ABSTRACT
Human polyomavirus 9 (HPyV9) is a closely related homologue of simian B-lymphotropic polyomavirus (LPyV). In order to define the architecture and receptor binding properties of HPyV9, we solved high-resolution crystal structures of its major capsid protein, VP1, in complex with three putative oligosaccharide receptors identified by glycan microarray screening. Comparison of the properties of HPyV9 VP1 with the known structure and glycan-binding properties of LPyV VP1 revealed that both viruses engage short sialylated oligosaccharides, but small yet important differences in specificity were detected. Surprisingly, HPyV9 VP1 preferentially binds sialyllactosamine compounds terminating in 5-N-glycolyl neuraminic acid (Neu5Gc) over those terminating in 5-N-acetyl neuraminic acid (Neu5Ac), whereas LPyV does not exhibit such a preference. The structural analysis demonstrated that HPyV9 makes specific contacts, via hydrogen bonds, with the extra hydroxyl group present in Neu5Gc. An equivalent hydrogen bond cannot be formed by LPyV VP1.
IMPORTANCE The most common sialic acid in humans is 5-N-acetyl neuraminic acid (Neu5Ac), but various modifications give rise to more than 50 different sialic acid variants that decorate the cell surface. Unlike most mammals, humans cannot synthesize the sialic acid variant 5-N-glycolyl neuraminic acid (Neu5Gc) due to a gene defect. Humans can, however, still acquire this compound from dietary sources. The role of Neu5Gc in receptor engagement and in defining viral tropism is only beginning to emerge, and structural analyses defining the differences in specificity for Neu5Ac and Neu5Gc are still rare. Using glycan microarray screening and high-resolution protein crystallography, we have examined the receptor specificity of a recently discovered human polyomavirus, HPyV9, and compared it to that of the closely related simian polyomavirus LPyV. Our study highlights critical differences in the specificities of both viruses, contributing to an enhanced understanding of the principles that underlie pathogen selectivity for modified sialic acids.
INTRODUCTION
Polyomaviruses (PyVs) are nonenveloped, double-stranded DNA viruses that can infect a large range of mammalian species and birds. Their viral genome typically encodes two regulatory proteins, the small and large T antigens, and three structural proteins, VP1, VP2, and VP3. The structural proteins assemble into a T=7d icosahedral shell that encapsidates the viral DNA (1).
Since their discovery 6 decades ago, polyomaviruses have emerged as highly useful models to understand the molecular events leading to cellular transformation and cancer. The human polyomaviruses JCPyV (2) and BKPyV (3) were discovered in 1971 and subsequently shown to be associated with progressive multifocal leukoencephalopathy and polyomavirus-associated nephropathy, respectively. Remarkably, 10 new human polyomaviruses have been identified in the last 6 years (4–14). One of these, Merkel cell polyomavirus (MCPyV), is the causative agent of Merkel cell carcinoma, a rare neuroendocrine tumor of the skin (6). Infection with another recently identified polyomavirus can lead to trichodysplasia spinulosa, which, while not life threatening, causes significant disfigurement (8). This disease is seen almost exclusively in transplant recipients immunosuppressed with drugs such as cyclosporine (15).
Human polyomavirus 9 (HPyV9) was identified in 2011 from the serum and urine of an asymptomatic kidney transplant patient (9) and a skin sample of a patient with Merkel cell carcinoma (10). HPyV9 has a high overall nucleotide sequence identity (76%) with B-lymphotropic polyomavirus (LPyV), which is also sometimes referred to as African green monkey polyomavirus (AGMPyV). Although, the role of HPyV9 in human disease has not yet been established, the isolation and identification of HPyV9 have resolved the puzzling finding of the serological cross-reactivity against LPyV in 20 to 30% of apparently healthy humans (16–20). Antibodies against polyomaviruses are mostly elicited against their apical surface, formed by the viral major capsid protein, VP1 (21–23). HPyV9 VP1 has 87% amino acid identity with LPyV VP1 (9, 10). It has therefore been hypothesized that these viruses share immunogenic epitopes for antibody engagement (9, 10, 19, 20) and perhaps also other properties.
VP1 is the major polyomavirus capsid protein, and a collection of available VP1 structures from different polyomaviruses shows that the protein adopts a jelly roll fold and assembles into a homopentamer around a central cavity (24–31). While the pentameric core structure is conserved in all VP1 proteins, these structures from different polyomaviruses exhibit substantial differences in the sequence, length, and conformation of their surface-exposed loops. These highly variable regions contain the sites for receptor engagement (25–27, 29) as well as antibody binding (21–23, 32). All human polyomavirus VP1 structures known to date engage glycans terminating in 5-N-acetyl neuraminic acid (Neu5Ac) as receptors (27, 29–31), with, in some cases, drastically different interactions taking place.
Whereas humans cannot synthesize a modified version of Neu5Ac, 5-N-glycolyl neuraminic acid (Neu5Gc), most other mammals can produce both Neu5Ac and Neu5Gc (33, 34). Humans can, however, acquire Neu5Gc from the red meat and milk in their diets. Assimilated Neu5Gc is therefore found in the human gut epithelium and kidney vasculature (35–37). Some viruses and bacteria can discriminate between the subtle differences of neuraminic acid variants and are able to specifically recognize only one of them, an ability which, in turn, influences their host range. For example, the polyomavirus simian virus 40 (SV40) preferentially binds to the Neu5Gc version of its GM1 ganglioside receptor (38), and this specificity is thought to limit SV40 infections in humans. On the other hand, the closely related human BKPyV exclusively recognizes receptors containing Neu5Ac but not those containing Neu5Gc (30). Similarly, animal rotavirus strains have variable preferences for Neu5Gc or Neu5Ac. The porcine and bovine rotaviruses prefer Neu5Gc as a component of their attachment receptor, whereas the rhesus monkey rotavirus apparently utilizes Neu5Ac-type receptors for infection (39). Finally, the subtilase cytotoxin secreted by Shiga-toxigenic Escherichia coli (STEC) specifically recognizes Neu5Gc-type glycan receptors in the human gut epithelium and kidney vasculature (35).
In order to define the receptor-binding specificity of HPyV9, we performed glycan microarray screening with recombinant HPyV9 VP1 and LPyV VP1. We also solved the crystal structure of HPyV9 VP1 in complex with relevant oligosaccharides containing Neu5Gc and Neu5Ac identified in the array, namely, Neu5Gc-α2,3-Gal-β1,4-GlcNAc (3GSLN) and Neu5Ac-α2,3-Gal-β1,4-GlcNAc (3SLN), and also with the sialyllactose Neu5Ac-α2,3-Gal-β1,4-Glc (3SL). A comparison of the structure of HPyV9 VP1 with that of LPyV VP1 when the proteins were in complex with Neu5Ac-containing glycan ligand (31) identified subtle differences in the binding site, which result in altered interactions with the different sialic acids present in the oligosaccharide structures. Furthermore, a comparison of the atomic structures of the VP1 proteins from HPyV9, LPyV, and MCPyV provides a basis for understanding properties such as antigenic variability and the serological cross-reactivity among these three viruses.
MATERIALS AND METHODS
DNA cloning and protein expression.
HPyV9 VP1 DNA (GenBank accession number HQ696595.1) was synthesized for expression in E. coli as described previously (19). The DNA fragment encoding protein residues 31 to 304 was amplified by PCR and was subsequently cloned into the pET 28a(+) expression vector (Novagen) downstream of a hexahistidine tag and a thrombin cleavage site. The protein was overexpressed in E. coli BL21(DE3) cells and purified by nickel affinity chromatography (GE Healthcare) and size exclusion chromatography on a Superdex 200 column (GE Healthcare). For crystallization, the untagged protein was obtained by incubation of the immobilized protein with thrombin. Recombinant VP1 contained four additional vector-encoded residues (Gly-Ser-His-Met) at the N terminus. Also, in the absence of a reducing agent and at a protein concentration of 0.5 mg/ml, the protein was allowed to form covalent dimers of pentamers, mediated by disulfide bonds involving the five cysteines at the bottom of the pentamer. Size exclusion chromatography on a Superdex 200 column (GE Healthcare) was used to obtain homogeneous dimers of pentamers that were concentrated to 4 mg/ml and stored in 20 mM HEPES (pH 7.5), 150 mM NaCl at −80°C. HPyV9 VP1 with an N282V point mutation was generated using mutagenic PCR and purified using the same protocol.
Synthesis of 3GSLN.
CMP-Neu5Gc was synthesized from Neu5Gc and CTP using the recombinant CMP-Neu5Ac synthetase (construct NSY-05) from Neisseria meningitidis (40). 3GSLN was synthesized from CMP-Neu5Gc and N-acetyllactosamine using the recombinant Cst-I α-2,3-sialyltransferase (construct CST-06) from Campylobacter jejuni (41). The synthesized 3GSLN was purified by size exclusion chromatography on a Superdex peptide column (10 mm by 30 cm; GE Healthcare) using 20 mM NH4CO3 as the eluent.
Glycan microarray screening.
Microarrays comprised lipid-linked oligosaccharide probes robotically printed on nitrocellulose-coated glass slides using a noncontact instrument (42, 43). For the present study, an array set of 28 glycan probes (most are ganglioside related; in-house designation, ganglioside dose-response microarray set 2) was used. The probes are listed in Table 1 and were arrayed at four levels: 0.3, 0.8, 1.7, and 5.0 fmol/spot. The microarray analysis was performed essentially as described previously (27, 31). In brief, microarrays were blocked in 5 mM HEPES (pH 7.4), 150 mM NaCl, 3% (wt/vol) bovine serum albumin (BSA; Sigma), and 5 mM CaCl2 (referred to as HBS-BSA). HPyV9 VP1 (the wild type [wt] and the N282V mutant) and LPyV VP1 were tested as protein-antibody complexes that were prepared by preincubating VP1 with mouse monoclonal antipolyhistidine and biotinylated anti-mouse IgG antibodies (both from Sigma) at a ratio of 4:2:1 (by weight) and diluted in HBS-BSA to provide a final VP1 concentration of 150 μg/ml. Binding was detected with Alexa Fluor 647-labeled streptavidin (Molecular Probes). Microarray data analysis was as described previously (44).
TABLE 1.
Probes included in ganglioside dose-response microarray set 2 and their sequencesa

The oligosaccharide probes are all lipid linked and are from the collection assembled in the course of research in Glycosciences Laboratory. Cer, natural glycolipids with various ceramide moieties; DH, neoglycolipids (NGLs) prepared from reducing oligosaccharides by reductive amination with the amino lipid 1,2-dihexadecyl-sn-glycero-3-phosphoethanolamine (DHPE).
Protein crystallization.
The dimers of HPyV9 VP1 pentamers were crystallized at 20°C by hanging-drop vapor diffusion against a reservoir of 20% (vol/vol) isopropanol, 0.2 M calcium chloride, 0.1 M sodium acetate, pH 5.15. The crystals were harvested into the respective reservoir solution supplemented with 25% (vol/vol) ethylene glycol as a cryoprotectant and directly flash frozen in liquid nitrogen. For the liganded structures, crystals were soaked in a reservoir solution including 40 mM sialyloligosaccharide ligands for 40 min. 3SL and 3SLN were purchased from Dextra, United Kingdom, and Carbosynth, United Kingdom, respectively. 3GSLN was custom synthesized as described above. The crystals were cryoprotected in soaking solution supplemented with 25% (vol/vol) ethylene glycol before freezing them in liquid nitrogen.
Structure determination.
All data were collected at −180°C using a wavelength of 1 Å at beam lines ID14-4 at ESRF (Grenoble, France) and X06DA at SLS (Villigen, Switzerland). The diffraction data were processed with the XDS program package (45), and the native structure was solved by molecular replacement with the MOLREP program (46) in the CCP4 suite using the core region of the LPyV VP1 pentamer (PDB accession number 4MBX) as a search model. Refinement was initially performed with PHENIX software (rigid body, simulated annealing, and restrained coordinate refinement) (47) and subsequently with the Refmac5 program (48). The structures were completed by iterating rounds of model building in the Coot program (49) and with restrained coordinate and B factor as well as TLS refinement in Refmac5. TLS parameters were generated using the TLSMD server (50). The asymmetric unit contains two complete VP1 pentamers, and noncrystallographic symmetry restraints were used throughout the refinement for the bulk of the protein. Ligands were fitted into weighted difference electron density maps at contour levels of 3σ and subsequently refined using the CCP4 library and user-defined restraints. Ligands were bound to either 4 or 5 binding sites per pentamer. The final models agree well with the experimental data and have good geometry (Table 2). Figures showing the X-ray structures were prepared with the PyMOL program (Schrödinger, LLC.).
TABLE 2.
Data collection and refinement statistics
| Parameter | Valuea |
|||
|---|---|---|---|---|
| VP1 | VP1-3SL | VP1-3SLN | VP1-3GSLN | |
| Data collection statistics | ||||
| Space group | P212121 | P212121 | P212121 | P212121 |
| Unit cell dimensions | ||||
| a, b, c (Å) | 97.33, 180.09, 199.39 | 97.01, 180.40, 199.66 | 96.91, 180.63, 199.42 | 103.40, 177.65, 198.65 |
| α, β, γ (°) | α = β = γ = 90 | α = β = γ = 90 | α = β = γ = 90 | α = β = γ = 90 |
| Resolution (Å) | 50–2.00 (2.05–2.00) | 50–2.09 (2.14–2.09) | 50–2.00 (2.05–2.00) | 50–2.10 (2.15–2.10) |
| Total no. of reflections | 802,267 (58,665) | 645,464 (43,686) | 737,257 (47,697) | 812,743 (62,074) |
| No. of unique reflections | 226,820 (16,603) | 204,785 (14,851) | 230,735 (16,974) | 211,585 (15,585) |
| Redundancy | 3.5 (3.5) | 3.2 (2.9) | 3.2 (2.8) | 3.8 (4.0) |
| 〈I〉/σ〈I〉 | 11.1 (2.1) | 8.4 (1.9) | 10.0 (2.0) | 10.5 (2.2) |
| Completeness (%) | 96.2 (95.9) | 99.0 (97.9) | 98.0 (98.0) | 99.4 (99.6) |
| Rmeas (%) | 11.0 (78.1) | 14.8 (77.9) | 12.4 (75.2) | 15.1 (80.7) |
| Wilson B factor (Å2) | 30.4 | 29.4 | 28.3 | 28.3 |
| Refinement statistics | ||||
| Rwork (%) | 19.6 | 17.8 | 17.9 | 18.4 |
| Rfreeb (%) | 23.6 | 21.6 | 21.3 | 22.4 |
| Coordinate errorc (Å) | 0.26 | 0.25 | 0.24 | 0.25 |
| No. of atoms: | ||||
| Protein | 20,822 | 20,644 | 20,734 | 20,686 |
| Oligosaccharide | 387 | 414 | 470 | |
| Water | 1,885 | 2,364 | 2,523 | 2,395 |
| Othersd | 94 | 182 | 170 | 166 |
| Avg B factors (Å2) | ||||
| Protein atoms | 27.4 | 27.3 | 26.6 | 25.8 |
| Oligosaccharide | 35.8 | 35.2 | 30.2 | |
| Water | 30.9 | 29.9 | 29.1 | 29.3 |
| Othersd | 38.9 | 35.6 | 36.8 | 35.5 |
| RMSD | ||||
| Bond length (Å) | 0.008 | 0.008 | 0.008 | 0.008 |
| Bond angle (°) | 1.26 | 1.23 | 1.21 | 1.22 |
| Ramachandran plote (%) | ||||
| Favorable | 95.76 | 96.10 | 96.32 | 96.07 |
| Allowed | 4.24 | 3.90 | 3.68 | 3.93 |
| Outlier | 0 | 0 | 0 | 0 |
Data for the highest-resolution shell are shown in parentheses. A single crystal was used for each data set.
Five percent of total reflections were used to calculate Rfree.
Coordinate error by Luzzati plot was calculated using the sfcheck program in the CCP4 suite.
Others include the substances used for crystallization, i.e., isopropanol, calcium, and ethylene glycol.
Calculated using the MolProbity server (http://molprobity.biochem.duke.edu/).
Protein structure accession numbers.
Coordinates and structure factor amplitudes have been deposited in the Protein Data Bank (PDB) under accession numbers 4POQ for unliganded VP1 and 4POR, 4POS, and 4POT for VP1 bound to 3SL, 3SLN, and 3GSLN, respectively.
RESULTS
HPyV9 preferentially binds short, linear glycan sequences terminating in Neu5Gc.
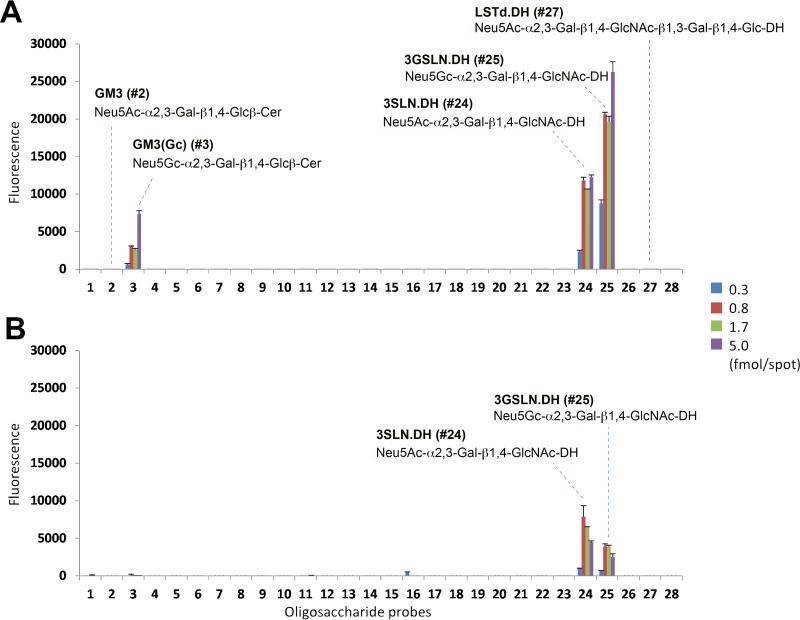
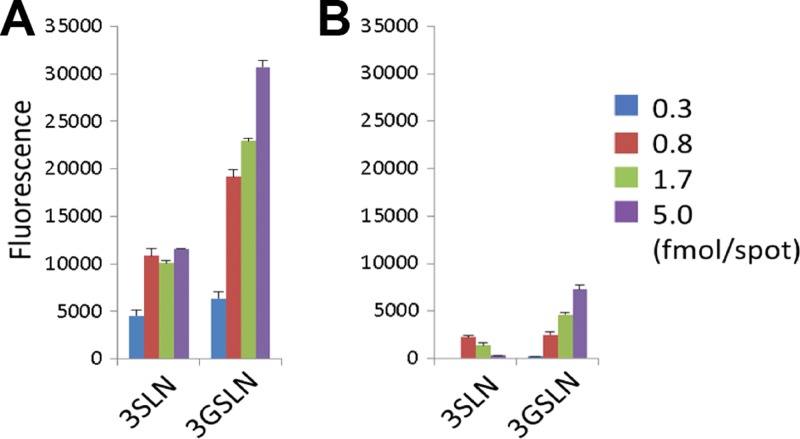
We hypothesized that HPyV9 might interact with sialic acids on the basis of its similarity to LPyV, which binds to sialylated oligosaccharides (31). However, the residues in the sialic acid binding region of the LPyV VP1 are less conserved in HPyV9 of VP1. This variability in the binding site would be especially interesting with respect to Neu5Ac and Neu5Gc binding by a pair of highly homologous viruses that have been isolated from humans and monkeys. We therefore performed comparative glycan microarray analysis using recombinantly expressed HPyV9 and LPyV VP1 pentamers using an array comprised of 28 lipid-linked oligosaccharide probes (Table 1). The microarray included 23 ganglioside-related probes (glycosylceramides and neoglycolipids), as well as neoglycolipids of sialyl-N-acetyllactosamine terminating in either Neu5Ac or Neu5Gc (designated 3SLN and 3GSLN, respectively) and three pentasaccharides from human milk (lactoseries sialyl tetrasaccharides LSTa, LSTd, and LSTc). The 3SL.DH probe, the neoglycolipid version of glycosylceramide GM3, was not included in the array. Each glycan probe was arrayed at four levels ranging from 0.3 to 5 fmol per spot in the form of liposomes (42).
The VP1 proteins of HPyV9 and LPyV were overlaid onto the microarrays after precomplexing with mouse anti-His and biotinylated anti-mouse IgG antibodies. This process resulted in increased multivalency of the protein and usually enabled detection of low-affinity ligands on the array surface. The VP1s of HPyV9 and LPyV showed similar binding profiles overall: highly selective binding to 3SLN.DH (probe 24) and 3GSLN.DH (probe 25) (Fig. 1). The signal intensities with HPyV9 VP1 were higher than those with LPyV VP1 under identical assay conditions. In parallel experiments, when the VP1s were analyzed without precomplexation, HPyV9 VP1 gave robust binding results that were almost identical to those obtained with the precomplexed protein (data not shown); in contrast, no binding was detected with LPyV VP1 under these conditions. These results suggest that HPyV9 VP1 has a higher binding avidity toward its glycan ligands than LPyV VP1.
FIG 1.
Glycan microarray screening of HPyV9 VP1 (A) and LPyV VP1 (B) using 28 lipid-linked oligosaccharide probes. Each oligosaccharide probe was arrayed at four levels (as indicated) in duplicate. The complete list of probes and their sequences are provided in Table 1. Numerical scores of the binding signals are shown as the means of the fluorescence intensities of duplicate spots with error bars, which represent half of the difference between the two values. Cer, natural glycolipids with various ceramide moieties; DH, neoglycolipids (NGLs) prepared from reducing oligosaccharides by reductive amination with the amino lipid 1,2-dihexadecyl-sn-glycero-3-phosphoethanolamine (DHPE).
Intriguingly, HPyV9 VP1 showed preferential binding to sialyloligosaccharides terminating in the nonhuman Neu5Gc over their Neu5Ac analogues. The probe 3GSLN.DH (probe 25) was more strongly bound than 3SLN.DH (probe 24), and weak binding to the glycosylceramide GM3(Gc) (probe 3) was detected. Neither LPyV nor HPyV9 VP1 showed detectable binding to GM3 (probe 2). LPyV VP1 gave only a slightly higher signal for 3SLN.DH than 3GSLN.DH (Fig. 1). Little or weak binding to glycosylceramides GM3 and GM3(Gc) by both proteins is consistent with the finding in our recent study of LPyV VP1 using a different array set (31). It should be noted that no signals of binding to longer oligosaccharide sequences with the trisaccharide 3SLN terminus, such as probe LSTd.DH (probe 27), were detected with HPyV9 VP1. This is also a feature shared with LPyV VP1, as we recently described (31). The 3SLN.DH and 3GSLN.DH probes were prepared by reductive amination and thus have ring-opened, reduced Glc moieties, which most likely confer greater flexibility of glycan presentation on the array, thereby facilitating engagement.
A further difference between HPyV9 VP1 and LPyV VP1 is the ligand density effect observed in the binding of the two proteins. With LPyV VP1, a higher signal intensity was observed at a lower density of ligand (1.7 fmol per spot) than at a high density of ligand (5 fmol per spot) (Fig. 1). This is consistent with our earlier analysis of LPyV VP1 binding using a different array set (31). Interestingly, such an effect was not observed with HPyV9 VP1, which showed the strongest binding (or a plateau) to probes tested at the highest ligand density, 5 fmol per spot (Fig. 1). Thus, where the avidity of binding of VP1 is high, the spacing of the ligands is not as critical as in the case of LPyV, perhaps also because the HPyV9 binding site is more accessible (see below).
Overall structure of HPyV9 VP1.
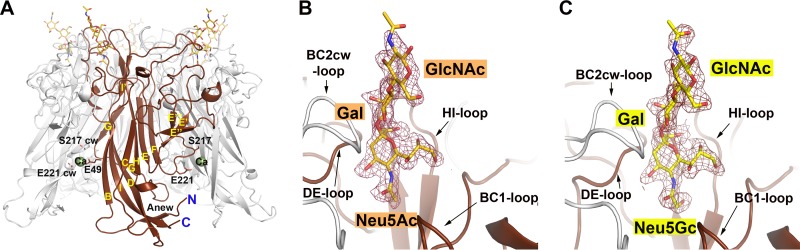
In order to rationalize the observed differences in ligand recognition, we solved high-resolution crystal structures of HPyV9 VP1 alone and in complex with the two most prominent ligands from the glycan microarray analysis, 3GSLN and 3SLN, as well as in complex with 3SL (Table 2; Fig. 2). The bulk of HPyV9 VP1 is constructed from two four-stranded β sheets (formed by strands B, I, D, and G and strands C, H, E, and F, respectively) that are stacked against each other (Fig. 2A). Additional smaller β strands from adjacent monomers are oriented such that they give rise to the cylindrical shape of the pentameric VP1 protein. Loops in VP1 are named according to the two β strands that they connect (e.g., the DE loop connects strands D and E), and these loops account for most of the exterior surface of the pentamer.
FIG 2.
Structure of HPyV9 VP1 in complex with sialyloligosaccharides. (A) HPyV9 VP1 pentamer structure in complex with 3SLN. The pentamer is shown in cartoon representation, with one VP1 monomer highlighted in chocolate color and the other monomers shown in gray. The β strands (Anew, B to I) are labeled. 3SLN is drawn as a stick model and colored according to atom type (blue, nitrogens; red, oxygens; gold, carbons). One calcium ion is bound to each VP1 monomer, and the ions bound to the colored VP1 monomer are shown as green spheres, with the protein residues contacting it shown in stick representation. (B and C) Close-up views of the 3SLN (B) and 3GSLN (C) binding sites. HPyV9 VP1 and 3SLN are drawn as described for panel A, while the carbon atoms are colored yellow for 3GSLN. The view is adjusted to show more clearly the trisaccharide with the associated VP1 loops (the BC1, DE, and HI loops) of one monomer and the BC2 loop of the clockwise (cw) monomer. Composite-annealed omit difference density maps are shown contoured at 2.5σ around glycan at a 2.1-Å radius.
Complexes with all three glycans were obtained by soaking crystals in the respective oligosaccharide solution (see Methods and Methods; Table 2). The glycans bound to the same binding site on HPyV9 VP1 (Fig. 2 and 3). Not all binding sites were fully occupied due to limited accessibility in the crystals. However, there was excellent electron density for each ligand in several binding sites in each structure, allowing us to assign contacts with confidence (Fig. 3). None of the interactions discussed below are influenced by crystal contacts.
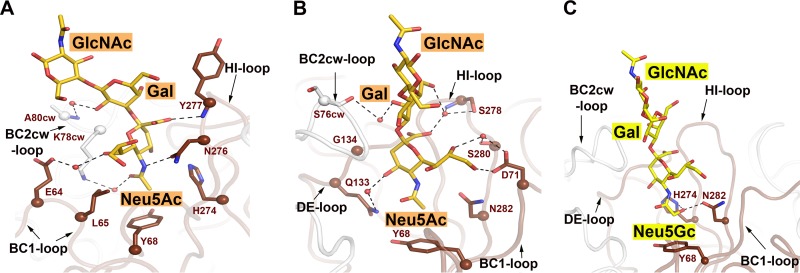
FIG 3.
Interactions between HPyV9 VP1 and sialyloligosaccharides. (A and B) Interactions between HPyV9 VP1 and 3SLN. The main chain of HPyV9 VP1 is shown in cartoon representation. One monomer is shown in chocolate color, while other monomers are shown in gray. The amino acid side chains interacting with 3SLN are shown as sticks, and the corresponding C-α atoms are shown as spheres. The backbone amide and carbonyl groups are shown only when engaging the oligosaccharide. 3SLN is shown as in Fig. 2B. Water molecules are represented as red spheres. Hydrogen bonds are shown as black dashed lines. The view in panel A was rotated by about 90° along a vertical axis to obtain the view presented in panel B. (C) Interactions between HPyV9 VP1 and 3GSLN. HPyV9 VP1 is drawn as described for panel A; the ligand 3GSLN is shown as in Fig. 2C. Residues H274 and N282, which interact specifically with the hydroxymethyl chain of Neu5Gc, are shown in stick representation.
Interactions of HPyV9 VP1 with Neu5Ac-containing sialyloligosaccharides.
The interactions of HPyV9 VP1 with 3SLN (Fig. 3A and B) are discussed first. As with other polyomavirus structures, the BC1, DE, and HI loops of one VP1 monomer and the BC2 loop of the clockwise (cw) neighboring monomer (BC2cw) contribute to the sialyloligosaccharide binding site. BC1 and BC2 are two subdivisions of the long BC loop that point in different directions. The sialic acid moiety of the ligand penetrates deeply into a cleft formed by these loops and makes the most contacts with the protein. The extended LacNAc disaccharide projects out of the binding pocket and makes few contacts with protein residues. HPyV9 VP1 does not undergo substantial conformational change after complexation, as native and liganded VP1s can be superimposed with a low root mean square deviation (RMSD) value of 0.2 Å for main-chain atoms. However, ligand engagement does rigidify the tip of the BC2 loop (i.e., residue 76 to residue 80), as demonstrated by a decrease in temperature factors from 40.8 Å2 (unbound state) to 29.6 Å2 (bound state).
Most contacts involve Neu5Ac, which forms extensive polar and nonpolar interactions with its carboxylate and N-acetyl groups. Neu5Ac is contacted by protein residues on both sides of the ring, with the side chain of K78cw contacting one face of the ring. The N-acetyl methyl group inserts deepest into the binding pocket, penetrating into a cavity lined with the hydrophobic parts of the side chains of residues L65, Y68, H274, and N282 (Fig. 3A). There is a hydrogen bond between the nitrogen of the N-acetyl group and N276. The carboxylate group of Neu5Ac is pointed toward the HI loop and makes one direct and two water-mediated hydrogen bonds with backbone atoms of Y277 as well as backbone and side chain atoms of S278, respectively. The glycerol chain of Neu5Ac faces toward the solvent and interacts with the protein via one direct hydrogen bond to D71 and three water-mediated hydrogen bonds. The interaction of Neu5Ac with the protein buries a surface of 319 Å2 (51).
The Gal moiety makes three water-mediated contacts with protein residues, burying an additional 133 Å2 of surface (Fig. 3B). Finally, the Glc(NAc) residue does not contact the protein directly and has elevated temperature factors. Nevertheless, this sugar can also be docked unambiguously into the electron density, probably because it is stabilized by the conformational preferences of the glycosidic bond.
We also solved the structure of HPyV9 VP1 in complex with 3SL (Table 2). The structural analysis of the two complexes shows that HPyV9 VP1 engages both 3SLN and 3SL with identical contacts to the Neu5Ac and Gal residues. Neither the GlcNAc in 3SLN nor the Glc in 3SL contributes any contacts to protein residues (data not shown).
Structural basis for preference of HPyV9 for 3GSLN.
Glycan microarray screening of HPyV9 VP1 revealed a preference for the Neu5Gc-containing 3GSLN. The structure of HPyV9 VP1 in complex with 3GSLN provides the structural basis of this preference (Fig. 3C). Whereas most of the interactions of 3GSLN and 3SLN with HPyV9 VP1 are conserved, the presence of an extra hydroxyl group in 3GSLN leads to additional contacts that are well-defined in the electron density maps (Fig. 2C). The Neu5Gc glycolyl chain faces toward a hydrophilic rim lining the hydrophobic cavity that accommodates the methylene (CH2) group through shape complementarity. This rim is formed by the hydrophilic parts of the H274 and N282 side chains. The hydroxyl group of Neu5Gc donates a hydrogen bond to the carbonyl oxygen of N282 and receives one from H274 (Fig. 3C). These polar contacts likely strengthen the interaction of HPyV9 VP1 with 3GSLN, especially because they occur in the relatively hydrophobic environment of a protein cavity.
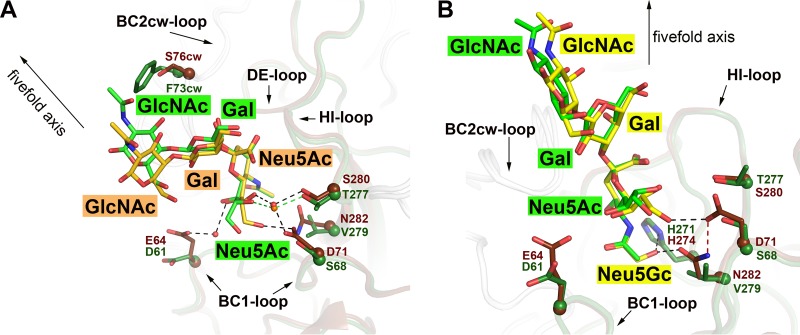
Comparison of structure and glycan specificity of HPyV9 and LPyV VP1.
As predicted from their high overall sequence identity of 87%, the structures of HPyV9 VP1 and LPyV VP1 (31) are very similar. Their C-α backbones can be superimposed with very low RMSD values of 0.62 Å (monomers) and 0.68 Å (pentamers). The two proteins also share a similar specificity for short, linear oligosaccharides containing α2,3-linked sialic acids. However, the binding sites are less conserved than the overall protein, sharing only 66% sequence identity. This difference is reflected in the signals in the glycan array, which are generally stronger for HPyV9 VP1 and also show that HPyV9 preferentially interacts with Neu5Gc-bearing glycans. The two binding regions differ at five amino acid positions (Table 3). One of these differences (HPyV9 VP1 S280 to LPyV VP1 T277) is unlikely to affect contacts, as both residues form a similar hydrogen bond to the ligand (Fig. 4A), whereas the other four substitutions subtly modulate the observed binding specificity of each viral protein and likely also its affinity.
TABLE 3.
Variable residues in the sialyloligosaccharide binding pocket of HPyV9 and LPyV VP1
| Loop | HPyV9 VP1 |
LPyV VP1 |
||
|---|---|---|---|---|
| Amino acid | Interaction | Amino acid | Interaction | |
| BC1 | E64 | Water-mediated H bond with O-7 of glycerol chain | D61 | None |
| D71 | Direct H bond with O-9 of glycerol chain | S68 | None | |
| D71 | Water-mediated H bond with O-8 of glycerol chain | S68 | None | |
| HI | S280 | Water-mediated H bond with O-8 of glycerol chain | T277 | Water-mediated H bond with O-8 of glycerol chain |
| N282 | Hydrophobic interaction with methyl moiety of N-acetyl group | V279 | Hydrophobic interaction with methyl moiety of N-acetyl group | |
| N282 | Direct H bond with extra hydroxyl of N-glycolyl group | V279 | None | |
| BC2cw | S76 | None | F73 | Hydrophobic interaction with methyl moiety of N-acetyl group of NAG |
FIG 4.
Comparison of the sialyloligosaccharide binding pockets of HPyV9 and LPyV VP1 proteins. (A) The structure of HPyV9 VP1-3SLN is superimposed onto the structure of LPyV VP1-3SLN (PDB accession number 4MBZ). HPyV9 VP1 and 3SLN are shown as in Fig. 3. For LPyV VP1, one monomer is shown in forest green and other monomers are shown in gray. The side chains are shown in stick representation, and the corresponding C-α atoms are shown as spheres. The 3SLN bound to LPyV VP1 is shown in stick representation; nitrogen and oxygen atoms are colored blue and red, respectively, and carbon atoms are colored forest green. Only residues forming contacts with 3SLN that differ between the two viruses are shown. Water molecules are represented as red and orange spheres for HPyV9 and LPyV, respectively. Hydrogen bonds are shown as black and green dashed lines for HPyV9 and LPyV, respectively. The 5-fold axis of the VP1 pentamer is indicated to depict the orientation of the superimposed pentamers. (B) The structure of HPyV9 VP1-3GSLN is superimposed onto the structure of LPyV VP1-3SLN (PDB accession number 4MBZ). HPyV9 VP1 and 3GSLN are shown as in Fig. 3C. LPyV VP1 and 3SLN are shown as described for panel A. Histidine is present in the binding sites of both viruses (H274 in HPyV9 and H271 in LPyV), but only in HPyV9 does it coordinate the hydroxyl group of Neu5Gc, which also interacts with the side chain of N282 of HPyV9 VP1. Hydrogen bonds are shown as black dashed lines. D71 is present in the proximal vicinity and stabilized the side chain of the N282 via a hydrogen bond (red dashed line).
The binding region has two additional substitutions in the BC1 loop, where E64 and D71 in HPyV9 VP1 are replaced with D61 and S68, respectively, in LPyV VP1. The two HPyV9 residues have longer side chains and contribute additional interactions with the sialic acid glycerol chain that result in a decrease in mobility (as evidenced by reduced temperature factors and better electron density) and a small change in the position of the glycerol chain (Fig. 4A). In particular, D71 forms a hydrogen bond with the glycerol O-9 hydroxyl group in HPyV9 VP1, while the corresponding S68 in LPyV VP1 does not. The extra contacts with the glycerol chain are likely responsible for the increased signals in the glycan arrays. This is in accordance with the higher B factor of the glycerol chain in the structure of the LPyV complex.
Additionally, the solvent-exposed F73 at the entrance of the binding pocket in LPyV is replaced with S76 in HPyV9. Although the significance of this substitution is not clear, the phenyl group of F73 contributes minor contacts to the third sugar's acetyl group in 3SLN in some copies of LPyV VP1 (31). S76 does not form any direct contacts with ligands, but its smaller side chain may facilitate access to the binding site, again in accord with the higher signals observed for HPyV9 in the glycan microarrays and the absence of the unusual ligand density effect observed with LPyV. Moreover, GlcNAc is located about ∼6 Å closer to VP1 in the LPyV complex than in the HPyV9 complex, which can likely be attributed to the presence of F73 (Fig. 4A).
Finally, HPyV9 and LPyV display key differences in the region that accommodates the glycolyl and acetyl groups of Neu5Gc and Neu5Ac, respectively, with implications for specificity. Of the two HPyV9 side chains that form hydrogen bonds to the glycolyl hydroxyl group, H274 is conserved in LPyV and thus does not help to discriminate between Neu5Ac and Neu5Gc. However, HPyV9 residue N282 is replaced with V279 in LPyV (Fig. 4B). The conformation of the N282 side chain is stabilized by a hydrogen bond with D71 (which is replaced with a serine in LPyV; Fig. 4B). The side chains of D71, S280, and N282 also stack against each other, thus stabilizing the overall conformation of this three-residue motif. The region surrounding the glycolyl group is significantly more polar in HPyV9. The hydrophobic V279 side chain in LPyV cannot favorably interact with the glycolyl oxygen of Neu5Gc.
Mutagenesis of HPyV9.
In order to test whether the substitution of a single residue in the HPyV9 binding site suffices to eliminate the binding preference for Neu5Gc and switch the specificity of HPyV9 to that of LPyV, we mutated HPyV9 residue N282 to valine (N282V), which is the amino acid found in LPyV VP1 at this position (Fig. 4B). Comparative glycan array analysis of wt and N282V HPyV9 VP1 showed that the mutant bound very poorly to the lipid-linked 3SLN and 3GSLN oligosaccharides, with a remaining slight preference for 3GSLN (Fig. 5). The most likely explanation for this finding is that the point mutation compromises the binding site. Since N282 lies directly next to D71 (Fig. 4), the N282V mutation places a hydrophobic side chain in the direct vicinity of D71, and it also removes favorable stacking interactions between N282 and D71. The mutation may thus alter the D71 side chain conformation and perhaps even cause other local structural rearrangements in the vicinity of the ligand. We conclude that specificity for Neu5Gc has to be attributed to the described set of three unique HPyV9 residues (D71, S280, and N282), which must be regarded as an entity.
FIG 5.
Comparison of binding of the wild-type HPyV9 VP1 (A) and the mutant HPyV9 VP1 (B) to lipid-linked oligosaccharide probes (3SLN.DH and 3GSLN.DH) in microarray analyses. Both probes were arrayed at four levels (as indicated) in duplicate. Numerical scores of the binding signals are shown as the means of the fluorescence intensities of duplicate spots with error bars, which represent half of the difference between the two values. The wt and mutant HPyV9 VP1 proteins were analyzed in parallel at 150 μg/ml precomplexed with anti-His and biotin–anti-mouse IgG antibodies.
Implications for serological cross-reactivity among polyomaviruses.
Serological cross-reactivity among closely related polyomaviruses has been investigated for decades, as reviewed by Moens et al. and references in their work (52). The high level of amino acid sequence identity is the most likely reason for serological cross-reactivity (18). HPyV9 and LPyV cross-react in immunodiagnostic assays (19, 20). Recently, a novel chimpanzee polyomavirus (PtrosPyV2) was also reported to cross-react with HPyV9 (53). Moreover, other novel polyomaviruses that also showed reactivity with human serum were also isolated from chimpanzees, while no human homologues of these viruses are known (53).
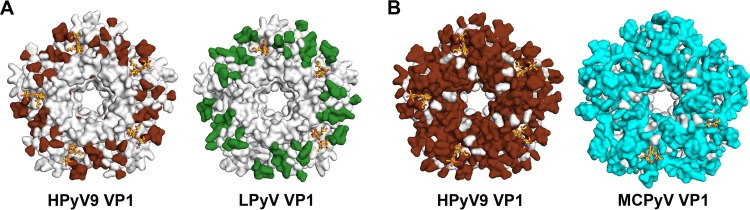
Pairwise comparisons of the structures of the closely related polyomavirus VP1 proteins provide an explanation for these findings. We highlighted the identical amino acid residues at the outermost apical surface of VP1 between two closely related viruses in gray, while the nonidentical residues were given a distinct color, as shown in Fig. 6. This analysis shows that cross-reactive polyomaviruses have significant degrees of amino acid conservation on the apical, solvent-exposed surface compared to non-cross-reactive polyomaviruses. Figure 6A shows a pairwise comparison of the apical surfaces of HPyV9 and LPyV, where extensive conserved areas (highlighted in gray) also include the bulk of the receptor binding region. In contrast, the comparison of the surface conservation between HPyV9 and MCPyV yields a very different picture (Fig. 6B). Here, the proportion of conserved residues on the apical surface is negligible, despite 58% overall sequence identity, providing the likely explanation for the distinct antigenicities of these two viruses (18, 20). This visualization provides support for the previous notion that HPyV9, rather than MCPyV, might be the cause for the serological reactivity of human serum against the monkey-derived LPyV (20).
FIG 6.
Structural representation of serological cross-reactivity and antigenicity of polyomaviruses. (A) Pairwise comparison of the apical surfaces of HPyV9 VP1 and LPyV VP1 (PDB accession number 4MBZ) pentamers. The apical surfaces of VP1 are shown in surface representation. Identical amino acids between these two viruses are colored gray. Nonidentical amino acids between the two viruses are colored chocolate and forest green for HPyV9 and LPyV, respectively. The glycan ligand (3SLN) is shown in stick representation, as in Fig. 3. (B) Pairwise comparison of the apical surfaces of HPyV9 VP1 and MCPyV VP1 (PDB accession number 4FMI) pentamers. The apical surfaces are shown in surface representation. Identical amino acids between the two viruses are colored gray. Nonidentical amino acids between the two viruses are colored chocolate and cyan for HPyV9 and MCPyV, respectively. The glycan ligand (3SLN) is shown in stick representation, as in Fig. 3.
DISCUSSION
A large number of viral and bacterial pathogens engage sialic acid-based compounds to establish their initial contact with a target cell (54). Although sialic acids often display modifications, the molecular principles that govern recognition of sialic acid variants are poorly understood, as the majority of structures have been solved using Neu5Ac-based compounds as ligands. Neu5Gc, which can be synthesized by nonhuman mammals but not by humans, has attracted much interest in recent years due to its role in understanding and defining host and tissue tropism (35, 38, 39). In order to investigate differences in sialic acid receptor binding in two closely related viruses from different species, we analyzed the ligand specificity and determined the structure of the major capsid protein VP1 from HPyV9, which was isolated from humans, and compared its properties with those of the VP1 protein from the monkey-derived virus LPyV.
We found that HPyV9 differs from LPyV with its unexpected preferential binding to the Neu5Gc glycan, which humans can acquire only through a diet containing red meat and milk. The observed preference for Neu5Gc can be explained by the architecture of the sialic acid binding site in HPyV9, which allows specific recognition of the extra Neu5Gc hydroxyl group via hydrogen bonds to side chains. Three residues unique to the HPyV9 binding site (D71, S280, and N282) act in concert to achieve this specificity. Replacement of one of these residues, N282, with the valine found in LPyV is not sufficient to alter the specificity and in fact compromises the binding pocket. Thus, the preference of HPyV9 for Neu5Gc is likely mediated by the set of three residues. A substitution at the entrance to the binding pocket (S76) likely enhances its accessibility in HPyV9 compared with that in LPyV (F76), in line with the observed binding profile of HPyV9 on glycan microarrays.
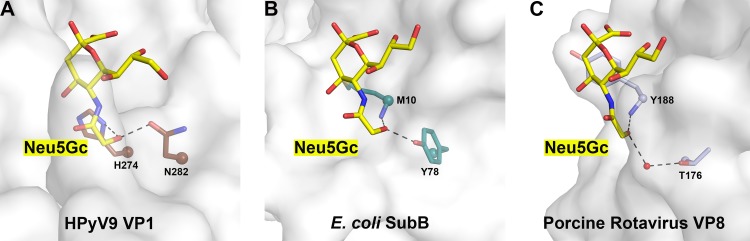
Only two other crystal structures of proteins in complex with Neu5Gc-based receptors have been reported: those of the porcine rotavirus strain CRW-8 spike protein domain VP8 (39) and the subtilase cytotoxin B subunit (SubB) produced by Shiga-toxigenic Escherichia coli (STEC) (35). Similar to HPyV9 VP1, CRW-8 VP8 can bind both Neu5Ac and Neu5Gc, but it prefers the latter compound, which shows a higher binding affinity and results in increased cellular infectivity (39). Likewise, SubB can also bind Neu5Ac and Neu5Gc in glycan microarrays, but structural studies were successful only for the Neu5Gc complex, again indicating a preference for Neu5Gc binding (35). A comparison of the Neu5Gc-binding sites of HPyV9 VP1, CRW-8 VP8, and STEC SubB shows that all three proteins acquire their preference for the glycolyl chain of Neu5Gc through a hydrophilic environment in its vicinity (Fig. 7). In all three cases, two hydrogen bonds are formed between protein residues and the extra hydroxyl group in Neu5Gc. Mutation of any of these residues results in a significant loss of activity of CRW-8 VP8 or SubB (35, 39). In contrast to HPyV9, the binding site for Neu5Gc is more surface exposed in CRW-8 VP8 and STEC SubB, with one side of the ring facing the protein and the other side facing the solvent. This is also in accordance with the larger buried surface area of Neu5Gc in the HPyV9 complex (314.5 Å2) than in the CRW-8 VP8 and STEC SubB complexes (229.7 Å2 and 234.6 Å2, respectively) (51).
FIG 7.
Comparison of Neu5Gc binding sites. Recognition of Neu5Gc by HPyV9 VP1 (A), Shiga-toxigenic E. coli SubB (PDB accession number 3DWP) (B), and porcine rotavirus strain CRW-8 VP8 (PDB accession number 3TAY) (C). The proteins are shown in surface representation. Neu5Gc is shown as in Fig. 4B. The critical amino acids making hydrogen bonds (black dashed lines) with Neu5Gc are shown in stick representation, and associated C-α atoms are shown as spheres. The water molecule in panel C is shown as a red sphere. The extra hydroxyl group of Neu5Gc makes two hydrogen bonds in each case.
Bacterial toxins similar to SubB, such as the pertussis toxin of Bordetella pertussis, do not exhibit a preference for Neu5Gc, as they lack the residue needed to coordinate the extra hydroxyl group of Neu5Gc, but they nevertheless accommodate Neu5Ac in a similar location and orientation (55). Similarly, in contrast to porcine rotavirus, the related rhesus monkey rotavirus VP8 preferentially binds Neu5Ac, also as a result of a single amino acid substitution (G187 to K187) that abolishes the ability of the virus to contact the hydroxyl group of Neu5Gc (39). The significance of these specificities for tropism is not always clear, although it has been suggested that the SubB has adapted to bind Neu5Gc so that it can use metabolically incorporated Neu5Gc as a receptor.
Our analysis shows that HPyV9 and LPyV form a closely related pair of viruses that differ in receptor specificity and, apparently, in species tropism. A similar pair of two closely related polyomaviruses is formed by BKPyV and SV40, which infect humans and simians, respectively. The BKPyV and SV40 VP1 proteins share a high degree of sequence identity and bind terminal sialic acid in a mostly conserved binding site that differs from the one observed in HPyV9 and LPyV (26, 30). However, SV40 displays a preference for receptors containing Neu5Gc over receptors containing Neu5Ac (38), whereas the sialic acid binding site of BKPyV exclusively binds to the smaller Neu5Ac and cannot accommodate the larger glycolyl chain of Neu5Gc (30). Thus, for these two viruses, the receptor specificities correlate well with the biosynthetically available variants of sialic acids in the respective hosts. However, an analogous correlation obviously does not exist in the case of the HPyV9 and LPyV pair of polyomaviruses.
Why, then, does the binding pocket of human HPyV9 favor binding to a ligand, Neu5Gc, that cannot be synthesized by humans? This question is difficult to answer at present, but the ability to engage Neu5Gc is likely advantageous for HPyV9. Humans can acquire Neu5Gc from their diet, as red meat and milk are rich sources of Neu5Gc. During digestion of such foods, Neu5Gc can be taken up by fast-growing body cells and is subsequently transferred to newly synthesized glycoproteins and glycolipids in the Golgi apparatus by a transferase that does not discriminate between Neu5Ac and Neu5Gc (35–37). Thus, the gut endothelium and kidney vasculature in particular display Neu5Gc on their surface, and this is exploited, for example, by the SubB toxin (35). Similarly, the expression of Neu5Gc on certain carcinomas has also been reported (56). It is therefore conceivable that the ability to bind to Neu5Gc confers an advantage to HPyV9 by directing it to specific tissues. HPyV9 may therefore have evolved to utilize a receptor that is provided by a diet. It will be interesting to test this hypothesis once tissue culture systems that allow propagation of HPyV9 are established. Whereas LPyV can propagate only in the cells of B lymphoblastoid origin (57), the reproductive niche for HPyV9 is not yet understood. We expect that our findings will facilitate a proper dissection of the attachment and entry mechanisms used by HPyV9.
ACKNOWLEDGMENTS
We thank members of the Glycosciences Laboratory for their collaboration in the establishment of the neoglycolipid-based microarray. We thank Marie-France Goneau for help with the synthesis of 3GSLN.
This project was supported by contract research Glykobiologie/Glykomik of the Baden-Württemberg Stiftung (to T.S.) as well as Wellcome Trust grants WT093378MA and WT099197MA, the United Kingdom Research Councils' Basic Technology Initiative Glycoarrays (GRS/79268), and an EPSRC translational grant (EP/G037604/1) (to T.F.).
Footnotes
Published ahead of print 19 March 2014
REFERENCES
- 1.Neu U, Stehle T, Atwood WJ. 2009. The Polyomaviridae: contributions of virus structure to our understanding of virus receptors and infectious entry. Virology 384:389–399. 10.1016/j.virol.2008.12.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Padgett BL, Walker DL, ZuRhein GM, Eckroade RJ, Dessel BH. 1971. Cultivation of papova-like virus from human brain with progressive multifocal leucoencephalopathy. Lancet i:1257–1260 [DOI] [PubMed] [Google Scholar]
- 3.Gardner SD, Field AM, Coleman DV, Hulme B. 1971. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet i:1253–1257 [DOI] [PubMed] [Google Scholar]
- 4.Allander T, Andreasson K, Gupta S, Bjerkner A, Bogdanovic G, Persson MA, Dalianis T, Ramqvist T, Andersson B. 2007. Identification of a third human polyomavirus. J. Virol. 81:4130–4136. 10.1128/JVI.00028-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gaynor AM, Nissen MD, Whiley DM, Mackay IM, Lambert SB, Wu G, Brennan DC, Storch GA, Sloots TP, Wang D. 2007. Identification of a novel polyomavirus from patients with acute respiratory tract infections. PLoS Pathog. 3:e64. 10.1371/journal.ppat.0030064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng H, Shuda M, Chang Y, Moore PS. 2008. Clonal integration of a polyomavirus in human Merkel cell carcinoma. Science 319:1096–1100. 10.1126/science.1152586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schowalter RM, Pastrana DV, Pumphrey KA, Moyer AL, Buck CB. 2010. Merkel cell polyomavirus and two previously unknown polyomaviruses are chronically shed from human skin. Cell Host Microbe 7:509–515. 10.1016/j.chom.2010.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Meijden E, Janssens RW, Lauber C, Bouwes Bavinck JN, Gorbalenya AE, Feltkamp MC. 2010. Discovery of a new human polyomavirus associated with trichodysplasia spinulosa in an immunocompromised patient. PLoS Pathog. 6:e1001024. 10.1371/journal.ppat.1001024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scuda N, Hofmann J, Calvignac-Spencer S, Ruprecht K, Liman P, Kuhn J, Hengel H, Ehlers B. 2011. A novel human polyomavirus closely related to the African green monkey-derived lymphotropic polyomavirus. J. Virol. 85:4586–4590. 10.1128/JVI.02602-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sauvage V, Foulongne V, Cheval J, Ar Gouilh M, Pariente K, Dereure O, Manuguerra JC, Richardson J, Lecuit M, Burguiere A, Caro V, Eloit M. 2011. Human polyomavirus related to African green monkey lymphotropic polyomavirus. Emerg. Infect. Dis. 17:1364–1370. 10.3201/eid1708.110278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buck CB, Phan GQ, Raiji MT, Murphy PM, McDermott DH, McBride AA. 2012. Complete genome sequence of a tenth human polyomavirus. J. Virol. 86:10887. 10.1128/JVI.01690-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Siebrasse EA, Reyes A, Lim ES, Zhao G, Mkakosya RS, Manary MJ, Gordon JI, Wang D. 2012. Identification of MW polyomavirus, a novel polyomavirus in human stool. J. Virol. 86:10321–10326. 10.1128/JVI.01210-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim ES, Reyes A, Antonio M, Saha D, Ikumapayi UN, Adeyemi M, Stine OC, Skelton R, Brennan DC, Mkakosya RS, Manary MJ, Gordon JI, Wang D. 2013. Discovery of STL polyomavirus, a polyomavirus of ancestral recombinant origin that encodes a unique T antigen by alternative splicing. Virology 436:295–303. 10.1016/j.virol.2012.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korup S, Rietscher J, Calvignac-Spencer S, Trusch F, Hofmann J, Moens U, Sauer I, Voigt S, Schmuck R, Ehlers B. 2013. Identification of a novel human polyomavirus in organs of the gastrointestinal tract. PLoS One 8:e58021. 10.1371/journal.pone.0058021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Izakovic J, Buchner SA, Duggelin M, Guggenheim R, Itin PH. 1995. Hair-like hyperkeratoses in patients with kidney transplants. A new cyclosporin side-effect. Hautarzt 46:841–846 (In German.) [DOI] [PubMed] [Google Scholar]
- 16.zur Hausen H, Gissmann L. 1979. Lymphotropic papovaviruses isolated from African green monkey and human cells. Med. Microbiol. Immunol. 167:137–153. 10.1007/BF02121180 [DOI] [PubMed] [Google Scholar]
- 17.Brade L, Muller-Lantzsch N, zur Hausen H. 1981. B-lymphotropic papovavirus and possibility of infections in humans. J. Med. Virol. 6:301–308 [DOI] [PubMed] [Google Scholar]
- 18.Kean JM, Rao S, Wang M, Garcea RL. 2009. Seroepidemiology of human polyomaviruses. PLoS Pathog. 5:e1000363. 10.1371/journal.ppat.1000363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trusch F, Klein M, Finsterbusch T, Kuhn J, Hofmann J, Ehlers B. 2012. Seroprevalence of human polyomavirus 9 and cross-reactivity to African green monkey-derived lymphotropic polyomavirus. J. Gen. Virol. 93:698–705. 10.1099/vir.0.039156-0 [DOI] [PubMed] [Google Scholar]
- 20.Nicol JT, Touze A, Robinot R, Arnold F, Mazzoni E, Tognon M, Coursaget P. 2012. Seroprevalence and cross-reactivity of human polyomavirus 9. Emerg. Infect. Dis. 18:1329–1332. 10.3201/eid1808.111625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murata H, Teferedegne B, Sheng L, Lewis AM, Jr, Peden K. 2008. Identification of a neutralization epitope in the VP1 capsid protein of SV40. Virology 381:116–122. 10.1016/j.virol.2008.07.032 [DOI] [PubMed] [Google Scholar]
- 22.Randhawa P, Viscidi R, Carter JJ, Galloway DA, Culp TD, Huang C, Ramaswami B, Christensen ND. 2009. Identification of species-specific and cross-reactive epitopes in human polyomavirus capsids using monoclonal antibodies. J. Gen. Virol. 90:634–639. 10.1099/vir.0.008391-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pastrana DV, Brennan DC, Cuburu N, Storch GA, Viscidi RP, Randhawa PS, Buck CB. 2012. Neutralization serotyping of BK polyomavirus infection in kidney transplant recipients. PLoS Pathog. 8:e1002650. 10.1371/journal.ppat.1002650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liddington RC, Yan Y, Moulai J, Sahli R, Benjamin TL, Harrison SC. 1991. Structure of simian virus 40 at 3.8-Å resolution. Nature 354:278–284. 10.1038/354278a0 [DOI] [PubMed] [Google Scholar]
- 25.Stehle T, Yan Y, Benjamin TL, Harrison SC. 1994. Structure of murine polyomavirus complexed with an oligosaccharide receptor fragment. Nature 369:160–163. 10.1038/369160a0 [DOI] [PubMed] [Google Scholar]
- 26.Neu U, Woellner K, Gauglitz G, Stehle T. 2008. Structural basis of GM1 ganglioside recognition by simian virus 40. Proc. Natl. Acad. Sci. U. S. A. 105:5219–5224. 10.1073/pnas.0710301105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neu U, Maginnis MS, Palma AS, Stroh LJ, Nelson CD, Feizi T, Atwood WJ, Stehle T. 2010. Structure-function analysis of the human JC polyomavirus establishes the LSTc pentasaccharide as a functional receptor motif. Cell Host Microbe 8:309–319. 10.1016/j.chom.2010.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neu U, Wang J, Macejak D, Garcea RL, Stehle T. 2011. Structures of the major capsid proteins of the human Karolinska Institutet and Washington University polyomaviruses. J. Virol. 85:7384–7392. 10.1128/JVI.00382-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neu U, Hengel H, Blaum BS, Schowalter RM, Macejak D, Gilbert M, Wakarchuk WW, Imamura A, Ando H, Kiso M, Arnberg N, Garcea RL, Peters T, Buck CB, Stehle T. 2012. Structures of Merkel cell polyomavirus VP1 complexes define a sialic acid binding site required for infection. PLoS Pathog. 8:e1002738. 10.1371/journal.ppat.1002738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neu U, Allen SA, Blaum BS, Liu Y, Frank M, Palma AS, Stroh LJ, Feizi T, Peters T, Atwood WJ, Stehle T. 2013. A structure-guided mutation in the major capsid protein retargets BK polyomavirus. PLoS Pathog. 9:e1003688. 10.1371/journal.ppat.1003688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neu U, Khan ZM, Schuch B, Palma AS, Liu Y, Pawlita M, Feizi T, Stehle T. 2013. Structures of B-lymphotropic polyomavirus VP1 in complex with oligosaccharide ligands. PLoS Pathog. 9:e1003714. 10.1371/journal.ppat.1003714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Corallini A, Mazzoni E, Taronna A, Manfrini M, Carandina G, Guerra G, Guaschino R, Vaniglia F, Magnani C, Casali F, Dolcetti R, Palmonari C, Rezza G, Martini F, Barbanti-Brodano G, Tognon MG. 2012. Specific antibodies reacting with simian virus 40 capsid protein mimotopes in serum samples from healthy blood donors. Hum. Immunol. 73:502–510. 10.1016/j.humimm.2012.02.009 [DOI] [PubMed] [Google Scholar]
- 33.Varki A. 2010. Colloquium paper: uniquely human evolution of sialic acid genetics and biology. Proc. Natl. Acad. Sci. U. S. A. 107(Suppl 2):8939–8946. 10.1073/pnas.0914634107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varki NM, Strobert E, Dick EJ, Jr, Benirschke K, Varki A. 2011. Biomedical differences between human and nonhuman hominids: potential roles for uniquely human aspects of sialic acid biology. Annu. Rev. Pathol. 6:365–393. 10.1146/annurev-pathol-011110-130315 [DOI] [PubMed] [Google Scholar]
- 35.Byres E, Paton AW, Paton JC, Lofling JC, Smith DF, Wilce MC, Talbot UM, Chong DC, Yu H, Huang S, Chen X, Varki NM, Varki A, Rossjohn J, Beddoe T. 2008. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature 456:648–652. 10.1038/nature07428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tangvoranuntakul P, Gagneux P, Diaz S, Bardor M, Varki N, Varki A, Muchmore E. 2003. Human uptake and incorporation of an immunogenic nonhuman dietary sialic acid. Proc. Natl. Acad. Sci. U. S. A. 100:12045–12050. 10.1073/pnas.2131556100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Banda K, Gregg CJ, Chow R, Varki NM, Varki A. 2012. Metabolism of vertebrate amino sugars with N-glycolyl groups: mechanisms underlying gastrointestinal incorporation of the non-human sialic acid xeno-autoantigen N-glycolylneuraminic acid. J. Biol. Chem. 287:28852–28864. 10.1074/jbc.M112.364182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Campanero-Rhodes MA, Smith A, Chai W, Sonnino S, Mauri L, Childs RA, Zhang Y, Ewers H, Helenius A, Imberty A, Feizi T. 2007. N-Glycolyl GM1 ganglioside as a receptor for simian virus 40. J. Virol. 81:12846–12858. 10.1128/JVI.01311-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu X, Dang VT, Fleming FE, von Itzstein M, Coulson BS, Blanchard H. 2012. Structural basis of rotavirus strain preference toward N-acetyl- or N-glycolylneuraminic acid-containing receptors. J. Virol. 86:13456–13466. 10.1128/JVI.06975-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karwaski MF, Wakarchuk WW, Gilbert M. 2002. High-level expression of recombinant Neisseria CMP-sialic acid synthetase in Escherichia coli. Protein Expr. Purif. 25:237–240. 10.1016/S1046-5928(02)00004-9 [DOI] [PubMed] [Google Scholar]
- 41.Pukin AV, Weijers CA, van Lagen B, Wechselberger R, Sun B, Gilbert M, Karwaski MF, Florack DE, Jacobs BC, Tio-Gillen AP, van Belkum A, Endtz HP, Visser GM, Zuilhof H. 2008. GM3, GM2 and GM1 mimics designed for biosensing: chemoenzymatic synthesis, target affinities and 900 MHz NMR analysis. Carbohydr. Res. 343:636–650. 10.1016/j.carres.2008.01.007 [DOI] [PubMed] [Google Scholar]
- 42.Liu Y, Childs RA, Palma AS, Campanero-Rhodes MA, Stoll MS, Chai W, Feizi T. 2012. Neoglycolipid-based oligosaccharide microarray system: preparation of NGLs and their noncovalent immobilization on nitrocellulose-coated glass slides for microarray analyses. Methods Mol. Biol. 808:117–136. 10.1007/978-1-61779-373-8_8 [DOI] [PubMed] [Google Scholar]
- 43.Palma AS, Feizi T, Zhang Y, Stoll MS, Lawson AM, Diaz-Rodriguez E, Campanero-Rhodes MA, Costa J, Gordon S, Brown GD, Chai W. 2006. Ligands for the beta-glucan receptor, Dectin-1, assigned using “designer” microarrays of oligosaccharide probes (neoglycolipids) generated from glucan polysaccharides. J. Biol. Chem. 281:5771–5779. 10.1074/jbc.M511461200 [DOI] [PubMed] [Google Scholar]
- 44.Stoll MS, Feizi T. 2009. Software tools for storing, processing and displaying carbohydrate microarray data. Beilstein Symposium on Glyco-Bioinformatics, Potsdam, Germany [Google Scholar]
- 45.Kabsch W. 2010. XDS. Acta Crystallogr. D Biol. Crystallogr. 66:125–132. 10.1107/S0907444909047337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vagin A, Teplyakov A. 2010. Molecular replacement with MOLREP. Acta Crystallogr. D Biol. Crystallogr. 66:22–25. 10.1107/S0907444909042589 [DOI] [PubMed] [Google Scholar]
- 47.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. 2010. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66:213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240–255. 10.1107/S0907444996012255 [DOI] [PubMed] [Google Scholar]
- 49.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126–2132. 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 50.Painter J, Merritt EA. 2006. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62:439–450. 10.1107/S0907444906005270 [DOI] [PubMed] [Google Scholar]
- 51.Krissinel E, Henrick K. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774–797. 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 52.Moens U, Van Ghelue M, Song X, Ehlers B. 2013. Serological cross-reactivity between human polyomaviruses. Rev. Med. Virol. 23:250–264. 10.1002/rmv.1747 [DOI] [PubMed] [Google Scholar]
- 53.Scuda N, Madinda NF, Akoua-Koffi C, Adjogoua EV, Wevers D, Hofmann J, Cameron KN, Leendertz SA, Couacy-Hymann E, Robbins M, Boesch C, Jarvis MA, Moens U, Mugisha L, Calvignac-Spencer S, Leendertz FH, Ehlers B. 2013. Novel polyomaviruses of nonhuman primates: genetic and serological predictors for the existence of multiple unknown polyomaviruses within the human population. PLoS Pathog. 9:e1003429. 10.1371/journal.ppat.1003429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Neu U, Bauer J, Stehle T. 2011. Viruses and sialic acids: rules of engagement. Curr. Opin. Struct. Biol. 21:610–618. 10.1016/j.sbi.2011.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stein PE, Boodhoo A, Armstrong GD, Heerze LD, Cockle SA, Klein MH, Read RJ. 1994. Structure of a pertussis toxin-sugar complex as a model for receptor binding. Nat. Struct. Biol. 1:591–596. 10.1038/nsb0994-591 [DOI] [PubMed] [Google Scholar]
- 56.Padler-Karavani V, Hurtado-Ziola N, Pu M, Yu H, Huang S, Muthana S, Chokhawala HA, Cao H, Secrest P, Friedmann-Morvinski D, Singer O, Ghaderi D, Verma IM, Liu YT, Messer K, Chen X, Varki A, Schwab R. 2011. Human xeno-autoantibodies against a non-human sialic acid serve as novel serum biomarkers and immunotherapeutics in cancer. Cancer Res. 71:3352–3363. 10.1158/0008-5472.CAN-10-4102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herrmann M, Oppenlander M, Pawlita M. 1995. Fast and high-affinity binding of B-lymphotropic papovavirus to human B-lymphoma cell lines. J. Virol. 69:6797–6804 [DOI] [PMC free article] [PubMed] [Google Scholar]