ABSTRACT
We report that UL133-UL138 (UL133/8), a transcriptional unit within the ULb′ region (ULb′) of the human cytomegalovirus (HCMV) genome, and UL97, a viral protein kinase encoded by HCMV, play epistatic roles in facilitating progression of the viral lytic cycle. In studies with HCMV strain TB40/E, pharmacological blockade or genetic ablation of UL97 significantly reduced the levels of mRNA and protein for IE2 and viral early and early-late genes during a second wave of viral gene expression that commenced at between 24 and 48 h postinfection. These effects were accompanied by significant defects in viral DNA synthesis and viral replication. Interestingly, deletion of UL133/8 likewise caused significant defects in viral DNA synthesis, viral gene expression, and viral replication, which were not exacerbated upon UL97 inhibition. When UL133/8 was restored to HCMV laboratory strain AD169, which otherwise lacks the locus, the resulting recombinant virus replicated similarly to the parental virus. However, during UL97 inhibitor treatment, the virus in which UL133/8 was restored showed significantly exacerbated defects in viral DNA synthesis, viral gene expression, and production of infectious progeny virus, thus recapitulating the differences between wild-type TB40/E and its UL133/8-null derivative. Phenotypic evaluation of mutants null for specific open reading frames within UL133/8 revealed a role for UL135 in promoting viral gene expression, viral DNA synthesis, and viral replication, which depended on UL97. Taken together, our findings suggest that UL97 and UL135 play interdependent roles in promoting the progression of a second phase of the viral lytic cycle and that these roles are crucial for efficient viral replication.
IMPORTANCE A unique feature of the herpesviruses, such as human cytomegalovirus (HCMV), is that they can undergo latency, a state during which the virus silences its gene expression, which allows lifelong viral persistence in immunocompetent hosts. We have uncovered an unexpected link between a cluster of HCMV genes involved in latency, UL133-UL138, and a virally encoded protein kinase, UL97, which plays crucial roles in manipulating the cell cycle during HCMV lytic replication. Although viral immediate early (IE) gene expression is essential for HCMV lytic replication, the activation of IE gene expression in latently infected cells is not sufficient to result in production of infectious virus. Our findings here and in an accompanying study (M. Umashankar, M. Rak, F. Bughio, P. Zagallo, K. Caviness, and F. D. Goodrum, J. Virol. 88:5987–6002, 2014) show that proteins expressed from the UL133-UL138 latency locus and UL97 play interdependent roles in overcoming checkpoints that restrict the viral lytic replication cycle, findings which suggest intriguing implications for establishment of and reactivation from HCMV latency.
INTRODUCTION
Human cytomegalovirus (HCMV) is the archetypal betaherpesvirus (reviewed in reference 1). Its genome, at over 235 kbp, is the largest of any virus known to infect humans. As with all herpesviruses, the HCMV lytic replication cycle is comprised of an orderly cascade of viral protein expression, which is classified into three phases: immediate early (IE; or α), early (E; or β; also called delayed early), and late (L; or γ). Viral genes are categorized into these kinetic classes on the basis of whether their expression shows sensitivity to inhibition of transcription (IE), translation (E), and/or viral DNA synthesis (L). The ability of herpesviruses such as HCMV to regulate their gene expression is important not only for lytic replication but also for viral latency, a state during which viral gene expression is minimized while the potential for reactivation is maintained.
Despite having such a large genome, HCMV encodes only one conventional protein kinase, UL97 (also referred to as pUL97), which is thought to act at multiple stages during infection to promote efficient viral replication (reviewed in reference 2). Despite its importance as an antiviral drug target (3), it remains unclear which roles of UL97 are the most crucial for infection and pathogenesis in vivo. The most well understood functions of the protein involve its mimicry of cellular cyclin-dependent kinase (CDK)–cyclin complexes: UL97 phosphorylates CDK substrates, including the retinoblastoma tumor suppressor protein (pRb) (4, 5), and A-type nuclear lamins (6–8). UL97 phosphorylates pRb at sites consistent with inactivation of its tumor suppressor function (4), which is thought to involve repression of cellular genes important for progression toward S phase (9). Moreover, UL97-mediated inactivation of pRb-family pocket proteins is required for efficient viral DNA synthesis in quiescent cells (10). UL97 phosphorylates A-type lamins on a site that regulates the disassembly of lamin polymers (7), which is hypothesized to facilitate the escape of newly assembled viral nucleocapsids from the nucleus (viral nuclear egress) (6–8, 11, 12). UL97 may also exert effects on the host cell cycle by phosphorylating Cdh1 (Fzr), a subunit of the anaphase-promoting complex/cyclosome (APC/C), which likewise represents a CDK substrate (13, 14).
In this study, we present evidence that UL97 is required during infection to stimulate progression of the viral lytic cycle. We found that UL97-deficient infections showed defects in a second phase of viral mRNA and protein expression, which involved IE2, a key transactivator of the viral lytic cycle, and viral early (E) and early-late (E-L) genes that participate in viral genome replication, which are likely regulated by IE2 (reviewed in reference 1). Consistent with these observations, we found that viral genome replication occurred at decreased levels when the second wave of viral gene expression was attenuated upon pharmacological inhibition or genetic ablation of UL97.
Furthermore, we demonstrate that progression of the viral lytic cycle is also strongly influenced by UL133-UL138 (UL133/8), a locus within the ULb′ region of the HCMV genome. Deletion of UL133/8 resulted in a basal defect in viral replication, which was accompanied by underlying defects in viral DNA synthesis, and in second-phase expression of IE2 and viral E and E-L genes, which were not further affected by UL97 inhibition. Conversely, reintroduction of UL133/8 to an HCMV strain that had spontaneously lost the locus during laboratory adaptation led to exacerbated defects during UL97 inhibition. Finally, we identify UL135, a constituent of UL133/8, to be necessary for these UL97-dependent effects on viral gene expression.
MATERIALS AND METHODS
Cells and virus.
The bacterial artificial chromosome (BAC) clone of HCMV strain AD169, AD169rv (15), was a gift of Ulrich Koszinowski. A BAC clone of TB40/E, TB40-BAC4 (16), was a gift of Christian Sinzger (University of Ulm, Ulm, Germany), and its UL97-null (Δ97) mutant derivative has been previously described (17). TB40/E with a deletion of the entire UL133/8 locus (TB40/E_UL133-UL138NULL [UL133/8NULL]), a TB40/E mutant unable to express UL138 (TB40/E_UL138STOP [UL138STOP]), and a TB40/E mutant that does not express either UL135 or UL138 (TB40/E_UL135STOP/UL138STOP [5/8STOP]) are described elsewhere (18, 19). AD169_UL133/8 was constructed using en passant Red mutagenesis in Escherichia coli (20, 21), as previously described (17). Briefly, a PCR product containing an I-SceI–aphAI cassette was generated using primers TB_UL135Kan_Fw (5′-GCT CTA CAT AGG AGA GGA TGG TCT GCC GAT AGA TAA ACC CGA GTT TCC TCC GGC GCG ATT TAG GGA TAA CAG GGT AAT CGA TTT-3′) and TB_UL135Kan_Rv (5′-TG GAT ACG TCG GGG ATC TCG AAT CGC GCC GGA GGA AAC TCG GGT TTA TCT ATC GGC AGA CGC CAG TGT TAC AAC CAA TTA ACC-3′) and inserted via Red recombination into the UL135 coding region of TB40/E. The resulting kanamycin-resistant (Kanr) BAC, TB40/E_UL135_aphAI, was used as the template to generate a PCR product of the entire UL133/8 region, including the I-SceI–aphAI cassette, using primers Ad_TBUL148A_Fw (5′-CTC CTC CAG GTA GTG GGT CTG ACT GCG ACG CAG CGT CCA GTT CAT GTA AAA CCT CCG TAG TCC TGT TCG C-3′) and Ad_TBUL139_Rv (5′-GAC TGG ATT AGT AAG CAG CCT CTT CGT GGC CGG ACA CGG CGA GAC CGA CTG TTT ATT ACT ATA TAA TTG A-3′). The 5′ ends of the primers were designed to promote a recombination event that would replace a 16-bp noncoding region between RL13 and UL132 of AD169, corresponding to AD169 nucleotide positions 42455 to 42470 (the numbering is according to that for the sequence with GenBank accession number AC146999.1), with the UL133/8 region of strain TB40/E. The inserted UL133/8 sequences corresponded to nucleotide positions 219231 to 223352 of the sequence with GenBank accession number EF999921.1 (TB40/E BAC). The PCR product was electroporated into E. coli GS1783 harboring the AD169rv BAC. After isolation of Kanr colonies, I-SceI endonuclease and bacteriophage λ Red recombinase activities were induced to remove the I-SceI–aphAI cassette. The resulting recombinant BAC, which we refer to as AD169_UL133/8, was verified by DNA sequencing of a 4.615-kb contiguous region that included the boundaries of sequences involved in en passant recombination events, as well as 4.122 kb of introduced UL133/8 sequences. The verified sequence spanned from nucleotide positions 42388 to 42882 of the AD169 genome (the numbering is according to that for the sequence with GenBank accession number AC146999.1 and does not reflect the insertion of UL133/8). The BAC was also analyzed by restriction enzyme digestion to confirm that spurious changes had not occurred and was reconstituted to generate infectious virus, as described previously (17). Viral stocks were prepared by ultracentrifuge concentration, and titers were determined by assays that measured the 50% tissue culture infective dose (TCID50) or the number of infectious units (IU)/ml, which have been described elsewhere (17). Replication kinetics experiments were performed as previously described (17).
Primary human foreskin fibroblasts (HFFs) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 20 μg/ml gentamicin sulfate, and 10 μg/ml ciprofloxacin (complete medium), and experimental infections were conducted in medium identical to complete medium, except that it contained 2% FBS instead of 10% FBS, all as described previously (17).
Pharmacological inhibitors.
1-(β-l-Ribofuranosyl)-2-isopropylamino-5,6-dichlorobenzimidazole (22, 23), which is also known by the trade name maribavir (MBV), was a generous gift of John Drach (University of Michigan, Ann Arbor, MI). Medium containing MBV at 2 μM or dimethyl sulfoxide (DMSO) at 0.1% (vol/vol) to control for the effects of the carrier was applied at 2 h postinfection (hpi) and maintained until samples were harvested.
Western blotting.
Western blotting procedures, including quantification of results on a LI-COR Odyssey system (LI-COR, Inc., Lincoln, NE), were carried out as described elsewhere (17). UL57, UL44, IE1, and human beta-actin were detected as previously described (17). IE2 was detected using mouse monoclonal antibody clone 5A8.2 (MAB8410; Millipore, Inc.). Rabbit polyclonal antibodies to UL138 and UL135 are described elsewhere (19, 24).
qPCR.
Real-time quantitative PCR (qPCR) to determine viral DNA levels was performed as previously described (8, 17, 25). For quantification of UL123, PCR efficiencies ranged from 96.8% to 109.9%, standard curve regression values (r2) ranged from 0.981 to 0.996, and the slopes of the calibration curve ranged from −3.321 to −3.4. For the 18S ribosomal DNA (rDNA), PCR efficiency ranged from 99.2% to 104.3%, r2 values were from 0.951 to 0.996, and the slopes of the calibration curve were from −3.106 to −3.519.
For quantification of viral RNA, total RNA was isolated from infected cells using an RNeasy minikit with an on-column DNase digestion step, as per the manufacturer's instructions (Qiagen, Inc., Valencia, CA). Two hundred nanograms of total RNA from infected cells was used as the template to produce oligo(dT)-primed cDNA with a ProtoScript II first-strand cDNA synthesis kit (New England BioLabs, Danvers, MA). Resulting cDNAs and control samples without reverse transcriptase (RT) were diluted 3-fold with water and used as the template in qPCRs to quantify the abundance of the transcripts of the indicated viral genes and of cellular GAPDH (glyceraldehyde-3-phosphate dehydrogenase) transcripts, which were used for normalization. Control samples in which RT was intentionally left out during the cDNA synthesis step uniformly produced qPCR results indistinguishable from the results of RT-qPCR on RNA samples from mock-infected cells (not shown). The primers used in RT-qPCR were specific for the GAPDH gene (5′-CTG TTG CTG TAG CCA AAT TCG T-3′ and 5′-ACC CAC TCC TCC ACC TTT GAC-3′) (26), UL69 (5′-CCT ACG ACT TTC GGT TCT TCT C-3′ and 5′-CGT CCA GTT CGT CGT CAA TAA-3′), UL57 (5′-TGT CAC CTA TGG GCA CAT TC-3′ and 5′-GAC ATC ACG AGC GAC TCT TT-3′), and UL44 (5′-GGC GTG AAA AAC ATG CGT ATC AAC-3′ and 5′-TAC AAC AGC GTG TCG TGC TCC G-3′). UL123 (IE1) was detected by the same primers referred to above for quantification of viral DNA (25), and UL122 (IE2) was detected using primers 5′-TGA CCG AGG ATT GCA ACG A-3′ and 5′-CGG CAT GAT TGA CAG CCT G-3′, which span an intron (27). Except for the primers specific for GAPDH, UL122, and UL123, all RT-qPCR primers were designed using PrimerQuest software from Integrated DNA Technologies, Inc. (Coralville, IA), and the specificity of their PCR products was confirmed by melting curve analysis (not shown). For newly designed primer pairs, the linearity of the results was validated across a series of dilutions of template (not shown).
Statistical analyses.
Analyses to determine statistical significance were performed using GraphPad Prism, version 6.0c (GraphPad Software, La Jolla, CA).
RESULTS
UL97 positively influences transcript levels for IE2 and for representative early and early-late genes during a second phase of viral gene expression.
We previously observed that pharmacological inhibition or genetic ablation of UL97 negatively influenced the expression of viral early (E; also called delayed-early) and early-late (E-L) proteins, such as UL57 and UL44, which directly participate in viral DNA synthesis (17). In the same study, we also found that the ULb′ region (ULb′) of the HCMV genome conferred an increased requirement for UL97 in viral DNA synthesis. These observations led us to hypothesize that UL97 regulates the expression of viral E and E-L proteins that participate in viral DNA synthesis and that ULb′ plays a role in this process. As a first step toward addressing our hypothesis, we sought to better define how UL97 might influence viral protein expression. Therefore, we conducted experiments with HCMV strain TB40/E, which contains full-length ULb′, to determine whether pharmacological inhibition or genetic ablation of UL97 would affect viral mRNA levels during infection.
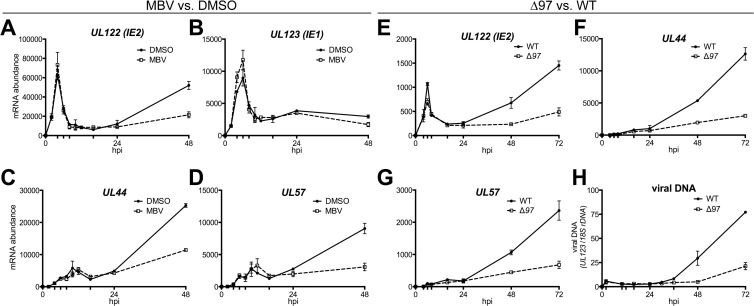
We infected fibroblasts at a multiplicity of infection (MOI) of 1 and incubated them from 2 hpi onwards in medium containing either the UL97 inhibitor maribavir (MBV) at 2 μM or 0.1% DMSO as a carrier-alone control. We quantified mRNA levels for UL123 (IE1) and UL122 (IE2), two viral genes driven by the viral major immediate early promoter (MIEP), and for UL44 and UL57, which represent E-L and E transcripts, respectively, over a time course of infection by reverse transcriptase quantitative PCR (RT-qPCR). For all four viral genes, we found that mRNA levels during infection exhibited a biphasic nature (Fig. 1A to D). As expected (28–30), the levels of IE1 (UL123) and IE2 (UL122) mRNAs rose to an initial peak at 6 hpi, after which their levels declined substantially, bottoming out at 10 to 16 hpi (Fig. 1A and B). IE1 (UL123) mRNA levels then rose again (though not nearly to their previous peak), plateaued at 24 hpi, and then exhibited a slight decline by 48 hpi (Fig. 1B). IE2 (UL122) mRNA levels, on the other hand, continued to increase from 24 to 48 hpi (Fig. 1A). Our observations of the biphasic accumulation of IE2 mRNA and the different patterns of transcript abundance between IE1 and IE2 are consistent with previous reports (28–30). However, we were intrigued that the second phase of IE2 (UL122) mRNA accumulation was markedly reduced during MBV treatment (Fig. 1A).
FIG 1.
Pharmacological inhibition or genetic ablation of UL97 leads to decreased late-phase mRNA accumulation for IE2 and for two viral early/early-late DNA synthesis factors. (A to D) mRNA abundance for the indicated viral genes was measured at 2, 4, 6, 8, 10, 12, 16, 24, and 48 hpi by RT-qPCR and normalized to GAPDH mRNA levels. Results are from infections at an MOI of 1 in the presence of MBV at 2 μM or DMSO at 0.1% (vol/vol) and are plotted relative to the signal from mock-infected samples. Control reactions excluding reverse transcriptase uniformly resulted in threshold cycle values indistinguishable from those of the RT-qPCR for mock-infected cells (not shown). (E to G) Viral mRNA levels for UL97-null TB40/E (Δ97) and parental wild-type TB40/E (WT) at an MOI of 1 were compared at 4, 6, 8, 16, 24, 48, and 72 hpi. (H) Viral DNA levels were monitored at 2, 10, 12, 16, 24, 30, 36, 48, and 72 hpi by qPCR following infection with the Δ97 mutant or WT. Error bars represent SEMs.
For viral E and E-L transcripts, represented by the UL57 and UL44 mRNAs, respectively, the first peak was detected at about 10 to 12 hpi and was modest in amplitude compared to the amplitudes of the two IE mRNAs (Fig. 1C and D). Subsequently, mRNA levels for UL57 and UL44 fell substantially, reaching a low point at 16 hpi (Fig. 1C and D). Congruent with the findings for IE2 (UL122), however, UL57 and UL44 mRNA levels began to rise again by 24 hpi and continued to increase through 48 hpi (Fig. 1C and D). As with IE2 (UL122), the first peak of viral E and E-L gene expression was unaffected during MBV treatment, while the second phase was strongly attenuated by it (Fig. 1C and D). Similar results were seen for other E and E-L mRNAs, including US28, UL97, UL54, UL69, and UL70 (G. Li and J. P. Kamil, unpublished results).
To exclude the possibility that off-target effects of MBV might have caused these effects, we compared wild-type virus (WT) and UL97-null (Δ97) mutant virus (Fig. 1E to G). Δ97 mutant virus infections exhibited 3-fold defects in the levels of IE2, UL44, and UL57 mRNA at 48 hpi and 72 hpi. Results from earlier time points, however, were for the most part indistinguishable between WT and the Δ97 mutant virus. Because these results were similar to those from MBV treatment, we concluded that the effects of MBV on viral mRNA levels were due to inhibition of UL97. Additionally, we observed that the onset of viral DNA synthesis in strain TB40/E, as indicated by qPCR detection of viral DNA, coincided with the second wave of viral mRNA expression (Fig. 1H). Moreover, the defect in viral DNA synthesis in the Δ97 mutant virus likewise coincided with its defects in mRNA expression (Fig. 1C to H). Together, the results suggest that UL97 influences the levels of mRNA for at least two viral genes involved in viral DNA synthesis and for IE2, a crucial transactivator of the HCMV lytic cycle (1).
Ablation of UL133/8 abolishes the effects of UL97 inhibition on viral DNA synthesis and viral replication.
Our mRNA results suggested that the roles of UL97 in viral gene expression might contribute to the requirement for UL97 in viral DNA synthesis, which we had previously found to involve ULb′ (17). To understand how ULb′ might influence viral DNA synthesis, we first needed to narrow down which particular gene(s) might be responsible. Therefore, we constructed three mutants with large deletions within ULb′ and observed that a virus ablated for UL133-UL141 exhibited attenuated defects in viral replication and viral DNA synthesis during treatment with a UL97 inhibitor (not shown). The UL133-UL138 (UL133/8) locus comprised the largest portion of UL133-UL141, and moreover, UL141 is nonfunctional in TB40/E (16, 31). We thus hypothesized that UL133/8 influences the requirement for UL97 during infection. To address our hypothesis, we tested the effects of UL97 inhibition on replication of a virus with a deletion of the entire UL133/8 locus, TB40/E_UL133/8NULL (UL133/8NULL).
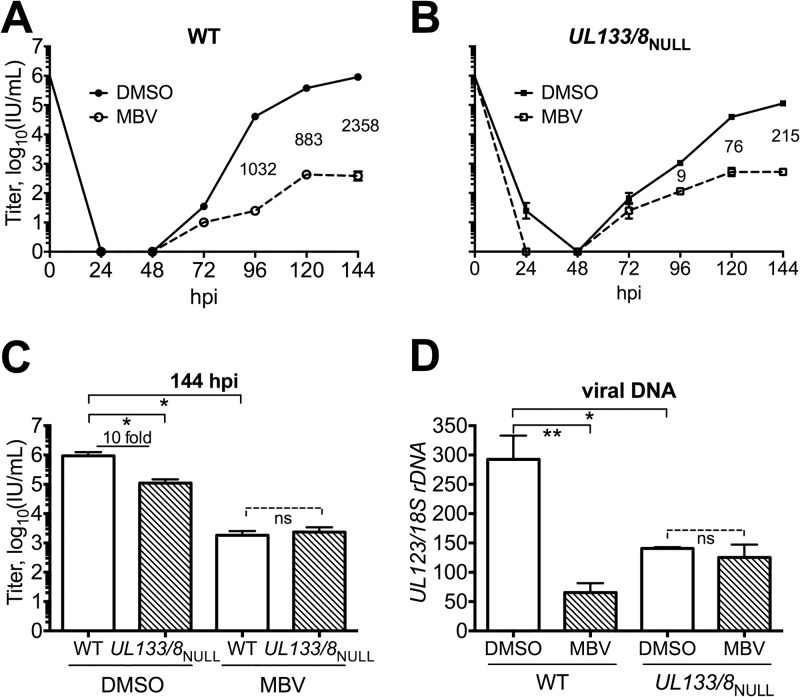
During infection at an MOI of 1, UL97 inhibitor treatment more drastically reduced replication in WT than in UL133/8NULL. At 120 hpi and 144 hpi, MBV caused defects in WT that were 10-fold larger than those seen in UL133/8NULL (Fig. 2A and B). However, under the DMSO control conditions, infectious virions were produced at approximately 10-fold lower levels in UL133/8NULL than in WT, and this difference was statistically significant (Fig. 2C). Despite the basal replication defect of UL133/8NULL, WT and UL133/8NULL replicated indistinguishably in the presence of MBV (Fig. 2A to C). These observations suggest (i) that UL133/8 provides a benefit to viral replication for strain TB40/E, leading to a viral replication defect in its absence, and (ii) that UL97 kinase activity is required for this benefit, such that UL133/8NULL and WT exhibit indistinguishable replication defects when UL97 is inhibited.
FIG 2.
Deletion of UL133/8 attenuates the effects of UL97 inhibition on viral replication and viral DNA synthesis. Fibroblasts were infected with WT or UL133/8NULL at an MOI of 1 in the presence of 2 μM MBV or 0.1% DMSO carrier alone. (A and B) Replication curves reflecting the production of cell-free virus are shown for WT (A) and UL133/8NULL (B). Numbers between curves indicate fold defects. Error bars, where perceptible, indicate standard deviations. (C) Results at 144 hpi from three independent biological replicates of the experiment shown in panels A and B are plotted side by side. (D) Levels of viral DNA at 96 hpi from DMSO control- and MBV-treated infections are shown as the number of copies of a viral gene, UL123, normalized to the number of copies of the cellular 18S rDNA determined by qPCR in two independent biological replicates. For panels C and D, error bars indicate SEMs. One-way analysis of variance, followed by Tukey's posttest, was used to determine statistical significance. ns, not significant; *, P < 0.05; **, P < 0.01.
We reasoned that if UL133/8 accounted for the influence of ULb′ on the requirement for UL97 (17), then the effects of a UL97 inhibitor on viral DNA synthesis should be attenuated in UL133/8NULL infections. Indeed, MBV failed to influence viral DNA levels in UL133/8NULL infections, while a significant, 5-fold decrease was observed for WT infections (Fig. 2D). Moreover, as with viral replication, basal levels of viral DNA synthesis were significantly lower in UL133/8NULL infections than in WT infections (Fig. 2D). Roles of UL97 in other processes, such as in viral nuclear egress (11), seem likely to explain why MBV still reduces the replication of UL133/8NULL (albeit to a reduced extent) while failing to affect its DNA synthesis. Overall, these results imply that UL97 and one or more genes encoded within UL133/8 must each be present for wild-type levels of viral DNA synthesis and, moreover, suggest an epistatic relationship between UL97 and UL133/8.
Timely expression of IE2 and high-level expression of viral early and early-late genes depend on UL97 and UL133/8.
The viral proteins that replicate the HCMV genome are expressed with E or E-L kinetics. For example, UL44, an E-L gene, encodes a viral DNA polymerase accessory factor, and UL57, an E gene, encodes a viral single-stranded DNA binding protein (reviewed in reference 1). Expression of the E and E-L genes is thought to be regulated by viral IE transactivators, most notably, IE2 (1). Because we had observed that the second phase of IE2 mRNA expression depended on UL97 (Fig. 1), it seemed plausible that UL97 and one or more functions encoded within UL133/8 were required to promote expression of IE2. Moreover, impaired expression of IE2 might be expected to result in the deficient expression of viral E and E-L products, which could explain the defects in viral DNA synthesis that we observed when either UL133/8 or UL97 was absent (Fig. 2D) (17). To investigate this possibility, we compared the effects of UL97 inhibition on viral gene expression during WT and UL133/8NULL infections.
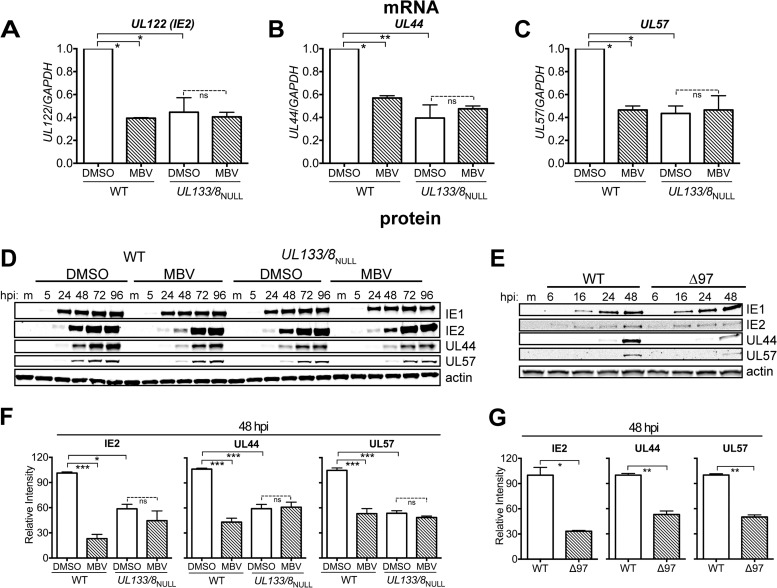
We found that UL133/8NULL infections showed significantly lower levels of mRNAs for IE2, UL44, and UL57 than WT infections at 48 hpi (Fig. 3A to C). Moreover, in UL133/8NULL infections, MBV failed to cause viral mRNA levels to decrease, while in WT infections, the drug caused statistically significant, ∼2- to 3-fold decreases in transcript levels for IE2 (UL122), UL44, and UL57 (Fig. 3A to C). From these results, it appears that inhibition of UL97 decreases the levels of mRNA for IE2 and for at least two viral E/E-L genes and that these effects depend on UL133/8.
FIG 3.
The UL133/8 locus is required for high-level accumulation of mRNA and protein for IE2 and for two viral proteins involved in DNA synthesis and for a UL97 inhibitor to influence their accumulation. (A to C) mRNA abundance at 48 hpi for UL122 (IE2), UL44, and UL57 is shown for infections of fibroblasts with either WT or UL133/8NULL at an MOI of 1 in the presence of 2 μM MBV or 0.1% DMSO. mRNA abundance is shown relative to that in DMSO control-treated WT infections, which were set to 1.0. The results represent the averages from two independent biological replicates; a one-way analysis of variance with Tukey's posttest was used to evaluate statistical significance. (D) Fibroblasts were infected with WT or UL133/8NULL at an MOI of 1 or mock infected (lanes m) and maintained in the presence or absence of MBV. A series of protein lysates was collected at the indicated time points, and expression of the specified viral proteins was monitored by Western blotting. (E) Fibroblasts were infected with WT or TB40/E Δ97 (Δ97) at an MOI of 1 or mock infected, and lysates were collected at 6, 16, 24, and 48 hpi. (F and G) Bands from detection of IE2, UL44, and UL57 at 48 hpi were quantified and normalized to the beta-actin signal. Results are shown relative to the signal intensity from DMSO-treated WT-infected samples, which was set to 100. (F) Results from three independent biological replicates of the conditions compared in panel D. (G) Results from two independent replicates of the experiment whose results are shown in panel E. A one-way analysis of variance with Tukey's posttest was used to determine statistical significance in panel F, and the Student t test was used for panel G. Error bars indicate SEMs. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Consistent with our mRNA results, MBV caused pronounced effects on expression of the proteins IE2, UL44, and UL57 but not IE1, and the effects were markedly attenuated in UL133/8NULL infections (Fig. 3D and F). At 48 hpi, MBV-treated WT infections showed defects in protein expression of 4-fold for IE2, of nearly 3-fold for UL44, and of 2-fold for UL57, all of which were statistically significant (Fig. 3D and F). In UL133/8NULL infections, however, MBV failed to affect the expression of any of these proteins (Fig. 3D and F). At 48 hpi, UL133/8NULL exhibited significant basal defects in the expression of IE2, UL44, and UL57 (Fig. 3F), which was consistent with its basal defect in viral DNA synthesis (Fig. 2D). By 72 to 96 hpi, however, IE2 levels in MBV-treated WT infections, as well as in UL133/8NULL infections, recovered to those seen under the DMSO-treated WT control condition, while defects in UL44 expression persisted (Fig. 3D). Thus, the absence of either UL97 or UL133/8 resulted in delayed IE2 expression and in prolonged defects in the expression of an E-L gene product.
To exclude the possibility that off-target effects of MBV could explain our results, we investigated protein expression during Δ97 mutant virus infections. We found that the levels of IE2, but not those of IE1, were appreciably lower in Δ97 mutant virus-infected cells than in WT-infected cells at 48 hpi but not at earlier time points (Fig. 3E and G). The IE2 expression defect for the Δ97 mutant virus was statistically significant and was accompanied by significant defects in the expression of UL44 and UL57. Because similar effects were seen in MBV-treated infections, it appeared that MBV faithfully recapitulated the phenotypic consequences of infections with UL97-null virus. From these experiments, we concluded that genetic deletion or pharmacological blockade of UL97 caused defects in mRNA levels and protein expression for IE2 and for at least two genes involved in viral DNA synthesis and, moreover, that the potential of a UL97 inhibitor to produce these defects was markedly attenuated in UL133/8NULL infections.
Reintroduction of UL133/8 into a laboratory strain.
Our results indicated that a 3.636-kb deletion in HCMV strain TB40/E that removed UL133/8 was sufficient to influence the requirement for UL97. The loss of UL133/8 appeared to preempt the ability of UL97 to influence viral gene expression and viral DNA synthesis by causing basal defects in these processes. Nonetheless, we could not yet rule out the possibility that effects of the UL133/8 deletion on neighboring genes might account for our findings. We also needed to exclude the possibility that the basal defects of the UL133/8NULL mutant somehow minimized the effects of MBV in a nonspecific manner. To address these issues, we decided to test whether the reintroduction of UL133/8 into strain AD169 would be sufficient to produce an increased requirement for UL97.
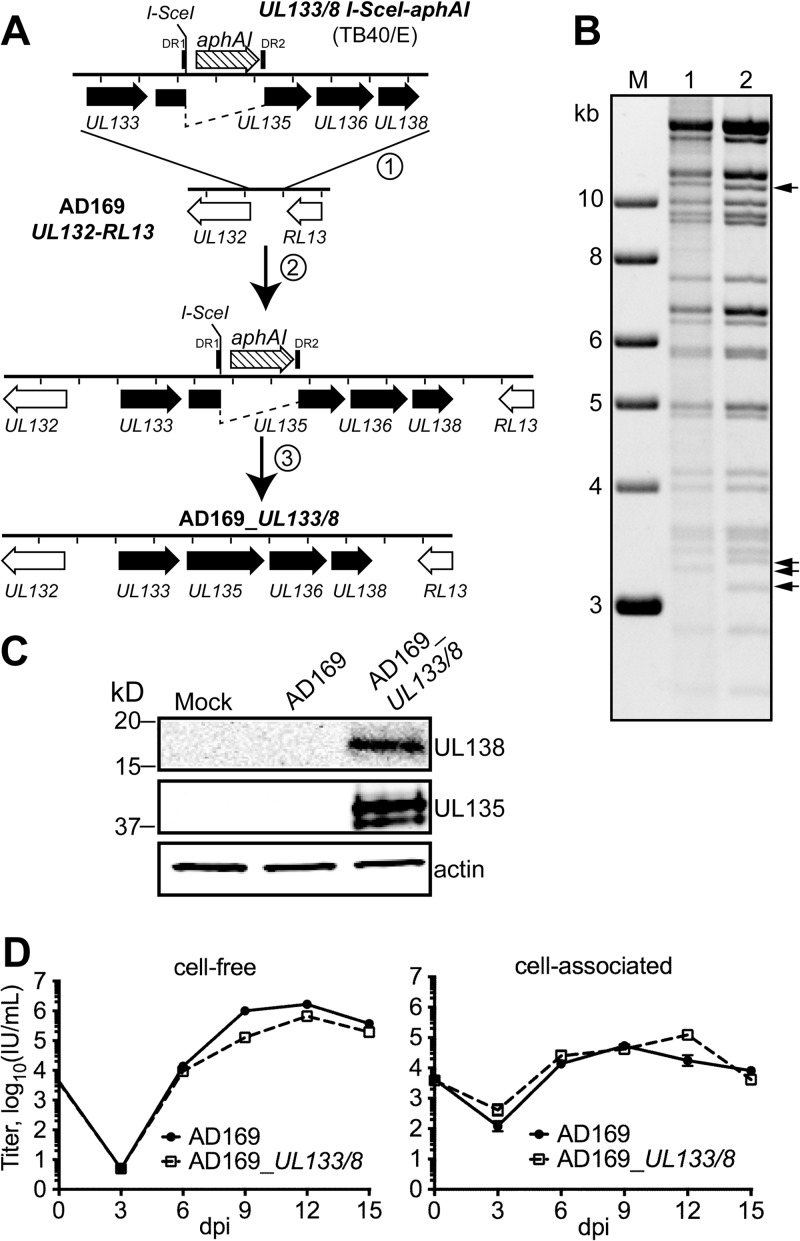
Strain AD169 lacks UL133/8, as well as 11 kb of additional ULb′ sequences present in strain TB40/E and in bona fide clinical HCMV isolates (16, 32–34). AD169 lost most of its ULb′ during serial passage in fibroblasts (32) and, in contrast to TB40/E, replicates robustly without UL133/8. We used en passant BAC mutagenesis to incorporate into the region between UL132 and RL13 of AD169 (arguably representing its remnant ULb′) a 4.122-kb segment of DNA from strain TB40/E encompassing a region from 281 bp upstream of UL133 up to and including a putative polyadenylation site 582 bp downstream of the UL138 stop codon (Fig. 4A). Our previous findings (24) implied that the sequences that we transferred into AD169 would be sufficient to drive the expression of UL133/8 proteins in the resulting recombinant virus, which we named AD169_UL133/8.
FIG 4.
Reintroduction of UL133/8 into laboratory-adapted HCMV strain AD169. (A) Methodology used to insert the UL133/8 locus of HCMV strain TB40/E into laboratory strain AD169. In steps 1 and 2, a PCR product encompassing UL133/8 disrupted by a kanamycin resistance marker (aphAI) was used to replace the noncoding region between UL132 and RL13 of AD169. The aphAI marker was coupled to an I-SceI site and flanked by direct repeats (DR1, DR2). In step 3, a recombination event was induced to remove the aphAI marker, resulting in AD169_UL133/8. (B) BAC DNA of parental strain AD169 (lane 1) and AD169_UL133/8 (lane 2) was EcoRI digested and analyzed by agarose gel electrophoresis. Black arrows, variations in the restriction pattern consistent with predicted differences between AD169_UL133/8 and parental strain AD169 BAC DNAs; lane M, molecular size marker. (C) UL135 and UL138 proteins were detected by Western blotting at 72 hpi of lysates of cells infected with the indicated virus at an MOI of 1, using rabbit polyclonal sera specific for the indicated proteins; the relative mobility of protein molecular mass standards is indicated at the left. (D) The multicycle replication of AD169_UL133/8 and parental wild-type AD169 was compared. Fibroblasts were infected at an MOI of 0.01, and titers of cell-free or cell-associated virus were determined at the indicated times. Error bars represent standard deviations.
We analyzed the EcoRI restriction digestion pattern of AD169_UL133/8 BAC DNA to verify the presence of the expected changes relative to the sequence of parental strain AD169 BAC DNA (Fig. 4B). We also confirmed by DNA sequencing that the incorporated UL133/8 sequences and neighboring sequences involved in our recombination strategy, spanning from 67 bp upstream of the UL133/8 insertion site to 412 bp downstream of it, did not contain spurious mutations (data not shown). Infectious virus was reconstituted in fibroblast cells, and virus stocks of AD169_UL133/8 were prepared.
Lysates of cells infected with AD169_UL133/8 at an MOI of 1 were evaluated to determine whether the incorporated UL133/8 locus could drive the expression of UL135 and UL138, the two proteins expressed from UL133/8 for which we had antibodies. In Western blotting experiments, immunoreactive bands were detected in cell lysates from AD169_UL133/8 infections but not those from parental strain AD169 infections or from mock infections (Fig. 4C). Moreover, the immunoreactive species exhibited relative mobilities consistent with the theoretical molecular masses of UL135 and UL138, which are 35.6 kDa and 19.3 kDa, respectively. We therefore concluded that the UL133/8 sequences that we had engineered into AD169_UL133/8 were sufficient to drive the expression of at least two of its encoded proteins.
To test whether incorporation of UL133/8 into strain AD169 would affect viral replication, we compared multicycle growth kinetics for parental strain AD169 and strain AD169_UL133/8 in fibroblasts following infection at an MOI of 0.01. Although AD169_UL133/8 exhibited ∼1-log-unit lower levels of cell-free virus at 9 days postinfection (dpi), viral yields from 12 to 15 dpi were indistinguishable between the two viruses (Fig. 4D). Cell-associated virus levels were modestly higher for AD169_UL133/8 at 12 dpi but were similar to those for parental strain AD169 at 9 dpi and 15 dpi (Fig. 4D). Overall, these results indicate that parental strain AD169 and AD169_UL133/8 replicate similarly in fibroblasts.
The results of our construction and initial characterization of AD169_UL133/8 confirmed that we had successfully introduced UL133/8 into strain AD169, that the resulting virus replicated similarly to parental strain AD169, and that it was capable of expressing at least two proteins from the incorporated locus (Fig. 4). For these reasons, we decided that AD169_UL133/8 was a suitable reagent to determine whether restoration of UL133/8 to a ULb′-deficient HCMV laboratory strain would influence its requirement for UL97.
Restoration of UL133/8 to a laboratory-adapted strain increases the requirement for UL97 in viral replication and viral DNA synthesis.
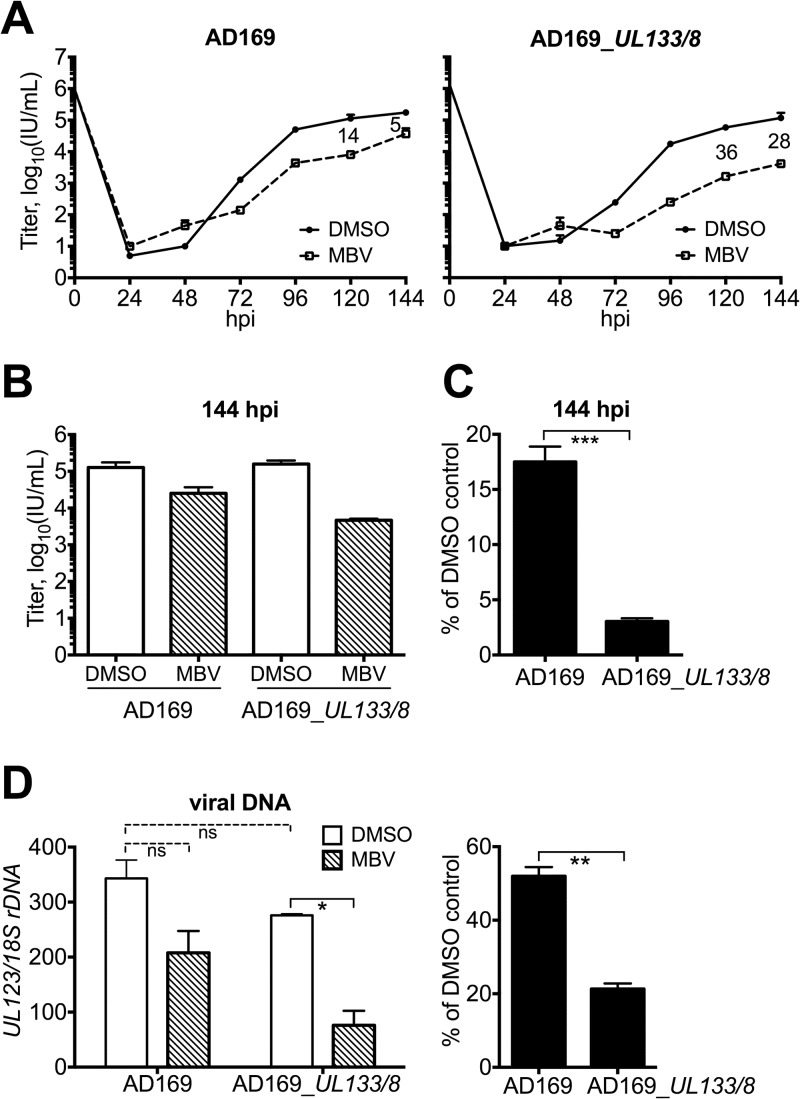
Our results thus far suggest that UL133/8 increased the requirement for UL97 in strain TB40/E. Therefore, we hypothesized that AD169_UL133/8 would show an increased requirement for UL97 in replication kinetics experiments. Although AD169_UL133/8 and its parental virus replicated indistinguishably in the absence of MBV, we found that treatment with the UL97 inhibitor caused larger defects in AD169_UL133/8 than in parental strain AD169 (Fig. 5A and B). In a representative result, we observed defects in AD169_UL133/8 of 36-fold and 28-fold at 120 hpi and 144 hpi, respectively, while parental strain AD169 showed only 14-fold and 5-fold defects at these time points (Fig. 5A and B). An ∼5-fold larger replication defect in AD169_UL133/8 at 144 hpi was repeatedly observed in multiple independent biological replicates comparing the two viruses (Fig. 5B and C), and the difference in the effect of the UL97 inhibitor on viral replication was statistically significant (P < 0.001) (Fig. 5C).
FIG 5.
A recombinant AD169 restored for UL133/8 shows exacerbated defects in replication and viral DNA synthesis when UL97 is inhibited. (A) Fibroblasts were infected with AD169 or AD169_UL133/8 at an MOI of 1 and maintained in the presence of 2 μM MBV or 0.1% DMSO. Cell-free virus was harvested at the indicated time points, and titers were determined. The fold difference in titer between DMSO control and MBV treatments at 120 hpi and 144 hpi is indicated in the space between the curves; error bars represent standard deviations. (B) Average viral yield results at 144 hpi for two independent biological replicates. (C) The viral yield at 144 hpi in the MBV condition is displayed as a percentage of that from the DMSO control condition, and the average results from three independent experiments are shown. Error bars indicate SEMs, and the Student t test was used to determine significance. (D) (Left) Absolute levels of viral DNA detected at 96 hpi from DMSO- and MBV-treated infections; (right) results from the MBV condition are also shown as a percentage of the values for the DMSO control condition. Error bars indicate SEMs. A one-way analysis of variance was used to determine statistical significance in the left panel, and the Student t test was used in the right panel. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
As expected, the phenotypic differences under MBV treatment extended to viral DNA synthesis. MBV significantly decreased viral DNA synthesis in AD169_UL133/8 but not in the parental virus, AD169 (Fig. 5D). At 96 hpi, MBV-treated AD169_UL133/8 infections exhibited a 5-fold defect in viral DNA synthesis, while parental strain AD169 showed a defect of less than 2-fold (Fig. 5D). The difference between these effects was likewise statistically significant. It therefore appeared to us that introduction of UL133/8 into strain AD169 significantly increased the effects of a UL97 inhibitor on viral replication and viral DNA synthesis.
Restoration of UL133/8 confers an increased requirement for UL97 in viral gene expression.
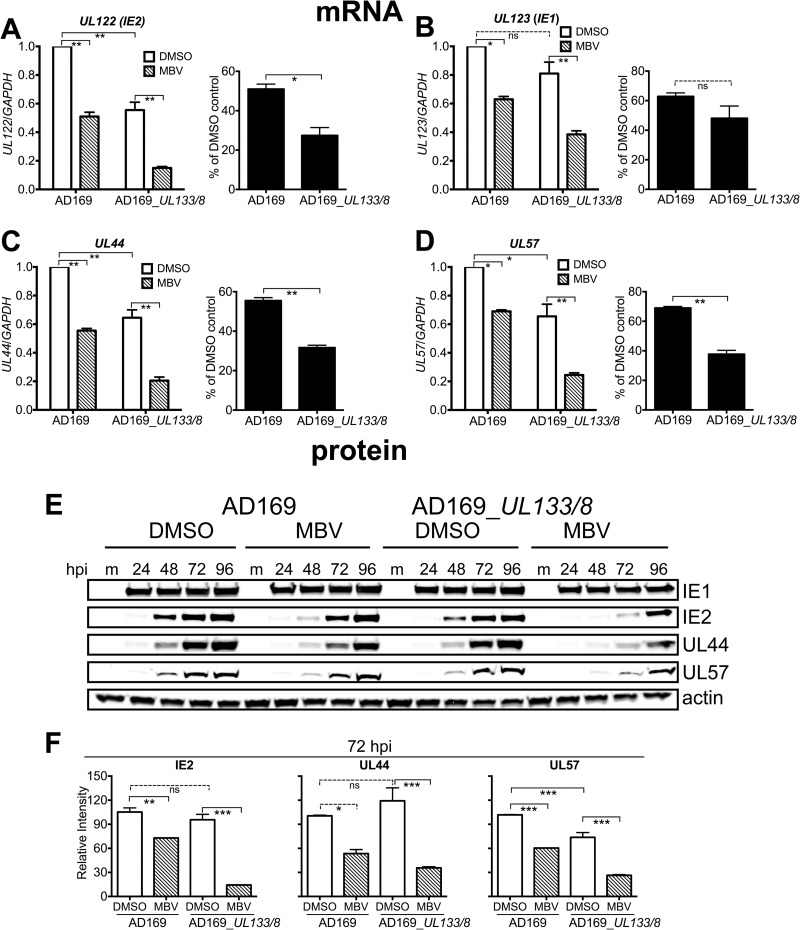
To determine whether differences in viral gene expression might underlie the increased requirement for UL97 in AD169_UL133/8, we quantified the effect of MBV on mRNA levels at 48 hpi for IE2 (UL122), IE1 (UL123), UL44, and UL57 by RT-qPCR. We observed a statistically significant 4-fold decrease in IE2 (UL122) mRNA levels in AD169_UL133/8 during MBV treatment but only a 2-fold decrease in parental strain AD169, which was also significant (Fig. 6A, left). Importantly, the difference between the viruses in the magnitude of their defects during MBV treatment was significant (Fig. 6A, right). Intriguingly, the basal level of IE2 transcripts was significantly lower in AD169_UL133/8 than in parental strain AD169, showing an ∼2-fold difference (Fig. 6A, left). The difference between the viruses in the effect of MBV on the levels of IE1 mRNA was less striking: a reduction of slightly over 2-fold was seen for AD169_UL133/8, while a 2-fold reduction was seen for parental strain AD169 (Fig. 6B). Although the effects of MBV on IE1 mRNA levels were significant for each virus, the difference between the effects was not (Fig. 6B).
FIG 6.
Incorporation of UL133/8 into laboratory strain AD169 exacerbates its defects in viral mRNA and protein expression during UL97 inhibition. (A to D) mRNA abundance at 48 hpi (MOI, 1) was quantified for UL122 (IE2), UL123 (IE1), UL44, and UL57, as indicated. (Left) Results are shown relative to those from DMSO-treated parental strain AD169 virus infections, which were set to 1.0, and one-way analysis of variance was used to determine statistical significance; (right) mRNA abundance from MBV-treated infections for each virus is shown as a percentage of that from the DMSO control condition, and the Student t test was used to determine statistical significance. (E and F) Fibroblasts were infected with AD169 or AD169_UL133/8 at an MOI of 1 and maintained in the presence of UL97 inhibitor (MBV) at 2 μM or 0.1% DMSO, to control for the effects of the carrier. (E) Expression of viral proteins at the indicated time points was detected by Western blotting. (F) Bands representing the expression of the IE2, UL44, and UL57 proteins at 72 hpi from three independent biological replicates were quantified and normalized to the level of beta-actin expression. Results relative to the signal from DMSO-treated infections with parental strain AD169 are shown. A one-way analysis of variance with Tukey's posttest was used to test for statistical significance. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
MBV significantly decreased UL44 and UL57 transcript levels in both viruses, showing effects of 3-fold or greater for AD169_UL133/8 and of 2-fold or less for AD169 (Fig. 6C and D). Thus, the presence of UL133/8 correlated with increased effects of the UL97 inhibitor (Fig. 6C and D). Moreover, the differences between the viruses in the effects of MBV on UL44 and UL57 transcript levels were significant (Fig. 6C and D, right). Finally, AD169_UL133/8-infected cells showed significantly lower basal levels of UL44 and UL57 mRNAs (Fig. 6C and D), as we had seen with IE2 (Fig. 6A). These results argue that UL133/8 is sufficient to significantly increase the effects of a UL97 inhibitor on the accumulation of viral mRNAs encoding IE2 and two E/E-L proteins.
AD169_UL133/8 also showed exacerbated defects in protein expression during MBV treatment (Fig. 6E). MBV caused a striking delay in IE2 protein accumulation in cells infected with AD169_UL133/8. This effect was markedly attenuated but not completely absent in parental strain AD169. Notably, we failed to observe appreciable effects of MBV on IE1 levels in either virus, which was consistent with our findings presented in Fig. 3 and with the results from our previous studies (8, 10, 17). Although IE2 negatively regulates the MIEP, which drives the expression of both IE1 and IE2, our failure to observe effects on IE1 comparable to those that we observed for IE2 are likely explained by their discordant expression kinetics (28, 29, 35).
The differences in IE2 expression during MBV treatment between AD169_UL133/8 and AD169 correlated with differences in the expression of UL44 and UL57, the most striking of which were seen at 48 and 72 hpi (Fig. 6E). To determine whether the effects were statistically significant, we quantified the levels of IE2, UL44, and UL57 protein expression at 72 hpi in three independent biological replicates (Fig. 6F). For AD169_UL133/8 infections, IE2 levels showed a statistically significant 7-fold decrease during MBV treatment. For parental strain AD169, IE2 showed only a 1.4-fold decrease, which nonetheless was also significant. AD169_UL133/8 also showed significant, ∼3-fold defects in UL44 and UL57 protein expression during MBV treatment. AD169 showed smaller, albeit significant, defects of ∼2-fold for UL44 and UL57. The difference between the viruses in the basal expression of viral proteins was found to be significant for UL57 but not for IE2 or UL44 (Fig. 6F).
Overall, our results demonstrate that restoration of UL133/8 to strain AD169 has significant effects on viral gene expression. First, the basal expression of transcripts for IE2, UL44, and UL57 was significantly decreased (Fig. 6A, C, and D). Second, the defects in their mRNA abundance were significantly exacerbated during inhibition of UL97 (Fig. 6A, C, and D). Third, UL97 inhibition produced exacerbated defects in protein expression, consistent with the mRNA results (Fig. 6E and F). These findings led us to conclude that UL133/8 is sufficient to influence the requirement for UL97 in viral gene expression.
In contrast to the UL133/8NULL mutant strain of TB40/E, the AD169_UL133/8 virus (i) did not exhibit a basal difference in viral replication relative to its parental virus (Fig. 4D and 5A) and (ii) showed less striking basal differences in viral protein expression—particularly for IE2—than its parental virus. Therefore, the results from our experiments comparing AD169_UL133/8 to its parental virus argue against the possibility that nonspecific replication defects of UL133/8NULL minimize the effects of UL97 inhibition by sheer coincidence. The results instead argue that UL133/8 and UL97 act in the same pathway to promote viral replication, likely via epistatic roles in viral gene expression.
The influence of the UL133/8 locus on viral sensitivity to a UL97 inhibitor requires UL135.
In an effort to identify which of the open reading frames within UL133/8 were necessary for its influence on the requirement for UL97, we screened null mutants of open reading frames within UL133/8 for their defects in the production of infectious virions during treatment with MBV (not shown). The initial screens, which included TB40/E mutants individually null for UL133, UL136, or UL138, failed to identify a mutant that showed reproducible differences relative to WT (not shown). As discussed in our companion paper in this issue, UL135 is required for viral replication when UL138 is expressed, and its expression is toxic to uninfected cells (19). Therefore, it has been a challenge to produce virus stocks containing disruptions only in UL135 (19).
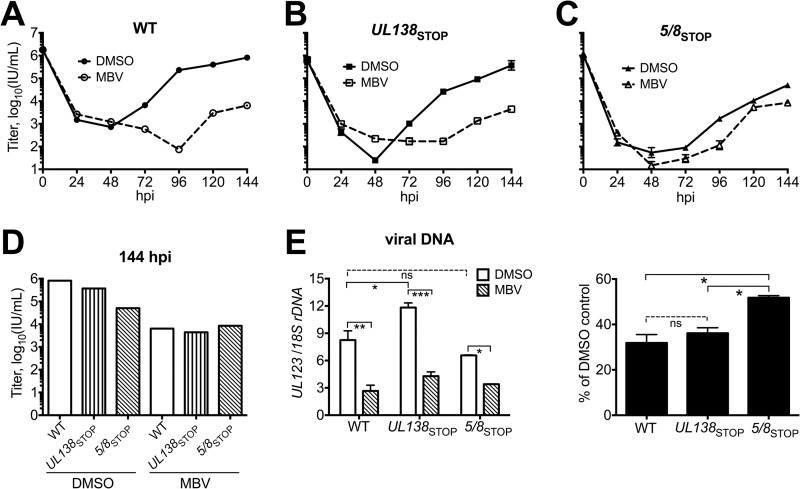
In order to address whether UL135 might influence the requirement for UL97, we assayed a TB40/E mutant that does not express either UL135 or UL138, which we subsequently refer to as 5/8STOP, for its ability to replicate in the presence of MBV. As controls, we included UL138STOP, a mutant unable to express UL138, and parental wild-type TB40/E (WT). At 144 hpi, MBV caused viral replication defects of ∼90-fold for UL138STOP and of over 100-fold for WT (Fig. 7A and B). In 5/8STOP, however, MBV caused a defect of only 6-fold (Fig. 7C). The attenuated effects of MBV on viral replication during 5/8STOP infections, coupled with the observation that basal replication of 5/8STOP exhibited a 1-log-unit defect relative to the effects on WT, suggested that the phenotype of 5/8STOP might resemble that of UL133/8NULL (Fig. 2A to C and 7A to D).
FIG 7.
Disruption of both UL135 and UL138, but not UL138 alone, attenuates the effects of UL97 inhibition on viral replication and viral DNA synthesis. Fibroblasts were infected with the indicated viruses at an MOI of 1 and maintained in the presence of MBV at 2 μM or 0.1% DMSO. The cell-free virus titer was determined for the indicated time points, and the resulting replication kinetics curves are shown for WT (A), UL138STOP (B), and 5/8STOP (C). Error bars indicate standard deviations. (D) Viral yield results at 144 hpi from panels A to C are compared side by side. (E) Viral DNA levels at 96 hpi were determined by qPCR in two independent biological replicates. (Left) Absolute levels of viral DNA; (right) results from MBV conditions as a percentage of the amount of viral DNA measured from the DMSO control conditions. For panel E, error bars indicate standard errors of the means, and a one-way analysis of variance was used to test for statistical significance. P values were determined by Tukey's posttest. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Viral DNA synthesis in 5/8STOP likewise resembled that in UL133/8NULL. In 5/8STOP infections, MBV reduced viral DNA levels by 2-fold, and the effect was not significant (Fig. 7E). In contrast, MBV significantly reduced viral DNA levels in WT and UL138STOP infections by 3.3-fold and 6-fold, respectively (Fig. 7E). Thus, MBV showed larger effects on viral DNA synthesis in WT and UL138STOP than in 5/8STOP (Fig. 7E). Although the basal levels of viral DNA synthesis in 5/8STOP infections decreased relative to those in WT infections, the difference was not found to be statistically significant (Fig. 7E). Intriguingly, UL138STOP infections showed significantly higher basal levels of viral DNA synthesis than WT infections, which is consistent with the results described in the accompanying article (19). During MBV treatment, however, all three viruses accumulated viral DNA to similar levels (Fig. 7E, left). These results suggest that UL135 and UL97 are each required for UL133/8 to influence viral replication and viral DNA synthesis.
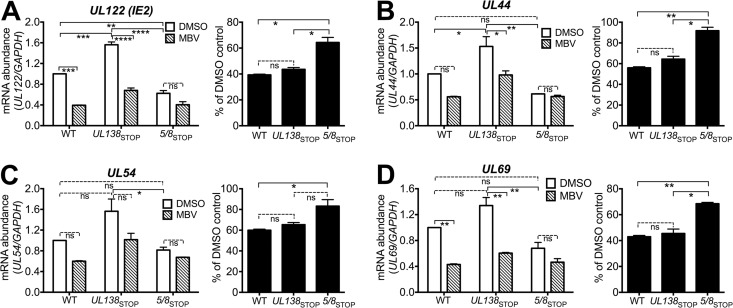
We next asked whether UL135 influenced viral mRNA levels and, if so, whether it would require UL97 to do so. We measured the effect of MBV on transcript levels at 48 hpi for IE2, UL44 (E), and two additional genes, UL54 (E) and UL69 (E-L), during infection with WT, UL138STOP, or 5/8STOP. In 5/8STOP infections, MBV failed to significantly reduce mRNA levels for any of the genes (Fig. 8A to D). In WT and UL138STOP infections, however, MBV caused significant decreases in mRNA levels for IE2 (UL122) and UL69 (Fig. 8A and D). The effects of MBV on transcript levels for UL44 and UL54, however, did not reach statistical significance in WT infections in this experiment. Nonetheless, the difference between WT and 5/8STOP in their mRNA defects during MBV treatment was significant for all four genes (Fig. 8A to D, right).
FIG 8.
A TB40/E mutant null for UL135 and UL138 exhibits viral early and early-late mRNAs at reduced levels that are insensitive to inhibition of UL97. Fibroblasts were infected with WT, UL138STOP, or 5/8STOP at an MOI of 1 and maintained in the presence of 2 μM MBV or 0.1% DMSO. Total RNA was prepared from infected cells at 48 hpi, and mRNA abundance was analyzed by RT-qPCR and normalized to that of GAPDH for UL122 (IE2) (A), UL44 (B), UL54 (C), and UL69 (D). (Left) The abundance of each viral transcript is shown relative to that of DMSO-treated WT infections, which was arbitrarily set to 1; (right) mRNA abundance from MBV-treated infections is shown as a percentage of that from the DMSO-treated condition. Results shown represent two independent biological replicates, and a one-way analysis of variance with Tukey's posttest was used to determine statistical significance. ns, not significant; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Interestingly, the basal viral mRNA levels seen during UL138STOP infections were consistently higher than those seen during WT or 5/8STOP infections, and this difference was found to be significant for IE2 and UL44 (Fig. 8). The latter observation is consistent with the viral DNA synthesis results from UL138STOP infections (Fig. 7D) and results described in the accompanying report (19). As UL97 inhibition significantly reduced viral mRNA levels during WT and UL138STOP infections but not during infections with a virus null for both UL135 and UL138, our findings imply that UL135 and UL97 are each required to promote the mRNA accumulation for IE2 and for at least a subset of E and E-L genes.
In summary, we have found that genetic ablation or pharmacological inhibition of UL97 led to decreased levels of mRNAs for IE2 and viral E and E-L genes during a second phase of viral mRNA expression, which commenced at between 24 and 48 hpi. Although 24 h is a considerable window, the onset of viral DNA synthesis in strain TB40/E appeared to occur during this time (Fig. 1H). We also observed that UL133/8-null mutants and mutants individually null for both UL138 and UL135 showed phenotypic profiles similar to those seen in UL97-deficient virus infections, with regard to defects in transcript and protein levels for IE2 and viral E and E-L proteins and in viral DNA synthesis (Fig. 2, 3, 7, and 8). Furthermore, we found that restoration of UL133/8 to laboratory strain AD169 recapitulated the differences in the effects of UL97 inhibitor treatment that we had observed between wild-type strain TB40/E and its mutant UL133/8NULL, while also causing decreased basal mRNA levels for IE2 and for certain viral E-L and E genes (Fig. 5 and 6). The defects that we detected in viral E and E-L transcript levels in infections with virus lacking either UL97 activity or UL133/8 are consistent with the roles of IE2 as a transactivator of viral E (and E-L) gene expression (reviewed in reference 1). Taken together, our study implicates the UL133/8 locus—in particular, UL135—and UL97 in regulating progression of the viral lytic cycle and, moreover, implies that UL135 and UL97 act epistatically (i.e., within the same pathway) to stimulate viral gene expression and viral DNA synthesis.
DISCUSSION
Eukaryotic cells are equipped with sophisticated mechanisms to regulate their gene expression. As an obligate intracellular parasite that establishes a lifelong persistent infection in a vertebrate host, HCMV might likewise be expected to possess intricate mechanisms to regulate the expression of its genes. The existence of latency in the herpesviruses is testament to the advantages conferred to intracellular pathogens capable of tightly regulating their gene expression. Conversely, inappropriate viral gene expression—during either latency or the lytic cycle—would almost certainly have devastating effects on viral fitness.
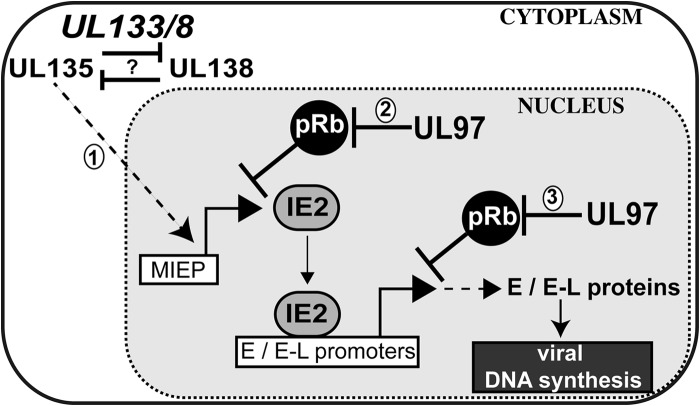
Model for the epistatic relationship between UL97 and UL133/8.
Among the key findings of this study is that progression of the HCMV lytic cycle is governed both by UL97, a viral protein kinase that inactivates the retinoblastoma tumor suppressor protein (pRb) (4, 5), and by the UL133/8 locus (UL133/8), which plays important roles in viral latency and in other cell type-specific phenotypes (18, 24, 36). Although the precise mechanisms are still unclear, our results implicate roles for UL97 and for two proteins expressed from UL133/8, UL135 and UL138, in regulating a second wave of expression of IE2, a key transactivator of the HCMV lytic cycle. Taken together with the findings presented in an accompanying study (19), our findings have led us to propose a model that suggests new hypotheses for understanding the genetic circuitry that governs HCMV lytic replication and latency (Fig. 9).
FIG 9.
Model for roles of UL97 and UL133/8 in the viral lytic cycle. In step 1, proteins expressed from UL133/8 promote transcription from the viral MIEP by an indirect mechanism that may involve a signal sent from the Golgi apparatus or cell surface, where UL133/8-encoded proteins localize during infection. The signal requires UL135, which is antagonized by UL138. In steps 2 and 3, pRb negatively regulates transcription from the MIEP and viral E and E-L promoters. In step 2, UL97 inactivates pRb, which enables the signal from UL135 to result in increased expression of IE2. Viral E and E-L promoters are positively regulated by IE2. In step 3, UL97-mediated inactivation of pRb may also be important for high-level expression of viral E and E-L proteins that participate in viral DNA synthesis.
By virtue of its nuclear localization (37) and its role in inactivation of pRb (4, 5, 10), we propose that UL97 promotes a second wave of transcription from the MIEP to upregulate IE2 expression; IE2, in turn, transactivates viral E and E-L genes, resulting in the biosynthesis of viral proteins that carry out viral DNA synthesis during the lytic cycle (Fig. 9). UL97's role in inactivating pRb may also be important for expression of viral E- and E-L genes, as observations from the literature, discussed below, suggest that IE2 may recruit pRb to silence viral gene expression. The mechanisms by which UL133/8 may influence IE2 expression are less clear. However, our results and those from the accompanying study (19) indicate that UL135 promotes the timely expression of IE2 during the second phase of its expression and that its influence is opposed by UL138. UL135, like other proteins expressed from UL133/8, is predominantly cytoplasmic, localizing to the cell surface and the Golgi apparatus during infection (18). Thus, it seems logical to propose that UL135 stimulates the MIEP by an indirect mechanism that involves transduction of a signal to the nucleus (Fig. 9). A crucial aspect of our findings here, which our model seeks to explain, is that UL135 cannot promote the viral lytic cycle without UL97, and vice versa.
Roles for UL133/8 during lytic replication.
While genes within UL133/8 have been found to be relevant during viral replication in endothelial cells (18, 36), our results presented here (Fig. 2, 3, and 5 to 8) and in an accompanying study (19) argue that UL133/8 also plays important roles during viral replication in fibroblasts, the cell type most frequently used in studies of HCMV lytic replication. Although members of our group previously reported that UL133/8 is dispensable for replication in fibroblasts, the same study documented an ∼5-fold replication defect in a UL133/8-null mutant of strain FIX (see Fig. S3 of reference 18). Moreover, UL133/8-null mutants also exhibited a delay in replication, which was more pronounced at lower MOIs (F. D. Goodrum, unpublished results).
Laboratory strains AD169 and Towne spontaneously accumulated deletions that encompass virtually the entire ULb′ region, including UL133/8 (32). This observation could be interpreted to suggest that ULb′ loci are dispensable. However, the robust replication of these laboratory strains in vitro likely belies the requirement for UL133/8 in HCMVs that have not undergone extensive laboratory adaptation. Notably, while restoration of UL133/8 to strain AD169 did increase its requirement for UL97, its UL97-dependent replication defect remained markedly less severe than the defects seen for strains such as TB40/E and TR, whose genomes more closely approximate those of HCMV clinical isolates (Fig. 2, 5, 6, and 7) (17). We thus hypothesize that AD169 and Towne have accumulated one or more mutations that suppress the requirement for UL133/8 and UL97 in replication, perhaps subverting controls that the virus imposes on itself in vivo.
Accordingly, it is worth noting that inhibition of UL97 was able to reduce viral mRNA levels and protein expression in parental strain AD169 but not in the TB40/E mutant UL133/8NULL, notwithstanding the fact that introduction of UL133/8 into AD169 exacerbated these defects (Fig. 3 and 6). Our model posits that the influence of UL135 is confined to the MIEP (Fig. 9), while UL97 acts both at the MIEP and at viral E/E-L promoters to stimulate viral gene expression. (Moreover, the literature, discussed below, suggests myriad indirect mechanisms by which UL97 might also affect viral gene expression.) We speculate that the expression of viral E and E-L genes is dysregulated in strain AD169 in a manner that fully relieves the requirement for UL133/8 while only partially alleviating the requirement for UL97. In contrast, because UL133/8 likely acts upstream of UL97 to regulate immediate early (IE) gene expression, the failure of UL97 inhibition to show an effect on viral E/E-L gene expression in UL133/8NULL may indicate that its upstream defect is sufficiently severe to obviate the influence of UL97 on E/E-L genes.
Implications of delayed IE2 expression.
Recently, Teng et al. demonstrated that HCMV's capacity for rapid expression of IE2 is crucial for viral fitness (38). We observed delayed accumulation of the IE2 protein when UL133/8 was ablated or when UL97 was either absent or inhibited (Fig. 3 and 6). Although IE2 levels ultimately recovered to wild-type levels, we routinely saw replication defects of ∼10-fold in viruses null for UL133/8 and of 100- to 1,000-fold in UL97-deficient HCMVs containing UL133/8 (Fig. 2 and 7) (17). While UL97-deficient viruses likely suffer from additional, functionally unrelated deficiencies, such as in viral nuclear egress (6–8), the findings of Teng et al. help explain how modest defects in IE2 expression, such as those observed in our study, might have outsized effects on the production of infectious particles (38).
How does UL97 influence viral gene expression and viral DNA synthesis?
The influence of UL97 on viral DNA synthesis appears to require its role in inactivation of pRb (10). However, our findings raise new questions as to whether roles for UL97 in the expression of IE2 and viral E/E-L genes might also impact viral DNA synthesis and whether such roles might also involve pRb. pRb is a tumor suppressor protein that regulates the cell cycle by binding to E2F transcription factors and restricting the expression of E2F-responsive (E2F-R) genes (reviewed in reference 39). E2F-R genes are important for progression toward S phase and, in certain examples, such as thymidylate synthase (10), are also necessary for viral DNA synthesis (40, 41). Interestingly, well before UL97 was found to phosphorylate pRb (4, 5), a physical interaction between IE2 and pRb was reported (42, 43). IE2 can functionally inactivate pRb in transfected cells (42), and its expression is sufficient to stimulate E2F-R genes (44). Despite this, UL97 is required for pRb inactivation during HCMV infection (10). Intriguingly, however, IE2 has also been hypothesized to recruit pRb to viral promoters to regulate their activity (42, 45). If so, UL97 perhaps influences such regulation by phosphorylating pRb.
Many HCMV proteins that participate in viral genome replication exhibit E-L expression kinetics, and the L component of E-L kinetics by definition depends on viral DNA synthesis (1). Therefore, it may be challenging to tease apart the roles of UL97 in directly regulating viral gene expression from the secondary effects of its roles in stimulating E2F-R cellular genes that support viral DNA synthesis, particularly since both may involve pRb. Furthermore, even though IE2 is an IE gene product, defects in IE2 expression have been observed in a temperature-sensitive HCMV mutant under conditions in which it could not synthesize viral DNA (29). Prima facie, the latter observation would raise the possibility that UL97-dependent defects in IE2 expression are in fact a consequence of UL97's influence on viral DNA synthesis and/or its roles in the expression of viral E and E-L genes, which play key roles in viral DNA synthesis. However, a number of observations argue against this interpretation. First, we observed UL97- and UL133/8-dependent defects in IE2 expression at 48 hpi, well before true late proteins (e.g., pp150, pp28, gB) could be detected in our system (Fig. 3 and 6) (17; Li and Kamil, unpublished). Second, we found significant UL97- and UL133/8-dependent effects on the expression of UL57, an E gene (Fig. 1, 3, 6, and 8). Because viral E genes are regulated by IE2 and do not require viral DNA synthesis for their expression, we hypothesize that the influence of UL97 on viral E and E-L genes is secondary to its effects on IE2 expression.
Many open questions remain as to how IE2, pRb, and UL97 might contribute to the intricacies of viral gene expression. Interestingly, of the two major repressive chromatin-modifying enzymes reported to functionally interact with pRb (46–49), the Suv39H1 histone methyltransferase, but not histone deacetylase 1 (HDAC1), was found to associate with IE2 in a pRb-dependent manner (50). Moreover, results from reporter assays imply that the pRb-Suv39H1 interaction contributes to IE2-mediated autorepression of the MIEP (50). Whether UL97 modulates viral gene expression by regulating the IE2-pRb complex remains to be determined. However, it has been noted that IE2, like E2Fs, fails to bind hyperphosphorylated, inactive forms of pRb (42). This suggests that UL97 might promote the dissociation of IE2-pRb complexes, much as it would be expected to cause the dissociation of E2F from pRb.
Bigley et al. recently reported that UL97 regulates the MIEP by phosphorylating HDAC1 and that this function of UL97 markedly influenced IE1 levels at immediate early and early times during infection, e.g., 6 to 12 hpi (51). Although we failed to observe substantial effects on mRNA or protein levels for IE1 at these times in this study (Fig. 1B and 3D) or in our previous studies (4, 8, 10), we did find modest effects on IE1 mRNA levels at later times during infection (Fig. 1B and 6B). Albeit we cannot fully reconcile the differences in our results, we do agree that UL97 may influence the MIEP. Nonetheless, our data argue that UL97's role in stimulating MIEP activity is confined to times subsequent to its de novo expression.
Biphasic viral gene expression.
Biphasic expression of IE2 during HCMV infection has been previously observed, and it appears that selective expression of IE2 at times subsequent to ∼24 hpi involves posttranscriptional mechanisms, such as alternative splice site selection, in addition to transcriptional regulation (28–30, 35). Interestingly, it has been reported that chromatin associated with the MIEP exhibits biphasic acetylation of histone H3 during the lytic cycle (52), and notably, the kinetics observed were similar to those seen in this study (Fig. 1) and in earlier work (29, 30, 35). Moreover, biphasic effects on viral chromatin during lytic replication were recently reported for Kaposi's sarcomas-associated herpesvirus, a gammaherpesvirus (53). Whether or not such biphasic phenomena turn out to be a general feature of the herpesvirus lytic cycle, the influence of chromatin on the viral genome is an area of active investigation (50–52, 54–59) and may provide new insights into how UL97 and UL133/8 impact viral gene expression.
Implications for studies of HCMV latency.
Taken together, our results suggest that UL97 and proteins expressed from UL133/8 play roles in regulating progression of the viral lytic cycle at times subsequent to the initial burst of viral IE gene expression. We hypothesize that UL97 plays an important role in this process by inactivating pRb (Fig. 9). Silencing of viral IE gene expression is an important hallmark of HCMV latency (60), and hence, detection of IE transcripts has been used as a proxy for reactivation during in vitro studies of latency. Nonetheless, studies on latently infected cells taken from healthy carriers suggest that viral IE gene expression is not sufficient to result in the production of infectious virions (61). Consistent with the findings reported here, UL135 has been shown to be critical for overcoming the suppressive effects of the UL138 latency determinant (19). Further investigation of the mechanisms by which UL135 and UL97 influence the viral lytic cycle may shed light on additional checkpoints that govern productive reactivation during HCMV latency.
ACKNOWLEDGMENTS
We thank Christian Sinzger (Institute for Virology, Ulm, Germany), Ulrich Koszinowski (Max von Pettenkofer Institute, Munich, Germany), Greg Smith (Northwestern University, Chicago, IL), Tom Shenk (Princeton University, NJ), and John Drach (University of Michigan, Ann Arbor, MI) for reagents. We are also grateful to Don Coen (Harvard Medical School, Boston, MA) for providing constructive feedback on the manuscript, to Lindsey Hutt-Fletcher (LSU Health Sciences Center, Shreveport, LA) for helpful advice, and to Jean Pesola (Harvard Medical School) for advice on statistics.
This project was supported by American Heart Association grant 12GRNT11890012 (to J.P.K.) and by the following grants from the U.S. Public Health Service: 8P20GM103433 (to J.P.K.) from the National Institute of General Medical Sciences and AI079059 (to F.D.G.) from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print 12 March 2014
REFERENCES
- 1.Mocarski ES, Shenk T, Griffiths PD, Pass RF. 2013. Cytomegaloviruses, p 1960–2014 In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B. (ed), Fields virology, 6th ed (electronic), vol 2 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 2.Prichard MN. 2009. Function of human cytomegalovirus UL97 kinase in viral infection and its inhibition by maribavir. Rev. Med. Virol. 19:215–229. 10.1002/rmv.615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biron KK. 2006. Antiviral drugs for cytomegalovirus diseases. Antiviral Res. 71:154–163. 10.1016/j.antiviral.2006.05.002 [DOI] [PubMed] [Google Scholar]
- 4.Hume AJ, Finkel JS, Kamil JP, Coen DM, Culbertson MR, Kalejta RF. 2008. Phosphorylation of retinoblastoma protein by viral protein with cyclin-dependent kinase function. Science 320:797–799. 10.1126/science.1152095 [DOI] [PubMed] [Google Scholar]
- 5.Prichard MN, Sztul E, Daily SL, Perry AL, Frederick SL, Gill RB, Hartline CB, Streblow DN, Varnum SM, Smith RD, Kern ER. 2008. Human cytomegalovirus UL97 kinase activity is required for the hyperphosphorylation of retinoblastoma protein and inhibits the formation of nuclear aggresomes. J. Virol. 82:5054–5067. 10.1128/JVI.02174-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marschall M, Marzi A, aus dem Siepen P, Jochmann R, Kalmer M, Auerochs S, Lischka P, Leis M, Stamminger T. 2005. Cellular p32 recruits cytomegalovirus kinase pUL97 to redistribute the nuclear lamina. J. Biol. Chem. 280:33357–33367. 10.1074/jbc.M502672200 [DOI] [PubMed] [Google Scholar]
- 7.Hamirally S, Kamil JP, Ndassa-Colday YM, Lin AJ, Jahng WJ, Baek MC, Noton S, Silva LA, Simpson-Holley M, Knipe DM, Golan DE, Marto JA, Coen DM. 2009. Viral mimicry of Cdc2/cyclin-dependent kinase 1 mediates disruption of nuclear lamina during human cytomegalovirus nuclear egress. PLoS Pathog. 5:e1000275. 10.1371/journal.ppat.1000275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reim NI, Kamil JP, Wang D, Lin A, Sharma M, Ericsson M, Pesola JM, Golan DE, Coen DM. 2013. Inactivation of retinoblastoma protein does not overcome the requirement for human cytomegalovirus UL97 in lamina disruption and nuclear egress. J. Virol. 87:5019–5027. 10.1128/JVI.00007-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyson N. 1998. The regulation of E2F by pRB-family proteins. Genes Dev. 12:2245–2262. 10.1101/gad.12.15.2245 [DOI] [PubMed] [Google Scholar]
- 10.Kamil JP, Hume AJ, Jurak I, Munger K, Kalejta RF, Coen DM. 2009. Human papillomavirus 16 E7 inactivator of retinoblastoma family proteins complements human cytomegalovirus lacking UL97 protein kinase. Proc. Natl. Acad. Sci. U. S. A. 106:16823–16828. 10.1073/pnas.0901521106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krosky PM, Baek MC, Coen DM. 2003. The human cytomegalovirus UL97 protein kinase, an antiviral drug target, is required at the stage of nuclear egress. J. Virol. 77:905–914. 10.1128/JVI.77.2.905-914.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharma M, Kamil JP, Coughlin M, Reim NI, Coen DM. 2014. Human cytomegalovirus UL50 and UL53 recruit viral protein kinase UL97, not protein kinase C, for disruption of nuclear lamina and nuclear egress in infected cells. J. Virol. 88:249–262. 10.1128/JVI.02358-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran K, Kamil JP, Coen DM, Spector DH. 2010. Inactivation and disassembly of the anaphase-promoting complex during human cytomegalovirus infection is associated with degradation of the APC5 and APC4 subunits and does not require UL97-mediated phosphorylation of Cdh1. J. Virol. 84:10832–10843. 10.1128/JVI.01260-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fehr AR, Gualberto NC, Savaryn JP, Terhune SS, Yu D. 2012. Proteasome-dependent disruption of the E3 ubiquitin ligase anaphase-promoting complex by HCMV protein pUL21a. PLoS Pathog. 8:e1002789. 10.1371/journal.ppat.1002789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hobom U, Brune W, Messerle M, Hahn G, Koszinowski UH. 2000. Fast screening procedures for random transposon libraries of cloned herpesvirus genomes: mutational analysis of human cytomegalovirus envelope glycoprotein genes. J. Virol. 74:7720–7729. 10.1128/JVI.74.17.7720-7729.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinzger C, Hahn G, Digel M, Katona R, Sampaio KL, Messerle M, Hengel H, Koszinowski U, Brune W, Adler B. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 89:359–368. 10.1099/vir.0.83286-0 [DOI] [PubMed] [Google Scholar]
- 17.Wang D, Li G, Schauflinger M, Nguyen CC, Hall ED, Yurochko AD, von Einem J, Kamil JP. 2013. The ULb′ region of the human cytomegalovirus genome confers an increased requirement for the viral protein kinase UL97. J. Virol. 87:6359–6376. 10.1128/JVI.03477-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Umashankar M, Petrucelli A, Cicchini L, Caposio P, Kreklywich CN, Rak M, Bughio F, Goldman DC, Hamlin KL, Nelson JA, Fleming WH, Streblow DN, Goodrum F. 2011. A novel human cytomegalovirus locus modulates cell type-specific outcomes of infection. PLoS Pathog. 7:e1002444. 10.1371/journal.ppat.1002444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umashankar M, Rak M, Bughio F, Zagallo P, Caviness K, Goodrum FD. 2014. Antagonistic determinants controlling replicative and latent states of human cytomegalovirus infection. J. Virol. 88:5987–6002. 10.1128/JVI.03506-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tischer BK, Smith GA, Osterrieder N. 2010. En passant mutagenesis: a two step markerless Red recombination system. Methods Mol. Biol. 634:421–430. 10.1007/978-1-60761-652-8_30 [DOI] [PubMed] [Google Scholar]
- 21.Tischer BK, von Einem J, Kaufer B, Osterrieder N. 2006. Two-step Red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197. 10.2144/000112096 [DOI] [PubMed] [Google Scholar]
- 22.McSharry JJ, McDonough A, Olson B, Talarico C, Davis M, Biron KK. 2001. Inhibition of ganciclovir-susceptible and -resistant human cytomegalovirus clinical isolates by the benzimidazole l-riboside 1263W94. Clin. Diagn. Lab. Immunol. 8:1279–1281. 10.1128/CDLI.8.6.1279-1281.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biron KK, Harvey RJ, Chamberlain SC, Good SS, Smith AA, III, Davis MG, Talarico CL, Miller WH, Ferris R, Dornsife RE, Stanat SC, Drach JC, Townsend LB, Koszalka GW. 2002. Potent and selective inhibition of human cytomegalovirus replication by 1263W94, a benzimidazole l-riboside with a unique mode of action. Antimicrob. Agents Chemother. 46:2365–2372. 10.1128/AAC.46.8.2365-2372.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petrucelli A, Rak M, Grainger L, Goodrum F. 2009. Characterization of a novel Golgi apparatus-localized latency determinant encoded by human cytomegalovirus. J. Virol. 83:5615–5629. 10.1128/JVI.01989-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nogalski MT, Chan G, Stevenson EV, Gray S, Yurochko AD. 2011. Human cytomegalovirus-regulated paxillin in monocytes links cellular pathogenic motility to the process of viral entry. J. Virol. 85:1360–1369. 10.1128/JVI.02090-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang X, Milyavsky M, Shats I, Erez N, Goldfinger N, Rotter V. 2004. Activated p53 suppresses the histone methyltransferase EZH2 gene. Oncogene 23:5759–5769. 10.1038/sj.onc.1207706 [DOI] [PubMed] [Google Scholar]
- 27.Fehr AR, Yu D. 2011. Human cytomegalovirus early protein pUL21a promotes efficient viral DNA synthesis and the late accumulation of immediate-early transcripts. J. Virol. 85:663–674. 10.1128/JVI.01599-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stamminger T, Puchtler E, Fleckenstein B. 1991. Discordant expression of the immediate-early 1 and 2 gene regions of human cytomegalovirus at early times after infection involves posttranscriptional processing events. J. Virol. 65:2273–2282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stenberg RM, Depto AS, Fortney J, Nelson JA. 1989. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate-early gene region. J. Virol. 63:2699–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stenberg RM, Witte PR, Stinski MF. 1985. Multiple spliced and unspliced transcripts from human cytomegalovirus immediate-early region 2 and evidence for a common initiation site within immediate-early region 1. J. Virol. 56:665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomasec P, Wang EC, Davison AJ, Vojtesek B, Armstrong M, Griffin C, McSharry BP, Morris RJ, Llewellyn-Lacey S, Rickards C, Nomoto A, Sinzger C, Wilkinson GW. 2005. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat. Immunol. 6:181–188. 10.1038/ni1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cha TA, Tom E, Kemble GW, Duke GM, Mocarski ES, Spaete RR. 1996. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J. Virol. 70:78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy E, Yu D, Grimwood J, Schmutz J, Dickson M, Jarvis MA, Hahn G, Nelson JA, Myers RM, Shenk TE. 2003. Coding potential of laboratory and clinical strains of human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 100:14976–14981. 10.1073/pnas.2136652100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cunningham C, Gatherer D, Hilfrich B, Baluchova K, Dargan DJ, Thomson M, Griffiths PD, Wilkinson GW, Schulz TF, Davison AJ. 2010. Sequences of complete human cytomegalovirus genomes from infected cell cultures and clinical specimens. J. Gen. Virol. 91:605–615. 10.1099/vir.0.015891-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puchtler E, Stamminger T. 1991. An inducible promoter mediates abundant expression from the immediate-early 2 gene region of human cytomegalovirus at late times after infection. J. Virol. 65:6301–6306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bughio F, Elliott DA, Goodrum F. 2013. An endothelial cell-specific requirement for the UL133-UL138 locus of human cytomegalovirus for efficient virus maturation. J. Virol. 87:3062–3075. 10.1128/JVI.02510-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michel D, Pavic I, Zimmermann A, Haupt E, Wunderlich K, Heuschmid M, Mertens T. 1996. The UL97 gene product of human cytomegalovirus is an early-late protein with a nuclear localization but is not a nucleoside kinase. J. Virol. 70:6340–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Teng MW, Bolovan-Fritts C, Dar RD, Womack A, Simpson ML, Shenk T, Weinberger LS. 2012. An endogenous accelerator for viral gene expression confers a fitness advantage. Cell 151:1569–1580. 10.1016/j.cell.2012.11.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burkhart DL, Sage J. 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 8:671–682. 10.1038/nrc2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gribaudo G, Riera L, Lembo D, De Andrea M, Gariglio M, Rudge TL, Johnson LF, Landolfo S. 2000. Murine cytomegalovirus stimulates cellular thymidylate synthase gene expression in quiescent cells and requires the enzyme for replication. J. Virol. 74:4979–4987. 10.1128/JVI.74.11.4979-4987.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lembo D, Gribaudo G, Riera L, Mondo A, Cavallo R, Angeretti A, Landolfo S. 2000. The thymidylate synthase inhibitor ZD1694 potently inhibits murine and human cytomegalovirus replication in quiescent fibroblasts. Antiviral Res. 47:111–120. 10.1016/S0166-3542(00)00096-6 [DOI] [PubMed] [Google Scholar]
- 42.Hagemeier C, Caswell R, Hayhurst G, Sinclair J, Kouzarides T. 1994. Functional interaction between the HCMV IE2 transactivator and the retinoblastoma protein. EMBO J. 13:2897–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sommer MH, Scully AL, Spector DH. 1994. Transactivation by the human cytomegalovirus IE2 86-kilodalton protein requires a domain that binds to both the TATA box-binding protein and the retinoblastoma protein. J. Virol. 68:6223–6231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Song YJ, Stinski MF. 2002. Effect of the human cytomegalovirus IE86 protein on expression of E2F-responsive genes: a DNA microarray analysis. Proc. Natl. Acad. Sci. U. S. A. 99:2836–2841. 10.1073/pnas.052010099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choi KS, Kim SJ, Kim S. 1995. The retinoblastoma gene product negatively regulates transcriptional activation mediated by the human cytomegalovirus IE2 protein. Virology 208:450–456. 10.1006/viro.1995.1175 [DOI] [PubMed] [Google Scholar]
- 46.Brehm A, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. 1998. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 391:597–601. 10.1038/35404 [DOI] [PubMed] [Google Scholar]
- 47.Magnaghi-Jaulin L, Groisman R, Naguibneva I, Robin P, Lorain S, Le Villain JP, Troalen F, Trouche D, Harel-Bellan A. 1998. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 391:601–605. 10.1038/35410 [DOI] [PubMed] [Google Scholar]
- 48.Nielsen SJ, Schneider R, Bauer UM, Bannister AJ, Morrison A, O'Carroll D, Firestein R, Cleary M, Jenuwein T, Herrera RE, Kouzarides T. 2001. Rb targets histone H3 methylation and HP1 to promoters. Nature 412:561–565. 10.1038/35087620 [DOI] [PubMed] [Google Scholar]
- 49.Vandel L, Nicolas E, Vaute O, Ferreira R, Ait-Si-Ali S, Trouche D. 2001. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol. Cell. Biol. 21:6484–6494. 10.1128/MCB.21.19.6484-6494.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reeves M, Murphy J, Greaves R, Fairley J, Brehm A, Sinclair J. 2006. Autorepression of the human cytomegalovirus major immediate-early promoter/enhancer at late times of infection is mediated by the recruitment of chromatin remodeling enzymes by IE86. J. Virol. 80:9998–10009. 10.1128/JVI.01297-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bigley TM, Reitsma JM, Mirza SP, Terhune SS. 2013. Human cytomegalovirus pUL97 regulates the viral major immediate early promoter by phosphorylation-mediated disruption of histone deacetylase 1 binding. J. Virol. 87:7393–7408. 10.1128/JVI.02825-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cuevas-Bennett C, Shenk T. 2008. Dynamic histone H3 acetylation and methylation at human cytomegalovirus promoters during replication in fibroblasts. J. Virol. 82:9525–9536. 10.1128/JVI.00946-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Toth Z, Brulois K, Lee HR, Izumiya Y, Tepper C, Kung HJ, Jung JU. 2013. Biphasic euchromatin-to-heterochromatin transition on the KSHV genome following de novo infection. PLoS Pathog. 9:e1003813. 10.1371/journal.ppat.1003813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abraham CG, Kulesza CA. 2013. Polycomb repressive complex 2 silences human cytomegalovirus transcription in quiescent infection models. J. Virol. 87:13193–13205. 10.1128/JVI.02420-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Knipe DM, Lieberman PM, Jung JU, McBride AA, Morris KV, Ott M, Margolis D, Nieto A, Nevels M, Parks RJ, Kristie TM. 2013. Snapshots: chromatin control of viral infection. Virology 435:141–156. 10.1016/j.virol.2012.09.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nitzsche A, Paulus C, Nevels M. 2008. Temporal dynamics of cytomegalovirus chromatin assembly in productively infected human cells. J. Virol. 82:11167–11180. 10.1128/JVI.01218-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nitzsche A, Steinhausser C, Mucke K, Paulus C, Nevels M. 2012. Histone H3 lysine 4 methylation marks postreplicative human cytomegalovirus chromatin. J. Virol. 86:9817–9827. 10.1128/JVI.00581-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Woodhall DL, Groves IJ, Reeves MB, Wilkinson G, Sinclair JH. 2006. Human Daxx-mediated repression of human cytomegalovirus gene expression correlates with a repressive chromatin structure around the major immediate early promoter. J. Biol. Chem. 281:37652–37660. 10.1074/jbc.M604273200 [DOI] [PubMed] [Google Scholar]
- 59.Zalckvar E, Paulus C, Tillo D, Asbach-Nitzsche A, Lubling Y, Winterling C, Strieder N, Mucke K, Goodrum F, Segal E, Nevels M. 2013. Nucleosome maps of the human cytomegalovirus genome reveal a temporal switch in chromatin organization linked to a major IE protein. Proc. Natl. Acad. Sci. U. S. A. 110:13126–13131. 10.1073/pnas.1305548110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sinclair J. 2008. Human cytomegalovirus: latency and reactivation in the myeloid lineage. J. Clin. Virol. 41:180–185. 10.1016/j.jcv.2007.11.014 [DOI] [PubMed] [Google Scholar]
- 61.Taylor-Wiedeman J, Sissons P, Sinclair J. 1994. Induction of endogenous human cytomegalovirus gene expression after differentiation of monocytes from healthy carriers. J. Virol. 68:1597–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]