ABSTRACT
Kaposi's sarcoma-associated herpesvirus (KSHV) establishes persistent latent infection in immunocompetent hosts. Disruption of KSHV latency results in viral lytic replication, which promotes the development of KSHV-related malignancies in immunocompromised individuals. While inhibitors of classes I and II histone deacetylases (HDACs) potently reactivate KSHV from latency, the role of class III HDAC sirtuins (SIRTs) in KSHV latency remains unclear. Here, we examined the effects of inhibitors of SIRTs, nicotinamide (NAM) and sirtinol, on KSHV reactivation from latency. Treatment of latently KSHV-infected cells with NAM or sirtinol induced transcripts and proteins of the master lytic transactivator RTA (ORF50), early lytic genes ORF57 and ORF59, and late lytic gene ORF65 and increased the production of infectious virions. NAM increased the acetylation of histones H3 and H4 as well as the level of the active histone H3 trimethyl Lys4 (H3K4me3) mark but decreased the level of the repressive histone H3 trimethyl Lys27 (H3K27me3) mark in the RTA promoter. Consistent with these results, we detected SIRT1 binding to the RTA promoter. Importantly, knockdown of SIRT1 was sufficient to increase the expression of KSHV lytic genes. Accordingly, the level of the H3K4me3 mark in the RTA promoter was increased following SIRT1 knockdown, while that of the H3K27me3 mark was decreased. Furthermore, SIRT1 interacted with RTA and inhibited RTA transactivation of its own promoter and that of its downstream target, the viral interleukin-6 gene. These results indicate that SIRT1 regulates KSHV latency by inhibiting different stages of viral lytic replication and link the cellular metabolic state with the KSHV life cycle.
IMPORTANCE Kaposi's sarcoma-associated herpesvirus (KSHV) is the causal agent of several malignancies, including Kaposi's sarcoma, commonly found in immunocompromised patients. While latent infection is required for the development of KSHV-induced malignancies, viral lytic replication also promotes disease progression. However, the mechanism controlling KSHV latent versus lytic replication remains unclear. In this study, we found that class III histone deacetylases (HDACs), also known as SIRTs, whose activities are linked to the cellular metabolic state, mediate KSHV replication. Inhibitors of SIRTs can reactivate KSHV from latency. SIRTs mediate KSHV latency by epigenetically silencing a key KSHV lytic replication activator, RTA. We found that one of the SIRTs, SIRT1, binds to the RTA promoter to mediate KSHV latency. Knockdown of SIRT1 is sufficient to induce epigenetic remodeling and KSHV lytic replication. SIRT1 also interacts with RTA and inhibits RTA's transactivation function, preventing the expression of its downstream genes. Our results indicate that SIRTs regulate KSHV latency by inhibiting different stages of viral lytic replication and link the cellular metabolic state with the KSHV life cycle.
INTRODUCTION
Kaposi's sarcoma-associated herpesvirus (KSHV) is a gammaherpesvirus associated with several AIDS-related malignancies, including Kaposi's sarcoma (KS), primary effusion lymphoma (PEL), and a subset of multicentric Castleman's disease (MCD). Like other herpesviruses, the life cycle of KSHV has latent and lytic replication phases. Following primary infection, KSHV establishes latent infection in the host cells, displaying a restricted latent replication program. During latency, KSHV expresses only a few viral latent genes, including latent nuclear antigen (LANA or LNA) encoded by ORF73, vFLIP encoded by ORF72, vCyclin encoded by ORF71, and more than two dozen microRNAs derived from 12 precursor microRNAs (1). Upon stimulation by specific signals, KSHV reactivates from latency, during which it expresses cascades of lytic genes and produces infectious virions. The KSHV switch from latent to lytic replication is initiated by the expression of an immediate early (IE) gene, RTA, encoded by ORF50, which is essential and sufficient for activating the entire viral lytic replication cycle (2, 3). In KS tumors, most KSHV-infected cells are in a latent state, indicating the importance of this phase of viral replication in tumor development. However, lytic replication also promotes tumor progression through an autocrine and paracrine mechanism (1). Indeed, clinical studies have shown that KSHV lytic replication is associated with disease incidence and progression (4–6). Thus, factors that disrupt KSHV latency and trigger viral lytic replication might contribute to the development of KSHV-related malignancies.
Histone deacetylases (HDACs) repress gene transcription by promoting highly condensed chromatin structures associated with histone deacetylation (7). Four groups of HDACs are involved in diverse cellular processes. Class I HDACs are homologous to the Saccharomyces cerevisiae yeast protein Rpd3 and consist of HDAC1, HDAC2, HDAC3, and HDAC8, while HDACs 4 to 7 and HDAC9, which correspond to the Hdal yeast protein, belong to the class II HDACs. Class III HDACs, also known as sirtuins (SIRTs), are a class of newly discovered HDACs (8). They have sequence similarity to Sir2, a transcriptional repressor of yeast. The seven members of SIRTs, named SIRTs 1 to 7, are unique in that they require NAD+ as a cofactor for their activity (8). In particular, SIRT1 is involved in the regulation of gene expression, cellular metabolism, and the stress response through interaction with a variety of proteins. SIRT1 preferentially deacetylates histone H3 at lysines 9 and 14 (H3K9 and H3K14) and histone H4 at lysine 16 (H4K16), leading to chromatin condensation and transcriptional repression (9). SIRT1 also regulates a number of nonhistone proteins, including p53 and FOXO3, thus linking cellular metabolism to apoptosis and the stress response (10–12). SIRTs exhibit biochemical features different from those of class I and II HDACs and are insensitive to their inhibitors, such as sodium butyrate (NaB) and trichostatin A (TSA). Similarly, specific inhibitors of SIRTs, such as nicotinamide (NAM), the amide of vitamin B3, and sirtinol, do not inhibit deacetylation of class I and II HDACs.
KSHV latent and lytic replication phases are tightly associated with histone posttranslational modifications. During latent infection, the RTA promoter is associated with hypoacetylated histones (13). Class I and II HDACs have been shown to regulate KSHV latent and lytic replication. NaB and TSA induce latently KSHV-infected cells into lytic replication (13–15). Treatment with NaB leads to histone hyperacetylation and subsequent chromatin remodeling in the RTA promoter, resulting in the induction of RTA expression (13). However, there is so far no report on the role of class III HDACs in the KSHV life cycle. In this study, we investigated the effect of inhibitors of SIRTs, NAM and sirtinol, on KSHV reactivation from latency in PEL cell lines. We demonstrate that treatment with NAM or sirtinol can efficiently reactivate KSHV from latency. In addition, we show SIRT1 binding to the RTA promoter. Knockdown of SIRT1 can increase the expression of KSHV lytic genes. Consistent with these results, we detected increased histone H3 trimethyl Lys4 (H3K4me3) mark and decreased histone H3 trimethyl Lys27 (H3K27me3) mark levels in the RTA promoter following SIRT1 knockdown. Finally, we show that SIRT1 inhibits RTA transactivation of its own promoter and that of its downstream target, the viral interleukin-6 (vIL-6; ORF-K2) gene. Together, our findings illustrate a role of SIRTs in multiple steps of KSHV replication and provide a link between cellular metabolism and the KSHV life cycle.
MATERIALS AND METHODS
Cell culture and reagents.
KSHV-positive PEL cell lines BCP-1, BC-3, and BCBL-1 were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS). BCBL-1–BAC36 cells harboring recombinant KSHV bacterial artificial chromosome 36 (BAC36) were previously described (16). 293T cells were grown in Dulbecco modified Eagle medium with 10% FBS. NAM, sirtinol, and NaB were purchased from Sigma (St. Louis, MO).
Plasmids and transfection.
Plasmids pcDNA3Flag-RTA and pcDNAMyc-His-RTA were previously described (17). Luciferase reporter constructs pRp-luc and pvIL-6-luc were kindly provided by Ren Sun of the University of California at Los Angeles, Los Angeles, CA. pRp-luc contains a 3-kb upstream RTA promoter sequence (18), while pvIL-6-Luc contains a 3.2-kb upstream vIL-6 promoter sequence (19). Plasmids pLTc-luc and pcDNA4ToSIRT1 were obtained from Angus C. Wilson and Danny Reiberg of New York University School of Medicine, New York, NY, respectively. Transfection was performed using Lipofectamine 2000 according to the instructions of the manufacturer (Invitrogen, Carlsbad, CA).
shRNA knockdown of SIRT1.
Knockdown with short hairpin RNAs (shRNAs) was performed as previously described (20). Briefly, cells infected with lentiviruses for 3 days were collected and used for examining the expression of proteins and mRNAs and chromatin immunoprecipitation (ChIP) assay. The SIRT1 shRNA knockdown sequences were GCGGGAATCCAAAGGATAATT for shRNA1 and GCTTGATGGTAATCAGTATCT for shRNA2, both of which were cloned into the pLKO.1 lentiviral vector (Addgene, Cambridge, MA). Lentivirus production and infection were carried out as previously described (20).
Luciferase reporter assay.
The luciferase reporter assay was performed as previously described (21). The luciferase reporter of RTA or vIL-6 or a latent LANA promoter, β-galactosidase expression plasmid pSV-β-gal (Promega, Madison, WI), a SIRT1 expression plasmid, or a control plasmid was cotransfected into 293T cells cultured in 6-well plates. To determine if NAM can reverse the effect of SRIT1, NAM was added to reach a concentration of 10 mM at 12 h posttransfection. Reporter activities were determined at 48 h posttransfection using a luciferase assay system (Promega, Madison, WI). The β-galactosidase expression plasmid was used in the cotransfections to normalize the transfection efficiency. All data are presented as averages and standard deviations from three independent experiments.
RNA isolation and RT-qPCR.
Total RNA extraction and cDNA synthesis were carried out as previously described (22). Reverse transcription (RT)-quantitative real-time PCR (qPCR) was performed with an ABI Prism 7900 system and SYBR green Taq polymerase mix (Applied Biosystems). Primers for RT-qPCR and the method for quantification of gene expression were previously described (22).
IFA.
KSHV lytic proteins ORF59 and ORF65 were detected by immunofluorescence assay (IFA) as previously described (23). The RTA protein was detected with an RTA polyclonal antibody kindly provided by Ke Lan at the Institut Pasteur of Shanghai, Chinese Academy of Sciences, Shanghai, China.
Confocal microscopy.
Confocal microscopy was used to examine the colocalization of RTA and SIRT1. The procedures for confocal microscopy were previously described (24). Myc-RTA and Flag-SIRT1 proteins coexpressed in 293T cells were detected using anti-Myc and anti-Flag mouse monoclonal antibodies (Sigma), respectively. Endogenous RTA and SIRT1 proteins in BCBL-1 cells were detected with a SIRT1 antibody purchased from EMD Millipore Corporation (Billerica, MA) and the RTA antibody described above.
Western blotting.
Western blotting was carried out as previously described (23). A monoclonal antibody was used to detect ORF65 (22), while the RTA and SIRT1 proteins were detected with the antibodies described above. Rabbit polyclonal antibodies were used to detect histone H3, histone H4, acetyl-histone H3, and acetyl-histone H4 (EMD Millipore Corporation). A monoclonal antibody to β-tubulin was from Sigma.
Titration of infectious virions and quantification of virus particles by qPCR.
Supernatants from uninduced BCBL-1 cells or BCBL-1 cells treated with NAM, sirtinol, and NaB for 4 days were collected and filtered through 0.45-μm-pore-size filters. The filtered supernatants were then used to infect rat mesenchymal precursor cells (MM cells) to quantify the relative virus titers as previously described (25, 26).
Virus particles were also measured by qPCR. The filtered supernatants were treated with DNase for 30 min at 37°C to remove the free DNA. Virus DNA was extracted and used for qPCR using primers 5′-TCCGGCGGATATACCGTCAC-3′ and 5′-GGTGCAGCTGGTACAGTGTG-3′ designed from ORF73. The relative virus genome copy number was calculated by use of the amount of DNA from uninduced cells set equal to 1.
Coimmunoprecipitation (co-IP).
To detect an RTA and SIRT1 interaction, 293T cells transfected with RTA and SIRT1 expression plasmids for 48 h were lysed in a lysis buffer containing 50 mM Tris-HCl, 150 mM NaCl, EDTA, 1% Triton X-100, and a cocktail of protease inhibitors (Sigma). The cleared lysate was precipitated with either mouse anti-Flag M2 agarose (Sigma) or the anti-Myc antibody (Sigma), followed by incubation with protein G agarose beads (Sigma) to capture the protein-antibody complexes. Western blotting was used to detect either Flag-tagged SIRT1 or Myc-tagged RTA.
Similar co-IP procedures were used to detect the RTA and SIRT1 interaction in BCBL-1 cells. In addition to the SIRT1 and RTA antibodies described above, two other RTA polyclonal antibodies were also used in these experiments. One was kindly provided by Charles Wood at the University of Nebraska—Lincoln, Lincoln, NE, while the second one was kindly provided by Gary Hayward at Johns Hopkins University, Baltimore, MD.
ChIP assay.
Untreated BCBL-1 cells and BCBL-1 cells treated with NAM and NaB for 12 h were collected for ChIP assays as previously described (27) using the antibodies to acetyl-histone H3 and acetyl-histone H4 (EMD Millipore Corporation). The immune complexes were eluted in an elution buffer containing 1% SDS and 0.1 M NaHCO3. The eluted and input samples were incubated at 65°C overnight to achieve reverse cross-linking and purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA). ChIP DNA was amplified by semiquantitative PCR using primers 5′-GGTACCGAATGCCACAATCTGTGCCCT-3′ and 5′-ATGGTTTGTGGCTGCCTGGACAGTATTC-3′ for locus 1 of the RTA promoter and primers 5′-GGTACCGAATGCCACAATCTGTGCCCT-3′ and 5′-ATGGTTTGTGGCTGCCTGGACAGTATTC-3′ for locus 2 of the RTA promoter (13). The LANA promoter was amplified using primers 5′-CCAGACTCTTCAACACCTATGCG-3′ and 5′-GGATGATCCCACGTAGATCGG-3′. Primers for GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were 5′-TACTAGCGGTTTTACGGGCG-3′ and 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′. The amplified PCR product was analyzed by agarose gel electrophoresis and ethidium bromide staining. The ChIP products were also analyzed by quantitative real-time PCR (ChIP-qPCR) as previously described (17).
ChIP assays were also carried out with antibodies to SIRT1, histone H3 trimethyl Lys4 (H3K4me3), and histone H3 trimethyl Lys27 (H3K27me3) (EMD Millipore Corporation). The products were analyzed by ChIP-qPCR using primers for the RTA promoter (5′-AAAGTCAACCTTACTCCGCAAG-3′ and 5′-GCTGCCTGGACAGTATTCTCAC-3′) and the LANA promoter (5′-CTTAACACAAATCATGTACA-3′ and 5′-GCAGCTTGGTCCGGCTGACT-3′).
RESULTS
NAM and sirtinol increase the expression of KSHV lytic transcripts.
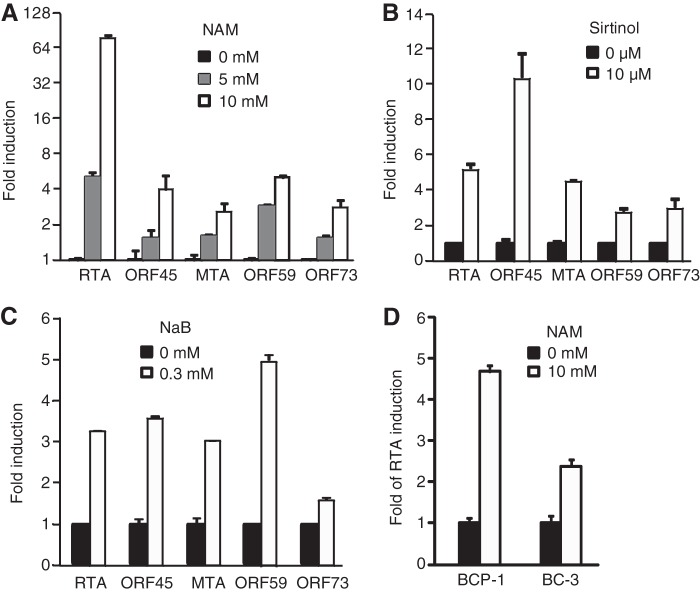
To examine the role of class III HDACs in KSHV lytic replication, we treated BCBL-1–BAC36 cells with 0, 5, and 10 mM NAM for 48 h. RT-qPCR showed that the expression of the KSHV IE lytic gene RTA was increased in a dose-dependent fashion, with the increase reaching 85-fold at 48 h posttreatment (Fig. 1A). As expected, KSHV lytic genes ORF45, MTA (ORF57), and ORF59 as well as latent gene LANA were also induced in a dose-dependent manner. Consistent with these results, the expression of KSHV lytic genes was increased when BCBL-1–BAC36 cells were treated with 10 μM sirtinol, another inhibitor of class III HDACs (Fig. 1B). These results were similar to those obtained with inhibitor of class I and II HDACs NaB (Fig. 1C). Examination of two other KSHV-infected cell lines, BCP-1 and BC-3, confirmed that NAM also induced the expression of the RTA transcript in these cells (Fig. 1D), indicating that the effects of inhibitors of class III HDACs on the expression of KSHV lytic genes is not cell line specific.
FIG 1.
Treatment with NAM or sirtinol increases the expression of KSHV lytic transcripts. (A to C) Expression of KSHV transcripts in BCBL-1 cells following treatment with NAM (A), sirtinol (B), and NaB (C). (D) Expression of the RTA transcript in BCP-1 and BC-3 cells following treatment with NAM. Cells were treated with the indicated concentrations of the chemicals for 48 h. Viral transcripts were detected by RT-qPCR, and their levels of expression were normalized to the level of expression of cellular GAPDH. Experiments were repeated three times, and results are shown as averages and standard deviations, with the value for untreated cells being set equal to 1.
NAM and sirtinol increase the expression of KSHV lytic proteins.
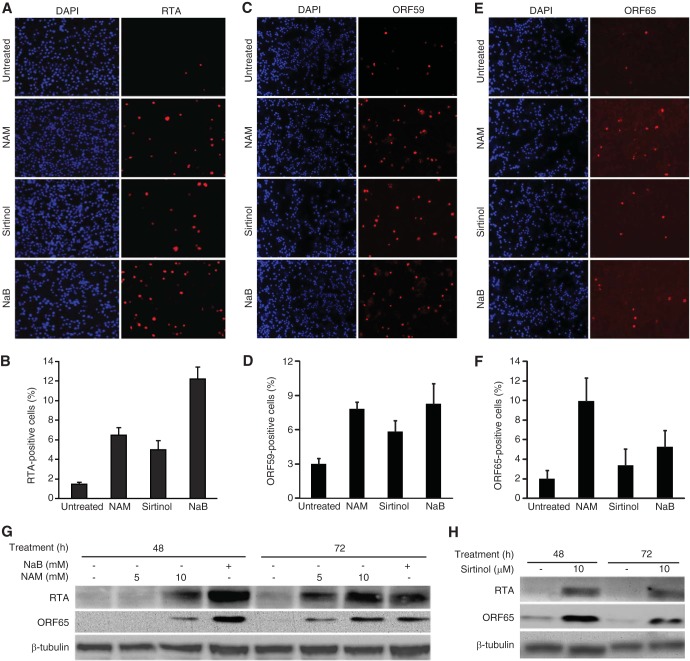
We further examined the effect of NAM on the expression of KSHV lytic proteins. Untreated BCBL-1–BAC36 cells had low levels of spontaneous lytic replication, with 1.6%, 3%, and 2% of the cells expressing the RTA, ORF59, and ORF65 proteins, respectively (Fig. 2A to F), as previously reported (28). A chemical inducer can increase the expression of lytic proteins and the production of infectious virions (29, 30). The expression kinetics of each viral gene is distinct but might vary depending on the potency of the inducer. This was evident following treatment with NAM, sirtinol, and NaB (Fig. 1). As a result, the percentage of cells expressing a lytic protein might vary from 2- to 10-fold, depending on when it is examined following the lytic induction (29, 30). Treatment with NAM and sirtinol for 48 h increased the proportion of RTA-positive cells to 6.5% and 5.7%, respectively (Fig. 2A and B), that of ORF59-positive cells to 7.6% and 5.8%, respectively (Fig. 2C and D), and that of ORF65-positive cells to 9.8% and 3.3%, respectively (Fig. 2E and F). These results were in the same ranges of those for cells treated with NaB, which had 11.9%, 8.1%, and 4.8% RTA-, ORF50-, and ORF65-positive cells, respectively (Fig. 2A to F). Western blotting showed that RTA and ORF65 proteins were robustly induced following treatment with NAM (Fig. 2G) and sirtinol (Fig. 2H). These results were in agreement with those for the viral transcripts, suggesting that SIRTs might be involved in controlling KSHV lytic replication. Interestingly, while NAM induced a higher level of the RTA transcript than sirtinol and NaB did, it did not entirely translate into higher levels of lytic proteins (Fig. 2).
FIG 2.
Treatment with NAM or sirtinol increases the expression of KSHV lytic proteins. (A to F) IFA detection of KSHV lytic proteins RTA (A and B), ORF59 (C and D), and ORF65 (E and F) following treatment with NAM, sirtinol, or NaB. BCBL-1 cells treated with 10 mM NAM, 10 μM sirtinol, or 0.3 mM NaB were subjected to IFA staining (A, C, and E) (DAPI, 4′,6-diamidino-2-phenylindole). The percentages of RTA-, ORF59-, and ORF65-positive cells are represented by the averages and standard deviations calculated from eight random fields (B, D, and F). (G and H) Western blot analysis of RTA and ORF65 proteins following treatment with NAM, sirtinol, or NaB. BCBL-1 cells treated with the indicated concentrations of NAM and 0.3 mM NaB (G) or 10 μM sirtinol (H) for the specified times were harvested and examined for the expression of RTA and ORF65 proteins using β-tubulin as a loading control.
NAM and sirtinol increase the production of KSHV infectious virions.
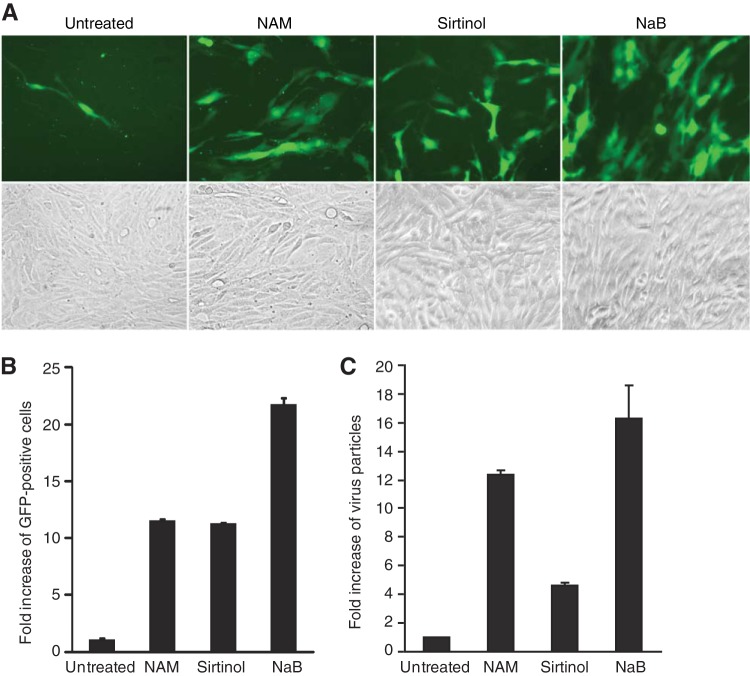
KSHV BAC36 contains a green fluorescent protein (GFP) cassette that can be used to track the infection of cells by the virus (16, 25). We estimated the relative yields of infectious virions by inoculating MM cells, which are highly susceptible to KSHV infection (26), with supernatants from cells producing infectious virions and subsequently counting the numbers of GFP-positive cells (Fig. 3A). Treatment with NAM and sirtinol increased the yields of infectious virions by 11.5- and 11-fold, respectively (Fig. 3B). We observed similar results by direct quantification of virus particles by qPCR (Fig. 3C). While cells induced with sirtinol produced about 2-fold fewer virus particles than those induced with NAM, they still produced 4.5-fold more virus particles than the uninduced cells did (Fig. 3C). These results indicate that the latently KSHV-infected cells induced by inhibitors of class III HDACs complete the full viral replication cycle, resulting in the increased production of KSHV infectious virions. Overall, the yields of infectious virions were consistent with the expression of all lytic proteins except the ORF65 protein (Fig. 2F).
FIG 3.
Treatment with NAM or sirtinol induces the production of KSHV infectious virions. (A) Titration of KSHV infectious virions by inoculating MM cells with supernatants from BCBL-1–BAC36 KSHV-infected cells induced with NAM, sirtinol, or NaB. Cells were induced with 10 mM NAM, 10 μM sirtinol, or 0.3 mM NaB for 4 days. Cell-free supernatants were used to infect MM cells for 48 h. Images of GFP-positive cells illustrate the presence of infectious virions in the supernatants. (B) Relative virus titers were determined on the basis of the numbers of GFP-positive cells from five random fields and are shown as averages and standard deviations. (C) The relative amounts of virus particles produced by the induced cells were determined by qPCR for virion DNA. Supernatants of induced cells collected at the end of induction were filtered and subjected to differential centrifugation to obtain virus particles. The pellets were treated with DNase to eliminate nonspecifically attached nonvirion DNA. Viral DNA was then extracted and analyzed by qPCR.
NAM alters the status of histone acetylation at promoters of KSHV genes.
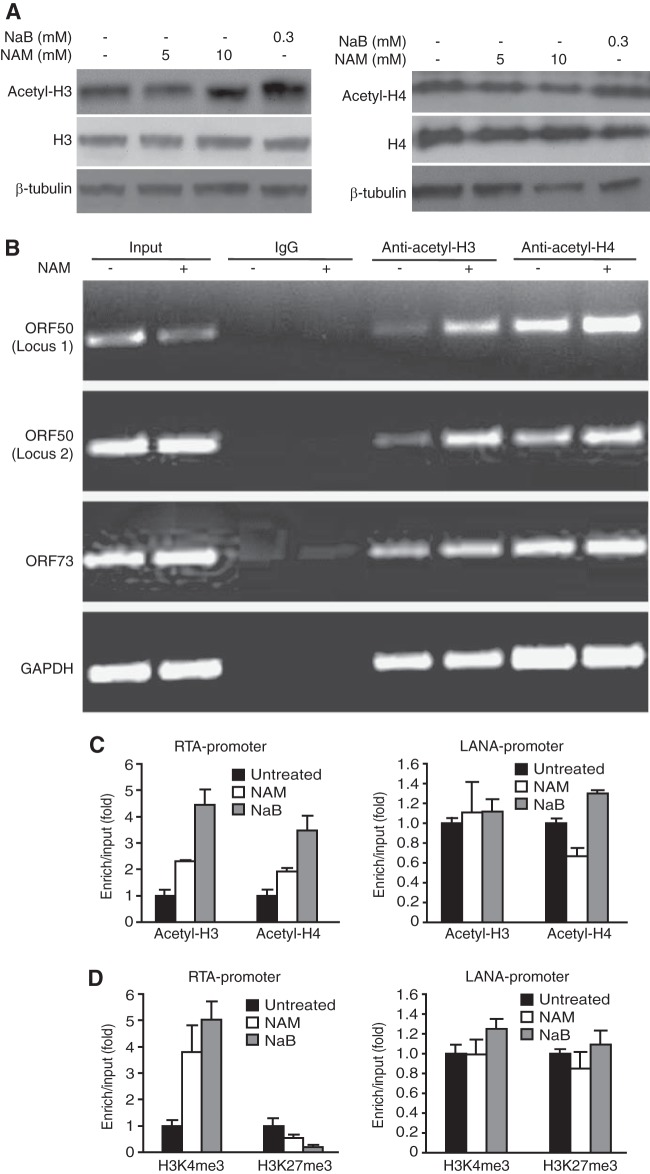
While histone acetylation is often correlated with activation of gene expression, histone deacetylation is linked to transcriptional silencing. To determine the mechanism of NAM induction of KSHV lytic genes, we first examined the total and acetylated forms of histone H3 and histone H4 proteins following treatment with NAM because NAM activates gene expression by inhibiting the deacetylase activity of SIRTs to directly affect histone acetylation status (31). Treatment with NaB slightly increased the acetylated forms of histone H3 and histone H4 proteins but had a minimal effect on the total histone H3 and histone H4 proteins (Fig. 4A). Treatment with NAM had almost no effect on the total and acetylated forms of histone H3 and histone H4 proteins (Fig. 4A). We then reasoned that SIRTs might be recruited to specific promoters and NAM's effect could be observed only at these promoters. Indeed, the levels of the acetylated forms of histones H3 and H4 at two loci of the RTA promoter were significantly increased (Fig. 4B). In contrast, the level of acetylated H3 was only slightly increased in the LANA promoter, which could be due to the slight higher level of input in the NAM-treated cells (Fig. 4B). There was no obvious change for acetylated H4 in the LANA promoter. The increased levels of the acetylated forms of histones H3 and H4 in the RTA promoter were also confirmed by ChIP-qPCR (Fig. 4C). Similarly, no obvious change in the levels of the acetylated forms of histones H3 and H4 in the LANA promoter was observed by ChIP-qPCR (Fig. 4C). NAM also did not affect the levels of binding of the acetylated forms of histones H3 and H4 to the promoter of the cellular GAPDH gene (Fig. 4B). The levels of the active histone mark H3K4me3 and the repressive histone mark H3K27me3 are directly correlated with the level of gene expression (32). NAM increased the level of the H3K4me3 mark but decreased the level of the H3K27me3 mark in the RTA promoter but not LANA promoter (Fig. 4D). Similar results were also obtained with NaB treatment. These results indicate that the enhanced RTA transcription is associated with the acetylation status of histone H3 and histone H4 at its promoter and SIRTs might regulate KSHV latency by maintaining a low level of the active histone H3K4me3 mark and a high level of the repressive histone H3K27me3 mark in the RTA promoter. In contrast, SIRTs were unlikely to be involved in the regulation of the LANA promoter.
FIG 4.
Treatment with NAM induces the hyperacetylation of histones, increases the level of the active histone H3K4me3 mark, and decreases the level of the repressive histone H3K27me3 mark in the RTA promoter. (A) BCBL-1 cells were treated with the indicated concentrations of NAM or 0.3 mM NaB for 24 h and examined for the total and acetylated forms of histone H3 and histone H4 proteins by Western blotting. (B) Quantification of acetylated histones H3 and H4 in RTA and LANA promoters by ChIP-PCR assay. BCBL-1 cells treated with 10 mM NAM for 8 h were subjected to ChIP with an antibody to acetylated histone H3 (anti-acetyl-H3) or histone H4 (anti-acetyl-H4) or a control antibody. The immunoprecipitated DNAs were amplified by semiquantitative PCR for two loci in the RTA promoter, one locus in the LANA promoter, or one locus in the GAPDH intergenic region. (C) Quantification of acetylated histones H3 and H4 in the RTA and LANA promoters by ChIP-qPCR assays. Cells were treated as described in the legend to panel B, and the ChIP DNAs were quantified by qPCR. (D) Quantification of the H3K4me3 and H3K27me3 marks in RTA and LANA promoters by ChIP-qPCR assays. ChIP assays were performed with cells treated as described in the legend to B using an antibody to H3K4me3 or H3K27me3 or a control antibody. The ChIP DNAs were quantified by qPCR.
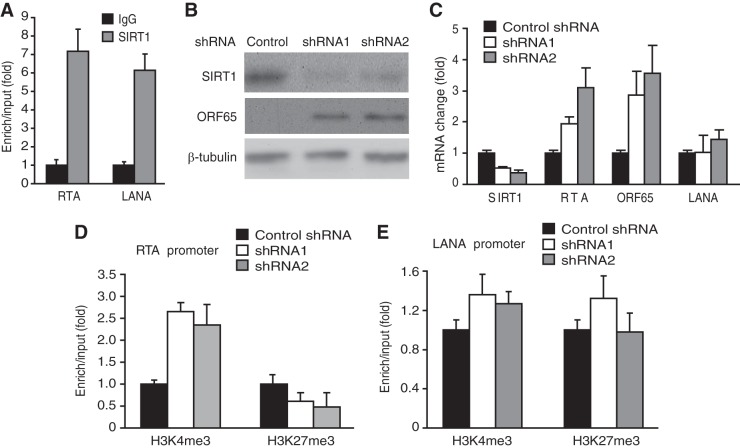
SIRT1 mediates KSHV latency.
We further searched for individual SIRTs that might regulate KSHV latency and identified SIRT1, which bound to both the RTA and LANA promoters (Fig. 5A). To determine if SIRT1 mediated epigenetic silencing of KSHV lytic genes, we performed shRNA knockdown of SIRT1 (Fig. 5B and C). Significantly, knockdown of SIRT1 alone was sufficient to increase the expression of KSHV lytic transcripts RTA and ORF65 but had a marginal effect on the expression of the LANA transcript (Fig. 5C). Knockdown of SIRT1 also increased the expression of the ORF65 protein (Fig. 5B). Consistent with these results, knockdown of SIRT1 increased the level of the H3K4me3 mark but decreased the level of the H3K27me3 mark in the RTA promoter (Fig. 5D) but had no effect on the level of the LANA promoter (Fig. 5E). Together, these results indicate that SIRT1 mediates KSHV latency by maintaining a low level of the active histone H3K4me3 mark and a high level of the repressive histone H3K27me3 mark in the RTA promoter. However, SIRT1 was not involved in the regulation of the LANA promoter.
FIG 5.
SIRT1 mediates KSHV latency. (A) SIRT1 binds to both RTA and LANA promoters. ChIP-qPCR was performed with BCBL-1 cells using an antibody to SIRT1 and a control antibody. (B) Short hairpin RNA knockdown of SIRT1 induces the expression of KSHV lytic protein ORF65. BCBL-1 cells infected with lentiviruses expressing shRNAs to SIRT1 or a scrambled control shRNA for 3 days were examined for the expression of SIRT1 and ORF65 proteins by Western blotting. (C) Expression of SIRT1, RTA, ORF65, and LANA transcripts in BCBL-1 cells following knockdown of SIRT1, as described in the legend to panel B. Viral transcripts were detected by RT-qPCR, and their levels were normalized to the level of cellular GAPDH. (D and E) Quantification of the H3K4me3 and H3K27me3 marks in RTA (D) and LANA (E) promoters by ChIP-qPCR assays following SIRT1 knockdown. Cells infected with lentiviruses expressing shRNAs to SIRT1 or a scrambled control shRNA as described in the legend to panel B were subjected to ChIP using an antibody to H3K4me3 or H3K27me3 or a control antibody. The ChIP DNAs were quantified by qPCR.
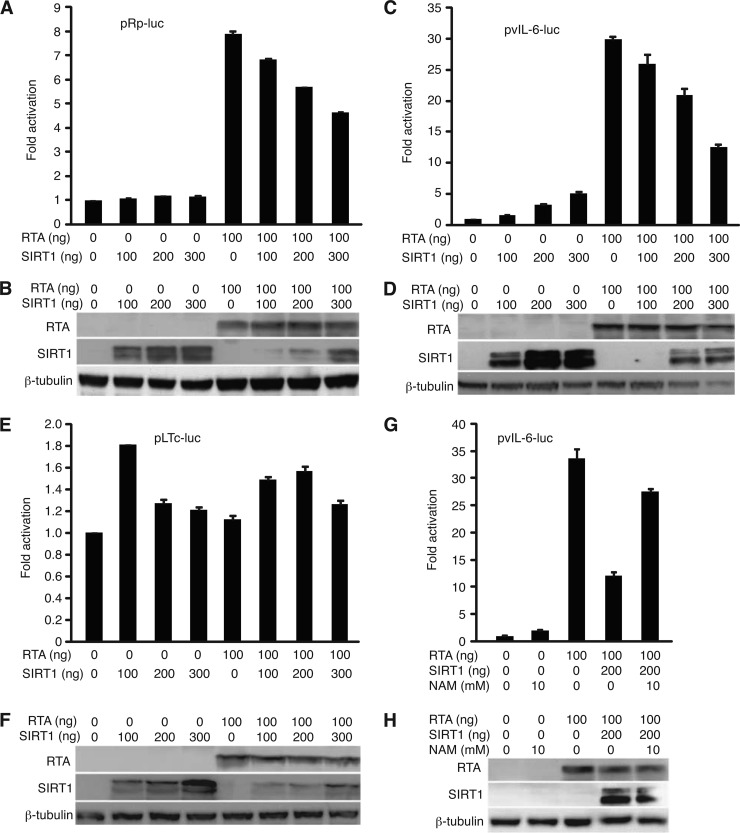
SIRT1 inhibits RTA-mediated transactivation, which is reversed by NAM.
Induction of RTA usually leads to the activation of other KSHV lytic genes and lytic replication (3, 33). RTA also activates its own promoter, resulting in the amplification of the lytic cascade. We examined if SIRT1 regulates RTA autoactivation of its own promoter and transactivation of its downstream genes in reporter assays. Expression of SIRT1 had no obvious effect on the reporter activity of the RTA promoter (Fig. 6A and B). As expected, expression of RTA increased the reporter activity of the RTA promoter by 7.8-fold. However, expression of SIRT1 repressed RTA transactivation of its own promoter in a dose-dependent fashion (Fig. 6A). Similar results were obtained with the vIL-6 promoter reporter, though SIRT1 increased the reporter activity without the presence of RTA (Fig. 6C and D). Expression of RTA increased the reporter activity of the vIL-6 promoter by 30-fold, which was repressed by SIRT1 in a dose-dependent fashion. At the highest dose used, SIRT1 repressed the activities of the RTA and vIL-6 promoters by 41% and 52%, respectively. Interestingly, coexpression of SIRT1 with RTA decreased the expression level of the SIRT1 protein (Fig. 6B and D). This was likely due to RTA's ubiquitin E3 ligase activity, which has been shown to promote the ubiquitination and degradation of cellular proteins (34). It is possible that any RTA-mediated degradation of the SIRT1 protein could alleviate SIRT1 repression of the RTA transactivation function. In contrast to the RTA and vIL-6 promoters, a reporter of the constitutively active LANA latent promoter pLTc, which was unresponsive to RTA, was only marginally affected by SIRT1 with or without the presence of RTA (Fig. 6E and F). Together, these results indicate that SIRT1 represses RTA-mediated transactivation of its downstream genes. Importantly, SIRT1 repression of RTA transactivation of the vIL-6 promoter reporter was relieved by NAM (Fig. 6G and H), further confirming that SIRT1's deacetylase activity is involved in inhibition of the RTA-mediated transactivation function.
FIG 6.
SIRT1 represses RTA transactivation in reporter assays. (A to F) SIRT1 represses RTA activation of its own promoter (A and B) and the vIL-6 promoter (C and D) but not LANA latent promoter LTc (E and F), as shown in reporter assays (A, C, and E) together with Western blots of the expressed RTA and SIRT1 proteins (B, D, and F). 293T cells were cotransfected with the luciferase reporter plasmid pRp-luc, pvIL-6-luc, or pLTc-luc together with RTA or without RTA in the presence of increased doses of a SIRT1 expression plasmid. A β-galactosidase expression construct was used to calibrate the transfection efficiency. The luciferase activity of the reporter without RTA and SIRT1 was given a value of 1. The protein extracts were also examined by Western blotting to detect the expression of the RTA and SIRT1 proteins. (G and H) NAM relieves SIRT1's inhibitory effect on RTA transactivation of the vIL-6 promoter. A reporter assay was carried out as described in the legend to panel A. NAM (10 mM) was added at 5 h posttransfection. The protein extracts were also examined by Western blotting to detect the expression of the RTA and SIRT1 proteins.
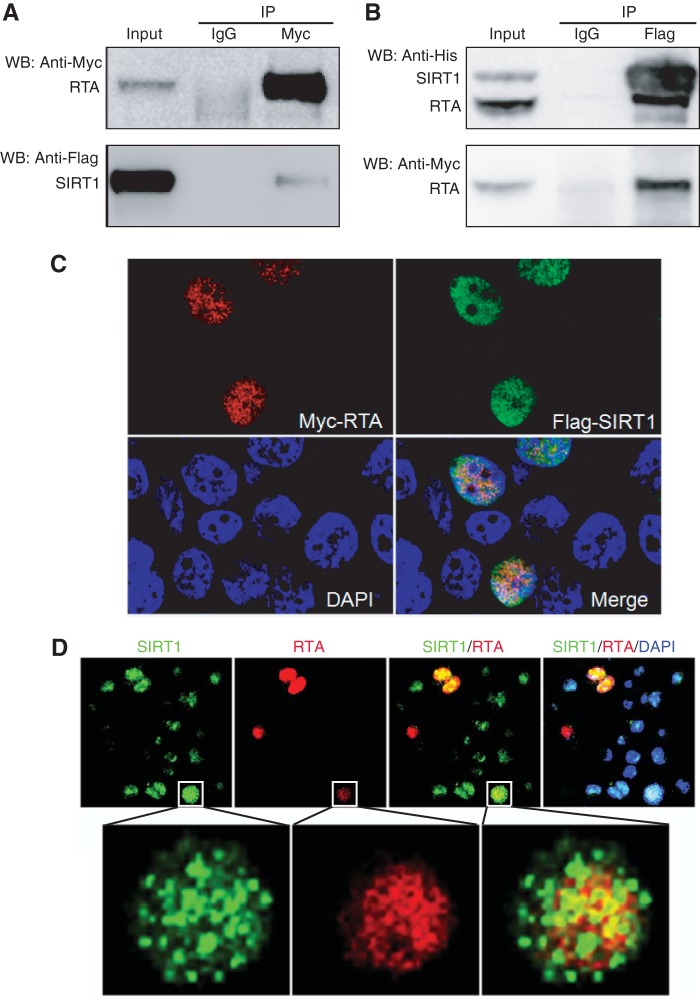
SIRT1 interacts with RTA.
RTA activation of its downstream genes requires acetylation of histones on their promoters and opening of their associated chromatins. To determine how SIRT1's deacetylation activity might inhibit the RTA transactivation function, we examined the SIRT1 and RTA protein interaction. RTA coimmunoprecipitated SIRT1 in 293T cells cotransfected with the expression plasmids of these two proteins (Fig. 7A). Similarly, SIRT1 efficiently coimmunoprecipitated RTA (Fig. 7B). Furthermore, SIRT1 and RTA were partially colocalized in the same nuclear compartment in 293T cells cotransfected with the SIRT1 and RTA expression plasmids (Fig. 7C). Unfortunately, we were not able to observe the SIRT1 and RTA interaction by co-IP in KSHV-infected BCBL-1 cells, albeit many attempts were made (data not shown). Nevertheless, SIRT1 and RTA colocalization was observed in BCBL-1 cells (Fig. 7D). NAM treatment did not affect either the staining patterns of RTA and SIRT1 or their interaction pattern (data not shown). These results indicate that SIRT1 may be associated with RTA to mediate its function. However, the mechanism of SIRT1 inhibition of RTA transactivation might be complex, possibly involving other unidentified viral and cellular factors.
FIG 7.
SIRT1 interacts with RTA. (A and B) Mutual coimmunoprecipitation of the SIRT1 and RTA proteins. (A) SIRT1 was coimmunoprecipitated by RTA; (B) RTA was coimmunoprecipitated by SIRT1. 293T cells were cotransfected with plasmids expressing Flag-His-SIRT1 and Myc-His-RTA. The whole-cell lysates were coimmunoprecipitated with agarose beads coated with an antibody to the Myc tag or Flag tag or control IgG. SIRT1 or RTA was detected by Western blotting (WB) with an antibody to the Myc tag, Flag tag, or His tag. IP, immunoprecipitation. (C) Detection of SIRT1 and RTA colocalization following transient coexpression in 293T cells. 293T cells cotransfected with plasmids expressing Flag-SIRT1 and Myc-RTA were stained with antibodies to the Flag tag and Myc tag and analyzed with a confocal microscope. Nuclei were counterstained with DAPI (4′,6-diamidino-2-phenylindole). (D) Detection of SIRT1 and RTA colocalization in KSHV-infected BCBL-1 cells. BCBL-1 cells were treated with 10 mM NAM, stained for SIRT1 and RTA proteins, and analyzed with a confocal microscope. Nuclei were counterstained with DAPI.
DISCUSSION
KSHV reactivation from latency is a complex process that requires fine-tuning of the sequential expression of viral lytic genes (1). During latency, KSHV lytic genes are epigenetically silenced (13, 35, 36). Several viral latent products are involved in the control of KSHV latency by directly or indirectly recruiting cellular repression complexes to the viral genome (22, 23, 37–40). The efficient switch of KSHV from latency to lytic replication requires the disruption of the repressive signals. While a numbers of factors, including hypoxia, coinfection by other viruses, activation of Toll-like receptors, proinflammatory cytokines, and the reactive oxidative species hydrogen peroxide, have been shown to disrupt viral latency and induce KSHV lytic replication (30, 41–46), additional physiological signals that can trigger KSHV replication remain to be elucidated. In this study, we have demonstrated that inhibitors of class III HDACs, NAM and sirtinol, can efficiently reactivate KSHV from latency. Furthermore, we have shown that SIRT1 binds to both RTA and LANA promoters. Knockdown of SIRT1 is sufficient to reactivate KSHV from latency. These results indicate that SIRTs, or at least SIRT1, are likely involved in the control of KSHV latency and efficient KSHV reactivation from latency would likely require the inhibition of its activity.
SIRTs are closely linked to gene regulation and involved in a broad range of cellular functions, including cell survival, the stress response, tumorigenesis, and metabolism of glucose and fat (47). The deacetylase activity of SIRTs is distinct from that of the conventional class I and II HDACs, in that it is dependent on NAD+ as a cofactor for its activity, which in turn links its enzymatic activity to the cellular abundance of NAD+. NAD+ and NADP+ along with their reduced forms, NADH and NADPH, respectively, which serve as electron-donating and -accepting coenzymes, respectively, are critical for many cellular processes related to cellular metabolism and energy production. It is not surprising that NAD+ has an essential role in glycolysis, fat utilization, and protein deacetylation (48). Our results, for the first time, link class III HDACs to the regulation of KSHV latency and reactivation. Because the deacetylase activity of SIRTs is regulated by cellular NAD+ concentrations, these results also link the cellular metabolic state to KSHV latency and reactivation.
Posttranslational modification of histones represents an important mechanism for regulating gene expression in mammalian cells. In general, chromatin remodeling as a result of histone acetylation is associated with transcription activation, while histone deacetylation often results in a repressive chromatin structure corresponding to transcriptional repression. RTA, as a transactivator of KSHV lytic genes and lytic replication, plays a central role in the switch of KSHV from latency to lytic replication. In latently KSHV-infected cells, the expression of RTA is highly controlled in order for the virus to maintain latent infection. The chemical inducer NaB, an inhibitor of class I and II HDACs, is able to activate KSHV lytic replication by inhibiting the action of HDACs, resulting in the increased acetylation of histones H3 and H4 in the RTA promoter and the disruption of KSHV latency (13). Since NAM inhibits the deacetylation activity of SIRTs, treatment with NAM should alter the levels of acetylated histones in specific gene promoters. While we did not detect any significant changes in total or acetylated histones H3 and H4 following NAM treatment, we indeed found that the levels of acetylated histones H3 and H4 were significantly increased in the RTA promoter, implying that SIRTs might exert preferential deacetylation activity in specific nuclear domains or chromatin regions. Importantly, we found that the level of the active histone mark H3K4me3 was increased in the RTA promoter, while that of the repressive histone mark H3K27me3 was decreased. However, there was no detectable change in the LANA promoter following NAM treatment. Furthermore, we showed that knockdown of SIRT1 increased the level of the H3K4me3 mark and decreased that of the H3K27me3 mark in the RTA promoter but had no effect on the LANA promoter. These results further confirm the involvement of SIRTs, particularly SIRT1, in the control of KSHV latency and imply the potential use of inhibitors of SIRTs as agents that can disrupt KSHV latency. Besides SIRT1, other SIRT members might also regulate KSHV latency. SIRTs usually do not directly bind to chromatins, and their recruitment to specific chromatin domains is likely mediated by other chromatin-associated transcription factors. LANA mediates epigenetic silencing of the KSHV genome by recruiting cellular repressors MECP2 and Dnmt3a (37–39). Thus, SIRTs, particularly SIRT1, can be recruited to KSHV chromatin by interacting with LANA or any of these cellular proteins.
It is interesting that treatment with NAM or knockdown of SIRT1 had differential effects on the promoters of RTA and LANA and the expression of their genes. However, this is not surprising, as there are distinct chromatin boundaries surrounding KSHV latent and lytic loci tightly regulated by cohesins and CTCF (49). These chromatin insulators may coordinate with active and repressive complexes, which likely include SIRTs, to regulate the expression of viral genes.
Efficient KSHV replication requires RTA's optimal transactivation function. RTA transactivates its downstream genes through direct or indirect binding to their promoters (1). RTA also autoregulates its own promoter through interaction with cellular factors, such as CBP, Oct-1, and RBPJK (50–52), resulting in the amplified expression of lytic genes. RTA's transactivation function would need to be repressed in order for KSHV to establish and maintain a successful latent infection. Indeed, a number of viral and cellular factors, such as LANA, K-bZIP (ORF-K8), poly(ADP-ribose) polymerase 1 (PARP-1), HDAC1, and Ste20-like kinase hKFC proteins, have been reported to repress RTA's transactivation function (52–55). In this report, we have shown that SIRT1 interacts and inhibits RTA's transactivation function in promoter reporter assays. Treatment with NAM reverses SIRT1's inhibitory effect on RTA's function, suggesting a specific and functional interaction between the two proteins. Therefore, our results indicate that SIRTs might repress KSHV lytic replication not only during viral latency but also during active viral lytic replication, such as during primary infection, hence promoting the establishment of viral latency. Nevertheless, SIRT1 repression of RTA transactivation of its own promoter was less robust than that of the vIL-6 promoter. Consistent with these results, the reversal of SIRT1 repression of RTA transactivation of its own promoter by NAM was not reproducible (data not shown), which could be due to the weak repression effect of SIRT1 on RTA transactivation of this promoter. RTA directly binds to its own promoter but not the vIL-6 promoter (17). Thus, the mechanisms of transactivation of these two promoters by RTA are distinct, which could determine their responses to SIRT1 and, hence, NAM treatment. Indeed, our results showed that the two promoters differentially responded to RTA and SIRT1 in the reporter assays under both basal and RTA transactivation conditions (Fig. 6A to D).
SIRT1 is known to interact with a wide variety of other chromatin-associated transcription factors and cofactors, including HIF-2α, PGC-1α, SUV39H1, p53, AP-1, and histone acetyltransferase p300 (10, 31, 56–59). Of interest, some of these proteins are implicated in regulating KSHV lytic replication and mediating RTA's transactivation function. SIRT1 can also inhibit RTA's function by binding to the KSHV chromatin through these factors. Recent studies have identified a number of nonhistone proteins, including p53, FOXO, SUV39H1, and MyoD, as SIRT1 substrates (10, 12, 31, 57). The observed SIRT1 and RTA interaction and RTA's increased acetylation upon treatment with NAM (unpublished data) have raised the possibility that RTA is a substrate of SIRT1.
Of interest was the observation of a lower level of the SIRT1 protein when it was coexpressed with the RTA protein. These results are not surprising, because RTA has ubiquitin E3 ligase activity, which can promote the ubiquitination and degradation of cellular proteins (34). Thus, the moderate repressive effect of SIRT1 on RTA's transactivation function shown in the reporter assays of RTA and vIL-6 promoters could be attributed to the degradation of the SIRT1 protein mediated by RTA ligase activity. It could be speculated that, during KSHV reactivation from latency, expression of the RTA protein could target SIRT1 for degradation, which would further break SIRT1's control of KSHV latency, leading to positive amplification of the viral lytic replication program. Given that LANA and RTA are key proteins in mediating KSHV latent and lytic replication, further characterization of the interaction of SIRT1 with LANA and RTA could provide a better understanding of how the cellular metabolic state might regulate the KSHV life cycle.
ACKNOWLEDGMENTS
We thank Ren Sun, Ke Lan, Danny Reiberg, Angus C. Wilson, Gary Hayward, and Charles Wood for providing plasmids and antibodies.
This work was supported by grants from NIH (CA096512, CA124332, CA132637, and CA177377) to S.-J. Gao.
Footnotes
Published ahead of print 26 March 2014
REFERENCES
- 1.Ye FC, Lei XF, Gao SJ. 2011. Mechanisms of Kaposi's sarcoma-associated herpesvirus latency and reactivation. Adv. Virol. 2011:193860. 10.1155/2011/193860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lukac DM, Renne R, Kirshner JR, Ganem D. 1998. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252:304–312. 10.1006/viro.1998.9486 [DOI] [PubMed] [Google Scholar]
- 3.Sun R, Lin SF, Gradoville L, Yuan Y, Zhu F, Miller G. 1998. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc. Natl. Acad. Sci. U. S. A. 95:10866–10871. 10.1073/pnas.95.18.10866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cattelan AM, Calabro ML, Gasperini P, Aversa SM, Zanchetta M, Meneghetti F, De Rossi A, Chieco-Bianchi L. 2001. Acquired immunodeficiency syndrome-related Kaposi's sarcoma regression after highly active antiretroviral therapy: biologic correlates of clinical outcome. J. Natl. Cancer Inst. Monogr. 28:44–49 http://jncimono.oxfordjournals.org/content/2000/28/44.long [DOI] [PubMed] [Google Scholar]
- 5.Engels EA, Biggar RJ, Marshall VA, Walters MA, Gamache CJ, Whitby D, Goedert JJ. 2003. Detection and quantification of Kaposi's sarcoma-associated herpesvirus to predict AIDS-associated Kaposi's sarcoma. AIDS 17:1847–1851. 10.1097/00002030-200308150-00015 [DOI] [PubMed] [Google Scholar]
- 6.Guadalupe M, Pollock BH, Westbrook S, Redding S, Bullock D, Anstead G, Agan BK, Marconi VC, Barbieri S, Sankar V, Rebeles J, Flahive Y, Schoolfield J, Wang L, Lei X, Dow D, Yeh CK, Dang H, Infante AJ, Gao SJ. 2011. Risk factors influencing antibody responses to Kaposi's sarcoma-associated herpesvirus latent and lytic antigens in patients under antiretroviral therapy. J. Acquir. Immune Defic. Syndr. 56:83–90. 10.1097/QAI.0b013e3181fdc928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alkhatib SG, Landry JW. 2011. The nucleosome remodeling factor. FEBS Lett. 585:3197–3207. 10.1016/j.febslet.2011.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michan S, Sinclair D. 2007. Sirtuins in mammals: insights into their biological function. Biochem. J. 404:1–13. 10.1042/BJ20070140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. 2004. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol. Cell 16:93–105. 10.1016/j.molcel.2004.08.031 [DOI] [PubMed] [Google Scholar]
- 10.Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. 2001. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 107:137–148. 10.1016/S0092-8674(01)00524-4 [DOI] [PubMed] [Google Scholar]
- 11.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. 2001. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149–159. 10.1016/S0092-8674(01)00527-X [DOI] [PubMed] [Google Scholar]
- 12.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. 2004. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303:2011–2015. 10.1126/science.1094637 [DOI] [PubMed] [Google Scholar]
- 13.Lu F, Zhou J, Wiedmer A, Madden K, Yuan Y, Lieberman PM. 2003. Chromatin remodeling of the Kaposi's sarcoma-associated herpesvirus ORF50 promoter correlates with reactivation from latency. J. Virol. 77:11425–11435. 10.1128/JVI.77.21.11425-11435.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miller G, Rigsby MO, Heston L, Grogan E, Sun R, Metroka C, Levy JA, Gao SJ, Chang Y, Moore P. 1996. Antibodies to butyrate-inducible antigens of Kaposi's sarcoma-associated herpesvirus in patients with HIV-1 infection. N. Engl. J. Med. 334:1292–1297. 10.1056/NEJM199605163342003 [DOI] [PubMed] [Google Scholar]
- 15.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. 1996. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat. Med. 2:342–346. 10.1038/nm0396-342 [DOI] [PubMed] [Google Scholar]
- 16.Zhou FC, Zhang YJ, Deng JH, Wang XP, Pan HY, Hettler E, Gao SJ. 2002. Efficient infection by a recombinant Kaposi's sarcoma-associated herpesvirus cloned in a bacterial artificial chromosome: application for genetic analysis. J. Virol. 76:6185–6196. 10.1128/JVI.76.12.6185-6196.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen J, Ye F, Xie J, Kuhne K, Gao SJ. 2009. Genome-wide identification of binding sites for Kaposi's sarcoma-associated herpesvirus lytic switch protein, RTA. Virology 386:290–302. 10.1016/j.virol.2009.01.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng H, Young A, Sun R. 2000. Auto-activation of the rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus. J. Gen. Virol. 81:3043–3048 http://vir.sgmjournals.org/content/81/12/3043.long [DOI] [PubMed] [Google Scholar]
- 19.Deng H, Song MJ, Chu JT, Sun R. 2002. Transcriptional regulation of the interleukin-6 gene of human herpesvirus 8 (Kaposi's sarcoma-associated herpesvirus). J. Virol. 76:8252–8264. 10.1128/JVI.76.16.8252-8264.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He M, Zhang W, Bakken T, Schutten M, Toth Z, Jung JU, Gill P, Cannon M, Gao SJ. 2012. Cancer angiogenesis induced by Kaposi sarcoma-associated herpesvirus is mediated by EZH2. Cancer Res. 72:3582–3592. 10.1158/0008-5472.CAN-11-2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moody R, Zhu Y, Huang Y, Cui X, Jones T, Bedolla R, Lei X, Bai Z, Gao SJ. 2013. KSHV microRNAs mediate cellular transformation and tumorigenesis by redundantly targeting cell growth and survival pathways. PLoS Pathog. 9:e1003857. 10.1371/journal.ppat.1003857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Q, Zhou F, Ye F, Gao SJ. 2008. Genetic disruption of KSHV major latent nuclear antigen LANA enhances viral lytic transcriptional program. Virology 379:234–244. 10.1016/j.virol.2008.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ye FC, Zhou FC, Xie JP, Kang T, Greene W, Kuhne K, Lei XF, Li QH, Gao SJ. 2008. Kaposi's sarcoma-associated herpesvirus latent gene vFLIP inhibits viral lytic replication through NF-kappaB-mediated suppression of the AP-1 pathway: a novel mechanism of virus control of latency. J. Virol. 82:4235–4249. 10.1128/JVI.02370-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greene W, Zhang W, He M, Witt C, Ye F, Gao SJ. 2012. The ubiquitin/proteasome system mediates entry and endosomal trafficking of Kaposi's sarcoma-associated herpesvirus in endothelial cells. PLoS Pathog. 8:e1002703. 10.1371/journal.ppat.1002703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao SJ, Deng JH, Zhou FC. 2003. Productive lytic replication of a recombinant Kaposi's sarcoma-associated herpesvirus in efficient primary infection of primary human endothelial cells. J. Virol. 77:9738–9749. 10.1128/JVI.77.18.9738-9749.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones T, Ye FC, Bedolla RG, Meng FHYJ, Qian LW, Pan HY, Zhou FC, Moody R, Wagner B, Arar M, Gao SJ. 2012. Direct and efficient cellular transformation of primary rat mesenchymal precursor cells by KSHV. J. Clin. Invest. 122:1076–1081. 10.1172/JCI58530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye FC, Blackbourn DJ, Mengel M, Xie JP, Qian LW, Greene W, Yeh IT, Graham D, Gao SJ. 2007. Kaposi's sarcoma-associated herpesvirus promotes angiogenesis by inducing angiopoietin-2 expression via AP-1 and Ets1. J. Virol. 81:3980–3991. 10.1128/JVI.02089-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng JH, Zhang YJ, Wang XP, Gao SJ. 2004. Lytic replication-defective Kaposi's sarcoma-associated herpesvirus: potential role in infection and malignant transformation. J. Virol. 78:11108–11120. 10.1128/JVI.78.20.11108-11120.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xie J, Ajibade AO, Ye F, Kuhne K, Gao SJ. 2008. Reactivation of Kaposi's sarcoma-associated herpesvirus from latency requires MEK/ERK, JNK and p38 multiple mitogen-activated protein kinase pathways. Virology 371:139–154. 10.1016/j.virol.2007.09.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye F, Zhou F, Bedolla RG, Jones T, Lei X, Kang T, Guadalupe M, Gao SJ. 2011. Reactive oxygen species hydrogen peroxide mediates Kaposi's sarcoma-associated herpesvirus reactivation from latency. PLoS Pathog. 7:e1002054. 10.1371/journal.ppat.1002054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. 2007. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature 450:440–444. 10.1038/nature06268 [DOI] [PubMed] [Google Scholar]
- 32.Patel DJ, Wang Z. 2013. Readout of epigenetic modifications. Annu. Rev. Biochem. 82:81–118. 10.1146/annurev-biochem-072711-165700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lukac DM, Kirshner JR, Ganem D. 1999. Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73:9348–9361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu Y, Wang SE, Hayward GS. 2005. The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation. Immunity 22:59–70. 10.1016/j.immuni.2004.11.011 [DOI] [PubMed] [Google Scholar]
- 35.Toth Z, Maglinte DT, Lee SH, Lee HR, Wong LY, Brulois KF, Lee S, Buckley JD, Laird PW, Marquez VE, Jung JU. 2010. Epigenetic analysis of KSHV latent and lytic genomes. PLoS Pathog. 6:e1001013. 10.1371/journal.ppat.1001013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gunther T, Grundhoff A. 2010. The epigenetic landscape of latent Kaposi sarcoma-associated herpesvirus genomes. PLoS Pathog. 6:e1000935. 10.1371/journal.ppat.1000935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krithivas A, Young DB, Liao G, Greene D, Hayward SD. 2000. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J. Virol. 74:9637–9645. 10.1128/JVI.74.20.9637-9645.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Krithivas A, Fujimuro M, Weidner M, Young DB, Hayward SD. 2002. Protein interactions targeting the latency-associated nuclear antigen of Kaposi's sarcoma-associated herpesvirus to cell chromosomes. J. Virol. 76:11596–11604. 10.1128/JVI.76.22.11596-11604.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shamay M, Krithivas A, Zhang J, Hayward SD. 2006. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi's sarcoma-associated herpesvirus LANA. Proc. Natl. Acad. Sci. U. S. A. 103:14554–14559. 10.1073/pnas.0604469103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lei X, Bai Z, Ye F, Xie J, Kim CG, Huang Y, Gao SJ. 2010. Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat. Cell Biol. 12:193–199. 10.1038/ncb2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang J, Renne R, Dittmer D, Ganem D. 2000. Inflammatory cytokines and the reactivation of Kaposi's sarcoma-associated herpesvirus lytic replication. Virology 266:17–25. 10.1006/viro.1999.0077 [DOI] [PubMed] [Google Scholar]
- 42.Haque M, Davis DA, Wang V, Widmer I, Yarchoan R. 2003. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) contains hypoxia response elements: relevance to lytic induction by hypoxia. J. Virol. 77:6761–6768. 10.1128/JVI.77.12.6761-6768.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gregory SM, West JA, Dillon PJ, Hilscher C, Dittmer DP, Damania B. 2009. Toll-like receptor signaling controls reactivation of KSHV from latency. Proc. Natl. Acad. Sci. U. S. A. 106:11725–11730. 10.1073/pnas.0905316106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin D, Feng N, Fan W, Ma X, Yan Q, Lv Z, Zeng Y, Zhu J, Lu C. 2011. Activation of PI3K/AKT and ERK MAPK signal pathways is required for the induction of lytic cycle replication of Kaposi's sarcoma-associated herpesvirus by herpes simplex virus type 1. BMC Microbiol. 11:240. 10.1186/1471-2180-11-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu X, Zhou F, Qin D, Zeng Y, Lv Z, Yao S, Lu C. 2011. Human immunodeficiency virus type 1 induces lytic cycle replication of Kaposi's-sarcoma-associated herpesvirus: role of Ras/c-Raf/MEK1/2, PI3K/AKT, and NF-kappaB signaling pathways. J. Mol. Biol. 410:1035–1051. 10.1016/j.jmb.2011.03.055 [DOI] [PubMed] [Google Scholar]
- 46.Lu C, Zeng Y, Huang Z, Huang L, Qian C, Tang G, Qin D. 2005. Human herpesvirus 6 activates lytic cycle replication of Kaposi's sarcoma-associated herpesvirus. Am. J. Pathol. 166:173–183. 10.1016/S0002-9440(10)62242-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haigis MC, Sinclair DA. 2010. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 5:253–295. 10.1146/annurev.pathol.4.110807.092250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Belenky P, Racette FG, Bogan KL, McClure JM, Smith JS, Brenner C. 2007. Nicotinamide riboside promotes Sir2 silencing and extends lifespan via Nrk and Urh1/Pnp1/Meu1 pathways to NAD+. Cell 129:473–484. 10.1016/j.cell.2007.03.024 [DOI] [PubMed] [Google Scholar]
- 49.Stedman W, Kang H, Lin S, Kissil JL, Bartolomei MS, Lieberman PM. 2008. Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 27:654–666. 10.1038/emboj.2008.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liang Y, Chang J, Lynch SJ, Lukac DM, Ganem D. 2002. The lytic switch protein of KSHV activates gene expression via functional interaction with RBP-Jkappa (CSL), the target of the Notch signaling pathway. Genes Dev. 16:1977–1989. 10.1101/gad.996502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sakakibara S, Ueda K, Chen J, Okuno T, Yamanishi K. 2001. Octamer-binding sequence is a key element for the autoregulation of Kaposi's sarcoma-associated herpesvirus ORF50/Lyta gene expression. J. Virol. 75:6894–6900. 10.1128/JVI.75.15.6894-6900.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gwack Y, Byun H, Hwang S, Lim C, Choe J. 2001. CREB-binding protein and histone deacetylase regulate the transcriptional activity of Kaposi's sarcoma-associated herpesvirus open reading frame 50. J. Virol. 75:1909–1917. 10.1128/JVI.75.4.1909-1917.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gwack Y, Nakamura H, Lee SH, Souvlis J, Yustein JT, Gygi S, Kung HJ, Jung JU. 2003. Poly(ADP-ribose) polymerase 1 and Ste20-like kinase hKFC act as transcriptional repressors for gamma-2 herpesvirus lytic replication. Mol. Cell. Biol. 23:8282–8294. 10.1128/MCB.23.22.8282-8294.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Izumiya Y, Lin SF, Ellison T, Chen LY, Izumiya C, Luciw P, Kung HJ. 2003. Kaposi's sarcoma-associated herpesvirus K-bZIP is a coregulator of K-Rta: physical association and promoter-dependent transcriptional repression. J. Virol. 77:1441–1451. 10.1128/JVI.77.2.1441-1451.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lan K, Kuppers DA, Verma SC, Robertson ES. 2004. Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency. J. Virol. 78:6585–6594. 10.1128/JVI.78.12.6585-6594.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dioum EM, Chen R, Alexander MS, Zhang Q, Hogg RT, Gerard RD, Garcia JA. 2009. Regulation of hypoxia-inducible factor 2alpha signaling by the stress-responsive deacetylase sirtuin 1. Science 324:1289–1293. 10.1126/science.1169956 [DOI] [PubMed] [Google Scholar]
- 57.Amat R, Planavila A, Chen SL, Iglesias R, Giralt M, Villarroya F. 2009. SIRT1 controls the transcription of the peroxisome proliferator-activated receptor-gamma co-activator-1alpha (PGC-1alpha) gene in skeletal muscle through the PGC-1alpha autoregulatory loop and interaction with MyoD. J. Biol. Chem. 284:21872–21880. 10.1074/jbc.M109.022749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dey S, Bakthavatchalu V, Tseng MT, Wu P, Florence RL, Grulke EA, Yokel RA, Dhar SK, Yang HS, Chen Y, St Clair DK. 2008. Interactions between SIRT1 and AP-1 reveal a mechanistic insight into the growth promoting properties of alumina (Al2O3) nanoparticles in mouse skin epithelial cells. Carcinogenesis 29:1920–1929. 10.1093/carcin/bgn175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Das C, Lucia MS, Hansen KC, Tyler JK. 2009. CBP/p300-mediated acetylation of histone H3 on lysine 56. Nature 459:113–117. 10.1038/nature07861 [DOI] [PMC free article] [PubMed] [Google Scholar]