Abstract
Smith–Lemli–Opitz syndrome (SLOS) is an inborn error of cholesterol synthesis resulting from a defect in 7-dehydrocholesterol reductase (DHCR7), the enzyme that produces cholesterol from its immediate precursor 7-dehydrocholesterol. Current therapy employing dietary cholesterol is inadequate. As SLOS is caused by a defect in a single gene, restoring enzyme functionality through gene therapy may be a direct approach for treating this debilitating disorder. In the present study, we first packaged a human DHCR7 construct into adeno-associated virus (AAV) vectors having either type-2 (AAV2) or type-8 (AAV2/8) capsid, and administered treatment to juvenile mice. While a positive response (assessed by increases in serum and liver cholesterol) was seen in both groups, the improvement was greater in the AAV2/8–DHCR7 treated mice. Newborn mice were then treated with AAV2/8–DHCR7 and these mice, compared to mice treated as juveniles, showed higher DHCR7 mRNA expression in liver and a greater improvement in serum and liver cholesterol levels. Systemic treatment did not affect brain cholesterol in any of the experimental groups. Both juvenile and newborn treatments with AAV2/8–DHCR7 resulted in increased rates of weight gain indicating that gene transfer had a positive physiological effect.
Keywords: Smith–Lemli–Opitz syndrome (SLOS), 7-Dehydrocholesterol reductase (DHCR7), Gene therapy, AAV, Cholesterol synthesis
1. Introduction
Smith–Lemli–Opitz syndrome (SLOS, OMIM 270400. locus 11q13.4) was first described in 1964 [1] and was much later shown to be an inborn error of cholesterol (C) biosynthesis [2], [3] that results in a broad range of physical dysmorphias and cognitive impairment [4]. The biochemical cause is deficient 3β-hydroxysterol-Δ7-reductase (EC 1.3.1.21, 7-dehydrocholesterol reductase, DHCR7), which reduces the Δ7 bond in 7-dehydrocholesterol (7DHC) to form cholesterol in the last step of the Kandutsch–Russell cholesterol synthetic pathway [5]. As a result, cholesterol production decreases, and amounts of dehydro precursors of cholesterol, such as 7DHC and 8-dehydrocholesterol (8DHC), increase in tissues and serum [2], [3], [6]. Comparing the concentration of 7DHC in plasma or serum to control levels, as measured by Gas Chromatography/Mass Spectrometry (GC/MS) [7], is used to diagnose SLOS. Prenatal diagnosis is also possible by measurement of the characteristic dehydrosterols in amniotic fluid or chorionic villus cells [7], [8], [9], and fetal-derived dehydrosteroid derivatives in maternal urine or serum [10].
SLOS predominantly presents in Caucasian populations, with the incidence reported to be between 1:10,000 and 1:70,000 in northern and central Europe [11], [12], [13], [14], and likely less than 1:100,000 in North America [15], [16]. The carrier frequency for the commonly found DHCR7 mutations predicts a much higher incidence rate, and it is possible that SLOS is under-diagnosed [15], [17] because the milder cases may be missed [18], [19], and the more severe cases may result in prenatal or neonatal death [13], [20], [21]. Currently, the only treatment is dietary cholesterol supplementation, and while this has been suggested to have positive biochemical and developmental benefits [22], [23], [24], [25], pre-existing dysmorphias are irreversible and neurological outcomes are unchanged. In contrast to previous anecdotal evidence, a short-term, placebo controlled study [26] showed that supplemental exogenous cholesterol did not improve neurological outcome in SLOS children. Likely, this is because the blood–brain barrier prevents circulating cholesterol from altering the sterol composition in the brain [27]. Because fetal development is highly dependent on endogenous cholesterol synthesis [28], [29], [30], [31], [32], treatment would ideally be administered prenatally in order to moderate otherwise irreversible symptoms; however, any such treatment remains to be developed.
Gene therapy is a largely unexplored treatment for this disorder that could address many of the shortcomings of dietary cholesterol supplementation. By importing a functional DHCR7 gene, patients could increase cholesterol and concomitantly decrease 7DHC. This should be doubly beneficial, firstly because sufficient cholesterol is needed for its multiple roles in metabolism, structure and regulation, and secondly because the accumulation of 7DHC can change the physical properties of cell membranes [27] and has deleterious effects when metabolized to oxidized derivatives [33]. Therefore, gene therapy has the potential to improve systemic and brain cholesterol synthesis, and if administered in utero, may even decrease the prevalence of fetal loss and ameliorate early deleterious effects.
Here we focus on systemic treatment of newborn and juvenile mice. In a previous study, we showed “proof of concept” for gene therapy in a mouse model for SLOS. Using a recombinant type-2 adeno-associated virus (AAV2) vector carrying a human DHCR7 cDNA and a heterologous promoter, we achieved the production of active DHCR7 enzyme and increased cholesterol synthesis, as demonstrated by lowered serum 7DHC/C ratio [34]. Because the biochemical improvement was modest and a complete normalization of cholesterol was not achieved, we hypothesized, and presently describe, that higher vector doses, a more efficient vector, and/or earlier treatment administration can yield a greater positive effect.
2. Materials and methods
2.1. Animal husbandry
Animal work conformed to NIH guidelines and was approved by the Institutional Animal Care and Use Committee. All animals were maintained in an AAALAC certified facility and were fed a normal, cholesterol-free chow (Teklad irradiated rodent diet 2918: Harlan, Madison, WI).
2.2. Generation of experimental animals
The use of mutant mice and their breeding protocol was as previously described [35]. In brief, study animals were generated by crossing two separate mouse models of SLOS: a null mutant containing a partial deletion of Dhcr7 (Δ) [36] and a hypomorphic mutant containing a point mutation (T93M) [37]. Δ/T93M mice were used for all experiments because they exhibit the most severe, yet still viable, phenotype. Mice with genotypes of T93M/T93M, Δ/T93M and Δ/Δ show SLOS characteristics of increasing biochemical severity, with Δ/Δ being lethal [36], [37]. Mice heterozygous for the wild type allele, Δ/+ and T93M/+, have a normal phenotype.
The Δ/T93M mice exhibit classic SLOS symptoms, such as elevated 7DHC/C ratios in the serum and tissues, decreased size compared to wild-type littermates, and sporadic syndactyly. Littermates having the Δ/T93M genotype were assigned to each experimental group (treated or control), and an effort was made to balance the two groups with respect to the initial average serum 7DHC/C ratio. Males and females were included in both groups, as no apparent correlation has been noted between gender and 7DHC/C ratios [34].
2.3. Preparation of AAV2 and AAV2/8 vectors
Vector construction, production and purification were as described previously [34]. Human DHCR7 cDNA was cloned into the EcoRI site of the pV4.1c plasmid, which contained a CMV promoter/enhancer and AAV2 inverted terminal repeats. A woodchuck hepatitis virus post-transcriptional regulatory element (WPRE) was included at the 3′ untranslated end of the DHCR7 to increase expression levels. AAV2–DHCR7 particles were produced in an adenovirus-free system by triple-transfecting HEK293 cells with plasmid pV4.1c-DHCR7, an AAV2 packaging plasmid, and a plasmid containing adenovirus helper genes [38]. For AAV2/8–DHCR7 particles, the procedure was the same except that the packaging plasmid (p5E18-VD2/8 kindly provided by James Wilson, University of Pennsylvania) contained an AAV2 rep gene fused to an AAV8 cap gene [39]. DNA packaged in capsid was purified by an Optiprep gradient (Iodixanol) and 2 cesium chloride gradient centrifugations [40]. Finally, the viral vector was dialyzed against normal saline and the titer in vector genomes (vg) per ml was established by quantitative PCR (AAV2–DHCR7 titer: 5.7x1012 vg/ml, AAV2/8–DHCR7 titer: 5.8 × 1012 vg/ml).
2.4. Treatment of SLOS mice and serum collection
In an effort to match initial 7DHC/C ratios between treated and control groups, sterols were analyzed from blood collected from Δ/T93M newborns and juveniles prior to injection. From newborns, blood was collected from the temporal vein by inserting the bevel of a 29-guage needle into the superficial temporal vein and lifting up slightly to allow a droplet of blood to pool. From mice 3 weeks and older, blood was collected from the retro-orbital sinus. Newborns were treated at 3 days of age (1 and 2 day old SLOS pups were too fragile for treatment) and, a single 60 μl dose of the AAV vector was injected intravenously via the superficial temporal vein. For juveniles (4 weeks old) a single 200 μl dose was injected via the tail vein. Sham treated controls received comparable injections of saline. In the juvenile cohorts, blood was collected every week following injection to monitor serum 7DHC/C. For newborns, blood was not collected again until the mice were 3 weeks old, but was collected weekly thereafter. This was done because the temporal vein is no longer visible after the first post-natal week, and retro-orbital bleeding is not feasible until 3 weeks of age. All mice were weighed at the times of retro-orbital blood collection.
2.5. Nucleic acid extraction and analysis
Anesthetized mice were euthanized at 13 or 14 weeks of age by exsanguination via cardiac puncture. Livers and brains were excised, immediately frozen in liquid nitrogen and stored at − 80 °C until analysis. DNA and RNA extraction, PCR, RT-PCR and quantitative RT-PCR (SYBR Green detection in an ABI 7900 instrument) were as previously described [34]. A commercial primer pair specific for human DHCR7 yielded a 123 base pair PCR product and a primer pair specific for mouse Dhcr7 yielded a 125 base pair PCR product (OriGene, Rockville, MD). For quantitative RT-PCR, mouse glyceraldehyde 3-phosphate dehydrogenase (mGAPDH) mRNA was also measured to provide an internal reference.
2.6. Analysis of cholesterol and dehydrocholesterol by GC/MS
Sample preparation for serum, liver, and brain was the same as previously described [34]. An internal standard (stigmasterol) was added to the samples, which were then saponified. Afterwards, the samples were extracted and reacted with BSTFA to form the trimethylsilyl (TMS) derivatives. To prevent the conversion of 7DHC to previtamin-D3, all tubes were protected from light by foil covers, and the entire procedure was conducted under minimum lighting conditions. Analysis was as previously described [34], [41] using an Agilent 5975C Gas Chromatography/Mass Spectrometry instrument with a HP-5MS column [35]. Calibration curves were prepared daily for each analytical batch by combining increasing amounts of the analytes with a fixed amount of stigmasterol, followed by derivatization [34]. In the liver and serum samples, 8DHC levels were never significantly above background, so only 7DHC levels were used to calculate ratios to cholesterol. In brain samples, levels of 8DHC were significant and ratios were calculated as (7DHCR + 8DHCR) / C.
2.7. Statistical analysis
Because data did not meet the criteria for a normal distribution, they were subjected to nonparametric statistical analysis using the Mann–Whitney rank-sum test for each comparison at a given time point. In addition to single time point comparisons, multiple time point comparisons between treated and untreated groups were also subjected to statistical analysis. This allowed comparison across whole time curves and added statistical power. To compare curves for serum and weight data over time, the mixed random effects model for longitudinal analysis was used as previously described [34]. Since the raw data showed that the pattern of covariances was not constant, the more traditional statistical analyses were not valid. On the other hand, mixed random models do not require this assumption and were therefore chosen for the analysis of our data. Differences having P values of 0.05 or less (two-tailed) were taken as significant.
3. Results
3.1. Administration of vectors
For the experiments described, all SLOS mice were compound heterozygotes carrying one null allele (Δ) and one hypomorphic allele (T93M) of Dhcr7[37]. This provided the most severe yet still viable SLOS phenotype currently available in an animal model. To treat these mice, two vectors were utilized. Both contained the same DNA, which included a human DHCR7 cDNA driven by a CMV promoter/enhancer, but one (AAV2–DHCR7) was packaged with type 2 capsid and the other (AAV2/8–DHCR7) with type 8 capsid. Experiments were conducted to compare the efficacy of AAV2 against AAV2/8, and to examine the effect of age on treatment. Three cohorts of experimental mice were established: 1) a cohort of 17 mice injected at 4 weeks of age (10 treated with 1.1 × 1012 vg AAV2–DHCR7 for an average dose of 1014 vg/kg; 7 with saline alone), 2) a cohort of 16 mice injected at 4 weeks of age (8 treated with 1.2 × 1012 vg AAV2/8–DHCR7 for an average dose of 1014 vg/kg; 8 with saline), and 3) a cohort of 15 mice injected at 3 days of age (8 treated with 3.5 × 1011 vg AAV2/8–DHCR7 for an average dose of 2.5 × 1014 vg/kg; 7 with saline).
3.2. DHCR7 gene detection and mRNA expression
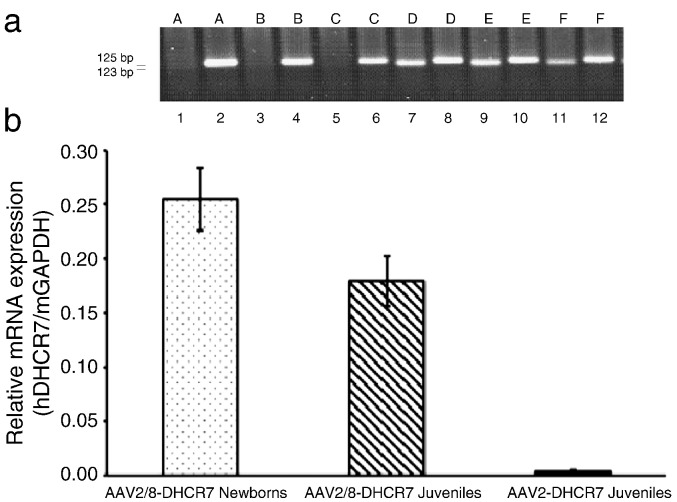
First, to confirm the presence or absence of vector DNA, liver DNA was subjected to PCR using primers specific for human DHCR7. In all three groups of experimental mice human DHCR7 DNA was detected in the livers of vector treated, but not control mice. Then, to measure DHCR7 mRNA, cDNA synthesized from liver mRNA was subjected to PCR using the human DHCR7 specific primers. As a positive control, RT-PCR was also done using primers specific to the mouse Dhcr7 gene transcript. Specificity of the human and mouse primer pairs and expression of human DHCR7 mRNA only in the treated mice were established by running RT-PCR products on a gel (Fig. 1a). Levels of human DHCR7 mRNA were then quantified by RT-qPCR using mouse GAPDH mRNA as an internal standard (Fig. 1b). Expression relative to the uniformly expressed mGAPDH gene transcript was calculated by dividing the concentration of hDHCR7 mRNA by the concentration of mGAPDH mRNA. The hDHCR7 gene expression in mice treated with AAV2/8–DHCR7 was significantly higher than in mice treated with AAV2–DHCR7 (P < 0.05). For AAV2/8–DHCR7, the expression in mice treated as newborns was greater than in mice treated as juveniles, although the criterion for statistical significance was not quite met (P = 0.07).
Fig. 1.
Expression of hDHCR7 mRNA in livers of treated mice. (a) RT-PCR was performed on liver RNA. Lanes 1 and 2 are from mouse A, lanes 3 and 4 are from mouse B etc. In odd numbered lanes primers to the hDHCR7 gene (123 base pair PCR product) were used, and in even numbered lanes primers to the mouse homologue (mDhcr7, 125 base pair product) were used. mDhcr7 was expressed in both control (A, B, C) and AAV2/8–DHCR7 treated (D, E, F) mice. hDHCR7 was expressed only in the treated mice. Qualitatively similar results were seen from all treated and control mice. (b) Levels of hDHCR7 mRNA expression determined by quantitative RT-PCR. Each bar represents the average relative mRNA expression ± SEM. Relative mRNA expression was calculated by dividing the concentration of hDHCR7 mRNA by the concentration of mGAPDH mRNA. The hDHCR7 expression in mice treated with AAV vector was significantly higher than background levels in sham-treated controls (AAV2–DHCR7 Juveniles: P = 0.03; AAV2/8–DHCR7 Juveniles: P < 0.01; AAV2/8–DHCR7 Newborns: P < 0.01).
3.3. 7DHC/C ratio in the serum, liver, and brain of treated mice
The ratio of 7DHC/C serves as a biochemical indication of SLOS severity. In the Δ/T93M mice this ratio was always elevated, whereas in normal mice (+/+, Δ/+ or T93M/+) it was not measurably above zero. Thus, a goal of treatment is to minimize this ratio.
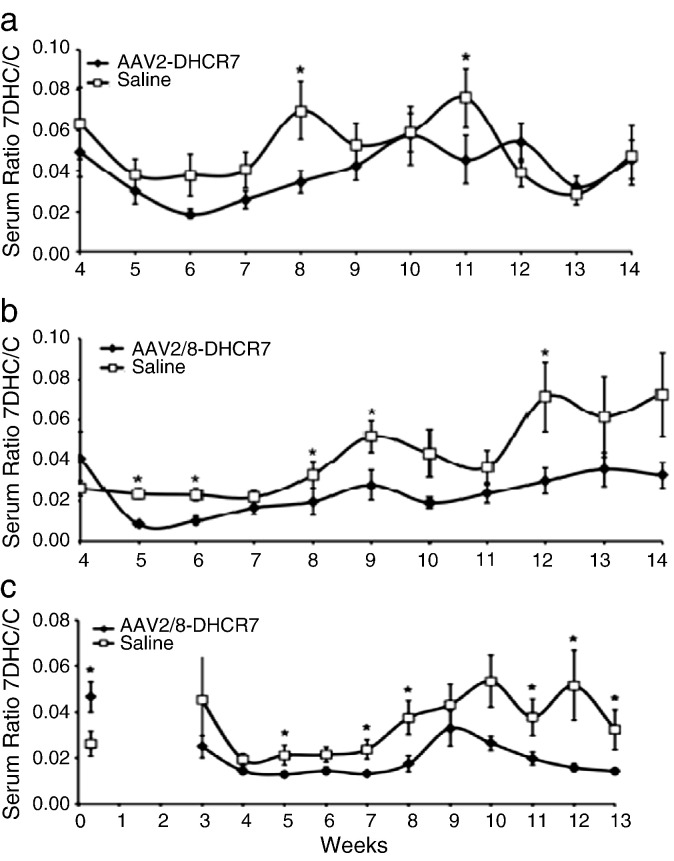
After treatment the ratio of 7DHC/C was measured in the sera of all mice at regular intervals. GC/MS was used to measure sterol concentrations and calculate ratios of 7DHC/C, and the results for the juvenile and newborn cohorts are shown in Fig. 2. As previously described [34], serum 7DHC/C ratios in treated and especially untreated SLOS mice varied considerably with age. In all three cohorts, the trend was for treated mice to have lower 7DHC/C ratios than their untreated littermates. In the AAV2–DHCR7 juvenile cohort the ratio of 7DHC/C in the serum of treated mice was apparently lower than in controls, but statistically significant at only 2 time points (Fig. 2a), whereas in the AAV2/8–DHCR7 juvenile cohort, treatment lowered the ratio with statistical significance at 5 time points (Fig. 2b). In the newborn treated cohort, the ratio of 7DHC/C was lower than in controls with statistical significance at 6 time points (Fig. 2c). To compare treated vs. control over the whole curve rather than at single time points, the mixed random effects model was employed. This more comprehensive analysis showed that each of the treatments had a statistically significant effect in lowering 7DHC/C ratios (AAV2 juveniles, P = 0.025; AAV2/8 juveniles, P < 0.001; AAV2/8 newborns, P < 0.001). Adjusting for sex did not affect these differences.
Fig. 2.
Weekly 7DHC/C ratios in serum comparing treated and sham-treated littermates. (a) AAV2–DHCR7 treated juvenile cohort. (b) AAV2/8–DHCR7 treated juvenile cohort. (c) AAV2/8–DHCR7 treated newborn cohort. *Denotes statistically significant (P < 0.05) differences in 7DHC/C ratio.
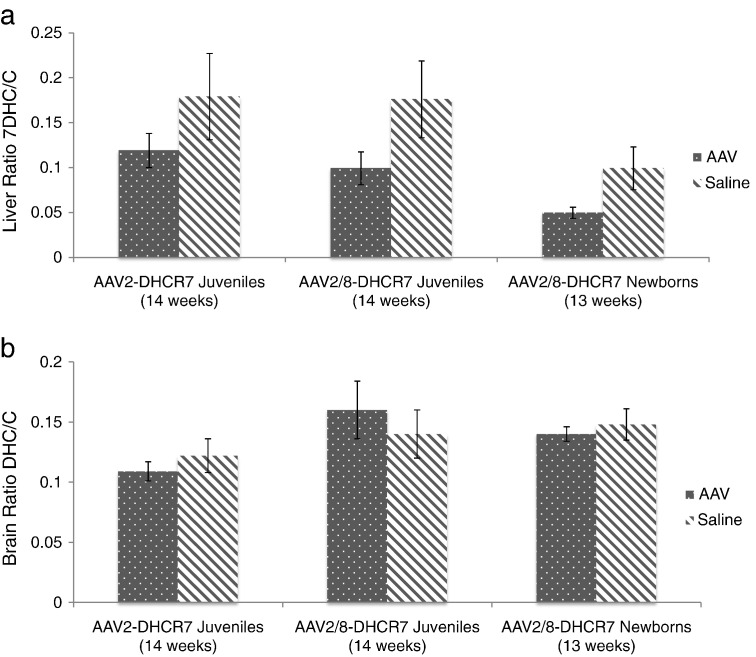
The ratio of 7DHC/C was also measured in the liver and brain after the animals were sacrificed. Although treatment resulted in an apparent decrease in liver ratios for all 3 cohorts, statistical significance in the AAV2–DHCR7 juvenile cohort was compromised by high variability among the untreated controls. In the juvenile and newborn cohorts treated with AAV2/8–DHCR7 the liver 7DHC/C ratios were significantly lower (P = 0.03 in each case) in the treated mice as compared to their littermate controls (Fig. 3a). Notably, there were no detectable differences in brain sterols between treated and untreated mice for any of the three cohorts (Fig. 3b).
Fig. 3.
Systemic treatment lowers DHC/C ratios in liver but not brain. (a) 7DHC/C ratios were determined for liver; (b) [7DHC + 8DHC]/C ratios were determined for brain. Because 8DHC levels in liver were not measurably above background, only 7DHC measurements were used to calculate DHC/C ratios. In brain 8DHC levels were significant and were therefore included in DHC/C ratios. Ratios shown are means ± SEM.
3.4. Gene therapy and growth rate
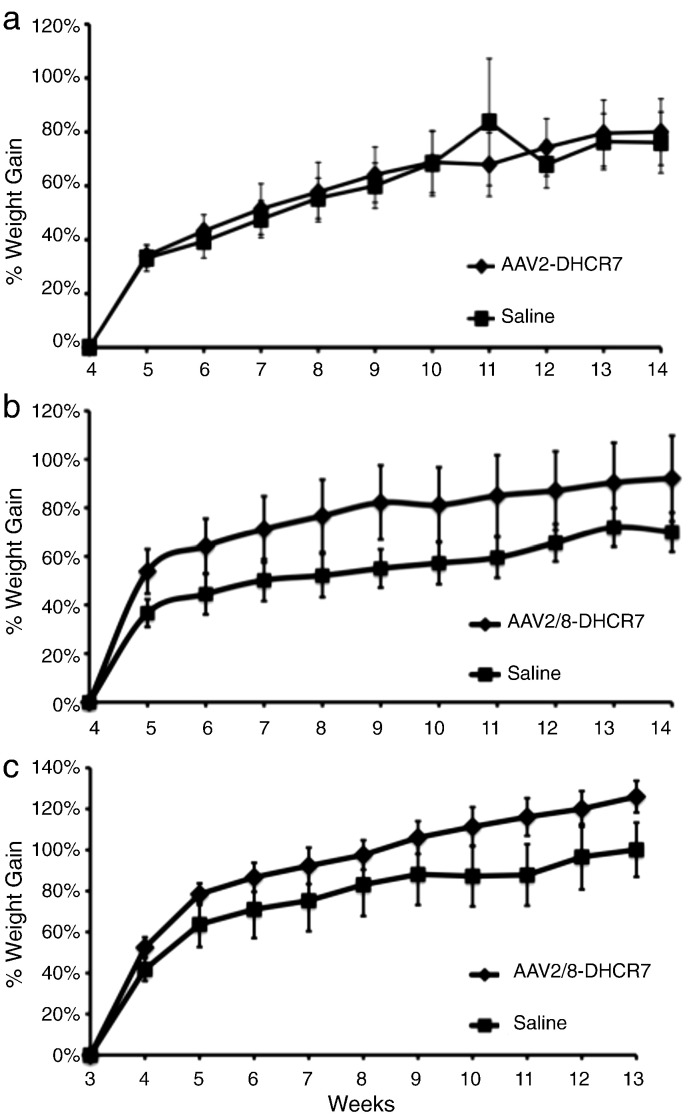
To determine if gene therapy was able to alter physical outcomes, all animals were weighed once a week at the time of serum sampling. Because the SLOS mice tend to be smaller than their wild-type siblings [37] and because there was considerable animal-to-animal variation, the relative rate of weight gain was used as an indicator of growth. This was calculated individually for each animal by dividing the weight at each week by the initial weight to yield the percent weight gain as a function of time (Fig. 4). The AAV2/8–DHCR7 treated mice, both newborns and juveniles, gained weight more rapidly than their sham-treated controls, while the AAV2–DHCR7 treated juveniles did not. Again, whole curves rather than individual time points were analyzed using the random effects model to make longitudinal comparisons. Both AAV2/8–DHCR7 treatments had highly significant effects on weight gain (P < 0.001). Adjusting for sex did not compromise the statistical significance of treatment even though males tended to grow faster than females (both treated and untreated).
Fig. 4.
Change in rate of weight gain following treatment. The weight of each animal at each time point was compared to its own weight at the time of injection or at the time of weaning. This provided cumulative % weight gains at weekly intervals. (a) Juveniles treated with AAV2–DHCR7 with weight gain indexed to the time of injection at 4 weeks of age (no statistical difference between treated and untreated). (b) Juveniles treated with AAV2/8–DHCR7 with weight gain indexed to time of injection at 4 weeks of age (treatment difference, P < 0.001). (c) Newborns treated with AAV2/8–DHCR7 with weight indexed to the time of weaning at 3 weeks of age (treatment difference, P < 0.001). Statistical comparisons of treated vs. littermate controls employed the mixed random effects model for longitudinal analysis.
For the AAV2/8–DHCR7 juvenile cohort, normal phenotype (T93M/+ genotype) littermates were also weighed weekly at 4 to 14 weeks of age. Although these mice were bigger than their SLOS littermates at both the beginning and throughout the time period, their relative rates of growth were indistinguishable from their untreated SLOS siblings (data not shown). That is, over this time period, the two groups grew in parallel. In contrast, the relative rate of growth for the treated SLOS mice was faster than normal, suggesting that treatment had a “catch up” effect on growth.
Compared to their respective littermate controls, newborn mice treated with AAV2/8–DHCR7 did not gain weight faster than juvenile mice treated with the same vector (compare Fig. 4b with 4c). It should be noted, however, that the newborn group might have already had an enhanced growth rate prior to its index weight at 3 weeks.
4. Discussion
Previously, we showed that gene transfer has the potential to alter cholesterol synthesis in a mouse model of SLOS, although the demonstrated effect was quite modest [34]. Here we report the following: 1) confirmation and extension of initial observations showing that gene therapy can significantly decrease the ratio of 7DHC/C in both the liver and serum; 2) comparison of the efficacy of two AAV vectors; 3) the effect of age at the time of treatment; 4) demonstration of a positive effect on growth rate.
Using both AAV2 and AAV2/8 vectors, the human DHCR7 gene was delivered and expressed in the liver, the primary target of these vectors following intravenous administration in mice [39], [42], [43], [44]. The dose used was sufficient to increase liver cholesterol and decrease 7DHC, which indicates that a functional enzyme was being produced and was actively converting 7DHC to cholesterol; this benefits the animal by reducing the dual burdens of cholesterol deficiency and 7DHC toxicity. (Direct measurements of DHCR7 enzyme activity were not done because cell disruption tends to inactivate this membrane-bound protein.) Since the liver is the principal systemic source of cholesterol, we hypothesized that local correction of cholesterol synthesis would have systemic effects, and this was evidenced by a strong correlation between the ratios of 7DHC/C in the serum and in the liver for each animal (r2 = 0.90, data not shown). Thus, we suggest that transduced liver cells might act as depots of DHCR7 activity and that 7DHC, either endogenous or imported, could be converted to cholesterol and then exported to other areas of the body as needed. Rigorous confirmation of this idea remains to be demonstrated. As expected, however, there was no significant change in brain sterols, because the blood–brain barrier limits the uptake of both viral vectors [45] and circulating cholesterol [46]. Preliminary (unpublished) results suggest that the blood–brain barrier can be circumvented by an alternate route of vector administration, namely intrathecal injection, which we have used successfully for other genetic diseases [45], [47]. This avenue is currently being pursued. Another possibility would be to use a different vector that is more effective in crossing the blood brain barrier, such as AAV 9 [48].
The greater reduction of the serum 7DHC/C ratio in juvenile mice treated with AAV2/8–DHCR7 suggests that the AAV2/8 vector was more efficient than the AAV2 vector for increasing systemic cholesterol. This is consistent with reports that AAV2/8 vectors better transduce hepatocytes than AAV2 vectors [39], [42], [49], [50]. Using AAV2/8–DHCR7, but not AAV2–DHCR7, the increase in systemic cholesterol was sufficient to alter growth outcomes, as both juvenile and newborn animals treated with the AAV2/8 vector grew faster. In the case of treated newborns, relative growth was indexed to the weight at 3 weeks of age (when regular blood collection began) rather than the time of treatment. Thus, these animals may have already partially overcome their delayed growth rates; nevertheless, they continued to grow more rapidly than their sham-treated littermates after weaning.
Administering gene therapy to both newborn mice and 4-week-old mice allowed us to evaluate the role of age in treatment effectiveness. When administered to either newborns or juveniles, gene therapy was persistent until at least 3 months of age. At the end of the experiments, treatment of newborns was 33% to 45% more effective by three biochemical criteria, but the differences were statistically marginal (P = 0.07 for hDHCR7 mRNA in liver; P = 0.06 for DHC/C ratio in liver; P = 0.03 for DHC/C in serum). Because of their size, newborns received a lower absolute dose of AAV2/8–DHCR7, but a higher relative dose (vector particles per body weight) than the mice treated as juveniles. This, in addition to age alone, could also contribute to the observed effectiveness of AAV2/8–DHCR7. The effectiveness of newborn treatment was somewhat surprising as it has been suggested that, for the long term, treating newborns is less effective than treating adults because episomal vector genomes in transduced cells are diluted by repeated cell replications [51]. But, it has also been observed that in treated newborns, a small fraction of vector genomes can integrate stably and result in persistent transgene expression [52]. Considering the potential advantages of earlier intervention, our results suggest that for SLOS there may be benefits from early administration in spite of vector genome dilution.
4.1. Conclusions
Gene therapy improved systemic cholesterol synthesis in juveniles and newborns of a mouse model for SLOS. Importantly, although complete normalization of sterol metabolism was not achieved, there was still a positive effect on physical outcome, as evidenced by improvements in growth rate. This supports two clinically relevant ideas: (1) Treatment does not have to completely normalize cholesterol in order to provide tangible benefit. (2) In spite of defects that arise early in development and which may not be reversible, treatment of SLOS newborns and even juveniles still has the potential to provide some physiological improvement. Perhaps with an even earlier treatment in utero, still greater normalization of cholesterol metabolism and more effective amelioration of SLOS symptoms can be achieved.
Conflict of interest
The authors have no conflict of interest to disclose.
Acknowledgments
This work was supported by the US National Institutes of Health Grant R01HD053036 to C. Shackleton and G. Watson. Dr. Serra was awarded a Beatriu de Pinós postdoctoral fellowship from Generalitat de Catalunya, Spain.
References
- 1.Smith D.W., Lemli L., Opitz J.M. A newly recognized syndrome of multiple congenital anomalies. J. Pediatr. 1964;64:210–217. doi: 10.1016/s0022-3476(64)80264-x. [DOI] [PubMed] [Google Scholar]
- 2.Irons M., Elias E.R., Salen G., Tint G.S., Batta A.K. Defective cholesterol biosynthesis in Smith–Lemli–Opitz syndrome. Lancet. 1993;341:1414. doi: 10.1016/0140-6736(93)90983-n. [DOI] [PubMed] [Google Scholar]
- 3.Tint G.S., Irons M., Elias E.R., Batta A.K., Frieden R., Chen T.S., Salen G. Defective cholesterol biosynthesis associated with the Smith–Lemli–Opitz syndrome. N. Engl. J. Med. 1994;330:107–113. doi: 10.1056/NEJM199401133300205. [DOI] [PubMed] [Google Scholar]
- 4.Porter F.D., Herman G.E. Malformation syndromes caused by disorders of cholesterol synthesis. J. Lipid Res. 2011;52:6–34. doi: 10.1194/jlr.R009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kandutsch A.A., Russell A.E. Preputial gland tumor sterols. 3. A metabolic pathway from lanosterol to cholesterol. J. Biol. Chem. 1960;235:2256–2261. [PubMed] [Google Scholar]
- 6.Batta A.K., Tint G.S., Shefer S., Abuelo D., Salen G. Identification of 8-dehydrocholesterol (cholesta-5,8-dien-3 beta-ol) in patients with Smith–Lemli–Opitz syndrome. J. Lipid Res. 1995;36:705–713. [PubMed] [Google Scholar]
- 7.Kelley R.I. Diagnosis of Smith–Lemli–Opitz syndrome by gas chromatography/mass spectrometry of 7-dehydrocholesterol in plasma, amniotic fluid and cultured skin fibroblasts. Clin. Chim. Acta. 1995;236:45–58. doi: 10.1016/0009-8981(95)06038-4. [DOI] [PubMed] [Google Scholar]
- 8.Tint G.S., Abuelo D., Till M., Cordier M.P., Batta A.K., Shefer S., Honda A., Honda M., Xu G., Irons M., Elias E.R., Salen G. Fetal Smith–Lemli–Opitz syndrome can be detected accurately and reliably by measuring amniotic fluid dehydrocholesterols. Prenat. Diagn. 1998;18:651–658. [PubMed] [Google Scholar]
- 9.Griffiths W.J., Wang Y., Karu K., Samuel E., McDonnell S., Hornshaw M., Shackleton C. Potential of sterol analysis by liquid chromatography–tandem mass spectrometry for the prenatal diagnosis of Smith–Lemli–Opitz syndrome. Clin. Chem. 2008;54:1317–1324. doi: 10.1373/clinchem.2007.100644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shackleton C.H., Marcos J., Palomaki G.E., Craig W.Y., Kelley R.I., Kratz L.E., Haddow J.E. Dehydrosteroid measurements in maternal urine or serum for the prenatal diagnosis of Smith–Lemli–Opitz syndrome (SLOS) Am. J. Med. Genet. A. 2007;143A:2129–2136. doi: 10.1002/ajmg.a.31901. [DOI] [PubMed] [Google Scholar]
- 11.Ryan A.K., Bartlett K., Clayton P., Eaton S., Mills L., Donnai D., Winter R.M., Burn J. Smith–Lemli–Opitz syndrome: a variable clinical and biochemical phenotype. J. Med. Genet. 1998;35:558–565. doi: 10.1136/jmg.35.7.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bzduch V., Behulova D., Skodova J. Incidence of Smith–Lemli–Opitz syndrome in Slovakia. Am. J. Med. Genet. 2000;90:260. doi: 10.1002/(sici)1096-8628(20000131)90:3<260::aid-ajmg17>3.3.co;2-i. [DOI] [PubMed] [Google Scholar]
- 13.Kelley R.I. A new face for an old syndrome. Am. J. Med. Genet. 1997;68:251–256. doi: 10.1002/(sici)1096-8628(19970131)68:3<251::aid-ajmg1>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 14.Lowry R.B., Yong S.L. Borderline normal intelligence in the Smith–Lemli–Opitz (RSH) syndrome. Am. J. Med. Genet. 1980;5:137–143. doi: 10.1002/ajmg.1320050205. [DOI] [PubMed] [Google Scholar]
- 15.Nowaczyk M.J., Zeesman S., Waye J.S., Douketis J.D. Incidence of Smith–Lemli–Opitz syndrome in Canada: results of three-year population surveillance. J. Pediatr. 2004;145:530–535. doi: 10.1016/j.jpeds.2004.06.045. [DOI] [PubMed] [Google Scholar]
- 16.Craig W.Y., Haddow J.E., Palomaki G.E., Kelley R.I., Kratz L.E., Shackleton C.H., Marcos J., Stephen Tint G., MacRae A.R., Nowaczyk M.J., Kloza E.M., Irons M.B., Roberson M. Identifying Smith–Lemli–Opitz syndrome in conjunction with prenatal screening for Down syndrome. Prenat. Diagn. 2006;26:842–849. doi: 10.1002/pd.1518. [DOI] [PubMed] [Google Scholar]
- 17.Battaile K.P., Battaile B.C., Merkens L.S., Maslen C.L., Steiner R.D. Carrier frequency of the common mutation IVS8-1G > C in DHCR7 and estimate of the expected incidence of Smith–Lemli–Opitz syndrome. Mol. Genet. Metab. 2001;72:67–71. doi: 10.1006/mgme.2000.3103. [DOI] [PubMed] [Google Scholar]
- 18.Langius F.A., Waterham H.R., Romeijn G.J., Oostheim W., de Barse M.M., Dorland L., Duran M., Beemer F.A., Wanders R.J., Poll-The B.T. Identification of three patients with a very mild form of Smith–Lemli–Opitz syndrome. Am. J. Med. Genet. A. 2003;122A:24–29. doi: 10.1002/ajmg.a.20207. [DOI] [PubMed] [Google Scholar]
- 19.Nowaczyk M.J., Whelan D.T., Hill R.E. Smith–Lemli–Opitz syndrome: phenotypic extreme with minimal clinical findings. Am. J. Med. Genet. 1998;78:419–423. doi: 10.1002/(sici)1096-8628(19980806)78:5<419::aid-ajmg5>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 20.Nowaczyk M.J., Waye J.S., Douketis J.D. DHCR7 mutation carrier rates and prevalence of the RSH/Smith–Lemli–Opitz syndrome: where are the patients? Am. J. Med. Genet. A. 2006;140:2057–2062. doi: 10.1002/ajmg.a.31413. [DOI] [PubMed] [Google Scholar]
- 21.Opitz J.M., Gilbert-Barness E., Ackerman J., Lowichik A. Cholesterol and development: the RSH (“Smith–Lemli–Opitz”) syndrome and related conditions. Pediatr. Pathol. Mol. Med. 2002;21:153–181. doi: 10.1080/15227950252852078. [DOI] [PubMed] [Google Scholar]
- 22.Nwokoro N.A., Mulvihill J.J. Cholesterol and bile acid replacement therapy in children and adults with Smith–Lemli–Opitz (SLO/RSH) syndrome. Am. J. Med. Genet. 1997;68:315–321. doi: 10.1002/(sici)1096-8628(19970131)68:3<315::aid-ajmg13>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 23.Elias E.R., Irons M.B., Hurley A.D., Tint G.S., Salen G. Clinical effects of cholesterol supplementation in six patients with the Smith–Lemli–Opitz syndrome (SLOS) Am. J. Med. Genet. 1997;68:305–310. doi: 10.1002/(sici)1096-8628(19970131)68:3<305::aid-ajmg11>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 24.Irons M., Elias E.R., Abuelo D., Bull M.J., Greene C.L., Johnson V.P., Keppen L., Schanen C., Tint G.S., Salen G. Treatment of Smith–Lemli–Opitz syndrome: results of a multicenter trial. Am. J. Med. Genet. 1997;68:311–314. [PubMed] [Google Scholar]
- 25.Linck L.M., Lin D.S., Flavell D., Connor W.E., Steiner R.D. Cholesterol supplementation with egg yolk increases plasma cholesterol and decreases plasma 7-dehydrocholesterol in Smith–Lemli–Opitz syndrome. Am. J. Med. Genet. 2000;93:360–365. doi: 10.1002/1096-8628(20000828)93:5<360::aid-ajmg4>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 26.Tierney E., Conley S.K., Goodwin H., Porter F.D. Analysis of short-term behavioral effects of dietary cholesterol supplementation in Smith–Lemli–Opitz syndrome. Am. J. Med. Genet. A. 2010;152A:91–95. doi: 10.1002/ajmg.a.33148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjorkhem I., Meaney S. Brain cholesterol: long secret life behind a barrier. Arterioscler. Thromb. Vasc. Biol. 2004;24:806–815. doi: 10.1161/01.ATV.0000120374.59826.1b. [DOI] [PubMed] [Google Scholar]
- 28.Jurevics H.A., Kidwai F.Z., Morell P. Sources of cholesterol during development of the rat fetus and fetal organs. J. Lipid Res. 1997;38:723–733. [PubMed] [Google Scholar]
- 29.Tint G.S., Yu H., Shang Q., Xu G., Patel S.B. The use of the Dhcr7 knockout mouse to accurately determine the origin of fetal sterols. J. Lipid Res. 2006;47:1535–1541. doi: 10.1194/jlr.M600141-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Edmond J., Korsak R.A., Morrow J.W., Torok-Both G., Catlin D.H. Dietary cholesterol and the origin of cholesterol in the brain of developing rats. J. Nutr. 1991;121:1323–1330. doi: 10.1093/jn/121.9.1323. [DOI] [PubMed] [Google Scholar]
- 31.Jurevics H., Morell P. Cholesterol for synthesis of myelin is made locally, not imported into brain. J. Neurochem. 1995;64:895–901. doi: 10.1046/j.1471-4159.1995.64020895.x. [DOI] [PubMed] [Google Scholar]
- 32.Turley S.D., Burns D.K., Rosenfeld C.R., Dietschy J.M. Brain does not utilize low density lipoprotein-cholesterol during fetal and neonatal development in the sheep. J. Lipid Res. 1996;37:1953–1961. [PubMed] [Google Scholar]
- 33.Gaoua W., Chevy F., Roux C., Wolf C. Oxidized derivatives of 7-dehydrocholesterol induce growth retardation in cultured rat embryos: a model for antenatal growth retardation in the Smith–Lemli–Opitz syndrome. J. Lipid Res. 1999;40:456–463. [PubMed] [Google Scholar]
- 34.Matabosch X., Ying L., Serra M., Wassif C.A., Porter F.D., Shackleton C., Watson G. Increasing cholesterol synthesis in 7-dehydrosterol reductase (DHCR7) deficient mouse models through gene transfer. J. Steroid Biochem. Mol. Biol. 2010;122:303–309. doi: 10.1016/j.jsbmb.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Serra M., Matabosch X., Ying L., Watson G., Shackleton C. Hair and skin sterols in normal mice and those with deficient dehydrosterol reductase (DHCR7), the enzyme associated with Smith–Lemli–Opitz syndrome. J. Steroid Biochem. Mol. Biol. 2010;122:318–325. doi: 10.1016/j.jsbmb.2010.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wassif C.A., Zhu P., Kratz L., Krakowiak P.A., Battaile K.P., Weight F.F., Grinberg A., Steiner R.D., Nwokoro N.A., Kelley R.I., Stewart R.R., Porter F.D. Biochemical, phenotypic and neurophysiological characterization of a genetic mouse model of RSH/Smith–Lemli–Opitz syndrome. Hum. Mol. Genet. 2001;10:555–564. doi: 10.1093/hmg/10.6.555. [DOI] [PubMed] [Google Scholar]
- 37.Correa-Cerro L.S., Wassif C.A., Kratz L., Miller G.F., Munasinghe J.P., Grinberg A., Fliesler S.J., Porter F.D. Development and characterization of a hypomorphic Smith–Lemli–Opitz syndrome mouse model and efficacy of simvastatin therapy. Hum. Mol. Genet. 2006;15:839–851. doi: 10.1093/hmg/ddl003. [DOI] [PubMed] [Google Scholar]
- 38.Matsushita T., Elliger S., Elliger C., Podsakoff G., Villarreal L., Kurtzman G.J., Iwaki Y., Colosi P. Adeno-associated virus vectors can be efficiently produced without helper virus. Gene Ther. 1998;5:938–945. doi: 10.1038/sj.gt.3300680. [DOI] [PubMed] [Google Scholar]
- 39.Gao G.P., Alvira M.R., Wang L., Calcedo R., Johnston J., Wilson J.M. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. U. S. A. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zolotukhin S., Byrne B.J., Mason E., Zolotukhin I., Potter M., Chesnut K., Summerford C., Samulski R.J., Muzyczka N. Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 1999;6:973–985. doi: 10.1038/sj.gt.3300938. [DOI] [PubMed] [Google Scholar]
- 41.Marcos J., Shackleton C.H., Buddhikot M.M., Porter F.D., Watson G.L. Cholesterol biosynthesis from birth to adulthood in a mouse model for 7-dehydrosterol reductase deficiency (Smith–Lemli–Opitz syndrome) Steroids. 2007;72:802–808. doi: 10.1016/j.steroids.2007.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakai H., Fuess S., Storm T.A., Muramatsu S., Nara Y., Kay M.A. Unrestricted hepatocyte transduction with adeno-associated virus serotype 8 vectors in mice. J. Virol. 2005;79:214–224. doi: 10.1128/JVI.79.1.214-224.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watson G.L., Sayles J.N., Chen C., Elliger S.S., Elliger C.A., Raju N.R., Kurtzman G.L., Podsakoff G.M. Treatment of lysosomal storage disease in MPS VII mice using a recombinant adeno-associated virus. Gene Ther. 1998;5:1642–1649. doi: 10.1038/sj.gt.3300775. [DOI] [PubMed] [Google Scholar]
- 44.Sharma V., Beckstead J.A., Simonsen J.B., Nelbach L., Watson G., Forte T.M., Ryan R.O. Gene transfer of apolipoprotein A-V improves the hypertriglyceridemic phenotype of apoa5 (−/−) mice. Arterioscler. Thromb. Vasc. Biol. 2013;33:474–480. doi: 10.1161/ATVBAHA.112.301078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Watson G., Bastacky J., Belichenko P., Buddhikot M., Jungles S., Vellard M., Mobley W.C., Kakkis E. Intrathecal administration of AAV vectors for the treatment of lysosomal storage in the brains of MPS I mice. Gene Ther. 2006;13:917–925. doi: 10.1038/sj.gt.3302735. [DOI] [PubMed] [Google Scholar]
- 46.Dietschy J.M. Central nervous system: cholesterol turnover, brain development and neurodegeneration. Biol. Chem. 2009;390:287–293. doi: 10.1515/BC.2009.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elliger S.S., Elliger C.A., Lang C., Watson G.L. Enhanced secretion and uptake of beta-glucuronidase improves adeno-associated viral-mediated gene therapy of mucopolysaccharidosis type VII mice. Mol. Ther. 2002;5:617–626. doi: 10.1006/mthe.2002.0594. [DOI] [PubMed] [Google Scholar]
- 48.Dayton R.D., Wang D.B., Klein R.L. The advent of AAV9 expands applications for brain and spinal cord gene delivery. Expert. Opin. Biol. Ther. 2012;12:757–766. doi: 10.1517/14712598.2012.681463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas C.E., Storm T.A., Huang Z., Kay M.A. Rapid uncoating of vector genomes is the key to efficient liver transduction with pseudotyped adeno-associated virus vectors. J. Virol. 2004;78:3110–3122. doi: 10.1128/JVI.78.6.3110-3122.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grimm D., Pandey K., Nakai H., Storm T.A., Kay M.A. Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J. Virol. 2006;80:426–439. doi: 10.1128/JVI.80.1.426-439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cunningham S.C., Dane A.P., Spinoulas A., Logan G.J., Alexander I.E. Gene delivery to the juvenile mouse liver using AAV2/8 vectors. Mol. Ther. 2008;16:1081–1088. doi: 10.1038/mt.2008.72. [DOI] [PubMed] [Google Scholar]
- 52.Inagaki K., Piao C., Kotchey N.M., Wu X., Nakai H. Frequency and spectrum of genomic integration of recombinant adeno-associated virus serotype 8 vector in neonatal mouse liver. J. Virol. 2008;82:9513–9524. doi: 10.1128/JVI.01001-08. [DOI] [PMC free article] [PubMed] [Google Scholar]