ABSTRACT
Enterovirus 71 (EV71) and coxsackievirus A16 (CA16) are the two most common etiological agents responsible for the epidemics of hand, foot, and mouth disease (HFMD), a childhood illness with occasional severe neurological complications. A number of vaccine candidates against EV71 or CA16 have been reported; however, no vaccine is currently available for clinical use. Here, we generated a secreted version of EV71 and CA16 virus-like particles (VLPs) using a baculovirus-insect cell expression system and reconstructed the three-dimensional (3D) structures of both VLPs by cryo-electron microscopy (cryo-EM) single-particle analysis at 5.2-Å and 5.5-Å resolutions, respectively. The reconstruction results showed that the cryo-EM structures of EV71 and CA16 VLPs highly resemble the recently published crystal structures for EV71 natural empty particles and CA16 135S-like expanded particles, respectively. Our cryo-EM analysis also revealed that the majority of previously identified linear neutralizing epitopes are well preserved on the surface of EV71 and CA16 VLPs. In addition, both VLPs were able to induce efficiently neutralizing antibodies against various strains of EV71 and CA16 viruses in mouse immunization. These studies provide a structural basis for the development of insect cell-expressed VLP vaccines and for a potential bivalent VLP vaccine against both EV71- and CA16-associated HFMD.
IMPORTANCE The recent outbreaks of hand, foot, and mouth disease (HFMD) in the Asia Pacific region spurred the search for effective vaccines against EV71 and CA16 viruses, the two most common etiological agents responsible for HFMD. In this paper, we show that secreted versions of EV71 and CA16 VLPs generated in the baculovirus-insect cell expression system highly resemble the crystal structures of their viral conterparts and that the majority of previously identified linear neutralizing epitopes are well preserved on the VLP surfaces. In addition, the generated VLPs can efficiently induce neutralizing antibodies against various strains of EV71 and CA16 viruses in mouse immunization. These studies provide a structural basis for the development of insect cell-expressed VLP vaccines and for a potential bivalent VLP vaccine against both EV71- and CA16-associated HFMD.
INTRODUCTION
Hand, foot, and mouth disease (HFMD) most frequently occurs in infants or children under 5 years old. Although most cases are mild and self-limited, HFMD can in some cases cause severe symptoms, such as myocarditis, pulmonary edema, and central nervous system complications ranging from flaccid paralysis to fatal encephalitis (1, 2). The recent epidemics of HFMD led to serious public health threats in the Asia Pacific region. In China, 1.16 million and 1.77 million cases of HFMD, including 353 and 905 deaths, were reported for 2009 and 2010, respectively (3). Etiologic studies revealed that enteroviruses, especially enterovirus 71 (EV71) and coxsackievirus A16 (CA16), are the most common causative agents responsible for HFMD epidemics (4–6). A clinical survey of the 2009 HFMD outbreak in mainland China revealed that EV71 and CA16 accounted for 50.4% and 38.3% of the 266 cases positive for human enteroviral infection (6). Another clinical study to investigate the relationship between pathogens and the ensuing nervous system complications in HFMD cases showed that EV71 and CA16 accounted for 70.7% and 20.7%, respectively, among 92 patients with neurological complications (7).
Continuing outbreaks of HFMD in recent years spurred the search for effective vaccines against enteroviruses, in particular, for EV71 and CA16 viruses. Various candidates against EV71 or CA16 virus, including inactivated vaccines (3, 8–22), live attenuated vaccines (23, 24), subunit vaccines based on the VP1 protein (25), virus-like particle (VLP) vaccines (26–34), and epitope-based vaccines (35–39), were shown to possess various levels of efficacy in animal studies or human clinical trials. In particular, inactivated EV71 vaccines showed promising results against EV71-associated diseases in recent phase 3 clinical studies conducted in mainland China (20–22). However, the exact efficacy of these vaccines remains to be identified before they can be used widely in the target population. In addition, these vaccines target only EV71 instead of both EV71 and CA16 viruses, the two most important causative agents of HFMD.
As an alternative choice for vaccine candidates, VLP vaccines have the advantages of being highly immunogenic, noninfectious, and accessible to quality control as well as to scaling-up during production. Expression systems based on baculovirus-insect cells are often used for the production of VLP vaccines. Coexpression in insect cells of the P1 (encoding viral structural proteins) and 3CD (encoding viral proteases) genes mimics the early steps of the enteroviral life cycle, including proteolysis of polyprotein, assembly into empty capsid, and generation of VLPs (26, 28, 29, 33). Both EV71 and CA16 VLPs consist of 60 copies for each of the capsid proteins, VP0, VP1, and VP3, which follow a P = 3 icosahedral arrangement. Previous studies showed that both EV71 and CA16 VLPs can elicit potent immune responses and are able to protect immunized mice against lethal challenges with EV71 or CA16 virus (27, 31, 33, 34). How VLP vaccines mimic their viral counterparts structurally and how the neutralizing epitopes are preserved on the surface of VLPs are issues of big interest for effective vaccine development. In the present study, we expressed a secreted version of EV71 and CA16 VLPs in Sf9 insect cells and determined the three-dimensional (3D) structure of both VLPs using cryo-electron microscopy (cryo-EM) single-particle analysis. We then compared the cryo-EM density maps of EV71 and CA16 VLPs with recently published crystal structures of authentic EV71 and CA16 viral particles (40, 41) and mapped the previously identified linear neutralizing epitopes (35–38) on both VLPs. In addition, we evaluated the immunogenicity and neutralizing antibody responses of both EV71 and CA16 VLPs by immunization of mice. These studies provide a structural basis for the production of VLP vaccines using an insect cell expression system, as well as for the potential development of a bivalent EV71 and CA16 VLP vaccine candidate against HFMD.
MATERIALS AND METHODS
Expression and purification of EV71 and CA16 virus-like particles.
The P1 and 3CD gene fragments of EV71 and CA16 were amplified by reverse transcription-PCR (RT-PCR) from the EV71 Fuyang strain (C4 genotype) (2) and CA16 09-7 strain (B1 genotype) (6), respectively. The P1 and 3CD genes were subsequently cloned into a pFastBac Dual vector (Invitrogen) under the control of the polyhedrin and p10 promoters (28). The p10 promoter was then replaced by the cytomegalovirus (CMV) immediately early promoter in order to produce the secreted version of VLPs (26). Recombinant baculoviruses were then generated using a Bac-to-Bac baculovirus expression system (Invitrogen) according to the instructions of the manufacturer. The expression of EV71 and CA16 recombinant proteins was confirmed by fluorescence-activated cell sorter (FACS) analysis. The EV71 and CA16 VLPs were produced by infecting Sf9 insect cells (4 × 106 to 6 × 106 cells/ml) at a multiplicity of infection (MOI) of 8 in a suspension culture with SFX-Insect medium (HyClone). The supernatants were collected 3 days postinfection and centrifuged at 20,000 × g for 20 min to remove cell debris. After filtration using a 0.22-μm-pore-size filter, the supernatants were then subjected to pelleting through a 30% sucrose cushion at 160,000 × g for 4 h at 4°C. Pellets were resuspended in phosphate-buffered saline (PBS) (pH 7.4) and cleared by centrifugation at 9,300 × g for 5 min. The samples were loaded onto 1.2 g/ml and 1.4 g/ml discontinuous cesium chloride density gradients and centrifuged at 220,000 × g for 20 h at 4°C. Fractions containing VLPs were collected and dialyzed against PBS (pH 7.4). After being concentrated, the VLPs were subjected to analytical ultracentrifugation (AUC) and SDS-PAGE analysis, cryo-EM study, and mouse immunization experiments. All bands visible after Coomassie staining of the SDS-PAGE gel were cut and processed for mass spectrometry analysis (LTQ Orbitrap XL).
Analytical ultracentrifugation.
The sedimentation coefficient distributions [c(s)] of EV71 VLPs (0.33 mg/ml) and CA16 VLPs (0.76 mg/ml) in 0.013 M PBS were determined using a Beckman Coulter ProteomeLab XL-I analytical ultracentrifuge with a 4-hole An-60Ti rotor. The absorbance at 280 nm was measured in a continuous scan mode during sedimentation at 6,532 × g in 12-mm double-sector cells at 20°C (measured cell radius from 5.9 cm to 7.2 cm). The program SedFit (42, 43) was used to analyze data in c(s) mode to give an apparent sedimentation coefficient distribution, and the 6th to 95th scans of both EV71 and CA16 VLPs were used for the c(s) calculations.
Cryo-EM imaging and three-dimensional reconstruction of EV71 and CA16 virus-like particles.
Aliquots of 3.5 μl of purified EV71 or CA16 VLPs were applied to glow-discharged Quantifoil grids (Quantifoil Micro Tools GmbH, Jena, Germany), blotted for 4.5 s in a 100% humidity chamber, and plunged into liquid ethane (cooled by liquid nitrogen) in an FEI Vitrobot Mark IV vitrification robot. Both VLPs were imaged in an FEI 300-kV Titan Krios cryo-electron microscope equipped with a Gatan UltraScan4000 (model 895) 16-megapixel charge-coupled device (CCD). Images were recorded at 300 kV at a calibrated magnification of ×160,770, corresponding to a pixel size of 0.933 Å, and the electron dose for each micrograph was approximately 20 electrons/Å2. The images of EV71 VLPs were collected by an automated acquisition system, Leginon (44), and the images of CA16 VLPs were acquired manually. The EV71 particles were selected by the program FindEM (45) embedded in the Appion system (46), and the CA16 particles were selected using the program boxer in EMAN1 (47). The initial model for CA16 VLPs was generated by IMIRS (48), and EMAN2 (49) was used for contrast transfer function (CTF) correction, 2D analysis and refinement, and final 3D reconstruction. For EV71 VLPs, the initial model was generated by filtering the reconstructed CA16 VLP structure to 30 Å, and the subsequent CTF correction, 2D analysis and refinement, and 3D reconstruction were similarly performed in EMAN2 (49). The resolutions of final maps were assessed by the Fourier shell correlation (FSC) method, with a 0.5-cutoff criterion (50).
Structure analysis of EV71 and CA16 virus-like particles.
UCSF Chimera software (51) was used to visualize the density maps of EV71 and CA16 VLPs, to segment the asymmetric units from the corresponding density maps, and to compare the structural similarities between VLPs and various reported structures of EV71 and CA16 viral particle counterparts (40, 41, 52–54). After docking the crystal structures of EV71 natural empty particles (PDB: 3VBU) (41) and CA16 135S-like expanded particles (PDB: 4JGZ) (40) into the EV71 and CA16 VLP density maps (σ = 1.00), the correlation coefficient (cc) values from comparisons of the cryo-EM density maps and crystal structures were calculated using “Fit in Map” in UCSF Chimera (51).
Mouse immunization and microneutralization assay.
Groups of 6 female BALB/c mice (6 to 8 weeks old, specific pathogen free [SPF]) were immunized intramuscularly (i.m.) with 2.5 μg EV71 or CA16 VLPs (2.5 μg EV71 VLPs plus 2.5 μg CA16 VLPs for the bivalent vaccine study) either alone or in combination with 100 μg alum or 10 μg CpG-oligodeoxynucleotides (CpG-ODN) (55) or both in a total volume of 100 μl. Mice were boosted with the same regimen 3 weeks later. Serum samples were collected 3 weeks postpriming and 2 weeks postboosting for serological analysis. The animal studies were carried out according to the guidelines of the Animal Care and Use Committee at the National Vaccine and Serum Institute in China. Neutralizing antibody titers were measured by a standard microneutralization assay in 96-well plates. Briefly, pooled sera from each group were diluted 10-fold in M-199 medium, filtered using a 0.22-μm-pore-size filter, and heated for 30 min at 56°C to inactivate complement. Serum samples were diluted in 96-well plates by 2-fold serial dilution with M-199 and incubated at room temperature for 1 h with 100 50% tissue culture infective doses (TCID50s) of EV71 or CA16 virus. Sera from naive mice at a 1:20 dilution were used as a negative control. Following incubation, 5,000 rhabdomyosarcoma (RD) cells were added to each well and incubated at 37°C with 5% CO2. After 7 (for EV71) or 8 (for CA16) days of culturing, neutralizing titers were determined as the highest dilution that could completely prevent cells from displaying cytopathogenic effects.
Cryo-EM density map accession numbers.
The cryo-EM density maps of EV71 and CA16 VLPs have been deposited in the Electron Microscopy Data Bank under accession numbers EMD-2607 and EMD-2608, respectively.
RESULTS AND DISCUSSION
Expression, purification, and characterization of recombinant EV71 and CA16 virus-like particles.
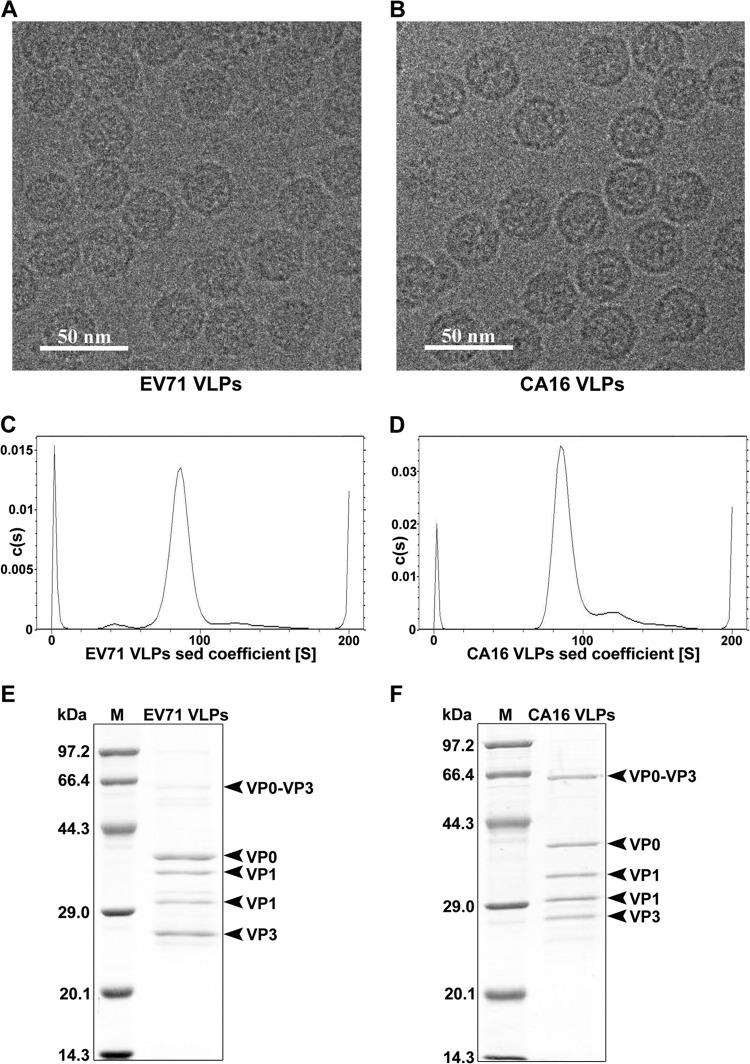
In an attempt to express EV71 and CA16 VLPs, the genes encoding P1 and 3CD were amplified by RT-PCR from strains EV71 Fuyang and CA16 09-7, respectively. The P1 and 3CD gene fragments were subsequently cloned into the pFastBac Dual shuttle plasmid, under the control of the polyhedrin and p10 promoters, respectively (28). In order to produce the secreted version of VLPs, the p10 promoter controlling the expression of 3CD was replaced by the CMV promoter (26). Recombinant baculoviruses were generated using a Bac-to-Bac expression system (Invitrogen). A suspension culture of Sf9 insect cells was infected with recombinant baculovirus, and the supernatants containing VLPs were harvested 3 days postinfection and subjected to a brief purification by cesium chloride density gradient centrifugation. The purified EV71 and CA16 VLPs were characterized by cryo-EM, analytical ultracentrifugation, SDS-PAGE, and mass spectrometry analysis. Cryo-EM images revealed that both EV71 and CA16 VLPs are about 30 nm in diameter, similar to the sizes of the authentic EV71 and CA16 viruses (Fig. 1A and B). Analytical ultracentrifugation (AUC) analysis showed that the sedimentation coefficients of both EV71 and CA16 VLPs are about 85S (Fig. 1C and D), a result that is consistent with previous measurements of EV71 and CA16 empty viral particles (40, 41), suggesting that both VLPs are empty particles devoid of nucleic acid. SDS-PAGE and mass spectrometry analysis showed that both EV71 and CA16 VLPs consisted mostly of VP0 (35 kDa), VP1 (33 and 30 kDa), and VP3 (27 kDa) capsid proteins and an additional, presumably incompletely processed band, VP0-VP3 (62 kDa) (Fig. 1E and F).
FIG 1.
Characterization of recombinant EV71 and CA16 virus-like particles. Purified EV71 VLPs (A, C, and E) and CA16 VLPs (B, D, and F) were characterized by cryo-electron microscopy (A and B), analytical ultracentrifugation (C and D), and SDS-PAGE analysis (E and F), respectively. sed, sedimentation; M, molecular mass marker.
Cryo-EM reconstructions of recombinant EV71 and CA16 virus-like particles.
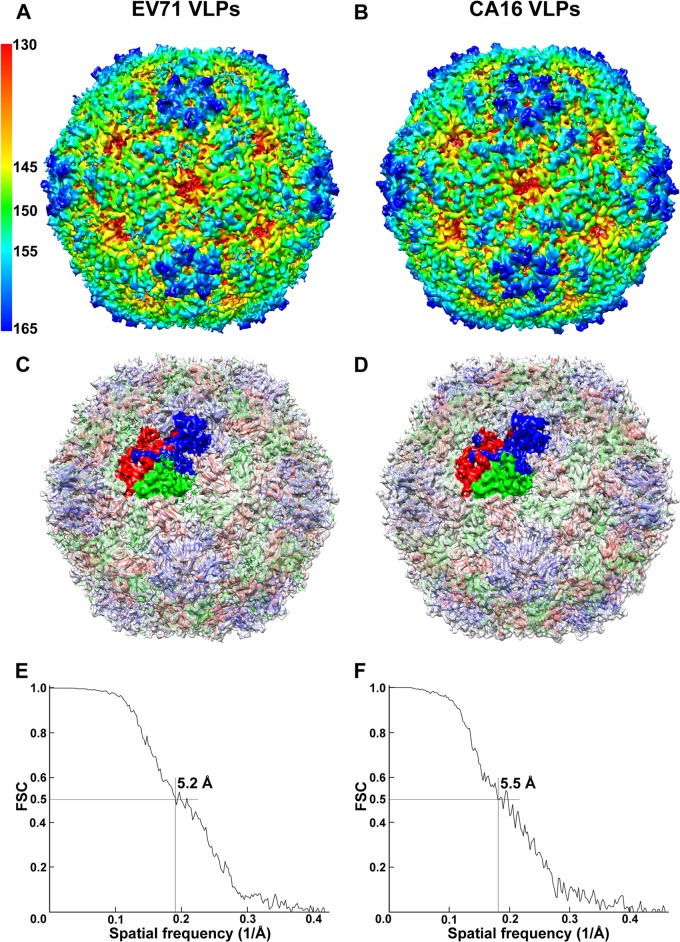
Cryo-EM images of EV71 and CA16 VLPs were recorded using an FEI Titan Krios electron microscope. A total of 30,386 EV71 VLPs and 21,403 CA16 VLPs were selected from the cryo-EM images and subjected to 2D alignment and 3D reconstruction by single-particle analysis using the EMAN2 package (49). The final resolutions for the 3D reconstructions of EV71 and CA16 VLP density maps (Fig. 2A and B) are 5.2 Å and 5.5 Å, respectively, as estimated by the Fourier shell correlation (FSC) at the 0.5-cutoff criterion (Fig. 2E and F) (50). After comparing various reported structures of authentic EV71 and CA16 viral particles (40, 41, 52–54) with our cryo-EM density maps of VLPs, we found that EV71 and CA16 VLPs produced from insect cells exhibit overall architectures highly similar to those of authentic EV71 and CA16 viral particles purified from the virus-infected cells. While the EV71 VLPs display a structure very similar to that of EV71 natural empty particles (PDB: 3VBU) (Fig. 2C) (cc = 0.88), which is slightly larger than that of EV71 mature virus (PDB: 3VBS) (41), the CA16 VLPs show a structure nearly identical to that of CA16 135S-like expanded particles (PDB: 4JGZ) (Fig. 2D) (cc = 0.91) (40). Both EV71 and CA16 VLP structures display a vertex around the 5-fold axis channel, and surrounding the vertex is a narrow depression known as a canyon (Fig. 2A and B). The structures of the 5-fold vertex and canyon in both EV71 and CA16 VLPs were well resolved in our cryo-EM density maps (Fig. 2A and B), into which the crystal structures of EV71 natural empty particles and CA16 135S-like expanded particles can be readily fitted (Fig. 2C and D). Like other enteroviruses, both EV71 and CA16 VLPs present a propeller feature (56) around the 3-fold axis of symmetry region (Fig. 2A and B). The crystal structures of the propellers from authentic EV71 and CA16 virions can also be readily fitted into the corresponding VLP density maps (Fig. 2C and D). At the 2-fold axis, both EV71 and CA16 VLPs present channels (Fig. 2A and B) measured as 9 × 26 Å for EV71 VLPs and 8 × 27 Å for CA16 VLPs, which are similar to those found in the crystal structures of EV71 natural empty particles (8 × 25 Å) and CA16 135S-like expanded particles (9 × 26 Å) (40, 41) (Fig. 2C and D). Together, these results demonstrate that the secreted versions of recombinant EV71 and CA16 VLPs have well-preserved structural characteristics of the EV71 natural empty particles and CA16 135S-like expanded particles, respectively (Fig. 2C and D).
FIG 2.
Cryo-EM reconstructions of EV71 and CA16 virus-like particles. (A and B) Cryo-EM structures of EV71 VLPs (A) and CA16 VLPs (B) viewed along a 2-fold axis of symmetry are shown at a contour level of 1σ and colored by radius from red to blue as indicated by the scale bar. (C and D) The crystal structures of EV71 natural empty particles (PDB: 3VBU) (41) and CA16 135S-like expanded particles (PDB: 4JGZ) (40) are docked into the cryo-EM density maps of EV71 VLPs (C) and CA16 VLPs (D). VP1, VP2, and VP3 in crystal structures are colored in blue, green, and red, respectively (VP4 protein was not resolved in crystal structures). Cryo-EM density maps are shown in gray at 50% transparency, and VP1, VP0 (precursor of VP2 and VP4), and VP3 of 1 asymmetric unit of VLPs are colored in blue, green, and red, respectively. (E and F) Fourier shell correlation curves suggested the reconstruction resolutions of EV71 VLPs (E) and CA16 VLPs (F) to be 5.2 Å and 5.5 Å, respectively, at a 0.5-cutoff criterion.
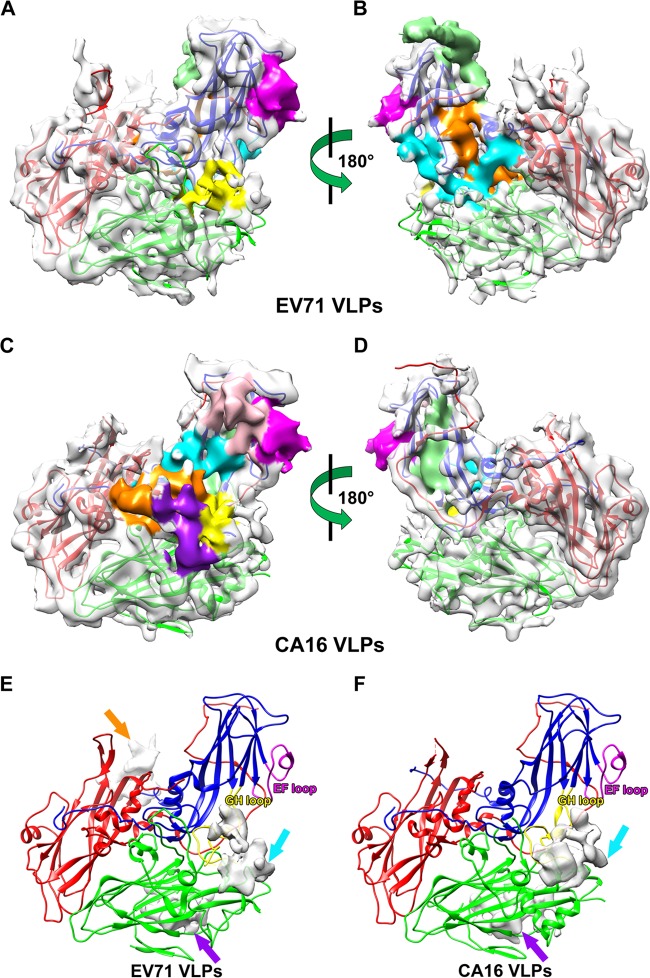
Interestingly, we identified three density blocks (extra contiguous densities) in the cryo-EM density map of EV71 VLPs (Fig. 3E) and two density blocks in that of CA16 VLPs (Fig. 3F) that are not occupied by the corresponding crystal structures of EV71 natural empty particles (PDB: 3VBU) (41) and CA16 135S-like expanded particles (PDB: 4JGZ) (40), respectively. Among these density blocks, two CA16 VLP density blocks partially overlapped with two of the three density blocks of EV71 VLPs (Fig. 3E and F, cyan and purple arrows). Comparing the cryo-EM density maps of EV71 and CA16 VLPs with the crystal structures of their counterparts, we found that one of the density blocks in the cryo-EM density of EV71 VLPs (Fig. 3E, cyan arrow) could be partially occupied by amino acids 211 to 217 of VP1 protein, which was missing in the crystal structure of EV71 natural empty particles (PDB: 3VBU) but was resolved in that of EV71 mature virus structure (PDB: 3VBS) (41). Therefore, we postulate that this density block is contributed by amino acids 211 to 217 of VP1 protein of EV71 VLPs (Fig. 3E, cyan arrow) and that the density block of CA16 VLPs located at the same position (Fig. 3F, cyan arrow) is contributed by amino acids 211 to 218 of VP1 protein of CA16 VLPs. The remaining two EV71 VLP density blocks (Fig. 3E, purple and orange arrows) and one CA16 VLP density block (Fig. 3F, purple arrow) are located near the N termini of VP1 protein according to the crystal structure of EV71 mature virus (41). We therefore hypothesize that these density blocks are contributed by parts of the VP1 N-terminal residues, which were not resolved in the crystal structures of EV71 natural empty particles (41) or CA16 135S-like expanded particles (40).
FIG 3.
Asymmetric units of EV71 and CA16 virus-like particles and epitope mapping. (A to D) Asymmetric units of EV71 VLPs (A and B) and CA16 VLPs (C and D) are viewed from the outer (A and C) and inner (B and D) surface of VLPs. Density maps of EV71 VLPs (A and B) and CA16 VLPs (C and D) are shown in gray at 50% transparency, and previous identified linear neutralizing epitopes (35–38) are shown in different colors (A to D). (E and F) Density blocks of EV71 VLPs (E) and CA16 VLPs (F) not resolved in the crystal structures of EV71 natural empty particles (41) and CA16 135S-like expanded particles (40) are shown in gray at 25% transparency. VP1, VP2, and VP3 of EV71 natural empty particle and CA16 135S-like expanded particle crystal structures docked into the asymmetric unit are colored in blue, green, and red, respectively (A to F). (A and B) EV71 neutralizing epitopes are colored as follows: SP55, 163 to 177 of VP1, magenta; SP70, 208 to 222 of VP1, yellow; K1, 253 to 267 of VP1, orange; K2, 28 to 42 of VP3, cyan; and K3, 1 to 15 of VP3, light green. (C and D) CA16 neutralizing epitopes are colored as follows: PEP32, 94 to 108 of VP1, pink; PEP37, 109 to 123 of VP1, cyan; PEP55, 163 to 177 of VP1, magenta; PEP63, 187 to 201 of VP1, light green; PEP71, 211 to 225 of VP1, yellow; PEP91, 271 to 285 of VP1, orange; and VP2-28, 136 to 150 of VP2, purple. (E and F) The EF and GH loops of VP1 are labeled in magenta and yellow, respectively.
Neutralizing epitopes in the recombinant EV71 and CA16 virus-like particles.
The preservation of neutralizing epitopes is critical for ensuring high efficacy for good vaccine candidates. We mapped various previously identified linear neutralizing epitopes of EV71 and CA16 viruses onto the density maps of our EV71 and CA16 VLPs. Foo et al. screened synthetic peptides covering the VP1 protein of EV71 and identified 2 linear neutralizing epitopes, SP55 and SP70 (35). These 2 epitopes are located in the EF and GH loops (Fig. 3E) of VP1, respectively, both of which are well exposed on the surface of EV71 VLPs (Fig. 3A). Using synthetic peptides spanning the entire EV71 capsid proteins, Liu et al. identified 2 linear neutralizing epitopes, VP1-43 and VP2-28 (37). The location of VP1-43 overlaps that of SP70, whereas the density of VP2-28 is missing in our cryo-EM structure of EV71 VLPs, possibly due to its high flexibility. Using an in silico approach, Kirk et al. identified 4 linear neutralizing epitopes of EV71, D1 (the same as SP70), K1, K2, and K3 (36). K1 covers the major part and several residue extension of the I β-strand of VP1 and is not exposed on the surface of VLPs (Fig. 3B). K2 and K3 are in the N-terminal loops of VP3, which are also buried inside the EV71 VLPs (Fig. 3B). Using a peptide screening method similar to Foo's, Shi et al. identified 6 linear neutralizing epitopes of CA16 VP1 protein (38). Among them, PEP32, PEP37, PEP55, and PEP71 are located in the BC, CD, EF, and GH loops of VP1, respectively, all of which are exposed on the surface of VLPs (Fig. 3C). PEP63 is present in the G β-strand and part of the GH loop, which is not exposed on the VLP surface (Fig. 3D). PEP91 is located on the VLP surface in the C-terminal loop of VP1 (Fig. 3C). In addition, the EV71 and CA16 VP2-28 cross-reactive neutralizing epitope (37) was found on the surface of CA16 VLPs, which is located in the EF loop of VP2 and spatially adjacent to PEP71 (Fig. 3C). Among the epitopes described above, K1, K2, and K3 of EV71 and PEP63 of CA16 are not exposed on the surface of VLPs. These epitopes are also buried inside the authentic viral particles, as shown in their crystal structures (40, 41), and may be exposed during conformational changes in the process of viral infection. Therefore, compared with the EV71 natural empty particles (PDB: 3VBU) and CA16 135S-like expanded particles (PDB: 4JGZ) (40, 41), the majority of identified linear neutralizing epitopes are well preserved on our secreted versions of EV71 and CA16 VLPs (Fig. 3A to D). Given the fact that the EV71 and CA16 VLPs resemble the EV71 natural empty particles and CA16 135S-like expanded particles, respectively, in 3D structures (Fig. 2C and D), it is reasonable to speculate that, besides the linear neutralizing epitopes, conformational epitopes could be well preserved in the recombinant insect cell-expressed EV71 and CA16 VLPs.
Immunogenicity evaluation of recombinant EV71 and CA16 virus-like particles.
To determine the immunogenicity of the secreted version of EV71 or CA16 VLPs, groups of mice were immunized with 2.5 μg EV71 or CA16 VLPs alone or in combination with alum or CpG-ODN or both (as a composite adjuvant). The serum samples were collected 3 weeks postpriming and 2 weeks postboosting and were analyzed by microneutralization assays against 4 strains of EV71 and 2 strains of CA16 viruses isolated from Chinese HFMD patients. The neutralizing antibody titers were read as the highest serum dilutions that resulted in 100% protection of RD cells from cytopathogenic effect at approximately 7 to 8 days postinfection. After priming with EV71 VLPs, the antigen group did not elicit neutralizing antibody titers above 1:20 (the minimum dilution tested) against 4 different strains of EV71 virus (Table 1). However, the neutralizing antibody titers were increased to approximately 1:20 to 1:80 after only one boosting (Table 1). In contrast to a single use of either alum or CpG-ODN showing no adjuvant effect, the composite adjuvant showed strong immunopotentiating effects after one or two immunizations (Table 1). After priming with CA16 VLPs, the antigen group elicited neutralizing antibody titers at 1:40 against 2 strains of CA16 virus (Table 2). After boosting, the neutralizing antibody titers were increased by 2-fold to 1:80 (Table 2). Alum, CpG-ODN, and the composite adjuvant showed different levels of enhancement on neutralizing antibody titers against both CA16 strains after one or two immunizations; of the three, the composite adjuvant was the most potent (Table 2).
TABLE 1.
Neutralizing antibody titers against 4 strains of EV71 virus
| Viral strain | Titer for group (antigen, 2.5 μg EV71 VLPs) |
|||||||
|---|---|---|---|---|---|---|---|---|
| VLPs |
VLPs + alum |
VLPs + CpG |
VLPs + alum + CpG |
|||||
| Prime | Boost | Prime | Boost | Prime | Boost | Prime | Boost | |
| EV71 0804232Y | <20 | 20 | <20 | 40 | <20 | 80 | 40 | 160 |
| EV71 08052303F | 20 | 80 | 20 | 40 | <20 | 40 | 40 | >2,560 |
| EV71 0804251Y | <20 | 20 | <20 | 20 | <20 | 20 | 20 | 640 |
| EV71 08061001Y | 20 | 80 | 20 | 40 | <20 | 320 | 40 | 640 |
TABLE 2.
Neutralizing antibody titers against 2 strains of CA16 virus
| Viral strain | Titer for group (antigen, 2.5 μg CA16 VLPs) |
|||||||
|---|---|---|---|---|---|---|---|---|
| VLPs |
VLPs + alum |
VLPs + CpG |
VLPs + alum + CpG |
|||||
| Prime | Boost | Prime | Boost | Prime | Boost | Prime | Boost | |
| CA16 0705212F | 40 | 80 | 40 | 320 | 40 | 160 | 20 | 640 |
| CA16 0705213F | 40 | 80 | 80 | 160 | 40 | 160 | 80 | 640 |
To evaluate whether a combination of the two VLPs can elicit a neutralizing antibody response to both viruses, groups of mice were immunized with 2.5 μg EV71 and 2.5 μg CA16 VLPs together or in combination with alum or CpG-ODN or both. After priming, the antigen group did not elicit detectable neutralizing antibody titers against any strain of EV71 or CA16 virus (Table 3). The single-adjuvant groups, i.e., those immunized with alum or CpG-ODN alone, showed weak or no immunopotentiating effects after priming, while the composite-adjuvant group elicited neutralizing antibody titers of from 1:160 to 1:320 against all strains of EV71 and CA16 viruses (Table 3). After boosting, the neutralizing antibody titers of the antigen alone and CpG-ODN adjuvant groups were increased to approximately 1:20 to 1:160 for all strains of EV71 virus but were still undetectable for both strains of CA16 virus (Table 3). In contrast, both the alum adjuvant group and the composite-adjuvant groups elicited neutralizing antibody titers of from 1:40 to 1:640 against all strains of EV71 and CA16 viruses; of the two, the composite adjuvant was the most potent (Table 3). These results suggest that a bivalent VLP vaccine against both EV71- and CA16-associated-HFMD, which is more desirable than a monovalent EV71 or CA16 VLP vaccine candidate, is feasible.
TABLE 3.
Neutralizing antibody titers against 4 strains of EV71 and 2 strains of CA16 virus
| Viral strain | Titer for group (antigen, 2.5 μg EV71 VLPs + 2.5 μg CA16 VLPs) |
|||||||
|---|---|---|---|---|---|---|---|---|
| VLPs |
VLPs + alum |
VLPs + CpG |
VLPs + alum + CpG |
|||||
| Prime | Boost | Prime | Boost | Prime | Boost | Prime | Boost | |
| EV71 0804232Y | <20 | 80 | <20 | 160 | <20 | 40 | 320 | 640 |
| EV71 08052303F | <20 | 80 | <20 | 80 | <20 | 80 | 320 | 640 |
| EV71 0804251Y | <20 | 40 | <20 | 160 | <20 | 20 | 320 | 640 |
| EV71 08061001Y | <20 | 80 | <20 | 80 | <20 | 160 | 320 | 640 |
| CA16 0705212F | <20 | <20 | 40 | 160 | <20 | <20 | 160 | 320 |
| CA16 0705213F | <20 | <20 | 20 | 40 | <20 | <20 | 160 | 320 |
Conclusion.
In summary, the present study demonstrated that the insect cell-expressed, secreted versions of recombinant EV71 and CA16 VLPs highly resemble EV71 natural empty particles and CA16 135S-like expanded particles, respectively, in their 3D structures and are able to elicit neutralizing antibodies in mouse immunization. These results provide a structural and immunological basis for the development of insect cell-expressed VLP vaccines and for a potential bivalent VLP vaccine against both EV71- and CA16-associated HFMD.
ACKNOWLEDGMENTS
This work was supported by grants from the National Basic Research Program of China (2010CB912400), the National Natural Science Foundation of China (31230018, 81261120418, and 21261130090), and the National Science and Technology Major Project for infectious disease of China (2013ZX10004606).
We thank Xiaomin Li and Feng Song for help in cryo-EM samples preparation, Ping Chen and Ting Yao for help in AUC analysis, Lunjiang Ling for technical assistance with the data processing at the HPC-Service Station at the Center for Biological Imaging, Institute of Biophysics, Chinese Academy of Sciences, Peng Xue for technique support of Mass Spectrometry, and Torsten Juelich for help with the manuscript editing and critical discussion.
Footnotes
Published ahead of print 26 March 2014
REFERENCES
- 1.McMinn PC. 2002. An overview of the evolution of enterovirus 71 and its clinical and public health significance. FEMS Microbiol. Rev. 26:91–107. 10.1111/j.1574-6976.2002.tb00601.x [DOI] [PubMed] [Google Scholar]
- 2.Solomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. 2010. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect. Dis. 10:778–790. 10.1016/S1473-3099(10)70194-8 [DOI] [PubMed] [Google Scholar]
- 3.Li YP, Liang ZL, Gao Q, Huang LR, Mao QY, Wen SQ, Liu Y, Yin WD, Li RC, Wang JZ. 2012. Safety and immunogenicity of a novel human Enterovirus 71 (EV71) vaccine: a randomized, placebo-controlled, double-blind, Phase I clinical trial. Vaccine 30:3295–3303. 10.1016/j.vaccine.2012.03.010 [DOI] [PubMed] [Google Scholar]
- 4.Rabenau HF, Richter M, Doerr HW. 2010. Hand, foot and mouth disease: seroprevalence of Coxsackie A16 and Enterovirus 71 in Germany. Med. Microbiol. Immunol. 199:45–51. 10.1007/s00430-009-0133-6 [DOI] [PubMed] [Google Scholar]
- 5.Wang Y, Feng Z, Yang Y, Self S, Gao Y, Longini IM, Wakefield J, Zhang J, Wang L, Chen X, Yao L, Stanaway JD, Wang Z, Yang W. 2011. Hand, foot, and mouth disease in China: patterns of spread and transmissibility. Epidemiology 22:781–792. 10.1097/EDE.0b013e318231d67a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang F, Zhang T, Hu Y, Wang X, Du J, Li Y, Sun S, Sun X, Li Z, Jin Q. 2011. Survey of enterovirus infections from hand, foot and mouth disease outbreak in China, 2009. Virol. J. 8:508. 10.1186/1743-422X-8-508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu W, Liu CF, Yan L, Li JJ, Wang LJ, Qi Y, Cheng RB, Xiong XY. 2012. Distribution of enteroviruses in hospitalized children with hand, foot and mouth disease and relationship between pathogens and nervous system complications. Virol. J. 9:8. 10.1186/1743-422X-9-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai Y, Liu Q, Huang X, Li D, Ku Z, Zhang Y, Huang Z. 2013. Active immunization with a Coxsackievirus A16 experimental inactivated vaccine induces neutralizing antibodies and protects mice against lethal infection. Vaccine 31:2215–2221. 10.1016/j.vaccine.2013.03.007 [DOI] [PubMed] [Google Scholar]
- 9.Cheng A, Fung CP, Liu CC, Lin YT, Tsai HY, Chang SC, Chou AH, Chang JY, Jiang RH, Hsieh YC, Su IJ, Chong PC, Hsieh SM. 2013. A Phase I, randomized, open-label study to evaluate the safety and immunogenicity of an enterovirus 71 vaccine. Vaccine 31:2471–2476. 10.1016/j.vaccine.2013.03.015 [DOI] [PubMed] [Google Scholar]
- 10.Hu YM, Wang X, Wang JZ, Wang L, Zhang YJ, Chang L, Liang ZL, Xia JL, Dai QG, Hu YL, Mao QY, Zhu FC, Song YF, Gao F, Chen JT. 2013. Immunogenicity, safety, and lot consistency of a novel inactivated enterovirus 71 vaccine in Chinese children aged 6 to 59 months. Clin. Vaccine Immunol. 20:1805–1811. 10.1128/CVI.00491-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li YP, Liang ZL, Xia JL, Wu JY, Wang L, Song LF, Mao QY, Wen SQ, Huang RG, Hu YS, Yao X, Miao X, Wu X, Li RC, Wang JZ, Yin WD. 2014. Immunogenicity, safety, and immune persistence of a novel inactivated human enterovirus 71 vaccine: a phase II, randomized, double-blind, placebo-controlled trial. J. Infect. Dis. 209:46–55. 10.1093/infdis/jit429 [DOI] [PubMed] [Google Scholar]
- 12.Liu CC, Lian WC, Butler M, Wu SC. 2007. High immunogenic enterovirus 71 strain and its production using serum-free microcarrier Vero cell culture. Vaccine 25:19–24. 10.1016/j.vaccine.2006.06.083 [DOI] [PubMed] [Google Scholar]
- 13.Liu L, Zhang Y, Wang J, Zhao H, Jiang L, Che Y, Shi H, Li R, Mo Z, Huang T, Liang Z, Mao Q, Wang L, Dong C, Liao Y, Guo L, Yang E, Pu J, Yue L, Zhou Z, Li Q. 2013. Study of the integrated immune response induced by an inactivated EV71 vaccine. PLoS One 8:e54451. 10.1371/journal.pone.0054451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mao Q, Cheng T, Zhu F, Li J, Wang Y, Li Y, Gao F, Yang L, Yao X, Shao J, Xia N, Liang Z, Wang J. 2013. The cross-neutralizing activity of enterovirus 71 subgenotype c4 vaccines in healthy Chinese infants and children. PLoS One 8:e79599. 10.1371/journal.pone.0079599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng FY, Li JX, Li XL, Chu K, Zhang YT, Ji H, Li L, Liang ZL, Zhu FC. 2012. Tolerability and immunogenicity of an inactivated enterovirus 71 vaccine in Chinese healthy adults and children: an open label, phase 1 clinical trial. Hum. Vaccin. Immunother. 8:668–674. 10.4161/hv.19521 [DOI] [PubMed] [Google Scholar]
- 16.Ong KC, Devi S, Cardosa MJ, Wong KT. 2010. Formaldehyde-inactivated whole-virus vaccine protects a murine model of enterovirus 71 encephalomyelitis against disease. J. Virol. 84:661–665. 10.1128/JVI.00999-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu SC, Liu CC, Lian WC. 2004. Optimization of microcarrier cell culture process for the inactivated enterovirus type 71 vaccine development. Vaccine 22:3858–3864. 10.1016/j.vaccine.2004.05.037 [DOI] [PubMed] [Google Scholar]
- 18.Zhu FC, Wang JZ, Li XL, Liang ZL, Ge HM, Meng FY, Mao QY, Zhang YT, Zhang ZY, Ji H, Gao F, Guo HJ, Zhu QY, Chu K, Wu X, Li JX, Chen QH, Chen XQ, Zhang WW, Hu YM, Li L, Li FX, Yao X, Liu P, Wang H, Shen XL. 2012. Reactogenicity and immunogenicity of an enterovirus 71 vaccine in Chinese healthy children and infants. Pediatr. Infect. Dis. J. 31:1158–1165. 10.1097/INF.0b013e31826eba74 [DOI] [PubMed] [Google Scholar]
- 19.Zhu F-C, Liang Z-L, Li X-L, Ge H-M, Meng F-Y, Mao Q-Y, Zhang Y-T, Hu Y-M, Zhang Z-Y, Li J-X, Gao F, Chen Q-H, Zhu Q-Y, Chu K, Wu X, Yao X, Guo H-J, Chen X-Q, Liu P, Dong Y-Y, Li F-X, Shen X-L, Wang J-Z. 2013. Immunogenicity and safety of an enterovirus 71 vaccine in healthy Chinese children and infants: a randomised, double-blind, placebo-controlled phase 2 clinical trial. Lancet 381:1037–1045. 10.1016/S0140-6736(12)61764-4 [DOI] [PubMed] [Google Scholar]
- 20.Zhu F-C, Meng F-Y, Li J-X, Li X-L, Mao Q-Y, Tao H, Zhang Y-T, Yao X, Chu K, Chen Q-H, Hu Y-M, Wu X, Liu P, Zhu L-Y, Gao F, Jin H, Chen Y-J, Dong Y-Y, Liang Y-C, Shi N-M, Ge H-M, Liu L, Chen S-G, Ai X, Zhang Z-Y, Ji Y-G, Luo F-J, Chen X-Q, Zhang Y, Zhu L-W, Liang Z-L, Shen X-L. 2013. Efficacy, safety, and immunology of an inactivated alum-adjuvant enterovirus 71 vaccine in children in China: a multicentre, randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 381:2024–2032. 10.1016/S0140-6736(13)61049-1 [DOI] [PubMed] [Google Scholar]
- 21.Zhu F, Xu W, Xia J, Liang Z, Liu Y, Zhang X, Tan X, Wang L, Mao Q, Wu J, Hu Y, Ji T, Song L, Liang Q, Zhang B, Gao Q, Li J, Wang S, Hu Y, Gu S, Zhang J, Yao G, Gu J, Wang X, Zhou Y, Chen C, Zhang M, Cao M, Wang J, Wang H, Wang N. 2014. Efficacy, safety, and immunogenicity of an enterovirus 71 vaccine in China. N. Engl. J. Med. 370:818–828. 10.1056/NEJMoa1304923 [DOI] [PubMed] [Google Scholar]
- 22.Li R, Liu L, Mo Z, Wang X, Xia J, Liang Z, Zhang Y, Li Y, Mao Q, Wang J, Jiang L, Dong C, Che Y, Huang T, Jiang Z, Xie Z, Wang L, Liao Y, Liang Y, Nong Y, Liu J, Zhao H, Na R, Guo L, Pu J, Yang E, Sun L, Cui P, Shi H, Wang J, Li Q. 2014. An inactivated enterovirus 71 vaccine in healthy children. N. Engl. J. Med. 370:829–837. 10.1056/NEJMoa1303224 [DOI] [PubMed] [Google Scholar]
- 23.Arita M, Nagata N, Iwata N, Ami Y, Suzaki Y, Mizuta K, Iwasaki T, Sata T, Wakita T, Shimizu H. 2007. An attenuated strain of enterovirus 71 belonging to genotype a showed a broad spectrum of antigenicity with attenuated neurovirulence in cynomolgus monkeys. J. Virol. 81:9386–9395. 10.1128/JVI.02856-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arita M, Shimizu H, Nagata N, Ami Y, Suzaki Y, Sata T, Iwasaki T, Miyamura T. 2005. Temperature-sensitive mutants of enterovirus 71 show attenuation in cynomolgus monkeys. J. Gen. Virol. 86:1391–1401. 10.1099/vir.0.80784-0 [DOI] [PubMed] [Google Scholar]
- 25.Wu CN, Lin YC, Fann C, Liao NS, Shih SR, Ho MS. 2001. Protection against lethal enterovirus 71 infection in newborn mice by passive immunization with subunit VP1 vaccines and inactivated virus. Vaccine 20:895–904. 10.1016/S0264-410X(01)00385-1 [DOI] [PubMed] [Google Scholar]
- 26.Chung CY, Chen CY, Lin SY, Chung YC, Chiu HY, Chi WK, Lin YL, Chiang BL, Chen WJ, Hu YC. 2010. Enterovirus 71 virus-like particle vaccine: improved production conditions for enhanced yield. Vaccine 28:6951–6957. 10.1016/j.vaccine.2010.08.052 [DOI] [PubMed] [Google Scholar]
- 27.Chung YC, Ho MS, Wu JC, Chen WJ, Huang JH, Chou ST, Hu YC. 2008. Immunization with virus-like particles of enterovirus 71 elicits potent immune responses and protects mice against lethal challenge. Vaccine 26:1855–1862. 10.1016/j.vaccine.2008.01.058 [DOI] [PubMed] [Google Scholar]
- 28.Chung YC, Huang JH, Lai CW, Sheng HC, Shih SR, Ho MS, Hu YC. 2006. Expression, purification and characterization of enterovirus-71 virus-like particles. World J. Gastroenterol. 12:921–927. 10.3748/wjg.v12.i6.921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu YC, Hsu JT, Huang JH, Ho MS, Ho YC. 2003. Formation of enterovirus-like particle aggregates by recombinant baculoviruses co-expressing P1 and 3CD in insect cells. Biotechnol. Lett. 25:919–925. 10.1023/A:1024071514438 [DOI] [PubMed] [Google Scholar]
- 30.Ku Z, Ye X, Huang X, Cai Y, Liu Q, Li Y, Su Z, Huang Z. 2013. Neutralizing antibodies induced by recombinant virus-like particles of enterovirus 71 genotype C4 inhibit infection at pre- and post-attachment steps. PLoS One 8:e57601. 10.1371/journal.pone.0057601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li HY, Han JF, Qin CF, Chen R. 2013. Virus-like particles for enterovirus 71 produced from Saccharomyces cerevisiae potently elicits protective immune responses in mice. Vaccine 31:3281–3287. 10.1016/j.vaccine.2013.05.019 [DOI] [PubMed] [Google Scholar]
- 32.Lin YL, Yu CI, Hu YC, Tsai TJ, Kuo YC, Chi WK, Lin AN, Chiang BL. 2012. Enterovirus type 71 neutralizing antibodies in the serum of macaque monkeys immunized with EV71 virus-like particles. Vaccine 30:1305–1312. 10.1016/j.vaccine.2011.12.081 [DOI] [PubMed] [Google Scholar]
- 33.Liu Q, Yan K, Feng Y, Huang X, Ku Z, Cai Y, Liu F, Shi J, Huang Z. 2012. A virus-like particle vaccine for coxsackievirus A16 potently elicits neutralizing antibodies that protect mice against lethal challenge. Vaccine 30:6642–6648. 10.1016/j.vaccine.2012.08.071 [DOI] [PubMed] [Google Scholar]
- 34.Zhao H, Li HY, Han JF, Deng YQ, Li YX, Zhu SY, He YL, Qin ED, Chen R, Qin CF. 2013. Virus-like particles produced in Saccharomyces cerevisiae elicit protective immunity against coxsackievirus A16 in mice. Appl. Microbiol. Biotechnol. 97:10445–10452. 10.1007/s00253-013-5257-3 [DOI] [PubMed] [Google Scholar]
- 35.Foo DG, Alonso S, Phoon MC, Ramachandran NP, Chow VT, Poh CL. 2007. Identification of neutralizing linear epitopes from the VP1 capsid protein of Enterovirus 71 using synthetic peptides. Virus Res. 125:61–68. 10.1016/j.virusres.2006.12.005 [DOI] [PubMed] [Google Scholar]
- 36.Kirk K, Poh CL, Fecondo J, Pourianfar H, Shaw J, Grollo L. 2012. Cross-reactive neutralizing antibody epitopes against Enterovirus 71 identified by an in silico approach. Vaccine 30:7105–7110. 10.1016/j.vaccine.2012.09.030 [DOI] [PubMed] [Google Scholar]
- 37.Liu CC, Chou AH, Lien SP, Lin HY, Liu SJ, Chang JY, Guo MS, Chow YH, Yang WS, Chang KH, Sia C, Chong P. 2011. Identification and characterization of a cross-neutralization epitope of Enterovirus 71. Vaccine 29:4362–4372. 10.1016/j.vaccine.2011.04.010 [DOI] [PubMed] [Google Scholar]
- 38.Shi J, Huang X, Liu Q, Huang Z. 2013. Identification of conserved neutralizing linear epitopes within the VP1 protein of coxsackievirus A16. Vaccine 31:2130–2136. 10.1016/j.vaccine.2013.02.051 [DOI] [PubMed] [Google Scholar]
- 39.Ye X, Ku Z, Liu Q, Wang X, Shi J, Zhang Y, Kong L, Cong Y, Huang Z. 2014. Chimeric virus-like particle vaccines displaying conserved enterovirus 71 epitopes elicit protective neutralizing antibodies in mice through divergent mechanisms. J. Virol. 88:72–81. 10.1128/JVI.01848-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ren J, Wang X, Hu Z, Gao Q, Sun Y, Li X, Porta C, Walter TS, Gilbert RJ, Zhao Y, Axford D, Williams M, McAuley K, Rowlands DJ, Yin W, Wang J, Stuart DI, Rao Z, Fry EE. 2013. Picornavirus uncoating intermediate captured in atomic detail. Nat. Commun. 4:1929. 10.1038/ncomms2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang X, Peng W, Ren J, Hu Z, Xu J, Lou Z, Li X, Yin W, Shen X, Porta C, Walter TS, Evans G, Axford D, Owen R, Rowlands DJ, Wang J, Stuart DI, Fry EE, Rao Z. 2012. A sensor-adaptor mechanism for enterovirus uncoating from structures of EV71. Nat. Struct. Mol. Biol. 19:424–429. 10.1038/nsmb.2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuck P, Rossmanith P. 2000. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers 54:328–341. [DOI] [PubMed] [Google Scholar]
- 43.Brown PH, Schuck P. 2006. Macromolecular size-and-shape distributions by sedimentation velocity analytical ultracentrifugation. Biophys. J. 90:4651–4661. 10.1529/biophysj.106.081372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, Stagg S, Potter CS, Carragher B. 2005. Automated molecular microscopy: the new Leginon system. J. Struct. Biol. 151:41–60. 10.1016/j.jsb.2005.03.010 [DOI] [PubMed] [Google Scholar]
- 45.Roseman A. 2004. FindEM—a fast, efficient program for automatic selection of particles from electron micrographs. J. Struct. Biol. 145:91–99. 10.1016/j.jsb.2003.11.007 [DOI] [PubMed] [Google Scholar]
- 46.Lander GC, Stagg SM, Voss NR, Cheng A, Fellmann D, Pulokas J, Yoshioka C, Irving C, Mulder A, Lau P-W, Lyumkis D, Potter CS, Carragher B. 2009. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J. Struct. Biol. 166:95–102. 10.1016/j.jsb.2009.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ludtke SJ, Baldwin PR, Chiu W. 1999. EMAN: semiautomated software for high-resolution single-particle reconstructions. J. Struct. Biol. 128:82–97. 10.1006/jsbi.1999.4174 [DOI] [PubMed] [Google Scholar]
- 48.Liang Y, Ke EY, Zhou ZH. 2002. IMIRS: a high-resolution 3D reconstruction package integrated with a relational image database. J. Struct. Biol. 137:292–304. 10.1016/S1047-8477(02)00014-X [DOI] [PubMed] [Google Scholar]
- 49.Tang G, Peng L, Baldwin PR, Mann DS, Jiang W, Rees I, Ludtke SJ. 2007. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 157:38–46. 10.1016/j.jsb.2006.05.009 [DOI] [PubMed] [Google Scholar]
- 50.Unser M, Trus BL, Steven AC. 1987. A new resolution criterion based on spectral signal-to-noise ratios. Ultramicroscopy 23:39–51. 10.1016/0304-3991(87)90225-7 [DOI] [PubMed] [Google Scholar]
- 51.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 52.Plevka P, Perera R, Cardosa J, Kuhn RJ, Rossmann MG. 2012. Crystal structure of human enterovirus 71. Science 336:1274. 10.1126/science.1218713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shingler KL, Yoder JL, Carnegie MS, Ashley RE, Makhov AM, Conway JF, Hafenstein S. 2013. The enterovirus 71 A-particle forms a gateway to allow genome release: a cryoEM study of picornavirus uncoating. PLoS Pathog. 9:e1003240. 10.1371/journal.ppat.1003240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cifuente JO, Lee H, Yoder JD, Shingler KL, Carnegie MS, Yoder JL, Ashley RE, Makhov AM, Conway JF, Hafenstein S. 2013. Structures of the procapsid and mature virion of enterovirus 71 strain 1095. J. Virol. 87:7637–7645. 10.1128/JVI.03519-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gong MQ, Zhou J, Yang CT, Deng Y, Zhao GY, Zhang YH, Wang Y, Zhou YS, Tan WJ, Xu HL. 2012. Insect cell-expressed hemagglutinin with CpG oligodeoxynucleotides plus alum as an adjuvant is a potential pandemic influenza vaccine candidate. Vaccine 30:7498–7505. 10.1016/j.vaccine.2012.10.054 [DOI] [PubMed] [Google Scholar]
- 56.Bubeck D, Filman DJ, Cheng N, Steven AC, Hogle JM, Belnap DM. 2005. The structure of the poliovirus 135S cell entry intermediate at 10-angstrom resolution reveals the location of an externalized polypeptide that binds to membranes. J. Virol. 79:7745–7755. 10.1128/JVI.79.12.7745-7755.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]