ABSTRACT
Epstein-Barr virus (EBV) encodes BPLF1, a lytic cycle protein with deubiquitinating activity that is contained in its N-terminal domain and conserved across the Herpesviridae. EBV replication is associated with cellular DNA replication and repair factors, and initiation of EBV lytic replication induces a DNA damage response, which can be regulated at least in part by BPLF1. The cellular DNA repair pathway, translesion synthesis (TLS), is disrupted by BPLF1, which deubiquitinates the DNA processivity factor, PCNA, and inhibits the recruitment of the TLS polymerase, polymerase eta (Pol eta), after damage to DNA by UV irradiation. Here we showed that the E3 ubiquitin ligase, which activates TLS repair by monoubiquitination of PCNA, is also affected by BPLF1 deubiquitinating activity. First, BPLF1 interacts directly with Rad18, and overexpression of BPLF1 results in increased levels of the Rad18 protein, suggesting that it stabilizes Rad18. Next, expression of functionally active BPLF1 caused relocalization of Rad18 into nuclear foci, which is consistent with sites of cellular DNA replication that occur during S phase. Also, levels of Rad18 remain constant during lytic reactivation of wild-type virus, but reactivation of BPLF1 knockout virus resulted in decreased levels of Rad18. Finally, the contribution of Rad18 levels to infectious virus production was examined with small interfering RNA (siRNA) targeting Rad18. Results demonstrated that reducing levels of Rad18 decreased production of infectious virus, and infectious titers of BPLF1 knockout virus were partially restored by overexpression of Rad18. Thus, BPLF1 interacts with and maintains Rad18 at high levels during lytic replication, which assists in production of infectious virus.
IMPORTANCE Characterization of EBV BPLF1's deubiquitinating activity and identification of its targets and subsequent functional effects remain little studied. All members of the Herpesviridae contain BPLF1 homologs with conserved enzymatic activity, and findings discovered with EBV BPLF1 are likely applicable to other members of the family. Discovery of new targets of BPLF1 will point to cellular pathways and viral processes regulated by the enzymatic activity of the EBV-encoded deubiquitinating enzyme. Here we determined the importance of the cellular ubiquitin ligase Rad18 in these processes and how it is affected by BPLF1. Our findings demonstrate that EBV can co-opt Rad18 as a novel accessory factor in the production of infectious virus.
INTRODUCTION
Epstein-Barr virus (EBV), a human herpesvirus, is one of the most common human viruses, infecting more than 90% of people worldwide. EBV is transmitted through saliva and initially infects the epithelial cells in the oropharyngeal cavity (1–3). As the initial lytic infection is brought under control, EBV, as do all members of the herpesviridae, establishes lifelong latency in secondary target cells, which in the case of EBV are memory B lymphocytes (1, 4). There are three types of EBV latency: I, II, and III, each characterized by expression of a limited set of viral RNAs and gene products. EBV latency types are associated with several distinct malignancies, including immunoblastic lymphomas (type III), Hodgkin lymphoma (type II), and Burkitt lymphoma (type I), as well as nasopharyngeal carcinoma (type II) (5–7). Latent virus can be reactivated periodically into the lytic state, with production of virus, which is asymptomatic except in persons with acquired or inborn immunodeficiency. Both during initial infection and after reactivation of the lytic cycle, the full complement of approximately 90 lytic proteins is expressed. Infectious mononucleosis is the classic disease caused by EBV lytic infection as a result of primary infection during adolescence and young adult years (8).
BPLF1 is the largest EBV protein (3,149 amino acids) and is expressed late in the lytic cycle. However, BPLF1 and its herpesviral homologs can function both early and late in viral infection, since the proteins are located in the tegument, and mRNA levels are detected as early as 6 to 8 h after infection (9–13). BPLF1 has deubiquitinating (DUB) as well as deneddylase activity expressed from its N-terminal domain (14–16). Both enzymatic activities are localized to a catalytic triad, composed of a His, Asp, and Cys residue, that is strictly conserved across the herpesvirus family. Mutation of the catalytic triad results in loss of both deubiquitinating and deneddylating activities (16–19). EBV BPLF1 knockout virus, as well as knockout of herpesvirus homolog genes, results in loss of infectivity (typically 90%) and/or reduces genome copy numbers (17, 20–24). BPLF1 has been shown to block proteasomal degradation of cytosolic and endoplasmic reticulum proteins by removal of ubiquitin from targeted substrates (25). Several major targets have been identified for BPLF1 deubiquitinating activity. The first identified was the large subunit of EBV ribonucleotide reductase, deubiquitination of which downregulates viral ribonucleotide reductase activity; this is the only viral target identified to date (17). We have also found that BPLF1 deubiquitinates the cellular processivity factor, PCNA, the first cellular target identified for BPLF1 deubiquitinating activity, and inhibits the DNA repair process, translesion synthesis (TLS), after UV damage (26). Saito et al. demonstrated that BPLF deubiquitinates TRAF6, which can inhibit NF-κB signaling during lytic infection (24). The Kaposi's sarcoma-associated herpesvirus (KSHV) homolog, Orf64, was shown to decrease RIG-I ubiquitination and reduce RIG-I-mediated interferon (IFN) signaling (27). The deneddylase activity of BPLF1 has been implicated in modulating the activity of cullin-RING ligases (16, 28). These various findings demonstrate important roles for BPLF1 and potentially its homologs in viral replication and infectivity.
Seven lytic gene products have been identified as necessary and sufficient for replication of EBV at oriLyt: BZLF1 (the EBV immediate early transactivator), BALF5 (DNA polymerase), BMRF1 (DNA processivity factor), BALF2 (single-stranded DNA binding protein), BBLF4 (helicase), BSLF1 (primase), and BBLF2/3 (helicase-primase accessory protein) (29–36). While EBV encodes its own proteins that are sufficient for viral DNA replication in vitro, many cellular DNA replication and repair proteins that regulate and enhance viral replication in vivo are associated with viral DNA replication. For example, the EBV DNA polymerase, BALF5, is dependent on chaperone Hsp90 for its localization to the nucleus (37). Kudoh et al. showed that many homologous recombinational repair factors, including Rad51, Rad52, RPA, and the MRN complex (MRE11-RAD50-NBS1), are located in replication complexes and are loaded onto newly synthesized viral genomes, implicating them in viral DNA synthesis (38). Additionally, the mismatch repair proteins MSH2, MSh6, MLH1, and PMS2, along with PCNA and the PCNA clamp-loader complex, are also localized to EBV replication compartments (39). Homologous recombination factors and the MRN complex have also been detected in replication compartments of other members of the herpesviridae (40–45). Our recent work identified TLS repair factors that are also associated with EBV and are specifically affected by the deubiquitinating activity of BPLF1 (26). PCNA, which is loaded onto viral DNA during EBV DNA replication (39), is monoubiquitinated in response to DNA damage and initiates postreplication repair (PRR) through recruitment of the specialized TLS polymerase, polymerase eta (Pol eta), to the site of damage. Deubiquitination of PCNA by BPLF1 abolishes recruitment of pol eta to sites of DNA damage, resulting in inhibition of TLS (26). Here we report on the contributions of the TLS factor Rad18, which is engaged and modulated by BPLF1.
Rad18 (the E3 ubiquitin-ligating enzyme) is part of the E2/E3 ubiquitin complex and is responsible for monoubiquitination of PCNA, which initiates several cellular DNA repair processes (46–50). Rad18 forms a complex with the E2 ubiquitin-conjugating enzyme Rad6 through a conserved RING finger motif (51). Ubiquitin is activated by the E1 ubiquitin-activating enzyme, Ube1, in an ATP-dependent fashion and transfers ubiquitin to the active site cysteine. E2 catalyzes the transfer of ubiquitin from the active site of E1 to the active site of E2. The E3 ubiquitin-ligating enzyme then transfers the ubiquitin through an isopeptide bond that typically links a lysine residue of the target protein to the C-terminal glycine of ubiquitin.
Given that Rad18 monoubiquitinates PCNA, is a member of the TLS DNA damage response pathway, and is itself ubiquitinated, we set out to determine if BPLF1 could directly affect Rad18 ubiquitination and confer a role for Rad18 in the EBV life cycle. Here we report that BPLF1 interacts with the E2/E3 ubiquitin complex (Rad6/18), upregulates Rad18 protein levels, and reorganizes Rad18 into focus-forming units in the nucleus. Finally, we have shown that Rad18 is important for efficient production of infectious virus. Thus, these findings uncover a new function for this cellular ubiquitin E3 ligase and its likely important accessory role in EBV replication and infectivity in the viral life cycle. In light of the conserved nature of the herpesvirus deubiquitinating enzymes, observations on the role of Rad18 in EBV infection are likely to extend to other members of the herpesviridae family.
MATERIALS AND METHODS
Cell line, growth, and transfection.
The H1299, 293T, 293EBV+, and 293EBV+ BPLF1 knockout cell lines (kindly provided by T. Tsurumi [24]) were cultured in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum and antibiotics (26). Lipofectamine 2000 (Invitrogen) and Effectene (Qiagen) were used for transfections according to the manufacturer's protocol.
Antibodies, immunoprecipitations, and immunoblotting.
FLAG-tagged BPLF1 (17) was immunoprecipitated with FLAG M2 antibody bound to magnetic Dynabeads (Invitrogen). Myc-tagged Rad18 was immunoprecipitated with Myc antibody (Santa Cruz), and His-tagged BPLF1 was precipitated with anti-BPLF1 antibody (26). PCNA was detected with PCNA antibody (Santa Cruz Biotechnology), Rad18 was detected with Rad18 antibody (Bethyl Laboratories), Rad6 was detected with anti-Rad6 antibody (Bethyl Laboratories), and FLAG-tagged ubiquitin was detected with FLAG M2 antibody (Sigma). Immunoblotting was performed as done previously (26).
BPLF1, PCNA, Ube1, FLAG-ubiquitin, and RAD6/RAD18 purification.
His-tagged BPLF1 and His-tagged PCNA were purified from Escherichia coli as previously described (26). BL21 Codon Plus DE3-RIPL cells were transformed with the pET22b-Rad6/Rad18 construct. Colonies were selected on LB agar plates containing 100 μg/ml ampicillin. Single colonies were inoculated into 5 ml LB broth containing ampicillin and cultured overnight at 37°C. Overnight preculture was used to inoculate 1 liter of LB broth at 37°C, cultured to an absorbance of 1.0 at 600 nm, and induced with isopropyl-β-d-thiogalactopyranoside (IPTG) with a final concentration of 0.2 mM Cells were grown for 15 to 17 h at 18°C. Cells were harvested by centrifugation and lysed by sonication in lysis buffer (50 mM NaH2PO4 [pH 8.0], 300 mM NaCl, 10 mM imidazole, 2 μM ZnCl2, 5 mM β-mercaptoethanol, and protease inhibitors). Cleared lysate was incubated with 1 ml nickel-nitrilotriacetic acid (Ni-NTA) resin and further purified as described previously (52). Human ubiquitin-activating enzyme (Ube1) and FLAG-ubiquitin were from R&D Systems.
In vitro competition assay.
BPLF1 (10 nM) was incubated with various amounts of PCNA (10 to 100 nM) and Rad6/Rad18 (10 to 100 nM) at room temperature for 2 h and then subjected to immunoprecipitation with BPLF1 antibody and probed with Rad18, BPLF1, and PCNA antibodies.
In vitro deubiquitination of Rad6.
Human Ube1 (100 μm) was preincubated with FLAG-ubiquitin (10 μM) in ubiquitination buffer (25 mM Tris-HCl [pH 8.0], 150 mM NaCl, 2 μM ZnCl2, 10 mM MgCl2, 5 mM beta-mercaptoethanol [β-ME], and 10 mM ATP) at room temperature for 10 min. Rad6/Rad18 (1 μM) and BPLF1 (100 nM) were added, and the samples were incubated at 30°C for 2 h. Samples were boiled in SDS-PAGE loading buffer and run on a 12% SDS-PAGE. Western blotting was performed and probed with FLAG, BPLF1, and Rad6 antibodies.
Immunofluorescence and visualization of yellow fluorescent protein-tagged Rad18 (YFP-Rad18) foci.
Rad18 was cloned into the monomeric pEYFP-N1 plasmid, and the construct was confirmed by DNA sequencing. H1299 cells were either transfected with pEYFP-N1-Rad18 or cotransfected with pEYFP-N1-Rad18 and the first 246 N-terminal amino acids of BPLF1, FLAG tagged (FLAG-BPLF1 1-246). Twenty-four hours later, cells were washed with ice-cold phosphate-buffered saline solution (PBS) and fixed with 4% paraformaldehyde for 5 min. The cells were washed three times with PBS, permeabilized with 0.1% Triton X-100 at room temperature for 2 min, and then washed three times with PBS. After blocking with normal donkey serum at room temperature for 2 h, cells were probed with primary FLAG antibody (1:1,000; mouse) (clone M2; Sigma), incubated with Alexa Fluor 594 mouse antibody, stained with 4′,6′-diamidino-2-phenylindole (DAPI), and examined under an Olympus IX 81-ZDC inverted fluorescence microscope (40× magnification).
Wild-type (WT) EBV and BPLF1 knockout cell lines (24, 53) were used to study Rad18 focus formation under lytic conditions. Cells were transfected with YFP-Rad18 and the EBV transactivator, BZLF1 (Z), to induce the lytic phase. Twenty-four hours later, cells were processed as described above and probed with primary EA-D antibody (1:100; mouse) to monitor viral induction.
Rad18 siRNA, viral reactivation, infectivity assays, and extracellular genome copies.
Small interfering RNA (siRNA), previously demonstrated to reduce endogenous protein levels (54, 55), against Rad18, was purchased from Thermo Fisher. siGENOME nontargeting siRNA no. 1 was used as a control. The 293EBV and 293EBV+ BPLF1 knockout cell lines contain a green fluorescent protein (GFP)-tagged viral genome and were transfected with siRNA twice: 24 h prior to viral induction and at the time of induction with BZLF1. 293 cells containing the EBV genome (293EBV+) and the BPLF1 knockout cell line (293EBV+ BPLF1) were reactivated into the viral replicative cycle by transfection of the viral transactivator BZLF1. Upon reactivation, these cells produce the full complement of EBV lytic genes. Forty-eight hours after lytic induction, supernatants and cells were collected for Western blotting, genome copy number determination, and infectivity studies. For infectivity assays, supernatant fluids were cleared and placed on Raji cells. Forty-eight and seventy-two hours after infection, GFP-positive Raji cells were detected by flow cytometry and used as a measure for viral infectivity. Supernatant fluids used for determining extracellular genome copies were treated with DNase before DNA extraction using the DNeasy Blood and Tissue kit (Qiagen) to eliminate DNA not encapsidated in DNase-resistant viral particles. Extracellular genome copies were determined by real time-PCR as described elsewhere, with probes targeting the BamHI repeat region (17).
RESULTS
BPLF1 interacts with Rad18 in cell culture.
In previous work, we demonstrated that BPLF1 physically interacts with and deubiquitinates PCNA and thereby downregulates translesion synthesis (TLS) (26). In this work, since the balance between ubiquitination and deubiquitination is a dynamic process, we examined if the E3 ubiquitin ligase, Rad18, responsible for monoubiquitin of PCNA, could itself be affected by BPLF1. We explored whether BPLF1 inhibits the function of the E2/E3 ubiquitin complex (Rad6/Rad18) responsible for PCNA ubiquitination and activation of TLS. Since cellular replication and repair proteins are detected at sites of EBV replication (38, 39), we explored whether Rad18 is involved in viral replication and infectivity through its association with BPLF1. First, to determine if there is physical interaction between BPLF1 and the Rad6/Rad18 complex, catalytically active FLAG-tagged BPLF1 1-246 (17) was overexpressed in H1299 cells, and lysates were immunoprecipitated with FLAG antibody (Sigma), separated on a 10% SDS-PAGE gel, and probed for endogenous Rad18. Rad18 coprecipitated with FLAG-tagged BPLF1 1-246, thus showing that interaction between BPLF1 1-246 and Rad18 could occur in cells (Fig. 1A). PCNA served as a positive control for interaction with BPLF1 (input levels are indicated in the lower panels of Fig. 1A).
FIG 1.
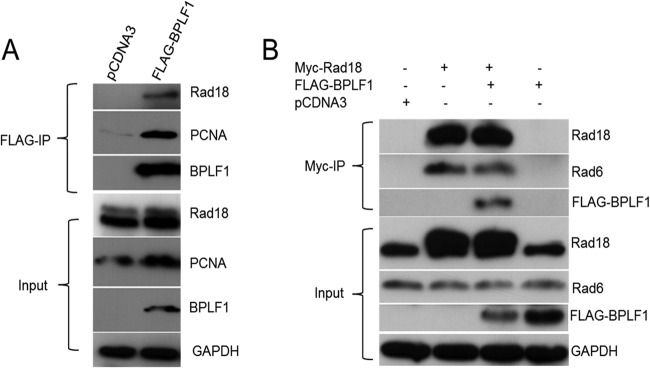
Rad18 interacts with BPLF1 in vivo. (A) H1299 cells were transfected with FLAG-tagged BPLF1 1-246. Lysates were immunoprecipitated (IP) with anti-FLAG antibody, followed by immunoblotting with FLAG, Rad18, and PCNA antibodies. Whole-cell lysates were also probed to indicate input levels. (B) Reverse immunoprecipitations were performed to confirm BPLF1 interaction with Rad18. H1299 cells were cotransfected with BPLF1 1-246 and Myc-tagged Rad18. Immunoprecipitations were performed with Myc antibody, followed by immunoblot analysis with Myc, FLAG, and HA antibodies. Whole-cell lysates were also probed.
In a reverse immunoprecipitation in which Myc-tagged Rad18 (56) was overexpressed in the presence of BPLF1 1-246 and hemagglutinin (HA)-tagged PCNA (57) in H1299 cells, Rad18 was immunoprecipitated with Myc antibody, and the immunoblot was probed for BPLF1 (Fig. 1B). BPLF1 was detected only when it and Myc-tagged Rad18 were coexpressed. Rad6, in addition to Rad18, was also detected in the immunoprecipitate, which was expected because physical association of Rad6 with Rad18 is required for stability of Rad18 (58). The lower panels of Fig. 1 indicate protein levels in lysates. These data demonstrate that BPLF1 interacts with the Rad6/18 complex either directly or through a complex involving other cellular proteins.
BPLF1 interacts with Rad18 in vitro.
We next investigated if interactions of Rad6/18 with BPLF1 are direct and independent of PCNA or are mediated by other cellular factors with the use of purified proteins in vitro. Since Rad18 is not stable in the absence of Rad6 (58), both proteins (48) were coexpressed and purified from E. coli. When purified BPLF1 1-246 (17) and Rad6/18 protein complex were coincubated, interaction of BPLF1 with both Rad6 and Rad18 was detected, demonstrating a direct interaction (Fig. 2A, lanes 1 and 3). PCNA interacted with BPLF1 as expected and served as a positive control (Fig. 2A, lane 1 and 2). The findings indicate not only that Rad6/18 complex and BPLF1 can interact directly but that the interaction can occur in the absence of PCNA or other cellular proteins.
FIG 2.
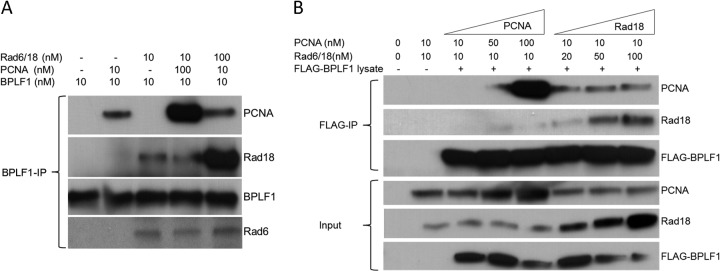
Rad18 interacts with BPLF1 but does not compete with PCNA for a common binding site on BPLF1. (A) BPLF1 interacts with Rad18 in vitro. BPLF1 and Rad6/Rad18 protein complex expressed and purified from E. coli were incubated together at equimolar concentrations (lanes 1 to 3) at room temperature for 1 h and then immunoprecipitated with anti-BPLF1 antibody, and Western blots were probed with Rad18, PCNA, and BPLF1 antibodies. Results showed that BPLF1 interacts with Rad18 independently of PCNA. Additionally, the BPLF1 protein (10 nM) was incubated with either Rad18 (10 nM) and 10-fold excess of PCNA (100 nM) (lane 4) or PCNA (10 nM) and RAD18 (100 nM) (lane 5) and subjected to immunoprecipitation with BPLF1 antibody. (B) PCNA and Rad18 do not compete for a common binding site on BPLF1. A semi-in vivo assay was used. FLAG-BPLF1 was expressed in H1299 cells, and whole-cell lysates were incubated with increasing concentrations of purified PCNA and Rad18 at room temperature for 1 h. Reaction mixtures were immunoprecipitated with FLAG antibody and probed for PCNA, Rad18, and BPLF1. Results indicate no competition between Rad18 and PCNA for BPLF1 binding.
PCNA and Rad18 do not compete for binding to BPLF1.
Since BPLF1 interacts with both PCNA and Rad18, we tested whether there is competition between PCNA and Rad18 for binding to BPLF1. Competition assays were performed with the concentration of BPLF1 held constant and Rad6/18 and PCNA concentrations either held constant or adjusted to 10-fold molar excess. Immunoprecipitations were performed as before (Fig. 2A, lanes 3 and 4). Molar excess of Rad6/18 did not decrease binding of PCNA to BPLF1 (Fig. 2A, lanes 2 and 5), nor did excess PCNA decrease binding of Rad18 to BPLF1 (Fig. 2A, lanes 3 and 4). These results indicate there is little or no competition between Rad18 and PCNA for binding to BPLF1. Had competition been observed, binding of the less-dominant partner to BPLF1 1-246 would have been reduced. Thus, the binding sites on BPLF1 for PCNA, which binds through the PIP (PCNA-interacting peptide) domain of BPLF1 (26), and Rad18 are different and function independently.
These results were substantiated by incubating BPLF1 1-246 containing whole-cell lysates from H1299 cells with graduated amounts of purified PCNA and Rad18 (Fig. 2B). An increase in the PCNA protein concentration did not affect binding of BPLF1 to Rad18, nor did an increase in the Rad18 protein concentration affect binding of BPLF1 to PCNA. Binding of PCNA to BPLF1 was not detected with a 10 nM concentration of PCNA, but an increase in the concentration of Rad18 from 10 nM to 20, 50, and 100 nM enhanced binding of PCNA to BPLF1 (Fig. 2B, compare the top panel, lane 3, to lanes 6 to 8). Together, these data not only substantiate the noncompetitive nature of binding of PCNA and Rad18 to BPLF1 but suggest cooperativity, since increasing Rad18 levels results in increased binding of PCNA to BPLF1.
BPLF1 deubiquitinates Rad6/18 complex.
The cascade of enzymatic reactions that culminates in ubiquitination of the target protein proceeds in an ATP-dependent manner. Binding of BPLF1 with the E2/E3-conjugating and -ligating enzymes (Rad6/Rad18 complex) offered the opportunity to probe the effects of BPLF1 deubiquitinating activity on this complex. We incubated purified Ube1, ubiquitin, and Rad6/Rad18 with or without BPLF1, used FLAG antibody to detect ubiquitinated products, and found that Ube1, Rad6, and Rad18 were ubiquitinated when BPLF1 was absent (Fig. 3A). Addition of purified BPLF1 to the reaction mixture resulted in deubiquitination of Rad6 and to a lesser extent Rad18 (Fig. 3A, lanes 3 and 4). Ube1 was not deubiquitinated, suggesting that deubiquitination of Rad6/18 by BPLF1 is specific (Fig. 3A, lanes 1 and 2). These results indicate that functional activity of the Rad6/18 complex is inhibited by BPLF1 and suggest that all downstream targets of Rad6/18 ubiquitination would be subsequently inhibited by the enzymatic activity of the viral DUB.
FIG 3.
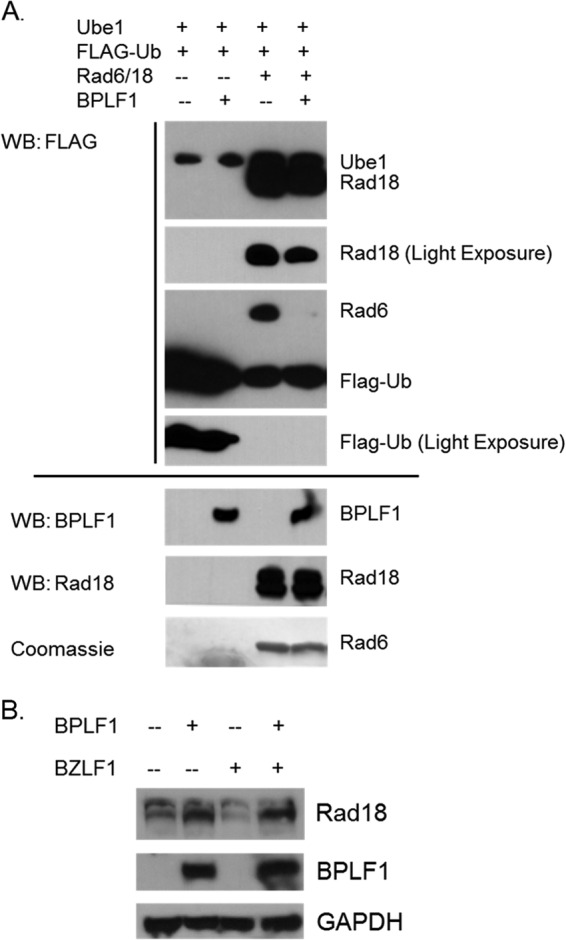
BPLF1 deubiquitinates the Rad6/Rad18 complex. (A) BPLF1 deubiquitinates Rad6/18 in vitro. Ube1 was incubated with Rad6/Rad18 and FLAG-ubiquitin in the presence or absence of BPLF1. Samples were run on a 12% SDS-PAGE gel, immunoblotted, and probed with FLAG, Rad18, and BPLF1 antibody. BPLF1 specifically deubiquitinates Rad6 but not Ube1. Slight deubiquitination of Rad18 was also observed. Input levels are shown at the bottom. (B) Rad18 is likely deubiquitinated by BPLF1 in vivo. 293EBV+ cells were transfected with BPLF1 1-246 and BZLF1 (to induce lytic replication) where indicated. The upper band in the Rad18 panel is consistent with the monoubiquitinated form of Rad18 and appears to be reduced in the presence of BPLF1. BPLF1 and GAPDH loading controls are shown in the middle and bottom panels.
Results shown in Fig. 3B suggest that BPLF1 partially deubiquitinates Rad18 in vivo. 293EBV+ cells are a GFP-tagged EBV-containing cell line in which viral replication can be reactivated by the EBV immediate early transactivator, BZLF1, to induce the lytic cycle and produce infectious virus. 293EBV+ cells were transfected with BPLF1 and BZLF1 as appropriate. When BPLF1 was expressed, either with or without induction of the lytic cycle, the amount of the ubiquitinated form (Fig. 3B, upper band) of Rad18 relative to that of the nonubiquitinated form was reduced (Fig. 3B, compare lane 1 to lane 2 and lane 3 to lane 4). These results suggest that BPLF1 1-246 partially reduces Rad18 ubiquitination in vivo, in agreement with in vitro results presented in Fig. 3A. Rad6 ubiquitination was not detected under the in vivo conditions tested.
BPLF1 increases Rad18 focus formation.
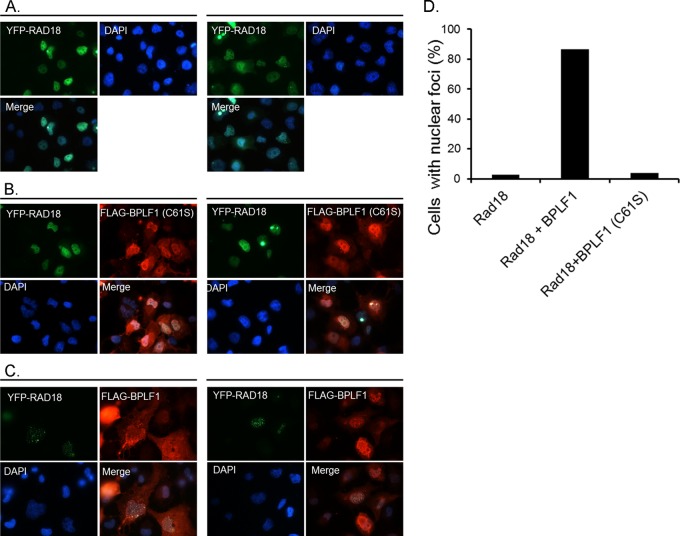
Normally, Rad18 is localized to the nucleus and is found in replication foci during the S phase of the cell cycle (59). To investigate the effect of BPLF1 expression on localization of Rad18, we transiently transfected H1299 cells with YFP-Rad18 alone or with BPLF1 1-246 and BPLF1 C61S (Fig. 4). Rad18 when expressed alone localized predominantly in the nucleus and distributed diffusely, as reported elsewhere (Fig. 4A) (59, 60). However, when YFP-Rad18 and BPLF1 were coexpressed, there was relocalization of Rad18 into distinct nuclear foci in about 85% of cells, compared with 2 to 3% of cells not expressing BPLF1 (Fig. 4B and D). We observed not only a BPLF1-dependent increase in formation of Rad18 foci but also a marked increase in the size of the nucleus in the BPLF1-expressing cells, in agreement with a previous report (16). When the active-site mutant of BPLF1 (C61S) was coexpressed with Rad18, neither Rad18 focus formation nor an increase in nuclear size was observed (Fig. 4C). We conclude that the enzymatic activity is responsible for the formation of Rad18 foci. The Rad18 foci observed are consistent with those seen at cellular replication forks, which suggests, interestingly, that BPLF1 induces localization of Rad18 to sites of active viral replication.
FIG 4.
Expression of BPLF1 increases Rad18 focus formation. Two separate fields of view are shown for each condition (left panels versus right panels [A-C]). (A) H1299 cells transfected with YFP-Rad18 are homogeneously distributed and are localized primarily in the nucleus. (B) H1299 cells coexpressing the inactive BPLF1 C61S and YFP-Rad18 do not affect focus formation. (C) H1299 cells coexpressing FLAG-BPLF1 and YFP-Rad18 show increased nuclear focus formation of Rad18. (D) Quantification of percentages of cells with focus formation in panels A, B, and C. One hundred cells were counted for each sample.
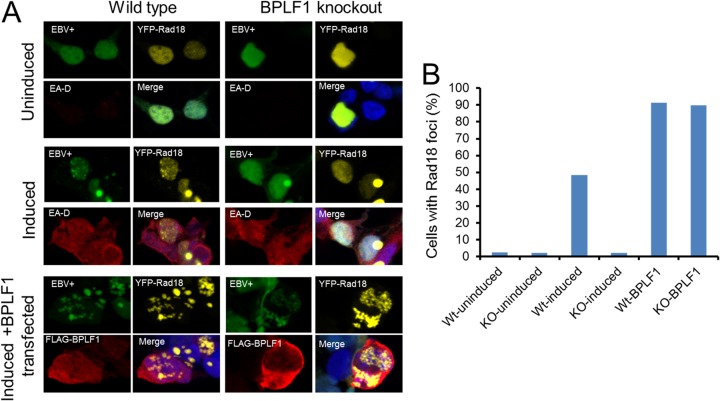
To confirm this important observation, we repeated these experiments with wild-type EBV and BPLF1 knockout cell lines (24, 53). The BPLF1 knockout cell line and virus, generously provided by Tsurumi and colleagues, was constructed by replacing BPLF1 nucleotides 1 through 975 with neomycin and streptomycin resistance genes, which effectively eliminates the deubiquitinating activity contained within its first 205 amino acids (24). Wild-type and DUB knockout EBV-containing 293 cells were induced into the viral lytic phase, in which the full complement of lytic genes is expressed, by transfection with the immediate early EBV transactivator, BZLF1 (Z), along with YFP-Rad18. After 24 h, immunofluorescence studies revealed that ∼50% of cells contained YFP-Rad18 nuclear foci (Fig. 5A) in cells in which the early lytic protein, EA-D, was also detected. The results showed that endogenous levels of BPLF1 can induce formation of Rad18 foci (Fig. 5A, upper and middle panels). Upon overexpression of BPLF1 in reactivated wild-type EBV-containing 293 cells, there was a ∼90% increase of YFP-Rad18 foci (Fig. 5A, lower panels). These results are consistent with those obtained in H1299 cells (Fig. 4). Finally, complementation experiments were performed with BPLF1 knockout virus. Reactivated BPLF1 knockout virus resulted in virtually no Rad18 focus formation; however, complementation with exogenously expressed BPLF1 resulted in ∼90% of cells containing Rad18 foci (Fig. 5A, middle- and lower-right panels). Moreover, complementation with BPLF1 C61S in BPLF1 knockout cells resulted in no appreciable increase in cells containing YFP-Rad18 foci (data not shown). Twenty cells under each condition were examined for the presence of Rad18 foci, and the percentages of cells containing Rad18 foci are quantitated in Fig. 5B. These data demonstrate that endogenous levels of BPLF1 can induce Rad18 nuclear focus formation and that functional enzymatic activity of BPLF1 is required, as well as suggesting that Rad18 localizes to sites of viral replication during lytic reactivation.
FIG 5.
Endogenous BPLF1 expression induces Rad18 focus formation. (A) 293EBV+ WT and 293EBV+ BPLF1 knockout cell lines were transfected with YFP-Rad18 (upper panels), additionally transfected with BZLF1 to induce lytic replication (middle panels), and induced and supplied with BPLF1 exogenously (lower panels). Induction of WT virus produced Rad18 foci but not BPLF1 knockout virus. Cell lines containing the EBV genomes are GFP tagged. EA-D (red) is an early lytic gene and demonstrates viral reactivation. (B) Quantitation of cells containing Rad18 foci. Twenty cells under each condition were examined for the presence of Rad18 foci.
BPLF1 expression increases levels of Rad18.
Since BPLF1 interacts with Rad18, we examined whether BPLF1 could affect Rad18 protein levels. 293EBV+ cells were transfected with BPLF1, and total lysates were probed for endogenous levels of Rad18. Overexpression of BPLF1 resulted in increased levels of Rad18 (Fig. 6A) whether or not the lytic cycle was induced with BZLF1 (top panel, lanes 2 and 4). These results indicate that BPLF1 regulates Rad18 levels. Interestingly, Rad6 levels remained largely unchanged. We also investigated protein levels in 293T cells (Fig. 6B) and again observed that the presence of functional BPLF1 resulted in increased Rad18 levels; however, enzymatically inactive BPLF1 C61S did not increase Rad18 levels. These results suggest that BPLF1 indirectly affects Rad18 protein levels through its deubiquitinating activity, either by stabilizing Rad18 or by increasing its expression.
FIG 6.
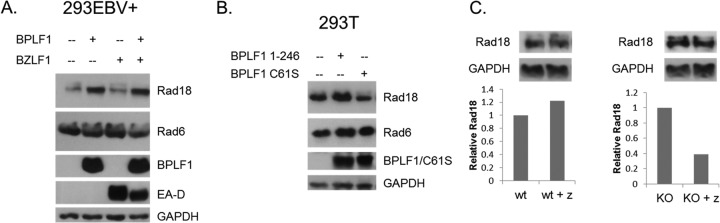
BPLF1 expression increases Rad18 protein levels. (A) 293EBV+ cells were transfected with BPLF1 1-246, and the lytic cycle was induced with BZLF1. Immunoblotting revealed a substantial increase in Rad18 protein levels when BPLF1 was expressed. Rad6 levels remained largely unchanged. EA-D is an EBV early protein that indicates induction of the lytic program. (B) 293T cells were transfected with functional (BPLF1 1-246) or nonfunctional (BPLF1 C61S) BPLF1. Rad18 levels were increased only in the presence of functional BPLF1. (C) Induction of BPLF1 knockout virus results in less Rad18 protein levels than with the WT. WT and BPLF1 knockout viruses were induced into the lytic replication cycle by transfection of BZLF1. Levels of Rad18 were normalized to GAPDH protein levels.
To determine if upregulation of Rad18 protein levels is important during viral replication, 293EBV+ cells, which express the full complement of lytic proteins upon reactivation, and 293EBV+ BPLF1 KO cells, where the sequence encoding the N-terminal region of BPLF1 was deleted (generous gift of T. Tsurumi) (24), were induced with the viral lytic transactivator BZLF1. Endogenous levels of BPLF1 expressed during EBV lytic replication keep protein levels of Rad18 high during viral reactivation (Fig. 6C). Upon reactivation of WT virus, Rad18 levels remained fairly constant in relation to cellular glyceraldehyde-3-phosphate dehydrogenase (GAPDH) protein levels. However, induction of the EBV BPLF1 KO virus decreased levels of the Rad18 protein by approximately 50% (Fig. 6C, upper panels). The graphs below the blots show the relative Rad18 protein levels when normalized to GAPDH levels in the cell lysates. These data suggest that endogenous levels of BPLF1 are sufficient and perhaps necessary to maintain high levels of Rad18 during induction of WT virus, which would otherwise be reduced in the absence of BPLF1. Miyase et al. (61) showed that Rad18 polyubiquitination could direct its degradation by the proteasome in vitro, which suggests that the increase in Rad18 levels could be due to rescue of Rad18 by BPLF1 from proteasomal degradation; however, in our studies, we did not detect polyubiquitinated forms of Rad18.
Silencing of endogenous Rad18 by siRNA does not affect extracellular genome copy number.
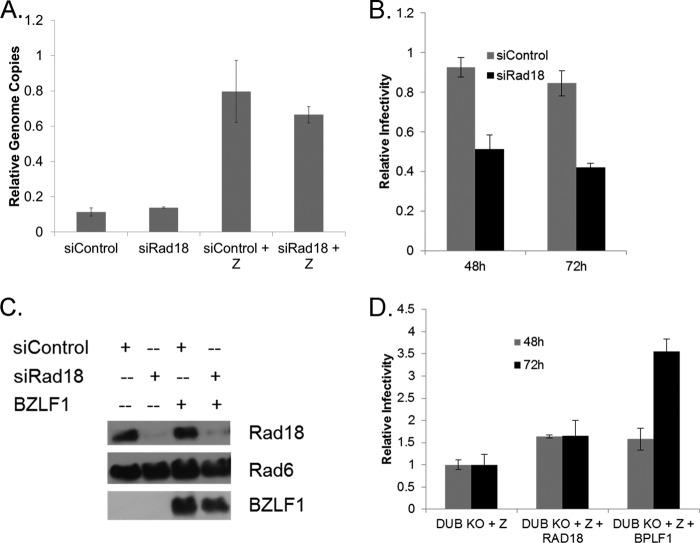
Finally, the effects of Rad18 on viral replication and infectivity were investigated by transfecting 293EBV+ cells with siRNA against Rad18 24 h prior to induction of the lytic cycle. Forty-eight hours after induction, supernatant fluids were collected, cleared, and used for determination of the number of extracellular genome copies. EBV infectious units were assayed by infection of Raji cells (62). Before extraction of DNA from supernatant fluids, samples were treated with DNase to eliminate nonencapsidated DNA. EBV particles are DNase resistant, and extracellular genome copy numbers represent viral genomes packaged into virions. The cell pellet was also collected and used for Western blotting. Levels of extracellular EBV genome copies were similar when Rad18 levels were reduced with siRNA (Fig. 7A). It should be noted that cellular DNA replication is halted upon induction of the EBV lytic cycle, so that effects on cellular DNA should be negligible, and the observed effects are likely to be specific to viral replication processes and production of viral genomes.
FIG 7.
Rad18 levels contribute to viral infectivity. Knockdown of Rad18 levels with siRNA reduces viral infectivity (A to C). (A) Extracellular genome copies. 293EBV+ cells were treated with siRNA against Rad18 and induced with BZLF1 for 48 h. DNA was extracted from supernatant fluids, and genome copies were detected with quantitative PCR (qPCR) targeting the BamHI repeat region. (B) Viral infectivity. The same supernatant fluids collected for assay of extracellular genome copy number were assayed for infectious titer on Raji cells. Infectivity was determined with flow cytometry 48 and 72 h after infection. (C) Immunoblots showing knockdown of Rad18. (D) Viral infectivity of BPLF1 knockout virus is partially restored by overexpression of Rad18. BPLF1 knockout virus was induced with Z and assayed for infectivity on Raji cells. Rad18 and BPLF1 were overexpressed as indicated. Experiments were performed in triplicate, and error bars represent the standard errors of means.
Rad18 is necessary for efficient production of infectious virus.
To assay for viral infectivity, Raji cells were inoculated with supernatant fluids containing EBV virions from 293EBV+ cells treated with control or Rad18 siRNA, and infection continued for 48 and 72 h. Infectivity was measured by detection of GFP-tagged EBV genomes in Raji cells by flow cytometry. Figure 7B shows that reduction of Rad18 protein levels by siRNA decreases viral infectivity both at 48 and 72 h after infection. While the amounts of genome-containing DNase-resistant EBV particles released into the supernatant fluid from siRNA Rad18-treated and untreated cell lines are similar, approximately 50% fewer virions produced from the siRNA Rad18-treated cell lines were infectious. This result suggests that viral genome copies, which are still produced in similar numbers and are encapsidated and egress efficiently as DNase-resistant viral particles, may contain damaged viral DNA due to the loss of Rad18 and result in decreased infectivity. Efficient knockdown of Rad18 was achieved with siRNA, and protein levels of Rad6 were largely independent of Rad18 levels (Fig. 7C). The data demonstrate that Rad18 has a significant impact on EBV infectivity.
Rad 18 overexpression partially restores loss of infectivity of the DUB activity knockout virus.
Since knockdown of Rad18 resulted in decreased WT EBV infectivity and the presence of BPLF1 increases Rad18 protein levels and focus formation, we investigated if overexpression of Rad18 could overcome deficiencies in infectivity observed with the BPLF1 knockout virus (24) (Fig. 7D). Cells containing the EBV BPLF1 knockout virus were induced with the viral transactivator BZLF1 alone or in the presence of exogenously supplied Rad18 or BPLF1. Figure 7D reveals that complementation with exogenous Rad18 during the productive cycle of the knockout virus increased infectious virus production by approximately 50%, a result that is quite consistent with the decrease in infectivity produced with knockdown of Rad18 with siRNA (Fig. 7B). Exogenous expression of BPLF1 resulted in similar levels of infectious titer rescue at 48 h and a marked further restoration of infectious titer at 72 h. These data illustrate that Rad18, through its interaction and regulation by BPLF1, partially compensates for deficiencies in infectivity produced by the loss of BPLF1 expression and further demonstrate that Rad18 enhances viral infectivity. The results identify an important and previously unknown role of the Rad6/18 complex in EBV replication and infectivity.
DISCUSSION
Deletion of conserved herpesvirus deubiquitinating genes or disruption of the catalytic triad results in a reduction of either viral genome copies or infectivity by approximately 90% (17, 20–24). The importance of BPLF1 during EBV infection is expanding as more of its targets and functional effects are discovered. The only viral target discovered to date for BPLF1 is the EBV ribonucleotide reductase (RR), in which deubiquitination of its large subunit results in a decrease of RR activity (17). In addition to PCNA, TRAF6 and cullin ring ligases have now been identified as cellular targets for BPLF1 (16, 24, 28).
Additionally, reactivation of EBV lytic replication elicits a cellular DNA damage response, specifically the ataxia telangiectasia-mutated (ATM) pathway (63, 64), which in part may be regulated by BPLF1 (26). The effects of BPLF1 on cellular DNA repair processes are significant. We demonstrated previously that BPLF1 aborts TLS repair by deubiquitinating the monoubiquitinated form of PCNA, which initiates DNA repair and results in a sharp decrease in recruitment of the repair polymerase, pol eta, to sites of cellular DNA damage (26). We hypothesized that these repair factors may also be involved in viral replication.
In this report, we examine another DNA repair factor, Rad18, which is responsible for monoubiquitinating PCNA and activating TLS, following observations that there was a modest increase in Rad18 levels when BPLF1 was overexpressed under conditions that damage DNA. We first evaluated effects of BPLF1 on Rad18 and then investigated Rad18's subsequent effects on infectivity in the absence of exogenously induced DNA damage.
We found that BPLF1 interacts physically with endogenous levels of Rad18 in vivo. However, since Rad18 and PCNA both interact with BPLF1, we then examined if the interaction between the Rad18 and BPLF1 proteins was direct with the use of purified proteins in vitro. These studies demonstrated that BPLF1 and Rad18 can interact directly, that the interaction is not dependent on the presence of PCNA, and that Rad18 must not compete with PCNA for binding to the conserved PIP (PCNA-interacting peptide) site (26) on BPLF1, since increasing the amounts of either Rad18 or PCNA did not inhibit binding of either protein to BPLF1. Conversely, when BPLF1-containing lysates were incubated with increasing concentrations of PCNA, an increase in Rad18 binding to BPLF1 was detected (Fig. 2B). Neither binding of PCNA nor Rad18 to BPLF1 was detected under conditions of short exposure when both Rad18 and PCNA were held at 10 nM concentrations. However, when Rad18 concentrations were increased to 20, 50, and 100 nM, binding of PCNA to BPLF1 increased, suggesting that some level of cooperativity may occur. Conversely, increased levels of PCNA did not result in increased binding of Rad18 to BPLF1.
The in vitro ubiquitination/deubiquitination experiments utilizing the E1-activating enzyme, Ube1, and the Rad6/18 E2- and E3-conjugating and ligating enzymes, respectively, demonstrated that the Rad6/18 complex, which is autoubiquitinated, is deubiquitinated by BPLF1. The deubiquitination of Rad6/18 appears to be specific, since the ubiquitinated form of Ube1 was not affected by BPLF1. While Rad18 was only partially deubiquitinated under the conditions tested, Rad6 was almost completely deubiquitinated. This is significant because the ubiquitin covalently linked to the active site of Rad6 is transferred by Rad18 to the target protein. In this manner, BPLF1 can inhibit PCNA ubiquitination by stripping the ubiquitin from the E2-conjugating enzyme, thereby making it unavailable for transfer to PCNA. These findings suggest a new mechanism by which BPLF1 can inhibit TLS, in addition to its inhibition by direct deubiquitination of PCNA, which we reported previously.
Apart from TLS, additional targets of Rad6 activity may be regulated by BPLF1's ability to remove monoubiquitin from Rad6. Rad6 can directly ubiquitinate substrates and regulate important cellular processes in the absence of Rad18. Rad6 is highly conserved in eukaryotic evolution and has two isoforms with approximately 95% identity, Rad6A and Rad6B (26). There is a requirement for at least one functional form in somatic cell types (65). Rad6 is responsible for ubiquitinating the histones H2A and H2B, as well as β-catenin, resulting in β-catenin stabilization in the absence of E3 ubiquitin ligases (66–69). Rad6 also forms a complex with p53 and p14ARF, both tumor suppressor genes, and mediates ubiquitination of p53 (70). Our finding of deubiquitination of Rad6 by BPLF1 introduces these targets of Rad6 activity as new avenues for study, and we are now beginning to explore the effects of BPLF1 deubiquitination of Rad6 on histone modification.
We also showed that BPLF1 expression caused relocalization of Rad18 to discrete nuclear foci, that the focus formation was dependent on enzymatically active BPLF1, and that endogenous levels of BPLF1 were sufficient to induce Rad18 focus formation This result is in agreement with published data that Rad18 foci are formed during S phase and is relevant because BPLF1 induces an S-phase-like state which results in increased nuclear size and DNA content (16). We also demonstrate that the presence of functional BPLF1 results in increased levels of Rad18 (Fig. 6). Significant changes in Rad18 protein levels have been observed during cell cycle progression, with levels highest during S phase (54). These results would be consistent with BPLF1 inducing an S-phase-like environment favorable for viral replication. Rad18 mRNA levels were found to fluctuate during cell cycle progression as well (60).
Here we have shown that protein levels of Rad18 are clearly increased in the presence of BPLF1, and endogenous levels themselves contribute to increased Rad18 expression. The data show that viral reactivation in the absence of BPLF1 utilizing the BPLF1 knockout virus results in decreased Rad18 protein levels. However, when BPLF1 is expressed endogenously in the context of WT virus, Rad18 protein levels remain high, suggesting that high levels of Rad18 are important for EBV replication. BPLF1 may influence Rad18 protein levels by contributing to its stability or by upregulation of transcriptional or translational activities or both. The E2F family of transcription factors is activated by a variety of intracellular and extracellular signals, and E2F levels increase as EBV lytic replication proceeds (64, 71). E2F3 associates with the Rad18 promoter and regulates its activity (71), and perhaps BPLF1 affects this activity during EBV infection.
Last, we have demonstrated that Rad18 is important for viral infectivity. Knockdown of Rad18 did not significantly reduce the number of extracellular genome copies; however, infectivity was significantly decreased, by ∼50%. Additionally, a decrease in production of infectious virus by reactivation of BPLF1 knockout virus could be partially restored by overexpression of Rad18. This result confirms the idea that Rad18 is indeed utilized by the virus for efficient replication of functional virus and that the levels of Rad18 are upregulated and maintained during lytic infection by BPLF1. The mechanism by which Rad18 levels are regulated is currently unclear and will require additional investigation, but the enzymatic function of BPLF1 seems to be required to maintain high levels of Rad18.
EBV itself is not known to encode genes necessary to repair viral DNA damage arising during lytic replication. Cellular repair factors, such as Rad18 and PCNA, may be necessary to maintain the integrity of viral DNA. If Rad18 does function to repair viral DNA damage, extracellular genomes released in its absence may contain more DNA errors and therefore lead to the decrease in infectious particles. Errors in viral DNA may contribute to faulty translation of proteins that are required for entry and establishment of a second round of replication. Results showing that infectivity is decreased by ∼50% when Rad18 is suppressed indicate that it is not strictly required yet is important for viral infectivity. This partial decrease in infectivity may be explained in that only a fraction of viral genomes are substantially damaged during normal replication. Future studies have been designed to evaluate the incidence of damage to EBV DNA during lytic replication. These studies are significant in that DNA damage is a major contributor to genetic instability, and they may provide evidence that BPLF1 deubiquitinating activity contributes to the tumorigenic properties of EBV.
Previously we showed that BPLF1 deubiquitinates PCNA and is inhibitory for TLS, and yet now we suggest that BPLF1 promotes an increase in Rad18 protein levels, which would be expected to stimulate TLS under DNA-damaging conditions. While these findings appear to be paradoxical, EBV may utilize these TLS factors in a nonclassical fashion for viral replication, and in addition, Rad18 also contributes to other effector pathways besides TLS. In our previous study, UV exposure largely shifted the function of Rad18 to activation of TLS, which would likely not be activated so strongly in the course of normal infection. Perhaps, therefore, BPLF1 shuts off TLS via PCNA deubiquitination under induced damage conditions yet promotes Rad18 stability and expression under infectious conditions, thereby contributing to ubiquitination of other Rad18 substrates, perhaps viral, and providing for an environment advantageous for viral replication and efficient production of infectious virus. It is not unusual for a deubiquitinating enzyme to target an E3 ligase, which could serve as a means of regulating the function of target proteins in a cell cycle-dependent fashion (72–75).
Exposure of latently EBV-infected cells to DNA damage stimuli, including chemotherapy and irradiation, can reactivate viral replication and induce the lytic cycle (76, 77). Induction of EBV lytic replication elicits a DNA damage response, specifically, activation of the ataxia telangiectasia-mutated (ATM) signal transduction pathway (64), which plays a key role in phosphorylating and activating DNA damage effectors, p53, γH2AX, and CHK2 (64, 78, 79). Additionally, the Kenney lab showed that ATM is involved in promoting EBV lytic reactivation (63). Knockdown of ATM inhibited EBV reactivation, and conversely, activation of ATM with Nutlin-3 increased lytic reactivation. Kulinski et al. demonstrated that chronic mouse gammaherpesvirus 68 (MHV68) infection is poorly controlled in mice that have insufficient ATM levels, suggesting that ATM is necessary for adaptive immune responses against this gammaherpesvirus (80). Nikitin et al. found that the DNA damage response is involved in tumor suppressor function: inhibition of ATM and Chk2 increased the transformation efficiency of EBV in primary B cells (81). Additionally, the EBV latency proteins LMP1, EBNA1, and EBNA3C also inhibit the DNA damage response (82, 83) but are not expected to affect viral replication. In summary, we report that the EBV lytic cycle protein BPLF1 mediates Rad18 function, and we have demonstrated the necessity for Rad18 for efficient production of infectious virus. BPLF1's deubiquitinating function may be a mechanism by which EBV co-opts cellular repair and replication factors in maintaining the fidelity of viral replication.
ACKNOWLEDGMENTS
This work was supported by NIH grants P01-CA19014-29 (to J.S.P.) and R21-AI095180 (to J.S.P.).
We thank T. Tsurumi for generously supplying EBV BPLF1 knockout virus, T. Murata for helpful discussions, and T. Sixma for the Rad6/18 construct. Thanks go to C. Vaziri and C. Moody for critical readings of the manuscript.
Footnotes
Published ahead of print 26 March 2014
REFERENCES
- 1.Anagnostopoulos I, Hummel M, Kreschel C, Stein H. 1995. Morphology, immunophenotype, and distribution of latently and/or productively Epstein-Barr virus-infected cells in acute infectious mononucleosis: implications for the interindividual infection route of Epstein-Barr virus. Blood 85:744–750 [PubMed] [Google Scholar]
- 2.Lemon SM, Hutt LM, Shaw JE, Li JL, Pagano JS. 1977. Replication of EBV in epithelial cells during infectious mononucleosis. Nature 268:268–270. 10.1038/268268a0 [DOI] [PubMed] [Google Scholar]
- 3.Niederman JC, Miller G, Pearson HA, Pagano JS, Dowaliby JM. 1976. Infectious mononucleosis. Epstein-Barr-virus shedding in saliva and the oropharynx. N. Engl. J. Med. 294:1355–1359 [DOI] [PubMed] [Google Scholar]
- 4.Babcock GJ, Decker LL, Volk M, Thorley-Lawson DA. 1998. EBV persistence in memory B cells in vivo. Immunity 9:395–404. 10.1016/S1074-7613(00)80622-6 [DOI] [PubMed] [Google Scholar]
- 5.Pagano J. 2009. EBV diseases, p 794 In Damania B, Pipas J. (ed), DNA tumor viruses. Springer, New York, NY [Google Scholar]
- 6.Raab-Traub N. 2007. EBV-induced oncogenesis. In Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, Yamanishi K. (ed), Human herpesviruses: biology, therapy, and immunoprophylaxis. Cambridge University Press, Cambridge, United Kingdom: [PubMed] [Google Scholar]
- 7.Saha A, Robertson ES. 2011. Epstein-Barr virus-associated B-cell lymphomas: pathogenesis and clinical outcomes. Clin. Cancer Res. 17:3056–3063. 10.1158/1078-0432.CCR-10-2578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niederman JC, McCollum RW, Henle G, Henle W. 1968. Infectious mononucleosis. Clinical manifestations in relation to EB virus antibodies. JAMA 203:205–209 [DOI] [PubMed] [Google Scholar]
- 9.Yuan J, Cahir-McFarland E, Zhao B, Kieff E. 2006. Virus and cell RNAs expressed during Epstein-Barr virus replication. J. Virol. 80:2548–2565. 10.1128/JVI.80.5.2548-2565.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abaitua F, Daikoku T, Crump CM, Bolstad M, O'Hare P. 2011. A single mutation responsible for temperature-sensitive entry and assembly defects in the VP1–2 protein of herpes simplex virus. J. Virol. 85:2024–2036. 10.1128/JVI.01895-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Batterson W, Furlong D, Roizman B. 1983. Molecular genetics of herpes simplex virus. VIII. Further characterization of a temperature-sensitive mutant defective in release of viral DNA and in other stages of the viral reproductive cycle. J. Virol. 45:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bottcher S, Granzow H, Maresch C, Mohl B, Klupp BG, Mettenleiter TC. 2007. Identification of functional domains within the essential large tegument protein pUL36 of pseudorabies virus. J. Virol. 81:13403–13411. 10.1128/JVI.01643-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Desai PJ. 2000. A null mutation in the UL36 gene of herpes simplex virus type 1 results in accumulation of unenveloped DNA-filled capsids in the cytoplasm of infected cells. J. Virol. 74:11608–11618. 10.1128/JVI.74.24.11608-11618.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schlieker C, Korbel GA, Kattenhorn LM, Ploegh HL. 2005. A deubiquitinating activity is conserved in the large tegument protein of the herpesviridae. J. Virol. 79:15582–15585. 10.1128/JVI.79.24.15582-15585.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schlieker C, Weihofen WA, Frijns E, Kattenhorn LM, Gaudet R, Ploegh HL. 2007. Structure of a herpesvirus-encoded cysteine protease reveals a unique class of deubiquitinating enzymes. Mol. Cell 25:677–687. 10.1016/j.molcel.2007.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gastaldello S, Hildebrand S, Faridani O, Callegari S, Palmkvist M, Di Guglielmo C, Masucci MG. 2010. A deneddylase encoded by Epstein-Barr virus promotes viral DNA replication by regulating the activity of cullin-RING ligases. Nat. Cell Biol. 12:351–361. 10.1038/ncb2035 [DOI] [PubMed] [Google Scholar]
- 17.Whitehurst CB, Ning S, Bentz GL, Dufour F, Gershburg E, Shackelford J, Langelier Y, Pagano JS. 2009. The Epstein-Barr virus (EBV) deubiquitinating enzyme BPLF1 reduces EBV ribonucleotide reductase activity. J. Virol. 83:4345–4353. 10.1128/JVI.02195-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzalez CM, Wang L, Damania B. 2009. Kaposi's sarcoma-associated herpesvirus encodes a viral deubiquitinase. J. Virol. 83:10224–10233. 10.1128/JVI.00589-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kattenhorn LM, Korbel GA, Kessler BM, Spooner E, Ploegh HL. 2005. A deubiquitinating enzyme encoded by HSV-1 belongs to a family of cysteine proteases that is conserved across the family Herpesviridae. Mol. Cell 19:547–557. 10.1016/j.molcel.2005.07.003 [DOI] [PubMed] [Google Scholar]
- 20.Bottcher S, Maresch C, Granzow H, Klupp BG, Teifke JP, Mettenleiter TC. 2008. Mutagenesis of the active-site cysteine in the ubiquitin-specific protease contained in large tegument protein pUL36 of pseudorabies virus impairs viral replication in vitro and neuroinvasion in vivo. J. Virol. 82:6009–6016. 10.1128/JVI.00280-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gredmark-Russ S, Isaacson MK, Kattenhorn L, Cheung EJ, Watson N, Ploegh HL. 2009. A gammaherpesvirus ubiquitin-specific protease is involved in the establishment of murine gammaherpesvirus 68 infection. J. Virol. 83:10644–10652. 10.1128/JVI.01017-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim ET, Oh SE, Lee YO, Gibson W, Ahn JH. 2009. Cleavage specificity of the UL48 deubiquitinating protease activity of human cytomegalovirus and the growth of an active-site mutant virus in cultured cells. J. Virol. 83:12046–12056. 10.1128/JVI.00411-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Loveland AN, Kattenhorn LM, Ploegh HL, Gibson W. 2006. High-molecular-weight protein (pUL48) of human cytomegalovirus is a competent deubiquitinating protease: mutant viruses altered in its active-site cysteine or histidine are viable. J. Virol. 80:6003–6012. 10.1128/JVI.00401-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saito S, Murata T, Kanda T, Isomura H, Narita Y, Sugimoto A, Kawashima D, Tsurumi T. 2013. Epstein-Barr virus deubiquitinase downregulates TRAF6-mediated NF-kappaB signaling during productive replication. J. Virol. 87:4060–4070. 10.1128/JVI.02020-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ernst R, Claessen JH, Mueller B, Sanyal S, Spooner E, van der Veen AG, Kirak O, Schlieker CD, Weihofen WA, Ploegh HL. 2011. Enzymatic blockade of the ubiquitin-proteasome pathway. PLoS Biol. 8:e1000605. 10.1371/journal.pbio.1000605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitehurst CB, Vaziri C, Shackelford J, Pagano JS. 2012. Epstein-Barr virus BPLF1 deubiquitinates PCNA and attenuates polymerase eta recruitment to DNA damage sites. J. Virol. 86:8097–8106. 10.1128/JVI.00588-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inn KS, Lee SH, Rathbun JY, Wong LY, Toth Z, Machida K, Ou JH, Jung JU. 2011. Inhibition of RIG-I-mediated signaling by Kaposi's sarcoma-associated herpesvirus-encoded deubiquitinase ORF64. J. Virol. 85:10899–10904. 10.1128/JVI.00690-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gastaldello S, Callegari S, Coppotelli G, Hildebrand S, Song M, Masucci MG. 2012. Herpes virus deneddylases interrupt the cullin-RING ligase neddylation cycle by inhibiting the binding of CAND1. J. Mol. Cell Biol. 4:242–251. 10.1093/jmcb/mjs012 [DOI] [PubMed] [Google Scholar]
- 29.Rooney CM, Rowe DT, Ragot T, Farrell PJ. 1989. The spliced BZLF1 gene of Epstein-Barr virus (EBV) transactivates an early EBV promoter and induces the virus productive cycle. J. Virol. 63:3109–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fixman ED, Hayward GS, Hayward SD. 1992. trans-acting requirements for replication of Epstein-Barr virus ori-Lyt. J. Virol. 66:5030–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cho MS, Milman G, Hayward SD. 1985. A second Epstein-Barr virus early antigen gene in BamHI fragment M encodes a 48- to 50-kilodalton nuclear protein. J. Virol. 56:860–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li JS, Zhou BS, Dutschman GE, Grill SP, Tan RS, Cheng YC. 1987. Association of Epstein-Barr virus early antigen diffuse component and virus-specified DNA polymerase activity. J. Virol. 61:2947–2949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsurumi T, Daikoku T, Kurachi R, Nishiyama Y. 1993. Functional interaction between Epstein-Barr virus DNA polymerase catalytic subunit and its accessory subunit in vitro. J. Virol. 67:7648–7653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsurumi T, Daikoku T, Nishiyama Y. 1994. Further characterization of the interaction between the Epstein-Barr virus DNA polymerase catalytic subunit and its accessory subunit with regard to the 3′-to-5′ exonucleolytic activity and stability of initiation complex at primer terminus. J. Virol. 68:3354–3363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daikoku T, Yamashita Y, Tsurumi T, Nishiyama Y. 1995. The US3 protein kinase of herpes simplex virus type 2 is associated with phosphorylation of the UL12 alkaline nuclease in vitro. Arch. Virol. 140:1637–1644. 10.1007/BF01322537 [DOI] [PubMed] [Google Scholar]
- 36.Fixman ED, Hayward GS, Hayward SD. 1995. Replication of Epstein-Barr virus oriLyt: lack of a dedicated virally encoded origin-binding protein and dependence on Zta in cotransfection assays. J. Virol. 69:2998–3006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kawashima D, Kanda T, Murata T, Saito S, Sugimoto A, Narita Y, Tsurumi T. 2013. Nuclear transport of Epstein-Barr virus DNA polymerase is dependent on the BMRF1 polymerase processivity factor and molecular chaperone Hsp90. J. Virol. 87:6482–6491. 10.1128/JVI.03428-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kudoh A, Iwahori S, Sato Y, Nakayama S, Isomura H, Murata T, Tsurumi T. 2009. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein-Barr virus replication compartments. J. Virol. 83:6641–6651. 10.1128/JVI.00049-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Daikoku T, Kudoh A, Sugaya Y, Iwahori S, Shirata N, Isomura H, Tsurumi T. 2006. Postreplicative mismatch repair factors are recruited to Epstein-Barr virus replication compartments. J. Biol. Chem. 281:11422–11430. 10.1074/jbc.M510314200 [DOI] [PubMed] [Google Scholar]
- 40.Lilley CE, Carson CT, Muotri AR, Gage FH, Weitzman MD. 2005. DNA repair proteins affect the lifecycle of herpes simplex virus 1. Proc. Natl. Acad. Sci. U. S. A. 102:5844–5849. 10.1073/pnas.0501916102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo MH, Rosenke K, Czornak K, Fortunato EA. 2007. Human cytomegalovirus disrupts both ataxia telangiectasia mutated protein (ATM)- and ATM-Rad3-related kinase-mediated DNA damage responses during lytic infection. J. Virol. 81:1934–1950. 10.1128/JVI.01670-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shirata N, Kudoh A, Daikoku T, Tatsumi Y, Fujita M, Kiyono T, Sugaya Y, Isomura H, Ishizaki K, Tsurumi T. 2005. Activation of ataxia telangiectasia-mutated DNA damage checkpoint signal transduction elicited by herpes simplex virus infection. J. Biol. Chem. 280:30336–30341. 10.1074/jbc.M500976200 [DOI] [PubMed] [Google Scholar]
- 43.Taylor TJ, Knipe DM. 2004. Proteomics of herpes simplex virus replication compartments: association of cellular DNA replication, repair, recombination, and chromatin remodeling proteins with ICP8. J. Virol. 78:5856–5866. 10.1128/JVI.78.11.5856-5866.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wilkinson DE, Weller SK. 2006. Herpes simplex virus type I disrupts the ATR-dependent DNA-damage response during lytic infection. J. Cell Sci. 119:2695–2703. 10.1242/jcs.02981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sugimoto A, Kanda T, Yamashita Y, Murata T, Saito S, Kawashima D, Isomura H, Nishiyama Y, Tsurumi T. 2011. Spatiotemporally different DNA repair systems participate in Epstein-Barr virus genome maturation. J. Virol. 85:6127–6135. 10.1128/JVI.00258-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durando M, Tateishi S, Vaziri C. 2013. A non-catalytic role of DNA polymerase eta in recruiting Rad18 and promoting PCNA monoubiquitination at stalled replication forks. Nucleic Acids Res. 41:3079–3093. 10.1093/nar/gkt016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Masuda Y, Suzuki M, Kawai H, Hishiki A, Hashimoto H, Masutani C, Hishida T, Suzuki F, Kamiya K. 2012. En bloc transfer of polyubiquitin chains to PCNA in vitro is mediated by two different human E2–E3 pairs. Nucleic Acids Res. 40:10394–10407. 10.1093/nar/gks763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hibbert RG, Huang A, Boelens R, Sixma TK. 2011. E3 ligase Rad18 promotes monoubiquitination rather than ubiquitin chain formation by E2 enzyme Rad6. Proc. Natl. Acad. Sci. U. S. A. 108:5590–5595. 10.1073/pnas.1017516108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Williams SA, Longerich S, Sung P, Vaziri C, Kupfer GM. 2011. The E3 ubiquitin ligase RAD18 regulates ubiquitylation and chromatin loading of FANCD2 and FANCI. Blood 117:5078–5087. 10.1182/blood-2010-10-311761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geng L, Huntoon CJ, Karnitz LM. 2010. RAD18-mediated ubiquitination of PCNA activates the Fanconi anemia DNA repair network. J. Cell Biol. 191:249–257. 10.1083/jcb.201005101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Masuda Y, Suzuki M, Kawai H, Suzuki F, Kamiya K. 2012. Asymmetric nature of two subunits of RAD18, a RING-type ubiquitin ligase E3, in the human RAD6A-RAD18 ternary complex. Nucleic Acids Res. 40:1065–1076. 10.1093/nar/gkr805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Notenboom V, Hibbert RG, van Rossum-Fikkert SE, Olsen JV, Mann M, Sixma TK. 2007. Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification. Nucleic Acids Res. 35:5819–5830. 10.1093/nar/gkm615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Delecluse HJ, Hilsendegen T, Pich D, Zeidler R, Hammerschmidt W. 1998. Propagation and recovery of intact, infectious Epstein-Barr virus from prokaryotic to human cells. Proc. Natl. Acad. Sci. U. S. A. 95:8245–8250. 10.1073/pnas.95.14.8245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang Y, Durando M, Smith-Roe SL, Sproul C, Greenwalt AM, Kaufmann W, Oh S, Hendrickson EA, Vaziri C. 2013. Cell cycle stage-specific roles of Rad18 in tolerance and repair of oxidative DNA damage. Nucleic Acids Res. 41:2296–2312. 10.1093/nar/gks1325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Day TA, Palle K, Barkley LR, Kakusho N, Zou Y, Tateishi S, Verreault A, Masai H, Vaziri C. 2010. Phosphorylated Rad18 directs DNA polymerase eta to sites of stalled replication. J. Cell Biol. 191:953–966. 10.1083/jcb.201006043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Palle K, Vaziri C. 2011. Rad18 E3 ubiquitin ligase activity mediates Fanconi anemia pathway activation and cell survival following DNA topoisomerase 1 inhibition. Cell Cycle 10:1625–1638. 10.4161/cc.10.10.15617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zlatanou A, Despras E, Braz-Petta T, Boubakour-Azzouz I, Pouvelle C, Stewart GS, Nakajima S, Yasui A, Ishchenko AA, Kannouche PL. 2011. The hMsh2-hMsh6 complex acts in concert with monoubiquitinated PCNA and Pol eta in response to oxidative DNA damage in human cells. Mol. Cell 43:649–662. 10.1016/j.molcel.2011.06.023 [DOI] [PubMed] [Google Scholar]
- 58.Bailly V, Lauder S, Prakash S, Prakash L. 1997. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272:23360–23365. 10.1074/jbc.272.37.23360 [DOI] [PubMed] [Google Scholar]
- 59.Inagaki A, van Cappellen WA, van der Laan R, Houtsmuller AB, Hoeijmakers JH, Grootegoed JA, Baarends WM. 2009. Dynamic localization of human RAD18 during the cell cycle and a functional connection with DNA double-strand break repair. DNA Repair (Amst.) 8:190–201. 10.1016/j.dnarep.2008.10.008 [DOI] [PubMed] [Google Scholar]
- 60.Masuyama S, Tateishi S, Yomogida K, Nishimune Y, Suzuki K, Sakuraba Y, Inoue H, Ogawa M, Yamaizumi M. 2005. Regulated expression and dynamic changes in subnuclear localization of mammalian Rad18 under normal and genotoxic conditions. Genes Cells 10:753–762. 10.1111/j.1365-2443.2005.00874.x [DOI] [PubMed] [Google Scholar]
- 61.Miyase S, Tateishi S, Watanabe K, Tomita K, Suzuki K, Inoue H, Yamaizumi M. 2005. Differential regulation of Rad18 through Rad6-dependent mono- and polyubiquitination. J. Biol. Chem. 280:515–524. 10.1074/jbc.M409219200 [DOI] [PubMed] [Google Scholar]
- 62.Hong GK, Delecluse HJ, Gruffat H, Morrison TE, Feng WH, Sergeant A, Kenney SC. 2004. The BRRF1 early gene of Epstein-Barr virus encodes a transcription factor that enhances induction of lytic infection by BRLF1. J. Virol. 78:4983–4992. 10.1128/JVI.78.10.4983-4992.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hagemeier SR, Barlow EA, Meng Q, Kenney SC. 2012. The cellular ataxia telangiectasia-mutated kinase promotes Epstein-Barr virus lytic reactivation in response to multiple different types of lytic reactivation-inducing stimuli. J. Virol. 86:13360–13370. 10.1128/JVI.01850-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kudoh A, Fujita M, Zhang L, Shirata N, Daikoku T, Sugaya Y, Isomura H, Nishiyama Y, Tsurumi T. 2005. Epstein-Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J. Biol. Chem. 280:8156–8163. 10.1074/jbc.M411405200 [DOI] [PubMed] [Google Scholar]
- 65.Roest HP, Baarends WM, de Wit J, van Klaveren JW, Wassenaar E, Hoogerbrugge JW, van Cappellen WA, Hoeijmakers JH, Grootegoed JA. 2004. The ubiquitin-conjugating DNA repair enzyme HR6A is a maternal factor essential for early embryonic development in mice. Mol. Cell. Biol. 24:5485–5495. 10.1128/MCB.24.12.5485-5495.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shekhar MP, Tait L, Gerard B. 2006. Essential role of T-cell factor/beta-catenin in regulation of Rad6B: a potential mechanism for Rad6B overexpression in breast cancer cells. Mol. Cancer Res. 4:729–745. 10.1158/1541-7786.MCR-06-0136 [DOI] [PubMed] [Google Scholar]
- 67.Jentsch S, McGrath JP, Varshavsky A. 1987. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329:131–134. 10.1038/329131a0 [DOI] [PubMed] [Google Scholar]
- 68.Sung P, Prakash S, Prakash L. 1988. The RAD6 protein of Saccharomyces cerevisiae polyubiquitinates histones, and its acidic domain mediates this activity. Genes Dev. 2:1476–1485. 10.1101/gad.2.11.1476 [DOI] [PubMed] [Google Scholar]
- 69.Shekhar MP, Gerard B, Pauley RJ, Williams BO, Tait L. 2008. Rad6B is a positive regulator of beta-catenin stabilization. Cancer Res. 68:1741–1750. 10.1158/0008-5472.CAN-07-2111 [DOI] [PubMed] [Google Scholar]
- 70.Lyakhovich A, Shekhar MP. 2003. Supramolecular complex formation between Rad6 and proteins of the p53 pathway during DNA damage-induced response. Mol. Cell. Biol. 23:2463–2475. 10.1128/MCB.23.7.2463-2475.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Varanasi L, Do PM, Goluszko E, Martinez LA. 2012. Rad18 is a transcriptional target of E2F3. Cell Cycle 11:1131–1141. 10.4161/cc.11.6.19558 [DOI] [PubMed] [Google Scholar]
- 72.Wu X, Yen L, Irwin L, Sweeney C, Carraway KL., III 2004. Stabilization of the E3 ubiquitin ligase Nrdp1 by the deubiquitinating enzyme USP8. Mol. Cell. Biol. 24:7748–7757. 10.1128/MCB.24.17.7748-7757.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Di Donato F, Chan EK, Askanase AD, Miranda-Carus M, Buyon JP. 2001. Interaction between 52 kDa SSA/Ro and deubiquitinating enzyme UnpEL: a clue to function. Int. J. Biochem. Cell Biol. 33:924–934. 10.1016/S1357-2725(01)00055-3 [DOI] [PubMed] [Google Scholar]
- 74.Jensen DE, Proctor M, Marquis ST, Gardner HP, Ha SI, Chodosh LA, Ishov AM, Tommerup N, Vissing H, Sekido Y, Minna J, Borodovsky A, Schultz DC, Wilkinson KD, Maul GG, Barlev N, Berger SL, Prendergast GC, Rauscher FJ., III 1998. BAP1: a novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 16:1097–1112. 10.1038/sj.onc.1201861 [DOI] [PubMed] [Google Scholar]
- 75.Soares L, Seroogy C, Skrenta H, Anandasabapathy N, Lovelace P, Chung CD, Engleman E, Fathman CG. 2004. Two isoforms of otubain 1 regulate T cell anergy via GRAIL. Nat. Immunol. 5:45–54. 10.1038/ni1017 [DOI] [PubMed] [Google Scholar]
- 76.Feng WH, Israel B, Raab-Traub N, Busson P, Kenney SC. 2002. Chemotherapy induces lytic EBV replication and confers ganciclovir susceptibility to EBV-positive epithelial cell tumors. Cancer Res. 62:1920–1926 http://cancerres.aacrjournals.org/content/62/6/1920.long [PubMed] [Google Scholar]
- 77.Westphal EM, Blackstock W, Feng W, Israel B, Kenney SC. 2000. Activation of lytic Epstein-Barr virus (EBV) infection by radiation and sodium butyrate in vitro and in vivo: a potential method for treating EBV-positive malignancies. Cancer Res. 60:5781–5788 http://cancerres.aacrjournals.org/content/60/20/5781.long [PubMed] [Google Scholar]
- 78.Smith J, Tho LM, Xu N, Gillespie DA. 2010. The ATM-Chk2 and ATR-Chk1 pathways in DNA damage signaling and cancer. Adv. Cancer Res. 108:73–112. 10.1016/B978-0-12-380888-2.00003-0 [DOI] [PubMed] [Google Scholar]
- 79.Zhang XP, Liu F, Wang W. 2011. Two-phase dynamics of p53 in the DNA damage response. Proc. Natl. Acad. Sci. U. S. A. 108:8990–8995. 10.1073/pnas.1100600108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kulinski JM, Leonardo SM, Mounce BC, Malherbe L, Gauld SB, Tarakanova VL. 2012. Ataxia telangiectasia mutated kinase controls chronic gammaherpesvirus infection. J. Virol. 86:12826–12837. 10.1128/JVI.00917-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Nikitin PA, Yan CM, Forte E, Bocedi A, Tourigny JP, White RE, Allday MJ, Patel A, Dave SS, Kim W, Hu K, Guo J, Tainter D, Rusyn E, Luftig MA. 2010. An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe 8:510–522. 10.1016/j.chom.2010.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cai Q, Guo Y, Xiao B, Banerjee S, Saha A, Lu J, Glisovic T, Robertson ES. 2011. Epstein-Barr virus nuclear antigen 3C stabilizes Gemin3 to block p53-mediated apoptosis. PLoS Pathog. 7:e1002418. 10.1371/journal.ppat.1002418 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 83.Gruhne B, Sompallae R, Masucci MG. 2009. Three Epstein-Barr virus latency proteins independently promote genomic instability by inducing DNA damage, inhibiting DNA repair and inactivating cell cycle checkpoints. Oncogene 28:3997–4008. 10.1038/onc.2009.258 [DOI] [PubMed] [Google Scholar]