ABSTRACT
Human respiratory syncytial virus (RSV) is the most common cause of bronchiolitis and pneumonia in infants and the elderly worldwide; however, there is no licensed RSV vaccine or effective drug treatment available. The RSV matrix (M) protein plays key roles in virus assembly and budding, but the protein interactions that govern budding of infectious virus are not known. In this study, we focus on M protein and identify a key phosphorylation site (Thr205) in M that is critical for RSV infectious virus production. Recombinant virus with a nonphosphorylatable alanine (Ala) residue at the site was markedly attenuated, whereas virus with a phosphomimetic aspartate (Asp) resulted in a nonviable virus which could only be recovered with an additional mutation in M (serine to asparagine at position 220), strongly implying that Thr205 is critical for viral infectivity. Experiments in vitro showed that mutation of Thr205 does not affect M stability or the ability to form dimers but implicate an effect on higher-order oligomer assembly. In transfected and infected cells, Asp substitution of Thr205 appeared to impair M oligomerization; typical filamentous structures still formed at the plasma membrane, but M assembly during the ensuing elongation process seemed to be impaired, resulting in shorter and more branched filaments as observed using electron microscopy (EM). Our data thus imply for the first time that M oligomerization, regulated by a negative charge at Thr205, may be critical to production of infectious RSV.
IMPORTANCE We show here for the first time that RSV M's role in virus assembly/release is strongly dependent on threonine 205 (Thr205), a consensus site for CK2, which appears to play a key regulatory role in modulating M oligomerization and association with virus filaments. Our analysis indicates that T205 mutations do not impair M dimerization or viruslike filament formation per se but rather the ability of M to assemble in ordered fashion on the viral filaments themselves. This appears to impact in turn upon the infectivity of released virus rather than on virus production or release itself. Thus, M oligomerization would appear to be a target of interest for the development of anti-RSV agents; further, the recombinant T205-substituted mutant viruses described here would appear to be the first RSV mutants affected in viral maturation to our knowledge and hence of considerable interest for vaccine approaches in the future.
INTRODUCTION
The human respiratory syncytial virus (RSV) is the most common cause of bronchiolitis and pneumonia in infants and the elderly worldwide. Despite the enormous burden of RSV disease, there is no efficacious vaccine or antiviral drug therapy yet available (1). RSV is a member of the Paramyxoviridae family. It is a pleomorphic, enveloped, single-strand RNA virus encoding 11 proteins, with the three glycoproteins, fusion (F), glycoprotein (G), and small hydrophobic (SH), present in the viral envelope. The virion itself contains an internal ribonucleoprotein (RNP) complex comprising the negative-sense genome encapsidated within the nucleoprotein (N), the phosphoprotein (P), and large (L) polymerase. The matrix (M) protein is present between the outer envelope and inner RNP and plays an important structural role as a key adaptor in the assembly process. In addition, the M2-1 and M2-2 proteins (both translated from the M2 gene) are associated with the nucleocapsid and have roles in RSV transcription and replication (2–4). Viral transcription and replication take place in cytoplasmic inclusions that contain the RNPs (5, 6). The M protein is believed to be the main driver of virus assembly on the plasma membrane through an adaptor role in interacting with the cytoplasmic tails of the glycoproteins and with the RNP complex in the cytoplasm (7, 8).
RSV assembles on the apical surface of polarized epithelial cells where viral filaments are formed on the plasma membrane (9, 10). These filaments are thought to be essential to cell-to-cell fusion and for syncytium formation. However, the virus has been shown to produce both filamentous and spherical forms during budding (11), with recent data suggesting that the filamentous particles rather than the spherical ones are infectious (12).
RSV viruslike filaments can be generated independently of viral infection, minimally requiring F, M, P, and N (13). Although little is known about the specific roles of P and N in budding, the cytoplasmic tail of F has been shown to have a critical role in filament formation and budding (13, 14). In the absence of the cytoplasmic tail of F, M remains relatively abundant in the cytoplasm and concentrated in inclusion bodies (IB), resulting in the complete absence of viral filaments and loss of infectivity (15). M's crucial role in viral filament maturation and elongation (16) probably relates to the transfer of RNP complexes from inclusion bodies to the sites of budding by interacting with the M2-1 protein (8). We have previously hypothesized that dimerization/oligomerization of M may drive the budding of infectious RSV at the membrane by bringing together the RNP and envelope glycoprotein complexes (17). RSV M, like other paramyxovirus matrix proteins, forms higher-order structures that are believed to be important in viral assembly on the plasma membrane (18–21). This is supported by electron micrographs showing a cylindrical matrix layer linked to the inner part of the viral envelope. Thus, M oligomerization is most probably crucial to maintain the filamentous shape of the budding virus (12). Based on structural alignment with the recently published structure of human metapneumovirus (hMPV) (22) and Newcastle disease virus (NDV) M protein (23), we hypothesized that RSV M forms structurally similar dimeric and oligomeric structures. Despite the sequence divergence of the NDV, hMPV, and RSV M proteins, their tertiary structure appears to be well conserved, and thus the mechanisms of M oligomerization may be similar (23).
In this study, we identify for the first time a key threonine residue (Thr205) within M that is critical for M phosphorylation and for infectivity of the released virus. Recombinant virus with a phosphomimetic Asp at the site could be recovered only with an additional mutation in M (Ser to Asn at position 220), strongly implying that Thr205 is critical for viral infectivity. Using a range of assays, we show that substitution of Thr205 impairs M distribution along viral filaments and modulates M oligomerization, with a nonphosphorylatable alanine residue at the site resulting in increased M oligomerization and negative charge at the site preventing infectious virus production; electron microscopic analysis shows that RSV with the phosphomimetic aspartate in M at the site forms viral filaments that are disordered compared to those formed by the wild-type (WT) virus. The results have important implications for M function in viral filament formation and its importance to RSV production.
MATERIALS AND METHODS
Cell culture and RSV preparation.
HEp-2 (ATCC CCL-23), 293T, A549, and Vero cells were maintained in Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum (FBS), 1% l-glutamine, and 1% penicillin-streptomycin. Transfections were performed using Lipofectamine 2000 (Invitrogen). All RSV recombinant (r) A2 strains were cultured in Vero cells. Cell-associated RSV was harvested when extensive cytopathic effect was observed. The culture supernatant was removed; cells were lysed in serum-free SPGA medium (218 mM sucrose, 7.1 mM K2HPO4, 4.9 mM sodium glutamate, 1% bovine serum albumin [BSA]), clarified by centrifugation (1,300 × g, 15 min, 4°C), and stored at −80°C until required. The RSV titer was determined on Vero cells using an immune plaque assay. Titers for each virus-containing cell lysate were determined on triplicate cultures.
Immune plaque assay.
Overnight cultures of HEp-2 or Vero cells (seeded at 4 × 104 cells/well on a 96-well plate) were infected with the virus stocks. On a separate 96-well plate, the virus stocks were diluted using doubling dilutions in serum-free DMEM starting at 1/10 for supernatant virus and 1/100 for cell-associated virus. Cells were washed once with serum-free DMEM, and 50 μl of each of the virus dilutions was added to triplicate wells and left for 2 h at 37°C. After 2 h, 100 μl of 2% FBS-DMEM was added and plates were incubated for a further 48 h at 37°C. In the case of immunostaining, medium was discarded, and cells were washed in 100 μl phosphate-buffered saline (PBS) and fixed for 20 min with 100 μl absolute methanol containing 2% hydrogen peroxide. Cells were washed in 200 μl PBS-1% BSA and stained with biotinylated anti-RSV antibody (1:500; AbD Serotec) for 2 h. Cells were washed again 3 times in PBS-1% BSA and stained with ExtrAvidin peroxidase (1:500; Sigma) for 2 h in the dark. Cells were washed 3 times, and plaques were visualized using Sigmafast diaminobenzidine (DAB) peroxidase substrate (Sigma).
Effect of protein kinase CK2 inhibitor or siRNA on RSV production/cell viability.
Overnight subconfluent monolayers of HEp-2 or Vero cells were infected with RSV rA2 at a multiplicity of infection (MOI) of 1 and then treated with the protein kinase CK2-specific inhibitor 4,5,6,7-tetrabromobenzo-triazole (TBB) (24, 25) added to the culture medium to a final concentration of 2.8 ng/ml for 12 h at 6 h or 18 h postinfection (p.i.); control cells were left untreated. Medium was then replaced with fresh medium without TBB, and cells were cultured further up to 48 h p.i. Culture supernatants and cell-associated virus were harvested, and viral titers were determined as described previously (26, 27) on monolayers of Vero cells followed by crystal violet staining to determine 50% tissue culture infective dose (TCID50)/ml. Virus titers were calculated as the percentage relative to the infected, nontreated sample. Alternatively, cells were transfected with 20 nM small inhibitory RNAs (siRNAs), control scrambled or CK2-specific SMARTpool siRNAs (Dharmacon), by using the transfection reagent DharmaFECT (according to the manufacturer's recommendations) 48 h prior to infection; viral infection was subsequently carried out as described above, and titration was determined as described above.
Cytotoxicity.
Release of lactate dehydrogenase (LDH) into the culture medium (28), as a measure of cell death, was estimated using the Promega nonradioactive LDH release assay as per the manufacturer's instructions. Data are depicted in terms of LDH activity relative to that in TBB-untreated cells and are calculated using the following formula: (OD490 of sample − OD490 of negative control)/(OD490 of infected, nontreated sample − OD490 of negative control), where OD490 is optical density at 490 nm.
In vivo RSV phosphorylation.
Subconfluent monolayers of HEp-2 cells in 6-well plates were infected without (mock) or with RSV rA2 at an MOI of 1. At 18 h p.i., culture medium was replaced with fresh medium containing 0.3m Ci/ml orthophosphate (32Pi) and incubated for 2 h. Cells were then lysed in RIPA buffer by incubating on ice for 30 min followed by removal of cellular debris by centrifugation for 20 min at 16,100 × g. Cleared cell lysates were used for immunoprecipitation with goat anti-RSV polyclonal antibody attached to protein G Sepharose beads (Merck Millipore). Bound proteins were eluted by boiling in Laemmli buffer and separated by SDS-PAGE; the gel was dried under vacuum and exposed to X-ray film.
Phosphorylation by purified CK2 or by cell lysates.
Recombinant His6-tagged M was expressed in bacteria as described below, T-ag-green fluorescent protein (GFP) (SV40 large tumor antigen amino acids 111 to 135, including the CK2 site at Ser111/112, fused N-terminal to the coding sequence for GFP) (29–31) and GFP alone were expressed in bacteria and purified as described previously (32). Phosphorylation was performed in 100-μl reaction samples in assay buffer (100 mM Tris-HCl [pH 7.5 at 30°C], 25 mM MgCl2, 2.5 mM dithiothreitol [DTT], 750 mM KCl), containing 2 ng recombinant CK2 (Millipore), 20 μg recombinant protein, 0.1 mM ATP, and 2 to 4 μCi [γ32]ATP, for 30 to 120 min at 30°C. Reactions were either stopped for SDS-PAGE by boiling in Laemmli buffer or for quantitative analysis by spotting onto Whatman P-81 phosphocellulose paper and washing in 0.5% phosphoric acid, with incorporated 32Pi determined by scintillation counting (29–31). Background measurements of phosphorylation in cell lysates in the absence of added CK2 peptide or recombinant M protein were made routinely, with the counts subtracted from those where CK2 peptide or M proteins were added, to enable determination of specific incorporation into the respective exogenous substrate. In the case of PAGE, gels were dried and analyzed by autoradiography on the Typhoon Trio (GE Healthcare).
Cell lysates for phosphorylation experiments were prepared from the indicated cell lines with or without prior infection with RSV for 24 h (MOI of 4) or treatment with siRNA (20 nM control scrambled or CK2α-specific SMARTpool siRNAs; Dharmacon) by washing the cells twice with PBS and then suspending the cell monolayers into lysate buffer (50 mM HEPES [pH 7.4], 100 mM NaCl, 50 mM sodium fluoride, 5 mM β-glycerophosphate, 2 mM EDTA, 2 mM EGTA, 1 mM sodium vanadate [Na3VO4], 1% [vol/vol] Triton X-100) prior to centrifugation (16,100 × g for 10 min) to remove debris. Phosphorylation reactions were essentially as indicated above for pure CK2 except that cyclic AMP-dependent protein kinase inhibitor (PKI) was included at 1 μM, with ca. 30 μg of lysate used as the source of kinase. Where used, TBB was included at a final concentration of 25 μM, while CK2 peptide substrate (Sigma Chemical Co.) was used as a positive control at 150 μM. Reactions were stopped by spotting onto phosphocellulose paper and washing in phosphoric acid, and incorporated 32Pi was determined by scintillation counting as described above.
Generation and recovery of mutant recombinant RSV.
M mutations were cloned into the RSV antigenomic cDNA (D53) using standard molecular biology techniques as described previously (26). Recovery of recombinant RSV from cloned DNA was performed as described previously (33). Briefly, monolayers of BSR-T7 cells in six-well plates were transfected using GeneJuice (Novagen) with a mixture of pTM1-based plasmids encoding the RSV N, P, L, and M2-1 proteins and either WT or mutant antigenome cDNA (1:1:0.5:0.5:1 μg each/well). The transfection mixture was removed after 20 h of incubation at 37°C and replaced with fresh medium (DMEM supplemented with 5% FBS; Life Technologies). After an additional 48 h, the supernatants were harvested, clarified by centrifugation, and passaged onto fresh Vero cells. The recovered viruses were harvested 4 days later. Viral titers were determined by plaque assay on Vero cells. Plaques were visualized by immunostaining using a cocktail of three murine anti-F monoclonal antibodies followed by horseradish peroxidase-coupled anti-mouse IgG antibodies and 4CN substrate (Kirkegaard and Perry Laboratories). The presence of the mutations was confirmed in recombinant virus by reverse transcription (RT)-PCR of viral genomic RNA and nucleotide sequencing.
RT-PCR.
Quantitative RT-PCR (qRT-PCR) was performed on viral RNA from purified recombinant RSV. A total of 106 PFU of each virus was purified by pelleting through a 30% sucrose cushion (20,000 × g, 90 min, 4°C). Viral pellets were resuspended and viral RNA was isolated by the QIAamp viral RNA minikit (QIAgen), according to the manufacturer's instructions. One fifth of the total volume of isolated viral RNA was subjected to random-primed reverse transcription (iScript reverse transcription kit; Bio-Rad). A total of 2 μl of each RT reaction product was used in duplicate qPCRs (SensiFAST SYBR and fluorescein mix; Bioline) with primers specific for the various intergenic regions to measure genomic viral RNA (SH-G, 5′-TTAACATCCCACCATGCAAA-3′ (forward) and 5′-GCATTTGCCCCAATGTTATT-3′ (reverse); leader-NS1, 5′-TGCGTACAACAAACTTGCAT-3′ (forward) and 5′-GCTGCCCATCTCTAACCAAG-3′ (reverse); L-trailer, 5′-CACAGGTAGTCTGTTATACAACTTTCA-3′ (forward) and 5′-ACAGTGTTAGTGTATAGCTATGGGAAT-3′) (reverse). Relative amounts of viral RNA in each sample were compared directly after normalization to rA2.
Viruslike filament/particle formation.
Overnight cultures of HEp-2 cells (seeded at 4 × 105 cells/well in 6-well plates on 16-mm microcover glass) were transfected to express plasmids (0.4 μg each) encoding the RSV A2 WT or the mutant M protein along with pcDNA3.1 codon-optimized plasmids encoding the RSV A2 N, P, and F proteins (a gift from Marty Moore), using Lipofectamine 2000 (Invitrogen) according to the manufacturer's recommendations. Cells were fixed at 24 h posttransfection (p.t.), immunostained, and imaged as described below. For viruslike particle (VLP) formation, released VLPs were harvested from the supernatant; the supernatant was clarified of cell debris by centrifugation (1,300 × g, 10 min, 4°C) and pelleted through a 20% sucrose cushion. Cells were lysed in RIPA buffer. Cellular lysates and VLP pellets were dissolved in Laemmli buffer and subjected to Western analysis.
RSV infection for infectivity assay and viral filament visualization.
Overnight cultures of HEp-2 cells seeded at 4 × 105 cells/well in 6-well plates on 16-mm coverslips were infected with recombinant RSV WT (rA2) or a mutant at an MOI of 3. Cells were fixed 24 h postinfection, immunostained, and imaged, as described below, to visualize filaments. For infectivity assay, cell-associated and released virus fractions were harvested, and virus titers of cell-associated and supernatant fractions were determined using immune plaque assay (see “Immune plaque assay” above).
Fixation and immunostaining.
Cells were fixed with 4% paraformaldehyde in PBS for 10 min, blocked with 3% BSA in 0.2% Triton X-PBS for 10 min, and coimmunostained with monoclonal anti-M (1:200; gift from Mariethe Ehnlund, Karolinska Institute, Sweden), polyclonal anti-RSV (1:200; Abcam), or monoclonal anti-F (1:500; Millipore) antibodies, followed by species-specific secondary antibodies conjugated to Alexa Fluor 488 and Alexa Fluor 568 (1:1,000; Invitrogen). Images were obtained on a Zeiss 5 PASCAL confocal laser scanning microscope (LSM) using a ×63/1.4 Plan-Apochromat oil lens. Images were acquired using Zeiss LSM Image Browser software (4.2.0.121; Zeiss).
M purification and size exclusion chromatography.
WT M and mutant derivatives were expressed as His6-fusion proteins in pCDF-Duet1 modified vector (Goedele Maertens) in Rosetta 2 Escherichia coli; cells were grown from fresh starters in 0.25 liters of Luria Bertani broth for 5 h at 30°C, followed by induction with 0.4 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h at 25°C. Cells were harvested, resuspended in 40 ml 50 mM Na phosphate buffer (pH 7.4) containing 300 mM NaCl plus protease inhibitors (Roche Diagnostics), and lysed with 1 mg/ml lysozyme followed by sonication (4 × 20 s) in the presence of RNase (25 μg/ml; Sigma), DNase (120 units; Sigma), and 0.25% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS). Lysates were clarified by centrifugation (23,425 × g, 30 min, 4°C), and the soluble His-M protein was purified using Ni-nitrilotriacetic acid (NTA) Sepharose affinity beads (Sigma) and eluted with 250 mM imidazole. Protein-containing fractions were collected, and the protein was concentrated using Vivaspin 4 columns (Sartorius Stedim Biotech) up to ∼3 mg/ml and analyzed by gel filtration chromatography (S200; GE Healthcare) in 50 mM sodium phosphate buffer (pH 7.4) containing 150 mM NaCl 1 day later.
Transmission electron microscopy.
Cells were fixed in 0.5% glutaraldehyde in 200 mM sodium cacodylate buffer for 30 min, washed in buffer, and secondarily fixed in reduced 1% osmium tetroxide and 1.5% potassium ferricyanide for 1 h. The samples were washed in distilled water and stained overnight at 4°C in 0.5% magnesium uranyl acetate, washed in distilled water, and dehydrated in graded ethanol. The samples were then embedded flat in the dish in Epon resin. Resin-filled stubs were placed on embedded cell monolayers and polymerized. Ultrathin sections (typically 50 to 70 nm) were cut parallel to the dish, stained with Reynolds' lead citrate, and examined in an FEI Tecnai electron microscope with a charge-coupled-device (CCD) camera image acquisition. Images were acquired using Olympus Image Browser software (siViewer; Olympus).
SDS-PAGE and Western analysis.
Protein samples were analyzed on 12% polyacrylamide gels and subjected to electrophoresis in Tris-glycine buffer. All samples were boiled for 3 min prior to electrophoresis. Gels were then transferred to a polyvinylidene difluoride (PVDF) membrane (Roche Diagnostics). The blots were blocked with 5% nonfat milk in Tris-buffered saline (pH 7.4) followed by incubation in polyclonal anti-RSV (1:1,000; Abcam) and horseradish peroxidase (HRP)-conjugated donkey anti-goat (1:5,000; Santa Cruz Biotechnology) antibody. Western blots were developed using freshly prepared chemiluminescent substrate (100 mM Tris-HCl [pH 8.8], 1.25 mM luminol, 0.2 mM p-coumaric acid, and 0.05% H2O2) and exposed to FUJI autoradiography films.
RESULTS
Threonine 205 within the RSV matrix protein is essential for virus production.
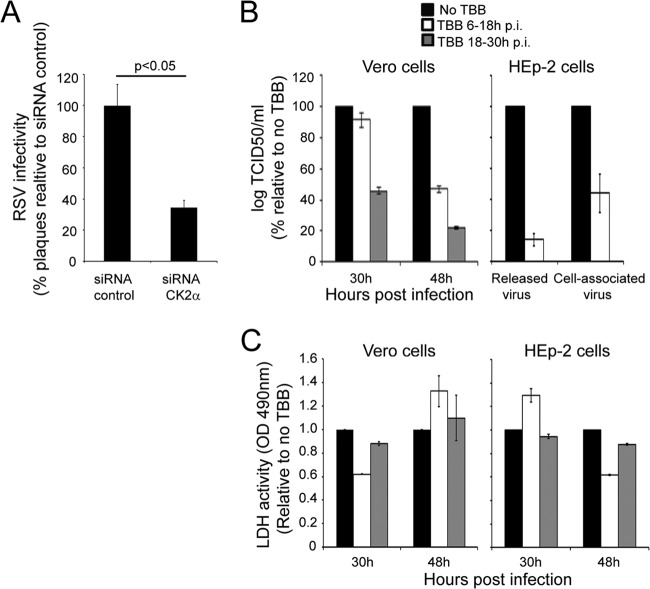
In similar fashion to a number of other paramyxoviruses (34–36), the M protein of RSV is known to be phosphorylated in infected cells (37). Since our previous analysis had implicated protein kinase CK2 as being involved in phosphorylation of M (7, 38), we firstly tested the effect on RSV production (MOI of 1) of siRNA-mediated knockdown of the CK2α subunit, with scrambled siRNA as a control (Fig. 1A). CK2α-depleted cells showed a consistent >60% decrease in the number of PFU, implying a role for CK2 in the RSV infectious cycle. We confirmed this by testing the effect on infection of the specific CK2 inhibitor 4,5,6,7-tetrabromobenzo-triazole (TBB) (24, 25) added at either 6 or 18 h postinfection. Strikingly, RSV production was reduced up to 80% (Fig. 1B, left); these inhibitory effects were also observed in HEp-2 cells (Fig. 1B, right), where a decrease in titer was observed for both cell-associated and released virus. Cell death, as monitored by measuring LDH release into the culture supernatant, was not significantly altered in TBB-treated cultures compared to that in untreated cultures (Fig. 1C), indicating clearly that the observed decrease in RSV titer was not due to induction of cell death by TBB.
FIG 1.
CK2 activity enhances RSV infectious virus production. (A) A549 cells were treated for 48 h with 20 nM siRNA specific for CK2α or control siRNA, followed by infection with RSV at an MOI of 1 for 24 h. Cells were harvested, and cell-associated virus was analyzed by plaque assay on Vero cells. Results shown are represented as the percentages of plaque compared to that of the negative control and are the means ± standard errors of the means (SEM) (n = 3 experiments); significant difference (Student's t test) is indicated by the P value. (B) Vero or HEp-2 cells were infected with RSV at an MOI of 1 and subsequently treated with the CK2-specific inhibitor TBB at 2.8 ng/ml (24, 25) for 12 h either 6 or 18 h postinfection (p.i.). In the case of Vero cells, cell-associated virus was harvested at 30 h and 48 h p.i. and titrated on Vero cells. In the case of HEp-2 cells, culture supernatants and cell-associated virus were collected at 48 h p.i. and infectious virus titers were determined on Vero cells. Results for log TCID50/ml (mean ± SEM, n = 3 experiments) are shown as percentages relative to the control (no TBB). (C) Vero and HEp-2 cells were infected with RSV A2 at an MOI of 1. Cells were treated without or with 2.8 ng/ml TBB for 12 h at 6 to 18 h p.i. or 18 to 30 h p.i. The medium was then replaced with fresh medium without TBB, and cells were cultured further up to 48 h p.i. Cell toxicity was determined by LDH release in culture supernatants as described in Materials and Methods. Data (means ± standard deviations [SD]) from two independent experiments are represented as LDH activity, determined in triplicate, relative to no treatment.
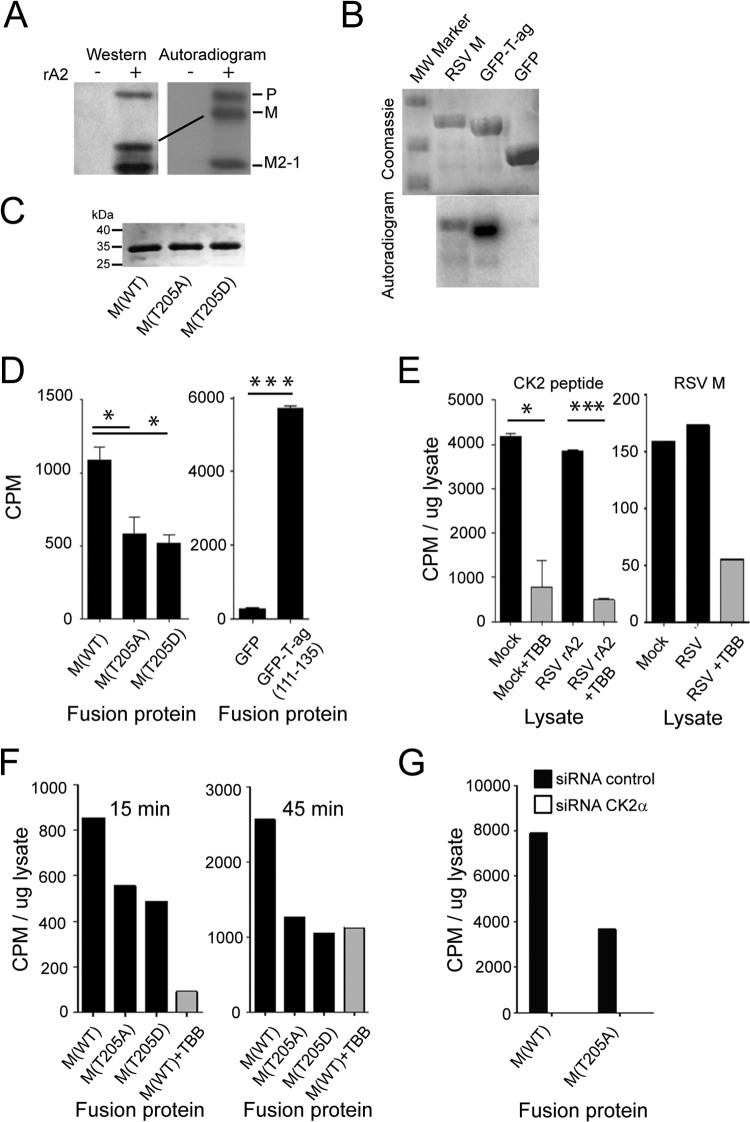
To confirm that M is phosphorylated in RSV-infected cells, as observed previously (37), HEp-2 cells were infected with RSV rA2 at an MOI of 1 and labeled with 32Pi for 2 h at 18 h p.i., and RSV proteins were immune-precipitated from the lysate using an anti-RSV antibody. Samples were analyzed by Western analysis/autoradiography, with phosphorylated RSV P, M, and M2-1 proteins able to be detected (see reference 37); in the case of M, the phosphorylated protein showed altered mobility on the SDS-PAGE gel (Fig. 2A). In the absence of suitable antibodies to perform immunoprecipitation of untagged M protein from infected cells, we set out to analyze potential CK2 phosphorylation sites responsible for the observed phosphorylation. We expressed and purified recombinant RSV M WT and mutant (see below) M proteins as described in Materials and Methods and then compared WT M protein to T-ag-GFP (SV40 large tumor antigen amino acids 111 to 135, including two strong sites for CK2 phosphorylation [39] fused in frame to the GFP coding sequence), with respect to its ability to be phosphorylated in vitro by purified CK2 (Fig. 2B). In contrast to GFP alone as a negative control, M was clearly able to be phosphorylated by CK2. We decided to focus on the consensus CK2 site at Thr205 within the nuclear export sequence of RSV M, which is critical for viral infection (7, 26), initially substituting Thr205 to either a nonphosphorylatable Ala (T205A) or phosphomimetic Asp (T205D) residue, expressing the resultant recombinant M proteins in bacteria (Fig. 2C), and quantifying phosphorylation by purified CK2 (29, 30, 39) (see Materials and Methods). Phosphorylation of WT M Thr205 was clearly evident (Fig. 2D), with the two mutant variants lacking a phosphorylatable Thr205 showing markedly (ca. 50%) reduced phosphorylation levels; results for T-ag-GFP and GFP alone are shown in the right panel of Fig. 2D for comparison. The clear implication is that Thr205 is able to be phosphorylated specifically by CK2.
FIG 2.
Thr205 within M protein is a key target for CK2. (A) RSV A2 (+)- or mock (−)-infected (MOI of 1) HEp-2 cells were metabolically labeled with 32Pi for 2 h at 18 h p.i. followed by lysis in RIPA buffer. RSV proteins were immunoprecipitated from the cleared lysates with goat anti-RSV antibody conjugated to protein G Sepharose beads (Merck Millipore), eluted by boiling in Laemmli buffer, and separated by SDS-PAGE. The gel was dried under vacuum and exposed to X-ray film. The P, M, and M2-1 proteins of RSV are indicated on the right. In parallel, samples were subjected to Western analysis using anti-RSV (Abcam) polyclonal antibody followed by donkey anti-goat HRP-conjugated secondary antibody (Santa Cruz Biotechnology). (B) M protein is phosphorylated by CK2 kinase in vitro. Recombinant His6-tagged M protein was expressed in E. coli and purified on Ni-NTA beads, along with His-tagged T-ag-GFP and GFP as controls. Purified proteins were incubated with 2 ng purified CK2 (Millipore) in the presence of γ32P[ATP] for 30 min at 30°C, and the reaction was stopped by boiling in Laemmli buffer. Reaction products were separated by SDS-PAGE and visualized by Coomassie blue staining (top), and the gel was dried and processed for autoradiography using the Typhoon Trio (GE Healthcare) (bottom). (C) M proteins used for phosphorylation experiments. Recombinant His6-tagged M protein derivatives were expressed in E. coli and purified on Ni-NTA beads. Proteins were run on SDS-PAGE and visualized by Coomassie blue staining. (D) M protein can be phosphorylated by CK2 at Thr205 in vitro. Purified proteins from panel C, along with His-tagged T-ag-GFP (positive control; see Materials and Methods) and GFP (negative control) were incubated with 2 ng purified CK2 (Millipore) in the presence of γ32P[ATP] for 120 min at 30°C, and the reaction was stopped by spotting onto P-81 paper/washing in phosphoric acid. Counts incorporated were determined by scintillation counting. Results represent the means ± SD (n = 2 experiments). (E) M protein can be phosphorylated in a TBB-sensitive manner by lysates prepared from RSV-infected cells in vitro. Phosphorylation was carried out for 15 min in the presence of ca. 24 μg cell lysate prepared as described in Materials and Methods from mock- or RSV-infected cells treated without or with TBB (2.8 ng/ml), prior to analysis of incorporated counts as described for panel D. Positive control (left) used a peptide CK2 substrate. Results represent the means ± SD (n = 2 experiments). (F) Thr205 within M is a key site for phosphorylation by cell lysates. M protein derivatives were incubated with cell lysates as described for panel E, without or with TBB (25 μM) treatment. Analysis was as described for panels D and E. (G) M protein phosphorylation by cell lysates is dependent on CK2. A549 cells were transfected with siRNA (scrambled or CK2α specific) at 20 nM, and then lysates were prepared 48 h later. Phosphorylation of recombinant purified M was carried out for 15 min in the presence of ca. 24 μg cell lysate prepared as described in Materials and Methods, prior to analysis of incorporated Pi as described for panel E; results are from a single typical experiment from a series of 2 experiments. Significant differences are denoted: *, P < 0.05; ***, P < 0.0001.
As a step toward relating the above-given results to infected cells, we prepared cell lysates from Vero cells in the absence or presence of RSV infection (MOI of 4) 24 h p.i. and assessed phosphorylation of a CK2 model peptide substrate and M (Fig. 2E, left and right, respectively) in the absence or presence of TBB. RSV-infected cell lysates were clearly able to phosphorylate the CK2 peptide in a TBB-sensitive manner, in comparable fashion to uninfected cells, consistent with robust CK2 activity in the infected cells. Importantly, M was also able to be phosphorylated by extracts from both uninfected and infected Vero cells to a comparable extent; the latter was able to be inhibited ca. 70% by TBB (Fig. 2E, right), implying the contribution of CK2 to this phosphorylation. Finally, M and the T205A and T205D mutant derivatives were tested for phosphorylation by cell lysates, results clearly indicating that WT M can be phosphorylated to levels about 2-fold higher than those for the T205A- or T205D-substituted proteins (Fig. 2F). This implies that Thr205 is a major site for phosphorylation of M in cells; importantly, phosphorylation of M was reproducibly able to be inhibited by TBB, implicating CK2 as the main kinase responsible. Consistent with this idea, knockdown of CK2α using specific siRNA reduced M phosphorylation by cell lysates (Fig. 2G), whereas nonspecific (scrambled) siRNA did not.
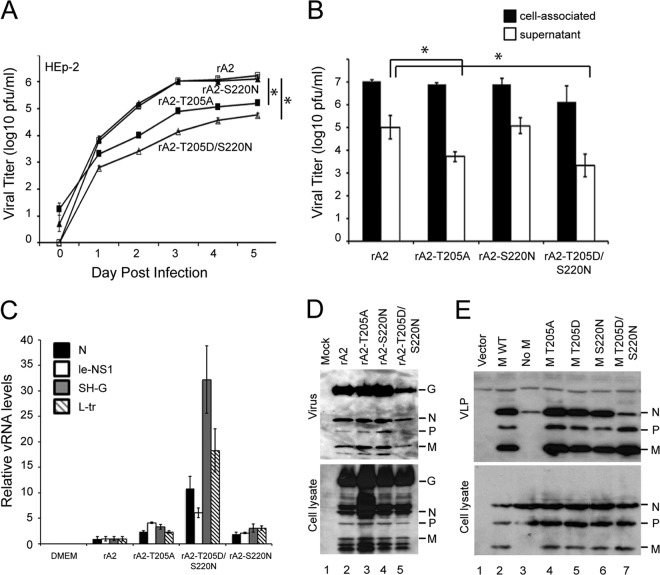
To test for the importance of Thr205 in RSV infection, we substituted Thr205 to either a nonphosphorylatable Ala (T205A) or phosphomimetic Asp (T205D) residue in the context of the full-length RSV A2 antigenome using reverse genetics (40). Although RSV with the T205A mutation (rA2–T205A) could be readily recovered, T205D-containing RSV could not be recovered in more than 10 attempts; however, a partial revertant virus was able to be recovered, which was found to retain a second-site suppressor mutation at position 220 (S220N) in M additional to T205D (rA2–T205D/S220N). Ser220 itself represents a putative phosphorylation site for the double-stranded DNA-dependent protein kinase, with examination of the published crystal structure for M (41) highlighting the fact that Thr205 and Ser220 are in relatively close proximity on the surface of the molecule and implying potential functional interaction between the two sites (see below). We subsequently generated and were able to recover the S220N mutant RSV (rA2–S220N).
The three mutant viruses were initially compared to WT rA2 virus in a multiple-step (MOI of 0.01) replication assay (Fig. 3A). The rA2–T205D/S220N virus showed markedly slower replication kinetics in HEp-2 cells (Fig. 3A), with titers 10- to 100-fold lower than those of the WT virus (rA2). The rA2–T205A virus also showed slower kinetics, with titers up to 10-fold lower than those of rA2, while rA2–S220N showed virus production kinetics indistinguishable from those of rA2. Both rA2–T205A and rA2–T205D/S220N viruses had significantly (P < 0.05) reduced infectivity compared to that of the WT for all time points measured. Similar results for virus production were obtained in A549 cells (not shown). We also compared the numbers of released infectious virus in a single-step (MOI of 3) infectivity assay in HEp-2 cells, finding that the titer of infectious RSV in the supernatant at 24 h postinfection was significantly (P < 0.05) lower for rA2–T205A virus (>10-fold) and up to 100-fold lower for rA2–T205D/S220N virus than for rA2 (Fig. 3B). To test whether this was attributable to impaired virus release or reduced infectivity of the virus released, we performed qRT-PCR to determine the copies of viral RNA per PFU (vRNA/PFU ratio) as normalized to the WT. Significantly (P < 0.01) higher copy numbers in the rA2–T205D/S220N virus (up to 30-fold higher than in rA2) were estimated (Fig. 3C), clearly implying that released rA2–T205D/S220N virus has reduced infectivity. It should be noted that although PCR results for the SH-G region were somewhat higher than those for the other regions/primer sets over three different experiments, presumably due to differences in primer binding/PCR efficiency, the rA2–T205D/S220N virus clearly showed higher levels than the other viruses tested for all RNA regions amplified, consistent with reduced infectivity of released virus.
FIG 3.
M protein Thr205 is required for optimal RSV production. (A) Monolayers of HEp-2 cells were infected by WT rA2 or the indicated M mutant rRSVs at an MOI of 0.01, and the supernatant was sampled every 24 h for 5 days prior to determination of infectious virus, performed by standard plaque assay on Vero cells as described previously (26). Results represent the means ± SEM (n = 3 experiments). *, P < 0.05. (B) HEp-2 cells were infected with the indicated viruses at an MOI of 3; samples were collected 24 h later, and titers of cell-associated and released virus were determined on HEp-2 cells using an immune plaque assay as described in Materials and Methods. Results represent the means ± SD (n = 3 experiments). *, P < 0.05. (C) Results of qRT-PCR analysis performed on viral RNA from 106 PFU of each virus (supernatant fraction) using primers specific for the N, leader-NS1, SH-G, and L-trailer regions of the genome. Results are for the means ± SEM (n = 3 experiments). (D) HEp-2 cells were infected without (mock) or with WT rA2 or the indicated M mutant rRSVs at an MOI of 3. Twenty-four hours p.i., virus was isolated from the supernatant after centrifugation (1,300 × g, 10 min, 4°C) to remove cell debris and pelleted through a sucrose cushion. Cell lysates were generated using RIPA buffer. Virus (top) and cell lysate (bottom) were then subjected to Western analysis using anti-RSV (Abcam) polyclonal antibody followed by donkey anti-goat HRP-conjugated secondary antibody (Santa Cruz Biotechnology). (E) HEp-2 cells were cotransfected with pcDNA3.1 vector plasmid (lane 1), with pcDNA3.1 plasmids encoding RSV P, N, and F proteins plus pcDNA3.1 RSV M WT (lane 2, positive control), with pcDNA3.1 plasmids encoding RSV P, N, and F proteins plus empty pcDNA3.1 vector (lane 3, negative control), or with the indicated RSV M mutant constructs (lanes 4 to 7). Forty-eight hours posttransfection, VLPs (top) were isolated from the supernatant after centrifugation (1,300 × g, 10 min, 4°C) to get rid of cell debris, and the clean supernatant was pelleted through a sucrose cushion. Cell lysates (bottom) were generated using RIPA buffer. VLPs and cell lysates were then subjected to Western analysis using anti-RSV (Abcam) polyclonal antibody followed by donkey anti-goat HRP-conjugated secondary antibody (Santa Cruz Biotechnology).
In order to examine further whether impaired fitness of the rA2–T205A and rA2–T205D/S220N mutants is due to reduced infectivity of released virus or a result of impaired virus release itself, we infected HEp-2 cells with the rA2, rA2–T205A, rA2–S220N, or rA2–T205D/S220N virus at an MOI of 3 and analyzed both cell-associated and supernatant (released) virus by Western analysis using RSV antibody (Fig. 3D). Viral protein expressions appeared to be comparable between all the strains in cell lysates (cell-associated virus); in the case of released virus, protein expressions were again comparable between all of the strains with the exception of the rA2–T205D/S220N mutant, which also showed somewhat reduced protein levels. Clearly, however, the mutant recombinant viruses are comparable to the wild type in terms of viral protein composition.
RSV viruslike particles (VLPs) can be generated independent of viral infection by transfecting cells with plasmids encoding the RSV M, N, P, and F proteins (13). We decided to use this assay to study M carrying the T205D mutation, which cannot otherwise be examined in the context of recombinant RSV. HEp-2 cells were transfected to express WT F, N, P, and various M constructs, and cell lysate and the VLPs in the supernatant fraction were analyzed by Western analysis using RSV antibody (Fig. 3E). All proteins were equally expressed in cells (Fig. 3E, bottom), indicating clearly that the M mutations do not impact M protein expression levels. In the presence of F, N, P, and WT M, RSV VLPs were formed (Fig. 3E, lane 2, top, positive control), whereas no VLPs could be detected at 24 h posttransfection in the absence of M (lane 3, top, negative control). M containing the T205A mutation did not prevent VLP release, nor did M containing the T205D or S220N mutation or the T205D/S220N double substitution (lanes 4 to 7, top). The clear implication is that T205 in M is critical for viral fitness, and point mutations in T205 do not reduce viral release per se but rather would appear to impact on the infectivity of the released viral particles.
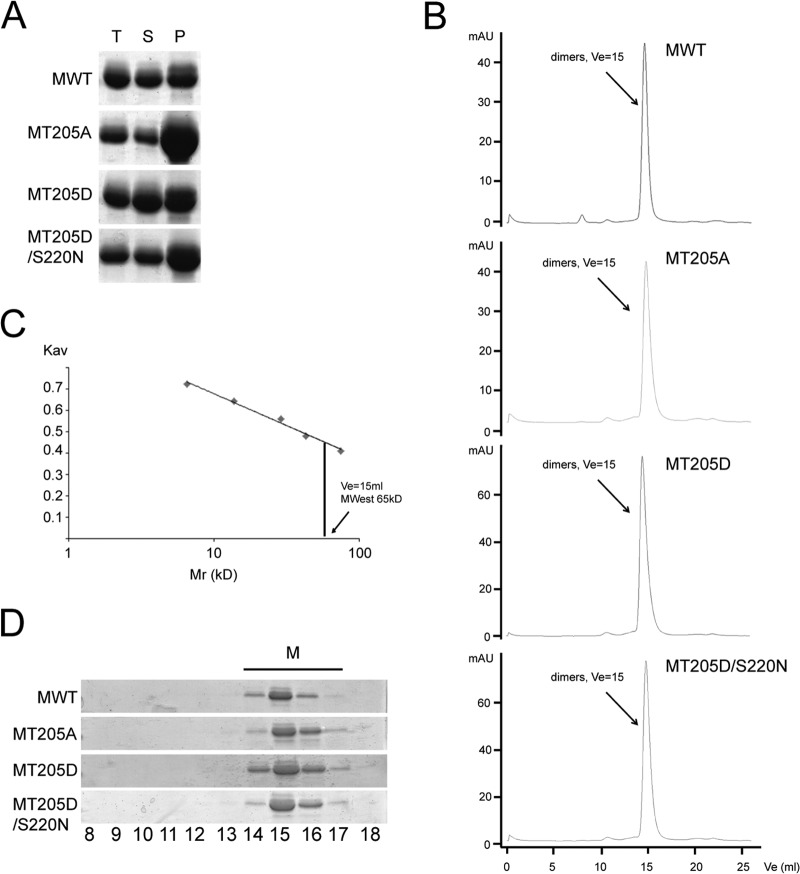
Substitution of M T205 does not impact on M dimerization.
RSV M, like other paramyxovirus matrix proteins, forms higher-order structures that are believed to be important in viral assembly on the plasma membrane (18–22). Although monomeric in the X-ray crystallographic model, recombinant RSV M is known to form higher-order oligomer structures (20, 41) and, based on similarities with NDV and hMPV M, speculated to form dimers as the basic biological unit for subsequent oligomerization (22, 23). We decided to test the ability of WT and mutant M recombinant proteins to form dimers/higher-order oligomers. Recombinant RSV M WT, T205A, T205D, and T205D/S220N proteins were expressed in bacteria, purified on nickel affinity chromatography, and concentrated to 3 mg/ml, all under conditions similar to those used for RSV M crystallization (41). Purified proteins were incubated for 24 h, and oligomerization was analyzed by measuring the ratio between the insoluble and soluble M protein after centrifugation to pellet-aggregated protein. As seen in Fig. 4A, all four proteins could be found distributed between the two fractions; insoluble protein, presumably largely comprising higher-order oligomers, was found in the pellet (P) fraction, with soluble protein in the supernatant (S). Total overall protein levels (T) were comparable for the different proteins, but while the WT and the T205D mutant were equally distributed between the P and S fractions, the T205A mutant was found predominantly in the P fraction, suggesting increased oligomerization/aggregation. The T205D/S220N double mutant, compared to the WT, showed slightly more protein in the P fraction. RSV M has previously been shown to form ordered oligomers in vitro when incubated with specific sets of lipids (20); we assume that the proteins found in the P fraction correspond to these higher-order oligomeric structures but aggregate in the absence of lipids. To analyze the oligomeric state of the soluble M proteins further, we ran the soluble fractions on size exclusion chromatography. WT M (molecular mass of 29 kDa) and the T205A, T205D, and T205D/S220N mutants all eluted as single dimeric species (estimated molecular mass of 65 kDa), implying that mutation of T205 does not impact on M's ability to dimerize (Fig. 4B and C). Eluted peaks were subjected to SDS-PAGE analysis and Coomassie staining to confirm the identity of the peak as M protein (Fig. 4D). Importantly, although recombinant T205A M appeared to form higher-order oligomeric aggregates with increased propensity, the results show that none of the mutations prevent stable dimer formation.
FIG 4.
Mutations of T205 do not abolish M's ability to form stable dimers. (A) Purification of bacterially expressed His6-tagged WT M and the T205A, T205D, and T205D/S220N mutant derivative proteins using nickel affinity chromatography. Purified proteins were concentrated to 3 mg/ml and pelleted to separate higher oligomers (P) from the soluble fraction (S), run on an SDS-PAGE gel, and stained with Coomassie. (B) Analytical S200 gel filtration chromatography was performed on the soluble fraction of WT M and the T205A, T205D, and T205D/S220N mutants. (C) Molecular masses were estimated by comparing the gel-phase distribution coefficient (Kav) of the M protein peaks with the values obtained for known calibration standards (GE Healthcare). Mw of M peaks was estimated by fitting to the calibration curve (plot Kav versus log Mr); the single peak was estimated to be a dimer (estimated Mw, 65 kDa) and is indicated by an arrow. (D) Fractions 8 to 18 were run on an SDS-PAGE gel and stained with Coomassie, identifying the peak as purified M protein.
T205D M can produce viruslike filaments but with altered distribution of M.
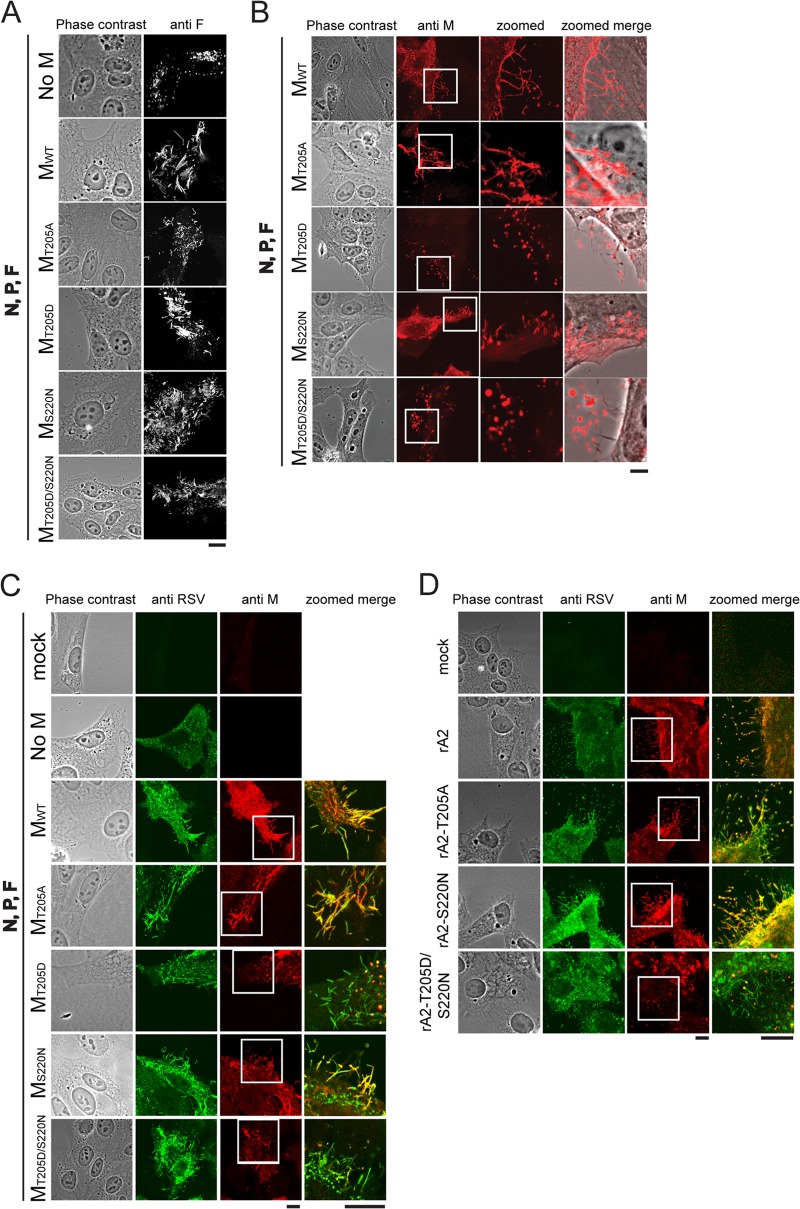
M is known to localize into RSV viral filaments, with oligomerization of M believed to be integral in filament elongation (12, 16, 20). To test whether the T205 mutations within M may impact on viruslike filament formation, we applied a transfection-based filament formation assay. No filaments were formed at 24 h after transfection in the absence of M (Fig. 5A, first row, negative control), in agreement with previous reports (13, 16). In the presence of F, N, P, and WT M, viruslike filaments were formed as detected by staining for F (Fig. 5A, second row, positive control). M proteins containing the T205A, T205D, S220N, and T205D/S220N mutations were all able to form filaments containing F (Fig. 5A), although they seemed to be shorter and not as regular as those observed for the WT (see also Fig. 6). Staining for M revealed clear differences between the WT and the T205 mutants in terms of distribution of M along the filaments (Fig. 5B). In the case of both WT M and M carrying the S220N mutation, M was clearly distributed along the whole length of the filaments. In contrast, T205D showed circular aggregates of M associated with the filaments (Fig. 5B, compare details with those for WT M); the T205D/S220N double mutant was similar. The T205A mutant, in contrast, showed increased staining for M associated with the filaments, including large aggregates. Clearly, oligomers/aggregates of M would appear to be localized to the filaments in the case of T205A and T205D M proteins, in contrast to the more ordered staining structures evident for WT and S220N M. Costaining for both M and RSV confirmed the results (Fig. 5C), revealing that viruslike filaments are formed in the case of the T205 mutants but that the distribution of M with respect to them is abnormal; the T205D and T205D/S220N mutants in particular showed circular aggregates that were mostly associated with the filaments. T205A in contrast resembled WT, with clear colocalization of M with the filaments and with aggregates not nearly as prominent. Importantly, viruslike filaments were evident in all cases, as detected by staining for RSV (Fig. 5C) or F (Fig. 5A), even for the T205D M mutant, implying that even in this case, sufficient M can be recruited to the plasma membrane to enable filament formation, in contrast to when M is absent. Despite this, M's localization/assembly on the filaments themselves appears to be irregular/altered in the case of the T205D M and T205D/S220N M mutants and increased in the case of the T205A M mutant.
FIG 5.
The T205D mutation alters M distribution on viruslike filaments. (A) HEp-2 cells were cotransfected with pcDNA3.1 plasmids encoding RSV P, N, and F proteins plus empty pcDNA3.1 vector (top row; negative control), pcDNA3.1 RSV M WT (second row; positive control), or the indicated RSV M mutant constructs (rows 3 to 6). Cells were fixed, permeabilized 24 h p.t., immunostained with anti-F (Millipore) monoclonal antibody and Alexa Fluor 568-coupled donkey anti-mouse secondary antibody (Invitrogen), and analyzed by confocal microscopy. (B) HEp-2 cells were transfected as described for panel A, fixed, permeabilized 24 h later, immunostained using monoclonal anti-M primary and Alexa Fluor 568-coupled donkey anti-mouse (Invitrogen) secondary antibodies, and analyzed by confocal microscopy. (C) HEp-2 cells were transfected as described for panel A, fixed, permeabilized 24 h later, immunostained using monoclonal anti-M and with polyclonal anti-RSV (Abcam) primary and Alexa Fluor 568-coupled donkey anti-mouse (Invitrogen) and Alexa Fluor 488-coupled rabbit anti-goat (Invitrogen) secondary antibodies, and analyzed by confocal microscopy. (D) HEp-2 cells were infected with the indicated recombinant viruses at an MOI of 3 and fixed and permeabilized 24 h later prior to immunostaining and confocal microscopy as described for panel C. Areas that are blown up in the details (zoomed column) are marked with white squares in the original panel. Scale bars represent 10 μm.
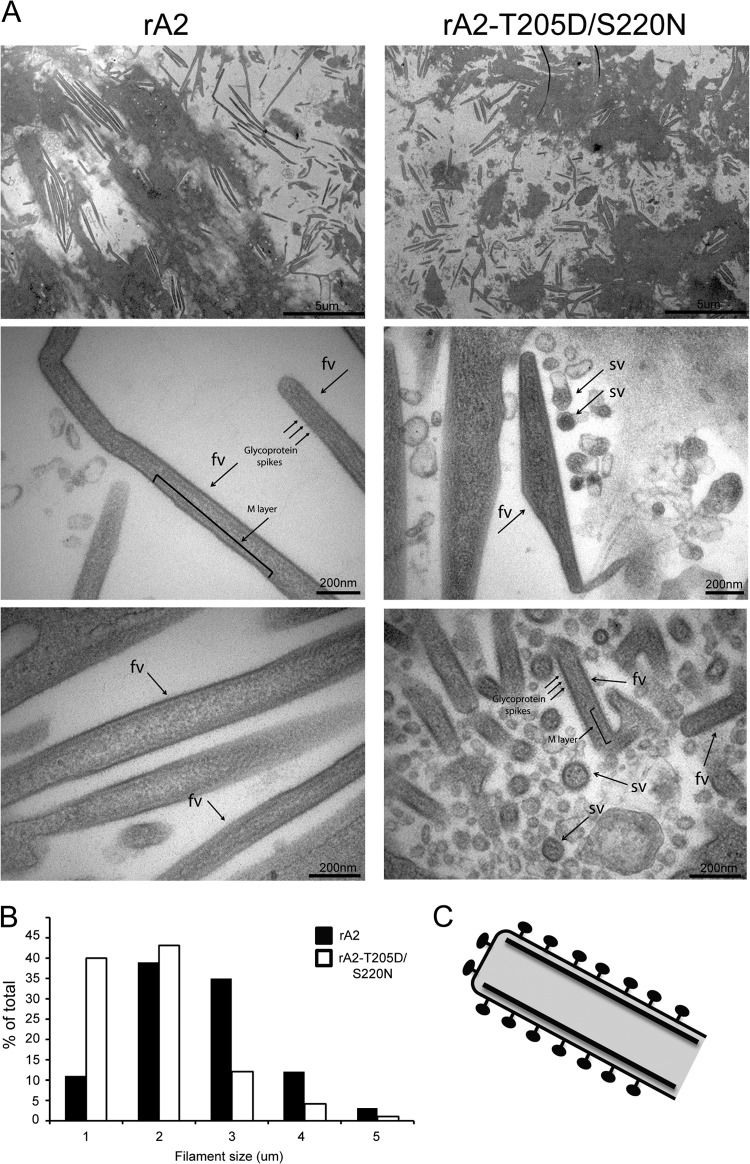
FIG 6.
(A) HEp-2 cells were infected with rA2 (left) or rA2–T205D/S220N (right) recombinant viruses at an MOI of 3. Cells were fixed 24 h later, ultrathin sections were cut parallel to the dish, and cells were stained and processed for transmission electron microscopy as described in Materials and Methods. Different magnifications shown are denoted by the scale bars; in the higher magnifications, fv marks filamentous and sv marks spherical virus particles, with the M layer and glycoprotein spikes indicated. (B) For each virus strain, 150 viral filaments were randomly selected and analyzed for filament dimensions, with the results shown as a percentage of the total. (C) A model diagram of viral filament is shown based on reference 12, with the indicated M layer below the membrane and glycoprotein spikes on the outside.
To extend the results to infected cells, we performed immunofluorescence on HEp-2 cells infected with the various recombinant viruses (Fig. 5D). Results were comparable to those described above, whereby cells infected with rA2 or rA2–S220N virus contained filaments with M largely distributed throughout the filaments (colocalization of anti-M and anti-RSV staining) and with few circular aggregates, while filaments formed in cells infected with rA2–T205D/S220N virus did not show extensive colocalization of M, except in the form of circular aggregates of M (Fig. 5D). Filaments in cells infected with rA2–T205A virus appeared to show more colocalization of M and RSV staining and increased levels of aggregates of M on the filaments. Viral filaments are thus clearly present in cells infected with virus carrying T205 mutations but appear to contain large regions of filaments free of M and/or containing associated circular aggregates of M. Since T205 mutations result in reduced RSV infectivity (Fig. 3), the implication is that ordered assembly of M within the viral filaments may be required for fully infectious RSV.
rA2–T205D/S220N virus produces disordered and shorter viral particles than rA2 WT.
To examine the viral filaments at higher resolution, we applied transmission electron microscopy (TEM) to HEp-2 cells infected with the rA2 and rA2–T205D/S220N viruses (Fig. 6); rA2–T205D/S220N virus was selected based on the fact that it showed the strongest phenotype in terms of M distribution along viral filaments (Fig. 5), as well as infectivity (Fig. 3), results confirming that the T205D mutation does not abolish cell-associated filament formation. rA2 (Fig. 6, left) produced viral filaments mostly 2 to 3 μm in length, with some >4 μm. Higher magnification revealed the filaments to be ordered, mostly linear structures and enabled identification of a layer beneath the filament membrane, probably representing the M protein, and spikes on the outside, probably representing the glycoproteins (12). In contrast, rA2–T205D/S220N (Fig. 6, right) filaments appeared shorter (80% at ≤2 μm, compared to 50% at ≥2 μm in the case of the WT virus), less regular, and with marked branching. In addition, larger numbers of spherical viral particles were seen, suggestive of the possibility that the cell-associated filaments may be less stable in this case and more easily disrupted (it should be noted that since serial sections were not analyzed, it is not formally possible to discern spherical particles and filament cross sections). Interestingly, although M was clearly within the filaments, the layer of M beneath the filament membrane in the rA2–T205D/S220N samples was less distinct than in those for rA2 (Fig. 6). Overall, our EM analysis indicates that rA2–T205D/S220N virus forms disordered viral filaments that are shorter than rA2 viral filaments, which presumably is the basis of the significantly reduced infectivity of rA2–T205D/S220N virus (Fig. 3) and the inability of rA2–T205D virus to be recovered. Together with recently published data (12), these results suggest an M-dependent mechanism for RSV filament morphogenesis during budding.
DISCUSSION
We show here for the first time that RSV M's role in virus assembly/release is strongly dependent on threonine 205 (Thr205), a consensus site for CK2, which appears to play a key regulatory role in modulating M oligomerization and association with virus filaments. Thr205 appears to be phosphorylated specifically by CK2, with mutant derivatives lacking the site being poorly phosphorylated, and the specific CK2 inhibitor TBB able to markedly reduce phosphorylation, strongly implying that CK2 is the major kinase targeting the site in infected cells; CK2 depletion or TBB treatment inhibits RSV production in infected cells (Fig. 1). Underlining the physiological importance of the site, virus containing the phosphomimetic T205D mutation residue could not be recovered except with an additional, compensating S220N substitution, which, however, could not compensate fully for proper M assembly. Our analysis using recombinant M protein showed that the T205A, T205D, and T205D/S220N mutations do not affect M's ability to dimerize but likely impact on M's ability to form ordered oligomeric structures. Importantly, although the T205D mutation does not preclude the formation of M dimers in vitro or impact on viruslike filament formation in cell-based systems, rA2–T205D-carrying virus could not be recovered, presumably due to the increased propensity of the T205D mutant to form circular aggregates (Fig. 5) rather than assemble in ordered fashion into viral filaments (Fig. 6), and that appears to be critical for high infectivity. In a fashion similar to that of the T205D mutant, the T205D/S220N mutant derivative forms circular aggregates (Fig. 5) but appears to be able to form viral filaments, albeit of reduced length (Fig. 6) and with resultant reduced infectivity of released virus (Fig. 3). M was shown, by using electron cryotomography analysis of released filamentous virus, to form higher-order structures beneath the viral membrane (12), and we conclude here that M oligomerization during RSV assembly is critical for infectious virus production. The exact mechanism by which the S220N mutation in M can compensate for the T205D mutation, which in isolation is lethal to the RSV life cycle, is unclear, but it should be noted that Thr205 and Ser220 are in close proximity in the crystal structure of M (41), implying that Ser220 may well impact on access to/phosphorylation at Thr205 (see below). Since the T205A mutation also impacts RSV production (Fig. 3), it seems reasonable to postulate that reversible phosphorylation at Thr205 is critical in the RSV life cycle, presumably to regulate in precise fashion the ordered oligomerization process of M with respect to the nascent virus filaments in the infected cell (see also reference 22).
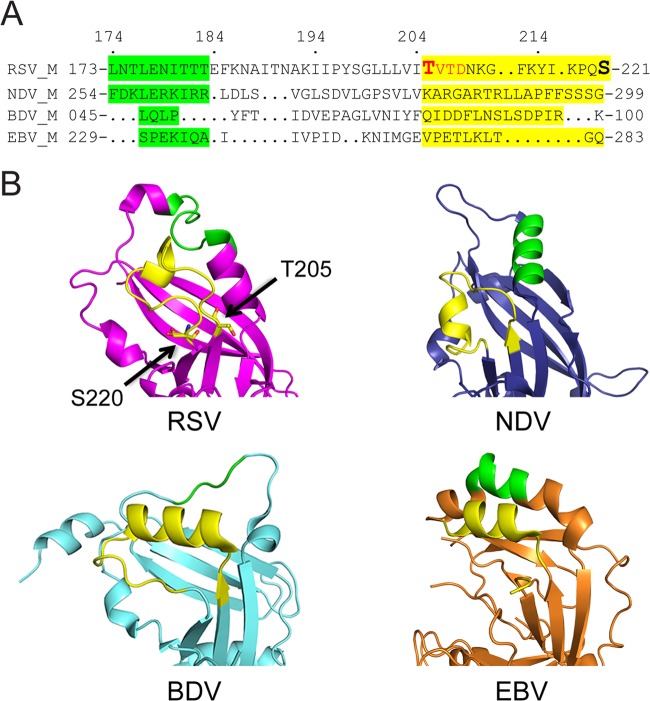
Structure-based sequence alignment (Fig. 7A) based on reference 23 indicates no real sequence conservation between RSV M (41) and most Mononegavirales M proteins (23, 42, 43). However, structural alignment of the area around the RSV M Thr205-Ser220 loop (Fig. 7B) indicates that all four proteins have an unstructured loop in that position, close to an α-helical region (green) that has been proposed to play a key role in higher-order oligomerization in the case of NDV M (23). Thr205 and Ser220 are in close proximity on the same surface of the RSV M molecule (Fig. 7B), with the region of RSV M postulated to be involved in oligomerization also in close proximity. Significantly, similar close proximity is also evident in the structures for NDV, Borna virus (BDV), and Ebola virus (EBV) between the sequences that align with the NDV oligomerization region (green) and the RSV M loop (yellow). The hMPV M structure (22) is not included in our structural alignment since the precise region of the loop was not resolved in the crystal structure, implying a highly flexible region. In the case of NDV, BDV, and EBV, a number of hydrogen bonds appear to hold these regions together; intriguingly, in this context, it can be speculated that phosphorylation at Thr205 with the loop region of RSV M may conceivably function to regulate/stabilize interaction of the loop and oligomerization region. Although this appears to be conserved in RSV strains, the mechanism by which interaction may be regulated in NDV, etc., is unclear at this stage.
FIG 7.
Structural conservation among different Mononegavirales matrix proteins. (A) Structure-based sequence (single-letter amino acid code) alignment (adapted from reference 23) around the loop from RSV M containing Thr205 to Ser220 (Thr205 and Ser220 marked), with residues proposed to play a key role in higher-order oligomerization based on structural alignment to NDV M highlighted in green, and the residues within the respective proteins comprising the loop highlighted in yellow based on alignment to RSV M. CK2 consensus site in RSV M indicated in red. Residues in each protein are numbered. (B) Structure-based alignment of known M structures, shown as a ribbon diagram; RSV M (pink) (41), NDV M (blue) (23), BDV M (cyan) (42), and EBV M (orange) (43), with the RSV M Thr205-Ser220 loop or comparable loops shown in yellow and oligomerization residues in green.
Our analysis indicates that T205 mutations do not impair M dimerization or viruslike filament formation per se, underlining the fact that the mutations do not cause gross misfolding of M or affect its stability but rather the ability of M to assemble in ordered fashion on the viral filaments themselves. This appears to impact in turn upon the infectivity of released virus rather than on virus production or release itself. Since RSV filaments are formed in rA2–T205A and rA2–T205D/S220N mutants but have quite distinct patterns of localization of M within them, it can be proposed that the ordered distribution of M along the entire viral filaments may be critical for the infectivity of released virus. That the defect in T205D-containing rA2 is unlikely to be related to the role of RSV F in viral filament formation (15) is indicated by the fact that F appeared to localize to normal extents to the RSV viral filaments in all cases (Fig. 5A). Based on detailed examination of RSV virion morphology (12), we concluded that RSV infectivity relates strongly to the fraction of filamentous as opposed to spherical virus particles; our EM analysis confirms that impaired oligomerization of M leads to production of more branched and less elongated viral filaments, correlated with reduced infectivity of the released virus. Thus, M oligomerization would appear to be a target of interest for the development of anti-RSV agents; further, the rA2–T205A and rA2–T205D/S220N mutant viruses described here would appear to be the first RSV mutants affected in viral maturation to our knowledge and hence of considerable interest for vaccine approaches in the future.
ACKNOWLEDGMENTS
We are indebted to Peter Collins for the reverse genetics system for RSV, to Marty Moore for codon-optimized expression plasmids, and to Mike Hollinshead for help with EM analysis.
This work was supported by institutional funds from Imperial College (identifier [ID] no. 707999), by a Marie Curie Career Integration grant (ID no. 321931) from the European Commission, by a fellowship from the Human Frontier Science Program Organization (M.B.), by project grants (ID no. 606407 and APP1043511) and a fellowship (ID no. APP1002486) from the National Health and Medical Research Council, Australia (D.A.J.), and by institutional funds from the University of South Florida and a grant (ID no. AI081977) from the NIH (M.N.T.).
Footnotes
Published ahead of print 26 March 2014
REFERENCES
- 1.Collins PL, Graham BS. 2008. Viral and host factors in human respiratory syncytial virus pathogenesis. J. Virol. 82:2040–2055. 10.1128/JVI.01625-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang YT, Collins PL, Wertz GW. 1985. Characterization of the 10 proteins of human respiratory syncytial virus: identification of a fourth envelope-associated protein. Virus Res. 2:157–173. 10.1016/0168-1702(85)90246-1 [DOI] [PubMed] [Google Scholar]
- 3.Fearns R, Collins PL. 1999. Role of the M2–1 transcription antitermination protein of respiratory syncytial virus in sequential transcription. J. Virol. 73:5852–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins PL, Crowe JE. 2007. Respiratory syncytial virus and human metapneumovirus, p 1601–1646 In Knipe H, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed, vol 2 Lippincott, Williams, and Wilkins, Baltimore, MD [Google Scholar]
- 5.Garcia J, Garcia-Barreno B, Vivo A, Melero JA. 1993. Cytoplasmic inclusions of respiratory syncytial virus-infected cells: formation of inclusion bodies in transfected cells that coexpress the nucleoprotein, the phosphoprotein, and the 22K protein. Virology 195:243–247. 10.1006/viro.1993.1366 [DOI] [PubMed] [Google Scholar]
- 6.Carromeu C, Simabuco FM, Tamura RE, Farinha Arcieri LE, Ventura AM. 2007. Intracellular localization of human respiratory syncytial virus L protein. Arch. Virol. 152:2259–2263. 10.1007/s00705-007-1048-4 [DOI] [PubMed] [Google Scholar]
- 7.Ghildyal R, Ho A, Jans DA. 2006. Central role of the respiratory syncytial virus matrix protein in infection. FEMS Microbiol. Rev. 30:692–705. 10.1111/j.1574-6976.2006.00025.x [DOI] [PubMed] [Google Scholar]
- 8.Li D, Jans DA, Bardin PG, Meanger J, Mills J, Ghildyal R. 2008. Association of respiratory syncytial virus M protein with viral nucleocapsids is mediated by the M2–1 protein. J. Virol. 82:8863–8870. 10.1128/JVI.00343-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roberts SR, Compans RW, Wertz GW. 1995. Respiratory syncytial virus matures at the apical surfaces of polarized epithelial cells. J. Virol. 69:2667–2673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gower TL, Pastey MK, Peeples ME, Collins PL, McCurdy LH, Hart TK, Guth A, Johnson TR, Graham BS. 2005. RhoA signaling is required for respiratory syncytial virus-induced syncytium formation and filamentous virion morphology. J. Virol. 79:5326–5336. 10.1128/JVI.79.9.5326-5336.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bachi T, Howe C. 1973. Morphogenesis and ultrastructure of respiratory syncytial virus. J. Virol. 12:1173–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liljeroos L, Krzyzaniak MA, Helenius A, Butcher SJ. 15 May 2013. Architecture of respiratory syncytial virus revealed by electron cryotomography. Proc. Natl. Acad. Sci. U. S. A. 10.1073/pnas.1309070110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shaikh FY, Cox RG, Lifland AW, Hotard AL, Williams JV, Moore ML, Santangelo PJ, Crowe JE., Jr 2012. A critical phenylalanine residue in the respiratory syncytial virus fusion protein cytoplasmic tail mediates assembly of internal viral proteins into viral filaments and particles. mBio 3:pii=e00270-11. 10.1128/mBio.00270-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oomens AG, Bevis KP, Wertz GW. 2006. The cytoplasmic tail of the human respiratory syncytial virus F protein plays critical roles in cellular localization of the F protein and infectious progeny production. J. Virol. 80:10465–10477. 10.1128/JVI.01439-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baviskar PS, Hotard AL, Moore ML, Oomens AG. 2013. The respiratory syncytial virus fusion protein targets to the perimeter of inclusion bodies and facilitates filament formation by a cytoplasmic tail-dependent mechanism. J. Virol. 87:10730–10741. 10.1128/JVI.03086-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mitra R, Baviskar P, Duncan-Decocq RR, Patel D, Oomens AG. 2012. The human respiratory syncytial virus matrix protein is required for maturation of viral filaments. J. Virol. 86:4432–4443. 10.1128/JVI.06744-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghildyal R, Jans DA, Bardin PG, Mills J. 2012. Protein-protein interactions in RSV assembly: potential targets for attenuating RSV strains. Infect. Disord. Drug Targets 12:103–109. 10.2174/187152612800100125 [DOI] [PubMed] [Google Scholar]
- 18.Russell PH, Almeida JD. 1984. A regular subunit pattern seen on non-infectious Newcastle disease virus particles. J. Gen. Virol. 65(Part 6):1023–1031 [DOI] [PubMed] [Google Scholar]
- 19.Heggeness MH, Smith PR, Choppin PW. 1982. In vitro assembly of the nonglycosylated membrane protein (M) of Sendai virus. Proc. Natl. Acad. Sci. U. S. A. 79:6232–6236. 10.1073/pnas.79.20.6232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McPhee HK, Carlisle JL, Beeby A, Money VA, Watson SM, Yeo RP, Sanderson JM. 2011. Influence of lipids on the interfacial disposition of respiratory syncytial virus matrix protein. Langmuir 27:304–311. 10.1021/la104041n [DOI] [PubMed] [Google Scholar]
- 21.Gaudin Y, Barge A, Ebel C, Ruigrok RW. 1995. Aggregation of VSV M protein is reversible and mediated by nucleation sites: implications for viral assembly. Virology 206:28–37. 10.1016/S0042-6822(95)80016-6 [DOI] [PubMed] [Google Scholar]
- 22.Leyrat C, Renner M, Harlos K, Huiskonen JT, Grimes JM. 2014. Structure and self-assembly of the calcium binding matrix protein of human metapneumovirus. Structure 22:136–148. 10.1016/j.str.2013.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Battisti AJ, Meng G, Winkler DC, McGinnes LW, Plevka P, Steven AC, Morrison TG, Rossmann MG. 2012. Structure and assembly of a paramyxovirus matrix protein. Proc. Natl. Acad. Sci. U. S. A. 109:13996–14000. 10.1073/pnas.1210275109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alvisi G, Jans DA, Guo J, Pinna LA, Ripalti A. 2005. A protein kinase CK2 site flanking the nuclear targeting signal enhances nuclear transport of human cytomegalovirus ppUL44. Traffic 6:1002–1013. 10.1111/j.1600-0854.2005.00331.x [DOI] [PubMed] [Google Scholar]
- 25.Sarno S, Papinutto E, Franchin C, Bain J, Elliott M, Meggio F, Kazimierczuk Z, Orzeszko A, Zanotti G, Battistutta R, Pinna LA. 2011. ATP site-directed inhibitors of protein kinase CK2: an update. Curr. Top. Med. Chem. 11:1340–1351. 10.2174/156802611795589638 [DOI] [PubMed] [Google Scholar]
- 26.Ghildyal R, Ho A, Dias M, Soegiyono L, Bardin PG, Tran KC, Teng MN, Jans DA. 2009. The respiratory syncytial virus matrix protein possesses a Crm1-mediated nuclear export mechanism. J. Virol. 83:5353–5362. 10.1128/JVI.02374-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mahy BWJ, Kangro HO. 1996. Virology methods manual, p 25–46 Harcourt Brace, New York, NY [Google Scholar]
- 28.Lappalainen K, Jaaskelainen I, Syrjanen K, Urtti A, Syrjanen S. 1994. Comparison of cell proliferation and toxicity assays using two cationic liposomes. Pharm. Res. 11:1127–1131. 10.1023/A:1018932714745 [DOI] [PubMed] [Google Scholar]
- 29.Xiao CY, Hubner S, Elliot RM, Caon A, Jans DA. 1996. A consensus cAMP-dependent protein kinase (PK-A) site in place of the CcN motif casein kinase II site simian virus 40 large T-antigen confers PK-A-mediated regulation of nuclear import. J. Biol. Chem. 271:6451–6457. 10.1074/jbc.271.11.6451 [DOI] [PubMed] [Google Scholar]
- 30.Xiao CY, Hubner S, Jans DA. 1997. SV40 large tumor antigen nuclear import is regulated by the double-stranded DNA-dependent protein kinase site (serine 120) flanking the nuclear localization sequence. J. Biol. Chem. 272:22191–22198. 10.1074/jbc.272.35.22191 [DOI] [PubMed] [Google Scholar]
- 31.Hubner S, Xiao CY, Jans DA. 1997. The protein kinase CK2 site (Ser111/112) enhances recognition of the simian virus 40 large T-antigen nuclear localization sequence by importin. J. Biol. Chem. 272:17191–17195. 10.1074/jbc.272.27.17191 [DOI] [PubMed] [Google Scholar]
- 32.Ghildyal R, Li D, Peroulis I, Shields B, Bardin PG, Teng MN, Collins PL, Meanger J, Mills J. 2005. Interaction between the respiratory syncytial virus G glycoprotein cytoplasmic domain and the matrix protein. J. Gen. Virol. 86:1879–1884. 10.1099/vir.0.80829-0 [DOI] [PubMed] [Google Scholar]
- 33.Tran KC, He B, Teng MN. 2007. Replacement of the respiratory syncytial virus nonstructural proteins NS1 and NS2 by the V protein of parainfluenza virus 5. Virology 368:73–82. 10.1016/j.virol.2007.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wechsler SL, Lambert DM, Galinski MS, Pons MW. 1985. Intracellular synthesis of human parainfluenza type 3 virus-specified polypeptides. J. Virol. 54:661–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wechsler SL, Lambert DM, Galinski MS, Heineke BE, Pons MW. 1985. Human parainfluenza virus 3: purification and characterization of subviral components, viral proteins and viral RNA. Virus Res. 3:339–351. 10.1016/0168-1702(85)90434-4 [DOI] [PubMed] [Google Scholar]
- 36.Sakaguchi T, Kiyotani K, Kato A, Asakawa M, Fujii Y, Nagai Y, Yoshida T. 1997. Phosphorylation of the Sendai virus M protein is not essential for virus replication either in vitro or in vivo. Virology 235:360–366. 10.1006/viro.1997.8701 [DOI] [PubMed] [Google Scholar]
- 37.Lambert DM, Hambor J, Diebold M, Galinski B. 1988. Kinetics of synthesis and phosphorylation of respiratory syncytial virus polypeptides. J. Gen. Virol. 69(Part 2):313–323 [DOI] [PubMed] [Google Scholar]
- 38.Alvisi G, Rawlinson SM, Ghildyal R, Ripalti A, Jans DA. 2008. Regulated nucleocytoplasmic trafficking of viral gene products: a therapeutic target? Biochim. Biophys. Acta 1784:213–227. 10.1016/j.bbapap.2007.08.021 [DOI] [PubMed] [Google Scholar]
- 39.Jans DA, Jans P. 1994. Negative charge at the casein kinase II site flanking the nuclear localization signal of the SV40 large T-antigen is mechanistically important for enhanced nuclear import. Oncogene 9:2961–2968 [PubMed] [Google Scholar]
- 40.Collins PL, Hill MG, Camargo E, Grosfeld H, Chanock RM, Murphy BR. 1995. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5′ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc. Natl. Acad. Sci. U. S. A. 92:11563–11567. 10.1073/pnas.92.25.11563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Money VA, McPhee HK, Mosely JA, Sanderson JM, Yeo RP. 2009. Surface features of a Mononegavirales matrix protein indicate sites of membrane interaction. Proc. Natl. Acad. Sci. U. S. A. 106:4441–4446. 10.1073/pnas.0805740106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neumann P, Lieber D, Meyer S, Dautel P, Kerth A, Kraus I, Garten W, Stubbs MT. 2009. Crystal structure of the Borna disease virus matrix protein (BDV-M) reveals ssRNA binding properties. Proc. Natl. Acad. Sci. U. S. A. 106:3710–3715. 10.1073/pnas.0808101106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dessen A, Volchkov V, Dolnik O, Klenk HD, Weissenhorn W. 2000. Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 19:4228–4236. 10.1093/emboj/19.16.4228 [DOI] [PMC free article] [PubMed] [Google Scholar]