Abstract
Dengue virus (DENV) and hepatitis C virus (HCV), members of the family Flaviviridae, are global human health concerns. As positive-strand RNA viruses, they each replicate in the cytoplasm of infected cells and induce distinct membranous replication compartments where most, if not all, steps of the viral life cycle occur. This Gem briefly reviews the most recent insights into the architecture and functional properties of membranous replication and assembly sites induced by DENV and HCV.
INTRODUCTION
Flaviviridae constitute a large virus family to which medically highly relevant human pathogens belong. Among those, hepatitis C virus (HCV), genus Hepacivirus, has infected around 170 million individuals worldwide and is a major cause of liver diseases, while dengue virus (DENV), genus Flavivirus, causes the most prevalent arthropod-borne viral disease, with an estimated ∼100 million symptomatic human infections per year. While direct-acting antivirals for treatment of HCV infections have recently been approved, it is expected that only a small proportion of infected individuals worldwide will benefit from this treatment. In case of DENV infection, neither selective therapy nor prophylactic vaccines exist. Thus, there are unmet medical needs for the development of novel treatment modalities and preventive strategies (1, 2).
While HCV and DENV belong to the same family and thus share many features of their life cycles, the molecular strategies underlying RNA replication and virus production differ substantially. Upon entry of HCV and DENV into the cell, the positive-strand RNA genome is delivered into the cytoplasm and used for RNA translation. The resulting polyprotein is cleaved by cellular and viral proteases, thus releasing several structural and nonstructural proteins (1, 2). Like all other positive-strand RNA viruses, HCV and DENV induce massive rearrangements of intracellular membranes to create in the cytoplasm a replication-favorable membranous microenvironment that we generically call “replication factories” or, in the case of HCV, “membranous web.” Analyses with electron tomography and three-dimensional (3D) reconstructions of remodeled membranes in HCV- and DENV-infected cells revealed endoplasmic reticulum (ER)-derived organelle-like structures whose morphologies differ strikingly (3, 4). These membranous structures are thought to serve several purposes: first, to coordinate the different steps of the viral life cycle by spatial segregation of replicating RNA from ribosomes and assembling capsids; second, to increase the local concentration of components required for efficient replication and assembly by reducing the diffusion space; third, to protect viral RNA against cellular nucleases and innate immunity-triggering pattern recognition receptors. Here, we will briefly summarize our current understanding of the HCV and DENV replication factories, focusing on their architecture and functional properties.
DENV-/HCV-INDUCED REPLICATION COMPARTMENTS
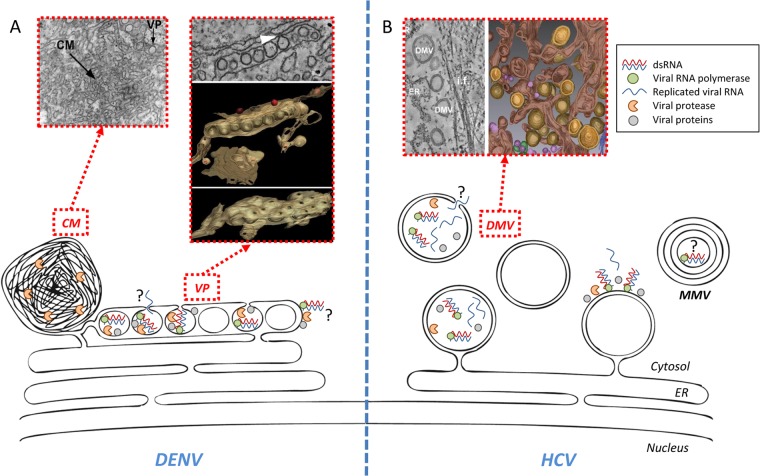
In DENV-infected cells, several membrane alterations can be observed (4) (Fig. 1A). First, there are invaginations of the ER membrane forming ∼90-nm-wide vesicles, which pack in-plane within ER sheets. These structures, termed vesicle packets (VPs), form a continuous membranous network in the ER and contain the viral RNA polymerase, nonstructural proteins 2B (NS2B), NS3, NS4A, and NS4B, and double-stranded RNA (dsRNA), the presumed replication intermediate, strongly suggesting that VPs are the site of RNA replication. In support of this model, an opening toward the cytosol could be observed in ∼50% of the vesicles. This 10-nm “pore” would allow metabolites (e.g., nucleoside triphosphates [NTPs]) to access replication complexes and newly synthesized viral RNA to exit VPs to be used for translation or virus particle assembly (see below). Second, in addition to VPs, large areas of convoluted membranes (CMs) were described and hypothesized to be the site of DENV RNA translation and polyprotein processing, because only the NS2B-3 viral protease complex, and no other DENV protein, was detected within these structures. Moreover, CMs are connected to the VP-containing ER network and, hence, might also constitute a “pool” of membranes required for de novo formation of replication factories. Nevertheless, CMs were undetectable in DENV-infected mosquito cells by electron microscopy (EM) (5), raising questions about their functional relevance or specific roles in the viral life cycle.
FIG 1.
DENV and HCV replication factories and associated membranes. (A) Schematic representation of DENV-induced endoplasmic reticulum (ER)-derived vesicle packets (VP) and convoluted membranes (CM). Corresponding transmission EM pictures (for VP and CM) and a 3D reconstruction from electron tomography (for VP) are also shown. The identified pore of each vesicle might act as an exit site for neosynthesized viral RNA. Note that DENV RNA replication on the cytosolic side of VPs is not excluded. (B) Schematic representation of HCV-induced membrane rearrangements. Note that the appearance of double-membrane vesicles (DMVs) coincides with the peak of RNA replication. At later time points after infection, multimembrane vesicles (MMV) as well as double-membrane tubes (not shown) are also observed, but their involvement in RNA replication remains to be determined. An opening toward the cytoplasm is observed in ∼10% of DMVs and might allow exit of viral RNA. Although enzymatically active HCV replicase is physically associated with DMVs, it is not known if RNA amplification occurs within DMVs and/or on their cytosolic side. Some of the insets are reprinted from reference 4 with permission from the publisher and from PLoS Pathogens (3).
Very little is known about the molecular mechanisms governing the formation of VPs and CMs. The sole expression of mature DENV NS4A was shown to induce membrane alterations reminiscent of CMs (6). Furthermore, this process appeared to depend on NS2B-3 protease-dependent maturation of NS4A, since no membrane alterations were observed for cells expressing the immature NS4A-2k precursor. This is consistent with the observation that the DENV protease partly localizes within CMs in infected cells. However, whether these NS4A-induced structures, or other DENV proteins crucial for RNA replication, are required for VP and CM formation remains to be elucidated; to our knowledge, DENV mutations specifically abrogating their biogenesis have not been described so far.
The architecture of HCV-induced membrane alterations differs strikingly from that of DENV VPs (Fig. 1B). The predominant membrane species in the case of HCV are double-membrane vesicles (DMVs), exhibiting an average diameter of ∼150 nm and accumulating in parallel to the peak of RNA replication (3, 7). At later time points of infection, double-membrane tubules and multimembrane vesicles appear, the latter presumably reflecting a stress-induced host cell response. By using electron tomography and 3D reconstructions, we found that with ∼50% of DMVs the outer membrane is connected to the ER membrane via a neck-like structure, arguing that DMVs are protrusions of the ER toward the cytoplasm (3). This topology strikingly contrasts with the invaginations observed for DENV VPs. A biochemical and morphological characterization of purified HCV DMVs demonstrated that these structures contain active viral replicase, suggesting that DMVs are bona fide replication sites (8). Reminiscent of DENV VPs, an opening toward the cytosol was found with ∼10% of DMVs (3). Whether this opening corresponds to “immature” DMVs, prior to complete closure of the membranes, or represents a distinct structure remains to be elucidated. Moreover, it remains also unclear whether RNA replication takes place within DMVs or on their outer surface.
For a long time, HCV NS4B, an integral membrane protein, was considered the main inducer of the membranous web since its sole expression can induce massive membrane rearrangements (9). Consistently, we found that mutations in NS4B inhibiting self-interaction affect DMV morphology and abrogate HCV RNA replication (10). Thus, via oligomerization, NS4B might contribute to DMV biogenesis by inducing ER membrane curvature. However, by using a replication-independent polyprotein expression approach, we found that NS5A was the only protein capable of inducing DMVs, while only single-membrane vesicles were observed in NS3/4A-, NS4B-, and NS5B-expressing cells (3, 11). Since none of the HCV proteins expressed on its own was capable of inducing a membranous web, these results suggest that web formation most likely requires a concerted action of the replicase proteins. In addition, several host cell factors are involved in this membrane remodeling. Notably, we have recently shown that de novo formation of DMVs was blocked by pharmacological inhibition of cyclophilin A (CypA), an NS5A interaction partner that is critical for RNA replication (reference 12 and references therein). This HCV cofactor might influence DMV biogenesis by inducing conformational changes in NS5A through its peptidyl-prolyl cis-trans isomerase activity. Interestingly, CypA inhibitors, such as alisporivir and NIM-811, show antiviral activity in HCV-infected patients and are currently being tested in phase II and III clinical trials. NS5A also associates with phosphatidyl-inositol-4-kinase-IIIα (PI4KIIIα) and induces the accumulation of PI4P within the membranous web. Knockdown of PI4KIIIα expression or NS5A mutations abrogating this interaction cause size reduction and aggregation of DMVs, strongly correlating with serious defects in RNA replication (reference 13 and references therein).
Immuno-gold labeling of purified DMVs revealed a drastic enrichment of cholesterol in these membranes compared to that in the isolated ER (8). DMVs depleted for cholesterol showed reduced size, highlighting the role of this lipid species in DMV morphology. The accumulation of cholesterol, and eventually other lipid species, might be mediated by the interplay between coopted cellular lipid-binding proteins, as recently described for PI4KIIIα and oxysterol-binding protein (14). These observations demonstrate that the membranous web is a highly specialized compartment.
DENV AND HCV ASSEMBLY COMPARTMENTS
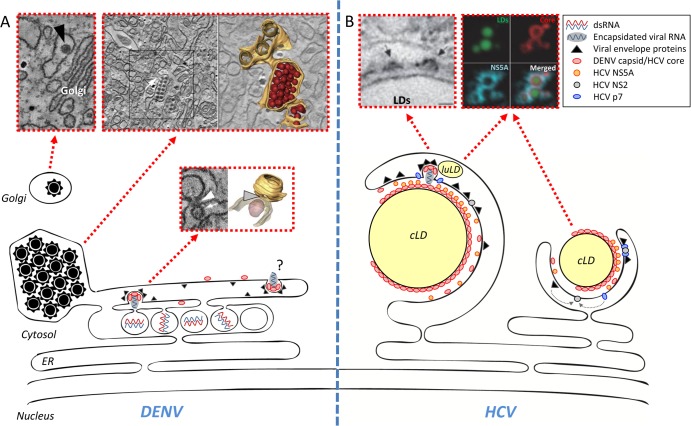
Assembly of infectious virus particles is a complex process requiring genome packaging into nucleocapsids and their subsequent envelopment. Although both viruses appear to use the conventional secretory pathway for particle release, the ways infectious virions are formed differ substantially. In case of DENV, putative virus budding sites at the ER in close proximity to VPs and VP pores have been found (Fig. 2A) (4). These results suggest that replicated DENV genomes released through the vesicle pore might be used directly for packaging into nucleocapsids that bud through the ER membrane in close vicinity. This close spatial relationship of replication and assembly sites might contribute to selectivity of DENV RNA encapsidation. It is also possible that at early time points after infection, when amounts of structural proteins are low, released viral RNA might be used preferentially for translation, but as the replication cycle progresses, capsid protein accumulates and sequesters viral RNA. This might shift the balance from RNA translation to genome encapsidation, implying a temporal regulation of the different steps of the DENV life cycle.
FIG 2.
DENV and HCV assembly sites. (A) Schematic model of DENV assembly. Three populations of virions are observed in the EM: first, budding viruses next to the pores of VPs (electron densities of detected structures presumably correspond to RNA); second, virus stacks within the lumen of the VP-containing ER network; third, viruses in Golgi-related vesicles. Note that virus budding might also occur in other areas of the ER and remote from VPs. Transmission EM images and 3D reconstructions of an electron tomogram are shown above the schematic. (B) Schematic model of HCV assembly. The majority of core protein is located on the surface of cytoplasmic lipid droplets (cLD), where a fraction of NS5A also resides. NS2 is thought to orchestrate the assembly process by recruiting on one hand p7, along with the two envelope glycoproteins, and on the other hand NS3 (not shown), eventually along with NS5A or the complete viral replicase (NS3 to NS5B). P7 is important for the envelopment of assembled capsids containing the HCV RNA genome. Assembly also involves components of the LDL synthesis machinery, including luminal lipid droplets (luLD) and apolipoproteins, most notably ApoE (not shown). Some of the insets are reprinted from reference 4 with permission from the publisher and from Nature Cell Biology (15).
The vast majority of budded DENV particles accumulate in regular arrays within dilated ER-derived cisternae, often connected to the VP-containing ER network (Fig. 2A). These densely packed virion stacks contained an electron-dense material and exhibited an envelope displaying a spike-like surface reminiscent of immature particles. This observation suggests that intracellular transport of assembled virions might be rate-limiting. Alternatively, these particles might be defective and retained within the cell. Importantly, a subset of single virions was found in distal portions of the ER or in vesicles of the Golgi apparatus, suggesting that these particles are trafficking through the secretory pathway to be released from the cell. It will be important to address the nature of the virion stacks and to identify the molecular switch(es) triggering their egress.
Unlike DENV, no intracellular reservoir of assembled HCV can be visualized by EM, and assembling, budding, or egressing virions are difficult to detect, suggesting that these processes are either rare or rapid. Nevertheless, biochemical and genetic approaches provided important insights into HCV particle assembly and release (Fig. 2B). The vast majority of the core protein is located on the surface of cytoplasmic lipid droplets (cLDs), which are responsible for neutral lipid storage and were found to be critical organelles for HCV assembly (15, 16). cLDs are tightly associated with the ER membrane within the membranous web, implying an HCV-induced microenvironment favorable for assembly. Save for NS5A that has an intrinsic LD targeting sequence, the core recruits other viral proteins as well as host cell factors involved in regulation of infectious particle production to the periphery of cLDs (17). Assembly of infectious HCV particles appears to be orchestrated by NS2 that interacts on one hand with p7 and the envelope glycoproteins (E1 and E2) and on the other hand with NS3, which is part of the viral replicase (18). Thus, NS2 might link the RNA-containing replicase with the machinery required for envelopment of HCV particles. In addition, p7 seems to be involved in capsid envelopment and protect virus particles against acidic pH during passage through the secretory pathway.
Ultrastructural studies revealed that in HCV-expressing cells, ∼30% of cLDs were closely juxtaposed to ER cisternae on which core- and E2-containing budding structures, reminiscent of virus particles, could be visualized (15). This suggests that HCV assembly occurs in this subcompartment of the membranous web. The close proximity of core-containing cLDs, DMVs, and E1/E2-containing ER membranes might facilitate coordination of replication and assembly. Finally, HCV assembly also relies on host cell machineries, such as components of the low-density lipoprotein pathway (19). This is consistent with the nature of HCV particles circulating in the blood as a lipoviroparticle (20).
The advent of RNA interference-mediated gene silencing and high-throughput screening approaches paved the way for the identification of numerous host dependency factors required for DENV and HCV life cycles. While these studies increased our knowledge about host cell pathways and machineries hijacked by these viruses, with a few exceptions, very little has been elucidated about which host factors are involved in de novo formation and/or maintenance of virus-induced replication factories. In this respect, proteomic and lipidomic analyses of highly purified replication factories will definitely contribute to answer this question. Moreover, compounds targeting the very first steps of virus-specific vesicle formation are expected to disrupt their biogenesis and, thus, to abrogate RNA replication. The proof of concept made with CypA inhibitors (see above) illustrates the idea that the biogenesis of replication factories induced by HCV, DENV, and more generally, positive-strand RNA viruses, represents a very attractive target for the development of highly potent broad-spectrum antiviral drugs.
ACKNOWLEDGMENTS
We thank Pietro Scaturro, David Paul, and Mirko Cortese for critical readings of the manuscript and Alessia Ruggieri for help with the figures. We apologize to all colleagues whose work could not be referenced because of length restrictions.
Work in our laboratory was supported by the Deutsche Forschungsgemeinschaft (SFB 638, TPA5 and TRR83, TP13) and the European Commission for Research and Innovation (FP7 HEALTH-2010 Collaborative Project SILVER; contract 260644).
Footnotes
Published ahead of print 12 March 2014
REFERENCES
- 1.Acosta EG, Kumar A, Bartenschlager R. 2014. Revisiting dengue virus-host cell interaction: new insights into molecular and cellular virology. Adv. Virus Res. 88:1–109. 10.1016/B978-0-12-800098-4.00001-5 [DOI] [PubMed] [Google Scholar]
- 2.Bartenschlager R, Lohmann V, Penin F. 2013. The molecular and structural basis of advanced antiviral therapy for hepatitis C virus infection. Nat. Rev. Microbiol. 11:482–496. 10.1038/nrmicro3046 [DOI] [PubMed] [Google Scholar]
- 3.Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, Walther P, Antony C, Krijnse-Locker J, Bartenschlager R. 2012. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 8:e1003056. 10.1371/journal.ppat.1003056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. 10.1016/j.chom.2009.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Junjhon J, Pennington JG, Edwards TJ, Perera R, Lanman J, Kuhn RJ. 2014. Ultrastructural characterization and three-dimensional architecture of replication sites in dengue virus-infected mosquito cells. J. Virol. 88:4687–4697. 10.1128/JVI.00118-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller S, Kastner S, Krijnse-Locker J, Buhler S, Bartenschlager R. 2007. The non-structural protein 4A of dengue virus is an integral membrane protein inducing membrane alterations in a 2K-regulated manner. J. Biol. Chem. 282:8873–8882. 10.1074/jbc.M609919200 [DOI] [PubMed] [Google Scholar]
- 7.Ferraris P, Blanchard E, Roingeard P. 2010. Ultrastructural and biochemical analyses of hepatitis C virus-associated host cell membranes. J. Gen. Virol. 91:2230–2237. 10.1099/vir.0.022186-0 [DOI] [PubMed] [Google Scholar]
- 8.Paul D, Hoppe S, Saher G, Krijnse-Locker J, Bartenschlager R. 2013. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J. Virol. 87:10612–10627. 10.1128/JVI.01370-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. 2002. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 76:5974–5984. 10.1128/JVI.76.12.5974-5984.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paul D, Romero-Brey I, Gouttenoire J, Stoitsova S, Krijnse-Locker J, Moradpour D, Bartenschlager R. 2011. NS4B self-interaction through conserved C-terminal elements is required for the establishment of functional hepatitis C virus replication complexes. J. Virol. 85:6963–6976. 10.1128/JVI.00502-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, Longerich T, Diehl S, Ramirez F, Balla T, Rohr K, Kaul A, Buhler S, Pepperkok R, Lengauer T, Albrecht M, Eils R, Schirmacher P, Lohmann V, Bartenschlager R. 2011. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe 9:32–45. 10.1016/j.chom.2010.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madan V, Paul D, Lohmann V, Bartenschlager R. Inhibition of HCV replication by cyclophilin antagonists is linked to replication fitness and occurs by inhibition of membranous web formation. Gastroenterology, in press. 10.1053/j.gastro.2014.01.055 [DOI] [PubMed] [Google Scholar]
- 13.Reiss S, Harak C, Romero-Brey I, Radujkovic D, Klein R, Ruggieri A, Rebhan I, Bartenschlager R, Lohmann V. 2013. The lipid kinase phosphatidylinositol-4 kinase III alpha regulates the phosphorylation status of hepatitis C virus NS5A. PLoS Pathog. 9:e1003359. 10.1371/journal.ppat.1003359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang H, Perry JW, Lauring AS, Neddermann P, De FR, Tai AW. Oxysterol-binding protein is a phosphatidylinositol 4-kinase effector required for HCV replication membrane integrity and cholesterol trafficking. Gastroenterology, in press. 10.1053/j.gastro.2014.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miyanari Y, Atsuzawa K, Usuda N, Watashi K, Hishiki T, Zayas M, Bartenschlager R, Wakita T, Hijikata M, Shimotohno K. 2007. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 9:1089–1097. 10.1038/ncb1631 [DOI] [PubMed] [Google Scholar]
- 16.Shavinskaya A, Boulant S, Penin F, McLauchlan J, Bartenschlager R. 2007. The lipid droplet binding domain of hepatitis C virus core protein is a major determinant for efficient virus assembly. J. Biol. Chem. 282:37158–37169. 10.1074/jbc.M707329200 [DOI] [PubMed] [Google Scholar]
- 17.Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. 2008. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 4:e1000035. 10.1371/journal.ppat.1000035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lindenbach BD. 2013. Virion assembly and release. Curr. Top. Microbiol. Immunol. 369:199–218. 10.1007/978-3-642-27340-7_8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Jr, Ye J. 2007. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. U. S. A. 104:5848–5853. 10.1073/pnas.0700760104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andre P, Komurian-Pradel F, Deforges S, Perret M, Berland JL, Sodoyer M, Pol S, Brechot C, Paranhos-Baccala G, Lotteau V. 2002. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 76:6919–6928. 10.1128/JVI.76.14.6919-6928.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]