ABSTRACT
The three subsets of virions that comprise the Brome mosaic virus (BMV) were previously thought to be indistinguishable. This work tested the hypothesis that distinct capsid-RNA interactions in the BMV virions allow different rates of viral RNA release. Several results support distinct interactions between the capsid and the BMV genomic RNAs. First, the deletion of the first eight residues of the BMV coat protein (CP) resulted in the RNA1-containing particles having altered morphologies, while those containing RNA2 were unaffected. Second, subsets of the BMV particles separated by density gradients into a pool enriched for RNA1 (B1) and for RNA2 and RNA3/4 (B2.3/4) were found to have different physiochemical properties. Compared to the B2.3/4 particles, the B1 particles were more sensitive to protease digestion and had greater resistivity to nanoindentation by atomic force microscopy and increased susceptibility to nuclease digestion. Mapping studies showed that portions of the arginine-rich N-terminal tail of the CP could interact with RNA1. Mutational analysis in the putative RNA1-contacting residues severely reduced encapsidation of BMV RNA1 without affecting the encapsidation of RNA2. Finally, during infection of plants, the more easily released RNA1 accumulated to higher levels early in the infection.
IMPORTANCE Viruses with genomes packaged in distinct virions could theoretically release the genomes at different times to regulate the timing of gene expression. Using an RNA virus composed of three particles, we demonstrated that the RNA in one of the virions is released more easily than the other two in vitro. The differential RNA release is due to distinct interactions between the viral capsid protein and the RNAs. The ease of RNA release is also correlated with the more rapid accumulation of that RNA in infected plants. Our study identified a novel role for capsid-RNA interactions in the regulation of a viral infection.
INTRODUCTION
Viruses use multiple strategies to maximize their fitness (1, 2). For viruses with genomes consisting of multiple RNA segments, their genomes can reassort with each infection to increase the ability to gain new combinations of traits (3, 4). A multipartite genome can also allow differential regulation for the expression of each segment of the genome. This could impact the timing of viral gene expression and limit exposure of the viral genome to attacks by host defenses (5, 6).
Brome mosaic virus (BMV) is an excellent model system for a virus with a multipartite genome. BMV has three positive-stranded genomic RNAs (RNA1, RNA2, and RNA3) and a subgenomic RNA (RNA4) (7, 8). The genomic RNAs are required for successful infection and are encapsidated into three separate virions. Subgenomic RNA4 is coencapsidated with RNA3. All three subsets of BMV virions contain 180 copies of the BMV coat protein (CP) and are indistinguishable in electron micrographs. Indeed, a mixture of three virions is used to grow crystals for structural studies (9, 10).
Contrary to the idea that subpopulations of BMV particles are structurally indistinguishable, several mutations in the BMV CP affected the specific encapsidation of one or more of the BMV RNAs, especially in the coencapsidation of RNA3 and RNA4 (reference 11 and references therein). In this work, we present evidence that the BMV virions have distinct capsid-RNA contacts that affect the stability of the virions, viral RNA release, and BMV infection.
MATERIALS AND METHODS
Construction of mutant BMV.
Mutant BMVs were made using an Agrobacterium-mediated gene delivery system to express the three BMV genomic RNAs (12). BMV RNA3 expressing the mutant CPs were made by PCR-mediated site-directed mutagenesis with appropriate primers, which will be available upon request. The sequence for the wild-type (WT) BMV CP has the GenBank accession number J02042.1. The virion containing the Δ8 had the normal initiation codon of the BMV CP changed from ATG deleted to allow translation initiation to start with the ATG at codon 9. The 1415AQ, 1920AA, and 1920KK mutants have pairs of codons changed to GCTCAA, GCTGCA, and AAGAAA, respectively. The sequences of the cDNA for BMV RNA3 were confirmed to have no other mutations. N. benthamiana plants used to produce virions were grown in a growth chamber kept at a constant 25°C, 70 to 75% humidity, and a 16/8 h light-dark cycle.
Virion purification.
The standard purification of one population of BMV virions used the method of Bujarski (13). To separate the particles containing RNA1 (B1) from those containing RNA2 and the coencapsidated RNA3 and RNA3 (B2.3/4), the total virions were separated in 45% cesium chloride (wt/vol) and centrifuged for 20 h at 50,000 rpm using a Beckman TLA110 rotor. Each band was collected separately and reconstituted in a 45% cesium chloride column for a second round of centrifugation for 20 to 24 h at 50,000 rpm. The top and bottom bands were collected and dialyzed with three changes of virus buffer (50 mM sodium acetate [NaOAc], 10 mM MgCl2 [pH 5.2]). The concentration of the virions was determined by spectrophotometric analysis and by comparing the Coomassie blue staining with known quantities of bovine serum albumin. The virions were stored at −80°C in aliquots.
BMV virions containing only RNA3 and RNA4 were produced using Agrobacterium engineered to express the two BMV replication proteins along with the corresponding RNA3 (11, 12). Due to the lack of BMV RNA1 and RNA2, only virions containing RNA3 and RNA4 can be produced in Nicotiana benthamiana (11).
Transmission electron microscopy.
BMV virions (50 μg/ml) were added to glow-discharged CF-400-Cu grids (Electron Microscopy Sciences). Where applicable, protease K was added to the virions for the prescribed amount of time and then adsorbed to the grid for 4 min. Residual solution was removed by blotting the edge of the grid with filter paper. The grids were washed by touching them to a drop of sample buffer and blotting to filter paper and then were immediately stained with 1% uranyl acetate for 60 s. Virions treated with RNase A were diluted in a Tris-HCl EDTA (pH 7.6) buffer. The samples were then diluted in virus buffer to the appropriate concentration for electron microscopy and prepared for staining by uranyl acetate as described above. The images were acquired with a JEM 1010 transmission electron microscope and a 4K-by-4K charge-coupled device (CCD) camera (TEM; JEOL, Inc.) at a ×60,000 magnification. Virion measurements used the Gatan imaging software.
Nanoindentation of BMV virions.
Imaging and mechanical indentation measurements were recorded in liquid and at room temperature using a Cypher atomic force microscope (Asylum Research). BMV virions were deposited on a freshly cleaved highly ordered pyrolytic graphite (HOPG) disk, mounted on a peizo scanner, and incubated for 10 min before analysis. A droplet holder was used to maintain a stable buffer environment.
Silicon cantilevers (BioLeverMini, Olympus) with a tip radius of ∼9 nm and spring constant of ∼0.1 N/m were used. The cantilever spring constant was calibrated using the thermal-oscillation method (14). Atomic force microscopy images were obtained in alternative contact mode using point amplitudes as low as possible (<10 mV) to minimize the tip-virus interaction. Samples were first imaged at low resolution (500 nm by 500 nm) to identify single virions. Higher-resolution topographic images (80 nm by 80 nm) were then recorded, followed by force mapping in contact mode. Force maps consisted of arrays of 6-by-6 force-distance curves on 80-nm by 80-nm areas, using a maximum applied force of 700 pN and a loading rate of 1 μm/s. After force mapping, a topographic image was collected to check the integrity of the virion. Data from virions that had suffered notable morphological damage during force mapping were rejected.
Instrumental response was estimated from force curves collected at the same position, the middle of the same virion, until a large deviation of the spring constant was observed, which was attributed to irreversible damage to the virus shell. The variance in the elastic constants characterizes the accuracy of measurements in the absence of heterogeneous broadening. Images and force maps were analyzed using Igor Pro 6.12 software (Wavemetrics).
For small indentations, compression of the virus by the tip can be represented as two springs connected in series representing by the virus and the cantilever (15). The elastic constant (Kv) of a virus is related to that of the cantilever as follows: Kv = (Kc · Keff)/(Kc − Keff), where Keff is the effective spring constant obtained by a least-square fit of the linear region in the force-distance curve, and Kc is the cantilever spring constant.
RNA extraction and analysis.
Virion-encapsidated RNAs were dissolved in an equal volume of 2× lysis buffer (200 mM Tris, 2 mM EDTA, 2% SDS [pH 8.5]) and extracted with a 1:1 mixture of phenol-chloroform. The buffer was adjusted to 0.3 M ammonium acetate, and RNA was precipitated with an equal volume of isopropanol. Total viral RNAs were extracted from 0.1 g of wheat leaves using TRIzol (11). The lysate was extracted with a 50% volume of chloroform solution followed by precipitation of the RNAs with isopropanol. All RNA concentrations were determined by spectrometry followed by analysis of RNA by agarose gel electrophoresis and staining with ethidium bromide.
Northern blotting was performed with glyoxal-treated RNA separated in a 1.2% agarose–BPTE gel (16). An in vitro-transcribed riboprobe specific to the 3′ untranslated region (UTR) of the positive-strand BMV RNAs was used as described by Hema et al. (16). Bands in the blots were quantified using ImageQuant software. Reverse transcription and quantitative PCR (RT-qPCR) to quantify minus-strand BMV RNA1 and RNA2 used 500 ng total RNA to synthesize the cDNA by strand-specific primers that recognized either nucleotides (nt) 100 to 120 on RNA1 or nt 371 to 392 on RNA2 as described by Subba-Reddy et al. (17). The cDNA was detected using pairs of primers that recognized nt 100 to 232 of RNA1 and nt 371 to 499 of RNA2.
Differential scanning fluorimetry.
Differential scanning fluorimetry (DSF) was performed in a 96-well plate in a real-time PCR machine (Stratagene Inc.). Each reaction of 25 μl contained 4 μg of purified virion, phosphate buffer adjusted to the appropriate pH, and SYPRO orange at a 2.5× final concentration from stock provided by the manufacturer (Molecular Probes). The temperature ramp was set at a rate of 1.0°C/min from 25 to 95°C. The fluorescence intensity was measured with an excitation wavelength of 470 nm and an emission wavelength of 550 nm (Ex470/Em550) and plotted as the first derivative of fluorescence versus temperature. The temperature with the fastest change in fluorescence intensity was interpreted as the apparent denaturation temperature (TMapp).
BMV partial proteolysis and MALDI-TOF detection.
Reaction mixtures containing 100 mM ammonium bicarbonate (pH 8.0), 50 μg virion, and 4 mM bradykinin fragment (Sigma) were preincubated for 5 min. Trypsin digestion (1 μg; Promega) was stopped at the indicated times by the addition of 1 μl of 10% trifluoroacetic acid. Peptide quantification was done by summation of the peptide ion intensities after normalization to the signal from the bradykinin fragment. Peptides were resolved by a Bruker Autoflex III matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometer in reflectron mode.
Reversible cross-linking peptide fingerprinting analysis (RCAP).
BMV virions were either treated or mock treated with formaldehyde and analyzed using the RCAP assay as previously described (16, 18). Mass spectrometric analysis used a Bruker Autoflex III mass spectrometer (Agilent Technologies) in positive ion mode. Assigned peaks all corresponded to within 0.5 Da of the theoretical masses of peptides from the BMV coat protein.
The database search used the program Mascot (Matrix Science), with the search directed against the BMV protein sequences from the NCBI. Molecular modeling used the crystal structure of the BMV coat protein (Protein Data Bank code 1JS9; retrieved using VIPERdb) and the Chimera program (19).
BMV capsid accumulation.
Independent samples were obtained from wheat seedlings grown in 4-in. pots. Multiple seedlings of the same treatment were harvested, cut into small pieces, and mixed thoroughly. Samples (150 mg) were homogenized using pestles that fit in 1.5-ml microcentrifuge tubes. The lysates were clarified by centrifugation at 10,000 × g for 10 min. Clarified lysate was separated by gel electrophoresis and subjected to Western blot analyses.
RESULTS
A deletion in the BMV CP can result in multiple virion morphologies.
The BMV CP contains an N-terminal tail that is enriched for positively charged residues (Fig. 1A). Several substitutions and insertions within this sequence resulted in virions that altered ratios of encapsidated BMV RNA3 and RNA4, although the mutations did not obviously affect virion morphologies (11). Rao and Grantham had previously deleted the first 7 residues of BMV and found that the resulting virus caused a hypersensitivity response in some plant hosts (20). Given that innate immune responses can act on nonself nucleic acids, these results suggest that the N-terminally truncated BMV CP results in defective RNA encapsidation. Therefore, we constructed and analyzed BMV virions named Δ8 that had a deletion of the first eight residues of the CP and initiate translation at the methionine codon at position 9.
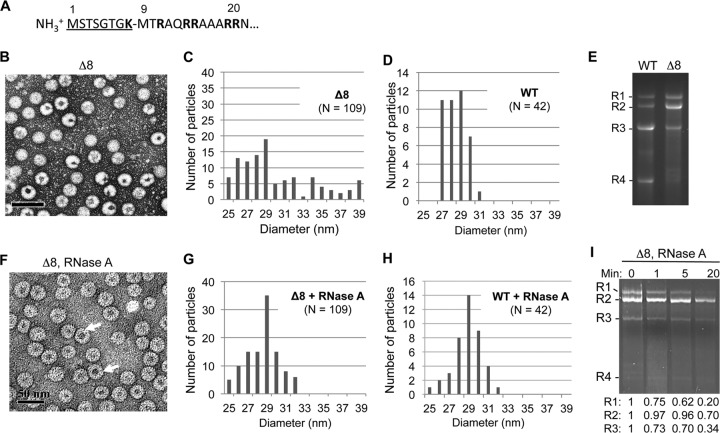
FIG 1.
Deletion of the N-terminal 8 residues of the BMV CP results in the formation of morphologically heterogeneous virions. (A) Sequence of the arginine-rich N-terminal tail of the BMV CP. The residues deleted in mutant Δ8 are underlined. (B) Δ8 virions stained with uranyl acetate exhibit a heterogeneous mixture of virions. (C) Diameters of Δ8 particles visualized by TEM. The virions diameters were measured from micrographs such as those in panel B using the Gatan digital micrograph program. (D) Diameters of WT BMV. (E) RNAs encapsidated by the WT or Δ8 virions. The RNAs were extracted from the virions with the use of phenol-chloroform and were separated on a 1.2% agarose gel in BPTE buffer. The RNAs were denatured by treatment with glyoxal. (F) Δ8 virions stained with uranyl acetate after RNase A treatment. Arrows indicate the locations of virions with larger inner cavities. (G) Diameters of Δ8 particles after treatment with RNase A. (H) Diameter of the WT BMV virions after treatment with RNase A. (I) Abundances of genomic RNAs encapsidated by Δ8 particles after RNase A treatment. The RNAs were extracted as described for panel E and quantified using programs in ImageQuant (Bio-Rad).
Unexpectedly, the Δ8 virions were a heterogeneous population (Fig. 1B). Some of the particles appeared to be wild type (WT), while others had altered staining with uranyl acetate within the viral particles (named lobed particles) (Fig. 1B). Random sampling of the Δ8 virions from multiple micrographs revealed that 29% ± 5% of the particles were lobed. The lobed particles also have more heterogeneous diameters than the WT BMV particle, which is 28 nm (Fig. 1C and D). Examination of the RNAs encapsidated by Δ8 revealed significant differences from those encapsidated by the WT BMV (Fig. 1E). BMV RNA2 was approximately half of the total RNA, while RNA1, RNA3, and RNA4 were underrepresented compared to the RNAs in WT BMV. These observations suggest that the deletion of the first eight residues of the BMV capsid negatively affected the formation of the virion containing BMV RNA1 (named B1) and RNA3/4 (B3/4) but had less of a negative impact on RNA2 encapsidation (B2).
Some of the Δ8 particles have diameters larger than 28 nm and also having altered staining with uranyl acetate (Fig. 1C); we wondered whether Δ8 virions have exposed RNAs in these virions. To test this, we treated the Δ8 virions with RNase A and imaged the resulting particles by negative-stained TEM. RNase A treatment changed the appearance of the particles so that several now had large hollow centers (Fig. 1F, arrows). The particles were also more homogeneous in diameter, with a mean diameter of ∼28 nm, suggesting that the removal of exposed RNA by RNase A resulted in rearrangements of the virus-like particles back to the diameter expected of WT BMV virions (Fig. 1G). Treatment of the WT BMV virions with RNase A resulted in a small proportion of virions of lower diameter, but the majority of the particles had diameters of 28 nm (Fig. 1H). Analysis of the RNAs extracted from RNase A-treated particles by gel electrophoresis revealed that RNA1 was preferentially degraded, while RNA2 was more resistant to digestion (Fig. 1I). RNA3 was degraded at an intermediate rate between that of RNA1 and RNA2. These results suggests that the lack of the first 8 residues of the CP left the RNA in the B1 virions more exposed, while the RNAs in the B2 particles were less affected.
The results with Δ8 led us to hypothesize that the three BMV virions have distinct capsid-RNA interactions that affect their physiochemical properties. We sought to test this hypothesis and to determine whether these properties affect BMV infection.
Protease digestion of subsets of BMV virions revealed different virion morphologies.
To demonstrate that the differences in capsid-RNA interactions also occur with wild-type BMV particles, we separated the subsets of the three WT BMV particles. The typical purification protocol results in one band in a cesium chloride density gradient that contains all three BMV virion types. However, we have observed that different virion preparations could form two bands with altered CsCl densities. Therefore, we manipulated the density of the cesium chloride gradients and the centrifugal force to consistently separate the virion preparations into two distinct bands, as described in Materials and Methods. RNAs extracted from the recovered virions revealed that the upper band (of lower density) was enriched for a mixture of the B2 and B3/4 virus particles (named B2.3/4), while the lower band (of higher density) was enriched for the B1 particles (Fig. 2A).
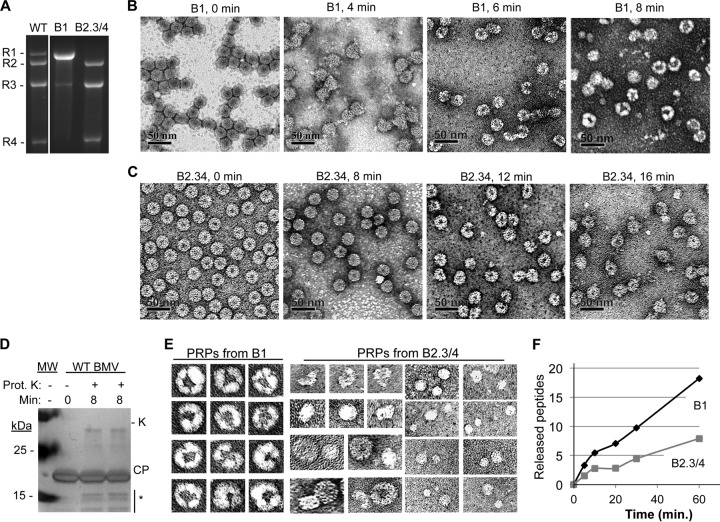
FIG 2.
Separation of BMV virions into subsets and partial proteolysis of the virions. (A) RNAs encapsidated by the B1 and the B2.3/4 virions. WT denotes a sample from a purified preparation that contains all three types of BMV virions. (B) Electron micrographs of B1 virions treated with protease K over an 8-min time course. The virions were stained with uranyl acetate. (C) Electron micrographs of B2.3/4 virions treated with protease K over a 16-min time course. (D) The state of the BMV CP after an 8-min treatment with protease K. The gel image was from an SDS-PAGE stained with Coomassie blue. The proteolyzed CPs are identified with an asterisk. Protease K added to the reaction mixture is labeled “K.” (E) Galleries of the four PRPs isolated from the B1 and the B2.3/4 virions. All of the images were adjusted to the same scale. (E) Rate of proteolysis of peptides from the B1 and B2.3/4 virions. Quantities of all of the peptides were determined relative to the amount of protease-resistant bradykinin fragment added to the virions prior to protease treatment. More than 90% of the peptides mapped to the N-terminal 46 residues of the BMV CP. All virions were digested overnight to ensure complete peptide fragmentation (data not shown).
The N-terminal tail of the bromovirus CP is susceptible to protease digestion, likely due to its lack of stable secondary structure and its movement in and out of the central cavity of the capsid (21; P. Ni, unpublished data). Given that the RNAs from the Δ8 virions are more susceptible to RNase digestion, it is likely that the capsid subunits may be less tightly associated with the encapsidated RNAs. To examine this directly, partial protease digestion of B1 and B2.3/4 particles was used to probe the capsid. Protease K digestion of the purified B1 over an 8-min time course resulted in particles that acquired the lobed structures previously observed with Δ8 (compare Fig. 1B with Fig. 2B). SDS-PAGE of the BMV CP treated with the same conditions revealed that less than 20% of the CP migrated to the position expected of the cleaved product and that the RNA was not fragmented (21) (Fig. 2C and 1B). Interestingly, B2.3/4 virions digested for the same amount of time did not result in the lobed structures of the B1 virions (2C). In addition, small rounded structures of ca. 8 to 20 nm were observed (Fig. 2C). A compilation of the protease-released particles (PRPs) from the B1 and the B2.3/4 particles is in Fig. 2E. These results suggest that the B1 particles are more sensitive to protease digestion than B2.3/4 and that disruption of the capsid shell results in the lobed particles.
To confirm the differential rates of proteolysis of the B1 versus B2.3/4 virions as well as to map the cleavage sites within the CP, we used MALDI-TOF to monitor the peptides released by proteolysis. A spiked bradykinin peptide was used as an internal quantification control. More peptides were released from the CP of the B1 virions than from the B2.3/4 virions. Furthermore, the majority of the peptides were from the N-terminal tail of the CP (Fig. 2). These results demonstrate that the capsid of the B1 virions is more accessible to proteolysis than that of the B2.3/4 virions.
B1 and B2.3/4 particles have distinct physiochemical properties.
We sought to obtain independent evidence that the B1 virions are more labile than the B2.3/4 virions. Populations of the virions were examined using differential scanning fluorimetry (DSF), which measures the thermodenaturation profiles of a protein-containing complex. Protein denaturation is measured by the binding (and induced fluorescence) of the SYPRO orange dye upon binding to hydrophobic regions of the protein exposed during denaturation. The temperature for the maximum change in fluorescence is designated the TMapp (22). In buffers with pH of 6.4 or below, the TMapp of the B1 virions was indistinguishable from that of the B2.3/4 virions. However, at pH 6.6 or above, the TMapp values for the B1 virions were 1 to 2.5°C lower than those for the B2.3/4 virions (Fig. 3A). This change occurred at a slightly higher pH than the expected midpoint of the pH-induced transition, pH 6.4 (23, 24), perhaps indicating that the B1 virions are affected in the rearrangements of the hydrophobic surfaces during or immediately after the transition to a swollen state.
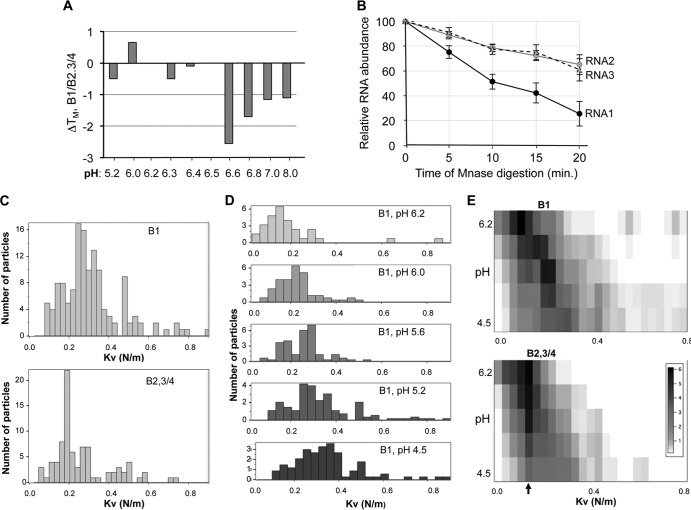
FIG 3.
B1 and B2.3/4 virions have distinct physiochemical properties. (A) Differential scanning fluorimetric analysis of the B1 and B2.3/4 virions. The difference in the TMapp of the B1 and the B2.3/4 virions is plotted. The virions were examined in phosphate buffer adjusted to the pH indicated on the horizontal axis. Each bar represents the mean of three independent measurements, and all standard errors were less than 5% of the mean. (B) The RNA in the B1 virion is more sensitive to micrococcal nuclease digestion than the RNAs in B2.3/4 virions. Virions (5 μg) at pH 7.0 were treated with 1 unit of micrococcal nuclease, and digestion was stopped by addition of 0.5 M EGTA at the indicated times. RNAs were extracted with TRIzol, precipitated, and subjected to electrophoresis in 1% agarose–TBE gels stained with ethidium bromide. The intact BMV RNAs were quantified with a Bio-Rad ImageQuant analyzer. The results are from four independent data sets. (C to E) Analysis of the B1 and B2.3/4 virions by nanoindentation using an atomic force microscope. (C) Histogram of elastic constants of B1 and B2.3/4 virions at pH 4.5. B1 virions exhibit a significantly broader distribution shifted toward stiffer elastic responses. (D) The effect of pH increase on B1 elasticity. The distribution shifts toward softer elastic responses. Note that the number of B1 virions resistant to nanoindentation decreased with increasing pH. (E) Comparison between the joint histograms of pH and elastic constants for B1 and B 2.3/4. For B2.3/4, the peak probability density at a K of 0.18 N/m is insensitive to pH changes, while for B1, it shifts gradually from a K of 0.38 N/m to 0.15 N/m.
A change in the swelling transition of the B1 virion should make the encapsidated RNA more susceptible to RNase digestion. To test this, we treated the B1 and the B2.3/4 virions at pH 7 with micrococcal nuclease over a time course. The full-length RNAs were then extracted and quantified after agarose gel electrophoresis. The BMV RNA1 was digested more rapidly from the B1 virions than RNA2 or RNA3 (Fig. 3E). These results suggest that RNA1 was more exposed than the other RNAs after the virions underwent a swelling transition. In the context of BMV infection, this could impact the accessibility of the different BMV RNAs for gene expression.
If certain RNAs expand more readily, the corresponding virions should exhibit higher mean elastic constants (i.e., be stiffer). We used nanoindentation by atomic force microscopy to measure the global elastic response of the virions to directional mechanical pressure (25). The B1 virions were indeed found to be stiffer than the B2.3/4 virions at pH 5.5, when the virions are in a contracted state (Fig. 3C).
With increasing pH, the elastic constant distributions for both the B1 and B2.3/4 virions shifted toward lower elasticity (Fig. 3C). This is to be expected from a swelling transition (24, 26). However, three observations are important to note: (i) the amount of B1 virions sufficiently stable to survive indentation after increasing the pH to 6.2 decreased 3-fold relative to B2 virions, (ii) B1 virions have significantly lower stability than B2.3/4 virions only at pH 6.4 or above when the virions are in a swollen state (Fig. 3D and E), and (iii) B2.3/4 virions exhibit a prominent, narrow elastic constant peak that is less sensitive to pH changes than that of B1 virions (Fig. 3E). The results from the nanoindentation experiments indicate that the stiffer B1 virions may become more easily disassembled than the B2.3/4 virions after they undergo the pH-dependent swelling transition. In other words, we found that B2.3/4 virions are soft and resilient, while B1 virions are stiff and brittle. Altogether, the results from DSF, RNase digestion, and nanoindentation support the thesis that the B1 virions exist in a different conformation than the B2.3/4 virions, especially after the pH-dependent transition.
The BMV virions have distinct CP-RNA interactions.
The difference in the physiochemical properties of the B1 and B2.3/4 virions could be affected by differences in capsid-RNA interaction. To map the site of RNA contact sites within the virions, we used the RCAP protocol, which combines reversible cross-linking, trypsin digestion, affinity RNA purification, and peptide fingerprinting (18). Peptides from the B1 capsid that coprecipitated with RNA1 were primarily from the N-terminal tail of the BMV CP and from the globular domain (Fig. 4A) (8). For the B2.3/4 sample, several peptides from the N-terminal tail of the CP differed from those from B1; there were fewer peptides from the globular domain of the CP and more from the C-terminal region of the BMV CP (residues 166 to 189) (Fig. 4A). The RNA contacts within the internal surfaces of the B1 and B2.3/4 are shown in Fig. 4B.
FIG 4.
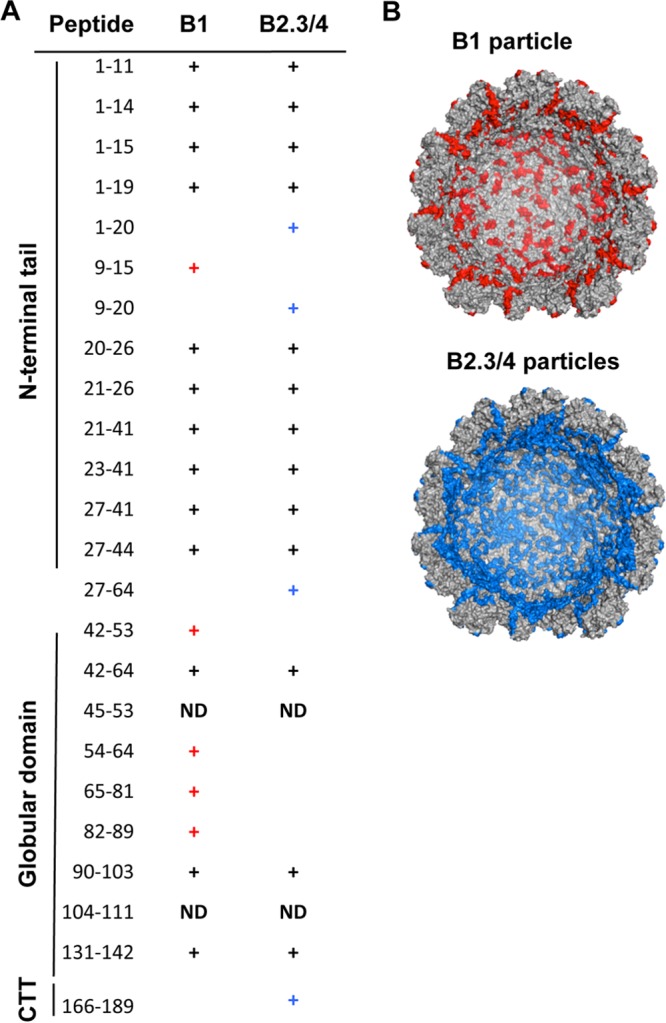
Distinct capsid-RNA contacts in the BMV virions. (A) Peptides from the BMV capsid that interact with RNA1 or RNA2 and RNA3/4. Mapping was performed with the RCAP assay. Peaks assigned to peptides within the BMV coat protein were based on theoretical tryptic digests that match calculated masses within 0.5 Da. (B) Molecular models of the peptides within the inner surface of the B1 and the B2.3/4 virions found to contact RNA. The structure was from PDB accession number 1JS9 and was retrieved as a half-capsid from VIPERdb (36). Peptides either common between the B1 and B2.3/4 particles or not found in the RCAP assay are gray, and unique peptides found to contact RNA1 or RNA 2.3/4 are red and blue, respectively.
The N-terminal tail of the BMV CP is required for specific encapsidation of RNA1.
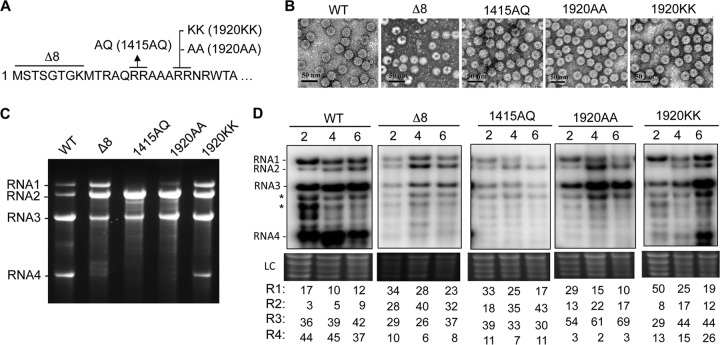
To examine whether the CP contacts affected RNA encapsidation, we made substitutions in the paired arginines at residues 14 and 15 (1415AQ mutant) and also at residues 19 and 20 (1920AA and 1920KK mutants) (Fig. 5A). These arginines contacted RNA1 but not RNA2.3/4. The Δ8 mutant that formed heterogeneous particles (Fig. 1B) was also analyzed. All of the mutant viruses were made by agroinfiltration of N. benthamiana. They produced virions at 10 to 60% of the WT and, except for Δ8, had virion morphologies similar to that of the WT (Fig. 5B). Interestingly, both the 1415AQ and the 1920AA mutant virions had reduced encapsidation of RNA1 and RNA4 (Fig. 5C). The 1920KK mutant virus encapsidated one RNA4 to every two RNA3 molecules, but it encapsidated RNA1 at levels similar to those of WT BMV (Fig. 5C). Northern blots of the total RNA from plants revealed that the defect in RNA1 encapsidation by the 1415AQ and 1920KK mutants was not due to reduced levels of RNA1 in plants (Fig. 5D). However, RNA4 accumulation was reduced in the Δ8, 1415AQ, and 1920AA mutants. These results suggest that residues in the N-terminal tail of the BMV CP can differentially affect encapsidation of RNA1. There is a second effect of the mutations on subgenomic RNA4 that we do not fully understand. However, it could be due to changes in the coding sequence of the CP affecting subgenomic RNA synthesis (27).
FIG 5.
BMV RNA1 has different encapsidation and replication requirements than other RNAs. (A) Mutations in the N-terminal tail of the BMV CP examined for BMV RNA encapsidation and RNA replication. (B) Transmission electron micrographs of the WT and the four mutant BMV virions. The black scale bar denotes 50 nm. (C) RNAs encapsidated by the WT and mutant BMV virions. All virions were prepared in N. benthamiana. The identities of the RNAs are shown to the left of the agarose gels containing glyoxal-treated RNAs. (D) The effects of the N-terminal tail mutations on BMV RNA accumulation in N. benthamiana plants at 2, 4, and 6 days after agroinfiltration. The RNAs were identified in Northern blots treated with a riboprobe that recognizes the conserved 3′ UTR of the BMV RNAs. Bands identified by the asterisks likely correspond to prematurely terminated RNA2 and/or degradation products (37). The relative amounts of the four full-length BMV RNAs within each sample are quantified below the image of the Northern blots. LC, rRNA used as a loading control.
RNA1 accumulates more rapidly in infected plants than other BMV RNAs.
The more facile interaction between BMV RNA1 and the capsid could allow higher levels of expression from RNA1 early in the infection process. To test this idea, we infected wheat seedlings with either B1 or B2.3/4 virions and detected the subsequently produced BMV RNAs over a time course. We note that the B1 and B2.3/4 virions contain small amounts of the other virions, which should allow the accumulation of all BMV RNAs.
Minus-strand RNA1 and RNA2 were detected by qRT-PCR using primer pairs having identical abilities to amplify transcripts of minus-strand RNA1 and RNA2. Minus-strand RNAs were detected at 16 h postinoculation, but their quantity could be assessed with better confidence at 24 h. As would be expected, plants inoculated with the enriched B1 virions had a higher accumulation of minus-strand RNA1 than minus-strand RNA2 (Fig. 6A). However, wheat inoculated with B2.3/4 that had only trace amounts of RNA1 also accumulated higher levels of minus-strand RNA1 than RNA2 (Fig. 6A). In wheat inoculated with an equal mixture of B1 and B2.3/4 virions, minus-strand RNA1 accumulation was again higher than that of minus-strand RNA2. This result was consistent in four independent samples.
FIG 6.
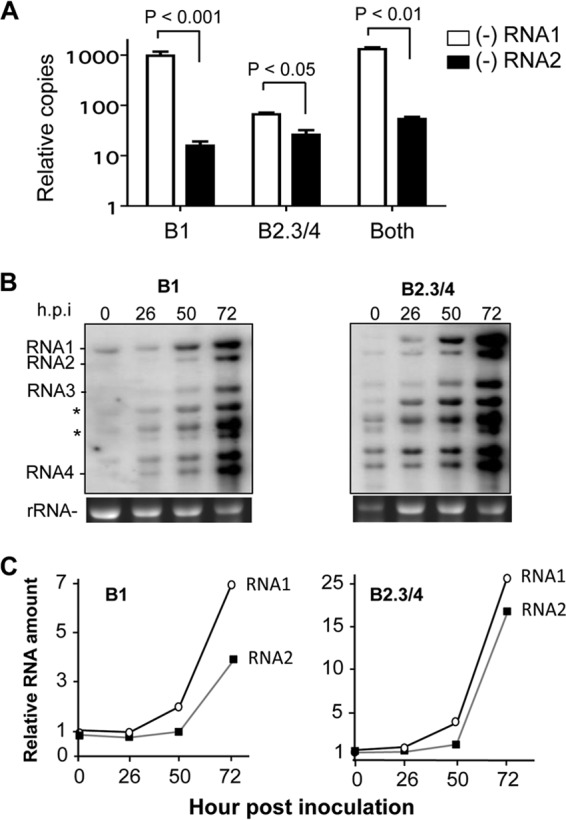
BMV RNA1 accumulated to higher levels than BMV RNA2 early in infection. (A) Minus-strand BMV RNA1 accumulated to higher levels than did RNA2 in wheat seedlings. Wheat seedlings were inoculated with 50 μg/ml of B1 or the B2.3/4 virions or an equal mixture of the two. Total RNAs were extracted with TRIzol reagent and quantified as described in Materials and Methods. Comparable amounts of input were confirmed by photospectrometry and gel electrophoresis. Abundances of the BMV RNAs were quantified using RT-PCR from three independent samples of the total RNA extracted from wheat seedlings inoculated with either B1, B2.3/4, or an equal molar ratio of the two sets of BMV virions. Total RNAs were harvested 24 h postinoculation (hpi). The range for one standard deviation is shown above the bars, as is the P value. The input RNA and performance of the primers against known amounts of BMV DNA were adjusted to allow accurate quantification of the BMV RNAs. (B) Plus-strand RNA1 replicates to a higher abundance than RNA2 after inoculation. The Northern blots were performed with total RNAs extracted from wheat seedlings after the number of hours postinoculation shown above the gel. rRNA served as a loading control in the RNA gel for Northern blot analysis. An image of the ethidium bromide-stained gels used for the Northern blot analysis is shown below the autoradiograms. The full-length BMV RNAs were identified by RNAs extracted from WT BMV virions. The bands highlighted with asterisks likely correspond to prematurely terminated RNA3 (37). (C) Quantification of the accumulation of plus-strand BMV RNA1 and RNA2 over time. The amount of RNA present was normalized to the amount present in the inoculum. The same results were observed in two independent experiments.
Northern blots were performed to detect plus-strand BMV RNAs. Consistent with the analysis of minus-strand BMV RNAs, RNA1 accumulated to higher levels than full-length BMV RNA2 or RNA3 in plants inoculated with either the B1 or the B2.3/4 virions (Fig. 6B). In addition, Northern blots of the plus-strand RNAs revealed an abundance of viral RNAs that are less than full length (Fig. 6B and 8C). Whether these were due to premature termination of RNA synthesis or degradation of the RNAs due to an imbalance of the input virions remains to be determined. However, between 50 and 72 h postinoculation, the abundance of the full-length RNA2 increased relative to that of full-length RNA1 (Fig. 6B and C). Altogether, the results in Fig. 6 suggest that early in the replication process, BMV RNA1 can replicate to higher levels than RNA2.
DISCUSSION
How the capsid releases RNA to initiate the infection process is not well understood for icosahedral viruses. For the tripartite BMV, which encapsidates its genomic RNAs into separate virions, we demonstrate that subsets of virions are physiochemically different. A deletion of the first 8 residues of the CP had different consequences for the encapsidation of BMV RNA1 versus RNA2. Wild-type virions separated into the B1 and the B2.3/4 particles exhibited different susceptibilities to protease digestion and thermodenaturation, responses to nanoindentation, and sensitivities of the encapsidated RNA to cleavage by RNase A. Notably, the differences between the B1 and the B2.3/4 virions are more pronounced after the virions undergo the swelling transition associated with the release of the encapsidated RNA. Mapping of the contacts made between the CP and the encapsidated RNA confirmed that RNA1 contacts the CP differently than RNA2 or RNA3/4. The functional significance of the identified contact sites was confirmed by reduced RNA1 encapsidation. Finally, we note that the more facile release of RNA1 from the B1 virions was correlated with more rapid accumulation of RNA1 in infected plants, even when it was underrepresented in the initial inoculum. These results suggest that different physiochemical properties of the BMV RNAs can impact the expression of the RNAs in a multipartite RNA virus.
An implication of these results is that the encapsidation of the three BMV RNAs may be more selective than previously believed. This idea is consistent with our previous observation that complementary electrostatic interactions between the BMV RNAs and capsids cannot fully account for the selective encapsidation of the BMV RNAs (11, 23). Multiple regions of the BMV CP contact RNA1 in the B1 virion, including arginine residues at positions 14, 15, 19, and 20 in the N-terminal tail (Fig. 5). The interaction between the CP and RNA1 likely accounts for the greater stiffness of the B1 particles as quantified by our nanoindentation experiments and the formation of larger PRP complexes when the capsid is breached by digestion with protease (Fig. 3C).
The physiochemical differences in the three virions could regulate the timing of BMV RNA release and subsequent gene expression during infection. The more facile release of RNA1 could ensure a higher level of expression of the 1a protein, which is responsible for the formation of the viral RNA replication factory. The 1a protein can also recruit the other BMV RNAs and possibly the RNA-dependent RNA polymerase (RdRp) to the replication factory (28). A mechanism to promote preferential expression of the 1a protein and the establishment of the replication factory could be advantageous in shielding BMV RNA replication from the host innate immune responses. This is consistent with the deletion of the first 8 residues of the BMV CP having been shown to cause necrosis, a plant innate immune response, in the alternative host plant Chenopodium quinoa (20). The stability of the virion is thus a potential regulator of the proper timing of gene expression. The proper balance of the BMV proteins and RNA could also ensure the integrity of the replicated RNAs, as evidenced by the increased amounts of truncated RNAs in plants inoculated with the B1 or the B2.3/4 virions (Fig. 6B). In addition, the BMV CP was recently reported to contain posttranslational modifications in a host-specific manner (29). These modifications could influence CP-RNA interaction to affect the stability and release of the BMV RNAs.
Viruses use a number of mechanisms to regulate the timing of gene expression. For example, proteolysis of alphaviruses and picornaviruses is used to regulate the transition from early to late gene expression (30, 31). The CPs of several viruses have been demonstrated to regulate activities of the viral RdRp as well as affect protein production and genome stability (32, 33). Perhaps most analogous to the selective release of RNAs from the BMV virion is the mechanism used by bacteriophage T7, which uses the selective expulsion of its DNA genome to release the portion of the genome that codes for early functions, while genes for the middle and late functions are released from the capsid due to the translocation of the T7 RNA polymerase during transcription (34, 35). It is possible that other viruses with multiple genomes encapsidated within the same virion or in separate virions could allow the selective release of their genomes from the capsids to regulate the kinetics of gene expression.
ACKNOWLEDGMENTS
We thank members of the Kao lab capsid subgroup for helpful discussions through the years and for being willing to do Friday afternoon experiments. We thank Laura Kao for editing the manuscript. We thank John Tragesser and Dan Calvin for the gift of spring wheat seeds.
This work was supported by the National Institutes of Health (1R01AI090280 to C.C.K.) and by the U.S. Department of Energy, Office of Science (DE-SC0010507 to B.D.).
Footnotes
Published ahead of print 26 March 2014
REFERENCES
- 1.Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 76:159–216. 10.1128/MMBR.05023-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gotte M. 2012. The distinct contributions of fitness and genetic barrier to the development of antiviral drug resistance. Curr. Opin. Virol. 2:644–650. 10.1016/j.coviro.2012.08.004 [DOI] [PubMed] [Google Scholar]
- 3.Van Reeth K. 2007. Avian and swine influenza viruses: our current understanding of the zoonotic risk. Vet. Res. 38:243–260. 10.1051/vetres:2006062 [DOI] [PubMed] [Google Scholar]
- 4.Reid AH, Taubenberger JK. 2003. The origin of the 1918 pandemic influenza virus: a continuing enigma. J. Gen. Virol. 84:2285–2292. 10.1099/vir.0.19302-0 [DOI] [PubMed] [Google Scholar]
- 5.Kawai T, Akira S. 2006. Innate immune recognition of viral infection. Nat. Immunol. 7:131–137. 10.1038/ni1303 [DOI] [PubMed] [Google Scholar]
- 6.Li K, Lemon SM. 2013. Innate immune responses in hepatitis C virus infection. Semin. Immunopathol. 35:53–72. 10.1007/s00281-012-0332-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Noueiry AO, Ahlquist P. 2003. Brome mosaic virus RNA replication: revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 41:77–98. 10.1146/annurev.phyto.41.052002.095717 [DOI] [PubMed] [Google Scholar]
- 8.Kao CC, Ni P, Hema M, Huang X, Dragnea B. 2011. The coat protein leads the way: an update on basic and applied studies with the Brome mosaic virus coat protein. Mol. Plant Pathol. 12:403–412. 10.1111/j.1364-3703.2010.00678.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larson SB, Lucas RW, McPherson A. 2005. Crystallographic structure of the T=1 particle of brome mosaic virus. J. Mol. Biol. 346:815–831. 10.1016/j.jmb.2004.12.015 [DOI] [PubMed] [Google Scholar]
- 10.Lucas RW, Larson SB, McPherson A. 2002. The crystallographic structure of brome mosaic virus. J. Mol. Biol. 317:95–108. 10.1006/jmbi.2001.5389 [DOI] [PubMed] [Google Scholar]
- 11.Ni P, Wang Z, Ma X, Das NC, Sokol P, Chiu W, Dragnea B, Hagan M, Kao CC. 2012. An examination of the electrostatic interactions between the N-terminal tail of the Brome mosaic virus coat protein and encapsidated RNAs. J. Mol. Biol. 419:284–300. 10.1016/j.jmb.2012.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gopinath K, Dragnea B, Kao C. 2005. Interaction between Brome mosaic virus proteins and RNAs: effects on RNA replication, protein expression, and RNA stability. J. Virol. 79:14222–14234. 10.1128/JVI.79.22.14222-14234.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bujarski JJ. 1998. Bromovirus isolation and RNA extraction. Methods Mol. Biol. 81:183–188 [DOI] [PubMed] [Google Scholar]
- 14.Proksch R, Schaffer TE, Cleveland JP, Callahan RC, Viani MB. 2004. Finite optical spot size and position corrections in thermal spring constant calibration. Nanotechnology 15:1344–1350. 10.1088/0957-4484/15/9/039 [DOI] [Google Scholar]
- 15.Michel JP, Ivanovska IL, Gibbons MM, Klug WS, Knobler CM, Wuite GJ, Schmidt CF. 2006. Nanoindentation studies of full and empty viral capsids and the effects of capsid protein mutations on elasticity and strength. Proc. Natl. Acad. Sci. U. S. A. 103:6184–6189. 10.1073/pnas.0601744103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hema M, Murali A, Ni P, Vaughan RC, Fujisaki K, Tsvetkova I, Dragnea B, Kao CC. 2010. Effects of amino-acid substitutions in the Brome mosaic virus capsid protein on RNA encapsidation. Mol. Plant Microbe Interact. 23:1433–1447. 10.1094/MPMI-05-10-0118 [DOI] [PubMed] [Google Scholar]
- 17.Subba-Reddy CV, Tragesser B, Xu Z, Stein B, Ranjith-Kumar CT, Kao CC. 2012. RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase in human cells reveals requirements for de novo initiation and protein-protein interaction. J. Virol. 86:4317–4327. 10.1128/JVI.00069-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaughan R, Running WE, Qi R, Kao C. 2012. Mapping protein-RNA interactions. Virus Adaptation Treat. 4:29–41. 10.2147/VAAT.S31299 [DOI] [Google Scholar]
- 19.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. 2004. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25:1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- 20.Rao AL, Grantham GL. 1995. Biological significance of the seven amino-terminal basic residues of brome mosaic virus coat protein. Virology 211:42–52. 10.1006/viro.1995.1377 [DOI] [PubMed] [Google Scholar]
- 21.Speir JA, Bothner B, Qu C, Willits DA, Young MJ, Johnson JE. 2006. Enhanced local symmetry interactions globally stabilize a mutant virus capsid that maintains infectivity and capsid dynamics. J. Virol. 80:3582–3591. 10.1128/JVI.80.7.3582-3591.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Niesen FH, Berglund H, Vedadi M. 2007. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat. Protoc. 2:2212–2221. 10.1038/nprot.2007.321 [DOI] [PubMed] [Google Scholar]
- 23.Running WE, Ni P, Kao CC, Reilly JP. 2012. Chemical reactivity of brome mosaic virus capsid protein. J. Mol. Biol. 423:79–95. 10.1016/j.jmb.2012.06.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bancroft JB, Hills GJ, Markham R. 1967. A study of the self-assembly process in a small spherical virus. Formation of organized structures from protein subunits in vitro. Virology 31:354–379 [DOI] [PubMed] [Google Scholar]
- 25.Roos WH, Gibbons MM, Arkhipov A, Uetrecht C, Watts NR, Wingfield PT, Steven AC, Heck AJ, Schulten K, Klug WS, Wuite GJ. 2010. Squeezing protein shells: how continuum elastic models, molecular dynamics simulations, and experiments coalesce at the nanoscale. Biophys. J. 99:1175–1181. 10.1016/j.bpj.2010.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee KK, Gan L, Tsuruta H, Moyer C, Conway JF, Duda RL, Hendrix RW, Steven AC, Johnson JE. 2008. Virus capsid expansion driven by the capture of mobile surface loops. Structure 16:1491–1502. 10.1016/j.str.2008.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Adkins S, Siegel RW, Sun JH, Kao CC. 1997. Minimal templates directing accurate initiation of subgenomic RNA synthesis in vitro by the brome mosaic virus RNA-dependent RNA polymerase. RNA 3:634–647 [PMC free article] [PubMed] [Google Scholar]
- 28.den Boon JA, Diaz A, Ahlquist P. 2010. Cytoplasmic viral replication complexes. Cell Host Microbe 8:77–85. 10.1016/j.chom.2010.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni P, Vaughan RC, Tragesser B, Hoover H, Kao CC. 2014. The plant host can affect the encapsidation of Brome mosaic virus (BMV) RNA; BMV virions are surprisingly heterogeneous. J. Mol. Biol. 426:1061–1076. 10.1016/j.jmb.2013.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bedard KM, Semler BL. 2004. Regulation of picornavirus gene expression. Microbes Infect. 6:702–713. 10.1016/j.micinf.2004.03.001 [DOI] [PubMed] [Google Scholar]
- 31.Strauss JH, Strauss EG. 1994. The alphaviruses: gene expression, replication, and evolution. Microbiol. Rev. 58:491–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subba-Reddy CV, Yunus MA, Goodfellow IG, Kao CC. 2012. Norovirus RNA synthesis is modulated by an interaction between the viral RNA-dependent RNA polymerase and the major capsid protein, VP1. J. Virol. 86:10138–10149. 10.1128/JVI.01208-12 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Ni P, Cheng Kao C. 2013. Non-encapsidation activities of the capsid proteins of positive-strand RNA viruses. Virology 446:123–132. 10.1016/j.virol.2013.07.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tzlil S, Kindt JT, Gelbart WM, Ben-Shaul A. 2003. Forces and pressures in DNA packaging and release from viral capsids. Biophys. J. 84:1616–1627. 10.1016/S0006-3495(03)74971-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia LR, Molineux IJ. 1995. Rate of translocation of bacteriophage T7 DNA across the membranes of Escherichia coli. J. Bacteriol. 177:4066–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shepherd CM, Borelli IA, Lander G, Natarajan P, Siddavanahalli V, Bajaj C, Johnson JE, Brooks CL, III, Reddy VS. 2006. VIPERdb: a relational database for structural virology. Nucleic Acids Res. 34:D386–389. 10.1093/nar/gkj032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wierzchoslawski R, Ubanowicz A, Dzianott A, Figlerowicz M, Bujarski JJ. 2006. Characterization of a novel 5′ subgenomic RNA3a derived from RNA3 of brome mosaic bromovirus. J. Virol. 80:12357–12366. 10.1128/JVI.01207-06 [DOI] [PMC free article] [PubMed] [Google Scholar]