ABSTRACT
Nuclear egress of herpesvirus capsids is mediated by a conserved heterodimeric complex of two viral proteins, designated pUL34 and pUL31 in herpes simplex virus and pseudorabies virus (PrV). pUL34, a tail-anchored membrane protein, is targeted to the nuclear envelope and recruits pUL31 to the inner nuclear membrane (INM) to provide the docking and envelopment machinery for the nascent capsid. While the less conserved C-terminal part of pUL34 is required for correct positioning of the nuclear egress complex (NEC) at the INM, the conserved N-terminal part functions as a docking site for pUL31. Since no crystal structure of NEC is available yet, structure-function studies depend on mutational analyses, with several approaches already being performed for different herpesvirus NECs. Here, we extended our studies on PrV pUL34 and identified two asparagine residues (N75, N103) and a dileucine motif (LL166/167), adjacent to an endoplasmic reticulum retention signal, which are absolutely required for NEC function. While the pUL34–N75A substitution mutant is unable to interact with pUL31, the pUL34–N103A mutant is nonfunctional, despite continuing complex formation. Surprisingly, mutant pUL34–G77A, which does not efficiently recruit pUL31 to the nuclear rim after cotransfection, nonetheless complements a UL34 deletion mutant, indicating that the NEC may be stabilized by additional viral factors during infection.
IMPORTANCE In the absence of a crystal structure of the nuclear egress complex (NEC) required for herpesvirus maturation, site-directed mutagenesis studies provide important information on critical amino acid residues. Here, we identify conserved amino acid residues in the membrane-bound component of the NEC which are relevant for its function.
INTRODUCTION
In herpesviruses, DNA replication and nucleocapsid formation occur in the host cell nucleus, while further virion maturation takes place in the cytosol. Thus, viral capsids have to cross the nuclear envelope (NE) to gain access to the final maturation compartment. In the meantime, it is well accepted that nuclear egress of herpesvirus capsids occurs by an envelopment-deenvelopment mechanism, in which capsids gain a transient lipid envelope by budding at the inner nuclear membrane (INM), resulting in primary enveloped virions being located in the perinuclear cleft. Subsequently, this primary envelope fuses with the outer nuclear membrane (ONM) or with the contiguous membrane of the endoplasmic reticulum (ER), resulting in the release of the capsids into the cytosol (1). This process can be regarded as a vesicle (primary envelope)-mediated transport of cargo, represented by the capsid, which was long believed to be unique in cell biology. However, recent studies on the nuclear export of large ribonucleoprotein complexes in Drosophila revealed a similar transport mechanism, leading to the hypothesis that herpesviruses could have adopted this cellular process for their replication (2–4). However, its molecular details remain enigmatic.
Nuclear egress of herpesvirus capsids is mediated by the nuclear egress complex (NEC), which is conserved within the Herpesviridae. It consists of a heterodimer of a type II membrane protein designated pUL34 in the prototypic alphaherpesvirus herpes simplex virus 1 (HSV-1) which interacts with nucleoplasmic pUL31. The NEC is important for efficient viral replication, and the absence of pUL31 and/or pUL34 leads to a drastic impairment of nuclear egress and of the formation of infectious progeny (5–11). In transfected cells, the tail-anchored pUL34 is targeted to the NE, whereas pUL31 localizes diffusely throughout the nucleus. During infection, pUL34 binds and recruits pUL31 to the INM. Subsequently, viral and cellular kinases are relocated, and these phosphorylate and thereby locally dissolve the nuclear lamina, so that intranuclear capsids can reach their budding sites at the INM (reviewed in references 2, 12, and 13). Interestingly, coexpression of pUL31 and pUL34 is sufficient to mediate the formation of primary envelopes without any additional viral factors (14, 15). However, for efficient fusion of the primary vesicle with the ONM, other viral and probably also cellular factors are required, as evidenced by the accumulation of NEC-containing primary envelopes in the perinuclear space, which resemble the accumulations of primary enveloped virions in cells infected with mutants lacking or expressing an inactive pUS3 kinase (16, 17).
Since no crystal structure of any herpesviral NEC has so far been obtained, elucidation of the molecular details of the pUL34-pUL31 interaction and NEC function continue to rely on random or targeted mutational analyses which were performed for several herpesvirus NECs (10, 18–20). Deletion/substitution analyses performed on the C terminus of pUL34 homologs comprising the transmembrane domain (10, 18, 20–22) revealed that this part is not required for pUL31 interaction but serves to anchor the protein to the nuclear membrane. The N-terminal part of pUL34 homologues shows significantly higher amino acid conservation within the Herpesviridae (10, 18–20), and the interaction domain with the pUL31 homologues was mapped to this region (10, 19, 23, 24). In pseudorabies virus (PrV), this interaction domain was delineated by N-terminal truncations and C-terminal substitutions/deletions to amino acids (aa) 5 to 161 (20). For further characterization of NEC formation, several mutational analyses within the interaction domain of pUL34 homologs revealed specific residues required for interaction and function during viral replication. In HSV-1, clusters of charged amino acids were mutagenized (18). The murine cytomegalovirus (MCMV) pUL34 homolog, M50, was analyzed using random transposon mutagenesis (10), whereas site-directed mutagenesis was performed on the human cytomegalovirus (HCMV) pUL50 (19). Interestingly, in all mutational analyses of pUL34 homologs in HSV-1, HCMV, MCMV, and PrV, a conserved pair of amino acids consisting of a glutamic acid and tyrosine (EY motif; E53 and Y54 in PrV) was shown to play a crucial role for function (10, 18–20). In addition, mutation of an RQR motif present in PrV pUL34 (aa 173 to 175), which was originally suggested to be an INM-targeting signal (25, 26) but more likely constitutes an ER retention signal, resulted in partial localization of pUL34 in the Golgi apparatus (20). Like the classical arginine-based ER retention signals for membrane proteins, this RQR is surrounded by a putative phosphorylation site and a characteristic dileucine motif (27) (Fig. 1).
FIG 1.

Amino acid sequence of PrV pUL34. Conserved amino acids (20) within the N terminus were mutagenized to alanine (highlighted by boxes). The pUL31 interaction domain is underlined, and the predicted transmembrane region is shaded. The complementation indexes for PrV-ΔUL34 by the cell lines expressing the mutated proteins are also given as +++, ++, +, or −, as defined in footnote b of Table 2.
In order to identify further residues within the N-terminal part of PrV pUL34 important for pUL31 interaction and NEC targeting and function, we extended our site-directed mutagenesis to 13 additional conserved amino acids within pUL34. This study identifies two asparagine residues (N75, N103) which, like the EY motif, are essential for pUL34 function. Mutation of the dileucine motif which separates the conserved N-terminal pUL31 interaction domain from the variable C-terminal membrane anchor region also abolished NEC function, despite unimpaired complex formation.
In addition to its function during nuclear egress, pUL34 also seems to be involved in direct viral cell-to-cell spread (20, 22, 28). However, although it was speculated that pUL34 affects trafficking of glycoprotein gE to cellular junctions, the molecular details remain elusive (28). To clarify the putative role of pUL34 in direct cell-to-cell transmission, we also analyzed pUL34 mutant-expressing cells for their capacity to complement plaque formation of our PrV pUL34 deletion mutant. Surprisingly, except for one protein, all mutant polypeptides, including those which quite efficiently supported replication of PrV-ΔUL34, showed a significant reduction in plaque size.
MATERIALS AND METHODS
Cells and viruses.
Rabbit kidney (RK13) cells were cultivated in Dulbecco's modified Eagle's minimum essential medium supplemented with 10% fetal calf serum. The wild-type PrV strain Kaplan (PrV-Ka) (29) was propagated on RK13 cells, while PrV-ΔUL34 and PrV-ΔUL34/US3, which express green fluorescent protein (GFP) instead of either UL34 or US3, respectively, were grown on RK13-UL34 cells as described previously (5, 16).
Generation of pUL34 mutants and stably expressing cell lines.
Site-directed mutagenesis was performed as described recently (20) using a QuikChange II XL site-directed mutagenesis kit (Agilent Technologies). Plasmid DNA carrying UL34 (pcDNA-UL34) (5) served as the template, and the mutant primers used are listed in Table 1 (only the forward primers are given, but they were used in combination with reverse complementary sequences). All constructs were verified by restriction enzyme cleavage and sequencing (data not shown).
TABLE 1.
Primers used for site-directed mutagenesis
| Name | Sequence (5′ to 3′)a | Location in PrV Ka (nt)b |
|---|---|---|
| UL34_L56A | CCGCTGGAGTACGTGCTGCGCCTCATGCGCAGC | 31548–31562 |
| UL34_N75A | CCTACGTGCGCGTGCAGGCCACGGGCGTGTCGGTGCTC | 31603–31649 |
| UL34_G77A | CGCGTGCAGAACACGGCCGTGTCGGTGCTCTTCC | 31611–31644 |
| UL34_V80A | CAGAACACGGGCGTGTCGGCGCTCTTCCAGGGCTTCTTCTTCC | 31617–31659 |
| UL34_P89A | GGCTTCTTCTTCCGGGCCGCCGACGCCCCCCTGG | 31647–31680 |
| UL34_N103A | CACGGCCGAGCACAACGCCGTGATCCTGGCCTCG | 31688–31721 |
| UL34_L106A | GCACAACAACGTGATCGCGGCCTCGACGCACAGC | 31697–31730 |
| UL34_L116A | GCACAGCACCGGCATGAGCGCCTCGGCGCTCGACGAC | 31724–31760 |
| UL34_K123A | GCGCTCGACGACATCGCGCGCGCCGGGGGCGTGG | 31749–31782 |
| UL34_P132A | GGCGTGGACACGCGGGCGCTGCGCGCCATGATGTCG | 31776–31811 |
| UL34_R144A | CGGTGAGCTGCTTCGTGGCCATGCCGCGCGTGCAGCTC | 31810–31847 |
| UL34_L167A | CAGACGCAGCGCCTCGCCGACCGCGCCGAGATG | 31881–31913 |
| UL34_L166/167A | CTCGCAGACGCAGCGCGCCGCCGACCGCGCCGAGATGC | 31877–31914 |
Only forward primers are listed, and the mutations introduced are underlined.
Positions correspond to those in the sequence with GenBank accession number BK001744. nt, nucleotide.
For generation of cell lines stably expressing the pUL34 mutants, RK13 cells were transfected with the pcDNA-UL34 constructs by calcium phosphate precipitation (30). After transfection, cells were selected in medium containing 0.5 mg/ml G418 (Invitrogen), and resistant clones were picked by aspiration. pUL34 expression was tested by Western blot analysis and immunofluorescence using polyclonal monospecific rabbit serum specific for PrV pUL34 (5).
Western blot analysis.
Western blot analysis was performed on cells expressing the pUL34 mutants by scraping the cells into medium and centrifugation. The cell pellets were washed twice with phosphate-buffered saline (PBS), resuspended in sodium dodecyl sulfate (SDS) sample buffer, and disrupted by supersonic lysis. Proteins were separated on an SDS-10% polyacrylamide gel. After transfer onto a nitrocellulose membrane, the blot was incubated with the PrV pUL34-specific polyclonal rabbit antiserum (5). Bound antibody was detected by peroxidase-coupled goat anti-rabbit antibody and visualized by enhanced chemiluminescence (Super Signal West Pico; Thermo Scientific). Images were recorded in an image analyzer (Bio-Rad).
Laser scanning confocal microscopy.
To analyze the localization and colocalization of mutated pUL34 with pUL31, RK13 cells were transfected with pcDNA3 constructs containing the corresponding genes for pUL31 or the pUL34 mutants. To investigate the colocalization of both proteins during infection, transgenic RK13-UL34 cell lines were infected in 6-well culture plates with PrV-Ka, PrV-ΔUL34, or PrV-ΔUL34/US3 under plaque assay conditions (300 PFU). Cells were fixed with 3% paraformaldehyde 2 days after transfection and labeled with polyclonal rabbit anti-pUL31 (8) and mouse anti-pUL34 (14) sera, which were diluted 1:500 and 1:300, respectively, in PBS. After incubation at room temperature for 1 h, bound antibody was detected with Alexa Fluor 488 goat antirabbit and Alexa Fluor 561 goat anti-mouse (Invitrogen) antibodies. In samples where GFP-expressing mutant viruses were used, the Alexa Fluor 634 anti-rabbit antibody (Invitrogen) was applied to detect bound anti-pUL31 antibodies. All secondary antibodies were diluted 1:1,000 in PBS. Fluorescence images were recorded with a laser scanning confocal microscope (SP5; Leica, Mannheim, Germany).
Complementation assays.
To test for functional complementation, cell lines expressing wild-type or mutant pUL34 were infected with PrV-Ka or PrV-ΔUL34 at a multiplicity of infection (MOI) of 3. After 1 h at 4°C, the inoculum was replaced by prewarmed medium and the cells were incubated at 37°C for an additional hour. Extracellular virus was inactivated by low-pH treatment (31). At 24 h after infection, cells and supernatant were harvested and progeny virus was titrated on RK13-UL34 cells. The mean values of three independent experiments were calculated and plotted with the corresponding standard deviations. The complementation index (CI) was calculated as the ratio of the titer of PrV-ΔUL34 and PrV-Ka on a given cell line and normalized to the ratio of PrV-ΔUL34/PrV-Ka on RK13-UL34 cells. The statistical significance of differences between PrV-Ka and PrV-ΔUL34 was evaluated by Student's t test.
To investigate the role of pUL34 mutants in viral cell-to-cell spread, transgenic cell lines were infected with PrV-Ka or PrV-ΔUL34 under plaque assay conditions and fixed with 5% formaldehyde 2 days after infection. The diameters of 30 plaques each were measured microscopically, and the plaque diameters of PrV-ΔUL34-infected samples were compared to those of PrV-Ka-infected plaques, which were set to 100%. The statistical significance of the differences between infection with PrV-Ka and PrV-ΔUL34 was evaluated by Student's t test.
Electron microscopy.
Cell lines expressing wild-type pUL34 or pUL34–N75A, -G77A, -N103A, -L116A, -P132A, -L167A, and -L166/167A were infected with PrV-ΔUL34 at an MOI of 1. At 1 h after infection, the inoculum was replaced by fresh medium, and at 16 h after infection, cells were fixed and processed for electron microscopy as described previously (5).
RESULTS
Mutation of residues within the pUL31 interaction domain of PrV pUL34.
We showed recently that the pUL31 interaction domain is contained within amino acids 5 to 161 of PrV pUL34 (20). Several amino acids which are conserved throughout the Herpesviridae can be identified in this region (19, 20). Recently, we showed that three conserved cysteine residues within this region could be functionally replaced by alanine, whereas the diresidue motif EY at positions 53 and 54 was crucial for interaction and function (20). To further identify essential amino acids within the pUL31 binding domain, 13 additional point mutations of conserved residues were generated (Fig. 1, boxed amino acids).
Localization and pUL31 interaction of mutant pUL34 after transient expression.
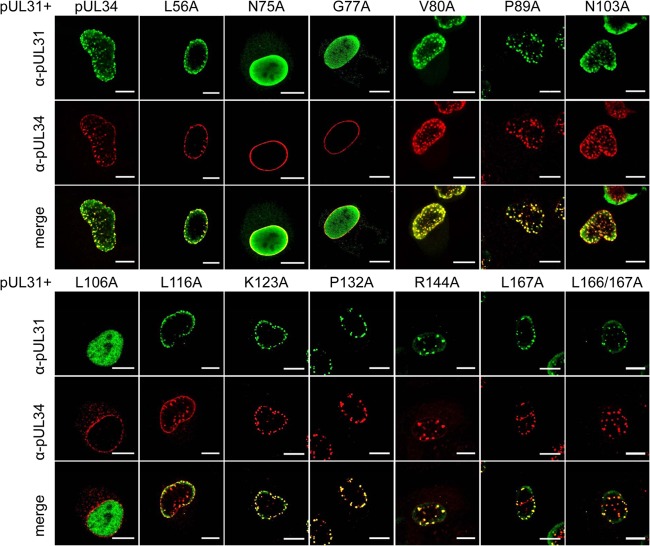
In transfected cells, all pUL34 mutants showed wild-type-like nuclear rim staining, indicating that none of the mutations interfered with targeting to the nuclear envelope (data not shown). After cotransfection with a pUL31 expression plasmid, pUL34 mutants pUL34–L56A, -V80A, -P89A, -N103A, -L116A, -K123A, -P132A, -R144A, and -L167A and the double dileucine mutant pUL34–L166/167A were able to induce speckles (Fig. 2; Table 2), i.e., accumulations of vesicles in which both proteins colocalize, reflecting successful pUL31 interaction (14). pUL34–N75A and pUL34–G77A failed to efficiently relocate pUL31, as evidenced by the diffuse nucleoplasmic staining of pUL31. This was also the case for cells cotransfected with pcDNA-UL34–L106A, but the detected amount of this mutated pUL34 was low, indicating either inefficient expression or instability of the protein. In summary, 10 of the 13 tested pUL34 mutants colocalized with pUL31 in the typical speckled pattern at the NE, indicating that they are able to interact with pUL31.
FIG 2.
Colocalization of mutated pUL34 with pUL31 after transient expression. In order to analyze the interaction between pUL31 and mutated pUL34, RK13 cells were cotransfected with the corresponding expression plasmids. Colocalization was studied using the monospecific murine anti-pUL34 (red) and rabbit anti-pUL31 (green) sera. Images were recorded by confocal laser scanning microscopy (Leica SP5). Speckles seen after coexpression of native pUL34 and pUL31 reflect the interaction of both proteins and the formation of primary envelopes (14), while diffuse nucleoplasmic pUL31 staining is due to failure of pUL31 recruitment. α, anti. Bars, 10 μm.
TABLE 2.
Summary of results
| Construct | Nuclear rim localization | Colocalization with pUL31 | Complementationb | Plaque diamc |
|---|---|---|---|---|
| pUL34 | + | + | +++ | 90 |
| pUL34–L56A | + | + | + | 30 |
| pUL34–N75A | + | − | − | 10 |
| pUL34–G77A | + | − | + | 30 |
| pUL34–V80A | + | + | ++ | 20 |
| pUL34–P89A | + | + | +++ | 40 |
| pUL34–N103A | + | + | − | 10 |
| pUL34–L116A | + | + | + | 20 |
| pUL34–K123A | + | + | ++ | 40 |
| pUL34–P132A | + | + | +++ | 80 |
| pUL34–R144A | + | + | ++ | 40 |
| pUL34–L167A | + | + | ++ | 40 |
| pUL34–L166/167A | + | + | − | 10 |
| Negative controla | NA | NA | − | 10 |
RK13 cells infected with PrV ΔUL34 served as a negative control. NA, not applicable.
Complementation is given as the CI: +++, CI ≥ 0.5; ++, 0.1 < CI < 0.3; +, 0.01 < CI < 0.09; −, CI < 0.009.
Data represent diameters as a percentage of that for PrV Ka, which was set to 100%.
Functional complementation by cell lines stably expressing mutated pUL34.
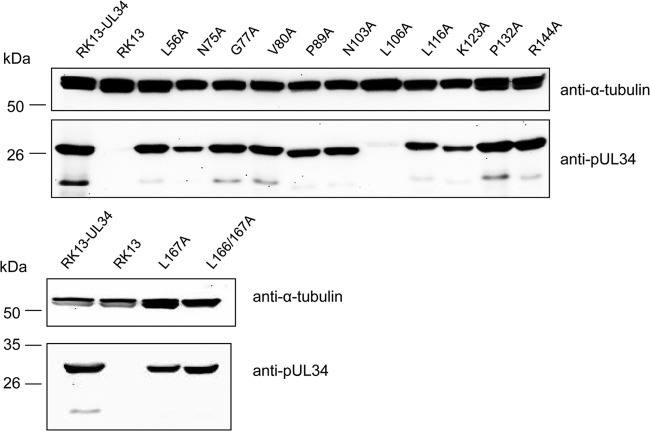
We next tested which of the pUL34 mutants were able to form a functional NEC and complement the defect of PrV-ΔUL34. To this end, cell lines stably expressing the mutated pUL34 were generated after transfection of RK13 cells and selection with G418. To verify correct protein expression, Western blot analysis was performed with cell lysates of the respective cell lines (Fig. 3). With the exception of RK13-UL34–L106A, all cell lines expressed pUL34 at levels comparable to the level of expression by RK13-UL34. Only a faint pUL34-specific signal was detectable in cell lysates of RK13-UL34–L106A, paralleling the results from transient-expression analyses. Therefore, this construct was excluded from further analyses.
FIG 3.
Western blot analysis of transgenic RK13 cell lines stably expressing mutated pUL34. Cell lysates of transgenic RK13 cell lines expressing the different pUL34 proteins were separated on an SDS-10% polyacrylamide gel, transferred onto a nitrocellulose membrane, and incubated with the polyclonal monospecific anti-pUL34 serum and monoclonal anti-α-tubulin antibody, which served as a loading control.
To test whether the pUL34 mutants were able to complement the defect of PrV-ΔUL34, the corresponding cell lines were infected with wild-type PrV-Ka or PrV-ΔUL34 at an MOI of 3, titers were determined after 24 h by titration on RK13-UL34 cells (Fig. 4), and the complementation index (CI) was calculated (Table 2). The proline substitution mutants pUL34–P89A and pUL34–P132A complemented PrV-ΔUL34 to titers similar to those of the wild type or decreased a maximum of 10-fold compared to the titer of the wild type. Infectious progeny titers derived from RK13-UL34–V80A, -K123A, -R144A, and -L167A cells were reduced approximately 50- to 100-fold. Progeny virus titers obtained from RK13-UL34–L56A and -L116A cells were even more drastically decreased, but they were still ca. 10-fold higher than those obtained from nontransfected RK13 cells, indicating a limited residual capacity for complementation. RK13-UL34–N75A, RK13-UL34–N103A, and RK13-UL34–L166/167A cells completely failed to produce titers above those from nontransgenic RK13 cells, demonstrating that they were nonfunctional. This was expected for RK13-UL34–N75A cells, since this protein failed to interact with pUL31. In contrast, despite a clear colocalization of pUL34–N103A with pUL31 (Fig. 2), the corresponding cell line failed to complement the defect of PrV-ΔUL34. On the other hand, viral progeny titers from infected RK13-UL34–G77A cells were only 50-fold lower than those from infected RK13-UL34 cells, although after transient expression, no interaction with pUL31 was detectable (Fig. 2). No significant difference in viral titers was found when PrV-Ka was propagated on the different pUL34 mutant-expressing cell lines, demonstrating that none of the mutant proteins exerted a dominant negative effect.
FIG 4.
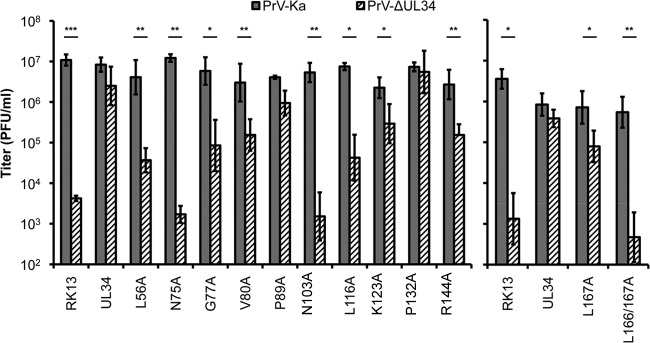
Functional complementation of RK13 pUL34-expressing cells. RK13 cells stably expressing the corresponding pUL34 mutants were infected with either PrV-Ka or PrV-ΔUL34 at an MOI of 3. Viral progeny was harvested at 24 h after infection, and the corresponding titers were determined by titration on RK13-UL34 cells. The mean values of three independent experiments and the corresponding standard deviations are given. Statistically significant differences between the titers of infection with PrV-Ka and PrV-ΔUL34 of each transgenic cell line are indicated: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
To investigate the phenotype of pUL34–G77A and pUL34–N103A in more detail, the respective cell lines, as well as RK13, RK13-UL34, and RK13-UL34–N75A cells as controls, were infected with PrV-ΔUL34/US3 and analyzed for colocalization of pUL31 and pUL34 by confocal microscopy (Fig. 5). A double deletion mutant, which was unable to express pUL34 and pUS3, was used to better visualize the formation of speckles which accumulate in the absence of the viral kinase pUS3 (16, 17). Colocalization of pUL34 and pUL31 in speckles was evident, as expected, after infection of RK13-UL34 cells but also after infection of RK13-UL34–G77A cells, indicating that complex formation between pUL34–G77A and pUL31 can occur during virus infection, probably by providing factors stabilizing the complex. Speckle formation and pUL31 colocalization were absent after infection of parental RK13 cells (data not shown) and RK13-UL34–N75A cells (Fig. 5). Unexpectedly, it was also missing in RK13-UL34–N103A cells, although cells with pUL34–N103A showed speckle formation and pUL34–N103A interacted with pUL31 after transient expression (compare Fig. 2 and 5).
FIG 5.
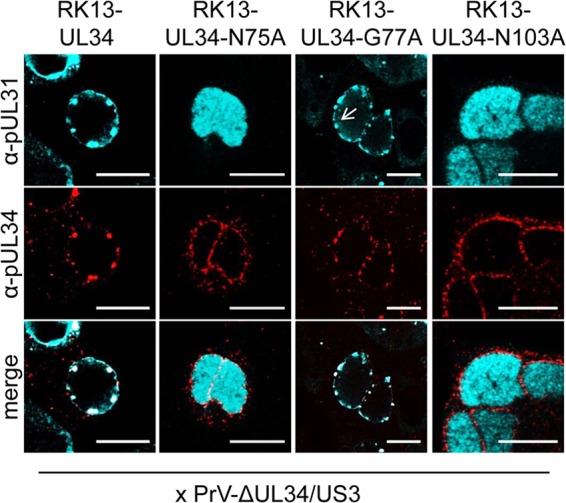
Colocalization of pUL31 and pUL34–N75A, pUL34–G77A, and pUL34–N103A in infected cells. To further investigate the divergent results obtained with pUL34–G77A and pUL34–N103A, the corresponding cell lines were infected with PrV-ΔUL34/US3 under plaque assay conditions. Two days after infection, cells were fixed, and colocalization of pUL31 and pUL34 was analyzed using monospecific pUL31 rabbit (turquoise) and pUL34 mouse (red) antisera. RK13-UL34 cells were used as a positive control, and RK13-UL34–N75A cells were used as a negative control. α, anti; arrow, speckle formation in infected RK13-UL34–G77A cells. Bars, 10 μm.
Ultrastructural analyses.
For further analysis, RK13-UL34 and RK13-UL34–N75A, -G77A, -N103A, -L116A, -P132A, -L167A, and -L166/167A cells were infected with PrV-ΔUL34 at an MOI of 1 and processed for transmission electron microscopy at 16 h after infection. Infected RK13-UL34 cells (Fig. 6A) served as a positive control. Immature capsid forms and DNA-filled nucleocapsids were observed in the nucleus as well as nucleocapsids in the cytosol and mature virions at the cell surface. The same was observed for infection of transgenic RK13-UL34–G77A, -L116A, -P132A, and -L167A cells (Fig. 6B to E). In contrast, RK13-UL34–N75A, -N103A, and -L166/167A cells (Fig. 6F to H) resembled nontransgenic RK13 cells, with capsids being present only in the nucleus. No budding stages at the INM or primary enveloped virions were observed in the perinuclear space, and capsids and virions were not observed in the cytosol or in the extracellular space, indicating that these pUL34 mutants do not support nuclear egress. Unfortunately, no differences between the pUL34 variants which were still able to interact with pUL31 (pUL34–N103A, pUL34–L166/167A) and the noninteracting mutant pUL34–N75A were evident in the ultrastructural analysis.
FIG 6.
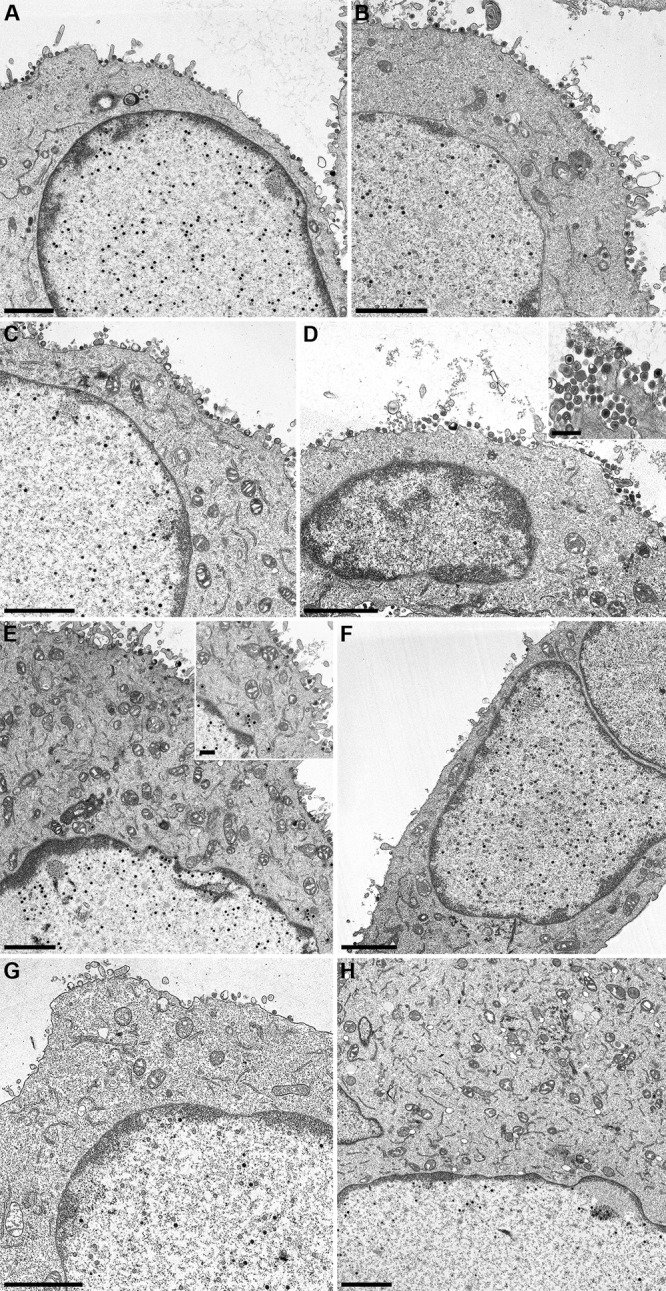
Ultrastructural analysis. Cell lines RK13-UL34 (A), RK13–G77A (B), RK13–L116A (C), RK13–P132A (D), RK13–L167A (E), RK13–N75A (F), RK13–N103A (G), and RK13–L166/167A (H) were infected with PrV-ΔUL34 at an MOI of 1 and processed for electron microscopy at 16 h after infection. Bars, 2 μm and 500 nm (insets).
Influence of pUL34 mutations on viral cell-to-cell spread.
Previous experiments on HSV-1 and PrV indicated that pUL34 might play a direct or indirect role in viral replication beyond nuclear egress (20, 22, 28). To investigate this further, transgenic cell lines expressing the mutated pUL34 constructs were infected with PrV-Ka and PrV-ΔUL34 under plaque assay conditions. Plaque diameters were measured microscopically 2 days after infection in three independent experiments and compared to those of PrV-Ka on the corresponding cell line. Except for RK13-UL34–P132A, the plaque diameters of PrV-ΔUL34 on all transgenic cells lines were 60 to 80% smaller than those of PrV-Ka (Fig. 7). This was not expected for mutants pUL34–V80A, -P89A, -K123A, and -R144A or mutants with cysteine substitutions, pUL34–C67A and pUL34–C141A, as shown previously (20), which replicated the UL34 deletion mutant to only slightly reduced viral titers compared with those for PrV-Ka. These data provide additional evidence that pUL34 plays a role in the efficiency of transfer of infectivity to neighboring cells independent of its role in nuclear egress. A summary of the results is shown in Table 2.
FIG 7.
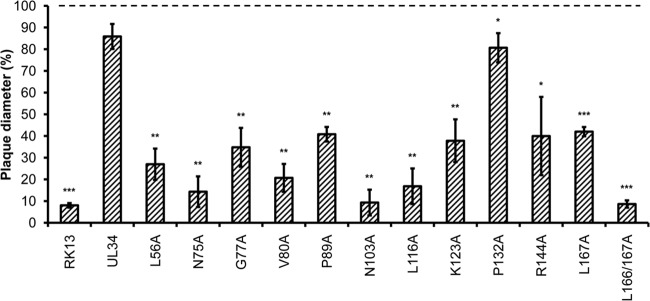
Complementation of direct cell-to-cell transmission. Direct viral cell-to-cell spread was analyzed by infecting the different pUL34-expressing RK13 cell lines with PrV-Ka or PrV-ΔUL34 under plaque assay conditions. After 2 days, samples were fixed and the diameters of 30 plaques of each cell line and virus were measured. Plaque diameters of PrV-Ka on a given cell line were set equal to 100%, and the diameters of the plaques induced by PrV-ΔUL34 were calculated accordingly. Values represent the means of three independent experiments. The corresponding standard deviations are indicated, and statistically significant differences of the infection with PrV-ΔUL34 compared to wild-type PrV-Ka infection of each transgenic cell line are indicated: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
DISCUSSION
In our previous study, we focused on three conserved cysteine residues which might influence the secondary structure of pUL34. However, mutations of these residues had only a moderate effect. In contrast, a highly conserved EY motif in PrV pUL34 was shown to be essential for NEC function, as had been demonstrated for the HCMV, MCMV, and HSV-1 homologs (10, 18, 19). We have now extended our mutational analysis to 13 additional conserved amino acids within the pUL31 interaction domain and the dileucine motif located outside but in the vicinity of the putative ER retention signal RQR, which was suggested to be relevant for correct targeting of pUL34 (20). All mutated pUL34 constructs were analyzed for interaction with pUL31 after transient transfection and for function after PrV-ΔUL34 infection of stably expressing cell lines.
All mutants described here localized to the nuclear rim, confirming that the N-terminal region does not seem to be involved in proper targeting. After cotransfection with a pUL31-expressing plasmid, most of the expressed mutant pUL34 proteins showed punctate, speckled structures at the nuclear rim comparable to those detectable after expression of the native proteins, indicating that the mutated amino acids are not necessary for pUL34 targeting, complex formation with pUL31, or, probably, the formation of NE-derived vesicles. Despite high expression levels, pUL34–N75A and pUL34–G77A failed to recruit coexpressed pUL31 to the NE. These residues are part of a conserved triad of amino acids which also includes a threonine residue (75-NTG-77). It might be speculated that this NTG motif and the essential upstream EY (53/54) motif form the major platform for pUL31 docking in PrV pUL34. pUL34–L106A did not show pUL31 recruitment either but was excluded from further analyses due to very low expression levels. In summary, 10 of the 13 tested PrV pUL34 mutants maintained an interaction with pUL31.
Six of the 10 pUL34 mutants which formed a complex with pUL31 after transient expression complemented the defect of the pUL34 deletion mutant to some extent. While RK13-UL34–P89A and -P132A complemented expression to wild-type levels, the titers of PrV-ΔUL34-infected RK13-UL34–V80A, -K123A, and -R144A cells were reduced a maximum of 100-fold, while those derived from RK13-UL34–L56A and -L116A cells were only slightly above the titers of infected RK13 cells. In contrast, RK13-UL34–N75A cells yielded very low titers similar to those of nontransgenic RK13 cells. Despite the use of high-resolution transmission electron microscopic images, no further conclusions on the different complementation abilities could be drawn. Surprisingly, pUL34–N103A was unable to complement the defect of PrV-ΔUL34, despite speckle formation indicative of pUL31 interaction after cotransfection. Since no pUL34/pUL31-positive speckles were found after infection of pUL34–N103A-expressing cells with PrV-ΔUL34/US3 and no cytosolic nucleocapsids or mature virions could be detected in ultrastructural analyses, the pUL34–N103A/pUL31 complex may either not form or be unstable during infection. In contrast, complex formation could not be observed after transient coexpression of pUL34–G77A with pUL31, whereas the cell line stably expressing pUL34–G77A complemented the defect of PrV-ΔUL34 to titers only approximately 50-fold lower than those of PrV-Ka, and nucleocapsids as well as mature virions were detectable in the cytosol in electron microscopic images. These data indicate that NEC formation may be influenced by other viral components besides the core NEC constituents.
While amino acids corresponding to P89 and K123 of PrV pUL34 were also found to be dispensable in HCMV UL50 (19), the roles of L116 and R144 were divergent. Whereas the residue corresponding to L116 could also be replaced in HCMV M50 (19), insertion of a linker in MCMV M50 at a site immediately preceding this residue resulted in a nonfunctional protein (10). It should be considered, however, that insertion of 6 amino acids by the transposon linker sequence might influence the protein structure more dramatically than a single amino acid substitution, and the results are therefore difficult to compare. On the other hand, the arginine corresponding to PrV pUL34–R144 was found to be exchangeable in HCMV M50 (19) and also had no relevance for pUL31 interaction or complementation in PrV (this study), while a charge cluster mutation including this residue (RMPR/AMPA; CL13) abrogated complementation by HSV-1 pUL34 (18). However, it was not further investigated whether a single alanine substitution would also result in a loss of function.
Since it has been proposed that pUL34 plays an additional role beyond nuclear egress affecting direct viral cell-to-cell spread (20, 22, 28), the transgenic pUL34 cell lines generated in this study were infected with PrV-Ka and PrV-ΔUL34, and plaque diameters were determined 2 days after infection. Surprisingly, except for pUL34–P132A, all tested cell lines supported cell-to-cell transmission of PrV-ΔUL34 with decreased efficiency. This also applied to those transgenic cell lines which complemented the production of infectious progeny of PrV-ΔUL34 rather efficiently. Although it has been proposed that HSV-1 pUL34 may influence targeting of gE to cell junctions, which might be a prerequisite for efficient cell-to-cell spread (28), this could not be shown for PrV (data not shown). Unfortunately, transmission electron microscopy did not provide further evidence of this defect. Therefore, it remains unclear which factors are involved or influenced directly or indirectly by pUL34 in direct herpesvirus cell-to-cell spread.
The dileucine motif L166/167 was speculated to be part of an arginine-based ER sorting motif (RQR) (20). Therefore, we mutated either the more highly conserved L167 or both leucine residues to alanine. Although a point mutation of RQR to RQG resulted in pUL34 accumulation in the Golgi apparatus (20), localization of pUL34–L167A or -L166/167A was indistinguishable from that of native pUL34, indicating that these residues are not required for nuclear rim localization or ER retention. Moreover, speckles could be observed after cotransfection with pUL31, correlating with the location of both residues outside the pUL31 interaction domain. Nevertheless, mutation of both leucine residues had a dramatic effect on NEC function, and RK13-UL34–L166/L167A cells completely failed to complement the UL34 deletion mutant, while the single mutation L167 had no effect. Therefore, this motif is apparently not involved in the proper translocation of pUL34 to the INM, as was previously hypothesized (20), but, rather, plays an important role in another step, e.g., in capsid recruitment or interaction with other factors involved in nuclear egress. Whether the L166A substitution is sufficient for this effect needs to be tested. Interestingly, both leucine residues are also present in MCMV and HCMV pUL34 homologs (10, 19), and a linker insertion at the corresponding site in MCMV UL50 also resulted in a nonfunctional protein. Thus, the location of the dileucine motif seems to separate the essential N-terminal pUL31 interaction domain from the replaceable C-terminal anchor domain.
In summary, our mutational analyses provide additional information on the role of different conserved amino acid residues of pUL34 in NEC formation and function. Together with the elucidation of the NEC structure, these data are crucial to further define the structure-function relations within the NEC and contribute to understanding vesicle-mediated nuclear egress.
ACKNOWLEDGMENTS
This study was supported by the Deutsche Forschungsgemeinschaft (DFG Me 854/12-1).
We thank Cindy Meinke and Petra Meyer for technical help and Mandy Jörn for photographical assistance.
Footnotes
Published ahead of print 19 March 2014
REFERENCES
- 1.Mettenleiter TC. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537–1547. 10.1128/JVI.76.4.1537-1547.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mettenleiter TC, Müller F, Granzow H, Klupp BG. 2013. The way out: what we know and do not know about herpesvirus nuclear egress. Cell. Microbiol. 15:170–178. 10.1111/cmi.12044 [DOI] [PubMed] [Google Scholar]
- 3.Speese SD, Ashley J, Jokhi V, Nunnari J, Barria R, Li Y, Ataman B, Koon A, Chang YT, Li Q, Moore MJ, Budnik V. 2012. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 149:832–846. 10.1016/j.cell.2012.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hatch EM, Hetzer MW. 2012. RNP export by nuclear envelope budding. Cell 149:733–735. 10.1016/j.cell.2012.04.018 [DOI] [PubMed] [Google Scholar]
- 5.Klupp BG, Granzow H, Mettenleiter TC. 2000. Primary envelopment of pseudorabies virus at the nuclear membrane requires the UL34 gene product. J. Virol. 74:10063–10073. 10.1128/JVI.74.21.10063-10073.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roller RJ, Zhou Y, Schnetzer R, Ferguson J, DeSalvo D. 2000. Herpes simplex virus type 1 U(L)34 gene product is required for viral envelopment. J. Virol. 74:117–129. 10.1128/JVI.74.1.117-129.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reynolds AE, Ryckman BJ, Baines JD, Zhou Y, Liang L, Roller RJ. 2001. U(L)31 and U(L)34 proteins of herpes simplex virus type 1 form a complex that accumulates at the nuclear rim and is required for envelopment of nucleocapsids. J. Virol. 75:8803–8817. 10.1128/JVI.75.18.8803-8817.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs W, Klupp BG, Granzow H, Osterrieder N, Mettenleiter TC. 2002. The interacting UL31 and UL34 gene products of pseudorabies virus are involved in egress from the host-cell nucleus and represent components of primary enveloped but not mature virions. J. Virol. 76:364–378. 10.1128/JVI.76.1.364-378.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neubauer A, Rudolph J, Brandmuller C, Just FT, Osterrieder N. 2002. The equine herpesvirus 1 UL34 gene product is involved in an early step in virus egress and can be efficiently replaced by a UL34-GFP fusion protein. Virology 300:189–204. 10.1006/viro.2002.1488 [DOI] [PubMed] [Google Scholar]
- 10.Bubeck A, Wagner M, Ruzsics Z, Lotzerich M, Iglesias M, Singh IR, Koszinowski UH. 2004. Comprehensive mutational analysis of a herpesvirus gene in the viral genome context reveals a region essential for virus replication. J. Virol. 78:8026–8035. 10.1128/JVI.78.15.8026-8035.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Popa M, Ruzsics Z, Lotzerich M, Dolken L, Buser C, Walther P, Koszinowski UH. 2010. Dominant negative mutants of the murine cytomegalovirus M53 gene block nuclear egress and inhibit capsid maturation. J. Virol. 84:9035–9046. 10.1128/JVI.00681-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mettenleiter TC, Klupp BG, Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234. 10.1016/j.virusres.2009.03.018 [DOI] [PubMed] [Google Scholar]
- 13.Johnson DC, Baines JD. 2011. Herpesviruses remodel host membranes for virus egress. Nat. Rev. Microbiol. 9:382–394. 10.1038/nrmicro2559 [DOI] [PubMed] [Google Scholar]
- 14.Klupp BG, Granzow H, Fuchs W, Keil GM, Finke S, Mettenleiter TC. 2007. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. U. S. A. 104:7241–7246. 10.1073/pnas.0701757104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desai PJ, Pryce EN, Henson BW, Luitweiler EM, Cothran J. 2012. Reconstitution of the Kaposi's sarcoma-associated herpesvirus nuclear egress complex and formation of nuclear membrane vesicles by coexpression of ORF67 and ORF69 gene products. J. Virol. 86:594–598. 10.1128/JVI.05988-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klupp BG, Granzow H, Mettenleiter TC. 2001. Effect of the pseudorabies virus US3 protein on nuclear membrane localization of the UL34 protein and virus egress from the nucleus. J. Gen. Virol. 82:2363–2371 http://vir.sgmjournals.org/content/82/10/2363.long [DOI] [PubMed] [Google Scholar]
- 17.Reynolds AE, Wills EG, Roller RJ, Ryckman BJ, Baines JD. 2002. Ultrastructural localization of the herpes simplex virus type 1 UL31, UL34, and US3 proteins suggests specific roles in primary envelopment and egress of nucleocapsids. J. Virol. 76:8939–8952. 10.1128/JVI.76.17.8939-8952.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bjerke SL, Cowan JM, Kerr JK, Reynolds AE, Baines JD, Roller RJ. 2003. Effects of charged cluster mutations on the function of herpes simplex virus type 1 UL34 protein. J. Virol. 77:7601–7610. 10.1128/JVI.77.13.7601-7610.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Milbradt J, Auerochs S, Sevvana M, Muller YA, Sticht H, Marschall M. 2012. Specific residues of a conserved domain in the N terminus of the human cytomegalovirus pUL50 protein determine its intranuclear interaction with pUL53. J. Biol. Chem. 287:24004–24016. 10.1074/jbc.M111.331207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paßvogel L, Trübe P, Schuster F, Klupp BG, Mettenleiter TC. 2013. Mapping of sequences in pseudorabies virus pUL34 that are required for formation and function of the nuclear egress complex. J. Virol. 87:4475–4485. 10.1128/JVI.00021-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ott M, Tascher G, Hassdenteufel S, Zimmermann R, Haas J, Bailer SM. 2011. Functional characterization of the essential tail anchor of the herpes simplex virus type 1 nuclear egress protein pUL34. J. Gen. Virol. 92:2734–2745. 10.1099/vir.0.032730-0 [DOI] [PubMed] [Google Scholar]
- 22.Schuster F, Klupp BG, Granzow H, Mettenleiter TC. 2012. Structural determinants for nuclear envelope localization and function of pseudorabies virus pUL34. J. Virol. 86:2079–2088. 10.1128/JVI.05484-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sam MD, Evans BT, Coen DM, Hogle JM. 2009. Biochemical, biophysical, and mutational analyses of subunit interactions of the human cytomegalovirus nuclear egress complex. J. Virol. 83:2996–3006. 10.1128/JVI.02441-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roller RJ, Bjerke SL, Haugo AC, Hanson S. 2010. Analysis of a charge cluster mutation of herpes simplex virus type 1 UL34 and its extragenic suppressor suggests a novel interaction between pUL34 and pUL31 that is necessary for membrane curvature around capsids. J. Virol. 84:3921–3934. 10.1128/JVI.01638-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer GA, Radsak KD. 2000. Identification of a novel signal sequence that targets transmembrane proteins to the nuclear envelope inner membrane. J. Biol. Chem. 275:3857–3866. 10.1074/jbc.275.6.3857 [DOI] [PubMed] [Google Scholar]
- 26.Meyer G, Gicklhorn D, Strive T, Radsak K, Eickmann M. 2002. A three-residue signal confers localization of a reporter protein in the inner nuclear membrane. Biochem. Biophys. Res. Commun. 291:966–971. 10.1006/bbrc.2002.6563 [DOI] [PubMed] [Google Scholar]
- 27.Michelsen K, Yuan H, Schwappach B. 2005. Hide and run. Arginine-based endoplasmic-reticulum-sorting motifs in the assembly of heteromultimeric membrane proteins. EMBO Rep. 6:717–722. 10.1038/sj.embor.7400480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haugo AC, Szpara ML, Parsons L, Enquist LW, Roller RJ. 2011. Herpes simplex virus 1 pUL34 plays a critical role in cell-to-cell spread of virus in addition to its role in virus replication. J. Virol. 85:7203–7215. 10.1128/JVI.00262-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaplan AS, Vatter AE. 1959. A comparison of herpes simplex and pseudorabies viruses. Virology 7:394–407. 10.1016/0042-6822(59)90068-6 [DOI] [PubMed] [Google Scholar]
- 30.Graham FL, van der Eb AJ. 1973. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52:456–467. 10.1016/0042-6822(73)90341-3 [DOI] [PubMed] [Google Scholar]
- 31.Mettenleiter TC. 1989. Glycoprotein gIII deletion mutants of pseudorabies virus are impaired in virus entry. Virology 171:623–625. 10.1016/0042-6822(89)90635-1 [DOI] [PubMed] [Google Scholar]