ABSTRACT
We have shown that glycoprotein K (gK) and its interacting partner, the UL20 protein, play crucial roles in virion envelopment. Specifically, virions lacking either gK or UL20 fail to acquire an envelope, thus causing accumulation of capsids in the cytoplasm of infected cells. The herpes simplex virus 1 (HSV-1) UL37 protein has also been implicated in cytoplasmic virion envelopment. To further investigate the role of UL37 in virion envelopment, the recombinant virus DC480 was constructed by insertion of a 12-amino-acid protein C (protC) epitope tag within the UL37 amino acid sequence immediately after amino acid 480. The DC480 mutant virus expressed full-size UL37 as detected by the anti-protC antibody in Western immunoblots, accumulated unenveloped capsids in the cytoplasm of infected cells, and produced very small plaques on African green monkey kidney (Vero) cells that were similar in size to those produced by the UL20-null and UL37-null viruses. The DC480 virus replicated nearly 4 log less efficiently than the parental wild-type virus when grown on Vero cells. However, DC480 mutant virus titers increased nearly 20-fold when the virus was grown on FRT cells engineered to express the UL20 gene in comparison to the titers on Vero cells, while the UL37-null virus replicated approximately 20-fold less efficiently than the DC480 virus on FRT cells. Coimmunoprecipitation experiments and proximity ligation assays showed that gK and UL20 interact with the UL37 protein in infected cells. Collectively, these results indicate that UL37 interacts with the gK-UL20 protein complex to facilitate cytoplasmic virion envelopment.
IMPORTANCE Herpes simplex viruses acquire their final envelopes by budding into cytoplasmic membranes derived from the trans-Golgi network (TGN). The tegument proteins UL36 and UL37 are known to be transported to the TGN sites of virus envelopment and to function in virion envelopment, since mutants lacking UL37 accumulate capsids in the cytoplasm that are unable to bud into TGN membranes. Viral glycoprotein K (gK) also functions in cytoplasmic envelopment, in a protein complex with the membrane-associated protein UL20 (UL20mp). This work shows for the first time that the UL37 protein functionally interacts with gK and UL20 to facilitate cytoplasmic virion envelopment. This work may lead to the design of specific drugs that can interrupt UL37 interactions with the gK-UL20 protein complex, providing new ways to combat herpesviral infections.
INTRODUCTION
The herpes simplex virus 1 (HSV-1) double-stranded DNA genome is enclosed in an icosahedral capsid and embedded within a proteinaceous tegument containing multiple viral proteins, all of which is packaged within an envelope decorated with multiple membrane proteins and glycoproteins (1–5). HSV-1 encodes at least 26 tegument proteins and 11 glycoproteins as well as several nonglycosylated membrane-associated proteins. The final steps in virion envelopment occur in the cytoplasm of infected cells and involve multiple interactions among viral proteins, including interaction between cytoplasmic portions of viral glycoproteins and tegument proteins bound to cytoplasmic capsids, as well as tegument-tegument protein interactions (1, 6–9).
HSV-1 UL37 is an approximately 120-kDa phosphorylated tegument protein expressed in both mature virions and light particles (10). The UL37 protein localizes to both the nucleus and cytoplasm in infected cells. The UL37 cytoplasmic localization is considered to be due to the presence of a nuclear export signal (NES) sequence within the UL37 protein that mediates transport of the UL37 protein to the cytoplasm in the absence of any other viral proteins (3, 11–13). The UL37 protein is capable of self-association and interacts via its carboxyl terminus with the UL36 major tegument protein, and this interaction is necessary and sufficient for transport of the UL36-UL37 protein complex to the trans-Golgi network (TGN), where virion envelopment is thought to occur (11, 14–16).
Deletion of the UL37 gene specified by either HSV-1 or pseudorabies virus (PRV) inhibits cytoplasmic virion envelopment, causing the accumulation of unenveloped capsids in the cytoplasm of infected cells (17–19). However, while the UL37 gene is absolutely essential for HSV-1 replication, deletion of the PRV UL37 gene allows the resultant mutant virus to replicate, albeit 100-fold less efficiently than the parental wild-type virus, thus demonstrating that certain PRV UL37 functions may be shared with other PRV proteins (17, 20). The pUL36 protein in the UL37-UL36 protein complex is thought to recruit the microtubule motors dynein, kinesin-1, and kinesin-2 onto capsids in the cytosol (21). These interactions are further supported by in vitro experiments in which capsids complexed with inner tegument proteins were able to associate with the microtubule motors, while the capsids lacking the tegument proteins did not (22). Thus, the UL37 protein plays important roles in capsid trafficking during virion entry, in a retrograde manner, i.e., toward the nucleus, and in conjunction with dynein motors and the microtubular network, while also playing an important role in capsid transport to the TGN (23–28).
The UL36-UL37 protein complex is part of a multiprotein complex within the virion particle and within infected cells. Specifically, the UL36 protein interacts with the outer tegument protein pUL48 (5) and the minor capsid protein UL25 (29). Additionally, the UL37 protein interacts with the UL46 and UL35 proteins (30). Also, it has been reported that UL37 is phosphorylated by cellular enzymes and that it interacts in the cytoplasm with ICP8, the major HSV-1 DNA binding protein, and is transported to the nuclei of infected cells (10). The HSV-1 UL37 protein contains a TRAF6 (tumor necrosis factor receptor-associated factor 6) binding domain that binds to TRAF6 and activates NF-κB expression, required for efficient viral replication, in a Toll-like receptor 2 (TLR2)-independent manner (31). These results suggest that the UL37 protein specifies additional functions in viral replication beyond its role in virion entry and cytoplasmic virion envelopment.
HSV-1 gK is a multimembrane-spanning viral glycoprotein encoded by the UL53 gene. It is expressed on virions, exists as a functional complex with the membrane-associated UL20 viral protein, and plays a role in both virion entry and cytoplasmic virion envelopment (32–34). The functions of the gK-UL20 protein complex in virion entry and cytoplasmic virion envelopment can be segregated genetically and physically from each other (34–39). Functions of gK-UL20 in virion entry involve direct interactions of the amino termini of both gK and UL20 with the fusogenic viral glycoprotein gB as well as with the membrane fusion regulator glycoprotein gH (35). These glycoproteins regulate fusion of the viral envelope with cellular membranes during virus entry, as well as virus-induced cell-to-cell fusion, i.e., formation of multinucleated cells (syncytia) which allow the virus to spread from cell to cell (40–43).
Here we demonstrate for the first time that the UL37 protein physically interacts with both gK and UL20 proteins in infected cells. Moreover, overexpression of the UL20 protein in FRT cells complements viral replication of the UL37 mutant DC480 virus, which is unable to acquire cytoplasmic envelopes, suggesting that the gK-UL20 interactions with UL37 facilitate cytoplasmic virion envelopment.
MATERIALS AND METHODS
Cell lines.
African green monkey kidney (Vero) cells were obtained from the American Type Culture Collection (Rockville, MD). The Vero-based UL37-complementing cell line BD45 was a gift from Prashant Desai (Johns Hopkins University, Baltimore, MD). The construction and use of the UL20-expressing FRT/UL20 cell line were detailed previously (39). The gK-transformed cell line VK302 was a gift from David C. Johnson (Oregon Health Sciences University, Portland, OR). All cells were maintained in Dulbecco's modified Eagle's medium (Gibco-BRL, Grand Island, NY) supplemented with 10% fetal calf serum and antibiotics.
Construction of HSV-1 mutant viruses.
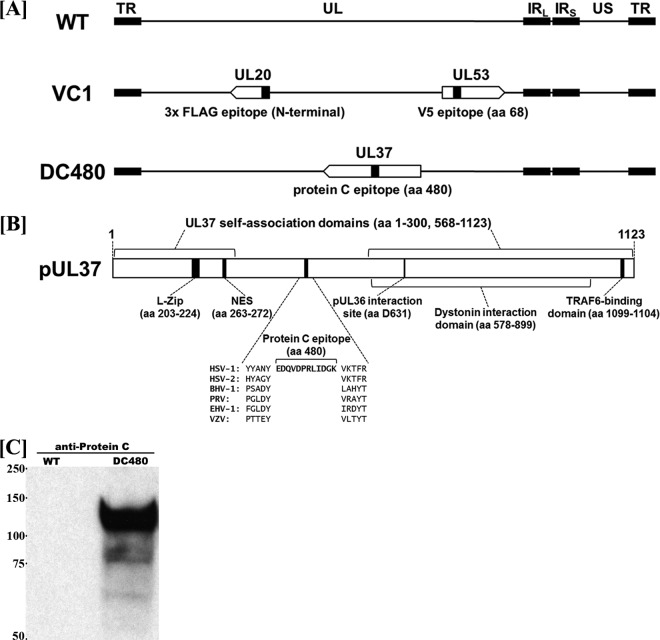
Mutagenesis was accomplished in Escherichia coli by using the markerless two-step Red recombination mutagenesis system and synthetic oligonucleotides (44, 45) (DC480 Forward, GGGTCGAACGTGTTTGGTCTGGCGCGGGAATACGGGTACTATGCCAACTACGATTACAAGGATGACGACGATAAG; and DC480 Reverse, CCGCCCGTGCGTGTGCTCGCTGGCGCCCTGGACCCGCCTGAAAGTTTTTACCTTGCCATCGATCAGTCGCGGATCCACCTGGTCTCGTAGTTGGCATAGTACCCGTACAACCAATTAACCAATTCTGATTAG) implemented on the bacterial artificial chromosome (BAC) plasmid pYEbac102 carrying the HSV-1(F) genome (46) (a gift from Y. Kawaguchi, University of Tokyo, Japan). Construction of the HSV-1 mutant virus ΔUL20 (UL20-null) was described previously (44). The DC480 recombinant virus was constructed by inserting a 12-amino-acid epitope tag (EDQVDPRLIDGK) for protein C (protC tag) into the HSV-1 UL37 gene immediately after the codon for amino acid 480 (Fig. 1). The recombinant mutant virus YE102-VC1 was modified to express gK and UL20 genes containing V5 and 3×FLAG antigenic epitopes, respectively, and was described previously (38). Construction of the HSV-1 UL37-null mutant virus (generously provided by Prashant Desai, Johns Hopkins University, Baltimore, MD) was described previously (17).
FIG 1.
Schematics of recombinant viruses and the UL37 gene. (A) Schematic arrangements of the wild-type (WT) virus and mutant viruses VC1 and DC480, with the unique long (UL) and unique short (US) regions flanked by the terminal repeat (TR) and internal repeat (IR) regions. The VC1 virus expresses UL20 and gK tagged with 3×FLAG and V5 epitope tags, respectively. The DC480 virus contains a 12-amino-acid (aa) protein C epitope tag inserted in frame immediately following amino acid 480. (B) Schematic of the UL37 protein showing its functional domains. (C) Western immunoblot of cell extracts obtained from Vero cells infected with the DC480 virus, using anti-protC antibody. Lane 1, wild-type virus-infected cellular extracts; lane 2, DC480 virus-infected cellular extracts.
Confirmation of targeted mutations, recovery of infectious virus, and plaque morphology analysis.
HSV-1 BAC DNAs were purified from 50-ml overnight BAC cultures by use of a Qiagen large-construct kit (Qiagen, Valencia, CA). All mutated DNA regions were sequenced to verify the presence of the desired mutations in BACs. Similarly, viruses recovered from infected Vero cells were sequenced to confirm the presence of the desired mutations. Viruses were recovered from cells transfected with BACs as described previously (44). Visual analysis of the plaque morphology of mutant viruses was performed as previously described (39, 42, 44, 47).
Viral growth kinetics.
Analysis of viral growth kinetics was performed as we described previously (32, 36). Briefly, nearly confluent Vero cell monolayers were infected with each virus at 4°C for 1 h at a multiplicity of infection (MOI) of 0.2. Thereafter, plates were incubated at 37°C and 5% CO2, and virus was allowed to penetrate for 1 h at 37°C. Any remaining extracellular virus was inactivated by low-pH treatment (pH 3.0), and cells were incubated at 37°C and 5% CO2.
Electron microscopy.
The ultrastructural morphology of virions within infected cells was examined by transmission electron microscopy essentially as described previously (37, 42, 44, 48, 49). All infected cells processed for electron microscopy were prepared at 18 h postinfection (hpi) and visualized on a JEOL transmission electron microscope.
Quantitative PCR (qPCR) analysis of cytoplasmic virions.
Cytoplasmic virions were separated by glycerol shock treatment essentially as originally described by Sarmiento and Batterson (50) and later modified by our laboratory (1). Real-time PCR was carried out on viral DNA from the cytoplasmic fractions of Vero cells. Specifically, the primers and probe (6-carboxytetramethylrhodamine [TAMRA]) for the real-time PCR were designed to detect HSV-1 US6 (gD) (gD forward primer, ACGTACCTGCGGCTCGTGAAGA; probe, 6-carboxyfluorescein [FAM]-GCCAAGGGCTCCTGTAAGTACGCCCT-TAMRA; and gD reverse primer, TCACCCCCTGCTGGTAGGCC). The cytoplasmic suspensions were treated with Turbo DNase I (Ambion, Inc.) for 1 h at 37°C. Viral DNA was extracted using a DNeasy blood and tissue kit (Qiagen, Inc.) per the manufacturer's instructions. Equal volumes of viral DNA were used for TaqMan PCR analysis. Purified HSV-1 bacterial artificial chromosome (YE102) DNA was used to generate the standard curve (1).
Immunoprecipitation and immunoblot assays.
Confluent Vero cells in T75 flasks were infected with the double-tagged recombinant virus YE102-VC1 (gK-V5 and UL20-FLAG) or the UL37-null virus at an MOI of 2. At 24 hpi, the infected cells were lysed with NP-40 cell lysis buffer (Life Technologies) supplemented with protease inhibitor tablets (Roche). The samples were centrifuged at 13,000 rpm for 10 min at 4°C. The supernatants were then used for immunoprecipitation. The proteins from virus-infected cells were immunoprecipitated using protein G magnetic Dynabeads according to the manufacturer's instructions (Invitrogen). Briefly, the beads were bound to their respective antibodies and left on a nutator for 10 min, followed by the addition of cell lysates. The lysate-bead mixture was kept on the nutator for 10 min at room temperature and subsequently washed three times with phosphate-buffered saline (PBS). The protein was eluted from the magnetic beads in 40 μl of elution buffer and used for immunoblot assays. Sample buffer containing 5% β-mercaptoethanol was added to the protein and heated at 55°C for 15 min. Proteins were resolved in a 4 to 20% SDS-PAGE gel and immobilized on nitrocellulose membranes. For Western immunoblots, subconfluent Vero cell monolayers were infected with the indicated virus at an MOI of 3. At 24 hpi, cells were collected by low-speed centrifugation, washed with PBS, and processed as described previously (34, 44). Immunoblot assays were carried out using monoclonal mouse anti-protein C antibody (Abcam, Cambridge, England), monoclonal mouse anti-VP5 antibody (Abcam, Inc., Cambridge, MA), monoclonal mouse anti-FLAG antibody (Sigma-Aldrich, Inc., St. Louis, MO), monoclonal mouse anti-V5 antibody (Invitrogen), horseradish peroxidase (HRP)-conjugated goat anti-mouse antibodies against the light chain (Fab) and heavy chain (Fc) (Abcam, Inc., Cambridge, MA), polyclonal rabbit anti-UL37 antibody (a gift from Frank J. Jenkins, University of Pittsburgh Cancer Institute), and HRP-conjugated goat anti-rabbit antibody (Abcam, Inc., Cambridge, MA).
In situ PLA of protein interactions in virus-infected cells.
Vero cells were grown on 8-well chamber slides (Nunc Lab-Tek II chamber slide system) and infected with F strain virus (untagged) or the tagged virus VC1 (gK-V5 and UL20-FLAG) at an MOI of 10. At 18 h postinfection, the cells were fixed with ice-cold methanol for 10 min at −20°C. After three washes with PBS, the samples were blocked for 2 h at 37°C with Duolink blocking buffer in a humidity chamber. Primary antibodies that were raised in two different species were diluted in antibody diluting buffer, added to the samples, and incubated overnight at 4°C. Mouse anti-V5 antibody (Invitrogen) and rabbit anti-FLAG antibody (Sigma-Aldrich, Inc., St. Louis, MO) were used for gK and UL20 detection, respectively (positive control). Mouse anti-FLAG and rabbit anti-UL37 antibodies were used for the HSV-1(F)-infected (virus with no epitope tags) and VC1-infected cells to detect UL20-UL37 interaction. Mouse anti-V5 and rabbit anti-UL37 antibodies were used for HSV-1(F)- and VC1-infected cells to detect gK-UL37 interaction. Mouse anti-ICP8 (Abcam) and rabbit anti-FLAG were used to detect ICP8-UL20 interaction. Unbound primary antibodies were removed by washing with 1× Tris-buffered saline (TBS)–Tween 20 (0.05%) three times for 5 min each. Duolink Anti-Rabbit Plus and Anti-Mouse Minus in situ proximity ligation assay (PLA) probes were added to the samples (1:5 dilution) and incubated at 37°C for 1 h. After the incubation step, washes were done with Duolink wash buffer A, twice for 5 min each. The ligation stock was diluted 1:5 in high-purity water and added to the wells (40 to 80 μl), and the slides were incubated at 37°C for 30 min. The slides were washed with buffer A three times for 5 min each, amplification solution (40 to 50 μl) was added, and the slides were incubated for 1.5 h at 37°C. The slides were then washed with wash buffer B twice for 10 min each, and once with 0.01% buffer B. The slides were mounted with mounting medium (Duolink II), stored at −20°C, and protected from light until confocal images were taken. The confocal images were taken using a 60× objective on an Olympus Fluoview FV10i confocal laser scanning microscope.
RESULTS
Construction and characterization of UL37 mutant viruses.
The recombinant virus DC480 was constructed by insertion of a protein C (protC) epitope tag immediately after the UL37 gene region encoding amino acid 480 (Fig. 1A). The protC epitope tag was inserted in frame immediately after a conserved tyrosine residue located within the central portion of UL37, distal to known functional domains of UL37 (Fig. 1B). Expression of the UL37-protC-tagged protein was confirmed by Western immunoblot analysis with anti-protC antibody. The UL37-protC construct was detected as a protein species migrating with an apparent electrophoretic mobility of approximately 120 to 130 kDa (Fig. 1C), in agreement with previously published reports (3, 11, 51).
Plaque morphology and replication kinetics of the DC480 recombinant virus.
The DC480 virus produced very small viral plaques on Vero and VK302 cells that were similar in appearance to those produced by the UL20-null virus and comprised, on average, 5 to 10 cells, but it formed nearly wild-type-like viral plaques on BD45 cells expressing the UL37 gene (Fig. 2). The UL37-null virus produced very small viral plaques on Vero, VK302, and FRT cells but formed wild-type-like plaques on BD45 cells. The UL20-null virus formed wild-type-like viral plaques on the FRT cell line expressing the UL20 gene, as expected. Interestingly, the DC480 virus formed plaques on FRT cells that were approximately 5 to 10 times larger than the plaques produced by the same virus on Vero or VK302 cells (Fig. 2). We reported previously that a lack of UL20 gene expression inhibits infectious virus production (39, 48). The DC480 virus replicated substantially less efficiently than the UL20-null virus in Vero cells, producing infectious virion titers that were nearly 4 log lower than those of the wild-type virus at late times postinfection, while the UL20-null titers were approximately 2 1/2 log lower than those of the wild-type virus (Fig. 3A). In contrast, the DC480 virus replicated nearly 20-fold more efficiently in FRT cells than in Vero cells, while the wild-type virus replicated equally efficiently in both FRT and Vero cells (Fig. 3B). The replication efficiency of the UL37-null virus on FRT cells was approximately 20-fold lower than that of the DC480 virus. As expected, the UL20-null virus was able to replicate in FRT cells substantially better than in Vero cells, although final titers remained lower than those of the wild-type virus (Fig. 3A and B).
FIG 2.
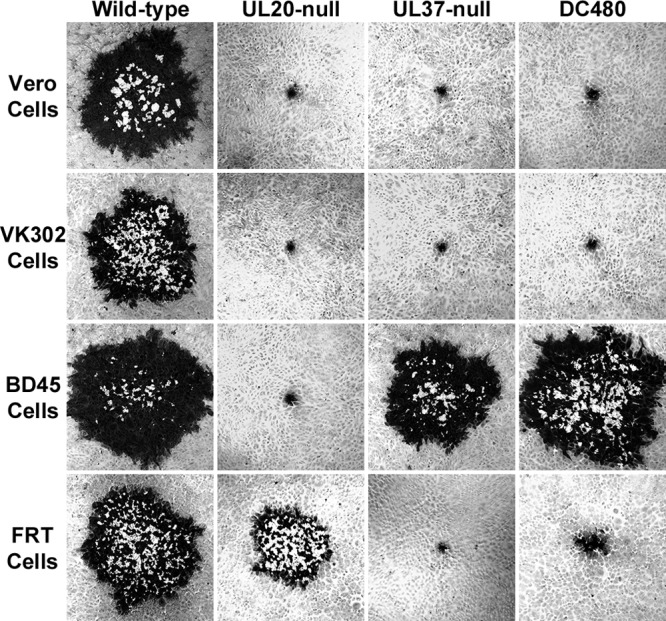
Representative plaque morphologies of wild-type and mutant viruses. Vero, BD45 (UL37 expressing), FRT (UL20 expressing), and VK302 (gK expressing) cell monolayers were infected at an MOI of 0.001, and viral plaques were visualized by immunohistochemistry using polyclonal rabbit anti-HSV-1 antibody at 48 hpi.
FIG 3.
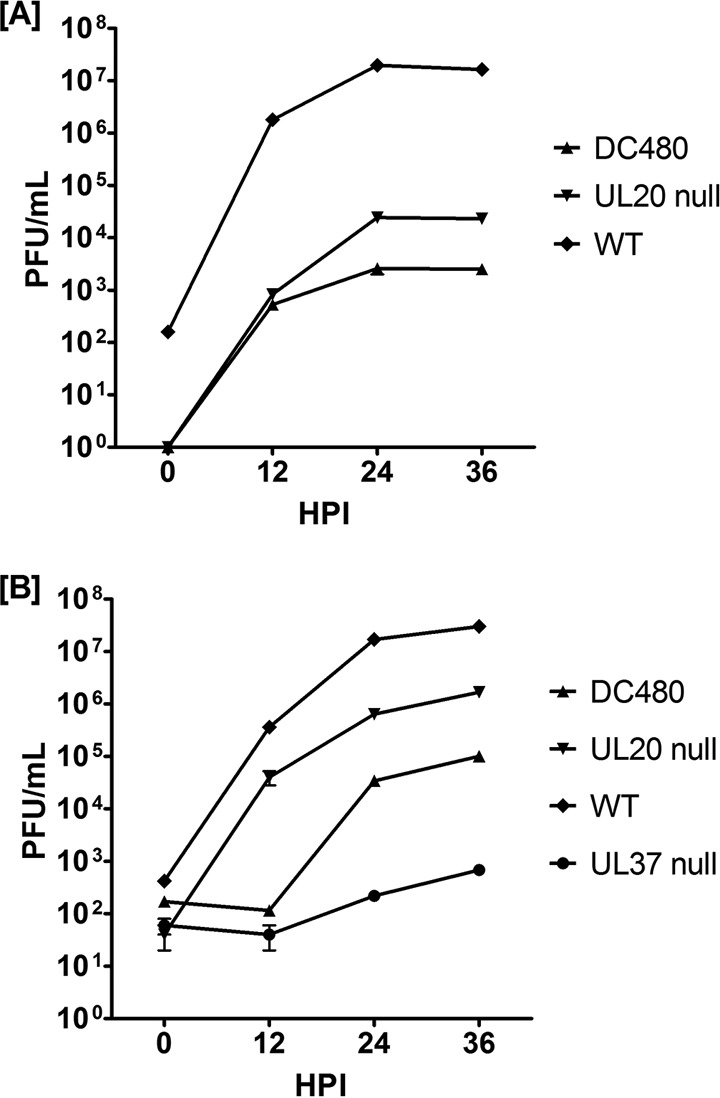
Replication kinetics of wild-type and mutant viruses. Confluent Vero (A) or FRT (B) cell monolayers were infected with each virus shown at an MOI of 0.2. Viral titers were obtained by plaque assay on the appropriate cell lines. The titers obtained were averaged, and the standard error of the mean was calculated for each time point.
Evaluation of envelopment efficiencies of the DC480 virus.
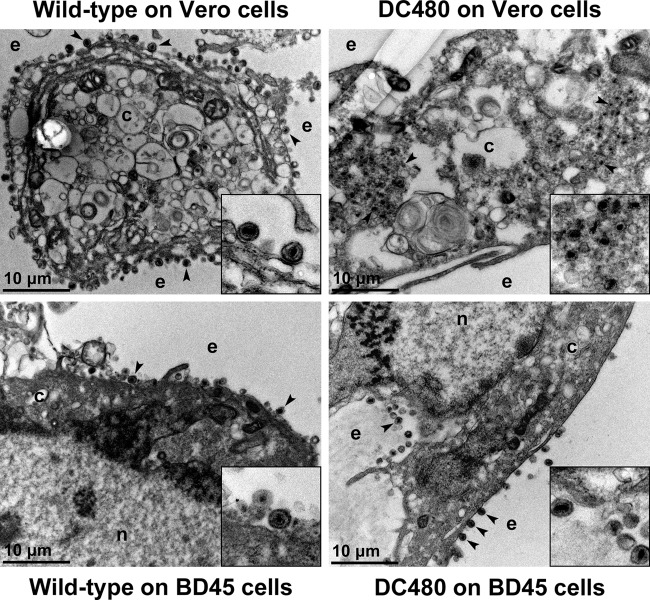
Ultrastructural visualization of infected cells revealed that the DC480 virus exhibited drastic defects in virion envelopment, as evidenced by the accumulation of unenveloped capsids in the cytoplasm of infected Vero cells and the absence of enveloped virions extracellularly, as previously reported for the UL37-null virus (17, 28), in comparison to wild-type virus infection. In contrast, the DC480 virions appeared to be egressing efficiently out of BD45 cells (Fig. 4).
FIG 4.
Ultrastructural morphology of wild-type and mutant viruses. Electron micrographs of Vero or BD45 cells infected at an MOI of 3 with wild-type or DC480 virus and processed for electron microscopy at 18 hpi are shown. Enlarged sections of the micrographs are included as insets. The nucleus (n), cytoplasm (c), and extracellular space (e) are marked. Representative virions are marked with black arrowheads.
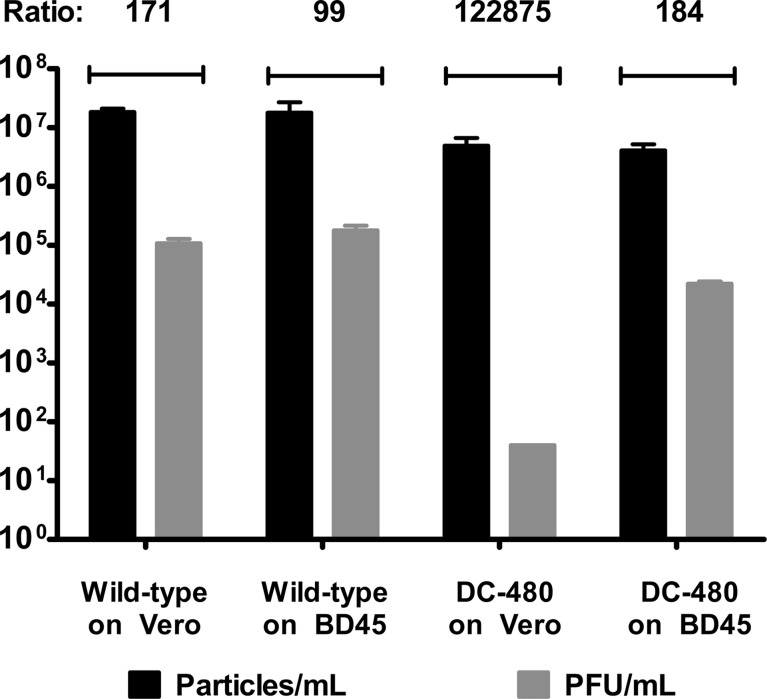
We previously described the use of qPCR for determination of the relative efficiency of viral cytoplasmic envelopment (1). In this assay, the total number of viral genomes found within DNase-resistant viral capsids is determined via qPCR and compared to the number of infectious virions obtained by plaque assay. The wild-type virus was efficiently enveloped and formed into infectious virions in both Vero and BD45 cells at an MOI of 1. In contrast, the viral envelopment and infectious virion production efficiency of the DC480 virus were inhibited >700-fold in Vero cells in comparison to BD45 cells. Ratios of the numbers of encapsidated DC480 viral genomes and PFU were only approximately 2-fold lower than that of the wild-type virus on BD45 cells (Fig. 5).
FIG 5.
Determination of relative efficiencies of virion envelopment of wild-type and mutant viruses in Vero or BD45 cells. The total number of viral genomes in the cytoplasm of infected Vero or BD45 cells at 24 hpi that were protected from DNase I treatment was obtained by qPCR. The total number of intracellular infectious virions was obtained by plaque assay. Ratios reflecting relative efficiencies of envelopment and infectious virion production were obtained by dividing the average number of capsid-protected viral genomes by the number of PFU. Error bars represent standard errors of the means.
Determination of UL37 interactions with gK and UL20.
To evaluate whether the UL37 protein interacts physically with either gK or the UL20 membrane protein, coimmunoprecipitation experiments were performed using the VC1 virus (Fig. 1A), which expresses gK tagged with a V5 epitope inserted in frame within the amino terminus of gK and UL20 tagged with a 3×FLAG epitope inserted in frame after amino acid 4 of the UL20 protein (38). Detection of the UL37 protein was achieved by using a rabbit anti-UL37 polyclonal antibody (3). The UL37 protein was detected as migrating with an apparent molecular mass of 130 kDa (Fig. 6A, arrow). Immunoprecipitation of cellular extracts obtained from wild-type-infected cells by use of the UL37 antibody and subsequent immunoblot detection by the UL37 antibody showed a UL37-specific protein species migrating with an apparent molecular mass of 130 kDa (Fig. 6A). However, this 130-kDa protein species was absent in the lysate and the immunoprecipitates from the UL37-null virus (Fig. 6A). Immunoblot probing of immunoprecipitates of the UL20 protein with the anti-3×FLAG antibody or immunoprecipitates of gK with the anti-V5 antibody readily detected the presence of the UL37 protein migrating at approximately 130 kDa (Fig. 6A). Immunoblot probing of infected lysate extracts with the anti-UL20 (anti-3×FLAG) antibody revealed the presence of UL20 migrating with an apparent molecular mass of approximately 28 kDa (Fig. 6B), as we reported previously (35). The UL20 protein was also readily detected in UL20, gK, and UL37 immunoprecipitates (Fig. 6B). The UL20 protein failed to immunoprecipitate VP5, while VP5 was readily detected in the lysate, migrating with an apparent molecular mass of 150 kDa (Fig. 6C) when probed with anti-VP5 antibody. Immunoblot probing with the anti-gK (anti-V5) antibody readily detected gK in VC1 virus-infected cell extracts (Fig. 6D) as well as gK, UL20, and UL37 immunoprecipitates. Although the apparent molecular mass of gK is approximately 38 kDa (32, 52), larger gK-related protein species were detected in UL20 and UL37 immunoprecipitates, presumably due to gK-UL20-UL37 protein complexes that were not fully resolved to individual protein species (Fig. 6D).
FIG 6.
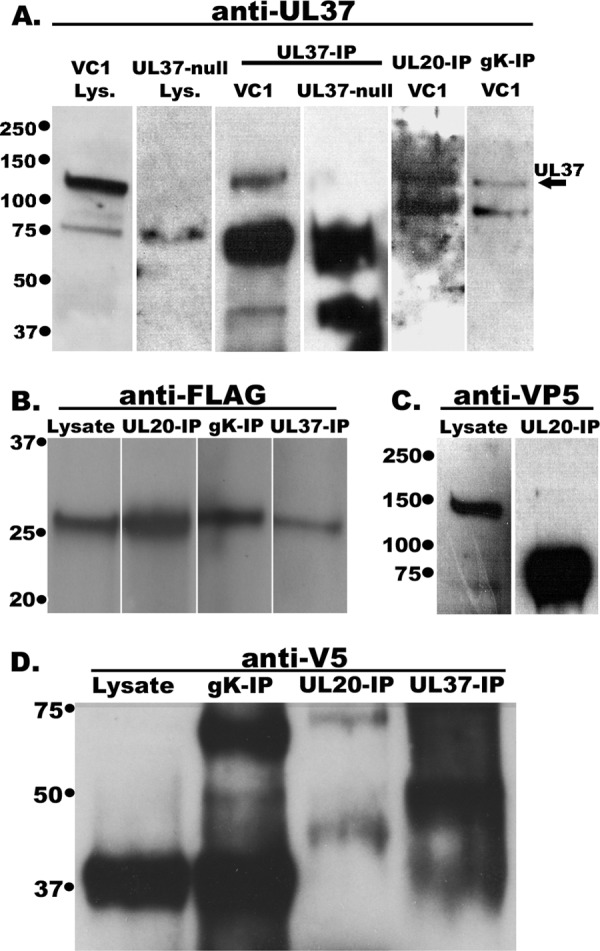
Western immunoblots of infected cell lysates and anti-UL37, anti-gK, and anti-UL20 immunoprecipitates. Vero cells were infected with the double-tagged virus YE102-VC1, expressing gK tagged with the V5 epitope tag and UL20 tagged with the 3×FLAG epitope, or with UL37-null virus. Infected cell lysates were obtained at 24 hpi, and immunoprecipitates were obtained using anti-gK (anti-V5; gK-IP), anti-UL20 (anti-FLAG; UL20-IP), and anti-UL37 (UL37-IP) antibodies in conjunction with magnetic beads. Immunoprecipitates were electrophoretically separated by SDS-PAGE and transferred to nitrocellulose membranes. The blots were probed with rabbit anti-UL37 antibody and HRP-conjugated secondary anti-rabbit antibody (A), mouse anti-FLAG antibody and HRP-conjugated goat anti-mouse antibody against heavy chain (Fc) (B), mouse anti-VP5 antibody and HRP-conjugated goat anti-mouse antibody (C), or mouse anti-V5 antibody and HRP-conjugated goat anti-mouse antibody against light chain (Fab) (D). In panels B, C, and D, lanes labeled “lysate” denote cellular extracts of VC1, and the other lanes represent immunoprecipitated samples from VC1-infected cells.
Determination of UL37 interactions with gK and UL20 by PLA.
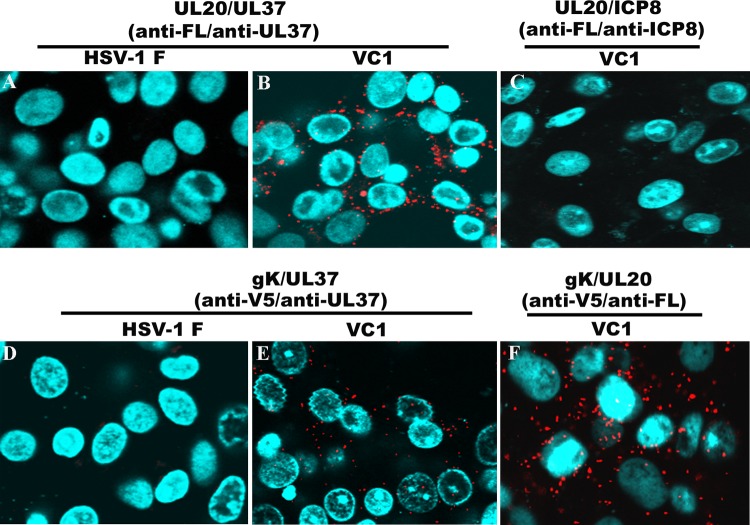
To further validate the observed interactions of the UL37 protein with gK and the UL20 membrane protein, we utilized PLA. In this assay, primary antibodies against UL37, gK (anti-V5), and UL20 (anti-3×FLAG) are first bound to their respective antigens. Species-specific secondary antibodies, called PLA probes, each having a unique short DNA primer covalently attached, are used to bind to the primary antibodies. When the PLA probes are in close proximity, the DNA primers can interact through two other circle-forming oligonucleotides that are added later. Enzymatic ligation of these two fluorescently labeled oligonucleotides followed by polymerase-dependent rolling circle amplification results in the generation of intense fluorescence, which is visualized as distinct bright spots by use of a fluorescence microscope. This assay can determine even transient or weak intracellular interactions (53, 54). PLA readily detected the known gK-UL20 interactions (Fig. 7F). Potential interactions were also detected between UL20 and UL37 (Fig. 7B) and between gK and UL37 (Fig. 7E), while wild-type virus-infected cells did not exhibit fluorescence signals (Fig. 7A and D). The fluorescence signals were also absent in our ICP8-UL20 control, suggesting the absence of potential interaction between these proteins (Fig. 7C).
FIG 7.
PLA to determine UL37 interactions with gK and UL20. PLA was performed using anti-UL37 (rabbit), anti-UL20 (anti-3×FLAG mouse antibody or anti-3×FLAG rabbit antibody), anti-ICP8 (mouse), and anti-gK (anti-V5 mouse antibody) antibodies in conjunction with appropriate secondary antibodies and oligonucleotides as described in Materials and Methods. Fluorescence images of cells infected with wild-type or VC1 virus were recorded using an Olympus confocal microscope at 18 hpi. (A and B) HSV-1(F)- and HSV-1(VC1)-infected Vero cells treated with anti-FLAG (anti-UL20) and anti-UL37 antibodies, respectively. (C) HSV-1(VC1)-infected Vero cells treated with anti-FLAG (anti-UL20) and anti-ICP8 antibodies. (D and E) HSV-1(F)- and HSV-1(VC1)-infected Vero cells treated with anti-V5 (anti-gK) and anti-UL37 antibodies, respectively. (F) HSV-1(VC1)-infected Vero cells treated with anti-V5 (anti-gK) and anti-FLAG (anti-UL20) antibodies.
DISCUSSION
HSV-1 cytoplasmic virion envelopment involves multiple interactions among viral glycoproteins and tegument proteins. The UL37 protein has been shown to be transported, in complex with the UL36 protein and in a capsid-independent manner, to the TGN, where it functions in cytoplasmic virion envelopment. Here we describe the phenotypic and replication properties of the UL37 mutant virus DC480, which contains a 12-amino-acid epitope tag inserted in frame at approximately the middle portion of the UL37 protein. The DC480 virus exhibits drastic defects in cytoplasmic virion envelopment, similar to those of mutant viruses lacking either the gK or UL20 gene. Partial complementation of the UL37 mutant virus in UL20-expressing FRT cells, along with coimmunoprecipitation experiments, revealed that the UL37 protein interacts with gK and UL20, suggesting that these interactions facilitate cytoplasmic virion envelopment.
Functional domains of the UL37 protein.
The UL37 protein contains multiple domains that appear to have distinct functions during virus replication. To facilitate detection of the UL37 protein, we inserted a protC epitope tag within the central portion of the UL37 protein, distal to the amino acid regions that are known to be involved in UL37 self-association, to bind to the UL36 protein, or to contain other known functional domains (Fig. 1). The mutant DC480 virus expressed a full-length UL37 protein that was readily detectable by Western immunoblots using anti-protC antibody. However, this epitope insertion resulted in a severe cytoplasmic envelopment defect similar to that of the UL37-null virus, as evidenced by ultrastructural examination of infected cells showing accumulation of unenveloped capsids in the cytoplasm. This apparent defect in cytoplasmic virion envelopment in the DC480 virus was further supported by highly elevated DNase-resistant particle-to-PFU ratios in comparison to those for wild-type virus infections. Alignment of a large subset of known UL37 amino acid sequences revealed the presence of highly conserved residues among the alphaherpesviruses, as noted previously (55). The protC epitope tag insertion was specifically targeted to be immediately adjacent to a highly conserved tyrosine (Y) residue at amino acid position 480. UL37 is also reported to be phosphorylated in infected cells (10). Reversible phosphorylation can result in conformational changes that can significantly alter the functions of proteins, as evidenced by the regulation of the p53 tumor suppressor protein via phosphorylation (56). Tyrosine sulfation is also a widespread posttranslational modification of eukaryotic proteins that modulate protein-protein interactions (57). Thus, it is possible that the protC epitope insertion alters Y-specific posttranslational modifications of the UL37 protein or alters protein-protein interactions that directly involve the amino acid region immediately adjacent to the conserved Y residue. Alternatively, disruption of this UL37 domain by the protC epitope insertion may change the overall conformation of the protein, thus affecting other functional domains of the UL37 protein. It has been shown that the UL37 protein is phosphorylated by cellular enzymes and that it interacts in the cytoplasm with the major DNA binding protein, ICP8. UL37 interactions with ICP8 enable transport of the UL37 protein to the nuclei of infected cells away from the cytoplasm; therefore, the UL37-ICP8 interactions are not likely to play a role in cytoplasmic virion envelopment (10).
As expected, both the DC480 and UL37-null viruses were efficiently complemented for virus replication and spread on BD45 cells transformed with an HSV-1 gene fragment containing the UL37 gene (17). Surprisingly, the DC480 virus was also complemented for virus replication and virus spread in FRT cells expressing the UL20 gene under the control of its native promoter, while the UL37-null virus was not. The observed DC480 virus complementation for virus growth on UL20-expressing FRT cells strongly suggests direct or indirect physical interactions between the UL37 protein and the UL20-gK protein complex. Coimmunoprecipitation experiments using the available anti-UL37 polyclonal antibody and either gK or UL20, tagged with a V5 or 3×FLAG epitope tag, respectively, showed that the UL37 protein interacted with both gK and UL20 in infected cells, thus providing additional evidence for physical interactions between UL37 and the gK-UL20 protein complex. It has been reported previously that the UL36-UL37 complex migrates to TGN membranes in the absence of capsids (11). There was no VP5 protein detected in UL20 immunoprecipitates, suggesting that the observed interactions between UL20 and gK involved a UL37 protein that was not attached to viral capsids. The UL37-UL36 protein complex may interact with gK-UL20 after its intracellular transport to TGN membranes. Alternatively, the UL36-UL37 protein complex may interact with gK-UL20 in the endoplasmic reticulum and be cotransported to the TGN sites of virion envelopment. Additional experiments are required to distinguish between these two possibilities.
We have shown that the gK-UL20 protein complex plays critical roles in cytoplasmic virion envelopment and egress from infected cells. Our current results as well as previous reports (11, 14–16) clearly indicate that the highly conserved UL37 protein is a crucial determinant for cytoplasmic virion envelopment. Thus, the facts that these two protein complexes (gK-UL20 and UL36-UL37) interact within infected cells and at TGN sites and are involved in cytoplasmic virion envelopment are not surprising. Similar interactions must occur within the virion particle, suggesting that during virion entry these interactions must be regulated to allow the capsid to enter into the cytoplasm, while virion envelopes and their glycoprotein content remain fused with cellular plasma membranes. Interactions of viral glycoproteins, including gK and its partner protein, UL20, with cellular receptors may trigger inhibition of the UL36-UL37 interactions with the gK-UL20 protein complex. Additional work is required to investigate these possibilities.
ACKNOWLEDGMENTS
Part of this work was supported by grant AI43000 from the National Institute of Allergy and Infectious Diseases to K.G.K., by a Louisiana Board of Regents Governor's Biotechnology Initiative grant to K.G.K., and by BioMMED Core Laboratory support by NIH:NIGMS COBRE grant P20GM103458. We acknowledge financial support by the LSU School of Veterinary Medicine.
We thank Y. Xiao and the Socolofsky Microscopy Center for their expert assistance with electron microscopy.
Footnotes
Published ahead of print 5 March 2014
REFERENCES
- 1.Chouljenko DV, Kim IJ, Chouljenko VN, Subramanian R, Walker JD, Kousoulas KG. 2012. Functional hierarchy of herpes simplex virus 1 viral glycoproteins in cytoplasmic virion envelopment and egress. J. Virol. 86:4262–4270. 10.1128/JVI.06766-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maringer K, Stylianou J, Elliott G. 2012. A network of protein interactions around the herpes simplex virus tegument protein VP22. J. Virol. 86:12971–12982. 10.1128/JVI.01913-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitz JB, Albright AG, Kinchington PR, Jenkins FJ. 1995. The UL37 protein of herpes simplex virus type 1 is associated with the tegument of purified virions. Virology 206:1055–1065. 10.1006/viro.1995.1028 [DOI] [PubMed] [Google Scholar]
- 4.Guo H, Shen S, Wang L, Deng H. 2010. Role of tegument proteins in herpesvirus assembly and egress. Protein Cell 1:987–998. 10.1007/s13238-010-0120-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vittone V, Diefenbach E, Triffett D, Douglas MW, Cunningham AL, Diefenbach RJ. 2005. Determination of interactions between tegument proteins of herpes simplex virus type 1. J. Virol. 79:9566–9571. 10.1128/JVI.79.15.9566-9571.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campadelli-Fiume G, Farabegoli F, Di Gaeta S, Roizman B. 1991. Origin of unenveloped capsids in the cytoplasm of cells infected with herpes simplex virus 1. J. Virol. 65:1589–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sugimoto K, Uema M, Sagara H, Tanaka M, Sata T, Hashimoto Y, Kawaguchi Y. 2008. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J. Virol. 82:5198–5211. 10.1128/JVI.02681-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turcotte S, Letellier J, Lippe R. 2005. Herpes simplex virus type 1 capsids transit by the trans-Golgi network, where viral glycoproteins accumulate independently of capsid egress. J. Virol. 79:8847–8860. 10.1128/JVI.79.14.8847-8860.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234. 10.1016/j.virusres.2009.03.018 [DOI] [PubMed] [Google Scholar]
- 10.Albright AG, Jenkins FJ. 1993. The herpes simplex virus UL37 protein is phosphorylated in infected cells. J. Virol. 67:4842–4847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Desai P, Sexton GL, Huang E, Person S. 2008. Localization of herpes simplex virus type 1 UL37 in the Golgi complex requires UL36 but not capsid structures. J. Virol. 82:11354–11361. 10.1128/JVI.00956-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Watanabe D, Ushijima Y, Goshima F, Takakuwa H, Tomita Y, Nishiyama Y. 2000. Identification of nuclear export signal in UL37 protein of herpes simplex virus type 2. Biochem. Biophys. Res. Commun. 276:1248–1254. 10.1006/bbrc.2000.3600 [DOI] [PubMed] [Google Scholar]
- 13.McLauchlan J. 1997. The abundance of the herpes simplex virus type 1 UL37 tegument protein in virus particles is closely controlled. J. Gen. Virol. 78:189–194 [DOI] [PubMed] [Google Scholar]
- 14.Bucks MA, Murphy MA, O'Regan KJ, Courtney RJ. 2011. Identification of interaction domains within the UL37 tegument protein of herpes simplex virus type 1. Virology 416:42–53. 10.1016/j.virol.2011.04.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kelly BJ, Diefenbach E, Fraefel C, Diefenbach RJ. 2012. Identification of host cell proteins which interact with herpes simplex virus type 1 tegument protein pUL37. Biochem. Biophys. Res. Commun. 417:961–965. 10.1016/j.bbrc.2011.12.044 [DOI] [PubMed] [Google Scholar]
- 16.Ko DH, Cunningham AL, Diefenbach RJ. 2010. The major determinant for addition of tegument protein pUL48 (VP16) to capsids in herpes simplex virus type 1 is the presence of the major tegument protein pUL36 (VP1/2). J. Virol. 84:1397–1405. 10.1128/JVI.01721-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desai P, Sexton GL, McCaffery JM, Person S. 2001. A null mutation in the gene encoding the herpes simplex virus type 1 UL37 polypeptide abrogates virus maturation. J. Virol. 75:10259–10271. 10.1128/JVI.75.21.10259-10271.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts AP, Abaitua F, O'Hare P, McNab D, Rixon FJ, Pasdeloup D. 2009. Differing roles of inner tegument proteins pUL36 and pUL37 during entry of herpes simplex virus type 1. J. Virol. 83:105–116. 10.1128/JVI.01032-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leege T, Granzow H, Fuchs W, Klupp BG, Mettenleiter TC. 2009. Phenotypic similarities and differences between UL37-deleted pseudorabies virus and herpes simplex virus type 1. J. Gen. Virol. 90:1560–1568. 10.1099/vir.0.010322-0 [DOI] [PubMed] [Google Scholar]
- 20.Klupp BG, Granzow H, Mundt E, Mettenleiter TC. 2001. Pseudorabies virus UL37 gene product is involved in secondary envelopment. J. Virol. 75:8927–8936. 10.1128/JVI.75.19.8927-8936.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sandbaumhuter M, Dohner K, Schipke J, Binz A, Pohlmann A, Sodeik B, Bauerfeind R. 2013. Cytosolic herpes simplex virus capsids not only require binding inner tegument protein pUL36 but also pUL37 for active transport prior to secondary envelopment. Cell. Microbiol. 15:248–269. 10.1111/cmi.12075 [DOI] [PubMed] [Google Scholar]
- 22.Pasdeloup D, McElwee M, Beilstein F, Labetoulle M, Rixon FJ. 2013. Herpesvirus tegument protein pUL37 interacts with dystonin/BPAG1 to promote capsid transport on microtubules during egress. J. Virol. 87:2857–2867. 10.1128/JVI.02676-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dodding MP, Way M. 2011. Coupling viruses to dynein and kinesin-1. EMBO J. 30:3527–3539. 10.1038/emboj.2011.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luxton GW, Haverlock S, Coller KE, Antinone SE, Pincetic A, Smith GA. 2005. Targeting of herpesvirus capsid transport in axons is coupled to association with specific sets of tegument proteins. Proc. Natl. Acad. Sci. U. S. A. 102:5832–5837. 10.1073/pnas.0500803102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antinone SE, Smith GA. 2010. Retrograde axon transport of herpes simplex virus and pseudorabies virus: a live-cell comparative analysis. J. Virol. 84:1504–1512. 10.1128/JVI.02029-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfstein A, Nagel CH, Radtke K, Dohner K, Allan VJ, Sodeik B. 2006. The inner tegument promotes herpes simplex virus capsid motility along microtubules in vitro. Traffic 7:227–237. 10.1111/j.1600-0854.2005.00379.x [DOI] [PubMed] [Google Scholar]
- 27.Radtke K, Kieneke D, Wolfstein A, Michael K, Steffen W, Scholz T, Karger A, Sodeik B. 2010. Plus- and minus-end directed microtubule motors bind simultaneously to herpes simplex virus capsids using different inner tegument structures. PLoS Pathog. 6:e1000991. 10.1371/journal.ppat.1000991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pasdeloup D, Beilstein F, Roberts AP, McElwee M, McNab D, Rixon FJ. 2010. Inner tegument protein pUL37 of herpes simplex virus type 1 is involved in directing capsids to the trans-Golgi network for envelopment. J. Gen. Virol. 91:2145–2151. 10.1099/vir.0.022053-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pasdeloup D, Blondel D, Isidro AL, Rixon FJ. 2009. Herpesvirus capsid association with the nuclear pore complex and viral DNA release involve the nucleoporin CAN/Nup214 and the capsid protein pUL25. J. Virol. 83:6610–6623. 10.1128/JVI.02655-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JH, Vittone V, Diefenbach E, Cunningham AL, Diefenbach RJ. 2008. Identification of structural protein-protein interactions of herpes simplex virus type 1. Virology 378:347–354. 10.1016/j.virol.2008.05.035 [DOI] [PubMed] [Google Scholar]
- 31.Liu X, Fitzgerald K, Kurt-Jones E, Finberg R, Knipe DM. 2008. Herpesvirus tegument protein activates NF-kappaB signaling through the TRAF6 adaptor protein. Proc. Natl. Acad. Sci. U. S. A. 105:11335–11339. 10.1073/pnas.0801617105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Foster TP, Rybachuk GV, Kousoulas KG. 2001. Glycoprotein K specified by herpes simplex virus type 1 is expressed on virions as a Golgi complex-dependent glycosylated species and functions in virion entry. J. Virol. 75:12431–12438. 10.1128/JVI.75.24.12431-12438.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foster TP, Melancon JM, Olivier TL, Kousoulas KG. 2004. Herpes simplex virus type 1 glycoprotein K and the UL20 protein are interdependent for intracellular trafficking and trans-Golgi network localization. J. Virol. 78:13262–13277. 10.1128/JVI.78.23.13262-13277.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foster TP, Chouljenko VN, Kousoulas KG. 2008. Functional and physical interactions of the herpes simplex virus type 1 UL20 membrane protein with glycoprotein K. J. Virol. 82:6310–6323. 10.1128/JVI.00147-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chouljenko VN, Iyer AV, Chowdhury S, Kim J, Kousoulas KG. 2010. The herpes simplex virus type 1 UL20 protein and the amino terminus of glycoprotein K (gK) physically interact with gB. J. Virol. 84:8596–8606. 10.1128/JVI.00298-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Foster TP, Alvarez X, Kousoulas KG. 2003. Plasma membrane topology of syncytial domains of herpes simplex virus type 1 glycoprotein K (gK): the UL20 protein enables cell surface localization of gK but not gK-mediated cell-to-cell fusion. J. Virol. 77:499–510. 10.1128/JVI.77.1.499-510.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Foster TP, Kousoulas KG. 1999. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J. Virol. 73:8457–8468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jambunathan N, Chowdhury S, Subramanian R, Chouljenko VN, Walker JD, Kousoulas KG. 2011. Site-specific proteolytic cleavage of the amino terminus of herpes simplex virus glycoprotein K on virion particles inhibits virus entry. J. Virol. 85:12910–12918. 10.1128/JVI.06268-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Melancon JM, Foster TP, Kousoulas KG. 2004. Genetic analysis of the herpes simplex virus type 1 UL20 protein domains involved in cytoplasmic virion envelopment and virus-induced cell fusion. J. Virol. 78:7329–7343. 10.1128/JVI.78.14.7329-7343.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eisenberg RJ, Atanasiu D, Cairns TM, Gallagher JR, Krummenacher C, Cohen GH. 2012. Herpes virus fusion and entry: a story with many characters. Viruses 4:800–832. 10.3390/v4050800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Atanasiu D, Saw WT, Cohen GH, Eisenberg RJ. 2010. Cascade of events governing cell-cell fusion induced by herpes simplex virus glycoproteins gD, gH/gL, and gB. J. Virol. 84:12292–12299. 10.1128/JVI.01700-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melancon JM, Luna RE, Foster TP, Kousoulas KG. 2005. Herpes simplex virus type 1 gK is required for gB-mediated virus-induced cell fusion, while neither gB and gK nor gB and UL20p function redundantly in virion de-envelopment. J. Virol. 79:299–313. 10.1128/JVI.79.1.299-313.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Browne H, Bruun B, Minson T. 2001. Plasma membrane requirements for cell fusion induced by herpes simplex virus type 1 glycoproteins gB, gD, gH and gL. J. Gen. Virol. 82:1419–1422 [DOI] [PubMed] [Google Scholar]
- 44.Lee HC, Chouljenko VN, Chouljenko DV, Boudreaux MJ, Kousoulas KG. 2009. The herpes simplex virus type 1 glycoprotein D (gD) cytoplasmic terminus and full-length gE are not essential and do not function in a redundant manner for cytoplasmic virion envelopment and egress. J. Virol. 83:6115–6124. 10.1128/JVI.00128-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. 10.2144/000112096 [DOI] [PubMed] [Google Scholar]
- 46.Tanaka M, Kagawa H, Yamanashi Y, Sata T, Kawaguchi Y. 2003. Construction of an excisable bacterial artificial chromosome containing a full-length infectious clone of herpes simplex virus type 1: viruses reconstituted from the clone exhibit wild-type properties in vitro and in vivo. J. Virol. 77:1382–1391. 10.1128/JVI.77.2.1382-1391.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fulmer PA, Melancon JM, Baines JD, Kousoulas KG. 2007. UL20 protein functions precede and are required for the UL11 functions of herpes simplex virus type 1 cytoplasmic virion envelopment. J. Virol. 81:3097–3108. 10.1128/JVI.02201-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Foster TP, Melancon JM, Baines JD, Kousoulas KG. 2004. The herpes simplex virus type 1 UL20 protein modulates membrane fusion events during cytoplasmic virion morphogenesis and virus-induced cell fusion. J. Virol. 78:5347–5357. 10.1128/JVI.78.10.5347-5357.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jayachandra S, Baghian A, Kousoulas KG. 1997. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J. Virol. 71:5012–5024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sarmiento M, Batterson WW. 1992. Glycerol shock treatment facilitates purification of herpes simplex virus. J. Virol. Methods 36:151–157. 10.1016/0166-0934(92)90146-5 [DOI] [PubMed] [Google Scholar]
- 51.McLauchlan J, Liefkens K, Stow ND. 1994. The herpes simplex virus type 1 UL37 gene product is a component of virus particles. J. Gen. Virol. 75:2047–2052. 10.1099/0022-1317-75-8-2047 [DOI] [PubMed] [Google Scholar]
- 52.Hutchinson L, Goldsmith K, Snoddy D, Ghosh H, Graham FL, Johnson DC. 1992. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J. Virol. 66:5603–5609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Soderberg O, Gullberg M, Jarvius M, Ridderstrale K, Leuchowius KJ, Jarvius J, Wester K, Hydbring P, Bahram F, Larsson LG, Landegren U. 2006. Direct observation of individual endogenous protein complexes in situ by proximity ligation. Nat. Methods 3:995–1000. 10.1038/nmeth947 [DOI] [PubMed] [Google Scholar]
- 54.Jarvius M, Paulsson J, Weibrecht I, Leuchowius KJ, Andersson AC, Wahlby C, Gullberg M, Botling J, Sjoblom T, Markova B, Ostman A, Landegren U, Soderberg O. 2007. In situ detection of phosphorylated platelet-derived growth factor receptor beta using a generalized proximity ligation method. Mol. Cell. Proteomics 6:1500–1509. 10.1074/mcp.M700166-MCP200 [DOI] [PubMed] [Google Scholar]
- 55.Kelly BJ, Mijatov B, Fraefel C, Cunningham AL, Diefenbach RJ. 2012. Identification of a single amino acid residue which is critical for the interaction between HSV-1 inner tegument proteins pUL36 and pUL37. Virology 422:308–316. 10.1016/j.virol.2011.11.002 [DOI] [PubMed] [Google Scholar]
- 56.Ashcroft M, Kubbutat MH, Vousden KH. 1999. Regulation of p53 function and stability by phosphorylation. Mol. Cell. Biol. 19:1751–1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kehoe JW, Bertozzi CR. 2000. Tyrosine sulfation: a modulator of extracellular protein-protein interactions. Chem. Biol. 7:R57–R61. 10.1016/S1074-5521(00)00093-4 [DOI] [PubMed] [Google Scholar]