ABSTRACT
Coronaviruses express a deubiquitinating protein, the papain-like protease-2 (PLP2), that removes both ubiquitin and the ubiquitin-like interferon (IFN)-stimulated gene 15 (ISG15) protein from target proteins. ISG15 has antiviral activity against a number of viruses; therefore, we examined the effect of ISG15 conjugation (ISGylation) in a model of acute viral hepatitis induced by the murine hepatitis virus strain 3 (MHV-3) coronavirus. Mice deficient in the ISG15 deconjugating enzyme, ubiquitin-specific peptidase-18 (USP18), accumulate high levels of ISG15-conjugated proteins and are hypersensitive to type I IFN. Infecting USP18−/− mice with MHV-3 resulted in extended survival (8 ± 1.2 versus 4 days) and in improved liver histology, a decreased inflammatory response, and viral titers 1 to 2 logs lower than in USP18+/+ mice. The suppression of viral replication was not due to increased IFN since infected USP18−/− mice had neither increased hepatic IFN-α, -β, or -γ mRNA nor circulating protein. Instead, delayed MHV-3 replication coincided with high levels of cellular ISGylation. Decreasing ISGylation by knockdown of the ISG15 E1 enzyme, Ube1L, in primary USP18+/+ and USP18−/− hepatocytes led to increased MHV-3 replication. Both in vitro and in vivo, increasing MHV-3 titers were coincident with increased PLP2 mRNA and decreased ISGylation over the course of infection. The pharmacologic inhibition of the PLP2 enzyme in vitro led to decreased MHV-3 replication. Overall, these results demonstrate the antiviral effect of ISGylation in an in vivo model of coronavirus-induced mouse hepatitis and illustrate that PLP2 manipulates the host innate immune response through the ISG15/USP18 pathway.
IMPORTANCE There have been a number of serious worldwide pandemics due to widespread infections by coronavirus. This virus (in its many forms) is difficult to treat, in part because it is very good at finding “holes” in the way that the host (the infected individual) tries to control and eliminate the virus. In this study, we demonstrate that an important host viral defense—the ISG15 pathway—is only partially effective in controlling severe coronavirus infection. Activation of the pathway is very good at suppressing viral production, but over time the virus overwhelms the host response and the effects of the ISG15 pathway. These data provide insight into host-virus interactions during coronavirus infection and suggest that the ISG15 pathway is a reasonable target for controlling severe coronavirus infection although the best treatment will likely involve multiple pathways and targets.
INTRODUCTION
Coronaviruses cause both common and severe clinical illness, as manifested by the severe acute respiratory syndrome (SARS) epidemic. During the 2002-2003 SARS epidemic, 8,422 people were affected, of whom 916 died from acute respiratory distress syndrome (1, 2). The episodic reemergence of severe coronavirus infections, most recently in the fall of 2012 and continuing into 2013, highlights the need for treatments against coronavirus infections (3).
Coronaviruses are positive-stranded enveloped RNA viruses with some of the largest viral genomes, ranging between 26 and 32 kDa. Coronaviruses target a myriad of distinct animal hosts and cause disease in a number of organs, such as the brain, liver, and lung. Organ damage from severe coronavirus infections is generally the result of an overexuberant activation of host innate immune mechanisms (4, 5). For example, murine hepatitis strain 3 (MHV-3) infection of susceptible mouse strains is a model of strong innate immune activation, resulting in fulminant viral hepatitis (6). Following infection by MHV-3, mice die in 3 to 4 days of hepatic parenchymal destruction mediated by a robust activation of local innate immunity (7–9). Understanding the limits of host immunity in the MHV-3 model may help identify novel targets for the treatment of severe coronavirus infections.
Interferon (IFN) stimulation as well as bacterial and viral infection induces the expression of IFN-stimulated genes (ISGs). One of the most abundantly expressed ISGs is ISG15, a 15-kDa protein (10). ISG15 is conjugated to target proteins, a process termed ISGylation, through consecutive interactions with an E1-activating enzyme (Ube1L), an E2-conjugating enzyme (UbcH6 or UbcH8), and an E3 ligase (Herc5, estrogen-responsive finger protein [EFP], or HHARI) (11). Over 160 proteins have been identified as targets of ISG15, including proteins involved in processes such as protein translation, cell cycle regulation, and immune regulation (11–13). Several ISG15 downstream targets are involved in the regulation of IFN signaling, including retinoic acid-inducible gene 1 (RIG-1), signal transducer and activator of transcription 1 (STAT-1), Janus-activated kinase-1 (JAK-1), and MxA (12–14). Although the ISG15 protease function has the potential to modulate IFN responses, this role has not been consistently demonstrated (15). The effect of ISGylation on downstream proteins is multifaceted as ISGylation has been reported to disrupt target protein function and/or alter cellular localization (16, 17).
The conjugation of ISG15 to protein targets is offset by the deconjugating activity of the ubiquitin (Ub)-specific peptidase-18 (USP18). USP18, like ISG15, is an ISG which is upregulated after stimulation by IFN or by viral and bacterial infection. USP18 is specific for ISG15 and strips ISG15 from target proteins through its isopeptidase activity (18). USP18−/− mice have markedly increased cellular ISGylation and are hypersensitive to the effects of type 1 IFN (19–21). The latter effect is independent of the isopeptidase activity and, instead, is due to the ability of USP18 to modulate IFN signaling by binding to the IFN-α receptor 2 (IFNAR2) (22). Thus, USP18 has several functions important for the host innate immune response: by binding to IFNAR2, USP18 can modulate the IFN response, and through its ISG15 isopeptidase function, it regulates the cellular ISGylation levels. However, the implications of the balance of ISGylation and USP18 isopeptidase activity require further elucidation.
The role of ISGylation in viral life cycles is specific to the virus involved. ISGylation can exert an antiviral pressure against some infections but can also stimulate viral replication in others. For example, USP18-deficient mice have increased ISGylation and are more resistant to lymphocytic choriomeningitis virus (LCMV) and herpes simplex virus (HSV) models of fatal viral encephalitis (20). During human immunodeficiency virus (HIV) infection, the conjugation of ISG15 to the HIV Gag protein arrests assembly of the Gag particle on the plasma membrane (23) and inhibits viral replication. On the other hand, we have found that ISGylation is necessary for robust production of hepatitis C virus (HCV) in human hepatocytes (24), and both ISG15 and USP18 are upregulated in the hepatocytes of patients with chronic HCV infection who do not respond to exogenous IFN treatment (25–27).
Previously, we demonstrated that coronavirus replication in vivo is held in check by host ubiquitination (28). Inhibition of the cellular proteasome leads to increased cellular ubiquitination levels and an early interruption in coronavirus replication. Coronaviruses have evolved countermeasures to such host cellular antiviral mechanisms. In this case, the deubiquitination protein papain-like protease-2 (PLP2) strips ubiquitin from target proteins (29). The PLP2 protein is not specific to ubiquitin: it acts on both ubiquitin and the ubiquitin-like protein ISG15. This suggests that ISG15 and its conjugation to cellular proteins may also exert an antiviral pressure effect against coronavirus infection. Moreover, several reports indicate that PLP2 has a 60-fold higher affinity for ISG15-conjugated than Ub-modified substrates (18, 30). However, it is unknown whether targeting PLP2 activity in an in vitro cell culture system or in vivo will inhibit coronavirus replication. Furthermore, questions remain as to whether ISGylation itself has an antiviral effect against coronavirus infection and/or whether the PLP2 machinery in vitro targets ISGylation directly to evade the host response.
In the present study, we infected USP18−/− mice with the coronavirus MHV-3 to evaluate the involvement of the ISG15/USP18 pathway in the virulence of MHV-3-induced hepatitis. We found that USP18−/− mice are more resistant to MHV-3 infection but that the virus gradually overcomes this protective effect. IFN type 1 and type 2 expression levels were not increased in USP18−/− mice following infection by MHV-3, allowing us to study the role of increased baseline ISGylation in the absence of IFN signaling. Silencing ISGylation reverses the antiviral milieu and leads to increased MHV-3 replication. Both in vitro and in vivo, viral persistence is accompanied by increased expression of the PLP2 protein. PLP2 expression is important for allowing viral replication; specific PLP inhibitors decreased viral protein expression. Overall, these results demonstrate both the role and the limits of the antiviral effect of ISGylation in severe coronavirus infection.
MATERIALS AND METHODS
Animals.
C57BL/6 USP18+/+ and USP18−/− mice between 6 and 8 weeks old were used for experiments. These mice were the kind gift of Dong Er Zhang (Scripps/University of California, San Diego [UCSD]). Animals were housed under specific-pathogen-free (SPF) conditions at the MaRS-Toronto Medical Discovery Tower (TMDT) Animal Resource Centre (Toronto, Canada) and were given chow and water ad libitum. Following MHV-3 infection, animals were housed in sterile cages in a level 2 facility in the Max Bell Research Center (MBRC; Toronto, Canada). Mice infected with MHV-3 were sacrificed at humane endpoints. Animals were treated according to the guidelines of the Canadian Council on Animal Care, and procedures were approved by the University Health Network Animal Care Committee.
Isolation of PEM and hepatocytes.
The isolation of peritoneal exudative macrophages (PEM) has been previously described (28). Briefly, mice were injected with 2 ml of sterile 3% thioglycolate. Animals were sacrificed after 3 days, and PEM were retrieved by washing the intraperitoneal cavity with 10 ml of ice-cold Hanks' balanced salt solution (HBSS) (Life Technologies). Cells were washed two times, spun down at 300 × g, and resuspended in Dulbecco's modified Eagle medium (DMEM) (Life Technologies) supplemented with l-Gln. Using this method, the purity of PEM was found to be >90% with a viability of >97% by trypan blue exclusion. For experiments, PEM were plated on polystyrene plates at a density of 1 × 106 cells/ml and incubated for 24 h at 37°C in 5% CO2. Nonadherent cells were then washed away.
Primary hepatocytes were isolated as previously described (31, 32). Briefly, mice were anesthetized with 50 mg/kg of pentobarbital intraperitoneally (i.p.). The portal vein was cannulated with a 21-gauge needle. The liver was flushed via the portal vena cava with perfusion solution (2.5 mM EGTA in HBSS without Ca2+ or Mg2+) at 37°C and a rate of 7 ml/min using an infusion pump for 3 min. The liver was then perfused with solution 2 (0.02% collagenase IV [Sigma-Aldrich] with calcium and magnesium in HBSS) at the same pressure. The liver was then carefully harvested into a petri dish containing DMEM supplemented with 15% FBS and 1 × 10−7 M insulin (Sigma-Aldrich) and minced. This solution was filtered through a 120-μm-pore-size nylon mesh and centrifuged for 2 min at 40 × g at room temperature (RT). Cells were washed two times with DMEM plus insulin. Cells were counted on a hemocytometer, and viability was determined by trypan blue exclusion. Hepatocytes were plated at a density of 1 × 105 cells/ml in DMEM supplemented with 1 × 10−7 M insulin on a six-well plate (Corning, Inc.). Hepatocytes were incubated for 2 h, and the medium was replaced with serum-free DMEM supplemented with 1 × 10−7 M insulin. Hepatocytes were inoculated with MHV-3 (multiplicity of infection [MOI] of 1, 0.1, or 0.01) and incubated for 1 h. Cells were then washed with serum-free medium plus 1 × 10−7 M insulin and cultured for the indicated time (Fig. 3 and 4).
FIG 3.
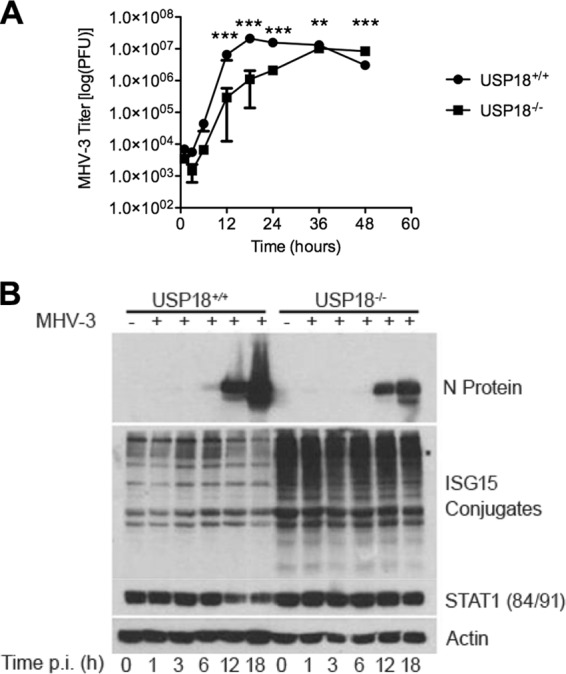
Elevated ISGylation in USP18−/− mice coincides with increased resistance to MHV-3 infection in vitro. (A) Primary hepatocytes of USP18−/− mice are more resistant to MHV-3 infection than those of USP18+/+ mice. Primary hepatocytes were infected with MHV-3 (MOI of 1), and viral titers were assayed using a standard plaque assay. Titers are represented as means ± SD (n = 3). (B) Elevated ISGylation coincides with decreased N protein production in USP18−/− hepatocytes. A time course of N protein production and ISG15 conjugate formation by WB is shown. Hepatocytes were inoculated at an MOI of 1 and harvested at the indicated time points. N protein production is a marker of viral replication. The WB is a representative blot of two independent experiments. ***, P < 0.001; **, P < 0.01; *P < 0.05.
FIG 4.
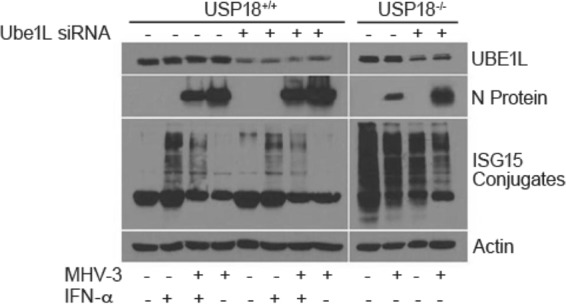
ISGylation inhibits MHV-3 replication. The E1 conjugation enzyme, Ube1L, is critical for ISG15 conjugation to target proteins. Ube1L expression was silenced to determine if increased expression of ISG15 or increased ISGylation had an antiviral effect on coronavirus. Primary hepatocytes were transfected with Ube1L or a control siRNA (Santa Cruz) and incubated for 24 h. Hepatocytes were infected with MHV-3 (MOI of 1), and cells were harvested at 12 h p.i. Samples were visualized on a WB for Ube1L expression, N protein production, and ISGylation. The blot is representative of two independent experiments.
Virus, viral infection, and viral titers.
MHV-3 virus was grown to titers of 10 × 106 to 50 × 106 PFU/ml RPMI medium on confluent 17CL cells. To determine viral titers, harvested cells or liver tissue was lysed or homogenized in ice-cold DMEM supplemented with 10% FBS using a TissueLyser (Qiagen). MHV-3 titers were assessed on monolayers of L2 cells in a standard plaque assay (6). For in vivo studies mice were infected i.p. with 50 PFU of MHV-3 and housed under sterile conditions. Animals were monitored daily and sacrificed at a humane endpoint.
Histology.
Tissues were harvested and preserved in 10% buffered formalin. Fixed tissues were paraffin embedded and cut into 5-μm sections by the Department of Pathology at the Hospital for Sick Children (Toronto, Canada). Sections were stained with hematoxylin and eosin (H&E) using standard protocols (33). Sections were scored in a blinded fashion by an experienced liver pathologist (M. J. Phillips). Sections were assessed for inflammation, parenchymal changes, and tissue necrosis.
Measurement of ALT and AST in the serum.
Blood was harvested by cardiac puncture and incubated for 10 min at RT. Samples were spun down at 2,000 × g, and the serum was collected for analysis. Alanine transaminase (ALT)/aspartate aminotransferase (AST) levels were measured using a Vitro DT60 II Chemistry System (Ortho Clinical Diagnostics, Neckargemund, Germany).
Serum protein levels of type I and type II IFN.
Blood was harvested from mice by cardiac puncture. Serum was incubated at 4°C overnight (O/N). The samples were centrifuged at 2,000 × g for 10 min, and the serum was removed to a fresh tube. Enzyme-linked immunosorbent assay (ELISA) kits for IFN-α and IFN-β were purchased from PBL Interferon Source (Piscataway Township, NJ, USA). The ELISA kit for IFN-γ was purchased from BioLegend, Inc. (San Diego, CA, USA). ELISAs were performed according to the manufacturers' instructions.
Real-time PCR.
Mice were infected with MHV-3 and sacrificed on the indicated day(s) (Fig. 1, 2, 6, and 7). Total RNA was extracted from liver tissue using TRIzol reagent (Invitrogen) according to the manufacturer's specifications. The isolated RNA was treated with DNase (Qiagen), and the quality of the RNA was assessed by measuring the 260 nm/280 nm absorption ratio on a Nanodrop spectrophotometer. One microgram of RNA was reverse transcribed using a First Strand cDNA Synthesis Kit (GE Health Care) using random hexamer oligonucleotides. The reverse transcriptase reaction was run as per the manufacturer's specifications. Primers used for real-time PCRs were based on published and primer bank sequences. The following primer sequences were used: mouse IFN-α forward primer, 5′-CTTTGGATTCCCGCAGGA-3′; mouse IFN-α reverse primer, 5′-TGTAGGACAGGGATGGCTTGA-3′; mouse IFN-β forward primer, 5′-TGAATGGAAAGATCAACCTCACCTA-3′; mouse IFN-β reverse primer, 5′-CTCTTCTGCATCTTCTCCGTCA-3′; mouse IFN-γ forward primer, 5′-CAGCAACAGCAAGGCGAAA-3′; mouse IFN-γ reverse primer, 5′-CTGGACCTGTGGGTTGTTGAC-3′; mouse hypoxanthine phosphoribosyltransferase (HPRT) forward primer, 5′-GTTGGATACAGGCCAGACTTTGTTG-3′ ; mouse HPRT reverse primer, 5′-GATTCAACTTGCGCTCATCTTAGGC-3′; mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH) forward primer, 5′-ACAACTTTGGCATTGTGGAA-3′; mouse GAPDH reverse primer, 5′-GATGCAGGGATGATGTTCTG-3′; MHV-3 PLP2 forward primer, 5′-AAATGTGGCTTGTTTGATGC-3′; and MHV-3 PLP2 reverse primer, 5′-CCTTCGCTAAGACCAAGGAC-3′.
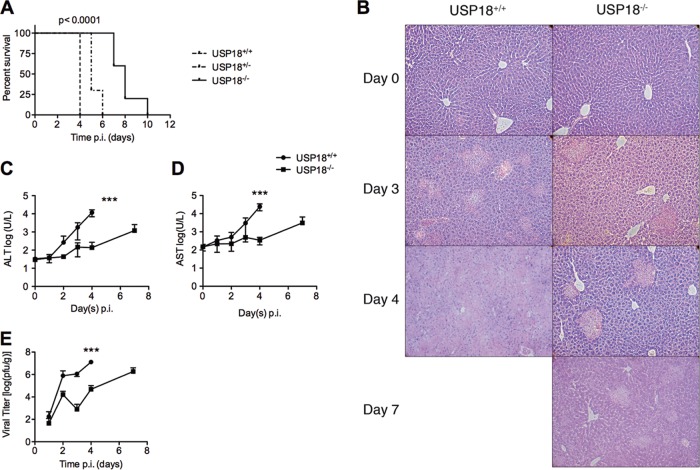
FIG 1.
USP18−/− mice showed delayed morbidity and death following MHV-3 infection. (A) Survival curve of USP18+/+ and USP18−/− mice following infection with 50 PFU of MHV-3 i.p. USP18−/− mouse survival was prolonged up to 10 days, with a median survival of 8 days compared to 4 days for control mice (n = 10 per group; P < 0.0001 using the log rank test for trend). (B) H&E staining of livers from USP18+/+ and USP18−/− mice at days 0, 3, 4, and 7 p.i. Black arrows point to areas of focal necrosis. Photomicrographs at day 4 and day 7 for USP+/+ and USP−/− mice, respectively, show confluent necrosis of the liver. Magnification, ×40. Photomicrographs are representative of each study group. (C and D) The increase in liver markers of injury, ALT and AST, was delayed in USP18−/− mice. ALT and AST were measured, and results are represented as log(U/liter) ± SD. (E) USP18−/− mice show decreased viral titers. Viral titers were measured daily until day 4 and on day 7 only in USP18−/− mice using a standard plaque assay. Data are represented as the mean log(PFU/g) ± SD (***, P < 0.001; **, P < 0.01; *, P < 0.05).
FIG 2.
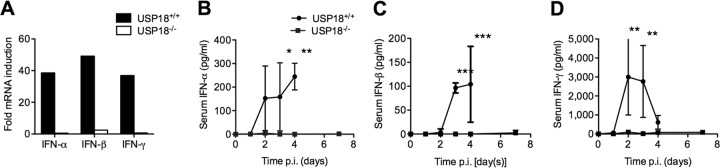
The IFN response to MHV-3 is blunted in USP18−/− mice. Animals infected with MHV-3 were sacrificed on days 0 through 4, with an additional measurement at day 7 for USP18−/− mice (USP18+/+ mice succumbed to disease by day 4 p.i.). (A) mRNA expression of both type I and type II IFN is inhibited in USP18−/− mice. mRNA was harvested from livers of USP18+/+ or USP18−/− mice infected with 50 PFU of MHV-3 and sacrificed at day 3 p.i. (B to D) Serum levels of IFNs were measured by ELISA, as indicated. IFN-α, IFN-β, and IFN-γ levels in the plasma were absent in the USP18−/− mice, whereas USP18+/+ mice were able to induce type I and type II IFN responses. Data are represented as means ± SD (A, n 2; B, n ≥ 2; C, n ≥ 3; D, n ≥ 3). ***, P < 0.001; **, P < 0.01; *, P < 0.05.
FIG 6.
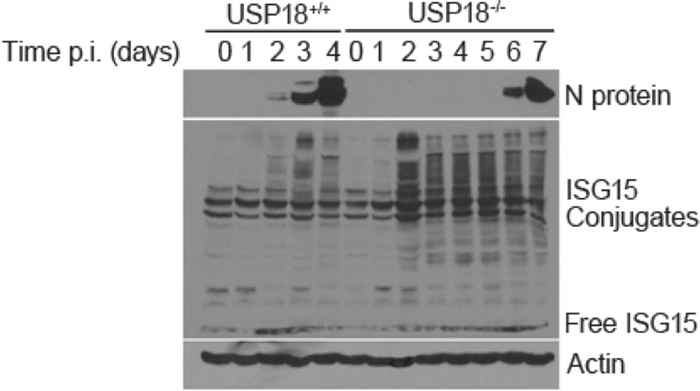
ISGylation delays MHV-3 replication. Increased ISGylation coincides with prolonged survival and delayed N protein accumulation. USP18+/+ or USP18−/− mice were infected with 50 PFU MHV-3/mouse. Mice (n = 4) from each group were sacrificed daily, and livers were harvested for WB analysis. N protein and ISG15 conjugates were visualized by WB.
FIG 7.
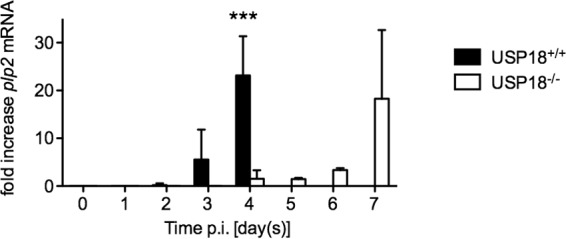
The onset of PLP2 expression is delayed in USP18−/− mice. N protein expression coincides with increased viral expression. To determine if PLP2 expression followed this expression pattern, we tested mRNA expression of PLP2 following MHV-3 expression. USP18+/+ or USP18−/− mice (n = 4/group) were infected with MHV-3 (50 PFU of MHV-3/mouse). Livers were harvested on the indicted days, and mRNA was extracted. Data are presented as the mean fold increase of PLP2 mRNA over that of the housekeeping gene GAPDH ± SD. ***, P < 0.001.
Real-time PCR was performed on an ABI Prism 7900H (Applied Biosystems) with SYBR green real-time PCR master mix (Applied Biosystems) according to the manufacturer's specifications using a 96-well plate format. Data were analyzed using SDS, version 2.2, software (Applied Biosystems) using the standard curve method. Samples were normalized to HPRT and GAPDH.
Western blotting (WB).
Harvested tissues were homogenized and lysed in ice-cold cell lysis buffer. Cells from in vitro cultures were lysed with 2× Laemmli buffer and 0.1 mM dithiothreitol (DTT). Protein concentrations were determined by the Bradford assay. Twenty micrograms of protein was visualized on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane (Pall). Membranes were probed with the following antibodies: anti-nucleocapsid (N) protein mouse monoclonal antibody (made in-house), anti-mouse β-actin mouse monoclonal antibody (Sigma), anti-mouse ISG15 rabbit polyclonal antibody, anti-mouse Ube1L goat polyclonal IgG antibody, or anti-mouse STAT-1 (p84/p91) rabbit polyclonal antibody (Santa Cruz Biotechnology). The corresponding horseradish peroxidase (HRP)-conjugated secondary antibodies were used: sheep anti-mouse IgG (GE Healthcare), donkey anti-rabbit IgG (Santa Cruz Biotechnology), or a donkey anti-goat IgG (Santa Cruz Biotechnology). Blots were developed using an enhanced chemiluminescence (ECL) system (Amersham Pharmacia Biotech).
Silencing Ube1L expression.
Isolated primary hepatocytes were plated at a density of 1 × 105 cells/ml in DMEM. Cells were incubated O/N at 37°C in 0.5% CO2, whereupon cells reached 60 to 80% confluence. Cells were transfected with Ube1L small interfering RNA (siRNA; Santa Cruz Biotechnology) as per the manufacturer's instruction. Briefly, cells were washed with transfection medium and then overlaid with transfection reagent solution and incubated for 6 h at 37°C. DMEM including 10% fetal bovine serum (FBS) was added, and the cells were incubated for a total of 24 h. The medium was replaced with normal growth medium, and cells were infected with MHV-3 (MOI of 1) with or without IFN-α (PBL Interferon Source). Samples were harvested at 12 h postinfection (p.i.) using 200 μl of protein lysis buffer (BD bioscience), run on a 10% SDS-PAGE gel, and visualized by WB.
PLP2 inhibition.
PLP2 inhibitors were generated in-house according to Ratia et al. (34). Isolated PEM were incubated in the presence of MHV-3 (MOI of 1) for 1 h. Medium was washed away, and fresh medium with or without 100 μM inhibitor 6, 100 μM inhibitor 7, 100 μM GRL0617, or 100 IU/ml IFN-α was added (PBL Interferon Source). Cells were harvested at 9 h p.i., run on a 10% SDS-PAGE gel, and visualized by WB.
Statistics.
Statistical analysis was performed using Prism software, version 5 (GraphPad Software, Inc.). Survival curves were calculated using a log rank (Mantel-Cox) test.
Data analysis was performed using two-way analysis of variance (ANOVA) followed by a Bonferroni post hoc test unless otherwise noted. Data are represented as means ± standard deviations (SD) (Fig. 1 to 3 and 7). Results with a P value below 0.05 were considered significant.
RESULTS
USP18−/− mice have increased survival following MHV-3 infection.
To test our hypothesis that ISGylation has an antiviral effect on murine hepatitis virus 3 (MHV-3) infection, we infected USP18−/− mice, which have high baseline levels of ISGylation, and control USP18+/+ mice with 50 PFU of MHV-3 (20). The infection of susceptible C57BL/6 mice with MHV-3 induces a fulminant hepatitis with hepatocellular necrosis, sinusoid thrombosis, and strong cytokine induction and leads to death within 3 to 4 days p.i. (6). USP18+/+ (C57BL/6) mice survived a median of 4 days and displayed the rapid development of symptoms characteristic of MHV-3 infection (Fig. 1A). In contrast, USP18−/− mice (C57BL/6) survived up to 10 days p.i., with a median survival of 8 days. The lengthened survival was accompanied by improved liver histology at all time points. On day 3, all USP18+/+ liver histology samples showed 40 to 50% focal necrosis, and by day 4, 70 to 90% of the liver showed evidence of confluent necrosis (Fig. 1B). In comparison, USP18−/− livers on day 4 showed only small areas of focal necrosis affecting 5 to 10% of the liver. MHV-3 infection of USP18−/− mice did gradually lead to widespread liver damage (40 to 50% focal necrosis) although it was never as pronounced as in USP18+/+ mice. Improved liver histology in USP18−/− mice coincided with a decrease in the levels of serum transaminases (markers of liver injury) and decreased viral titers compared to those in USP18+/+ mice (Fig. 1C to E). Histology, viral titers, and liver function in USP18−/− mice were improved at all time points compared with USP18+/+ mice. However, at day 7 p.i., pathogenesis in USP18−/− mice approached levels comparable to those in USP18+/+ mice at day 3 p.i. for all three of the measured criteria, including histology, serum transaminase levels, and viral titers (Fig. 1B to E). Therefore, the absence of USP18 confers an early antiviral advantage against MHV-3 infection. However, USP18−/− mice ultimately succumbed to infection.
MHV-3 infection does not induce type I IFN in USP18−/− mice.
USP18−/− mice have increased baseline cellular ISGylation levels, and they have increased sensitivity to type I IFN signaling (20). Either of the two aspects of the USP18−/− phenotype may explain the suppression of viral replication and decreased inflammatory response seen in response to MHV-3 infection. In order to determine whether increased sensitivity to type 1 IFN contributed to the suppression of MHV-3 replication in USP18−/− mice, we measured type I IFN (IFN-α and IFN-β) induction by MHV-3 infection. In parallel, we measured type II IFN (IFN-γ) as a control proinflammatory cytokine. MHV-3 infection of USP18+/+ mice induced strong mRNA and protein expression of IFN-α, IFN-β, and IFN-γ in the liver and serum, respectively. In USP18−/− mice, there was no induction of IFN-α, IFN-β, or IFN-γ hepatic mRNA or protein levels in response to MHV-3 infection at any time point studied (Fig. 2A to D). Similarly, MHV-3 infection did not induce type I or type II IFN induction in USP18−/− primary hepatocytes (data not shown). Finally, USP18−/− peritoneal exudative macrophages (PEM) that were infected with MHV-3 did not show increased type I IFN signaling, as seen by the absence of STAT-1 phosphorylation (data not shown). These data imply that the increased antiviral activity against MHV-3 observed in the USP18−/− mice is not due to increased IFN production or signaling.
Cellular ISGylation inhibits MHV-3 replication.
Since the decrease in MHV-3 replication and mortality following infection of USP18−/− mice is not due to increased IFN or type I IFN signaling, we asked whether decreased MHV-3 replication and mortality were due to increased levels of ISGylation (21). First, we examined whether the decreased proliferation of MHV-3 in USP18−/− cells in vivo could be replicated in vitro in primary hepatocytes. Hepatocytes isolated from USP18+/+ and USP18−/− mice were infected with MHV-3 (multiplicity of infection [MOI] of 1), and viral titers were measured up to 48 h p.i. (Fig. 3A). USP18+/+ hepatocytes reached peak MHV-3 titers, 2.09 × 107 ± 1.29 × 106 PFU, between 12 and 18 h p.i. Viral titers remained elevated until 36 h p.i., after which the levels started to decrease, presumably due to cell death. Viral titers increased in USP18−/− hepatocytes with slower kinetics than in USP18+/+ mice, with nearly a log decrease in MHV-3 titers at almost all time points. However, USP18−/− mice reached peak viral titers at 36 h at 1.03 × 107 ± 2.31 × 105 PFU, more than 18 h after peak titers were reached in USP18+/+ hepatocytes. These in vitro results support the in vivo finding that MHV-3 viral titers are suppressed in the presence of increased ISGylation. Although the viral titers in USP18−/− hepatocytes are suppressed initially, the virus eventually overwhelms the antiviral response. This is similar to the extended survival seen in USP18−/− mice as these mice ultimately succumb to MHV-3 infection. Of note, at early time points following MHV-3 inoculation, namely, 1 h, 3 h, and 6 h, viral titers of USP18+/+ and USP18−/− mice do not differ significantly (two-way ANOVA with Bonferroni post hoc test). This suggests that virus binding and entry into the cell may not be affected by antiviral activity of interferon-stimulated gene 15 (ISG15); however, further studies are required to differentiate the effects of ISGylation on virus binding and entry and virus replication.
The amount of ISG15-conjugated proteins is markedly higher in USP18−/− hepatocytes than in USP18+/+ hepatocytes at all time points following MHV-3 infection (Fig. 3B). The expression of viral nucleocapsid (N) protein is detectable by WB starting at 6 h p.i. in USP18+/+ hepatocytes and continues to accumulate until 18 h (Fig. 3B) when peak N protein expression coincides with peak viral titers (Fig. 3A). In contrast, in USP18−/− hepatocytes, overall N protein accumulation is decreased and is delayed compared to that in USP18+/+ hepatocytes. These in vitro data mirror the in vivo results and suggest that high ISGylation levels are an intrinsic antiviral force against coronavirus in USP18−/− mice.
To determine whether ISGylation is causally linked to MHV-3 virus production, we knocked down the ISG15 pathway E1 enzyme Ube1L to block ISGylation. Ube1L is the unique activating enzyme of the ISG15/USP18 pathway and forms an ATP-dependent thioester bond with ISG15 (11, 35). Ube1L knockdown in primary USP18+/+ murine hepatocytes decreased Ube1L protein and attenuated the increase in ISGylation that follows exposure of cells to IFN-α (Fig. 4). Ube1L silencing in USP18+/+ primary cells infected with MHV-3 abrogates ISGylation and leads to increased N protein expression. As shown previously (Fig. 3B), N protein levels are markedly decreased in USP18−/− compared to USP18+/+ cells at 12 h p.i. The effect of Ube1L knockdown is more prominent in USP18−/− primary cells; Ube1L silencing markedly decreased protein ISGylation in USP18−/− primary cells from high baseline levels of ISGylation although ISGylation levels remained at elevated levels following MHV-3 infection compared to control hepatocytes. Following both MHV-3 infection and Ube1L knockdown, the levels of ISGylated proteins dropped further compared to Ube1L knockdown alone (Fig. 4), and N protein expression, the surrogate of MHV-3 replication, reached levels of expression higher than those induced by MHV-3 infection alone. MHV-3 infection decreased ISGylation in both USP18+/+ and USP18−/− mice. In MHV-3-infected USP18+/+ hepatocytes, however, decreased ISGylation was observed only following treatment with IFN-α which strongly induces ISGylation. In the absence of IFN-α, no ISGylation is detected by WB. These data suggest that the Ube1L conjugation of ISG15 to target proteins and the subsequent accumulation of ISGylated proteins mediate an antiviral effect against MHV-3.
The effect of PLP inhibitors on MHV-3 infection.
Since there is an observed decrease in ISGylation in USP18−/− cells following MHV-3 infection, we next asked whether the MHV-3 PLP2 protein is the mechanism through which MHV-3 ultimately escapes the effect of host ISGylation. To this end, we treated MHV-3-infected cells with synthetic PLP inhibitors. The PLP inhibitors are noncovalent cysteine inhibitor compounds generated against the catalytic residues of the SARS coronavirus PLP (34). PLP inhibitors increased ISGylation levels in USP18+/+ PEM. No observable change in ISGylation was observed in USP18−/− PEM even in less saturated blots (data not shown) due, perhaps, to already high baseline levels of ISGylation in USP18−/− PEM. USP18+/+ PEM infected with MHV-3 showed strong expression of N protein at 9 h p.i. Treatment of USP18+/+ PEM with PLP inhibitors resulted in decreased levels of N protein production (Fig. 5). Treatment with GRL0617, the PLP inhibitor with the highest binding capacity (34), resulted in the greatest decrease in viral N protein production. In MHV-3-infected USP18−/− PEM, where baseline levels of ISGylated proteins are high (21), the expression of N protein was significantly decreased compared to that in wild-type PEM. The expression of N protein was even more inhibited following treatment with all PLP inhibitors. By WB analysis, GRL0617 completely inhibited the expression of N protein. Consistent with these data, viral titers were decreased in the presence of PLP2 inhibitors in both USP18+/+ and USP18−/− PEM (data not shown). Similar data were obtained for hepatocytes (data not shown). Overall, these results suggest that MHV-3 proliferation and evasion from the cellular antiviral ISGylation milieu are dependent on PLP2 activity.
FIG 5.
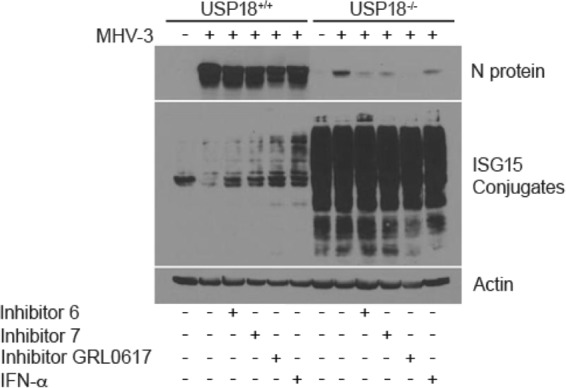
PLP2 inhibitors and ISG15 antiviral activity inhibit MHV-3 N protein production. PEM from USP18+/+ or USP18−/− mice were inoculated with MHV-3 (MOI of 0.1) for 1 h; the virus was then washed away, and PEM were treated with PLP inhibitors (100 μM inhibitor 6, 7, or GRL0617) or IFN-α (100 IU/ml) (34). Cells were harvested at 9 h p.i. N protein and ISG15 conjugates were visualized by WB.
ISGylation delays onset of MHV-3 production and death in vivo.
Above, we showed that ISGylation in vitro exerts an antiviral pressure on MHV-3 coronavirus replication. However, this antiviral pressure is overcome as the virus replicates and expresses the deubiquitinase protein (DUB) PLP2. In order to test whether this holds true in vivo, we inoculated USP18+/+ and USP18−/− animals with 50 PFU of MHV-3 i.p. and measured ISGylation and PLP2 levels in whole-liver tissue. As before, USP18+/+ mice became moribund and were euthanized by day 4 p.i. Liver protein ISGylation increased and peaked at day 3 p.i. (Fig. 6); N protein was first detectable at day 2 and increased steadily until the animals were euthanized on day 4. In contrast, USP18−/− mice had higher baseline liver ISGylation levels throughout the course of infection, and MHV-3 replication as seen by N protein expression was first detected at day 6 p.i. Concurrently, we measured viral expression of plp2 mRNA. plp2 mRNA expression increased as N protein expression increased, and plp2 expression was delayed in USP18−/− mice compared to expression in control mice (Fig. 7). These data are consistent with the ability of increased PLP2 to gradually overwhelm the antiviral effect of ISGylation in vivo by stripping ISG15 from target proteins.
DISCUSSION
During the innate immune response, ISGylation is an important barrier against viral infection. ISGs are upregulated following the activation of type I IFN signaling and have various functions in the innate immune response, including the feedback regulation of the host immune response and antiviral activities (36–41). ISG15 is one of the most abundantly upregulated ISGs. In this study, we show that ISG15 conjugation (ISGylation) has antiviral activity in a model of severe coronavirus infection and that this antiviral activity is independent of type I IFN signaling.
Studies have shown that ISGylation has an antiviral effect against several viruses, including influenza B virus, Sindbis virus, Sendai virus, and vaccinia virus (15, 41–44). ISG15 and IFN-α/β receptor double-deficient mice, which lack ISG15 and have an impaired ability to induce ISG following IFN-α/β stimulation, were rescued against Sindbis virus-induced lethality by expressing ISG15 (42, 43). The antiviral activity associated with ISG15 is dependent on the conjugation of ISG15 to target proteins as the mutation of the ISG15 C-terminal isopeptidase target motif rendered ISGylation ineffective against Sindbis virus (42). However, ISGylation is permissive for certain viruses. We along with others have demonstrated that cellular ISGylation is necessary for the efficient replication of hepatitis C virus (25, 45). Our data demonstrate that ISGylation has antiviral activity against MHV-3 infection. In USP18−/− mice, where ISGylation levels are high, there was a significant decrease in viral N protein expression in vitro and in vivo that coincided with prolonged survival. We attribute the prolongation in survival to the antiviral effect of ISGylation since we did not observe the induction of a type I or type II IFN response in the USP18−/− animals. The role of ISGylation was confirmed by Ube1L knockdown experiments, in which silencing Ube1L in primary hepatocytes reversed the antiviral activity of ISGylation (Fig. 4). These data are in accordance with studies in which both Ube1L−/− mice, which lack the ability to conjugate ISG15 to target proteins, and ISG15−/− mice were more sensitive to Sindbis virus infection (43, 46) than wild-type animals. Therefore, our data provide another example of the antiviral nature of ISGylation activity but in a novel context, coronavirus infection.
Coronaviruses evade the host innate immune response by interfering with the induction of IFN (47–52). MHV-3 infection of USP18+/+ mice resulted in markedly increased type I IFN mRNA and protein (Fig. 2A to D). In contrast, MHV-3 infection of USP18−/− mice resulted in little, if any, IFN production at either the mRNA or protein level. At first glance this is surprising since USP18−/− mice are hypersensitive to IFN, and we expected that any IFN-β produced upon exposure to MHV-3 virus would stimulate IFN-α production in a positive feedback loop (53). Furthermore, the increased ISGylation observed in USP18−/− cells would be expected to lead to increased IFN production since the direct ISGylation of interferon regulatory transcription factor 3 (IRF-3) prolongs the activation of IRF-3 and results in increased expression of IFN-β and other ISGs (41). However, we observed an almost complete absence of type I IFN production in response to MHV-3. While it is tempting to ascribe the lack of IFN production to the increased ISGylation seen in USP18−/− cells, this is unlikely. First, USP18 was shown to regulate type I IFN induction independent of isopeptidase activity, and it was shown that ISG15 antiviral activity is independent of IFN signaling (22, 43). Second, the observed lack of a type I IFN response may also be due to differences in the immune cell phenotype of USP18−/− mice. Cong et al. described a USP18-dependent but ISGylation-independent difference in dendritic cell maturation (54). Similarly, we have observed a fundamental difference in the phenotype of USP18−/− macrophages and Kupffer cells, with a shift from the M1 proinflammatory phenotype in USP18+/+ mice to an M2 regulatory phenotype in USP18−/− mice (unpublished data). This result may suggest that the fact that MHV-3 does not induce IFN is not specific to the virus but the phenotype of the USP18−/− macrophages and dendritic cells, both of which are key producers of hepatic IFN (23, 53, 55). Differences in immune cell phenotype and the consequent lack of IFN production in USP18−/− mice raise the possibility of confounding immune effects in our in vivo data. However, the lack of a type I IFN response in the USP18−/− mice would be expected to promote viral replication, but we observed the reverse effect. The lack of IFN signaling thus allows us to attribute our results to ISGylation as opposed to IFN signaling in USP18−/− mice.
The mechanism by which ISGylation exerts antiviral activity against MHV-3 is unclear. ISGylation can modulate virus production in one of two ways: by being conjugated to viral proteins in a manner that disrupts the viral life cycle or by conjugating to host proteins that play a role in the viral life cycle or in the host innate immune response (11). In the context of the ISGylation of viral proteins, the conjugation of ISG15 to the influenza A virus NS1 protein has an antiviral effect on influenza A virus infection in humans (39). ISGylation of NS1 prevents its association with importin-α and its subsequent translocation to the nucleus, ultimately inhibiting viral replication (38). In an example of host protein modification, the ISGylation of host 4E homologous protein (4EHP) protein exerts an antiviral effect by interfering with viral (and host) protein translation. 4EHP is a 5′ mRNA cap binding protein that competes with eukaryotic initiation factor 4 (eIF4) to suppress translation. The ISGylation of 4EHP increases its RNA binding capacity and strongly suppresses translation of both host and viral proteins during the innate immune response (37). Lastly, another group of investigators reported a mechanism that incorporates both host and viral protein ISGylation: the antiviral pressure of ISGylation observed in this model is exerted by the indiscriminate ISGylation of newly synthesized proteins that follows IFN stimulation or USP18 deletion (36). The nonspecific ISGylation of the nascent subset of each protein population present in the cell has been suggested to disrupt viral proliferation through a dominant negative effect. Such an effect has been shown in models of human papillomavirus (HPV) and HIV infection (36, 56) and is consistent with our finding that the effect of ISGylation blunts viral replication but does not abrogate it altogether. Further studies are necessary to determine the mechanism by which ISGylation specifically delays MHV-3 replication.
Although ISGylation exerts an antiviral pressure on the MHV-3 coronavirus, the virus ultimately overcomes this pressure both in vitro and in vivo. Viral titers in states of increased ISGylation (USP18−/−) approach peak levels seen in states of low ISGylation (USP18+/+) after a 3-day delay. Additionally, N protein production and PLP2 expression levels increase while ISGylation levels in USP18−/− mice decrease (Fig. 6 and 7). All coronaviruses express a DUB that removes both ubiquitin and ISG15 from conjugated proteins (29, 57, 58). PLPs are multifunctional viral proteins and have several functions relevant to coronavirus production within the host cell, including the processing of the viral polyprotein. In our study, some of the inhibitory effect on MHV-3 production that follows treatment with PLP inhibitors may be due to this effect (34, 51). However, treatment of wild-type PEM and hepatocytes with PLP inhibitors increased cellular ISGylation following infection and inhibited viral N protein expression (Fig. 5). Our data, therefore, may be the result of both nsp3 cleavage and ISGylation, but PLP2 definitely contributes to the stripping of ISG15 from target proteins. Regardless of the relative contributions of each mode of action, these data support the use of PLP2 inhibitors as one arm of treatment against coronavirus infection. The DUB activity may thus contribute to the evasion of the host antiviral response by altering immune responses through de-ISGylation or by reactivating ISGylated viral proteins.
We have previously shown that increasing ubiquitination exerts an antiviral effect on coronavirus replication (28), and in this study we have shown that ISGylation exerts a similar effect. Both DUB functions of PLP2 would favor MHV-3 replication although PLP2 has more affinity for ISGylated proteins than for ubiquitinated proteins and therefore may be more relevant as an antiviral mechanism (29, 30, 59).
Overall, our data are consistent with the ability of ISGylation to exert an antiviral pressure in a model of severe MHV-3 coronavirus infection. This effect is most marked in the setting of USP18 deletion, suggesting that targeting USP18 might have a clinical benefit in treating severe coronavirus infections. However, our data also strongly argue that treatment of a coronavirus infection should be multifaceted since the virus has evolved mechanisms to overwhelm the antiviral effect of ISGylation, one of the chief effector mechanisms of IFN. As illustrated by this study, coronavirus can escape the host antiviral response in a number of ways that are independent of IFN signaling. IFN therapy, the current treatment for coronavirus infection, should be considered in conjunction with other therapies, such as the inhibition of PLP activity and proteasome inhibition, to comprehensively counter the effects of the viral DUB (28).
ACKNOWLEDGMENTS
We especially thank Dong Er Zhang for the gift of the USP18−/− mice.
This research was supported by the Canadian Institute of Health Research (grant 200622).
Footnotes
Published ahead of print 19 March 2014
REFERENCES
- 1.Rota PA, Oberste MS, Monroe SS, Nix WA, Campagnoli R, Icenogle JP, Penaranda S, Bankamp B, Maher K, Chen MH, Tong S, Tamin A, Lowe L, Frace M, DeRisi JL, Chen Q, Wang D, Erdman DD, Peret TC, Burns C, Ksiazek TG, Rollin PE, Sanchez A, Liffick S, Holloway B, Limor J, McCaustland K, Olsen-Rasmussen M, Fouchier R, Gunther S, Osterhaus AD, Drosten C, Pallansch MA, Anderson LJ, Bellini WJ. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399. 10.1126/science.1085952 [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. 15 August 2003, posting date Summary table of SARS cases by country, 1 November 2002–7 August 2003. WHO, Geneva, Switzerland: http://www.who.int/csr/sars/country/2003_08_15/en/. [Google Scholar]
- 3.World Health Organization. 16 February 2013, posting date Novel coronavirus infection-update. World Health Organization, Geneva, Switzerland: http://www.who.int/csr/don/2013_02_16/en/ [Google Scholar]
- 4.Perlman S, Dandekar AA. 2005. Immunopathogenesis of coronavirus infections: implications for SARS. Nat. Rev. Immunol. 5:917–927. 10.1038/nri1732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bergmann CC, Lane TE, Stohlman SA. 2006. Coronavirus infection of the central nervous system: host-virus stand-off. Nat. Rev. Microbiol. 4:121–132. 10.1038/nrmicro1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy GA, Leibowitz JL, Edgington TS. 1981. Induction of monocyte procoagulant activity by murine hepatitis virus type 3 parallels disease susceptibility in mice. J. Exp. Med. 154:1150–1163. 10.1084/jem.154.4.1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marsden PA, Ning Q, Fung LS, Luo X, Chen Y, Mendicino M, Ghanekar A, Scott JA, Miller T, Chan CW, Chan MW, He W, Gorczynski RM, Grant DR, Clark DA, Phillips MJ, Levy GA. 2003. The Fgl2/fibroleukin prothrombinase contributes to immunologically mediated thrombosis in experimental and human viral hepatitis. J. Clin. Invest. 112:58–66. 10.1172/JCI200318114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pope M, Rotstein O, Cole E, Sinclair S, Parr R, Cruz B, Fingerote R, Chung S, Gorczynski R, Fung L. 1995. Pattern of disease after murine hepatitis virus strain 3 infection correlates with macrophage activation and not viral replication. J. Virol. 69:5252–5260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McGilvray ID, Lu Z, Wei AC, Dackiw AP, Marshall JC, Kapus A, Levy G, Rotstein OD. 1998. Murine hepatitis virus strain 3 induces the macrophage prothrombinase fgl-2 through p38 mitogen-activated protein kinase activation. J. Biol. Chem. 273:32222–32229. 10.1074/jbc.273.48.32222 [DOI] [PubMed] [Google Scholar]
- 10.Farrell PJ, Broeze RJ, Lengyel P. 1979. Accumulation of an mRNA and protein in interferon-treated Ehrlich ascites tumour cells. Nature 279:523–525. 10.1038/279523a0 [DOI] [PubMed] [Google Scholar]
- 11.Lenschow DJ. 2010. Antiviral properties of ISG15. Viruses 2:2154–2168. 10.3390/v2102154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giannakopoulos NV, Luo JK, Papov V, Zou W, Lenschow DJ, Jacobs BS, Borden EC, Li J, Virgin HW, Zhang DE. 2005. Proteomic identification of proteins conjugated to ISG15 in mouse and human cells. Biochem. Biophys. Res. Commun. 336:496–506. 10.1016/j.bbrc.2005.08.132 [DOI] [PubMed] [Google Scholar]
- 13.Zhao C, Denison C, Huibregtse JM, Gygi S, Krug RM. 2005. Human ISG15 conjugation targets both IFN-induced and constitutively expressed proteins functioning in diverse cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 102:10200–10205. 10.1073/pnas.0504754102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malakhov MP. 2003. High-throughput immunoblotting. Ubiquitin-like protein ISG15 modifies key regulators of signal transduction. J. Biol. Chem. 278:16608–16613 [DOI] [PubMed] [Google Scholar]
- 15.Osiak A, Utermohlen O, Niendorf S, Horak I, Knobeloch KP. 2005. ISG15, an interferon-stimulated ubiquitin-like protein, is not essential for STAT1 signaling and responses against vesicular stomatitis and lymphocytic choriomeningitis virus. Mol. Cell. Biol. 25:6338–6345. 10.1128/MCB.25.15.6338-6345.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeon YJ, Choi JS, Lee JY, Yu KR, Kim SM, Ka SH, Oh KH, Il Kim K, Zhang D-E, Bang OS, Ha Chung C. 2009. ISG15 modification of filamin B negatively regulates the type I interferon-induced JNK signalling pathway. EMBO Rep. 10:374–380. 10.1038/embor.2009.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zou W, Papov V, Malakhova O, Kim KI, Dao C, Li J, Zhang D-E. 2005. ISG15 modification of ubiquitin E2 Ubc13 disrupts its ability to form thioester bond with ubiquitin. Biochem. Biophys. Res. Commun. 336:61–68. 10.1016/j.bbrc.2005.08.038 [DOI] [PubMed] [Google Scholar]
- 18.Malakhov MP. 2002. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 277:9976–9981. 10.1074/jbc.M109078200 [DOI] [PubMed] [Google Scholar]
- 19.Ritchie KJ. 2002. Dysregulation of protein modification by ISG15 results in brain cell injury. Genes Dev. 16:2207–2212. 10.1101/gad.1010202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie KJ, Hahn CS, Kim KI, Yan M, Rosario D, Li L, de la Torre JC, Zhang D-E. 2004. Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat. Med. 10:1374–1378. 10.1038/nm1133 [DOI] [PubMed] [Google Scholar]
- 21.Malakhova OA. 2003. Protein ISGylation modulates the JAK-STAT signaling pathway. Genes Dev. 17:455–460. 10.1101/gad.1056303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Malakhova OA, Kim KI, Luo JK, Zou W, Kumar KG, Fuchs SY, Shuai K, Zhang DE. 2006. UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 25:2358–2367. 10.1038/sj.emboj.7601149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woods MW, Kelly JN, Hattlmann CJ, Tong JG, Xu LS, Coleman MD, Quest GR, Smiley JR, Barr SD. 2011. Human HERC5 restricts an early stage of HIV-1 assembly by a mechanism correlating with the ISGylation of Gag. Retrovirology 8:95. 10.1186/1742-4690-8-95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen L, Sun J, Meng L, Heathcote J, Edwards AM, McGilvray ID. 2010. ISG15, a ubiquitin-like interferon-stimulated gene, promotes hepatitis C virus production in vitro: implications for chronic infection and response to treatment. J. Gen. Virol. 91:382–388. 10.1099/vir.0.015388-0 [DOI] [PubMed] [Google Scholar]
- 25.Chen L, Borozan I, Sun J, Guindi M, Fischer S, Feld J, Anand N, Heathcote J, Edwards AM, McGilvray ID. 2010. Cell-type specific gene expression signature in liver underlies response to interferon therapy in chronic hepatitis C infection. Gastroenterology 138:1123–1133.e3. 10.1053/j.gastro.2009.10.046 [DOI] [PubMed] [Google Scholar]
- 26.Chen L, Borozan I, Feld J, Sun J, Tannis L-L, Coltescu C, Heathcote J, Edwards AM, McGilvray ID. 2005. Hepatic gene expression discriminates responders and nonresponders in treatment of chronic hepatitis C viral infection. Gastroenterology 128:1437–1444. 10.1053/j.gastro.2005.01.059 [DOI] [PubMed] [Google Scholar]
- 27.McGilvray I, Feld JJ, Chen L, Pattullo V, Guindi M, Fischer S, Borozan I, Xie G, Selzner N, Heathcote EJ, Siminovitch K. 2012. Hepatic cell-type specific gene expression better predicts HCV treatment outcome than IL28B genotype. Gastroenterology 142:1122–1131.e1. 10.1053/j.gastro.2012.01.028 [DOI] [PubMed] [Google Scholar]
- 28.Ma XZ, Bartczak A, Zhang J, Khattar R, Chen L, Liu MF, Edwards A, Levy G, McGilvray ID. 2010. Proteasome inhibition in vivo promotes survival in a lethal murine model of severe acute respiratory syndrome. J. Virol. 84:12419–12428. 10.1128/JVI.01219-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barretto N, Jukneliene D, Ratia K, Chen Z, Mesecar AD, Baker SC. 2005. The papain-like protease of severe acute respiratory syndrome coronavirus has deubiquitinating activity. J. Virol. 79:15189–15198. 10.1128/JVI.79.24.15189-15198.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lindner HA, Lytvyn V, Qi H, Lachance P, Ziomek E, Ménard R. 2007. Selectivity in ISG15 and ubiquitin recognition by the SARS coronavirus papain-like protease. Arch. Biochem. Biophys. 466:8–14. 10.1016/j.abb.2007.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Selzner N, Liu H, Boehnert MU, Adeyi OA, Shalev I, Bartczak AM, Xue-Zhong M, Manuel J, Rotstein OD, McGilvray ID, Grant DR, Phillips MJ, Levy GA, Selzner M. 2012. FGL2/fibroleukin mediates hepatic reperfusion injury by induction of sinusoidal endothelial cell and hepatocyte apoptosis in mice. J. Hepatol. 56:153–159. 10.1016/j.jhep.2011.05.033 [DOI] [PubMed] [Google Scholar]
- 32.McQueen CA, Williams GM. 1987. The hepatocyte primary culture/DNA repair test using hepatocytes from several species. Cell Biol. Toxicol. 3:209–218. 10.1007/BF00058457 [DOI] [PubMed] [Google Scholar]
- 33.De Albuquerque N, Baig E, Ma X, Zhang J, He W, Rowe A, Habal M, Liu M, Shalev I, Downey GP, Gorczynski R, Butany J, Leibowitz J, Weiss SR, McGilvray ID, Phillips MJ, Fish EN, Levy GA. 2006. Murine hepatitis virus strain 1 produces a clinically relevant model of severe acute respiratory syndrome in A/J. mice. J. Virol. 80:10382–10394. 10.1128/JVI.00747-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ratia K, Pegan S, Takayama J, Sleeman K, Coughlin M, Baliji S, Chaudhuri R, Fu W, Prabhakar BS, Johnson ME, Baker SC, Ghosh AK, Mesecar AD. 2008. A noncovalent class of papain-like protease/deubiquitinase inhibitors blocks SARS virus replication. Proc. Natl. Acad. Sci. U. S. A. 105:16119–16124. 10.1073/pnas.0805240105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan W, Krug RM. 2001. Influenza B virus NS1 protein inhibits conjugation of the interferon (IFN)-induced ubiquitin-like ISG15 protein. EMBO J. 20:362–371. 10.1093/emboj/20.3.362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durfee LA, Lyon N, Seo K, Huibregtse JM. 2010. The ISG15 conjugation system broadly targets newly synthesized proteins: implications for the antiviral function of ISG15. Mol. Cell 38:722–732. 10.1016/j.molcel.2010.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Okumura F, Zou W, Zhang DE. 2007. ISG15 modification of the eIF4E cognate 4EHP enhances cap structure-binding activity of 4EHP. Genes Dev. 21:255–260. 10.1101/gad.1521607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao C, Hsiang TY, Kuo RL, Krug RM. 2010. ISG15 conjugation system targets the viral NS1 protein in influenza A virus-infected cells. Proc. Natl. Acad. Sci. U. S. A. 107:2253–2258. 10.1073/pnas.0909144107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsiang TY, Zhao C, Krug RM. 2009. Interferon-induced ISG15 conjugation inhibits influenza A virus gene expression and replication in human cells. J. Virol. 83:5971–5977. 10.1128/JVI.01667-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu SY, Sanchez DJ, Cheng G. 2011. New developments in the induction and antiviral effectors of type I interferon. Curr. Opin. Immunol. 23:57–64. 10.1016/j.coi.2010.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi HX, Yang K, Liu X, Liu XY, Wei B, Shan YF, Zhu LH, Wang C. 2010. Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol. Cell. Biol. 30:2424–2436. 10.1128/MCB.01466-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lenschow DJ, Giannakopoulos NV, Gunn LJ, Johnston C, O'Guin AK, Schmidt RE, Levine B, Virgin HW., IV 2005. Identification of interferon-stimulated gene 15 as an antiviral molecule during Sindbis virus infection in vivo. J. Virol. 79:13974–13983. 10.1128/JVI.79.22.13974-13983.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenschow DJ, Lai C, Frias-Staheli N, Giannakopoulos NV, Lutz A, Wolff T, Osiak A, Levine B, Schmidt RE, Garcia-Sastre A, Leib DA, Pekosz A, Knobeloch KP, Horak I, Virgin HW., IV 2007. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. U. S. A. 104:1371–1376. 10.1073/pnas.0607038104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guerra S, Caceres A, Knobeloch KP, Horak I, Esteban M. 2008. Vaccinia virus E3 protein prevents the antiviral action of ISG15. PLoS Pathog. 4:e1000096. 10.1371/journal.ppat.1000096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Broering R, Zhang X, Kottilil S, Trippler M, Jiang M, Lu M, Gerken G, Schlaak JF. 2010. The interferon stimulated gene 15 functions as a proviral factor for the hepatitis C virus and as a regulator of the IFN response. Gut 59:1111–1119. 10.1136/gut.2009.195545 [DOI] [PubMed] [Google Scholar]
- 46.Giannakopoulos NV, Arutyunova E, Lai C, Lenschow DJ, Haas AL, Virgin HW. 2009. ISG15 Arg151 and the ISG15-conjugating enzyme UbE1L are important for innate immune control of Sindbis virus. J. Virol. 83:1602–1610. 10.1128/JVI.01590-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou H, Perlman S. 2007. Mouse hepatitis virus does not induce Beta interferon synthesis and does not inhibit its induction by double-stranded RNA. J. Virol. 81:568–574. 10.1128/JVI.01512-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Versteeg GA, Bredenbeek PJ, van den Worm SH, Spaan WJ. 2007. Group 2 coronaviruses prevent immediate early interferon induction by protection of viral RNA from host cell recognition. Virology 361:18–26. 10.1016/j.virol.2007.01.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun L, Xing Y, Chen X, Zheng Y, Yang Y, Nichols DB, Clementz MA, Banach BS, Li K, Baker SC, Chen Z. 2012. Coronavirus papain-like proteases negatively regulate antiviral innate immune response through disruption of STING-mediated signaling. PLoS One 7:e30802. 10.1371/journal.pone.0030802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spiegel M, Pichlmair A, Martinez-Sobrido L, Cros J, Garcia-Sastre A, Haller O, Weber F. 2005. Inhibition of beta interferon induction by severe acute respiratory syndrome coronavirus suggests a two-step model for activation of interferon regulatory factor 3. J. Virol. 79:2079–2086. 10.1128/JVI.79.4.2079-2086.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Clementz MA, Chen Z, Banach BS, Wang Y, Sun L, Ratia K, Baez-Santos YM, Wang J, Takayama J, Ghosh AK, Li K, Mesecar AD, Baker SC. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 84:4619–4629. 10.1128/JVI.02406-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Devaraj SG, Wang N, Chen Z, Tseng M, Barretto N, Lin R, Peters CJ, Tseng CT, Baker SC, Li K. 2007. Regulation of IRF-3-dependent innate immunity by the papain-like protease domain of the severe acute respiratory syndrome coronavirus. J. Biol. Chem. 282:32208–32221. 10.1074/jbc.M704870200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Theofilopoulos AN, Baccala R, Beutler B, Kono DH. 2005. Type I interferons (alpha/beta) in immunity and autoimmunity. Annu. Rev. Immunol. 23:307–336. 10.1146/annurev.immunol.23.021704.115843 [DOI] [PubMed] [Google Scholar]
- 54.Cong XL, Lo MC, Reuter BA, Yan M, Fan JB, Zhang DE. 2012. Usp18 promotes conventional CD11b+ dendritic cell development. J. Immunol. 188:4776–4781. 10.4049/jimmunol.1101609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rose KM, Weiss SR. 2009. Murine coronavirus cell type dependent interaction with the type i interferon response. Viruses 1:689–712. 10.3390/v1030689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee SK, Harris J, Swanstrom R. 2009. A strongly transdominant mutation in the human immunodeficiency virus type 1 gag gene defines an Achilles heel in the virus life cycle. J. Virol. 83:8536–8543. 10.1128/JVI.00317-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lindner HA, Fotouhi-Ardakani N, Lytvyn V, Lachance P, Sulea T, Menard R. 2005. The papain-like protease from the severe acute respiratory syndrome coronavirus is a deubiquitinating enzyme. J. Virol. 79:15199–15208. 10.1128/JVI.79.24.15199-15208.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ratia K. 2006. Severe acute respiratory syndrome coronavirus papain-like protease: structure of a viral deubiquitinating enzyme. Proc. Natl. Acad. Sci. U. S. A. 103:5717–5722. 10.1073/pnas.0510851103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen Z, Wang Y, Ratia K, Mesecar AD, Wilkinson KD, Baker SC. 2007. Proteolytic processing and deubiquitinating activity of papain-like proteases of human coronavirus NL63. J. Virol. 81:6007–6018. 10.1128/JVI.02747-06 [DOI] [PMC free article] [PubMed] [Google Scholar]