Abstract
Hepatitis C virus (HCV) nonstructural protein 2 (NS2) is required for HCV polyprotein processing and particle assembly. It comprises an N-terminal membrane domain and a C-terminal, cytosolically oriented protease domain. Here, we demonstrate that the NS2 protease domain itself associates with cellular membranes. A single charged residue in the second α-helix of the NS2 protease domain is required for proper membrane association, NS2 protein stability, and efficient HCV polyprotein processing.
TEXT
Hepatitis C virus (HCV) nonstructural protein 2 (NS2) is required for HCV polyprotein processing and particle formation (1–3). It comprises an N-terminal membrane domain (amino acids [aa] 1 to 93) and a C-terminal, cytosolically oriented protease domain (NS2pro; aa 94 to 217). The membrane domain is composed of three predicted transmembrane segments and serves as a molecular platform for the coordination of HCV structural and nonstructural proteins in viral assembly (4–7). NS2pro forms a dimeric cysteine protease with two composite active sites (8). It is composed of two subdomains connected by an extended linker. The N-terminal subdomain includes two α-helices, H1 and H2, and the C-terminal subdomain, a four-stranded antiparallel β-sheet. As the crystal structure of NS2pro contained detergent molecules interacting with H1 and H2, it was proposed that NS2pro itself may associate with membranes (8). However, this hypothesis has not been tested experimentally.
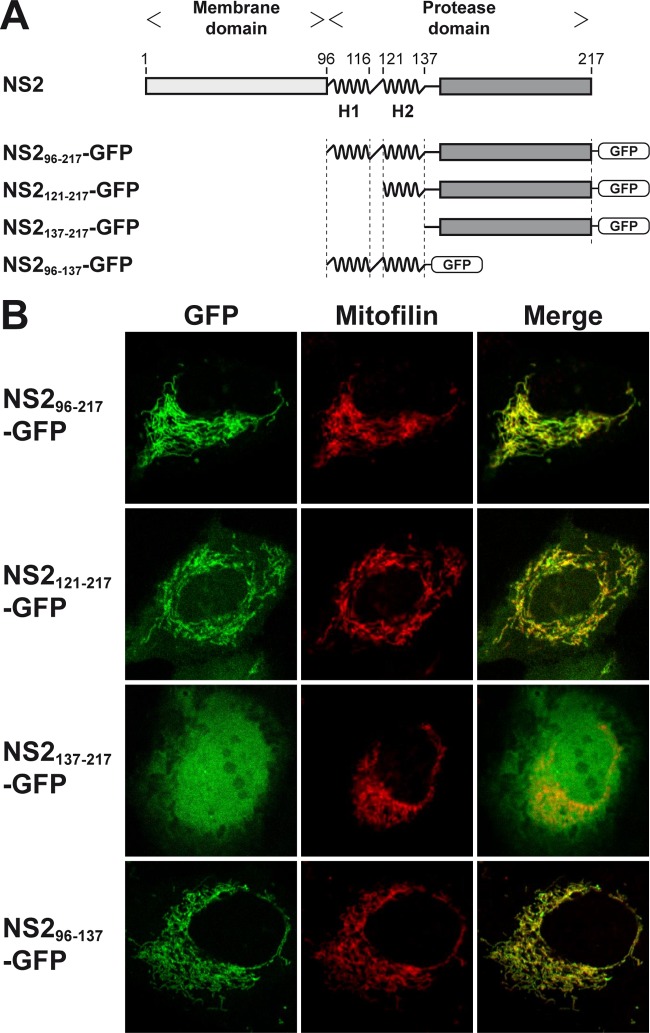
Therefore, a panel of green fluorescent protein (GFP) fusion constructs comprising NS2 segments derived from HCV genotype 1a clone H77 (9) (kindly provided by Charles M. Rice, The Rockefeller University, New York, NY) was prepared and transiently expressed in U-2 OS human osteosarcoma cells, as described previously (4, 10) (Fig. 1A). We have previously shown that GFP alone is diffusely distributed in the cytoplasm and nucleus of these cells whereas full-length NS2 expressed as a fusion protein with GFP localizes mainly to membranes of the endoplasmic reticulum (ER) (4). As shown in Fig. 1B, NS2pro localized to cellular membranes and, more specifically, to mitochondria, as revealed by colocalization with mitofilin. Membrane association of NS2pro was confirmed by membrane flotation analyses (data not shown). Interestingly, deletion of H1 and H2 (NS2137–217-GFP) abolished membrane association whereas fusion of H1 and H2 (NS296–137-GFP) or of H2 alone (NS2121–137-GFP) to GFP resulted in its membrane association. Collectively, these observations indicate that NS2pro itself associates with membranes through the α-helices within NS2 aa segment 96 to 137.
FIG 1.
Membrane association of the HCV NS2 protease domain. (A) Schematic representation of HCV NS2 and of GFP fusion constructs. The two α-helices located at the N terminus of the protease domain (comprising aa 96 to 116 [H1] and 121 to 137 [H2]) are highlighted. (B) Confocal laser scanning microscopy analyses of U-2 OS human osteosarcoma cells transiently transfected with cytomegalovirus (CMV) promoter-driven GFP fusion constructs, as illustrated in panel A. A polyclonal antibody against mitofilin (Sigma-Aldrich, St. Louis, MO) was used as a marker for mitochondria.
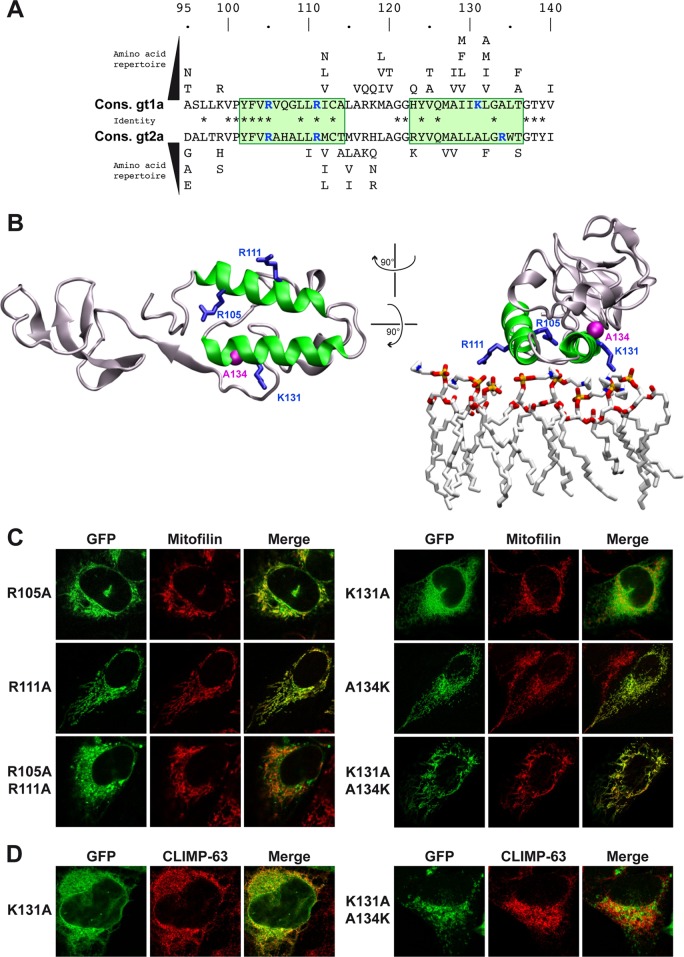
H1 and H2 display a number of surface-exposed hydrophobic residues as well as conserved positively charged residues with putative accessibility to cellular membrane lipids (Fig. 2A and B). Two arginine residues are found at positions 105 and 111 within H1. Within H2, a positively charged residue is found in all HCV isolates referenced in the European HCV Database (http://euhcvdb.ibcp.fr/), at either position 131 or position 134. A lysine is found in position 131 in HCV genotype 1 (as well as in genotypes 4 and 6; data not shown) whereas an arginine is found in position 134 in HCV genotype 2 (as well as in genotypes 3 and 5; data not shown), at a distance that corresponds to one α-helix turn (Fig. 2A). The positively charged side chains of these residues are laterally oriented on the helices (Fig. 2B).
FIG 2.
A single positively charged residue in H2 affects membrane association of NS2pro. (A) Sequence analysis of the two α-helices (H1 and H2, highlighted by green boxes) of NS2pro identifies conserved positively charged residues (highlighted in blue) at positions 105 and 111 within H1 and at position 131 (genotype 1) or 134 (genotype 2) within H2. The distance between positions 131 and 134 corresponds to one α-helix turn. Asterisks indicate residues that are identical between genotypes. Cons., consensus. (B) Structure of an NS2pro monomer (PDB entry 2HD0, H77 isolate, genotype 1a) (8) drawn as a ribbon using VMD 1.9 software (14). H1 and H2 are colored in green and the positively charged residues highlighted in blue. The right panel shows the presumed close proximity of H1 and H2 to the membrane surface represented as a simulated model of a POPC (1-palmitoyl-2-oleoyl-2-sn-glycero-3-phosphocholine) layer obtained from reference 15 (a stick representation with phosphorus, oxygen, nitrogen, and carbon atoms is colored in yellow, red, blue, and light gray, respectively). Positively charged amino acid residues 105, 111, and 131 are expected to interact with the negative charges of phosphate groups of POPC. The alanine residue at position 134 is highlighted in pink by its Van der Waals surface. (C and D) Confocal laser scanning microscopy analyses of U-2 OS human osteosarcoma cells transiently transfected with CMV promoter-driven NS296-217-GFP fusion constructs (see Fig. 1A) harboring the indicated mutations. A polyclonal antibody against mitofilin (Sigma-Aldrich, St. Louis, MO) and monoclonal antibody G1/296 against CLIMP-63 (Enzo Life Sciences, Farmingdale, NY) were used as markers for mitochondria and the endoplasmic reticulum in panels C and D, respectively.
Single or double alanine substitutions of the multiple conserved hydrophobic residues in H1 and H2 did not alter membrane association of NS2pro (data not shown). Likewise, alanine substitution of the arginine residues within H1 did not affect membrane association (R105A, R111A, and R105A/R111A; Fig. 2C). In contrast, alanine substitution of Lys 131 resulted in redistribution of the protease domain from mitochondria to the ER, indicating a change in the character of membrane association, likely due to altered protein-protein and/or protein-lipid interactions (K131A; Fig. 2C and D). Indeed, the mitochondrial membrane contains phospholipids, such as cardiolipin, whose negatively charged phosphate residues are accessible to positively charged basic amino acid residues. Remarkably, the membrane association phenotype could be restored by introduction of a lysine residue at position 134 (K131A/A134K; Fig. 2C and D). Conversely, alanine substitution of the naturally occurring arginine in position 134 of NS2pro from HCV genotype 2a (isolate JFH1) resulted in the same change in subcellular distribution that was rescued by introduction of an arginine in position 131 (data not shown). Taken together, these observations reveal a significant contribution of a single positively charged amino acid residue at position 131 (or 134) to the membrane association of NS2pro.
The mutations described above were introduced into full-length mono- and bicistronic HCV constructs to examine their impact on polyprotein processing and viral particle assembly. Of note, the bicistronic constructs, harboring NS2 from HCV genotype 1a isolate H77 (11) (kindly provided by Charles M. Rice), allow investigation of mutations independently of an effect on NS2 protease-mediated polyprotein processing. Analyses were performed 72 h after electroporation of RNA transcribed from these constructs into Huh-7.5 human hepatocellular carcinoma cells (12) (kindly provided by Charles M. Rice).
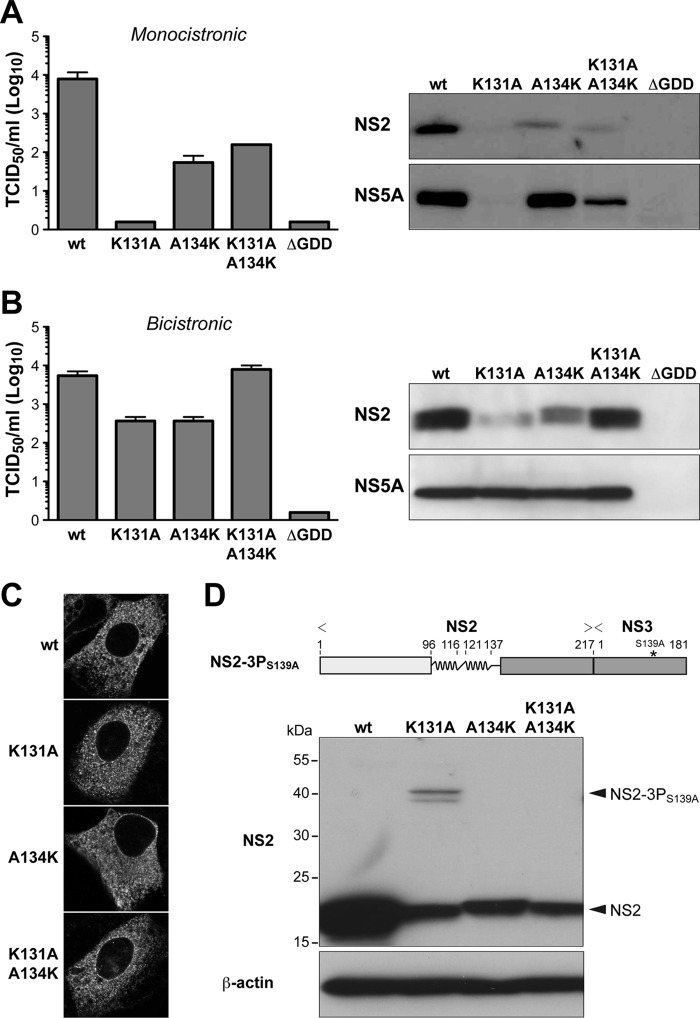
In the monocistronic context, mutation K131A completely abolished virus production (Fig. 3A, left panel). This defect could be partially rescued by introduction of a lysine in position 134 (K131A/A134K). Immunoblot analyses revealed a strong reduction of NS2 expression by mutant K131A, which could be partially restored by a lysine in position 134 (K131A/A134K) (Fig. 3A, right panel).
FIG 3.
The positively charged residue in H2 is important for HCV polyprotein processing. (A and B) The indicated mutations were introduced into a monocistronic full-length construct harboring NS2 from HCV genotype 1a isolate H77 (11) (A) or a bicistronic full-length construct harboring an encephalomyocarditis virus (EMCV) internal ribosome entry site (IRES) between NS2 from HCV genotype 1a isolate H77 and NS3 (11) (B). Virus infectivity, quantified shown on the left, was assessed by 50% tissue culture infectious dose (TCID50) determination at 72 h postelectroporation, as described previously (16). Immunoblot analyses using monoclonal antibodies (MAbs) 6H6 against NS2 (8, 11) and 9E10 against NS5A (16) (both kindly provided by Charles M. Rice) are shown on the right. (C) NS2 expressed in the context of the different bicistronic viruses was examined 48 h postelectroporation by immunofluorescence microscopy using MAb 6H6. (D) A CMV promoter-driven construct allowing expression of NS2 and the protease domain of NS3 with an inactivating S139A mutation in its catalytic site (NS2-3PS139A) was derived from HCV genotype 1a isolate H77 and is illustrated on the top. Processing of the wild-type (wt) construct as well as of constructs harboring the indicated mutations in NS2 H2 was analyzed by transient transfection into U-2 OS cells and immunoblotting using MAb 6H6 against NS2. Mouse monoclonal AC-15 against β-actin was used as a loading control. The positions of NS2-3PS139A and of processed NS2 are indicated on the right.
In the bicistronic context, mutation K131A also reduced virus production (Fig. 3B, left panel), but this could be completely restored by introduction of a lysine in position 134 (K131A/A134K). Mutation K131A had no effect on NS5A expression, i.e., did not affect HCV RNA replication (Fig. 3B, right panel). However, NS2 expression by this mutant was reduced. This defect was fully rescued by introduction of a lysine in position 134 (K131A/A134K). Hence, lack of a positively charged residue at position 131 (or 134) in H2 appears to strongly impact on NS2 protein stability, possibly by affecting proper protein-lipid interactions at the membrane surface. However, mutation K131A within NS2pro did not affect the overall subcellular localization of full-length NS2, which, as shown previously (4), is determined by its N-terminal membrane domain (Fig. 3C).
The effect of the mutations described above on NS2 protease activity was analyzed further in a construct comprising NS2 and the N-terminal one-third of NS3, whose serine protease activity was inactivated by an S139A mutation in its catalytic site (NS2-3PS139A). As shown in Fig. 3D, the efficiency of NS2-mediated cleavage at the NS2/NS3 junction is impaired by the K131A mutation and restored in construct K131A/A134K. Taken together, these observations indicate that positively charged amino acid residue 131 (or 134) in H2 is required not only for NS2 protein stability but also for efficient HCV polyprotein processing. In contrast, this residue appears to play only a minor role in HCV assembly, the second key function of NS2.
In conclusion, NS2pro associates with intracellular membranes independently from the N-terminal membrane domain of NS2. A single positively charged residue in H2 is required for proper membrane association, NS2 protein stability, and efficient HCV polyprotein processing. In more general terms, based on the findings reported here and previous work on the HCV NS3-4A complex (13), proper positioning of HCV nonstructural proteins on intracellular membranes is ensured by multiple determinants, including membrane domains as well as determinants for membrane association within their catalytic domains.
ACKNOWLEDGMENTS
We gratefully acknowledge Ivo Lorenz for critical review of the manuscript and Rosa Castillo and Audrey Kennel for expert technical assistance as well as Thomas Dentzer and Charles M. Rice for reagents.
This work was supported by the Swiss National Science Foundation (31003A-138484 to D.M.), the Novartis Foundation (09C53 to D.M.) and the French National Agency for Research on AIDS and Viral Hepatitis (ANRS, to F.P.). C.M.L. is supported by the Deutsche Forschungsgemeinschaft (LA 2806/1-1 and LA 2806/2-1).
Footnotes
Published ahead of print 19 March 2014
REFERENCES
- 1.Bartenschlager R, Lohmann V, Penin F. 2013. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat. Rev. Microbiol. 11:482–496. 10.1038/nrmicro3046 [DOI] [PubMed] [Google Scholar]
- 2.Lindenbach BD, Rice CM. 2013. The ins and outs of hepatitis C virus entry and assembly. Nat. Rev. Microbiol. 11:688–700. 10.1038/nrmicro3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moradpour D, Penin F. 2013. Hepatitis C virus proteins: from structure to function. Curr. Top. Microbiol. Immunol. 369:113–142 [DOI] [PubMed] [Google Scholar]
- 4.Jirasko V, Montserret R, Lee JY, Gouttenoire J, Moradpour D, Penin F, Bartenschlager R. 2010. Structural and functional studies of nonstructural protein 2 of the hepatitis C virus reveal its key role as organizer of virion assembly. PLoS Pathog. 6:e1001233. 10.1371/journal.ppat.1001233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma Y, Anantpadma M, Timpe JM, Shanmugam S, Singh SM, Lemon SM, Yi M. 2011. Hepatitis C virus NS2 protein serves as a scaffold for virus assembly by interacting with both structural and nonstructural proteins. J. Virol. 85:86–97. 10.1128/JVI.01070-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Popescu C-I, Callens N, Trinel D, Roingeard P, Moradpour D, Descamps V, Duverlie G, Penin F, Héliot L, Rouillé Y, Dubuisson J. 2011. NS2 protein of hepatitis C virus interacts with structural and non-structural proteins towards virus assembly. PLoS Pathog. 7:e1001278. 10.1371/journal.ppat.1001278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stapleford KA, Lindenbach BD. 2011. Hepatitis C virus NS2 coordinates virus particle assembly through physical interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 85:1706–1717. 10.1128/JVI.02268-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lorenz IC, Marcotrigiano J, Dentzer TG, Rice CM. 2006. Structure of the catalytic domain of the hepatitis C virus NS2-3 protease. Nature 442:831–835. 10.1038/nature04975 [DOI] [PubMed] [Google Scholar]
- 9.Kolykhalov AA, Agapov EV, Blight KJ, Mihalik K, Feinstone SM, Rice CM. 1997. Transmission of hepatitis C by intrahepatic inoculation with transcribed RNA. Science 277:570–574. 10.1126/science.277.5325.570 [DOI] [PubMed] [Google Scholar]
- 10.Gouttenoire J, Castet V, Montserret R, Arora N, Raussens V, Ruysschaert JM, Diesis E, Blum HE, Penin F, Moradpour D. 2009. Identification of a novel determinant for membrane association in hepatitis C virus nonstructural protein 4B. J. Virol. 83:6257–6268. 10.1128/JVI.02663-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dentzer TG, Lorenz IC, Evans MJ, Rice CM. 2009. Determinants of the hepatitis C virus nonstructural protein 2 protease domain required for production of infectious virus. J. Virol. 83:12702–12713. 10.1128/JVI.01184-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J. Virol. 76:13001–13014. 10.1128/JVI.76.24.13001-13014.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brass V, Berke JM, Montserret R, Blum HE, Penin F, Moradpour D. 2008. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc. Natl. Acad. Sci. U. S. A. 105:14545–14550. 10.1073/pnas.0807298105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Humphrey W, Dalke A, Schulten K. 1996. VMD: visual molecular dynamics. J. Mol. Graph. 14:33–38, 27–28. 10.1016/0263-7855(96)00018-5 [DOI] [PubMed] [Google Scholar]
- 15.Chandler DE, Penin F, Schulten K, Chipot C. 2012. The p7 protein of hepatitis C virus forms structurally plastic, minimalist ion channels. PLoS Comput. Biol. 8:e1002702. 10.1371/journal.pcbi.1002702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626. 10.1126/science.1114016 [DOI] [PubMed] [Google Scholar]