Abstract
The TBC1D1 plays a key role in body energy homeostasis by regulating the insulin-stimulated glucose uptake in skeletal muscle. The present study aimed to identify the association between genetic polymorphisms of TBC1D1 and body weight (BW) in rabbits. Among the total of 12 SNPs detected in all 20 exons, only one SNP was non-synonymous (c.214G>A. p.G72R) located in exon 1. c.214G>A was subsequently genotyped among 491 individuals from two rabbit breeds by the high-resolution melting method. Allele A was the predominant allele with frequencies of 0.7780 and 0.6678 in European white rabbit (EWR, n = 205) and New Zealand White rabbit (NZW, n = 286), respectively. The moderate polymorphism information content (0.25<PIC<0.50) was present in both breeds. The association analysis revealed that genotypes GA and AA had higher 35 d body weight (BW) than genotype GG in both EWR (p<0.01) and NEW (p<0.05). For the 56 d BW and 70 d BW traits, genotypes AA and GA were higher than genotype GG in both two breeds, the difference was not significant (p>0.05). Our results implied that the c.214G>A of TBC1D1 gene might be one of the candidate loci affecting the trait of 35 d BW in the rabbit.
Keywords: TBC1D1, Polymorphism, High-resolution Melting, Growth Traits, Rabbit
INTRODUCTION
In skeletal muscle cells, the highest ability for glucose transport is accomplished via the insulin-responsive glucose transporter 4 (GLUT4) between intracellular storage compartments with the plasma membrane; therefore this pathway is crucial for maintaining glucose homeostasis (Felber and Golay, 2002; Watson et al., 2004). As the members of TBC1 domain protein family, AS160 (also known as TBC1D4) and TBC1D1 have been implicated in regulating glucose transport as the Akt substrates in skeletal muscle (Taylor et al., 2008; Funai and Cartee, 2009). The TBC1D1 holds two partial or full phosphotyrosine-binding (PTB) domains at the N-terminus, which is typically involved in regulation of cellular location post-translation (Marchler et al., 2013). Near the C-terminus, a complete TBC (Tre-2/BUB2/Cdc16) domain is known for its Rab (a cellular co-factor) GAP activity and regulation of Rab family small molecular weight GTPases (Will and Gallwitz, 2001). Previous studies showed that TBC1D1 regulated GLUT4 translocation through its GAP activity as AS160, in which, the insulin-stimulated phosphorylation of this protein on Akt identified sites led to suppression of its GAP activity and induced the GTP form elevating of the critical Rab, the activation of the Rab triggers then moved to, and/or docked with the plasma membrane of GLUT4-containing vesicles (Sakamoto and Holman, 2008; Zaid et al., 2008). Other related studies also suggested that TBC1D1 may regulate glucose transporter 1 (GLUT1) expression through the mammalian target of rapamycin (mTOR)-p70 S6 kinase pathway in adipocytes (Zhou et al., 2008).
The polymorphisms of TBC1D1 gene may affect the insulin-stimulated GLUT4 translocation, which could potentially result in disorder of glucose homeostasis and energy metabolism in skeletal muscle and blood. A mutation of this gene had been identified to be involved in severe obesity disease and human type 2 diabetes (Stone et al., 2006). A lean SJL (Swiss Jim Lambert mouse strain)-specific mutation in the TBC1D1 gene was found to protect mice from diet induced obesity (Chadt et al., 2008). The genotype GA located in intron 2 (g.219G>A) of TBC1D1 had a significant effect on ham weight in Italian Large White pigs (Fontanesi et al., 2011). In chicken, whole-genome resequencing of several domestic chickens revealed a divergent selection pattern of TBC1D1 haplotypes, which would suggest there was direct connection between TBC1D1 and important production traits (Rubin et al., 2010).
In summary, it would be expected that TBC1D1 can be considered as a candidate gene significantly associated with the obesity-related diseases in human as well as the natural growth pace in farm animals. At present, no study has been carried out to identify its variability and the association analysis of TBC1D1 with rabbit growth traits. This paper focused on finding out the genetic polymorphisms of TBC1D1 gene and its association with growth traits in the European white rabbit (EWR) and New Zealand White rabbit (NZW).
MATERIALS AND METHODS
Growth traits collection and genomic DNA extraction
A total of 491 commercial meat rabbits were used in this study, including 205 EWR and 286 NZW. The nutritional levels and feeding management were described in our previous study (Zhang et al., 2011). The growth traits dataset included body weight (BW) of 35 (BW35), 56 (BW56), and 70 (BW70) day of age. Ear tissues were collected and the genomic DNA was extracted using AxyPrep Genomic DNA Miniprep Kit (Axygen, USA), the DNA was stored at −20°C.
Screening SNP of TBC1D1
To scan the genetic polymorphisms of TBC1D1 gene coding region, five primer pairs were designed according to the TBC1D1 gene mRNA sequence (GenBank accession NO. XM_002709384.1) using Premier 5.0 (Table 1). For each primer pair, 20 rabbits were randomly selected from 491 individuals for mutational analysis. The 30 μL PCR reaction volume included 15 μL 2× Taq PCR MasterMix (TIANGEN, Beijing, China), 3 μL cDNA template (20 ng/μL), 9.6 μL ddH2O, 1.2 μL of each primer (10 pmol/μL). The PCR reaction was performed with the following conditions: one initial denaturation cycle at 95°C for 4 min, followed by 34 cycles of 95°C for 30 s, 50°C to 61°C for 30 to 45 s and 72°C for 40 to 80 s, end with an extension cycle at 72°C for 10 min. The purified PCR products were directly sequenced on a 3700 DNA sequencer in both directions using the BigDye Terminator sequencing kit (Applied Biosystems, Foster City, CA, USA), according to the manufacturer’s instructions.
Table 1.
Primer sequences, PCR product sizes, Tm value and location
| Primers | Primer sequences (5′→3′) | Product (bp) | Annealing (°C) | Location |
|---|---|---|---|---|
| Seq1 | F:TGGGCAGACCTCCAGAAAG | 1,342 | 62.0 | Exon1–9 |
| R:GATGTCCAGCCTAAAGCGAGT | ||||
| Seq2 | F:GAGCATCCTGTCCCGAGGTAATA | 1,436 | 60.0 | Exon9–18 |
| R:CGAAGACTCTGGCGACAAATC | ||||
| Seq3 | F:GATGTGCCATACAAAGAACTCC | 806 | 59.2 | Exon15–20 |
| R:TCAAGGAGGTCAAGGTTCTGT | ||||
| Seq4 | F:CACCATTTGTAGCCTTTCTATG | 716 | 57.0 | Exon1 |
| R:CCCTCCGCAACTTACTTTT | ||||
| Seq5 | F:GAGCCCATCCCTGAAGTGTT | 697 | 57.0 | Exon20 |
| R:CGTGTCCATGCCATCTAAAGC | ||||
| HRM214 | F:TTCCAGAAAGGAGCCCGTGAC | 90 | 62.1 | c.214G>A |
| R:GGTTGACTCTTGCCCAGGT |
Genotyping using high-resolution melting (HRM)
Primer pair of HRM214F and HRM214R (Table 1) was designed to amplify the 90 bp fragment containing the target SNP of c.214G>A, which was further subjected to HRM analysis. HRM analysis was performed on the Rotor-Gene Q series real-time PCR detection system (Corbett Research Pty Ltd, Sydney, Australia). The 10 μL reaction volume contained 5 μL 1×SsoFast EvaGreen supermix (Bio-Rad, USA), 1 μL DNA template (20 ng/μL), 3.2 μL ddH2O, 0.4 μL of each primer (10 pmol/μL), which was pipetted into each well on the 72-well PCR plate. The PCR profile was as following: one initial denaturation cycle at 98°C for 3 min, 35 cycles of 98°C for 5 s, 62.1°C for 30 s, subsequently 95°C for 10 s. Each run contained a non-template control (NTC) and three known genotype individuals including wild-type, heterozygote, homozygote from sequencing verified reference sequence types as the standards. Genotyping of each sample was repeated twice in the same run. The high-resolution melting data was analyzed with the precision melt analysis software (Corbett Research Pty Ltd, Sydney, Australia).
Statistical analysis
The DNA sequences were assembled and analyzed with the DNAstar program (DNAS Inc, Madison, WI, USA). The localization prediction of rabbit TBC1D1 gene mutations was obtained with SMART web server (Letunic et al., 2009). HRM curve data was analyzed with the Analysis-only Rotor-Gene Q series software VIRTUAL MODE software (Corbett Research Pty Ltd, Sydney, Australia). Allele frequencies, genotype frequencies, heterozygosity (He), effective number of alleles (Ne) and Polymorphic Information Content (PIC) were estimated based on the previous study (Botstein et al., 1980). The x2 analysis was used to test for deviation of genotype from Hardy Weinberg Equilibrium (HWE). The associations of genotypes with the growth traits were carried out by the least-squares method as applied in the General Linear Model (GLM) procedure of SAS 9.2 program. The statistical model as follows:
Where Yijkl was a record of the trait, μ was the overall mean of observations, Gi was the gender effect, Mj was the fixed genotype effect, Pk was the fixed born time effect, and eijkl was the residual error.
RESULTS AND DISCUSSION
Identification of SNP in rabbit TBC1D1
Genetic polymorphism of TBC1D1 in rabbit has not been reported before, we first scanned the coding region of the TBC1D1 to detect potential SNPs in rabbit. A total of 12 SNPs were detected and all of them were distributed over five exons region (Figure 1). Among them, there was only one non-synonymous mutation (c.214G>A) which was detected in exon 1 and resulted in the amino acid change from glycine to arginine (G72R). In porcine TBC1D1 gene, three synonymous mutations (g.40A>G, g.151C>T, and g.172G>A) were also observed, which were located in exon 2; other two mutations (g.219G>A and g.252G>A) were located in intron 2 and the g.219G>A mutation was correlated with some growth traits and slaughter traits (Fontanesi et al., 2011). In the human TBC1D1 gene, a non-synonymous mutation (R125W, rs35859249) was observed which showed an association with higher risk of obesity (Stone et al., 2006; Meyre et al., 2008).
Figure 1.

Schematic illustration of rabbit TBC1D1 gene organization and the identified genetic variants. Black bars represent the 20 exons. The inverted arrows indicate location of identified variants in the present study. Bold arrow at c.214 represents non-synonymous mutation, whereas the others indicate the synonymous mutations.
The prediction of rabbit TBC1D1 domains showed that the two PTB domains spanned the amino acids 16 to 164 and 168 to 387, respectively; the TBC domain spanned from the amino acids 800 to 1020; a coiled coil spanned amino acids 1077 to 1132, the remains were lower conserved sequences. Among the 12 SNPs, seven synonymous mutations (c.2451G>A, c.2514A>G, c.2535G>A, c.2589G>A, c.2763T>C, c.2844T>C, c.2937G>T) were located in the TBC domain of TBC1D1 in rabbit. The TBC domain is known for its Rab GAP activity and regulation of Rab family small molecular weight GTPases (Will and Gallwitz, 2001). In a previous study, a mutation in the tbc1d1 gene in SJL strain mice resulted in a truncated protein absence of the TBC Rab-GTPase-activative protein domain which suppressed high-fat diet-induced obesity (Chadt et al., 2008). It suggested that the functional mutation in TBC domain may play a potential role in regulating energy metabolism. We detected only seven synonymous mutations in this region; more individuals need to be sequenced to explore the extent of non-synonymous mutations in the TBC domain. The non-synonymous mutation c.214G>A (p.G72R) and other four synonymous mutations (c.153T>C, c.252A>G, c.564G>A, c.591G>A) were located in two PTB domains, respectively. As mentioned previously, the two PTB domains located in N-terminus of TBC1D1 were conserved in AS160. The PTB domain is typically involved in targeting protein to the appropriate cellular location or interacting with a binding partner (Marchler-Bauer et al., 2013). The protein holding PTB domain possesses powerful functions including the ability to bind inositol phosphates and/to other proteins which is dependent on the phosphorylation status of tyrosine residues on the binding partner (Yan et al., 2002). In previous studies, a R125W mutation in TBC1D1 located in PTB domain was associated with obesity susceptibility in humans (Stone et al., 2006; Meyre et al., 2008). Some other studies have proved that altering an R in TBC1D1 at a position corresponding G72 affected the binding properties of the PTB domain (Zhou et al., 1995; Yaich et al., 1998). Thus, we could presume that G72R mutation may be linked to the binding of TBC1D1 to the binding partner known as Akt in a PI3-kinase-dependent manner (Ishiki and Klip, 2005) which would have immediate effect on growth traits. To prove the hypothesis, c.214G>A was selected to carry out the association analysis with growth traits in the rabbit.
The evidence accumulated over the past decade also suggests that synonymous mutations could have functional consequences that can result in an aberrant mRNA splice, affecting mRNA stability and thus protein expression and enzymatic activity. The synonymous mutations were also demonstrated to affect protein conformation and have function and clinical consequence. In future studies regarding TBC1D1 and growth traits, the effects of synonymous mutations should be considered in our research (Sauna and Kimchi-Sarfaty, 2011).
Genotyping of SNP in rabbit TBC1D1 by HRM
High-resolution melting (HRM) analysis is a relatively new post-PCR analysis method used to identify variations in nucleic acid sequences. This method is based on detecting small differences in PCR melting curves and is enabled by improved dsDNA-binding dyes used in conjunction with real-time PCR instrumentation. In the presence of the fluorescent DNA intercalating dye and unlabeled oligonucleotide probes, the change in stability of DNA duplexes induced by subtle sequence variation is monitored as the temperature of the solution increases (Martino et al., 2010). Therefore, the shape of the melting curve changes with the sequence variation (Montgomery et al., 2007). Since different genotypes have unique melt curves and Tm, it is possible to identify the two homozygotes according to Tm directly and to discern the heterozygotes based on Tm and the changes in the shapes of the melt curves as compared to homozygotes (Nguye-Dumont et al., 2009). In previous studies, genotyping by HRM showed high sensitivity and specificity (Wittwer et al., 2003; Garritano et al., 2009; Martino et al., 2010). To assess the sensitivity and specificity of HRM in the present study, we undertook direct sequencing of 36 samples on the same plate. The sensitivity and specificity of HRM for c.214G>A mutation detection were 0.97 [35 true positive/(35 true positive+1 fasle negative)], and 0.98 [71 true negative/(1 false positive+71 true negative)], respectively. The results were consistent with previous studies. Therefore, HRM analysis was used to genotype the c.214 G>A of TBC1D1 in two rabbit breeds and the normalized graphs produced by analyzing amplified fragment of the three different genotypes are presented in Figure 2.
Figure 2.
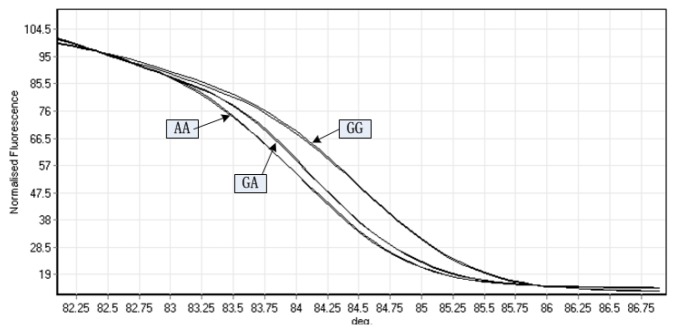
Different graphs produced with high resolution melting analyses of amplified fragments of the three different TBC1D1 c.214G>A SNP genotypes.
Genotype and allele frequencies in exon 1 of TBC1D1 in rabbit
The genotype and allele frequencies of c.214G>A in exon 1 of TBC1D1 in two rabbit breeds are shown in Table 2. Allele A was the predominant allele with frequencies of 0.78 in EWR and 0.67 in NZW, respectively. The frequencies of genotype AA for EWR and NZW were the highest (0.64 and 0.49, respectively). The polymorphic site showed low heterozygosity (He = 0.35) and moderate polymorphism information content (0.25<PIC<0.50) in EWR, high heterozygosity (He = 0.44) and moderate polymorphism information content (0.25<PIC<0.50) in NZW. The genetic diversity indicated higher genetic variation and a selective potentiality in NZW which could be expected to gain more genetic progress, while the lower genetic variation in EWR was probably due to the long-term artificial selection which made the negative allele frequency reduction (Andersson, 2001; Darwin, 2009). Genotype frequencies of c.214G>A were not in HWE in either EWR (x2 = 7.790, p = 0.020) or NZW (x2 = 11.002, p = 0.004), the number of AA homozymous subjects was higher than expected in both breeds, the potential cause would be the population dynamics resulted from the inbreeding of livestock that led to an increased frequency of homozygotes (Schaid and Jacobsen, 1999). In order to efficiently analyze genetic variation, more unrelated individuals and breeds need to be included in the study.
Table 2.
The frequencies of allele and genotype of the c.214G>A variation site
| Breeds | Sample (n) | Genotype frequency | Allele frequency | Genetic characteristic | |||||
|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|||||||
| AA | GA | GG | A | G | He | PIC | Ne | ||
| EWR | 205 | 131 (0.64) | 57 (0.28) | 17 (0.08) | 0.78 | 0.22 | 0.35 | 0.29 | 1.53 |
| NEW | 286 | 140 (0.49) | 102 (0.36) | 44 (0.15) | 0.67 | 0.33 | 0.44 | 0.35 | 1.80 |
The numbers in the brackets are the genotype frequency for the respective genotypes.
He = Heterozygosity. PIC = Polymorphism information content. Ne = Effective number of alleles.
Association between TBC1D1 genotypes and growth traits in rabbits
In the present study, the association analysis suggested that different genotypes had significant effects on the BW35, rabbits with genotypes GA and AA had higher 35 d BW than those with genotype GG in both EWR (p<0.01) and NEW (p<0.05). But the TBC1D1 genotypes had no significant association with other growth traits (p>0.05) (Table 3). In previous studies, a single nucleotide polymorphism (g.219A) located in intron 2 of porcine TBC1D1 had a significant association with higher ham weight and average daily gain (ADG) in Italian Large White pigs, the less frequent genotype g.219GA showed a lower ham weight and ADG (Fontanesi et al., 2011). In chicken, the TBC1D1 haplotype was significant for body weight at 12 weeks of age in broilers (Rubin et al., 2010; Gu et al., 2011). The TBC1D1 with R125W mutation was associated with obesity susceptibility in childhood and adult humans (Stone et al., 2006; Meyre et al., 2008). These results indicated that the TBC1D1 may be involved in rapid postnatal growth resulted from the energy metabolism in muscle and fat as genotypes at some of SNPs in TBC1D1 were associated with growth traits. Due to the negative genotype GG for c.214 G>A variant in TBC1D1 having a low frequency, groups with genotype GG had lower weight during postnatal growth than groups with AA and GA in both EWR and NZW. These effects indicated that the c.214G>A polymorphism of TBC1D1 was involved in regulating growth traits in rabbit. The groups with allele A had a positive effect on caloric shift which could lead advantageous growth in rabbit.
Table 3.
Least square means of growth traits of different genotypes
| Traits | Breeds | Genotype | p | ||
|---|---|---|---|---|---|
|
| |||||
| AA | GA | GG | |||
| 35-d-weight (g) | EWR | 1,026±10.67A | 1,012±16.18A | 938.8±29.36B | <0.01 |
| NEW | 619.2±11.80a | 636.4±13.87a | 561.1±21.07b | <0.01 | |
| 56-d-weight (g) | EWR | 1,888±20.71 | 1,893±32.66 | 1,776 ±55.70 | 0.13 |
| NEW | 1,081±17.56 | 1,109±20.81 | 1,062±31.54 | 0.21 | |
| 70-d-weight (g) | EWR | 2,381±19.70 | 2,369±29.32 | 2,363±95.76 | 0.72 |
| NEW | 1,360±22.14 | 1,359±25.91 | 1,358±40.27 | 0.96 | |
Different lowercase letters and capital letters mean significant difference at 0.05 and 0.01 levels, respectively.
NEW = New Zealand white rabbit; EWR = European white rabbit.
The proposed function for TBC1D1 was reasonable for its role involved in systemic energy balance by regulating the transport of GLUT4 or GLUT1 into cells. We predicted that the c.214G>A occurring in the protein interaction domain altered interaction of TBC1D1 with a binding partner known as Akt in insulin-induced glucose metabolism. The abnormal interaction may alter caloric disposal in insulin sensitive tissues, furthermore, the genotypes AA and GA had a positive effect on this process. When a high energy source is required for a rapid postnatal growth carried out around weaning, the groups with genotypes AA and GA have higher ability to uptake glucose for predominant binding of TBC1D1 to a binding partner and accumulate higher WB35 in two breeds. This speculation is supported in New Zealand obese (NZO) mice which present a feature of early-onset obesity and are highly susceptible to weight gain when fed a high-fat diet. Studies regarding NZO identified some dietary fat-responsive genetic loci and TBC1D1 was one of the genes in the Nob1.24 segment which was a suppressor of high-fat diet-induced obesity and the TBC1D1 gene mRNA expression was upregulated in high-fat diet energy metabolism (Crofford and Davis, 1965; West et al., 1992; Wuschke et al., 2006). Therefore, when a well-distributed growth is carried out in early development before 70 d of age for rabbit, the binding of TBC1D1 to Rab targets in glucose metabolism induced by energy source keeps a relatively stable state that had no significant effect on WB56 and WB70 in the present study. These results were also consistent with the function of TBC1D1 that is a regulator of either glucose or fatty acid transporter to achieve systemic energy balance by regulating the calories in cells (Kahn and Klip, 2000; Raz et al., 2005). Obviously, further experiments would be carried out to prove the precise molecular mechanism of TBC1D1 as well as the PTB domains of TBC1D1 involving in insulin-stimulated glucose metabolism, including the comparison of enzymatic activity among the alternative genotypes.
CONCLUSION
In the present study, one non-synonymous c.214G>A in TBC1D1 gene was first revealed to be significantly associated with early postnatal growth traits in rabbit. This would indicate that TBC1D1 is positively involved in the growth regulation. However, further functional investigation is necessary to explore the biological implication caused by or linked to this mutation.
Acknowledgments
This work was financially supported by the Earmarked Fund for China Agricultural Research System (Grant No: CARS-44-A-2).
REFERENCES
- Andersson L. Genetic dissection of phenotypic diversity in farm animals. Nat Rev Genet. 2001;2:130–138. doi: 10.1038/35052563. [DOI] [PubMed] [Google Scholar]
- Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Chadt A, Leicht K, Deshmukh A, Jiang LQ, Scherneck S, Bernhardt U, Dreja T, Vogel H, Schmolz K, Kluge R, Zierath JR, Hultschig C, RC Hoeben, Schürmann A, Alhasani H. Tbc1d1 mutation in lean mouse strain confers leanness and protects from diet-induced obesity. Nat Genet. 2008;40:1354–1359. doi: 10.1038/ng.244. [DOI] [PubMed] [Google Scholar]
- Crofford OB, Davis CK., Jr Growth characteristics, glucose tolerance and insulin sensitivity of New Zealand obese mice. Metabolism. 1965;14:271–280. doi: 10.1016/0026-0495(65)90068-5. [DOI] [PubMed] [Google Scholar]
- Darwin C. On the origin of species: by means of natural selection, or the preservation of favored races in the struggle for life. 2nd edition. Valde Books; New York, New York: 2009. [Google Scholar]
- Felber J, Golay A. Pathways from obesity to diabetes. International journal of obesity and related metabolic disorders: journal of the International Association for the Study of Obesity. 2002;26:S39–S45. doi: 10.1038/sj.ijo.0802126. [DOI] [PubMed] [Google Scholar]
- Fontanesi L, Colombo M, Tognazzi L, Scotti E, Buttazzoni L, Dall’Olio S, Davoli R, Russo V. The porcine TBC1D1 gene: mapping, SNP identification, and association study with meat, carcass and production traits in Italian heavy pigs. Mol Biol Rep. 2011;38:1425–1431. doi: 10.1007/s11033-010-0247-3. [DOI] [PubMed] [Google Scholar]
- Funai K, Cartee GD. Inhibition of contraction-stimulated AMP-activated protein kinase inhibits contraction-stimulated increases in PAS-TBC1D1 and glucose transport without altering PAS-AS160 in rat skeletal muscle. Diabetes. 2009;58:1096–1104. doi: 10.2337/db08-1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garritano S, Gemignani F, Voegele C, Nguyen-Dumont T, Calvez-Kelm FL, Silva DD, Lesueur F, Landi S, Tavtigian SV. Determining the effectiveness of high resolution melting analysis for SNP genotyping and mutation scanning at the TP53 locus. BMC Genet. 2009;10:5. doi: 10.1186/1471-2156-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Feng C, Ma L, Song C, Wang Y, Da Y, Li H, Chen K, Ye S, Ge C, Hu X, Li N. Genome-wide association study of body weight in chicken F2 resource population. PlOS ONE. 2011;6:e21872. doi: 10.1371/journal.pone.0021872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiki M, Klip A. Minireview: recent developments in the regulation of glucose transporter-4 traffic: new signals, locations, and partners. Endocrinology. 2005;146:5071–5078. doi: 10.1210/en.2005-0850. [DOI] [PubMed] [Google Scholar]
- Kahn BB, Flier JS. Obesity and insulin resistance. J Clin Invest. 2000;106:473–481. doi: 10.1172/JCI10842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucl Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: conserved domains and protein three-dimensional structure. Nucl. Acids Res. 2013;41(D1):D348–D352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino A, Mancuso T, Rossi AM. Application of high-resolution melting to large-scale, high-throughput SNP genotyping A comparison with the TaqMan. method. J. Biomol. Screen. 2010;15:623–629. doi: 10.1177/1087057110365900. [DOI] [PubMed] [Google Scholar]
- Meyre D, Farge M, Lecoeur C, Proenca C, Durand E, Allegaert F, Tichet J, Marre M, Balkau B, Weill J, Delplanque J, Froguel P. R125W coding variant in TBC1D1 confers risk for familial obesity and contributes to linkage on chromosome 4p14 in the French population. Hum Mol Genet. 2008;17:1798–1802. doi: 10.1093/hmg/ddn070. [DOI] [PubMed] [Google Scholar]
- Montgomery J, Wittwer CT, Palais R, Zhou L. Simultaneous mutation scanning and genotyping by high-resolution DNA melting analysis. Nat Protoc. 2007;2:59–66. doi: 10.1038/nprot.2007.10. [DOI] [PubMed] [Google Scholar]
- Nguyen-Dumont T, Calvez-Kelm FL, Forey N, McKay-Chopin S, Garritano S, Gioia-Patricola L, Silva DD, Weigel R, Sangrajrang S, Lesueur F, Tavtigian SV. Description and validation of high-throughput simultaneous genotyping and mutation scanning by high-resolution melting curve analysis. Hum Mutat. 2009;30:884–890. doi: 10.1002/humu.20949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raz I, Eldor R, Cernea S, Shafrir E. Diabetes: insulin resistance and derangements in lipid metabolism. Cure through intervention in fat transport and storage. Diabetes/Metabolism Res. Rev. 2005;21:3–14. doi: 10.1002/dmrr.493. [DOI] [PubMed] [Google Scholar]
- Rubin C-J, Zody MC, Eriksson J, Meadows JRS, Sherwood E, Webster MT, Jiang L, Ingman M, Sharpe T, Ka S, Hallöök F, Besnier F, Carlborg Ö, Bed’hom B, Tixier-Boichard M, Jensen P, Siegel P, Lindblad-Toh K, Andersson L. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Holman GD. Emerging role for AS160/TBC1D4 and TBC1D1 in the regulation of GLUT4 traffic. Am. J. Physiol-Endocrinol. Metab. 2008;295:E29–E37. doi: 10.1152/ajpendo.90331.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonmous mutations to human disease. Nat Rev Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, Jacobsen SJ. Blased tests of association: comparisons of allele frequencies when departing from hardy-weinberg proportions. Am J Epidemiol. 1999;149:706–711. doi: 10.1093/oxfordjournals.aje.a009878. [DOI] [PubMed] [Google Scholar]
- Stone S, Abkevich V, Russell DL, Riley R, Timms K, Tran T, Trem D, Frank D, Jammulapati S, Neff CD, Lliev D, Gress R, He G, Frech GC, Adams TD, Skolnick MH, Lanchbury JS, Gutin A, Hunt SC, Shattuck D. TBC1D1 is a candidate for a severe obesity gene and evidence for a gene/gene interaction in obesity predisposition. Hum Mol Genet. 2006;15:2709–2720. doi: 10.1093/hmg/ddl204. [DOI] [PubMed] [Google Scholar]
- Taylor EB, An D, Kramer HF, Yu H, Fujii NL, Roeckl KSC, Bowles N, Hirshman MF, Xie J, Feener EP, Goodyear LJ. Discovery of TBC1D1 as an insulin-, AICAR-, and contraction-stimulated signaling nexus in mouse skeletal muscle. J Biol Chem. 2008;283:9787–9796. doi: 10.1074/jbc.M708839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson RT, Kanzaki M, Pessin JE. Regulated membrane trafficking of the insulin-responsive glucose transporter 4 in adipocytes. Endocrinol Rev. 2004;25:177–204. doi: 10.1210/er.2003-0011. [DOI] [PubMed] [Google Scholar]
- West DB, Boozer CN, Moody DL, Atkinson RL. Dietary obesity in nine inbred mouse strains. Am. J. Physiol-Regul. Inetgr Comp Physiol. 1992;262:R1025–R1032. doi: 10.1152/ajpregu.1992.262.6.R1025. [DOI] [PubMed] [Google Scholar]
- Will E, Gallwitz D. Biochemical characterization of Gyp6p, a Ypt/Rab-specific GTPase-activating protein from yeast. J Biol Chem. 2001;276:12135–12139. doi: 10.1074/jbc.M011451200. [DOI] [PubMed] [Google Scholar]
- Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clin Chem. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- Wuschke S, Dahm S, Schmidt C, Joost HG, Al-Hasani H. A meta-analysis of quantitative trait loci associated with body weight and adiposity in mice. Int J Obes. 2006;31:829–841. doi: 10.1038/sj.ijo.0803473. [DOI] [PubMed] [Google Scholar]
- Yaich L, Ooi J, Park M, Borg JP, Landry C, Bodmer R, Margolis B. Functional analysis of the Numb phosphotyrosine-binding domain using site-directed mutagenesis. J Biol Chem. 1998;273:10381–10388. doi: 10.1074/jbc.273.17.10381. [DOI] [PubMed] [Google Scholar]
- Yan KS, Kuti M, Zhou MM. PTB or not PTB–that is the question. FEBS Lett. 2002;513:67–70. doi: 10.1016/s0014-5793(01)03305-1. [DOI] [PubMed] [Google Scholar]
- Zaid H, Antonescu C, Randhawa V, Klip-Biochem A. Insulin action on glucose transporters through molecular switches, tracks and tethers. Biochem J. 2008;413:201–215. doi: 10.1042/BJ20080723. [DOI] [PubMed] [Google Scholar]
- Zhang GW, Wang HZ, Chen SY, Li ZC, Zhang WX, Lai SJ. A reduced incidence of digestive disorders in rabbits is associated with allelic diversity at the TLR4 locus. Vet Immunol Immunop. 2011;144:482–486. doi: 10.1016/j.vetimm.2011.08.009. [DOI] [PubMed] [Google Scholar]
- Zhou M-M, Ravichandran KS, Olejniczak ET, Petros AM, Meadows RP, Sattler M, Harlan JE, Wade WS, Burakoff SJ, Fesik SW. Structure and ligand recognition of the phosphotyrosine binding domain of Shc. Nature. 1995;378:584–592. doi: 10.1038/378584a0. [DOI] [PubMed] [Google Scholar]
- Zhou QL, Jiang ZY, Holik J, Chawla A, Hagan GN, Leszyk J, Czech MP. Akt substrate TBC1D1 regulates GLUT1 expression through the mTOR pathway in 3T3-L1 adipocytes. Biochem J. 2008;411:647–655. doi: 10.1042/BJ20071084. [DOI] [PMC free article] [PubMed] [Google Scholar]


