ABSTRACT
Infectious salmon anemia (ISA) is a severe disease that affects farmed Atlantic salmon (Salmo salar), causing outbreaks in seawater in most salmon-producing countries worldwide, with particular aggressiveness in southern Chile. The etiological agent of this disease is a virus belonging to the Orthomyxoviridae family, named infectious salmon anemia virus (ISAV). Although it has been suggested that this virus can be vertically transmitted, even in freshwater, there is a lack of compelling experimental evidence to confirm this. Here we demonstrate significant putative viral loads in the ovarian fluid as well as in the eggs of two brood stock female adult specimens that harbored the virus systemically but without clinical signs. The target virus corresponded to a highly polymorphic region 3 (HPR-3) variant, which is known to be virulent in seawater and responsible for recent and past outbreaks of this disease in Chile. Additionally, the virus recovered from the fluid as well as from the interior of the eggs was fully infective to a susceptible fish cell line. To our knowledge, this is the first robust evidence demonstrating mother-to-offspring vertical transmission of the infective virus on the one hand and the asymptomatic transmission of a virulent form of the virus in freshwater fish on the other hand.
IMPORTANCE The robustness of the data presented here will contribute to a better understanding of the biology of the virus but most importantly will constitute a key management tool in the control of an aggressive agent constantly threatening the sustainability of the global salmon industry.
INTRODUCTION
Infectious salmon anemia (ISA) is the most destructive and challenging viral disease of farmed Atlantic salmon (1, 2). Although this disease has been observed in most salmon-producing countries that report to the International Veterinary Sanitary Authority (3, 4), the aggressiveness displayed in the extremely cold seawater of southern Chile, with cumulative mortality rates sometimes exceeding 90%, is an Achilles' heel for a blooming and promising industry (5–7). The disease was first described in Norway in 1984 (8), and devastating outbreaks have also been reported in other locations since the late 1990s, such as the Faroe Islands (9, 10), Scotland (11), the United States (12), and several locations in Canada (13, 14). The sole etiological agent of the disease has been identified as a novel RNA virus of the Orthomyxoviridae family (infectious salmon anemia virus [ISAV]) and classified as the only member of a new genus, Isavirus (2). Since the only known hosts and reservoirs for the virus are salmonid fish from the North Atlantic and North Pacific areas and because of the fact that ISAV populations consist of a mixture of avirulent and virulent strains, the epidemiology of ISA is still in its infancy, and substantial research is essential to better understand the biology of the virus and, as a result, to develop proper strategies for its control and ideally to prevent new epizootic outbursts.
One key issue that needs prompt and clear elucidation is whether, in addition to horizontal transmission, the virus is also transmitted vertically. Indeed, the readiness of horizontal transmission has been clearly demonstrated in tanks and net pens and in cohabitation experiments, suggesting that it constitutes the major pathway for the spread of this disease (15). The possibility of vertical transmission as an alternative remains highly controversial. On the one hand, the absence of the vertical route has been reported for quantitative real-time PCR (qRT-PCR)-positive brood fish (16), while on the other hand, positive detection in fertilized eggs from clinically ill brood fish, also via RT-PCR, indicated only that viral sequences were detectable, but the infective potential of a putative viral particle was not demonstrated (17). Additionally, a large real-time PCR-based screening of smolt production sites in Norway was positive for the virus, suggesting that most marine production sites might be carriers, with the virus circulating during the production cycle, based on which some sort of vertical transmission cannot be ruled out (18). Considering that all salmonid specimens in the Southern Hemisphere were introduced primarily from Europe (19), natural hosts for ISAV did not exist in Chile, and since the Chilean ISAV has been assigned to the EU genotype (6), the most likely explanation is that the virus arrived via the importation of embryos (20). Although this interpretation supports the possibility of vertical transmission, since it is not clear how the eggs were originally harvested, horizontal transmission cannot be excluded.
In the present study, we analyzed two brood stock adult female specimens from a freshwater farm which systemically harbored HPR-3, a virulent variant of ISAV, but in the absence of any clinical signs. The ovarian fluid and the eggs of the two specimens were screened, confirming the presence of low threshold cycle (CT) values for viral RNA via quantitative RT-PCR (qRT-PCR). Viral particles recovered from both sources were demonstrated to infect a sensitive fish cell line. The target virus was the HPR-3 variant, which is known to be highly virulent in seawater and is responsible for recent and past outbreaks of this disease in Chile (21). To our knowledge, this is the first robust evidence that demonstrates mother-to-offspring vertical transmission of the infective virus on the one hand and the asymptomatic transmission of a virulent form of the virus in freshwater fish on the other hand.
MATERIALS AND METHODS
Samples and processing.
Ovarian fluid and eggs were obtained from two asymptomatic Atlantic salmon (Salmo salar) female brood fish individually identified as ISAV positive by standardized qRT-PCR procedures using pooled organs (gills, heart, and kidney) in two independent laboratories, both of which are officially recognized by SERNAPESCA, the official Chilean state agency responsible for health management in the salmon production industry. The assayed specimens were reared in a freshwater farm in the Región de los Lagos, Chile.
RNA characterization in ovarian fluid.
Fifty milliliters of ovarian fluid recovered from each female was clarified by low-speed centrifugation (3,200 × g for 15 min), and 250 μl of the clarified supernatant and the resulting pellet were resuspended in an equivalent volume of 1× phosphate-buffered saline (PBS). RNA was then extracted by using the TRIzol LS procedure as specified by the manufacturer (TRIzol LS reagent; Invitrogen USA). The resulting RNA was recovered in a final volume of 30 μl of nuclease-free deionized water. Triplicates of 5 μl each were used to confirm ISAV by standard qRT-PCR procedures (19, 22).
Virus recovery.
Fifteen milliliters of ovarian fluid was clarified by centrifugation (3,200 × g for 15 min), and the supernatant was further centrifuged for 1 h at 18,000 × g. The pelleted virus was resuspended in 100 μl of serum-free L-15 tissue culture medium. Individual eggs were washed three times with L-15 medium and thoroughly homogenized in the presence of glass pearls (Glassperlen 0.10 to 0.11 mm; B. Braun Melsungen Apparatebau, Germany). The homogenate was clarified and processed as described above. A supernatant aliquot from infected SHK-1 cells was used as a positive control.
Ultrastructural analysis of ovarian fluid.
Fifteen milliliters of ovarian fluid was clarified by centrifugation (3,200 × g for 15 min), and the supernatant was further centrifuged for 30 min at 18,000 × g. The pellet was fixed in 2.5% glutaraldehyde (0.1 M cacodylate buffer, pH 7.2, for 48 h at room temperature), postfixed with aqueous 1% osmium tetroxide for 2 h, extensively rinsed with double-distilled water (ddH2O), and stained with aqueous 1% uranyl acetate for a further 2 h. Dehydration was obtained by consecutive exposure to 50%, 70%, 2× 95%, and 3× 100% acetone solutions for 30 min each. Following this, preinclusion with Epon-acetone (1:1) was performed overnight in order to incorporate pure Epon polymerized at 60°C for 24 h. Thin sections (60 to 70 nm) were obtained with a Sorvall ultramicrotome (MT-5000), layered over copper grills, stained with 4% uranyl acetate in methanol for 2 min and lead citrate (23) for 5 min, and observed under a Philips Tecnai electron microscope at a range of 12 to 80 kV.
RNA characterization of eggs.
One hundred sixty eggs from each female were thoroughly washed three times with PBS followed by a further three washes with PBS containing 0.05% NP-40 detergent to release potential viral particles attached to the surface of the eggs. Half of the eggs were ground and homogenized by using a MagNALyser instrument (Roche, USA) in the presence of TRIzol LS, and total RNA was further purified by means of an RNeasy Minikit (Qiagen, MD, USA). The remaining half of the eggs were individually placed into a 96-well enzyme-linked immunosorbent assay (ELISA) plate, each one was carefully punctured with a tuberculin needle, and the egg interior was recovered. As a first approach, these were pooled and similarly extracted with TRIzol LS.
Ultrastructural analysis of eggs.
Thoroughly washed eggs (see above) were fixed with 2.5% glutaraldehyde in cacodylate buffer (0.1 M; pH 7.2) for 24 h at room temperature. At hour 24, the eggs were sliced in half with a scalpel, and fixation continued for a further 24 h at 4°C, followed by postfixation in 1% aqueous osmium tetroxide for 2 h. After extensive washing with deionized water, block staining was performed with 1% aqueous uranyl acetate for 2 h. Acetone dehydration was obtained by exposure to increasing concentrations of acetone (50%, 70%, 2× 95%, and 3× 100%) for 30 min each. Overnight preinclusion with Epon-acetone (1:1) was then performed in order to finally incorporate pure Epon for polymerization at 60°C for 24 h. Thin sections (60 to 70 nm) were obtained on a Sorvall RT MT-5000 ultramicrotome, loaded onto copper grids, stained with 4% uranyl acetate in methanol for 2 min and lead citrate for 5 min (23), and analyzed under a Philips Tecnai electron microscope (12 to 80 kV).
Direct negative staining.
For negative staining, 15 μl of ASK-1 cells infected with a standard HPR-3 variant of ISAV and a virus-enriched fraction of ovarian fluid were independently loaded onto Formvar-coated carbon grids for 1 min, excess liquid was carefully withdrawn with Whatman no. 1 filter paper, and the cells were stained with 10 μl of 1% aqueous uranyl acetate followed by a 10-min incubation at 37°C before visualization with an electron microscope.
Evaluation of viral infectivity.
The SHK-1 Salmo salar macrophage-like continuous cell line (24) was maintained at 18°C in Leibovitz medium (L-15; Gibco, Invitrogen, United Kingdom) supplemented with penicillin (100 IU ml−1), streptomycin (100 g ml−1), and 15% fetal calf serum (FCS; Gibco). Cells were grown at a high cell density and infected with ISAV-containing clarified ovarian fluid in triplicate. The RNA was extracted 24 h later for evaluation.
Immunofluorescence staining for confocal microscopy.
SHK-1 cells were grown in 24-well tissue culture plates (Orange Scientific) containing sterile coverslips and infected with the putative virus recovered from the ovarian fluid and eggs of one fish as well as with a standard ISAV as a positive control. The cultures were incubated at 17°C, and at 24 h postinfection, cells were fixed with 4% formaldehyde in 0.13 M phosphate buffer at pH 7.4 and at room temperature for 15 min and washed with PBS. Cells were then permeabilized for 1 h at room temperature with 0.1% Triton X-100 in 1× PBS. The cells were fixed, and cell sections were blocked with 3% bovine serum albumin (BSA) containing 0.5% Tween 20 for 30 min at 37°C (25). A monoclonal antibody against the ISAV nucleoprotein (clone 2C2/H4; Grupo Bios, BiosChile) diluted 1:500 in 1× PBS and 3% BSA (26) was incubated overnight at 4°C, detected by using a goat anti-mouse antibody conjugated with Alexa Fluor 568 (Life Technologies, USA) diluted 1:1,000 in 1× PBS and 3% BSA, and incubated for 1 h at room temperature. For wheat germ agglutinin (WGA) depiction, samples were stained with 5 μg/ml Alexa Fluor 488-conjugated WGA (Invitrogen, USA) for 1 h at room temperature. Slides were prepared by using Vectashield mounting medium. Image acquisition was performed with a Leica TCSSP5 II confocal microscope, and samples were observed under a 100× lens.
High-resolution melting analysis.
Samples were analyzed as described previously by our laboratory (27). Briefly, PCR-amplified samples and controls were analyzed by using high-resolution melting (HRM)-PCR carried out with a dilution (1:1,000) of Melt DoctorHRM Master Mix (Applied Biosystems, USA) and our previously validated primers. The resulting amplicons were resolved in a 2% agarose gel in 1% Tris-borate-EDTA (TBE) buffer at 100 V for 30 min and visualized with a Photocapture system (Bio-Imaging System, Ltd., Israel). Amplification and melt curve analysis were then performed on an Applied Biosystems Step One instrument. The HRM curves, the shape of the curve, and the melting temperature (Tm) differences were internally normalized with High Resolution Melt v2.0 software.
Denaturant gradient gel electrophoresis (DGGE) analysis.
Samples were analyzed as described previously by our laboratory (28). Briefly, GC-clamped and validated primers were used to obtain specific amplicons, which were further resolved by their migration on an 8% polyacrylamide–30 to 60% denaturant (100% denaturant equal to 7 M urea and 40% deionized formamide) gradient gel in 1× Tris-acetate EDTA (TAE) buffer run at 150 V and 56°C for 90 min. The gel was stained with 3× GelRed (Biotium, Inc., CA, USA) and photographed with the Photo Capture system. Amplicon bands were recovered and sequenced.
Ethical considerations.
Tissue samples from fish were obtained from the surveillance program for fish disease in Chile. Fish were not killed for the purpose of this study. All sampling was performed according regulations of SERNAPESCA (Chilean government institution in charge of fish health) and carried out in strict compliance with the recommendations in chapter 7.4 of the Aquatic Animal Health Code of the World Organization for Animal Health (OIE, Paris, France) (3). Every effort was made to minimize animal suffering in all procedures.
RESULTS
In order to validate the information provided by SERNAPESCA that two asymptomatic female brood fish from a freshwater farm tested positive for ISAV with CT values indicative of high and low viral loads, respectively, we decided to further analyze both fish regarding the viral sequence content in their ovarian fluid and eggs by means of qRT-PCR. The results are summarized in Table 1.
TABLE 1.
qRT-PCR analysis of genomic segment 8 and cellular ELFa
| Sample | RNA concn (ng/μl) |
CT value |
|
|---|---|---|---|
| Segment 8b | Cellular ELFc | ||
| Organ pool,d fish 1 | ND | 15.0 | 21.6 |
| Organ pool,d fish 2 | ND | 36.16 | 21.3 |
| Clarified ovarian fluid, fish 1e | 12.3 | 29.37 | 31.34 |
| Clarified ovarian fluid, fish 2 | 70.6 | 37.87 | >38 |
| Pellet ovarian fluid, fish 1 | 13.4 | 21.47 | ND |
| Pellet ovarian fluid, fish 2 | 102.6 | 31.34 | ND |
| Whole egg, fish 1 | 9.8 | 32.4 | 26.54 |
| Internal egg fluid, fish 1 | 2.2 | 31.55 | 35.17 |
| Whole egg, fish 2f | 4.5 | 38.16 | 29.78 |
| Remaining infective viruse (1 h p.i.) | 17.8 | 28.8 | 29.1 |
| SHK-1 cells (24 h p.i.) | 31.1 | 23.7 | 26.97 |
| SHK-1 cellsf (24 h p.i.) | 1.3 | 25.18 | 24.3 |
Considering that both the ovarian fluid and the eggs tested positive for ISAV, although with different quantitative results for each fish (Table 1), we decided to continue working with the fish that presumptively contained a higher viral load in order to evaluate the possibility of detecting the virus intact in one or both compartments. Thus, samples were prepared for transmission electron microscopy analysis, and the results are shown in Fig. 1.
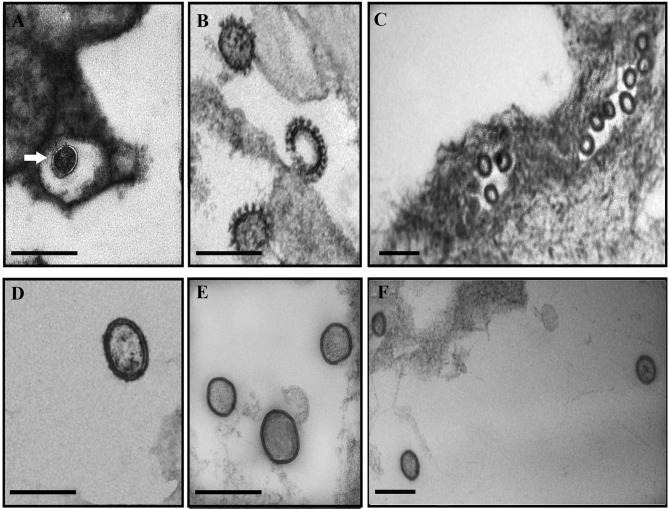
FIG 1.
Electron microscopy analysis of ISAV from eggs and ovarian fluid. (A to C) Sectioned eggs; (D) supernatant from an in vitro infection with an ISAV HPR-3 variant; (E and F) ovarian fluid. (A) The arrow shows a membranous connection between a virus particle and the membrane of the egg. (B) Magnification of three clearly distinctive viral particles inside the egg. (C) General view of numerous viral particles inside the egg. (D) Typical viral structure of an in vitro-grown ISAV. (E) Pelleted virus from ovarian fluid. (F) Direct staining of 15 μl of ovarian fluid. Scale bars, 200 nm.
Once we visualized putative ISAV in both compartments, we decided to examine the infectivity potential of the observed structures. In order to do so, an aliquot of the clarified ovarian fluid from fish 1 (Table 1) was used to infect ASK-1 cells to evaluate the fate of infection 24 h later, timing that we routinely use in the laboratory to evaluate infectivity from in vivo samples. A parallel infection with a highly aggressive HPR-3 ISAV variant was used as a positive control. Potential infection was assessed by confocal microscopy using a monoclonal antibody elicited against the nucleocapsid protein of ISAV. The results are shown in Fig. 2.
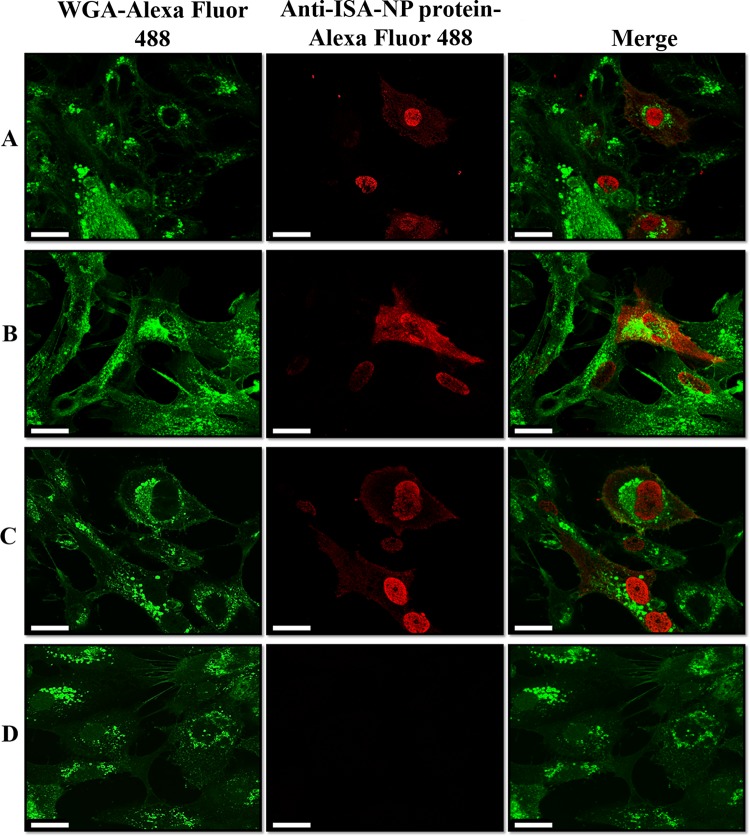
FIG 2.
Confocal microscopy analysis of infectious ISAV over SHK-1 tissue culture cells (magnification, ×100). Detection was obtained after staining of fixed cells with an anti-ISAV NP protein monoclonal antibody and an Alexa Fluor 647-conjugated secondary antibody at 24 h postinfection for infection with an ovarian fluid-enriched viral fraction (A), infection with an intracellular egg-enriched viral fraction (B), standard ISAV infection (C), and noninfected cells (D). Nuclear membranes are depicted in green with Alexa Fluor 488-conjugated wheat germ agglutinin (WGA), and reactive ISAV NP is shown in red with Alexa Fluor 568. Scale bars, 25 μm.
After confirmation that both the ovarian fluid and the eggs were enriched with ISAV and that the virus recovered from the ovarian fluid was infective in vitro, we aimed to further characterize the RNA type of the ISAV variant(s). We decided to use two complementary and highly sensitive procedures to accurately and thoroughly determine the potential variability of the highly polymorphic region in viral (29–31) genomic segment 6, which defines ISAV variants.
Figure 3 shows the high-resolution melting (HRM) profiles for targeted amplicons derived from the HPR region of viral genomic segment 6 obtained from the ovarian fluid RNA as well as that obtained from the RNA recovered from the interior of the eggs. In both cases, a perfect match was obtained with the established profiles for the ISAV HPR-3 variant.
FIG 3.
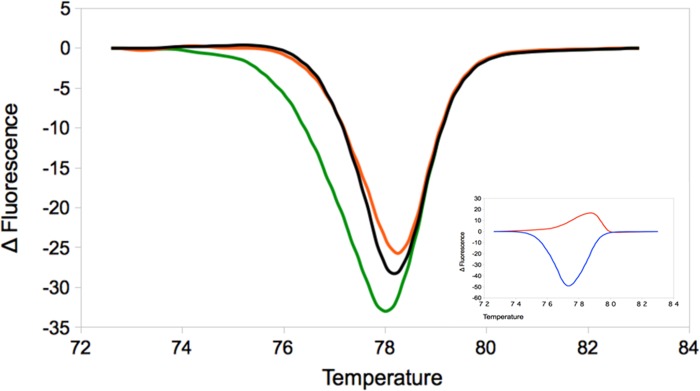
High-resolution melting profiles for ISAV genomic segment 6. Orange, ovarian fluid; green, intraegg fluid; black, reference ISAV HPR-3 variant. The inset shows the reference HRM standard variants HPR-0 (red) and HPR-7b (blue).
As a confirmatory experiment, the same amplicons were processed for denaturant gradient gel electrophoresis (DGGE). Figure 4 clearly shows a unique band for both samples which matches that of the ISAV HPR-3 variant.
FIG 4.
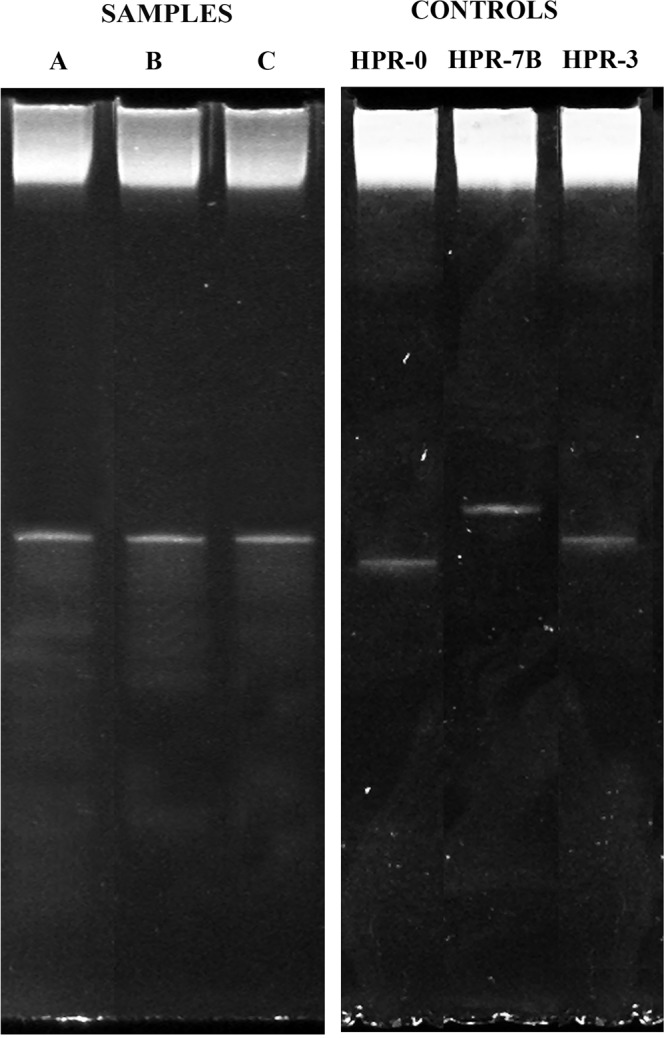
Denaturant gradient gel electrophoresis characterization of the HPR region of ISAV genomic segment 6. (A) Ovarian fluid; (B) intraegg fluid; (C) tissue culture-grown HPR-3 variant. The control panel shows profiles for the highly infective HPR-7b variant, the avirulent HPR-0 variant, and the virulent HPR-3 variant.
DISCUSSION
The biological mechanisms involved in the transmission of infectious salmon anemia virus (ISAV) remain largely unclear. Although horizontal transmission seems to be highly accepted among the scientific community and a well-understood fact under experimental conditions, general controversy surrounds the possibility that vertical transmission may exist under natural conditions. Unfortunately, as of today, robust experimental data are lacking to confirm and/or rule out this possibility. Nevertheless, there are a number of distinctive features associated with the biology of this virus that support its existence. First, subclinical infections with an apparently noninfective variant of the virus (HPR-0) seem to play an important although puzzling role in the pathogenesis of the virus worldwide (7, 29, 32); second, all species of salmonid fish cultivated in Chile were introduced primarily from northern Europe, thus ruling out a putative local source for the virus incubated in a local reservoir (29); and third, all eggs initially imported to Chile came from Norway, where the first outbreaks and description of the disease were reported (8).
In this study, we report that freshwater-reared Atlantic salmon (Salmo salar) brood stock fish, two randomly chosen phenotypically healthy female specimens, turned out to be systemic carriers of an epizootic virulent strain of infectious salmon anemia virus (HPR-3). Not only does our analysis of the ovarian fluids as well as the eggs of both fish demonstrate the presence of the virus in both compartments, it was also seen that the viruses recovered from both fractions were highly infective to an established fish cell line (Fig. 2), which is suggestive of functional vertical transmission, a cryptic fear shared by most scientists and producers. Interestingly enough, virus recovered from eggs of fish 2, whose organs displayed a high CT value, which is indicative of a low viral load (Table 1), was demonstrated to be highly infective, as unequivocally shown in Fig. 2B. Moreover, the comparatively higher NP protein labeling observed in the cytoplasm of infected cells might even be suggestive of a higher degree of aggressiveness of the egg-recovered virus. We have also demonstrated that the viral variant encountered in these two compartments perfectly matches the one detected in the initial analysis of the pooled organs. Sequences of all samples indicate that the variant recovered is HPR-3 (Fig. 5), and since we have grown and cloned this variant virus, we used the corresponding amplicon as a marker in the DGGE analysis (Fig. 4).
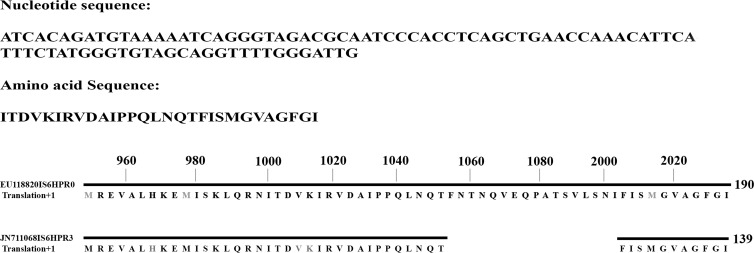
FIG 5.
Sequencing and alignment of the HPR-3 variant sequence. The length of the sequenced amplicon is 137 bp and matches the available GenBank sequence reported under accession number JN711068, which lies between bp 994 and 1080, corresponding to HPR-3.
Nevertheless, there is an intrinsic puzzling contradiction with these results. The sole variant detected in these two fully asymptomatic members of a brood stock corresponded to the HPR-3 type, which is known to be an unquestionably epizootic ISAV variant in vivo and which is also highly aggressive when passaged in vitro. We do not have a solid explanation for this interesting fact, and it therefore requires further analysis. Nonetheless, a selected number of observations gathered from the reference literature on the behavior of the virus offer alternatives that may help to elucidate this dilemma. First, the targeted brood fish from this study came from a freshwater unit, an environment in which ISAV has been extensively detected both in natural environments as well as under confined rearing conditions and without clinical expression. This may suggest that ISAV, independent of its potential aggressiveness, persists in a carrier stage under freshwater conditions. Second, most diagnostic procedures for ISAV rely on PCR alternatives concentrating primarily on the amplification of a conserved sequence of genomic segment 8 (19) or, alternatively, on the selective amplification of the HPR region of the genomic segment which defines infective variants (6). However, ISAV is a regular orthomyxovirus with 8 subgenomic segments, prone to reassortment and recombinatorial events of more than one variant in the same organism. This alternative has not been covered by our analysis, and it cannot be ruled out, as it might occur and may attenuate viral expression under different environmental conditions. Last but not least, the putative avirulent HPR-0 variant, which represents the “complete” virus with an intact HPR region in segment 6, has been proposed to be the master virus from which all virulent variants are derived by specific deletions in the HPR region (30). HPR-0 is the most prevalent and an almost commonplace ISAV form in all farms worldwide, particularly after viral outbreaks. It is our impression that it represents a clever strategy for viral self-regulation, and the coexistence of more than one viral variant in the same fish, organ, or cell might contribute to defining the eventual infectivity. In conclusion, for the first time, unequivocal vertical transmission is demonstrated for ISAV in vivo, and this is suggestive of a persistent stage of the virus in fish maintained under freshwater conditions.
ACKNOWLEDGMENTS
We gratefully acknowledge the Chilean National Fisheries and Aquaculture Service (SERNAPESCA) for their participation and provision of samples. We also acknowledge Constanza Cardenas for careful review of the manuscript and Dannia Giménez for keen technical assistance.
This research was supported by grants from SERNAPESCA Chile and CORFO Project Code 09CEII-6991 to S.H.M.
Footnotes
Published ahead of print 12 March 2014
REFERENCES
- 1.Mjaaland S, Rimstad E, Falk K, Dannevig BH. 1997. Genomic characterization of the virus causing infectious salmon anemia in Atlantic salmon (Salmo salar L.): an orthomyxo-like virus in a teleost. J. Virol. 71:7681–7686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawaoka Y, Cox NJ, Haller O, Hongo S, Kaverin N, Klenk H-D, Lamb RA, McCauley J, Palese P, Rimstad E, Webster RG. 2005. Infectious salmon anemia virus, p 681–693 In Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. (ed),Virus taxonomy. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press, New York, NY [Google Scholar]
- 3.World Organization for Animal Health. 2013. Aquatic animal health code. Infection with infectious salmon anemia virus, 16th ed. OIE, Paris, France: http://www.oie.int/es/normas-internacionales/codigo-acuatico/acceso-en-linea/ [Google Scholar]
- 4.Rimstad E, Dale OB, Dannevig BH, Falk K. 2011. Infectious salmon anemia, p 143–165 In Woo PTK, Bruno DW. (ed), Fish diseases and disorders, 2nd ed, vol 3 Viral, bacterial and fungal infections. Centre for Agricultural Bioscience International, Wallingford, United Kingdom [Google Scholar]
- 5.Asche F, Hansen H, Tveteras R, Tveteras S. 2009. The salmon disease crisis in Chile. Mar. Resour. Econ. 24:405–411. 10.5950/0738-1360-24.4.405 [DOI] [Google Scholar]
- 6.Godoy M, Aedo A, Kibenge MJ, Groman DB, Yason CV, Grothusen H, Lisperguer A, Calbucura M, Avendaño F, Imilan M, Jarpa M, Kibenge FS. 2008. First detection, isolation and molecular characterization of infectious salmon anemia virus associated with clinical disease in farmed Atlantic salmon (Salmo salar L) in Chile. BMC Vet. Res. 4:28–33. 10.1186/1746-6148-4-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cottet L, Cortez-San Martin M, Tello M, Olivares E, Rivas-Aravena A, Vallejos E, Sandino AM, Spencer E. 2010. Bioinformatic analysis of the genome of infectious salmon anemia virus associated with outbreaks of high mortality in Chile. J. Virol. 84:11916–11928. 10.1128/JVI.01202-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thorud K, Djupvik HO. 1988. Infectious anemia in Atlantic salmon (Salmo salar L). Bull. Eur. Assoc. Fish Pathol. 8:109–111 [Google Scholar]
- 9.Anonymous. 2000. ISA hits the Faroes. Fish Farming Int. 27:47 [Google Scholar]
- 10.Christiansen DH, Ostergaard PS, Snow M, Dale OB, Falk K. 2011. A low-pathogenic variant of infectious salmon anemia virus (ISAV-HPR0) is highly prevalent and causes a non-clinical transient infection in farmed Atlantic salmon (Salmo salar L) in the Faroe Island. J. Gen. Virol. 92:909–918. 10.1099/vir.0.027094-0 [DOI] [PubMed] [Google Scholar]
- 11.Cunningham CO, Gregory A, Black J, Simpson J, Raynard RS. 2002. A novel variant of the infectious salmon anemia virus (ISAV) hemagglutinin gene suggests mechanisms for virus diversity. Bull. Eur. Assoc. Fish Pathol. 22:366–374 [Google Scholar]
- 12.Bouchard DA, Brockway K, Giray C, Keleher W, Merrill PL. 2001. First report of infectious salmon anemia (ISA) in the United States. Bull. Eur. Assoc. Fish Pathol. 21:86–88 [Google Scholar]
- 13.McBeath AJA, Bain N, Snow M. 2009. Surveillance for infectious salmon anemia virus HPR0 in marine Atlantic salmon farms across Scotland. Dis. Aquat. Organ. 87:161–169. 10.3354/dao02128 [DOI] [PubMed] [Google Scholar]
- 14.Murray AG, Munro LA, Wallace IS, Berx B, Pendrey D, Fraser D, Raynard RS. 2010. Epidemiological investigation into the re-emergence and control of an outbreak of infectious salmon anemia in the Shetland Islands, Scotland. Dis. Aquat. Organ. 91:189–200. 10.3354/dao02262 [DOI] [PubMed] [Google Scholar]
- 15.EFSA Panel on Animal Health and Welfare. 2012. Scientific opinion on infectious salmon anemia. EFSA J. 10:2971. 10.2903/j.efsa.2012.2971 [DOI] [Google Scholar]
- 16.Melville KJ, Griffiths SG. 1999. Absence of vertical transmission of infectious salmon anemia virus (ISAV) from individually infected Atlantic salmon (Salmo salar L). Dis. Aquat. Organ. 38:231–234 [DOI] [PubMed] [Google Scholar]
- 17.Søfteland E. 2005. Feltforsøk med ILA-infisertstamfiskavlaks-erdetmulig å overføre ILA-virus via rognogmelke? Report Mattilsynet. Universitetet Bergen, Akvaforsk, Fomas, Bergen, Norway [Google Scholar]
- 18.Nylund A, Plarre H, Karlsen M, Fridell F, Ottem KF, Bratland A, Saether PA. 2007. Transmission of infectious salmon anemia virus (ISAV) in farmed populations of Atlantic salmon (Salmo salar). Arch. Virol. 152:151–179. 10.1007/s00705-006-0825-9 [DOI] [PubMed] [Google Scholar]
- 19.Snow M, McKay P, McBeath AJA, Black J, Doig F, Kerr R, Cunningham CO, Nylund A, Devold M. 2006. Development, application and validation of a Taqman real-time RT-PCR assay for the detection of infectious salmon anemia virus (ISAV) in Atlantic salmon (Salmo salar L). Dev. Biol. 126:133-145 [PubMed] [Google Scholar]
- 20.Vike S, Nylund S, Nylund A. 2009. ISA virus in Chile: evidence of vertical transmission. Arch. Virol. 154:1–8. 10.1007/s00705-008-0251-2 [DOI] [PubMed] [Google Scholar]
- 21.Godoy M, Kibenge M, Suarez R, Lazo E, Heisinger A, Aguinaga J, Bravo D, Mendoza J, Llegues K, Avendaño-Herrera R, Vera C, Mardones F, Kibenge F. 2013. Infectious salmon anemia virus (ISAV) in Chilean Atlantic salmon (Salmo salar) aquaculture: emergence of low pathogenic ISAV-HPR0 and re-emergence of virulent ISAV-HPRs: HPR3 and HPR14. Virol. J. 10:344. 10.1186/1743-422X-10-344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sepúlveda D, Bohle H, Labra A, Grothusen H, Marshall S. 2013. Design and evaluation of a unique RT-qPCR assay for diagnostic quality control assessment that is applicable to pathogen detection in three species of salmonid fish. BMC Vet. Res. 9:183. 10.1186/1746-6148-9-183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reynolds ES. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212. 10.1083/jcb.17.1.208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dannevig BH, Falk K, Namork E. 1995. Isolation of the causal virus of infectious salmon anemia (ISA) in a long-term cell line from Atlantic salmon head kidney. J. Gen. Virol. 76(Part 6):1353–1359. 10.1099/0022-1317-76-6-1353 [DOI] [PubMed] [Google Scholar]
- 25.Goić B, Bustamante J, Miquel A, Alvarez M, Vera MI, Valenzuela PD, Burzio LO. 2008. The nucleoprotein and the viral RNA of infectious salmon anemia virus (ISAV) are localized in the nucleolus of infected cells. Virology 379:55–63. 10.1016/j.virol.2008.05.036 [DOI] [PubMed] [Google Scholar]
- 26.Rivas-Aravena A, Vallejos-Vidal E, Cortez-San Martin M, Reyes-López F, Tello M, Mora P, Sandino AM, Spencer E. 2011. Inhibitory effect of a nucleotide analog on infectious salmon anemia virus infection. J. Virol. 85:8037–8045. 10.1128/JVI.00533-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sepúlveda D, Cárdenas C, Carmona M, Marshall SH. 2012. Novel strategy to evaluate infectious salmon anemia virus variants by high resolution melting. PLoS One 7:e37265. 10.1371/journal.pone.0037265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carmona M, Sepúlveda D, Cárdenas C, Nilo L, Marshall SH. 2012. Denaturing gradient gel electrophoresis (DGGE) as a powerful novel alternative for differentiation of epizootic ISA virus variants. PLoS One 7:e37353. 10.1371/journal.pone.0037353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nylund A, Devold M, Plarre H, Isdal E, Arseth M. 2003. Emergence and maintenance of infectious salmon anemia virus (ISAV) in Europe: a new hypothesis. Dis. Aquat. Organ. 56:11–24. 10.3354/dao056011 [DOI] [PubMed] [Google Scholar]
- 30.Markussen T, Jonassen CM, Numanovic S, Braen S, Hjortaas M, Nilsen H, Mjaaland S. 2008. Evolutionary mechanisms involved in the virulence of infectious salmon anemia virus (ISAV), a piscine orthomyxovirus. Virology 374:515–527. 10.1016/j.virol.2008.01.019 [DOI] [PubMed] [Google Scholar]
- 31.Devold M, Falk K, Dale B, Krossøy B, Biering E, Aspehaug V, Nilsen F, Nylund A. 2001. Strain variation, based on the hemagglutinin gene, in Norwegian ISA virus isolates collected from 1987 to 2001: indications of recombination. Dis. Aquat. Organ. 47:119–128. 10.3354/dao047119 [DOI] [PubMed] [Google Scholar]
- 32.SERNAPESCA. 2010. Informe técnico Programa Sanitario Específico de Vigilancia y Control de la Anemia Infecciosa del Salmón (PSEC-ISA), 2008-2010. Unidad de Acuicultura, SERNAPESCA, Valparaíso, Chile: http://mail.anfitrion.cl/GobiernoTransparente/pesca/NE/RES/2011/08/73385.html [Google Scholar]