ABSTRACT
The microRNA miR-122 is highly expressed in the liver and stimulates hepatitis C virus (HCV) replication in vitro. IFNL3 (lambda-3 interferon gene) polymorphisms and the expression of miR-122 have been associated with sustained virological response (SVR) to treatment with pegylated interferon plus ribavirin in patients with chronic hepatitis C (CHC). We investigated, in vivo, the relationship between miR-122 expression, IFNL3 polymorphism, fibrosis, and response to PEG-IFN plus ribavirin. Pretreatment liver biopsy specimens and serum samples from 133 patients with CHC were included. Sixty-six patients achieved SVR, and 64 failed to respond to the treatment (43 nonresponders [NR] and 21 relapsers [RR]). All stages of fibrosis were represented, with 39, 50, 23, and 19 patients, respectively, having Metavir scores of F1, F2, F3, and F4. miR-122 expression was assessed by real-time quantitative PCR (RT-qPCR) and IFNL3 rs12979860 by direct sequencing. Hepatic miR-122 expression was higher in patients with the IFNL3 CC genotype than in those with the IFNL3 CT or TT genotype, in all patients (P = 0.025), and in NRs plus RRs (P = 0.013). Increased hepatic miR-122 was more strongly associated with complete early virological response (cEVR) (P = 0.003) than with SVR (P = 0.016). In multivariate analysis, increased hepatic miR-122 was only associated with the IFNL3 CC genotype. miR-122 was decreased in patients with advanced fibrosis (Metavir scores of F3 and F4) compared to its levels in patients with mild and moderate fibrosis (F1 and F2) (P = 0.01). Serum and hepatic expression of miR-122 were not associated. The association between miR-122 and IFNL3 was stronger than the association between miR-122 and response to treatment. miR-122 may play a role in the early viral decline that is dependent on IFNL3 and the innate immune response.
IMPORTANCE miR-122 plays a crucial role during HCV infection. Moreover, it was reported that miR-122 binding within the HCV genome stimulates its replication. Moreover, miR-122 is highly expressed within hepatocytes, where it regulates many cellular pathways. A reduction of miR-122 expression has been suggested to be associated with responsiveness to IFN-based therapy in patients with chronic hepatitis C. Several independent genome-wide association studies reported a strong association between IFNL3 polymorphism and responsiveness to IFN-based therapy. We report here a strong association between the expression of miR-122 and IFNL3 polymorphism that is independent of the response to the treatment. Our data suggest that modification of miR-122 expression may play an important role in the molecular mechanism associated with IFNL3 polymorphism. Moreover, we report a reduction of miR-122 at more advanced stages of fibrosis in patients with chronic hepatitis C.
INTRODUCTION
Hepatitis C virus (HCV) is a major cause of chronic liver disease, with an estimated prevalence of 150 million cases worldwide (1). The severity of the disease varies from asymptomatic chronic infection to cirrhosis and hepatocellular carcinoma (2, 3).
The treatment of chronic hepatitis C (CHC) for HCV genotype 1-infected patients has greatly improved with the use of direct-acting antivirals (4, 5). However, in many parts of the world, pegylated-interferon (PEG-IFN)-plus-ribavirin dual therapy remains an option for patients with HCV genotype 1 and the standard of care for patients with all other HCV genotypes (6).
Two main single-nucleotide polymorphisms (SNPs), rs12979860 and rs8099917, located upstream from the lambda-3 interferon gene, IFNL3, were identified as predicting sustained virological response (SVR) in patients with CHC (7). To date, the molecular mechanism controlling the response to IFN therapy associated with IFNL3 SNPs has not been fully elucidated. Recently, a spliced form of the IFNL3 protein, IFNL4, has been reported to be associated with responsiveness to IFN-based therapy. IFNL4 is generated by individuals who carry the ΔG allele of the ss469415590 variant (IFNL4-ΔG). Linkage disequilibrium is strong between the IFNL4-ΔG allele and the unfavorable rs12979860-T allele (8). The expression of IFNL4 is associated with HCV persistence and a poorer response to PEG-IFN plus ribavirin (8). IFNL4 was suggested to regulate the antiviral activity against HCV (9). A strong decline of HCV viral RNA during the first weeks of treatment in patients with an IFNL3 rs12979860 CC genotype compared to the HCV viral RNA levels in patients with the CT or TT (CT/TT) genotype has been described (10). Patients with CT/TT were suggested to have aberrantly enhanced baseline induction of innate immune response pathways, resulting in impaired virologic response (11). An IFNL3 SNP was suggested to play a role in the innate immune response responsible for this early viral decline (11).
MicroRNAs (miRNAs) regulate the expression of up to 60% of cellular mRNAs. With the potential for one miRNA to regulate several mRNAs, miRNAs play key roles in diverse regulatory pathways (12, 13). Modification of the expression of the miRNA miR-122 during HCV expression has been reported in cells, in animal models, and in humans (14–17). miR-122 is highly expressed in hepatocytes, where it represents 70% of total miRNAs. In the liver, miR-122 regulates hepatocyte growth, neoplastic transformation, and lipid metabolism (18–20). miR-122 binds the HCV genome within two seed sites located upstream from the HCV internal ribosome entry site (IRES) and stimulates its replication in vitro. Miravirsen, an inhibitor of miR-122, has been reported to induce a decrease of HCV replication in humanized mice and in chimpanzees (21, 22). The antiviral effect of miravirsen has been assessed more recently in 27 patients with CHC (23). Moreover, the expression of miR-122 in HCV-nonpermissive cells facilitates the replication of HCV (24, 25). A recent study suggested that miR-122 regulates collagen production via suppressing P4HA1 expression in hepatic stellate cells (25).
Interestingly, a reduction of hepatic and serum miR-122 has been associated with primary nonresponse (pNR) and nonresponse (NR) in patients with CHC (26, 27). However, in further studies, these results were not statistically confirmed (17). Since both IFNL3 polymorphism and the expression of miR-122 are associated with response to treatment, we investigated in this study the relationship between the expression of miR-122 and IFNL3. Moreover, in patients with CHC, the monitoring of fibrosis is mandatory, because it reflects the progression of the disease. The progression from mild (F1) to moderate (F2) fibrosis (Metavir score) is a strong argument to initiate a treatment. Although the IFNL3 polymorphisms seem not to be associated with fibrosis (28, 29), the IFNL3 CC genotype was associated with a state of enhanced immunity that can promote necroinflammation (30). Moreover, reports suggested a role of miR-122 in liver fibrosis (31, 32). Since miR-122 regulates HCV replication, it is particularly interesting to investigate its role during fibrosis. We therefore also assessed the association of miR-122 with fibrosis in our cohort of patients.
MATERIALS AND METHODS
Patients.
A total of 133 treatment-naive patients with CHC were consecutively included in the study. The study conformed to the ethical guidelines of the 1975 Declaration of Helsinki. Assistance Publique des Hôpitaux de Paris granted approval for our research. All participants gave their informed consent, and the following criteria were met: (i) all patients had an established diagnosis of CHC; (ii) all patients had an absence of another cause of chronic liver disease and an absence of viral coinfection; and (iii) all patients received treatment with either PEG–IFN-α2b (Viraferonpeg; Schering Plough) at a dose of 1.5 μg/kg of body weight/week and weight-based ribavirin at a dose of 800 to 1,200 mg/day (Rebetol; Schering Plough) or PEG–IFN-α2a at a dose of 180 μg/week (Pegasys; Roche) and weight-based ribavirin at 1,000 to 1,200 mg/day (Copegus; Roche). The duration of treatment was 48 weeks for patients with HCV genotypes 1 and 4 and 24 weeks for those with genotypes 2 and 3. For patients with HCV genotypes 1 and 4, if HCV RNA was detectable at week 24, the treatment was stopped. For those with HCV genotype 1, if HCV RNA decreased less than 2 log10 from baseline to week 12 of therapy, treatment was stopped.
SVR was defined as undetectable HCV RNA 24 weeks after completion of treatment and NR as detectable serum HCV RNA at the end of the treatment (4). Relapse (RR) was defined as the reappearance of detectable serum HCV RNA within 24 weeks after cessation of treatment (4). Complete early virological response (cEVR) and rapid virological response (RVR) were defined as undetectable HCV RNA at week 12 and week 4, respectively, whereas primary nonresponse (pNR) was characterized as a reduction of HCV RNA of less than 2 log at week 12 (4).
Both serum samples and liver biopsy specimens were collected before the initiation of the treatment. Patients underwent treatment 1 week to 6 months after the liver biopsy was performed. From the 133 patients included, paired liver and serum samples were available for 83 patients.
Fibrosis was staged according to the Metavir score system. F0 characterized a liver with no fibrosis, F1 a portal fibrosis, F2 a portal fibrosis with few septa, F3 a septal fibrosis, and F4 a cirrhosis. Five patients had two liver biopsy specimens taken during their follow-up, with the time between biopsies varying from 2 years and 5 months to 8 years and 7 months.
Histologically normal controls.
Nine percutaneous liver biopsy specimens were histologically normal, with absence of inflammation, any other cause of liver disease, fibrosis, or pathological pattern and less than 5% steatosis. A previous study demonstrates the importance of collecting healthy controls under the same conditions as the other samples (33).
Total RNA extraction.
Frozen liver biopsy specimens were crushed and dissolved in 1 ml of iced RNAble (Eurobio). Chloroform was added, and the upper phase was collected after centrifugation and added to the same volume of isopropanol. RNA was precipitated and washed in 80% ethyl alcohol. The pellet was dissolved in RNase- and DNase-free H2O. The optical density of total RNA was measured at 260 nm. Total RNAs were extracted from 400 μl of serum (mirVana; Life Technologies). Since there is no miRNA stably expressed in the serum, we added 5.6 × 108 copies of miR-39 (Caenorhabditis elegans; Qiagen) to each serum sample before the extraction. miR-122 expression was then normalized to the miR-39 expression values.
Real-time RT-qPCR.
miR-122 was detected by reverse transcription-quantitative PCR (RT-qPCR). PCRs were performed using the Light Cycler 480 real-time PCR system and the LightCycler 480. The theoretical and practical aspects of RT-qPCR using the light cycler LC480 (Roche) have been already described in detail (34, 35).
One nanogram of cDNA from reverse transcription of miRNAs was added to each reaction mixture. The total amount of hepatic miR-122 was normalized to the amount of the control small RNA SNORD44, whereas the total amount of miR-122 in serum was normalized to the amount of Caenorhabditis elegans miR-39 (Qiagen). To choose the best noncoding small RNA to normalize the expression data of miR-122, 7 noncoding RNAs (miR-25_1, SNORD44, miR-191_1, miR-103_1, SNORD48, RNU6B_2, and RNU5A_1) were analyzed in 80 liver biopsy specimens from our cohort of patients. The expression levels of each of these 7 noncoding RNAs were compared and normalized to each other to identify the most accurate candidate to normalize the results. When the values were normalized to SNORD44, the expression of the other noncoding RNAs was the most stable among the 80 liver biopsy specimens tested. Therefore, SNORD44 was chosen to normalize the expression values of miR-122 in the biopsy specimens (data not shown, available on request).
For the patients who had 2 biopsies at different stages of fibrosis, the miR-122 content was analyzed by TaqMan RT-qPCR (Life Technologies).
All of the results were expressed as the n-fold difference in target gene expression relative to the expression of SNORD44 and are termed Ntarget. Ntarget was determined as Ntarget = 2ΔCpsample, where Cp is the crossing point. The ΔCp value was determined by subtracting the average Cp value of the target miRNA from the average Cp value of SNORD44 (liver biopsy samples) and miR-39 (serum samples).
IFNL3 genotyping.
In our series, the patients were mainly of Caucasian ancestry and infected with HCV genotype 1. The SNP rs12979860 was selected for this study because it has been the main polymorphism associated with response to treatment in patients with Caucasian ancestry and HCV genotype 1 (7). The 133 patients included in the cohort were genotyped for rs12979860 using direct sequencing (AmpliTaq gold and BigDye Terminator; Life Technologies). Free, circulating DNA was extracted from 500-μl serum samples (QIAamp circulating nucleic acid kit; Qiagen) as described previously (36). The PCR products were separated on an ABI 3130 sequencer and analyzed with SeqScape 2.6 (Life Technologies).
Statistical analysis.
We evaluated the statistical significance of the relationships between the expression of miR-122 in the liver and serum and bioclinical characteristics. The following variables were investigated: age (<45 years or ≥45 years), gender, stage of fibrosis (Metavir F1 versus F2 to 4), viral genotype (HCV genotype I versus other genotypes), body mass index (<25 or ≥25 kg/m2), alcohol intake (<30 g/day or ≥0.30 g/day), serum levels of aspartate amino transferase (ALAT) and alanine amino transferase (ASAT) (IU/ml), serum level of gamma glutamyl transferase (GGT), viral load at baseline (log10 IU), response to treatment (SVR versus NR plus RR [NR/RR]), and response at week 12 (cEVR versus pNR). For investigating binary variables, we performed Student's t tests. We report the means and standard errors together with the P values associated with the t tests. For comparing differences between liver tissues and histologically normal controls, we used the nonparametric Wilcoxon Mann-Whitney test. For investigating continuous variables, we performed linear regression analyses and reported the slope of the regression line (with its standard error), together with the P value associated with the corresponding Wald test. We performed a regularized (least absolute shrinkage and selection operator [LASSO]) linear regression analysis for variable selection with the log expression values of miR-122 as the dependent variable (glmnet package). We used multiple imputations by chained equations (mice package) to handle missing data. The regularization parameter for LASSO regression was chosen by 5-fold cross validation. A final multivariate linear regression model with selected variables (those having nonzero parameter estimates) is reported.
We tested the null hypothesis that the Pearson product moment correlation coefficient is zero using the classical test statistic which follows a Student's t distribution.
For all these tests, statistical significance was considered to be a P value of less than 0.05. All these analyses were carried out using R software and related packages (http://cran.r-project.org/index.html).
RESULTS
Patients and baseline characteristics.
Patient's characteristics are presented in Table 1. A total of 133 patients were included, of whom 50 were women and 83 men. Patients were mainly infected by HCV genotypes 1 (n = 80) and 4 (n = 25). The diagnosis of fibrosis was performed by liver biopsy and scored using Metavir. All stages of fibrosis were represented, with 39, 50, 23, and 19 patients scored as F1, F2, F3, and F4, respectively. Sixty-six patients had SVR, 43 had NR, and 21 had RR. At week 12 of the treatment, 47 patients had pNR and 63 had cEVR. rs12979860 and rs8099917 are both associated with SVR and are in strong linkage disequilibrium (37, 38). We selected the rs12979860 polymorphism because this polymorphism was reported to be more informative than rs8099917 in a cohort of patients with Caucasian ancestry (39). rs8099917 was analyzed by direct sequencing in 28 patients of the cohort as described in reference 40. Of the 28 patients tested, all who were rs12979860 CC were rs8099917 TT, and the 21 patients with the unfavorable allele rs12979860 CT/TT carried the rs8099917 unfavorable genotype TG or GG (see Table S2 in the supplemental material).
TABLE 1.
Characteristics of the patients
| Parametera | Mean value ± SD (range) or no. (%) of patients, unless otherwise indicated |
|---|---|
| No. of patients | 133 |
| Male/female | 83 (62.4)/50 (37.6) |
| Age (yr) | 47.8 ± 8.9 (28–73) |
| BMI (kg/m2) | 25.3 ± 4.0 (17.64–37.83) |
| ALAT (IU/liter; ULN, 35 IU/liter) | 109.7 ± 74.3 (18–459) |
| ASAT (IU/liter; ULN, 30 IU/liter) | 68.3 ± 47.6 (15–323) |
| GGT (IU/liter; ULN, 60 IU/liter) | 84.1 ± 81.8 (7–411) |
| Total cholesterol (mmol/liter) | 4.6 ± 1.2 (1.5–7.4) |
| Glycemia (mmol/liter) | 5.2 ± 1.2 (3.2–13.6) |
| Median viral load (log IU/ml) ± SD (range) | 5.5.± 0.7 (3.3–6.6) |
| HCV genotype | |
| 1 | 80 (60.2) |
| 2 | 12 (9.1) |
| 3 | 14 (10.5) |
| 4 | 25 (18.8) |
| 5 | 1 (0.7) |
| 6 | |
| Unknown | 1 (0.7) |
| IFNL3 rs12979860 genotype | |
| C/C | 38 (28.5) |
| C/T | 51 (38.3) |
| T/T | 29 (21.8) |
| Unknown | 15 (11.4) |
| Stage of fibrosisb | |
| F1 | 39 (29.3) |
| F2 | 50 (37.6) |
| F3 | 23 (17.3) |
| F4 | 19 (14.3) |
| unknown | 2 (1.5) |
| Treatment response | |
| SVR | 66 (49.6) |
| NR | 43 (32.3) |
| RR | 21 (15.8) |
| pNR | 47 (35.3) |
| cEVR | 63 (47.4) |
ALAT, alanine aminotransferase; ASAT, aspartate aminotransferase; GGT, gamma-glutamyl-transferase; BMI, body mass index; ULN, upper limit of normal; SVR, sustained virological response; NR, nonresponse; RR, relapse; pNR, primary nonresponse; cEVR, complete early virological response.
The Metavir system was used to determine fibrosis.
In the univariate analysis, we found no relationship between the log expression of hepatic miR-122 and age at therapy, gender, body mass index (BMI), alcohol use, HCV genotype, cholesterol, triglycerides, or glycemia (Table 2). A decrease in the mean log expression of hepatic miR-122 was associated with increased viral load at baseline (P = 0.025), the serum level of GGT (P < 0.001), and the ASAT and the ALAT values (P = 0.003 and P = 0.018, respectively) (Table 3). We performed a regularized (LASSO) linear regression analysis with log expression of hepatic miR-122 as the dependent variable. This analysis showed that only GGT and IFNL3 polymorphism had nonzero parameter estimates. The multivariate model, which included GGT and IFNL3, showed that patients with the CC genotype had higher log expression of hepatic miR-122 than patients with CT and TT (Table 4).
TABLE 2.
Univariate analysis of relationship between log expression of hepatic miR-122 and binary clinicobiological variablesa
| Variable | No. of patients | Mean (SE) | P value | Mean difference (SE) |
|---|---|---|---|---|
| Age (yr) | ||||
| <45 | 48 | −0.01 (0.59) | ||
| ≥45 | 84 | −0.13 (0.62) | 0.28 | −0.12 (0.01) |
| Gender | ||||
| Female | 50 | −0.08 (072) | ||
| Male | 83 | −0.10 (0.54) | 0.90 | −0.01 (0.01) |
| Obesity (BMI) | ||||
| ≤25 kg/m2 | 63 | −0.03 (0.58) | ||
| >25 kg/m2 | 63 | −0.15 (0.61) | 0.28 | −0.12 (0.01) |
| Alcohol use | ||||
| <30 g/day | 85 | −0.029 (0.65) | ||
| ≥30 g/day | 16 | −0.102 (0.53) | 0.67 | −0.07 (0.03) |
| Fibrosis (Metavir score) | ||||
| F1 | 39 | −0.06 (0.59) | ||
| ≥F2 | 92 | −0.15 (0.60) | 0.06 | −0.21 (0.01) |
| IFNL3 genotype | ||||
| CC | 38 | 0.08 (0.59) | ||
| CT/TT | 80 | −0.246 (0.63) | 0.01 | 0.33 (0.01) |
| Viral genotype | ||||
| HCV genotype 1 | 80 | −0.145 (0.64) | ||
| Other subtypes | 53 | −0.017 (0.59) | 0.25 | −0.13 (0.01) |
The Student t test was used to test the lack of statistically significant differences for miR-122 log expression between categories of each variable investigated. The P value for each variable is reported. We also report the observed mean difference with the associated standard error.
TABLE 3.
Relationship between log expression of hepatic miR-122 and continuous biological variablesa
| Variable | Slope | SE | P value | R2 coefficient |
|---|---|---|---|---|
| Viral load | −0.20 | 0.08 | 0.025 | 0.052 |
| Log(ASAT) | −0.28 | 0.08 | 0.003 | 0.073 |
| Log(ALAT) | −0.21 | 0.09 | 0.018 | 0.045 |
| Log(GGT) | −0.29 | 0.06 | 3.10−6 | 0.173 |
The continuous biological variables that are significantly (P < 0.05) associated with log expression of hepatic miR-122 are reported. The slope coefficient (with standard error) of the linear regression model, together with the associated P value (and R2 coefficient), are reported.
TABLE 4.
Multivariate analysis with log expression of hepatic miR-122 as the dependent variable and log(ASAT) and IFNL3 genotype as the independent variables
| Variable | Slope | SE | P value |
|---|---|---|---|
| IFNL3 (CC vs CT/TT) | 0.25 | 0.11 | 0.03 |
| Log (GGT) | −0.27 | 0.06 | <0.0001 |
Relationship between miR-122 expression, IFNL3 polymorphism, and response to therapy.
In univariate analysis, the level of expression of hepatic miR-122 before the initiation of treatment was reduced in patients with pNR and those with NR or RR (NR/RR) compared to, respectively, patients with cEVR (P = 0.003) (Fig. 1A) and SVR (P = 0.016) (Fig. 1B). There was no significant relationship between hepatic miR-122 and response to treatment at week 4.
FIG 1.
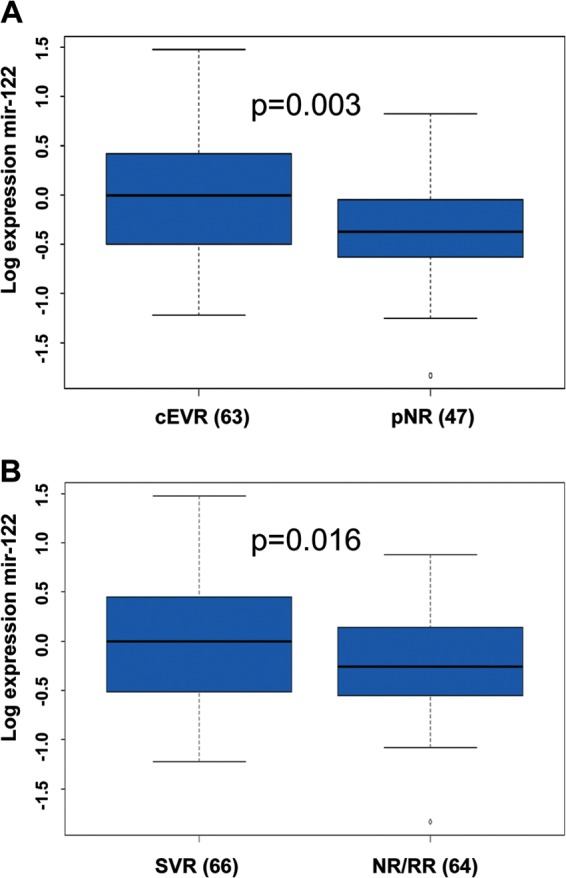
The hepatic expression of miR-122 is associated with early virological response. The expression of miR-122 was assessed by RT-qPCR. The ΔCp (ΔCpt = 2ΔCpsample) of miR-122 was calculated and normalized to the ΔCp value of SNORD44 in each biopsy specimen. The histograms represent the mean log expression of miR-122/SNORD44 within the groups of patients, comprising those with sustained virological response (SVR), nonresponse or relapse (NR/RR), primary nonresponse (pNR), and complete early virological response (cEVR), normalized to the expression of miR-122 within the group of histologically normal liver biopsy specimens. (A) The hepatic expression of miR-122 was compared in patients with pNR and cEVR. (B) The hepatic expression of miR-122 was compared in patients with NR/RR and SVR. The nonparametric Wilcoxon Mann-Whitney test was used to compare miR-122 expression. The numbers in parentheses show the number of patients in each group.
Patients carrying the IFNL3 CC genotype had significantly higher expression of hepatic miR-122 (P = 0.01) (Table 2) than patients with the CT/TT genotype (Fig. 2A). When analyzing the relationship between hepatic miR-122 and IFNL3 genotypes (CC versus CT/TT) stratified by response (SVR versus NR/RR), we showed that, for NRs, the genotype CC is associated with an increase of miR-122 (P = 0.005) (Fig. 2B), whereas for SVR, the genotype is not associated with miR-122 levels (P = 0.4) (Fig. 2C). When analyzing the relationship between miR-122 and response (SVR versus NR/RR) stratified by IFNL3 genotypes (CC versus CT/TT), we showed that for CT/TT genotypes, the response is associated with an increase of hepatic miR-122 (P = 0.04), whereas for the CC genotype, the response is not associated with hepatic miR-122 levels (P = 0.8) (Fig. 2D).
FIG 2.
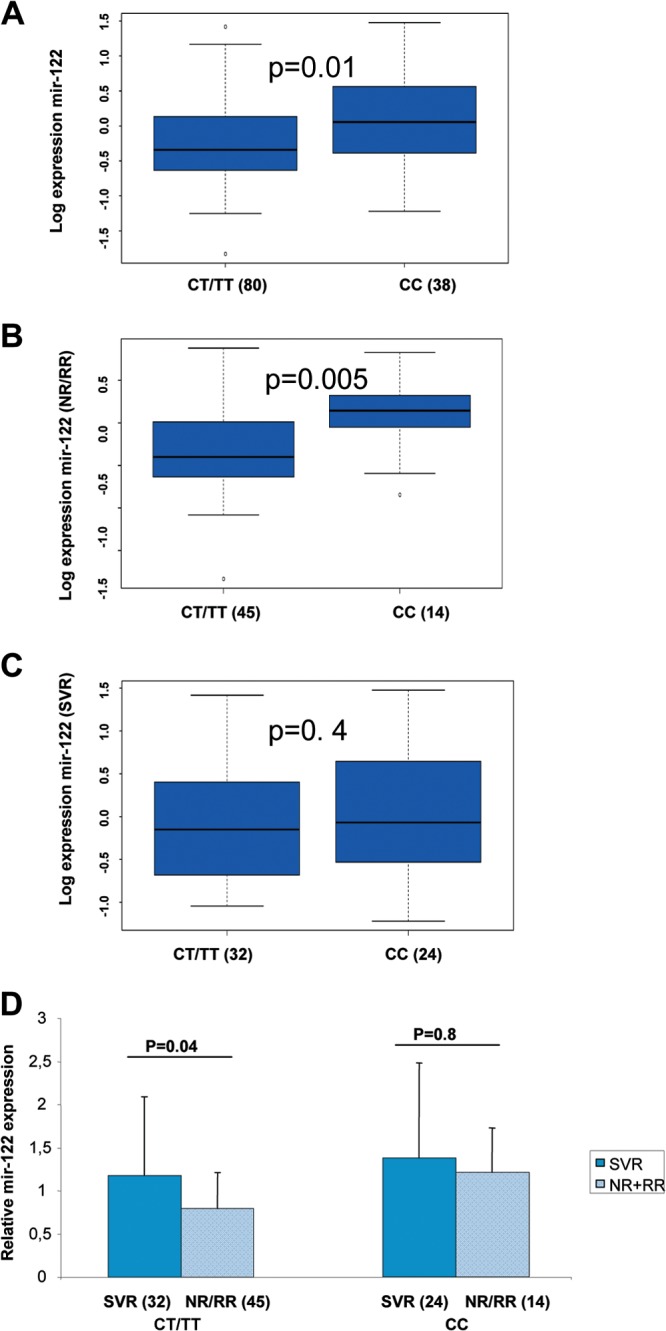
The IFNL3 CC genotype is associated with accumulation of hepatic miR-122. The expression of miR-122 was assessed by RT-qPCR. The ΔCp (ΔCpt = 2ΔCpsample) of miR-122 was calculated and normalized to the ΔCp value of SNORD44 in each biopsy specimen. (A to D) The histograms represent the mean log expression of miR-122/SNORD44 for all patients (A), patients who failed to respond to the treatment (NR/RR) (B), and SVR patients (C). The boxes and whiskers are as described in the legend to Fig. 1. We used the nonparametric Wilcoxon Mann-Whitney test to compare the expression of miR-122 in IFNL3 CC and CT/TT patients. (D) Reduction of hepatic expression of miR-122 in IFNL3 CT/TT patients who failed to respond to the treatment. The histograms represent the mean expression of miR-122/SNORD44 in IFNL3 CC and CT/TT patients with either SVR or NR/RR, normalized to the expression of miR-122 within the group of histologically normal liver biopsy specimens. The errors bars represent the standard deviations. The numbers in parentheses show the number of patients within each group.
The response (NR/RR versus SVR) and response at week 12 (cEVR versus pNR) variables did not significantly improve the fit of the multivariate model which included GGT and IFNL3.
The raw data for the expression of miR-122 in the liver and in the serum are presented in Table S3 in the supplemental material.
Relationship between miR-122 expression and fibrosis.
Hepatic miR-122 was reduced in patients infected with HCV (F1 to F4) compared to its level in healthy controls. Hepatic miR-122 was decreased in F3 and F4 patients compared to its levels in F1 and F2 patients (P = 0.01) (Fig. 3A). However, when we compared the F1 patients to those with Metavir scores of F2 to F4, the difference was not significant within all HCV genotypes (P = 0.06) (Fig. 3A).
FIG 3.
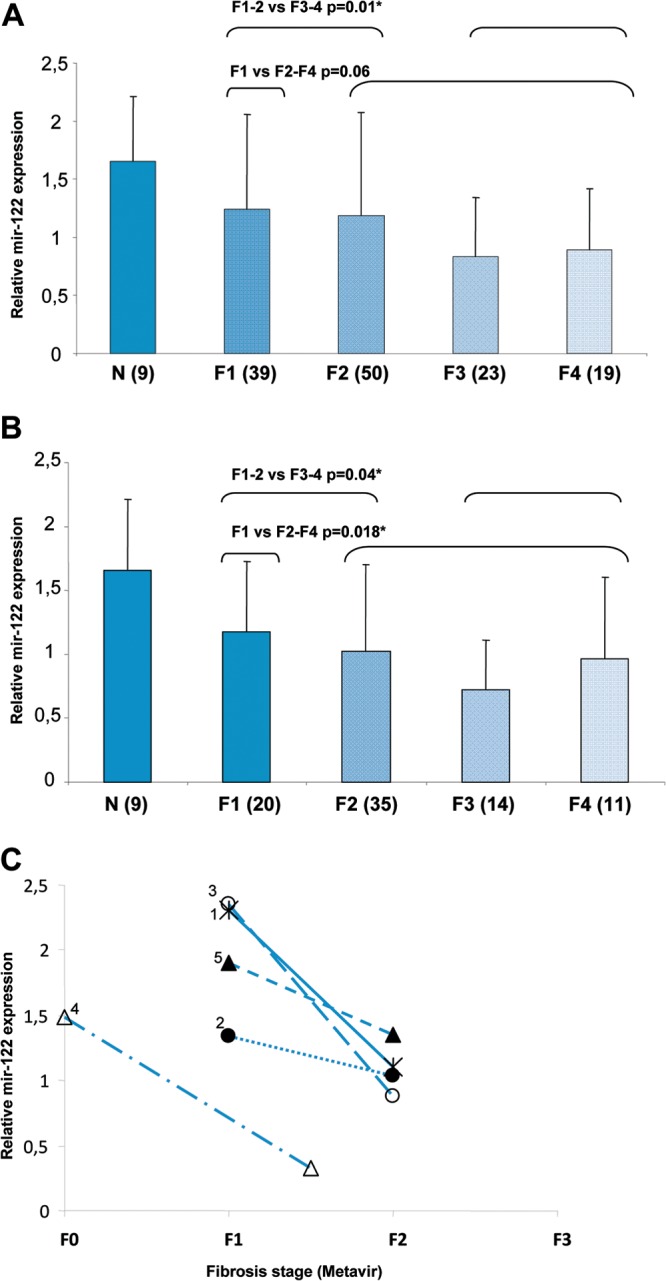
Modification of hepatic miR-122 expression at different stages of fibrosis in chronic hepatitis C. The expression of miR-122 was assessed by RT-qPCR. The ΔCp (ΔCpt = 2ΔCpsample) of miR-122 was calculated and normalized to the ΔCp value of SNORD44. (A and B) The histograms represent the mean expression of miR-122/SNORD44 within the groups of patients at different stages of fibrosis (F1 to F4). Modification of miR-122 expression at the different stages of fibrosis is shown for all patients with any HCV genotype (A) and for HCV genotype 1 patients only (B). The expression of mir-122 was compared in F1–F2 versus F3–F4 and in F1 versus F2–F4 in patients with all genotypes (A) and only genotype 1 (B). The number above the brackets is the P value for each association. We performed the nonparametric Wilcoxon Mann-Whitney test to compare miR-122 in each group of patients, including those with histologically normal liver biopsy samples (N) and those with stage F1 to F4 fibrosis. The numbers in parentheses show the number of patients within each group. The errors bars represent the standard deviations. (C) Reduction of miR-122 expression during early fibrosis progression. Five patients received 2 liver biopsies at different stages of fibrosis (x axis). The paired samples were obtained at intervals varying from 2 years and 5 months to 8 years and 7 months. The number next to each line is the identification number for each patient, also used in Table S1 in the supplemental material.
When we focused on patients with HCV genotype 1, we observed increased miR-122 levels in both F1 patients only and F1 and F2 patients compared to, respectively, those with Metavir scores of F2 to F4 or those with F3 and F4 (P = 0.018 and P = 0.04, respectively) (Fig. 3B).
Within HCV genotype 1 patients, hepatic expression of miR-122 was still associated with the viral load at baseline (P = 0.01), ASAT (P = 0.012), ALAT (P = 0.02), and GGT (P < 0.0001). HCV genotype 1 patients carrying the IFNL3 CC genotype had higher expression of hepatic miR-122 than CT/TT patients at baseline (P = 0.047).
Among the patients studied, 5 untreated patients with mild disease had two liver biopsies at time intervals ranging from 2 years and 5 months to 8 years and 7 months. All 5 patients showed a progression of the fibrosis stage at the second biopsy (F0 to F1–F2 [n = 1] and F1 to F2 [n = 4]). One patient who showed progression from F1 to F2 was infected by HCV genotype 2, whereas the other 4 patients had HCV genotype 1. The patient who showed progression from F0 to F1–F2 was a man, and 3 of the 4 patients who showed progression from F1 to F2 were women (see Table S1 in the supplemental material). In each of the 5 patients, the hepatic expression of miR-122 was reduced at the time of the second liver biopsy with its higher stage of fibrosis compared to the hepatic miR-122 expression at the time of the first liver biopsy at F0 or F1 (Fig. 3C).
Association with serum miR-122.
No relationship was found between hepatic and serum miR-122 expression (P = 0.21) (Fig. 4).
FIG 4.
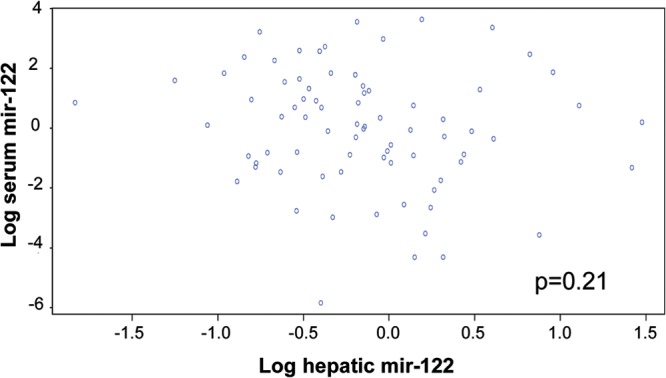
Lack of association between hepatic and serum miR-122. Log expression of hepatic and serum miR-122 was compared in patients with CHC. We tested the null hypothesis that the Pearson product moment correlation coefficient is zero using the classical statistical test which follows Student's t distribution.
In univariate analysis, we found no relationship of the log expression of serum miR-122 serum with age at therapy, BMI, alcohol use, fibrosis stage, HCV genotype, IFNL3, cholesterol, and glycemia. A 2.5-fold increase in the mean log expression of miR-122 in serum was found in men compared to women (P = 0.05). Moreover, an increase of serum miR-122 expression was associated with increased HCV viral load at baseline (P = 0.01), ASAT (P = 0.009), ALAT (P = 0.004), and GGT (P = 0.005).
We found no relationship of the log expression of serum miR-122 with response (NR/RR versus SVR) or response at week 12 (cEVR versus pNR).
Within patients with HCV genotype 1, we confirmed the association between expression of serum miR-122 and gender (P = 0.05), viral load at baseline (P = 0.02), and GGT (P = 0.025).
Using multivariate analysis, we found that men had higher log expression of serum miR-122 than women (P = 0.009). Moreover, an increase in ASAT was associated with an increase in the mean log expression of serum miR-122 (P = 0.004).
DISCUSSION
miR-122 is highly abundant within hepatocytes and plays a fundamental role in regulating hepatic functions (18–20, 31). Moreover, miR-122 expression is critical for HCV replication (24, 25, 41). However, so far, only a few studies have investigated the modification of miR-122 expression during CHC infection, especially in vivo. Interestingly, both miR-122 expression and IFNL3 polymorphisms have been associated with responsiveness to PEG-IFN plus ribavirin (7, 26). Therefore, we provided an analysis of miR-122 expression and its association with IFNL3 polymorphism, fibrosis progression, and viral load in a large and well-phenotyped cohort of 133 patients with CHC.
Taken together, our results suggest that the association between miR-122 and IFNL3 polymorphism is stronger than the association between miR-122 and both SVR and cEVR. The relationship we observed between miR-122 expression and responsiveness to IFN may be due to the strongest association between the expression of miR-122 and IFNL3 polymorphism. An IFNL3 CC genotype, SVR, and cEVR were associated with increased hepatic expression of miR-122 in univariate analysis. However, using multivariate linear regression analysis with log expression of hepatic miR-122 as the dependent variable, only IFNL3 CC polymorphism was independently associated with miR-122. Moreover, we observed increased expression of miR-122 in IFNL3 CC patients at baseline, whatever the response status. A previous study that included 61 patients with CHC reported a trend of upregulation of miR-122 in IFNL3 CC patients compared to its expression in TT patients (P = 0.08) (42). The larger number of patients in our study may explain the significant relationship we observed between the increased expression of miR-122 and IFNL3 CC.
Interestingly, during PEG-IFN–ribavirin therapy, patients with the IFNL3 CC genotype had a more rapid early HCV viral decline than CT/TT patients (10). Moreover, carriers of the IFNL3 CC genotype were more likely to have a superior innate immune response to IFN therapy (11). Patients with IFNL3 CT/TT were suggested to have an aberrantly enhanced baseline induction of innate immune response pathways, resulting in an impaired virological response (11). Therefore, in our study, the stronger association of miR-122 with cEVR (P = 0.003) than with SVR (P = 0.016) and the relationship between miR-122 and IFNL3 polymorphism suggest that miR-122 expression may play an important role during early viral decline induced by the innate immune response.
The expression of miR-122 is reduced in Huh-7 cells treated with IFN-β (43). Interestingly, our results correlate with this finding. Indeed, patients with IFNL3 CT/TT, who have high innate stimulation at baseline, had significantly lower expression of miR-122 than patients with the CC genotype, who have low innate stimulation. In IFNL3 CT/TT patients, a higher expression of IFN at baseline could explain both the lower miR-122 expression and high stimulation of the innate response.
Surprisingly, in our study, an increase in hepatic miR-122 expression was associated with a decrease in the HCV viral load at baseline, whereas miR-122 stimulates HCV replication in vitro (21). Moreover, patients with low viral decreases (pNR and NRs) presented the lowest hepatic expression of miR-122 (Fig. 1). In our series, we did not observe dramatic modification of miR-122 expression; the expression varied from 2- to 3-fold. Interestingly, in patients treated with miravirsen, the lowest dose, 3 mg/kg, had very limited antiviral effect (23), suggesting that a stringent inhibition of miR-122 is required to efficiently inhibit HCV replication.
In our study, the expression of miR-122 was increased in IFNL3 CC carriers, who had stronger activation of the innate immune response. Interestingly, even if miR-122 stimulates the expression of HCV, the activation of innate immune activity may be predominant and may result in a reduction of HCV replication.
miR-122 was suggested to inhibit, in vitro, the IFN-stimulated response element activity that controls the expression of interferon-stimulated genes (ISGs) (44). In patients with NR and in IFNL3 CT/TT carriers, ISGs are overexpressed at baseline. It was suggested that, in these patients, ISGs were already maximally induced and the addition of exogenous IFN was not able to activate it further. Therefore, in these patients, reduction of the expression of miR-122 at baseline may participate in increased expression of ISGs (37).
We found no association between hepatic miR-122 and either total cholesterol or triglycerides, whereas miR-122 regulates lipids and cholesterol metabolism (19, 41). In serum, the increased expression of miR-122 was associated with an increase of viral load and ALAT and ASAT values at baseline. Since miR-122 is highly abundant in hepatocytes, the lysis of hepatocytes may result in the release of ASAT and ALAT together with miR-122. However, if miR-122 release was only a consequence of hepatocyte lysis, we would expect to see a strong correlation between hepatic and serum expression of miR-122. In the 80 paired samples, however, hepatic and serum miR-122 were not associated (Fig. 4), and therefore, hepatocyte lysis may not be the only mechanism responsible for the presence of miR-122 in the serum samples.
We found no relationship between the expression of serum miR-122, IFNL3 polymorphism, and response to IFN therapy. However, Su and colleagues described a reduction of baseline serum miR-122 expression in NRs (27). They studied 126 serum samples of patients of Asian ethnicity infected with genotype 1b or 2 (27). Moreover, most of the patients included were SVR (98 versus 28 NR) and carried the IFNL3 CC polymorphism (97 CC versus 21 CT/TT) (27). In our series, different ethnicities (mainly Caucasian and African) were represented, with all HCV genotypes. Moreover, 53% of HCV genotype 1-infected patients were infected by subtype 1b. However, in accordance with our results, Su and colleagues found no significant association between both SVR and cEVR and serum miR-122 in the specific group of HCV genotype 1 patients.
Both serum and hepatic expression of miR-122 were strongly associated with GGT values (P = 0.005 and P = 3.10−6, respectively). Serum miR-122 has been associated with GGT in a previous report (27). Moreover, GGT was reported as an independent predictor of both SVR and clinical outcomes among patients with chronic hepatitis C (45).
Since miR-122 is very abundant in the liver, we tested its association with fibrosis in patients with CHC. In our series of patients, the expression of miR-122 was reduced in patients with F3 and F4 fibrosis compared to its expression in patients with F1 and F2 within all patients with any HCV genotype and HCV genotype 1-infected patients (P = 0.01 and P = 0.04). Moreover, within HCV genotype 1 patients, we found higher hepatic expression of miR-122 in patients with F1 fibrosis than in those with F2 to F4 (P = 0.018) (Fig. 3B). Interestingly, we observed a reduction of the expression of miR-122 during the progression of early stages of fibrosis. The patients tested all showed reduction of miR-122 expression when they progressed to F2, whereas the level of expression of miR-122 in the 5 patients was different at F0 or F1 (Fig. 3C). These 5 patients with historical paired liver biopsy specimens are highly informative, even though the results should be confirmed in a larger population.
The progression of fibrosis induces dramatic modification of the architecture of the liver, and the activation of hepatic stellate cells (HSCs) plays a crucial role during this process (46). A recent report described a reduction of miR-122 in both serum and liver of patients with CHC (32). miR-122 is mainly expressed by hepatocytes, and during fibrosis progression, the number of hepatocytes decreases and the numbers of lymphocytes, Kuppfer cells, and HSCs increase. In our experiment, we studied the expression of miR-122 in total liver cells. Therefore, in the samples from patients with more advanced fibrosis, the amount of miR-122 may be reduced more dramatically than the amount of the reporter miRNA (SNORD44). However, a reduction of miR-122 has been reported in trans-activated HSCs and in the liver of mice treated with CCl4 (31), suggesting that miR-122 plays an important role during fibrosis and that the reduced amount of miR-122 may not be just a consequence of hepatocyte loss at more advanced stages of fibrosis.
In conclusion, we provide an analysis of miR-122 expression in patients with CHC. We describe an increase in the expression of miR-122 in carriers of the IFNL3 CC polymorphism, independent of the response to PEG-IFN plus ribavirin. Our results suggest that miR-122 may be associated with molecular mechanisms regulating responsiveness to PEG-IFN plus ribavirin. Moreover, our results suggest that the reduction of miR-122 expression may play a role during the progression of fibrosis in patients with CHC. Interestingly, our study supports the idea that, in patients infected with HCV, miR-122 activity is not limited to stimulating the replication of the virus but may also regulate molecular mechanisms of IFN responsiveness and fibrosis progression.
Supplementary Material
ACKNOWLEDGMENTS
This project was supported by the French National Agency for Research on AIDS and Viral Hepatitis (ANRS) (grant no. 2011-447). E.E. received a fellowship funded by ANRS (grant NM/NM/no. 3117/CSS7/AO 2011-1).
Footnotes
Published ahead of print 26 March 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JVI.00016-14.
REFERENCES
- 1.Anonymous. 2004. Global burden of disease (GBD) for hepatitis C. J. Clin. Pharmacol. 44:20–29. 10.1177/0091270003258669 [DOI] [PubMed] [Google Scholar]
- 2.Marcellin P, Asselah T, Boyer N. 2002. Fibrosis and disease progression in hepatitis C. Hepatology 36:S47–S56. 10.1053/jhep.2002.36993 [DOI] [PubMed] [Google Scholar]
- 3.Seeff LB. 2002. Natural history of chronic hepatitis C. Hepatology 36:S35–S46. 10.1053/jhep.2002.36806 [DOI] [PubMed] [Google Scholar]
- 4.Craxì A, Pawlotsky JM, Wedemeyer H, Bjoro K, Flisiak R, Forns X, Mondelli M, Peck-Radosavljevic M, Rosenberg W, Sarrazin C, Jacobson I, Dusheiko G, European Association for the Study of the Liver 2011. EASL clinical practice guidelines: management of hepatitis C virus infection. J. Hepatol. 55:245–264. 10.1016/j.jhep.2011.02.023 [DOI] [PubMed] [Google Scholar]
- 5.Asselah T, Marcellin P. 2013. Interferon free therapy with direct acting antivirals for HCV. Liver Int. 33(Suppl 1):93–104. 10.1111/liv.12076 [DOI] [PubMed] [Google Scholar]
- 6.Asselah T, Marcellin P. 2014. Second-wave IFN-based triple therapy for HCV genotype 1 infection: simeprevir, faldaprevir and sofosbuvir. Liver Int. 34(Suppl 1):60–68 [DOI] [PubMed] [Google Scholar]
- 7.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, Urban TJ, Heinzen EL, Qiu P, Bertelsen AH, Muir AJ, Sulkowski M, McHutchison JG, Goldstein DB. 2009. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461:399–401. 10.1038/nature08309 [DOI] [PubMed] [Google Scholar]
- 8.Prokunina-Olsson L, Muchmore B, Tang W, Pfeiffer RM, Park H, Dickensheets H, Hergott D, Porter-Gill P, Mumy A, Kohaar I, Chen S, Brand N, Tarway M, Liu L, Sheikh F, Astemborski J, Bonkovsky HL, Edlin BR, Howell CD, Morgan TR, Thomas DL, Rehermann B, Donnelly RP, O'Brien TR. 2013. A variant upstream of IFNL3 (IL28B) creating a new interferon gene IFNL4 is associated with impaired clearance of hepatitis C virus. Nat. Genet. 45:164–171. 10.1038/ng.2521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamming OJ, Terczynska-Dyla E, Vieyres G, Dijkman R, Jorgensen SE, Akhtar H, Siupka P, Pietschmann T, Thiel V, Hartmann R. 2013. Interferon lambda 4 signals via the IFNlambda receptor to regulate antiviral activity against HCV and coronaviruses. EMBO J. 32:3055–3065. 10.1038/emboj.2013.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thompson AJ, Muir AJ, Sulkowski MS, Ge D, Fellay J, Shianna KV, Urban T, Afdhal NH, Jacobson IM, Esteban R, Poordad F, Lawitz EJ, McCone J, Shiffman ML, Galler GW, Lee WM, Reindollar R, King JW, Kwo PY, Ghalib RH, Freilich B, Nyberg LM, Zeuzem S, Poynard T, Vock DM, Pieper KS, Patel K, Tillmann HL, Noviello S, Koury K, Pedicone LD, Brass CA, Albrecht JK, Goldstein DB, McHutchison JG. 2010. Interleukin-28B polymorphism improves viral kinetics and is the strongest pretreatment predictor of sustained virologic response in genotype 1 hepatitis C virus. Gastroenterology 139:120–129. 10.1053/j.gastro.2010.04.013 [DOI] [PubMed] [Google Scholar]
- 11.Naggie S, Osinusi A, Katsounas A, Lempicki R, Herrmann E, Thompson AJ, Clark PJ, Patel K, Muir AJ, McHutchison JG, Schlaak JF, Trippler M, Shivakumar B, Masur H, Polis MA, Kottilil S. 2012. Dysregulation of innate immunity in hepatitis C virus genotype 1 IL28B-unfavorable genotype patients: impaired viral kinetics and therapeutic response. Hepatology 56:444–454. 10.1002/hep.25647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartel DP. 2004. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 13.Meister G, Tuschl T. 2004. Mechanisms of gene silencing by double-stranded RNA. Nature 431:343–349. 10.1038/nature02873 [DOI] [PubMed] [Google Scholar]
- 14.Gong BD, Xie Q, Xiang XG, Wang L, Zhao GD, An FM, Wang H, Lin LY, Yu H, Bao SS. 2010. Effect of ribavirin and interferon beta on miRNA profile in the hepatitis C virus subgenomic replicon-bearing Huh7 cells. Int. J. Mol. Med. 25:853–859 [DOI] [PubMed] [Google Scholar]
- 15.Peng X, Li Y, Walters KA, Rosenzweig ER, Lederer SL, Aicher LD, Proll S, Katze MG. 2009. Computational identification of hepatitis C virus associated microRNA-mRNA regulatory modules in human livers. BMC Genomics 10:373. 10.1186/1471-2164-10-373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ura S, Honda M, Yamashita T, Ueda T, Takatori H, Nishino R, Sunakozaka H, Sakai Y, Horimoto K, Kaneko S. 2009. Differential microRNA expression between hepatitis B and hepatitis C leading disease progression to hepatocellular carcinoma. Hepatology 49:1098–1112. 10.1002/hep.22749 [DOI] [PubMed] [Google Scholar]
- 17.Murakami Y, Tanaka M, Toyoda H, Hayashi K, Kuroda M, Tajima A, Shimotohno K. 2010. Hepatic microRNA expression is associated with the response to interferon treatment of chronic hepatitis C. BMC Med. Genomics 3:48. 10.1186/1755-8794-3-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elmen J, Lindow M, Silahtaroglu A, Bak M, Christensen M, Lind-Thomsen A, Hedtjarn M, Hansen JB, Hansen HF, Straarup EM, McCullagh K, Kearney P, Kauppinen S. 2008. Antagonism of microRNA-122 in mice by systemically administered LNA-antimiR leads to up-regulation of a large set of predicted target mRNAs in the liver. Nucleic Acids Res. 36:1153–1162. 10.1093/nar/gkm1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R, Subramaniam A, Propp S, Lollo BA, Freier S, Bennett CF, Bhanot S, Monia BP. 2006. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 3:87–98. 10.1016/j.cmet.2006.01.005 [DOI] [PubMed] [Google Scholar]
- 20.Girard M, Jacquemin E, Munnich A, Lyonnet S, Henrion-Caude A. 2008. miR-122, a paradigm for the role of microRNAs in the liver. J. Hepatol. 48:648–656. 10.1016/j.jhep.2008.01.019 [DOI] [PubMed] [Google Scholar]
- 21.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. 2005. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science 309:1577–1581. 10.1126/science.1113329 [DOI] [PubMed] [Google Scholar]
- 22.Lanford RE, Hildebrandt-Eriksen ES, Petri A, Persson R, Lindow M, Munk ME, Kauppinen S, Orum H. 2010. Therapeutic silencing of microRNA-122 in primates with chronic hepatitis C virus infection. Science 327:198–201. 10.1126/science.1178178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez-Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA, Hodges MR. 2013. Treatment of HCV infection by targeting microRNA. N. Engl. J. Med. 368:1685–1694. 10.1056/NEJMoa1209026 [DOI] [PubMed] [Google Scholar]
- 24.Fukuhara T, Kambara H, Shiokawa M, Ono C, Katoh H, Morita E, Okuzaki D, Maehara Y, Koike K, Matsuura Y. 2012. Expression of microRNA miR-122 facilitates an efficient replication in nonhepatic cells upon infection with HCV. J. Virol. 86:7918–7933. 10.1128/JVI.00567-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kambara H, Fukuhara T, Shiokawa M, Ono C, Ohara Y, Kamitani W, Matsuura Y. 2012. Establishment of a novel permissive cell line for the propagation of hepatitis C virus by expression of microRNA miR122. J. Virol. 86:1382–1393. 10.1128/JVI.06242-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W. 2009. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat. Med. 15:31–33. 10.1038/nm.1902 [DOI] [PubMed] [Google Scholar]
- 27.Su TH, Liu CH, Liu CJ, Chen CL, Ting TT, Tseng TC, Chen PJ, Kao JH, Chen DS. 2013. Serum microRNA-122 level correlates with virologic responses to pegylated interferon therapy in chronic hepatitis C. Proc. Natl. Acad. Sci. U. S. A. 110:7844–7849. 10.1073/pnas.1306138110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bochud PY, Bibert S, Kutalik Z, Patin E, Guergnon J, Nalpas B, Goossens N, Kuske L, Mullhaupt B, Gerlach T, Heim MH, Moradpour D, Cerny A, Malinverni R, Regenass S, Dollenmaier G, Hirsch H, Martinetti G, Gorgiewski M, Bourliere M, Poynard T, Theodorou I, Abel L, Pol S, Dufour JF, Negro F. 2012. IL28B alleles associated with poor hepatitis C virus (HCV) clearance protect against inflammation and fibrosis in patients infected with non-1 HCV genotypes. Hepatology 55:384–394. 10.1002/hep.24678 [DOI] [PubMed] [Google Scholar]
- 29.Marabita F, Aghemo A, De Nicola S, Rumi MG, Cheroni C, Scavelli R, Crimi M, Soffredini R, Abrignani S, De Francesco R, Colombo M. 2011. Genetic variation in the interleukin-28B gene is not associated with fibrosis progression in patients with chronic hepatitis C and known date of infection. Hepatology 54:1127–1134. 10.1002/hep.24503 [DOI] [PubMed] [Google Scholar]
- 30.Noureddin M, Wright EC, Alter HJ, Clark S, Thomas E, Chen R, Zhao X, Conry-Cantilena C, Kleiner DE, Liang TJ, Ghany MG. 2013. Association of IL28B genotype with fibrosis progression and clinical outcomes in patients with chronic hepatitis C: a longitudinal analysis. Hepatology 58:1548–1557. 10.1002/hep.26506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li J, Ghazwani M, Zhang Y, Lu J, Fan J, Gandhi CR, Li S. 2013. miR-122 regulates collagen production via targeting hepatic stellate cells and suppressing P4HA1 expression. J. Hepatol. 58:522–528. 10.1016/j.jhep.2012.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Trebicka J, Anadol E, Elfimova N, Strack I, Roggendorf M, Viazov S, Wedemeyer I, Drebber U, Rockstroh J, Sauerbruch T, Dienes HP, Odenthal M. 2013. Hepatic and serum levels of miR-122 after chronic HCV-induced fibrosis. J. Hepatol. 58:234–239. 10.1016/j.jhep.2012.10.015 [DOI] [PubMed] [Google Scholar]
- 33.Asselah T, Bieche I, Laurendeau I, Martinot-Peignoux M, Paradis V, Vidaud D, Valla DC, Bedossa P, Marcellin P, Vidaud M. 2008. Significant gene expression differences in histologically “Normal” liver biopsies: implications for control tissue. Hepatology 48:953–962. 10.1002/hep.22411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bieche I, Asselah T, Laurendeau I, Vidaud D, Degot C, Paradis V, Bedossa P, Valla DC, Marcellin P, Vidaud M. 2005. Molecular profiling of early stage liver fibrosis in patients with chronic hepatitis C virus infection. Virology 332:130–144. 10.1016/j.virol.2004.11.009 [DOI] [PubMed] [Google Scholar]
- 35.Asselah T, Bieche I, Laurendeau I, Paradis V, Vidaud D, Degott C, Martinot M, Bedossa P, Valla D, Vidaud M, Marcellin P. 2005. Liver gene expression signature of mild fibrosis in patients with chronic hepatitis C. Gastroenterology 129:2064–2075. 10.1053/j.gastro.2005.09.010 [DOI] [PubMed] [Google Scholar]
- 36.Asselah T, De Muynck S, Broet P, Masliah-Planchon J, Blanluet M, Bieche I, Lapalus M, Martinot-Peignoux M, Lada O, Estrabaud E, Zhang Q, El Ray A, Vidaud D, Ripault MP, Boyer N, Bedossa P, Valla D, Vidaud M, Marcellin P. 2012. IL28B polymorphism is associated with treatment response in patients with genotype 4 chronic hepatitis C. J. Hepatol. 56:527–532. 10.1016/j.jhep.2011.09.008 [DOI] [PubMed] [Google Scholar]
- 37.Estrabaud E, Vidaud M, Marcellin P, Asselah T. 2012. Genomics and HCV infection: progression of fibrosis and treatment response. J. Hepatol. 57:1110–1125. 10.1016/j.jhep.2012.05.016 [DOI] [PubMed] [Google Scholar]
- 38.Afdhal NH, McHutchison JG, Zeuzem S, Mangia A, Pawlotsky JM, Murray JS, Shianna KV, Tanaka Y, Thomas DL, Booth DR, Goldstein DB. 2011. Hepatitis C pharmacogenetics: state of the art in 2010. Hepatology 53:336–345. 10.1002/hep.24052 [DOI] [PubMed] [Google Scholar]
- 39.Beinhardt S, Aberle JH, Strasser M, Dulic-Lakovic E, Maieron A, Kreil A, Rutter K, Staettermayer AF, Datz C, Scherzer TM, Strassl R, Bischof M, Stauber R, Bodlaj G, Laferl H, Holzmann H, Steindl-Munda P, Ferenci P, Hofer H. 2012. Serum level of IP-10 increases predictive value of IL28B polymorphisms for spontaneous clearance of acute HCV infection. Gastroenterology 142:78–85. 10.1053/j.gastro.2011.09.039 [DOI] [PubMed] [Google Scholar]
- 40.Melis R, Fauron C, McMillin G, Lyon E, Shirts B, Hubley LM, Slev PR. 2011. Simultaneous genotyping of rs12979860 and rs8099917 variants near the IL28B locus associated with HCV clearance and treatment response. J. Mol. Diagn. 13:446–451. 10.1016/j.jmoldx.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elmen J, Lindow M, Schutz S, Lawrence M, Petri A, Obad S, Lindholm M, Hedtjarn M, Hansen HF, Berger U, Gullans S, Kearney P, Sarnow P, Straarup EM, Kauppinen S. 2008. LNA-mediated microRNA silencing in non-human primates. Nature 452:896–899. 10.1038/nature06783 [DOI] [PubMed] [Google Scholar]
- 42.Urban TJ, Thompson AJ, Bradrick SS, Fellay J, Schuppan D, Cronin KD, Hong L, McKenzie A, Patel K, Shianna KV, McHutchison JG, Goldstein DB, Afdhal N. 2010. IL28B genotype is associated with differential expression of intrahepatic interferon-stimulated genes in patients with chronic hepatitis C. Hepatology 52:1888–1896. 10.1002/hep.23912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pedersen IM, Cheng G, Wieland S, Volinia S, Croce CM, Chisari FV, David M. 2007. Interferon modulation of cellular microRNAs as an antiviral mechanism. Nature 449:919–922. 10.1038/nature06205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshikawa T, Takata A, Otsuka M, Kishikawa T, Kojima K, Yoshida H, Koike K. 2012. Silencing of microRNA-122 enhances interferon-alpha signaling in the liver through regulating SOCS3 promoter methylation. Sci. Rep. 2:637. 10.1038/srep00637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Everhart JE, Wright EC. 2013. Association of gamma-glutamyl transferase (GGT) activity with treatment and clinical outcomes in chronic hepatitis C (HCV). Hepatology 57:1725–1733. 10.1002/hep.26203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedman SL. 2010. Evolving challenges in hepatic fibrosis. Nat. Rev. Gastroenterol. Hepatol. 7:425–436. 10.1038/nrgastro.2010.97 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


