ABSTRACT
Marek's disease (MD) is a lymphoproliferative disease of chickens caused by the oncogenic Gallid herpesvirus 2, commonly known as Marek's disease virus (MDV). MD vaccines, the primary control method, are often generated by repeated in vitro serial passage of this highly cell-associated virus to attenuate virulent MDV strains. To understand the genetic basis of attenuation, we used experimental evolution by serially passing three virulent MDV replicates generated from an infectious bacterial artificial chromosome (BAC) clone. All replicates became completely or highly attenuated, indicating that de novo mutation, and not selection among quasispecies existing in a strain, is the primary driving force for the reduction in virulence. Sequence analysis of the attenuated replicates revealed 41 to 95 single-nucleotide variants (SNVs) at 2% or higher frequency in each population and several candidate genes containing high-frequency, nonsynonymous mutations. Five candidate mutations were incorporated into recombinant viruses to determine their in vivo effect. SNVs within UL42 (DNA polymerase auxiliary subunit) and UL46 (tegument) had no measurable influence, while two independent mutations in LORF2 (a gene of unknown function) improved survival time of birds but did not alter disease incidence. A fifth SNV located within UL5 (helicase-primase subunit) greatly reduced in vivo viral replication, increased survival time of birds, and resulted in only 0 to 11% disease incidence. This study shows that multiple genes, often within pathways involving DNA replication and transcriptional regulation, are involved in de novo attenuation of MDV and provides targets for the rational design of future MD vaccines.
IMPORTANCE Marek's disease virus (MDV) is a very important pathogen in chickens that costs the worldwide poultry industry $1 billion to $2 billion annually. Marek's disease (MD) vaccines, the primary control method, are often produced by passing virulent strains in cell culture until attenuated. To understand this process, we identified all the changes in the viral genome that occurred during repeated cell passage. We find that a single mutation in the UL5 gene, which encodes a viral protein necessary for DNA replication, reduces disease incidence by 90% or more. In addition, other candidate genes were identified. This information should lead to the development of more effective and rationally designed MD vaccines leading to improved animal health and welfare and lower costs to consumers.
INTRODUCTION
Marek's disease virus (MDV; also called Gallid herpesvirus 2) is an oncogenic alphaherpesvirus that causes Marek's disease (MD) in chickens, which is characterized by T-cell lymphomas, nerve lesions, and death in affected birds. MD costs the worldwide commercial poultry industry $1 billion to $2 billion in losses annually (1); therefore, control of MD is vital for economic viability. Vaccines have been successfully used to control MD since 1970 with the introduction of HVT, a related turkey herpesvirus (2). Since then, additional vaccines with better efficacy have been introduced to combat field strains that evolved to overcome existing vaccinal protection (3). This continuous need to improve MD vaccines is likely driven by the fact that MD vaccines are nonsterilizing and, thus, do not prevent vaccinated birds from becoming infected with virulent field strains that collectively replicate and evolve within the same bird (4). Therefore, new vaccines must be periodically developed and introduced to combat the evolution of more-virulent strains of MDV. Currently, the most protective vaccine, known as Rispens or CVI988, is an attenuated MDV strain developed from in vitro serial passage of a virulent virus until the resulting isolate became avirulent (5).
The process of attenuation via in vitro serial passage has been used to successfully generate candidate vaccines against MD since 1969 (6). Despite this widespread and well-established history of use, the underlying mechanism(s) behind attenuation remains poorly understood. Expansion of 132-bp repeats within the inverted repeat regions flanking the unique long region of the MDV genome has been consistently observed during serial passage of MDV strains and, therefore, was once postulated to be a driving cause for attenuation. Despite the ubiquitous nature of this 132-bp repeat region expansion, this phenomenon is correlated with, but not causal for, attenuation because recombinant viruses completely lacking the 132-bp repeat region are still virulent and able to become attenuated via serial passage in vitro (7). Recently, next-generation sequencing (NGS) has led to a massive expansion in sequencing power, allowing characterization at the molecular genetic level of the complete genomes for many viruses and strains, including serially passed MDV strains such as 648A (8) and several classical MD vaccines, including Rispens and SB-1 (9, 10). This increase in sequencing power allows comparisons for identification of a range of polymorphisms among strains that might account for differences in virulence, and yet connecting genetic variation with phenotypic differences still remains an additional and necessary step to understand attenuation at the genetic level.
To determine the genetic basis of attenuation, viral populations generated from the same infectious Md5 bacterial artificial chromosome (BAC) clone of MDV were serially passed in vitro to generate three attenuated replicates. For each replicate, the lowest attenuated passage was sequenced to ∼250 to 1,000× depth of coverage via NGS. The resulting sequence information allowed us to accurately identify even low-frequency mutations and track their occurrence within the viral populations as they evolved from the virulent parental virus and increased in frequency throughout the population. Furthermore, an additional benefit of using a BAC-derived MDV, besides ensuring initially uniform replicates before passage, is that candidate mutations identified in the serially passed attenuated replicates could be incorporated via Red-mediated recombination into the cloned viral genome, allowing us to generate near-isogenic viruses. These recombinant viruses were then tested in bird trials in order to measure the phenotypic effects of candidate mutations. This approach led to the identification of several mutations that increased survival time of infected birds and, most notably, a mutation within UL5, the helicase-primase subunit, which reduced MDV replication levels in vivo and lessened virulence, resulting in a reduction in disease incidence by over 90%.
MATERIALS AND METHODS
Tissue culture.
Chicken embryo fibroblasts (CEFs) and duck embryo fibroblasts (DEFs) were used to culture viruses. Cultures were maintained in a 1:1 mixture of Leibovitz's L-15 and McCoy's 5A (LM) medium supplemented with fetal bovine serum (FBS), 200 U/ml penicillin, 20 μg/ml streptomycin, and 2 μg/ml amphotericin B in a 37°C, 5% CO2 incubator. Cells were plated with 4% FBS LM medium and maintained in 1% FBS LM medium. For storage as viral stocks, infected cells were suspended in freezing medium composed of 10% dimethyl sulfoxide (DMSO), 45% FBS, and 45% LM medium and kept in liquid nitrogen.
Viruses.
A virulent Md5 BAC-derived virus stock was generated by transfecting Md5B40BAC, an infectious pBeloBAC11 clone that contains the complete MDV Md5 genome (11), into CEFs. Md5B40BAC-derived virus clone 1 (Md5B40BAC-c1) at passage 4 (p4) was used as the parental virus to infect three separate CEF plates with 200 PFU of virus. These three replicates were designated Rep 1 p5, Rep 2 p5, and Rep 3 p5. The original, uncloned Md5 strain p11 was also used to infect an additional plate of CEFs with 200 PFU of the virus and passed as a positive control. Plates containing a confluent monolayer of CEFs in 4% LM medium were infected with the appropriate viral stock, and after 24 h, the medium was changed and cells were maintained in 1% medium. When cytopathic effects developed on the entire monolayer, the viruses were passed by trypsinizing the monolayer and collecting infected cells in 1% LM medium before coculturing infected cells with fresh CEFs. Ten percent of the total cells harvested were transferred to a new plate containing a confluent monolayer of CEFs in 4% LM medium to complete one passage. This process was conducted repeatedly on the three Md5B40BAC-c1 replicates and the Md5 strain for 100 passages. At every 10 passages (i.e., p15, p25, p35, etc., for BAC-derived replicates and p21, p31, p41, etc., for the Md5 strain), viral stocks were saved for in vivo bird trials to determine virulence levels and identify the earliest passage at which attenuation occurred in each replicate.
In vivo virulence trials.
In order to determine virulence, viral stocks were used to challenge ADOL 15I5 × 71 maternal-antibody-negative chicks. A minimum of 17 day-old chicks were challenged intra-abdominally with 500 PFU of the designated virus and housed for 8 weeks in Horsfall-Bauer (HB) units. Moribund birds, or those that survived up to 8 weeks postchallenge, were terminated and examined via necropsy for signs of MD, including tumors and nerve enlargement. All experiments were approved by the USDA Avian Disease and Oncology Laboratory (ADOL) Animal Care and Use Committee (ACUC). The ACUC guidelines established and approved by the ADOL ACUC (April 2005) and the Guide for the Care and Use of Laboratory Animals published by the Institute for Laboratory Animal Research (2011) were followed throughout the experiments.
Virulence trials of mixed virulent and avirulent MDV.
Viral mixtures containing various known quantities of virulent Md5B40BAC-c1 p5 and avirulent ΔMeqBAC, a recombinant virus lacking both copies of the Meq (R-LORF7) oncogene (12), were used to infect birds. Mixtures containing 5, 10, 25, or 50 PFU of the virulent Md5B40BAC-c1 p5 were mixed with ΔMeqBAC to yield a total of 500 PFU of virus to challenge each bird. An additional series of birds were infected with the same quantity of Md5B40BAC-c1 p5 as before (5, 10, 25, and 50 PFU) but without the addition of ΔMeqBAC. Birds were challenged and disease incidence was measured as described previously.
Sequencing of viral stocks.
To identify changes in both the genome and gene expression of the attenuated replicates relative to the virulent progenitor virus, DNA and RNA of the lowest attenuated passage for each replicate, as well as the Md5 strain p11 and Md5B40BAC-c1 p5 parental viruses, were sequenced using the Illumina GAIIx platform (San Diego, CA). DNA from heavily infected plates was extracted and enriched for viral DNA using a micrococcal nuclease protocol (13), while RNA was extracted using the Stratagene Absolutely RNA kit (Agilent Technologies, Santa Clara, CA). Library preparation and sequencing were conducted by the Michigan State University Research Technology and Support Facility (www.rtsf.msu.edu).
Identification of mutations.
After processing, filtering, and trimming of reads based on quality scores, Illumina reads were mapped to the sequenced Md5B40BAC reference genome (HQ149526.1) using BWA (14). Single-nucleotide variants (SNVs) present in attenuated replicates were called using VarScan (15) with a P value of <0.005. SNVs were identified from RNA sequence data according to the same procedure described above, with analysis for differential expression utilizing BWA for mapping, followed by processing with Cufflinks (16).
Tracking SNV kinetics.
Primers were designed to amplify eight regions of the MDV genome containing 16 candidate SNVs identified via sequencing present at >20% in the attenuated replicates (Table 1). Template DNA was obtained from viral stocks of the serially passed replicates saved every 10 passages (from p15 to final attenuated passage of p65, p75, or p85 depending on the replicate). SNV-containing regions were amplified using Phusion High-Fidelity DNA polymerase (New England BioLabs, Ipswich, MA), and the resulting amplicons were barcoded and pooled for Illumina GAIIx sequencing. Sequenced reads were processed as previously described to determine SNV frequency at each passage level in the replicates.
TABLE 1.
Genomic regions and PCR primers used for targeted resequencing of SNVs
| Genomic region | Primer name | Primer sequence | Genome region amplified | SNV nucleotide position: amino acid changea |
|---|---|---|---|---|
| Block C | C-f | CAACTTCGCGGGTATGAATC | LORF2 (MDV012) | 17359: promoter |
| C-r | ACGCTCCCTAGATCGACTCC | 17521: synonymous | ||
| 17545: intronic | ||||
| Block D | D-f | ACGGCCATTTTTAGTTCGTG | UL5 (MDV017) | 23201: I682R |
| D-r | TGCGAAATTAGAAGCCAAC | |||
| Block Pre-E | Pre-E-f | CCGTATGACAGCGAACAAAG | UL41 (MDV054) | 98690: R17stop |
| Pre-E-r | CATCTGGCTAAGCTATGTGCAA | |||
| Block E | E-f | CATTAAACAAACAACAATTGAGC | UL42 (MDV055) and UL43 (MDV056) | 100014: D207G |
| E-r | GCCAAGATGTGAACAACGATT | 101786: A375T | ||
| Block F | F-f | AGAGAAACCAACGCGACAAT | UL46 (MDV059) | 106639: Q117R |
| F-r | ATAATGAAGCGGCTCAGCTC | |||
| Block B/G | G-f | AAATTCGCACTTGAGTGTTGG | Between R-LORF11 and R-LORF12 | 12990/128612: intergenic |
| G-r | TCTCATGAAACATACCCCTTCC | 12986/128616: intergenic | ||
| 12114/129489: intergenic | ||||
| Block I | I-f | TCAATCACATATATGGGTCTCAGG | ICP4 (MDV084/MDV100) | 145864/178590: S1630P |
| I-r | TCGCGTGGTATCACTGATTG | |||
| Block J | J2-f | CAGGTGAGGTGGAAGTAGG | ICP4 (MDV084/MDV100) | 149985/174469: L256S |
| J2-r | CTGTTCATGTCGGAGGTCTG | 150169/174285: T195A | ||
| 150563/173891: Q63H | ||||
| 150567/173887: G62V |
Nucleotide position in Md5B40BAC-c1 reference genome HQ149525.1 followed by amino acid change and its position in protein. SNVs in repeat genes have both equivalent positions listed within internal repeat long (IRL)/terminal repeat long (TRL) or internal repeat short (IRS)/terminal repeat short (TRS) indicated.
Pathway analysis of mutated genes.
The Database for Annotation, Visualization and Integrated Discovery (DAVID) (17, 18) was used to identify gene pathways that were enriched for mutations in the attenuated viruses. A list of genes that contained mutations at 20% or greater frequency in any of the four serially passed replicates (Rep 1 to Rep 3 and Md5 strain) was submitted to DAVID for analysis using the default parameters.
Generation of defined recombinant viruses.
Single point mutations in UL42, UL46, and UL5 and two different mutations within LORF2 (also called MDV012) were incorporated individually into the parental BAC using two-step Red-mediated recombineering (19). Phusion High-Fidelity DNA polymerase (New England BioLabs) and primer sets (Table 2) were used to mutate Md5B40BAC, yielding five independent recombinants: Mut UL42-D207G, Mut UL46-Q117R, and Mut UL5-I682R and two involving LORF2, referred to as LORF2-Promoter and LORF2-Intron. Mutations in the BAC were verified in the bacterial stocks via Sanger sequencing before transfecting purified BAC DNA into DEFs using the calcium phosphate method (20) to create viral stocks, which were again sequenced to confirm the desired mutation.
TABLE 2.
Mutational primers for generation of recombinant viruses
| Recombinant virus | Primer name | Primer sequencea |
|---|---|---|
| Mut UL42-D207G | MDV055 D207G-f | TTGAAAGCTGAAGGAGGTTTTTATGCCGGAACGATTTGTGgTGTGATAAGTTTTGATATAGtagggataacagggtaatcgattt |
| MDV055 D207G-r | TTGGACCATTGCGCTTCCATCTATATCAAAACTTATCACAcCACAAATCGTTCCGGCATAAgccagtgttacaaccaattaacc | |
| Mut UL46-Q117R | MDV059 Q117R-f | ACACCTGCGGTGGTAAAAGACTACTACACAGACTCGTATCgGCGCTATGTCTGTAAGCGGCtagggataacagggtaatcgattt |
| MDV059 Q117R-r | ATCAACACAAGGTATACGCCGCCGCTTACAGACATAGCGCcGATACGAGTCTGTGTAGTAGgccagtgttacaaccaattaacc | |
| Mut UL5- I682R | MDV017 I682R-f | GATAGTTATGTCGATAATGTGAGTTCGAGAGGATGTGAGAgATTCATAAACAACATGCGAGtagggataacagggtaatcgattt |
| MDV017 I682R-r | AGCAAGGGATAACATTCCCCCTCGCATGTTGTTTATGAATcTCTCACATCCTCTCGAACTCgccagtgttacaaccaattaacc | |
| Revt UL5-R682I | MDV017 revertant R682I-f | GATAGTTATGTCGATAATGTGAGTTCGAGAGGATGTGAGAtATTCATAAACAACATGCGAGtagggataacagggtaatcgattt |
| MDV017 revertant R682I-r | AGCAAGGGATAACATTCCCCCTCGCATGTTGTTTATGAATaTCTCACATCCTCTCGAACTCgccagtgttacaaccaattaacc | |
| LORF2-Promoter | Rep2 LORF2/17359-f | TCTCGCACAATCTATGCAGAAACAGATAATCTAGGGTGTGtGCGGTGCTTTGTACTTCCTAtagggataacagggtaatcgattt |
| Rep2 LORF2/17359-r | TTTTTATAACTGATCCGACGTAGGAAGTACAAAGCACCGCaCACACCCTAGATTATCTGTTgccagtgttacaaccaattaacc | |
| LORF2-Intron | Rep3 LORF2/17545-f | TTTCCGTCGTGAATTTGTACGCCAAATTTTACAACGTGAGaTTATATTGTTTGAAATTTCAtagggataacagggtaatcgattt |
| Rep3 LORF2/17545-r | ACGATAAAGATAATATACATTGAAATTTCAAACAATATAAtCTCACGTTGTAAAATTTGGCgccagtgttacaaccaattaacc |
Nucleotides in uppercase denote the portion of the primer designed to target the MDV genome. The desired point mutation to be incorporated is designated by a bold, underlined lowercase letter. Nucleotides in lowercase indicate the portion of the primer annealing to the plasmid template region containing the kanamycin cassette.
In vivo characterization of mutant viruses.
Day-old, 15I5 × 71 maternal-antibody-negative chicks were infected with 500 PFU of the recombinant MDV. Eighteen infected chicks were housed in one HB isolator to measure MD incidence, while an additional 21 infected chicks were housed separately in another HB isolator with six uninfected contact chicks to test for horizontal transmission of recombinant viruses between birds. Additionally, one HB isolator containing uninfected negative-control birds and another containing positive controls challenged with 500 PFU of the parental Md5B40BAC-c1 virus were included for each trial. For each recombinant virus, five birds were sacrificed at 7, 14, and 21 days postinfection (dpi) to collect spleen samples for quantitative PCR (qPCR). All surviving birds after 10 weeks postinfection were euthanized and examined via necropsy. Recombinant viruses that showed altered disease incidence in preliminary trials were used to challenge two more isolators each with a minimum of 17 day-old, 1515 × 71 maternal-antibody-negative chicks with 500 PFU of the virus, as previously described, in order to replicate measures of disease incidence.
In vivo replication of MDV.
DNA extracted from spleens of infected birds was used to assay the in vivo replication levels of serially passed replicates and recombinant viruses relative to control virulent Md5B40BAC-c1 p5. Extracted DNA was used to quantify the relative copy number of MDV genomes present via qPCR using primers for chicken glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and MDV gB with the TaqMan Fast Universal PCR kit (Applied Biosciences, Foster City, CA) as described by Gimeno et al. (21).
RESULTS
Attenuation of serially passed replicates.
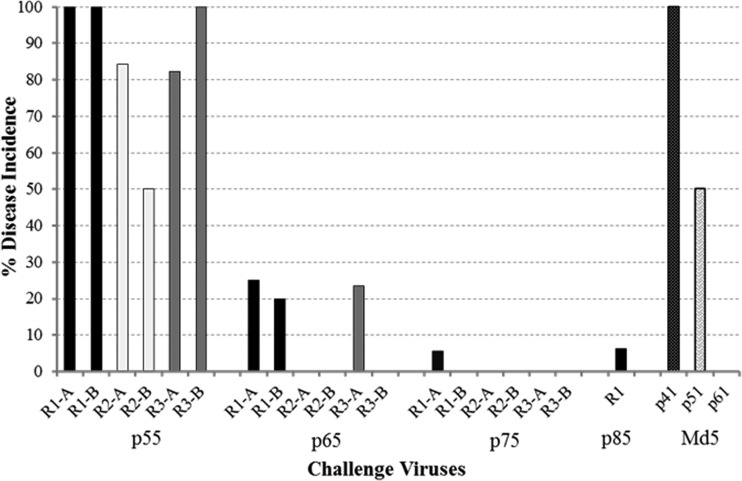
To assess the ability to identify de novo mutations in the MDV genome, three replicates derived from the virulent Md5B40BAC-c1 virus and one from the Md5 parental strain (p11), the strain originally used to clone the MDV genome, were serially passed in vitro. To determine the earliest passage at which viral attenuation occurred, birds were challenged with the passed MDV replicates and MD incidence was measured. As expected, the genetically heterogeneous Md5 p11 strain became attenuated at p61, with no virulence observed after 50 serial passages (Fig. 1). In addition, all passed replicates became attenuated, with complete attenuation occurring at passages 65 and 75 for Rep 2 and Rep 3, respectively, while Rep 1 retained low levels of virulence (6% MD) even at passage 85, indicating that the rate of attenuation is a variable process even among initially identical replicates (Fig. 1).
FIG 1.
Virulence of MDV replicates derived from a serially passed, BAC-cloned virus. Two bird trials (A and B) were conducted for Reps 1, 2, and 3 each at p55, p65, and p75, e.g., R1-A is replicate 1, trial A, at the indicated passages. In addition, the control strain Md5 was tested at p41, p51, and p61.
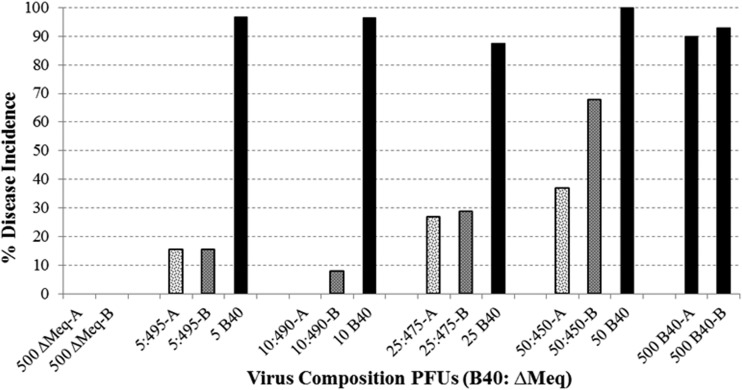
MD incidence in defined mixtures.
Defined mixtures containing known quantities of virulent Md5B40BAC-c1 p5 and avirulent ΔMeqBAC were used to simulate intermediate time points of the serially passed replicates, which were mixed populations composed of both attenuated and virulent viruses at various frequencies. Additionally, PFU of virulent Md5B40BAC-c1 p5 equal to that used in the mixed populations, but without addition of ΔMeqBAC, were used as a comparison to determine if virulence was simply a matter of the raw quantity of virulent MDV used to infect a bird, or if infectivity would be influenced by the presence of additional avirulent ΔMeqBAC virions. Increasing the percentage of virulent virus within a total dose of 500 PFU increased the MD induced by mixtures of Md5B40BAC-c1 and ΔMeqBAC, as expected, but the same pattern did not hold for birds infected with identical levels of Md5B40BAC-c1 alone (Fig. 2). For example, infection of birds with 5 PFU of the virulent Md5B40BAC-c1 along with 495 PFU of the avirulent ΔMeqBAC resulted in ∼15% of the birds developing MD, yet challenge of birds with 5 PFU of Md5B40BAC-c1 by itself resulted in over 90% of birds developing MD (Fig. 2). Thus, our test for virulence is very sensitive, being virtually saturated at 5 PFU of Md5B40BAC-c1. While the quantity of additional avirulent virus reduces disease incidence following challenge with a mixed viral population, we still detect MD, even after infection with a population in which 1% of the PFU derives from virulent MDV. Therefore, mutations that lead to attenuation in our samples should exist in the population at substantial frequencies in order to explain the loss of virulence for the population as a whole.
FIG 2.
Disease incidence in chickens challenged with defined mixtures of virulent and avirulent MDV. Bird trials conducted in duplicate are indicated by “A” and “B,” e.g., 5:495-A was trial A where the inoculation consisted of 5 PFU of B40-derived MDV and 495 PFU of rMd5ΔMeq.
Next-generation sequencing analysis of attenuated replicates.
Based on the results of tests above, the completely attenuated Rep 2 p65, Rep 3 p75, and Md5 strain p61, as well as the >90% attenuated Rep 1 at p75 and p85, were chosen for NGS to identify and quantify mutations in the attenuated viral genomes. The parental Md5B40BAC-c1 p5 virus used to generate the three replicate strains was also sequenced to determine any differences from the previously published Md5B40BAC reference sequence (GenBank accession no. HQ149525.1). This screening also allowed us to identify mutations already present in the progenitor virus prior to passage to eliminate preexisting SNVs from consideration as causative mutations for attenuation. Analysis of the data identified six SNVs fixed at 100% in the sequenced Md5B40BAC-c1 p5 viral stocks that differed from the reference. Due to the presence of these variations in the progenitor stock and, consequently, all resulting BAC-derived replicates, these SNVs were excluded from further analysis. While the number of SNVs identified in the sequenced viruses varied among replicates, 19 to 68% of called SNVs were present at frequencies of less than 2% in the viral populations (Table 3). To screen for the optimal candidate mutations involved in attenuation and eliminate false SNV calls due to sequencing error, mutations present at less than 2% were excluded from further analysis; the standard error of the frequency estimate was 0.9% and 0.4% for 250× and 1,000× depths of coverage, respectively. The total number remaining ranged from 41 to 95 SNVs, depending on the replicate (Table 4). These SNVs occurred within both coding and noncoding regions of the MDV genome, and of those within coding regions, over 60% were nonsynonymous mutations.
TABLE 3.
Mutations in sequenced viral stocks
| Mutation frequency in total viral population (%) | No. of SNVs in virus: |
||||
|---|---|---|---|---|---|
| Md5B40BAC-c1 p5 | Rep 1 p75 | Rep 1 p85 | Rep 2 p65 | Rep 3 p75 | |
| ≤2 | 33 | 23 | 21 | 16 | 36 |
| 3–10 | 3 | 53 | 55 | 23 | 45 |
| 11–20 | 2 | 9 | 11 | 4 | 7 |
| 21–30 | 1 | 4 | 3 | 4 | 1 |
| 31–40 | 2 | 2 | 1 | 2 | 0 |
| 41–50 | 0 | 4 | 4 | 1 | 4 |
| 51–60 | 0 | 4 | 5 | 3 | 0 |
| 61–70 | 1 | 6 | 2 | 1 | 3 |
| 71–80 | 1 | 6 | 1 | 0 | 1 |
| 81–90 | 0 | 5 | 1 | 2 | 3 |
| 91–100 | 6 | 2 | 2 | 1 | 3 |
| Total no. of mutations identified | 49a | 118 | 106 | 57 | 103 |
Six mutations in Md5B40BAC-c1 p5 were fixed at 100% in our viral stocks relative to the reference Md5B40BAC sequence. Considering that these mutations were present in the parental virus preparations, and therefore all serially passed replicates, these mutations were not considered in any further SNV analysis in the attenuated replicates.
TABLE 4.
SNVs identified in serially passed Md5B40BAC replicates
| Virus | No. of SNVs |
% nonsynonymous mutations | ||||
|---|---|---|---|---|---|---|
| >2% | Noncoding | Coding | Synonymous | Nonsynonymous | ||
| Rep 1 p75 | 95 | 67 | 28 | 7 | 21 | 75 |
| Rep 1 p85 | 85 | 57 | 28 | 10 | 18 | 64 |
| Rep 2 p65 | 41 | 22 | 19 | 7 | 12 | 63 |
| Rep 3 p75 | 67 | 43 | 24 | 5 | 19 | 79 |
Among attenuated replicates, eight identical nucleotide changes were present at frequencies greater than 2% in at least two of the four attenuated viruses (data not shown). Five of the eight identical nucleotide changes were instances in which one viral replicate had the mutation at a high frequency while other replicates containing the same mutation were at low frequencies, <10%. Only three mutations were present at moderate or high frequencies in two or more of the attenuated viral replicates. One mutation at moderate frequencies in two replicates was present at nucleotide 171661, which is located downstream of SORF2a. This mutation occurred at frequencies of 22% and 27% in Rep 1 and Rep 3, respectively. Of the two high-frequency mutations, one was identified within the three regions of the a-like sequence at positions 897, 140705, and 183749, occurring at rates between 50 and 89% in the four attenuated replicates, while the final point mutation at nucleotide 100014 in UL42 was found at rates of 84% and 85% in Rep 1 and Md5 p61, respectively. Due to the logistical complexity of incorporating mutations within the three a-like regions present within herpesvirus genomes, that mutation was not considered for creation as a recombinant virus, while the mutation within UL42 was deemed an excellent candidate.
Besides SNV mutations, we looked for changes in gene copy number variation reflected by differences in the depths of sequenced coverage over the MDV genome of attenuated replicates versus virulent virus. Previous studies of the MDV 132-bp repeats have shown this region to greatly expand in copy number during serial passage, although this increase in copy number is not causative for the loss of virulence (7). In the virulent Md5B40BAC-c1 p5 virus before passage, the depth of coverage across the entire MDV genome was fairly consistent, with the only exception being a larger number of reads mapping to the inverted repeat region containing the a-like sequences and telomeric repeat regions (mTMR) (Table 5). This expansion involving the repetitive a-like sequence and mTMR was also present in the three attenuated replicates. After serial passage, a 7- to 58-fold increase in depth of coverage corresponding to the 132-bp repeat regions was observed in the attenuated viruses, as would be expected (Table 5).
TABLE 5.
Depth of coverage for sequenced viruses
| MDV region | Fold depth of coverage |
||||||
|---|---|---|---|---|---|---|---|
| Md5B40BAC-c1 | Rep 1 p75 | Rep 1 p85 | Rep 2 p65 | Rep 3 p75 | Md5 p11 | Md5 p61 | |
| Whole genome | 246 | 1,011 | 909 | 357 | 600 | 406 | 213 |
| 132 bp in TRL | 200 | 11,855 | 11,460 | 5,796 | 3,549 | 365 | 2,556 |
| 132 bp in IRL | 208 | 11,892 | 11,505 | 5,817 | 3,542 | 353 | 2,578 |
| a-like sequence | 796 | 2,390 | 3,256 | 3,667 | 2,358 | 2,246 | 2,259 |
| 132-bp repeats | 204 | 11,874 | 11,482 | 5,806 | 3,546 | 359 | 2,567 |
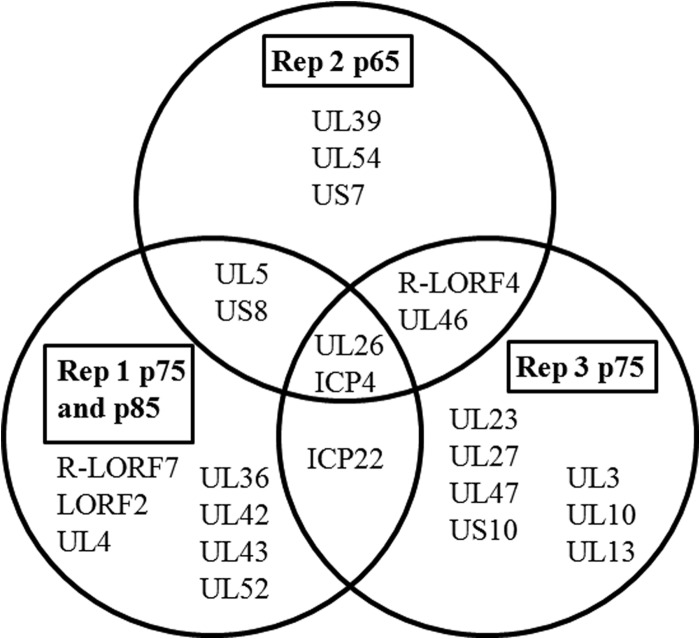
Candidate attenuation-causal genes were identified by screening for genes mutated more than once among the serially passed populations. Seventy-four percent of genes containing nonsynonymous mutations were mutated exclusively within only one of the three Md5B40BAC-c1 replicates (Fig. 3). Five genes had nonsynonymous mutations in two of the three replicates: UL5 (helicase-primase helicase subunit), US8 (gE), US1 (ICP22), R-LORF4 (unknown function), and UL46 (VP11/VP12). There were only two genes with missense mutations in all three replicates: UL26 (VP24), which had an identical but low-frequency (<5%) mutation in all three replicates at nucleotide 59317, and RS1 (ICP4). A third gene, LORF2, also contained unique mutations within all three BAC-derived replicates, although these mutations were not all nonsynonymous changes affecting protein sequence. Generally, mutated genes contained only one to two unique polymorphisms within a gene per replicate. Only ICP4 contained multiple, nonsynonymous mutations in all replicates (Table 6). In the completely attenuated Rep 2 and Rep 3, both replicates had multiple, high-frequency, nonsynonymous ICP4 SNVs such as G62V (Rep 2, 83.8%), T195A (Rep 2, 39.8%), Q63H (Rep 3, 100%), L256S (Rep 3, 65.9%), and S1630P (Rep 3, 85.7%). These mutations suggest ICP4 as a top candidate gene. The three Md5B40BAC-c1 replicates were not the only attenuated viruses containing high-frequency mutations within ICP4. The serially passed Md5 strain also possessed the SNV Y60C at 93%. All fully attenuated virus stocks except Rep 1 contained a high-frequency SNV in ICP4 within bases that encode amino acids 60 to 63, including the completely fixed mutation in Rep 3 encoding amino acid 62, suggesting that mutations within this region in particular may play an important role in attenuation.
FIG 3.
Genes with nonsynonymous mutations within attenuated MDV replicates. Serially passed MDV replicates were compared to identify commonly mutated genes shared among attenuated replicates.
TABLE 6.
ICP4 mutations in attenuated viruses
| Virus | Amino acid change | % frequency |
|---|---|---|
| Rep 1 p75 | L47P | 2.02 |
| I228V | 2.16 | |
| C844R | 7.13 | |
| L2163P | 4.71 | |
| Rep 1 p85 | L47P | 2.99 |
| I228V | 3.70 | |
| C844R | 9.26 | |
| L2163P | 9.25 | |
| Rep 2 p65 | G62V | 83.76 |
| T195A | 39.76 | |
| Rep 3 p75 | Q63H | 100.00 |
| L256S | 65.94 | |
| Y271C | 3.14 | |
| D1483V | 2.22 | |
| S1630P | 85.70 | |
| Md5 strain p61 | Y60C | 96.70 |
| Y271C | 6.87 | |
| 1631a | 8.14 | |
| H1670R | 2.77 |
Indicates synonymous mutation.
Following sequencing of DNA to identify mutations in the attenuated replicates, SNVs were also called in the sequenced RNA isolated from infected plaques grown on CEFs. While SNVs were first called in genomic DNA data, those mutations may not necessarily be expressed in mRNA, so the frequency of SNVs in DNA was compared to their frequency in RNA. Frequencies of mutations first identified in DNA were extremely consistent and comparable to their frequencies in RNA (Table 7). On the whole, SNV frequencies in RNA did not differ by more than 5% from their measured frequency called in DNA. Of the top candidate mutations identified, the sole exception to this trend was LORF2 in Rep 2 p65, which had the greatest difference in SNV frequency between RNA and DNA, 8.8%. This provides confirmation of the accuracy of the sequencing and variant calling, as well as indicating that there was no apparent preferential expression of wild-type or mutated alleles within a viral population.
TABLE 7.
Top candidate mutation frequencies in DNA versus RNA
| Mutation location | Mutation | DNA (%) | RNA (%) |
|---|---|---|---|
| UL5, Rep 2 p65 | I682R | 66.7 | 71.0 |
| UL42, Rep 1 p85 | D207G | 84.7 | 83.3 |
| UL46, Rep 2 p65 | Q117R | 99.5 | 98.4 |
| LORF2, Rep 2 p65 | 17359 | 56.0 | 64.8 |
| LORF2, Rep 3 p75 | 17545 | 94.8 | 97.3 |
| ICP4, Rep 2 p65 | T195A | 39.4 | 41.0 |
| ICP4, Rep 2 p65 | G62V | 83.8 | 81.3 |
| ICP4, Rep 3 p75 | S1630P | 85.7 | 81.4 |
| ICP4, Rep 3 p75 | L256S | 65.9 | 63.8 |
| ICP4, Rep 3 p75 | Q63H | 100 | 100 |
RNA data were then analyzed to determine if there was differential gene expression between attenuated and virulent viruses. Comparing each attenuated replicate to the virulent parental virus identified 5 to 14 genes that had significantly differential expression (Table 8). Analyzing genes that are differentially expressed among the attenuated replicates showed that one gene, UL45 (envelopment protein), was differentially expressed in all attenuated replicates with at least a 1.5- to 2.9-fold increase in all attenuated viruses. In addition to UL45, R-LORF2 (viral interleukin-8 [vIL-8]) was also differentially expressed in all replicates, although for Rep 1 there was differential expression only at p85. Despite the differential expression of these genes, there were no mutations within coding regions or nearby promoter regions of the genes themselves that occurred at frequencies exceeding 20%.
TABLE 8.
Significantly differentially expressed genes in attenuated MDV replicates versus virulent MDV
| Virus and passage | Genea | Fold change | P value |
|---|---|---|---|
| Rep 1 p75 | LORF3 | 1.01 | 2.68E−03 |
| UL8 | −1.06 | 1.80E−03 | |
| UL26.5 | −1.43 | 4.57E−04 | |
| UL40 | −1.33 | 2.64E−03 | |
| UL44 | 1.91 | 6.37E−13 | |
| UL45* | 2.89 | 1.16E−07 | |
| UL47 | 1.52 | 2.22E−04 | |
| Rep 1 p85 | LORF3 | 0.80 | 7.50E−03 |
| UL5 | −0.90 | 7.63E−03 | |
| UL10 | 1.22 | 1.59E−03 | |
| UL26.5 | −1.19 | 1.79E−03 | |
| UL33 | −0.94 | 6.00E−03 | |
| UL39 | −1.42 | 1.19E−03 | |
| UL40 | −1.29 | 6.37E−04 | |
| UL44* | 2.16 | 3.75E−13 | |
| UL45* | 2.83 | 1.06E−10 | |
| UL47 | 1.37 | 1.04E−04 | |
| R-LORF2* | 2.16 | 1.42E−05 | |
| US1 | −1.86 | 1.07E−06 | |
| US10 | −1.79 | 1.88E−03 | |
| US2 | −1.24 | 4.80E−04 | |
| Rep 2 p65 | R-LORF2* | 2.07 | 2.29E−07 |
| UL45 | 1.52 | 2.50E−04 | |
| US1* | −2.78 | 8.89E−12 | |
| US10* | −2.81 | 0.00E+00 | |
| US2 | −1.55 | 1.19E−04 | |
| Rep 3 p75 | R-LORF2 | 1.95 | 2.79E−98 |
| UL10 | 1.09 | 5.73E−04 | |
| UL45* | 2.00 | 1.77E−10 | |
| UL47 | 1.14 | 3.57E−04 | |
| SORF3 | 1.25 | 4.99E−04 | |
| R-LORF4 | 0.76 | 3.09E−03 |
Genes marked with an asterisk display more than a 2-fold significant difference in expression between attenuated and virulent viruses.
Analysis for gene pathways that were enriched for mutations among the attenuated replicates using DAVID found only one annotated cluster of functional genes that was enriched (DAVID enrichment score, 0.56) for the attenuated replicates. This cluster contained genes involved in transcriptional regulation and regulation of RNA metabolic processes (GO:0006355 and GO:0051252), including the genes UL46 and ICP4, previously identified as candidate genes for attenuation.
Candidate mutations and characterization of recombinant viruses.
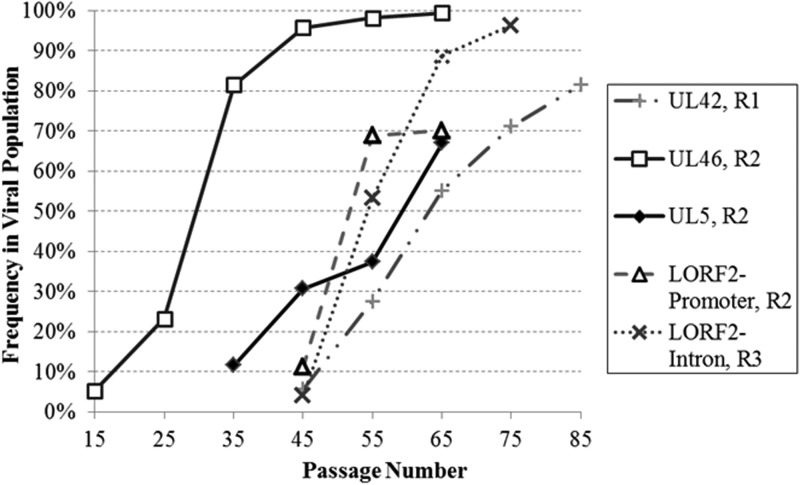
Given that sequence analysis identified only a modest number of nonsynonymous mutations in genes and the fact that the isolates were either completely or very highly attenuated, we further narrowed our focus to those mutations whose frequency exceeded 20% in the attenuated populations. We conducted targeted resequencing of 16 candidate SNVs identified in the attenuated replicates at consecutive 10-passage intervals from p15 until attenuation. This allowed us to estimate when these mutations first occurred and correlate increases in candidate SNV frequencies with decreasing virulence in vivo. Mutations that were at high frequencies in the final, attenuated population and whose increase in frequency occurred roughly around passages 55 to 65, when a drop in virulence of the serially passed populations was most predominant when tested in vivo (Fig. 1), were identified as top candidates, particularly if they were nonsynonymous mutations (Fig. 4). Of the candidate SNVs included in targeted resequencing, five mutations were picked as the top candidates to first test as recombinant viruses due to their location within either the unique long or the unique short region of the MDV genome (Fig. 4). Based on mutation frequencies and a concentration of high-frequency nonsynonymous mutations clustered around amino acids 60 to 63 in ICP4, ICP4 appears to be a top candidate for involvement in attenuation. However, ICP4 was not included among the five mutations initially tested using recombinant viruses due to its location within the repeat regions of the herpesvirus genome, as additional recombineering steps would have been required for mutating both copies of ICP4, instead of a single mutation necessary for genes located in unique regions. Despite this complication, characterization of ICP4 mutations via recombinant viruses is under way.
FIG 4.
Kinetics of candidate single-nucleotide polymorphism frequencies over serial passage. Targeted resequencing of candidate single-nucleotide polymorphisms allowed for the frequency of each mutation to be determined from the earliest passage in which the mutation occurred until attenuation.
These five mutations within four genes were incorporated into Md5B40BAC to generate the following recombinant viruses: Mut UL42-D207G, Mut UL46-Q117R, Mut UL5-I682R, Mut LORF2-Promoter, and Mut LORF2-Intron. The first three recombinant virus mutations altered amino acids within the double-stranded DNA (dsDNA)-binding protein/DNA polymerase processivity subunit, the VP11/VP12 tegument protein, and the helicase-primase subunit, respectively, while the two final mutations both are noncoding mutations that may affect expression of a fourth gene of unknown function, known as LORF2.
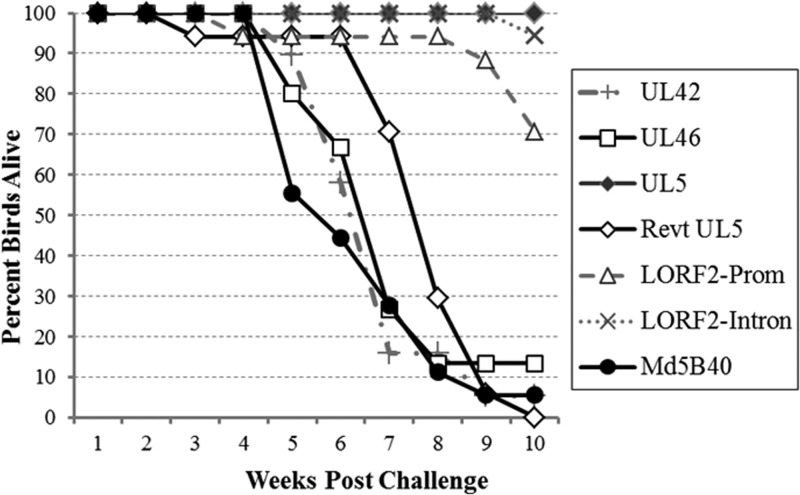
In vivo bird trials of the five recombinant viruses indicated that two of the mutations, UL42-D207G and UL46-Q117R, had no effect on survival or MD incidence (Fig. 5 and Table 9). Three recombinant viruses, Mut LORF2-Promoter, Mut LORF2-Intron, and Mut UL5-I682R, all increased length of survival in birds challenged with the recombinant viruses relative to the virulent Md5B40BAC-c1, but upon termination and examination via necropsy, both LORF2 mutants exhibited 100% MD incidence, although, interestingly, Mut LORF2-Intron did not appear to transmit horizontally. Only Mut UL5-I682R not only influenced survival time but also resulted in an almost complete reduction in virulence. Birds infected with Mut UL5-I682R survived to the end of the 10-week experiment, and upon termination, only 11% of infected birds developed any symptoms of MD, in comparison to 100% of control Md5B40BAC-c1-infected birds (Table 9). Further independent trials measuring disease incidence of Mut UL5-I682R resulted in 0% (0/17 birds) and 6% (1/18 birds) of birds developing MD.
FIG 5.
Survival of birds challenged with recombinant MDVs. Mortality due to MD over the course of the 10 weeks was determined for the five recombinant viruses and the virulent B40-derived MDV positive control.
TABLE 9.
MD incidence due to candidate recombinant viruses
| Virus | No. of MD+ birds/total no. of birds in group: |
|
|---|---|---|
| Challenged | Contact | |
| Mut UL42 | 20/20 | 6/6 |
| Mut UL46 | 15/15 | 5/6 |
| Mut UL5 | 2/18 | 0/6 |
| Revt UL5 | 17/17 | 5/5 |
| LORF2-Promoter | 17/17 | 5/5 |
| LORF2-Intron | 19/19 | 0/4 |
| Md5B40-c1 | 18/18 | 3/6 |
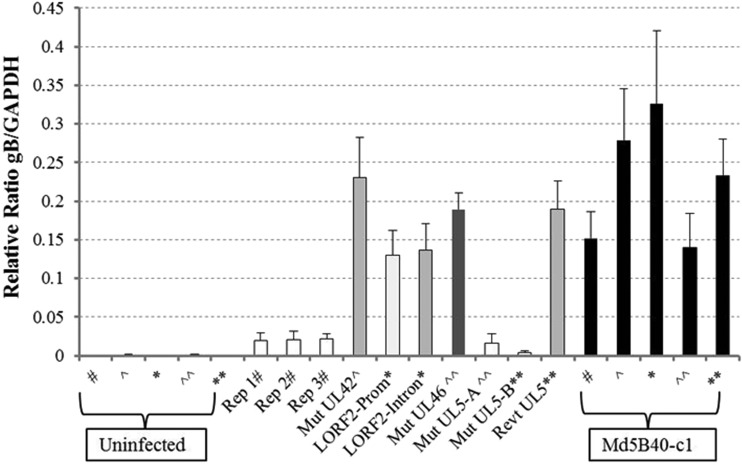
To determine if attenuated replicates and recombinant viruses retained the ability to replicate in vivo or if loss of oncogenicity resulted from loss of viral replication in vivo, splenic DNA from infected birds was extracted to measure MDV replication in birds using qPCR ratios comparing copy numbers of viral gB DNA versus copies of chicken GAPDH genes averaged over the three collection time points (Fig. 6). The average relative number of gB/GAPDH copies in Md5B40BAC-c1 ranged from 0.14 to 0.33, based on four separate bird trials. The three serially passed Md5B40BAC-c1 replicates exhibited about a 10-fold reduction in viral replication, while the recombinant Mut UL5-I682R also had a ratio comparable to those of these attenuated viruses of 0.016 (Fig. 6). The two virulent recombinant viruses, Mut UL42-D207G and Mut UL46-Q117R, had ratios comparable to that of the virulent virus (Fig. 6). The LORF2 mutations, which resulted in greater survival for challenged birds yet still exhibited 100% MD incidence, exhibited values of 0.13 to 0.14, which were toward the lower range of values observed with Md5B40BAC-c1. Virulent viruses, such as Md5B40BAC-c1, Mut UL42-D207G, Mut UL46-Q117R, and both Mut LORF2 recombinants, all exhibited significantly higher levels of MDV than did the three serially passed attenuated viruses or Mut UL5-I682R. While attenuated viruses had low levels of replication compared to virulent viruses, their levels of replication were higher than the background of uninfected controls, indicating that they still replicated in vivo. Not only was a low level of MDV detectable via qPCR in attenuated viruses, but viable viruses were able to be reisolated from peripheral blood lymphocytes of infected, but MDV-negative, birds challenged with the attenuated viruses when plated on DEFs to produce plaques (data not shown), further supporting the idea that these attenuated viruses replicated in vivo.
FIG 6.
Comparison of in vivo replication rates for attenuated replicates and recombinant viruses. Due to housing and chick availability, recombinant viruses were tested incrementally; therefore, positive (Md5B40-c1) and negative (uninfected) controls are matched to the respective recombinant viruses tested collectively by matched symbols (i.e., *, ∧, #, etc.). Trials for Mut UL5 were conducted in duplicate, and each trial indicated is by “A” or “B.”
Based on the dramatic decrease in virulence due to the UL5 mutation, a revertant of Mut UL5-I682R called Revt UL5-R682I was created. This revertant virus not only replicated at high levels in vivo but also restored levels of virulence comparable to that of the wild-type, Md5B40BAC-c1 virus (Fig. 6 and Table 9).
DISCUSSION
This work shows that attenuation of a genetically homogeneous MDV reproducibly occurs after repeated serial passage in vitro, indicating that de novo mutations arising during passage are sufficient to generate new, avirulent viral populations. A variety of mutations were identified within each attenuated MDV replicate relative to the parental virulent virus, of which several candidate mutations were found within genes of interest, including ICP4 (transcriptional transactivator), UL42 (DNA polymerase subunit), UL46 (VP11/VP12 tegument protein), UL5 (helicase-primase subunit), and LORF2 (unknown function). In particular, no single mutation rose to high frequency in every attenuated replicate, nor was any single gene mutated at high frequencies in every replicate. Two genes, ICP4 and UL26, were mutated in all attenuated replicates, but at least in Rep 1, the frequencies of the respective mutations suggest that they are not solely responsible for attenuation. Furthermore, we experimentally identified individual mutations in UL5 and, to a lesser extent, in LORF2, which resulted in a phenotypic change that contributed to attenuation, while mutations within UL5 have been shown to result in a reduction in virulence in other herpesviruses, suggesting that UL5 is a factor in virulence of herpesviruses (22, 23).
While attenuation of a virulent virus via serial passage in vitro is not a new or unusual phenomenon unique to MDV, the use of virus generated from a BAC-cloned MDV in this study allowed for a more detailed insight into the mechanism(s) of attenuation than that in previous studies (6, 8). When trying to understand the driving mechanism that causes a virulent viral population to become avirulent, the two most probable explanations are selection and mutation. Like many viruses, MDV strains, even those that are plaque purified, are often described as quasispecies composed of genetically diverse genotypes existing within a strain population (8, 11). Thus, while the viral strain may be described as a virulent, there can be unique subpopulations that do not share the same degree of virulence compared to the whole population. Therefore, viral attenuation could be driven by selection for preexisting variation of low-frequency, avirulent genotypes within a strain. A second explanation proposes that de novo mutations that occur during viral replication over serial passage generate new avirulent genotypes to attenuate the virus. Therefore, by passing both a traditional virulent MDV strain and viruses from a BAC-derived clone, we could distinguish whether attenuation requires preexisting variation within a strain, or if normal mutation rates occurring during serial passage are sufficient to generate avirulent viruses de novo from the genotypically homogenous virus. Not only did the Md5 strain become attenuated, as expected, but the three serially passed Md5B40BAC-c1 replicates became attenuated as well. This result shows that, despite beginning from a single virulent genotype, de novo mutation is sufficient to generate avirulent viruses, and attenuation is not simply selection upon preexisting variation within quasispecies. However, as the Md5 strain became attenuated at earlier passages, it is possible that selection of preexisting variants in the population may also be a contributing mechanism.
To better understand attenuation, we then compared durations of time before attenuation between the serially passed replicates, and the first noticeable difference was that time until attenuation varied between replicates. While a reduction in virulence occurred in all replicates, attenuation did not occur simultaneously in all replicates despite being derived from identical BAC-derived virus stocks before passage. Rep 2 was completely attenuated at p65, Rep 3 was completely attenuated at p75, and the virulence of Rep 1 was greatly reduced, but not completely attenuated, at p85. Despite these differences, all replicates showed dramatic decreases in virulence, with a 50 to 80% decrease in MD incidence within a span of ∼10 passages. This rapid change suggests that a small number of mutations with strong effects are likely responsible for attenuation. In contrast, if attenuation were the result of an accumulation of many small, additive mutations that progressively accrued over time, we would have expected a slower, more gradual decline in virulence (24, 25).
From a simple look at the total number of mutations that occurred at >2% in the viral population, it is seen that no replicate had greater than 100 mutations, with the number of SNVs in the three replicates ranging from 41 to 95. While this is a relatively small number of mutations within a genome of ∼184 kb, the actual number of candidate mutations is reduced even further to yield a combined list of only 16 candidate SNVs from the three Md5B40BAC-c1 replicates when considering only mutations occurring at high frequencies exceeding 20% in the population, which is still a conservative cutoff value. As a result, we conclude that there are only a few probable candidate mutations occurring at high frequency to cause attenuation of the population. Further filtering of these SNVs based on frequency kinetics in relation to decreasing virulence identified five candidate mutations and genes underlying attenuation.
Further comparisons among the three attenuated replicates to find commonly mutated genes identified ICP4 as a prime candidate gene. ICP4 contained several nonsynonymous candidate SNVs at frequencies greater than 60% in the completely attenuated MDV replicates, including one mutation completely fixed at 100% in a replicate. As ICP4 is an immediate early transcriptional regulator that controls expression of other immediate early, early, and late genes, it is reasonable that mutations altering normal ICP4 function could have a cascade effect, altering regulation of many downstream genes. Transcriptome sequencing analysis of attenuated viruses identified 5 to 14 genes that were differentially expressed, but compared to the list of SNVs identified, there were no mutations within the genes themselves to explain their differential expression. Instead, mutations within upstream regulators, such as ICP4, could explain this observation in which mutations within ICP4 could lead to larger, widespread effects within the population resulting from relatively few mutations occurring within ICP4.
The results of in vivo trials of recombinant viruses involving a single SNV within UL5 encoding the helicase-primase subunit showed that a single mutation can have a substantial impact on the phenotype of recombinant viruses. Unlike the other four recombinant viruses that retained 100% disease incidence in vivo, the UL5 I682R mutation reduced MD incidence by at least 89% or more, depending on the trial. This mutant virus also had much lower levels of viral replication in vivo than did the virulent Md5B40BAC-c1. All serially passed replicates had comparable levels of MDV replication in vivo, as did the Mut UL5-I682R. As a result of this dramatic phenotypic change due to the mutation within UL5, we created a revertant virus to verify that the results of in vivo trials could be attributed to the desired single point mutation and not extraneous changes that may have occurred unbeknownst to us during recombineering and generation of viral stocks. Challenging birds with this virus clearly showed that the reversion restored a wild-type phenotype for survival, levels of in vivo replication, and disease incidence, confirming our conclusion that the single point mutation within Mut UL5-I682R was responsible for the observed phenotypic changes. While this single mutation did not cause complete attenuation of the virus, it does show that even a single nucleotide change is capable of causing dramatic changes in virulence. This result clearly warrants further study to determine if introduction of additional candidate mutations, such as the next-highest-frequency candidate mutation within UL46 (VP11/VP12 tegument protein) identified within the same replicate, is able to have an additive or epistatic influence to cause complete attenuation in a recombinant virus.
This dramatic decrease in disease incidence exhibited by the mutation within UL5-helicase-primase within this study indicates the importance of this gene as a factor in virulence. Sequence comparison of Gallid herpesvirus 1, an alphaherpesvirus also known as infectious laryngotracheitis virus (ILTV), identified a nonsynonymous mutation within UL5 that was unique to the vaccine strain and enabled the attenuated virus to be distinguished from four other virulent ILTV strains (26). Mutations within UL5 have been shown to cause a reduction in virulence in herpesviruses other than Gallid herpesviruses as well. Studies involving herpes simplex virus 1 (HSV-1) have identified several mutations within UL5 that have been determined to cause a reduction in virulence (27). Biochemical analysis of the UL5 Gly 815 mutant shows a decrease in binding affinity of single-stranded DNA and reduced turnover rate (28). This mutation in HSV-1 was unable to replicate in vivo alone, but when additional purified components of the heterotrimeric complex were provided, the mutant was able to perform normal helicase-primase functions. While the three components of the functional complex, UL5, UL8, and UL52, are all necessary for normal function in vivo, during in vitro replication UL8 is not. Growth curves measuring the in vitro replication for our Mut I682R relative to the Md5B40BAC-c1 are yet to be conducted, but it is clear that the recombinant Mut UL5 replicates in vitro to form plaques and generates high viral titers. Despite this ability to replicate in vitro, its ability to replicate in vivo is severely reduced compared to that of the virulent wild type, which has been observed in other UL5 mutants. Since serial passage of MDV in tissue culture drives selection for improved in vitro replication, it would be anticipated that the mutation within UL5 would not detrimentally affect in vitro replication or even that, as a result of selection for improved in vitro growth, the observed Mut I682R could potentially improve in vitro growth. Whether in vitro replication is altered by the UL5 mutation compared to the wild-type BAC, growth curves quantifying replication of the Mut UL5-I682R virus may offer an explanation for how this mutation rose to a high frequency in the attenuated populations.
While this high-frequency SNV in UL5 resulting in greatly reduced virulence was present only in Rep 2, three additional mutations within UL5 were also found within Rep 1, although these SNVs were present at much lower frequencies than was the mutation found within Rep 2. As seen in ICP4, it appears that identical mutations shared among replicates are rare, but there were several commonly mutated genes between attenuated replicates, such as with ICP4, UL5, and LORF2. Experimental evolution studies involving over 100 replicates of Escherichia coli have shown high degrees of convergent evolution between replicates at the level of genes and pathways, while identical nucleotide mutations were rarely shared among the replicates (29). Based on these conclusions, we looked for commonly mutated pathways among attenuated replicates to determine if there were key pathways involved in attenuation. After screening all mutations for the most probable candidates, we identified five top candidate genes, two of which were genes involved in DNA replication: a mutation within UL5 and a second one within UL42. Considering that mutations within genes involving DNA replication are probable targets for selection to cause adaptation of viruses during in vitro growth, wider inspection of all mutations within attenuated replicates of the seven genes required for DNA replication in herpesviruses (UL5, UL8, UL9, UL29, UL30, UL42, and UL52) revealed multiple mutations within genes involved in DNA replication, especially within Rep 1. Of these genes, we found eight mutations at frequencies of >2% within all seven genes required for DNA replication within Rep 1 p85. Clustering of mutations within DNA replication genes, such as the UL5 (helicase-primase) and UL42 (DNA polymerase subunit), as well as the significant phenotypic effect of the mutation tested within UL5, suggests that mutations in the genes associated with the DNA replication pathway are targeted for selection during serial passage, leading to attenuation of the virus. To further identify commonly mutated pathways, we used DAVID to determine pathways that were enriched for mutants in the attenuated replicates. Considering only high-frequency, nonsynonymous mutations at >20% in the four attenuated viral populations, we found that pathways containing genes involving transcriptional regulation were enriched for mutations. It is possible that pathways involving transcriptional regulation, at both the DNA-dependent and RNA metabolism levels, are mechanisms which, when disturbed, can lead to attenuation. The finding of enrichment for pathways involving transcriptional regulation due to multiple mutations within genes involved in DNA replication, such as Mut I682R in UL5, supports our hypothesis that serial passage in vitro selects for increased replication and adaptation for growth in tissue culture. Mutations altering normal interactions of these gene products would impact viral replication and lead to the changes in phenotype observed in attenuated viruses.
In conclusion, our results show that it is possible to attenuate viral BAC-derived clones as a result of de novo mutation and that it is apparent that there is variation among viruses in the manner in which they become attenuated. All serially passed replicates contained unique candidate mutations, and while there may be commonly mutated genes among attenuated viruses, such as ICP4, it appears that there is no single route that all viruses must follow to become attenuated. Multiple candidate mutations have been identified and tested to determine their effect within genes of known and unknown function, such as LORF2, DNA polymerase subunit, VP11/VP12 tegument protein, and helicase-primase subunits. Singly, no one mutation was able to cause complete attenuation, yet one SNV within the helicase-primase subunit had a dramatic decrease in virulence, indicating an important role for this gene in MDV attenuation. Additional work to further characterize candidate mutations identified in this study may identify a minimum number of mutations required to create a fully avirulent virus through changing only a few, perhaps 2 to 3, key mutations. This could allow the precise engineering of avirulent viruses for production of candidate vaccines via mutation of critical genes involved in attenuation, without relying on blind serial passage and random chance to generate the next superior MD vaccine candidate.
ACKNOWLEDGMENTS
We thank Laurie Molitor and Lonnie Milam for technical assistance and Jerry Dodgson for critical input in the manuscript.
This project was supported in part by National Research Initiative competitive grant no. 2010-65119-20505 from the USDA National Institute of Food and Agriculture.
Footnotes
Published ahead of print 19 March 2014
REFERENCES
- 1.Morrow C, Fehler F. 2004. Marek's disease: a world-wide problem, p 49–61 In Davison F, Nair V. (ed), Marek's disease. Academic Press, San Diego, CA [Google Scholar]
- 2.Purchase H, Okazaki W. 1971. Effect of vaccination with herpesvirus of turkeys (HVT) on horizontal spread of Marek's disease herpesvirus. Avian Dis. 15:391–397. 10.2307/1588710 [DOI] [PubMed] [Google Scholar]
- 3.Witter R. 1997. Increased virulence of Marek's disease virus field isolates. Avian Dis. 41:149–163. 10.2307/1592455 [DOI] [PubMed] [Google Scholar]
- 4.Atkins KE, Read AF, Savill NJ, Renz KG, Islam AF, Walkden-Brown SW, Woolhouse ME. 2013. Vaccination and reduced cohort duration can drive virulence evolution: Marek's disease virus and industrialized agriculture. Evolution 67:851–860. 10.1111/j.1558-5646.2012.01803.x [DOI] [PubMed] [Google Scholar]
- 5.Rispens BH, Vloten H, Mastenbroek N, Maas HJ, Schat KA. 1972. Control of Marek's disease in the Netherlands. 1. Isolation of an avirulent Marek's disease virus (strain CVI988) and its use in laboratory vaccination trials. Avian Dis. 16:108–125 [PubMed] [Google Scholar]
- 6.Churchill AE, Chubb RC, Baxendale W. 1969. The attenuation, with loss of oncogenicity, of the herpes-type virus of Marek's disease (strain HPRS-16) on passage in cell culture. J. Gen. Virol. 4:557–564. 10.1099/0022-1317-4-4-557 [DOI] [PubMed] [Google Scholar]
- 7.Silva RF, Reddy SM, Lupiani B. 2004. Expansion of a unique region in the Marek's disease virus genome occurs concomitantly with attenuation but is not sufficient to cause attenuation. J. Virol. 78:733–740. 10.1128/JVI.78.2.733-740.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spatz SJ. 2010. Accumulation of attenuating mutations in varying proportions within a high passage very virulent plus strain of Gallid herpesvirus type 2. Virus Res. 149:135–142. 10.1016/j.virusres.2010.01.007 [DOI] [PubMed] [Google Scholar]
- 9.Spatz SJ, Petherbridge L, Zhao Y, Nair V. 2007. Comparative full-length sequence analysis of oncogenic and vaccine (Rispens) strains of Marek's disease virus. J. Gen. Virol. 88:1080–1096. 10.1099/vir.0.82600-0 [DOI] [PubMed] [Google Scholar]
- 10.Spatz SJ, Schat KA. 2011. Comparative genomic sequence analysis of the Marek's disease vaccine strain SB-1. Virus Genes 42:331–338. 10.1007/s11262-011-0573-0 [DOI] [PubMed] [Google Scholar]
- 11.Niikura M, Kim T, Silva RF, Dodgson J, Cheng HH. 2011. Virulent Marek's disease virus generated from infectious bacterial artificial chromosome clones with complete DNA sequence and the implication of viral genetic homogeneity in pathogenesis. J. Gen. Virol. 92:598–607. 10.1099/vir.0.026864-0 [DOI] [PubMed] [Google Scholar]
- 12.Silva RF, Dunn JR, Cheng HH, Niikura M. 2010. A MEQ-deleted Marek's disease virus cloned as a bacterial artificial chromosome is a highly efficacious vaccine. Avian Dis. 54:862–869. 10.1637/9048-090409-Reg.1 [DOI] [PubMed] [Google Scholar]
- 13.Volkening JD, Spatz SJ. 2009. Purification of DNA from the cell-associated herpesvirus Marek's disease virus for 454 pyrosequencing using micrococcal nuclease digestion and polyethylene glycol precipitation. J. Virol. Methods 157:55–61. 10.1016/j.jviromet.2008.11.017 [DOI] [PubMed] [Google Scholar]
- 14.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koboldt DC, Chen K, Wylie T, Larson DE, McLellan MD, Mardis ER, Weinstock GM, Wilson RK, Ding L. 2009. VarScan: variant detection in massively parallel sequencing of individual and pooled samples. Bioinformatics 25:2283–2285. 10.1093/bioinformatics/btp373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28:511–515. 10.1038/nbt.1621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang BDW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4:44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- 18.Huang DW, Sherman BT, Lempicki RA. 2009. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37:1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tischer BK, von Einem JV, Kaufer B, Osterrieder N. 2006. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. 10.2144/000112096 [DOI] [PubMed] [Google Scholar]
- 20.Moriuchi H, Moriuchi M, Smith HA, Straus SE, Cohen JI. 1992. Varicella-zoster virus open reading frame 61 protein is functionally homologous to herpes simplex virus type 1 ICP0. J. Virol. 66:7303–7308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gimeno IM, Cortes AL, Silva RF. 2008. Load of challenge Marek's disease virus DNA in blood as a criterion for early diagnosis of Marek's disease tumors. Avian Dis. 52:203–208. 10.1637/8089-081407-Reg.1 [DOI] [PubMed] [Google Scholar]
- 22.Biswas N, Weller SK. 2001. The UL5 and UL52 subunits of the herpes simplex virus type 1 helicase-primase subcomplex exhibit a complex interdependence for DNA binding. J. Biol. Chem. 276:17610–17619. 10.1074/jbc.M010107200 [DOI] [PubMed] [Google Scholar]
- 23.Jarosinski KW, Osterrieder N, Nair VK, Schat KA. 2005. Attenuation of Marek's disease virus by deletion of open reading frame RLORF4 but not RLORF5a. J. Virol. 79:11647–11659. 10.1128/JVI.79.18.11647-11659.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrick JE, Lenski RE. 2013. Genome dynamics during experimental evolution. Nat. Rev. Genet. 14:827–839. 10.1038/nrg3564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Elena SF, Lenski RE. 2003. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat. Rev. Genet. 4:457–469. 10.1038/nrg1088 [DOI] [PubMed] [Google Scholar]
- 26.Spatz SJ, Volkening JD, Keeler CL, Kutish GF, Riblet SM, Boettger CM, Clark KF, Zsak L, Afonso CL, Mundt ES, Rock DL, Garcia M. 2012. Comparative full genome analysis of four infectious laryngotracheitis virus (Gallid herpesvirus-1) virulent isolates from the United States. Virus Genes 44:273–285. 10.1007/s11262-011-0696-3 [DOI] [PubMed] [Google Scholar]
- 27.Biswas S, Miguel RN, Sukla S, Field HJ. 2009. A mutation in helicase motif IV of herpes simplex virus type 1 UL5 that results in reduced growth in vitro and lower virulence in a murine infection model is related to the predicted helicase structure. J. Gen. Virol. 90:1937–1942. 10.1099/vir.0.011221-0 [DOI] [PubMed] [Google Scholar]
- 28.Graves-Woodward KL, Weller SK. 1996. Replacement of gly815 in helicase motif V alters the single-stranded DNA-dependent ATPase activity of the herpes simplex virus type 1 helicase-primase. J. Biol. Chem. 271:13629–13635. 10.1074/jbc.271.23.13629 [DOI] [PubMed] [Google Scholar]
- 29.Tenaillon O, Rodríguez-Verdugo A, Gaut RL, McDonald P, Bennett AF, Long AD, Gaut BS. 2012. The molecular diversity of adaptive convergence. Science 335:457–460. 10.1126/science.1212986 [DOI] [PubMed] [Google Scholar]