ABSTRACT
Like most positive-strand RNA viruses, infection by plant tombusviruses results in extensive rearrangement of specific host cell organelle membranes that serve as the sites of viral replication. The tombusvirus Tomato bushy stunt virus (TBSV) replicates within spherules derived from the peroxisomal boundary membrane, a process that involves the coordinated action of various viral and cellular factors, including constituents of the endosomal sorting complex required for transport (ESCRT). ESCRT is comprised of a series of protein subcomplexes (i.e., ESCRT-0 -I, -II, and -III) that normally participate in late endosome biogenesis and some of which are also hijacked by certain enveloped retroviruses (e.g., HIV) for viral budding from the plasma membrane. Here we show that the replication of Carnation Italian ringspot virus (CIRV), a tombusvirus that replicates at mitochondrial membranes also relies on ESCRT. In plant cells, CIRV recruits the ESCRT-I protein, Vps23, to mitochondria through an interaction that involves a unique region in the N terminus of the p36 replicase-associated protein that is not conserved in TBSV or other peroxisome-targeted tombusviruses. The interaction between p36 and Vps23 also involves the Vps23 C-terminal steadiness box domain and not its N-terminal ubiquitin E2 variant domain, which in the case of TBSV (and enveloped retroviruses) mediates the interaction with ESCRT. Overall, these results provide evidence that CIRV uses a unique N-terminal sequence for the recruitment of Vps23 that is distinct from those used by TBSV and certain mammalian viruses for ESCRT recruitment. Characterization of this novel interaction with Vps23 contributes to our understanding of how CIRV may have evolved to exploit key differences in the plant ESCRT machinery.
IMPORTANCE Positive-strand RNA viruses replicate their genomes in association with specific host cell membranes. To accomplish this, cellular components responsible for membrane biogenesis and modeling are appropriated by viral proteins and redirected to assemble membrane-bound viral replicase complexes. The diverse pathways leading to the formation of these replication structures are poorly understood. We have determined that the cellular ESCRT system that is normally responsible for mediating late endosome biogenesis is also involved in the replication of the tombusvirus Carnation Italian ringspot virus (CIRV) at mitochondria. Notably, CIRV recruits ESCRT to the mitochondrial outer membrane via an interaction between a unique motif in the viral protein p36 and the ESCRT component Vps23. Our findings provide new insights into tombusvirus replication and the virus-induced remodeling of plant intracellular membranes, as well as normal ESCRT assembly in plants.
INTRODUCTION
Tombusviruses are positive-strand RNA [(+)RNA] viruses that infect a wide range of plant species and replicate at host cell membranes derived specifically from either peroxisomes (e.g., Tomato bushy stunt virus [TBSV]) or mitochondria (e.g., Carnation Italian ringspot virus [CIRV]) (1). Upon infection and depending on the tombusvirus, the peroxisomal or mitochondrial (outer) membranes progressively proliferate and invaginate, resulting in the formation of hundreds of spherules that serve to concentrate viral and host cell factors required for synthesis of the viral RNA genome and to protect nascent viral RNAs from degradation by host cell defenses (2, 3). Concomitant with these morphological changes, the modified organelles also form large appendages and coalesce, yielding aggregated structures that no longer resemble the organelles from which they were derived (1, 4).
The morphological transformation of peroxisomes or mitochondria in tombusvirus-infected cells involves two viral replication proteins: an auxiliary viral RNA-binding protein and an RNA-dependent RNA polymerase, referred to as p33 and p92, respectively, in TBSV, or p36 and p95, respectively, in CIRV (5). Both sets of replicase proteins are essential for viral genome replication (6, 7) and are encoded by overlapping open reading frames (ORFs), and p92 and p95 are products of translational read-through of an amber stop codon in p33 and p36, respectively (8, 9). Consequently, the N-terminal portion of p92/p95 is identical to p33/p36. Both sets of replicase proteins are also integral membrane proteins, each possessing two transmembrane domains (TMDs), as well as unique targeting signals that mediate their specific sorting to either peroxisomes or mitochondria (4, 10, 11) and thus dictate the intracellular site for viral replication.
Numerous host cell factors involved in tombusvirus replication have been identified as part of several large-scale genomic and proteomic studies performed with TBSV and Saccharomyces cerevisiae as a model host (12). Among these factors are several components of endosomal sorting complex required for transport (ESCRT). ESCRT is a network of ∼20 soluble proteins that, in noninfected cells, are sequentially recruited from the cytosol and assembled into several multiprotein subcomplexes (ESCRT-0, -I, -II, and -III) at the late endosomal surface, where they participate in sorting of ubiquitinated membrane-bound cargo proteins into intraluminal vesicles derived from the endosomal boundary membrane during multivesicular body (MVB) biogenesis. According to models based primarily on studies with yeasts and mammalian cells (13), ESCRT assembly begins with ESCRT-0 recognizing ubiquitinated cargo in the endosomal membrane and recruiting ESCRT-I to the endosomal surface. ESCRT-I also participates in ubiquitinated cargo sorting and recruits ESCRT-II, which subsequently recruits ESCRT-III, which forms polymeric filaments that drive membrane vesiculation. Thereafter, the AAA-ATPase Vps4 (vacuolar protein sorting 4) catalyzes the disassembly of ESCRT-III, a process that is coupled with membrane fission and results in the recycling of ESCRT(III) subunits to the cytosol, while the mature MVB fuses with the lysosome/vacuole, where its contents are degraded. Interestingly, while the ESCRT machinery and its interaction network are relatively well conserved and have similar cellular functions in plants compared to yeasts and mammals (14, 15), some key differences exist, which are not well understood. For example, homologs of mammalian and yeast ESCRT-0 do not exist in plants (16), and instead, recent work has identified the TOL family of proteins to be involved in early recognition of ubiquitinated cargo and their sorting to MVBs (17). However, the relationship between cargo recognition by TOL proteins and subsequent ESCRT recruitment and assembly has yet to be investigated.
In addition to MVB biogenesis, certain ESCRT components also participate in other important cellular processes involving membrane deformation, including scission of the midbody during cytokinesis or budding of enveloped retroviruses (18, 19). In cells infected with human immunodeficiency virus (HIV), for example, ESCRT is initially redirected or “hijacked” to the plasma membrane via interactions between peptide late-domain motifs in the HIV structural protein, Gag p6, and the ESCRT-I protein Tsg101 and/or the ESCRT accessory protein Alix (20). Among the most common and best characterized of these late-domain motifs is a proline-rich sequence (i.e., P[S/T]AP), which interacts with the ubiquitin E2 variant (UEV) domain of Tsg101 via a binding mechanism that mimics the interaction of certain ESCRT-0 proteins with Tsg101 (21, 22).
Analogous to retroviral budding, the proposed model for the role of ESCRT during TBSV infection is that ESCRT is recruited to the surface of the peroxisome to facilitate invagination of the peroxisomal membrane, as well as to concentrate and assemble the viral replicase complexes within nascent spherules (23). Evidence in support of this model includes the identification of several ESCRT proteins in a genome-wide screen for factors involved in TBSV replication in yeast (24) and the inhibition of TBSV replicase activity in yeast and plant cells upon overexpression of mutant versions of various ESCRT proteins (23). In addition, coexpression of the TBSV replicase protein p33 and the yeast ortholog of mammalian Tsg101, Vps23p, in yeast cells results in the relocalization of Vps23p to peroxisomes (23). TBSV p33 also interacts with yeast Vps23p in a manner that depends on the N-terminal UEV domain of Vps23p and two (mono)ubiquitinated lysine residues in p33, as well as a peptide sequence in p33 (i.e., PSVP) that resembles the PSAP late-domain motif of HIV Gag p6 (25).
Given the ultrastructural similarities of the modified peroxisomal and mitochondrial membranes in TBSV- and CIRV-infected cells, respectively, as well as the ability of chimeric versions of these two viruses to produce the corresponding organelle-specific membrane rearrangements (5, 11, 26), it seems likely that CIRV relies on ESCRT in a manner similar to TBSV. However, CIRV p36 does not possess a late-domain-like motif resembling that identified in TBSV p33 (Fig. 1) (23) (and in mammalian enveloped retroviruses [20]), implying that CIRV uses a distinct mechanism to recruit ESCRT to mitochondrial membranes. Consistent with this premise, in this study, we show that CIRV p36 binds to and recruits Vps23 to mitochondria in plant cells via a unique N-terminal sequence in p36 that is not present in TBSV p33. We show also that, in contrast to TBSV p33 (and mammalian enveloped retroviruses), the interaction between p36 and Vps23 does not require the UEV domain of Vps23 but, instead, requires the C-terminal steadiness box domain (StBox) of Vps23, which in yeasts and mammals is important for the assembly of ESCRT-I (27–30).
FIG 1.
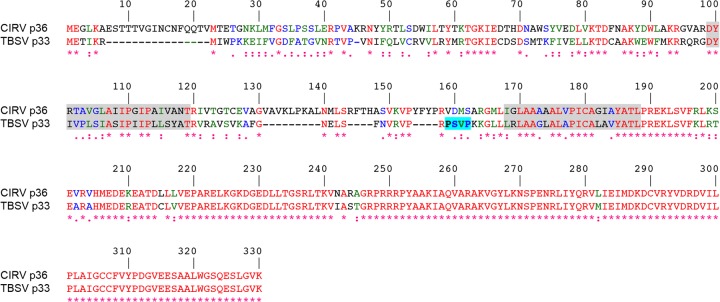
Alignment of the deduced amino acid sequences of CIRV p36 and TBSV p33. Sequences were obtained from GenBank (accession numbers CAA59477.2 and NP_062898.1) and aligned using ClustalW. Identical and similar amino acids in each protein are colored red and green or blue, respectively, and indicated also with asterisks and colons or periods, respectively. The numbers represent specific amino acid residues in full-length p36 (330 residues). Putative TMDs (shown on a gray background) were determined using TOPCONS and visual inspection, and the late-domain-like motif identified in the intervening loop sequence of p33 (23) but absent in p36 is shown on a bright blue background.
Collectively, our results highlight a unique mechanism for ESCRT recruitment by CIRV that utilizes a Vps23-interacting motif that appears to be distinct from other viruses that co-opt ESCRT, such as TBSV and HIV. This novel mechanism may be reflective of the absence of an ESCRT-0 complex, which raises new mechanistic questions about general features of ESCRT assembly in plants.
MATERIALS AND METHODS
Recombinant DNA procedures and reagents.
Molecular biology reagents were purchased either from New England BioLabs, Promega, PerkinElmer Life Sciences Inc., Stratagene, or Invitrogen. Custom oligonucleotides were synthesized by Sigma-Aldrich Ltd. All DNA constructs were verified using automated sequencing performed at the University of Guelph Genomics Facility. PCR-based mutagenesis was carried out using the QuikChange site-directed mutagenesis kit (Stratagene).
Plasmid construction.
The majority of the plasmids used in this study have been described previously, including the following binary plasmids used in Agrobacterium infiltration and rub inoculation of Nicotiana benthamiana leaves: pHST20/TBSV and pHST20/CIRV, encoding the entire TBSV and CIRV genomes, respectively (8, 31); pTRV1 and pTRV2, encoding RNA1 and RNA2 of the bipartite Tobacco rattle virus (TRV) genome (32), which were obtained from the Arabidopsis Biological Resource Center (ABRC); pMO4-AtSKD1[E232Q], encoding the green fluorescent protein (GFP) fused to the N terminus of a mutant version (i.e., a glutamic acid at position 232 replaced with glutamine) of Arabidopsis thaliana Vps4 (also referred to as SKD1 [33]), which was kindly provided by M. Otegui (University of Wisconsin); and pRCS2, serving as an “empty” binary vector control (34). All binary plasmids, as well as all other plant expression plasmids based on pRTL2, pSAT, and pUC18 vectors (see below), contain the cauliflower mosaic virus 35S promoter (CaMV 35S).
Plant expression plasmids used in transient transformations of Nicotiana tabacum Bright Yellow-2 (BY-2) cells include the following plasmids that have been described previously (11, 35): pRTL2/p36, encoding CIRV p36; pRTL2/Myc-p36, encoding p36 with an N-terminal Myc epitope tag; pRTL2/p95, encoding CIRV p95, in which the p36 amber stop codon was mutated to a tyrosine codon; pRTL2/Rep, encoding the overlapping CIRV open reading frame 1 (ORF1) and ORF2 that encode both p36 and p95; p361-90-CAT and p3690-190-CAT, encoding the N-terminal 90 or 90 to 190 amino acid residues of p36 fused to the N terminus of chloramphenicol acetyltransferase (CAT), respectively; pRTL2/Myc-Vps23, pRTL2/GFP-Vps23, pUC18/Vps23-GFP, and pRTL2/HA-Vps23, encoding either the Myc or hemagglutinin (HA) epitope tag or the green fluorescent protein fused to the N or C terminus of isoform A of Arabidopsis Vps23 (referred to as Vps23 in this study); pRLT2/Myc-Vps28 and pRTL2/Myc-Vps25, encoding N-terminal Myc-tagged Arabidopsis Vps28 isoform A and Arabidopsis Vps25, respectively. Other previously described plant expression vectors include the following plasmids: pRTL2/RFP, encoding the red fluorescent protein (RFP) (36); pUC18/GFP-Syp21 and pUC18/GFP-Syp52, which encode GFP fused to the N termini of the Arabidopsis membrane-bound Qa-SNAREs (soluble N-ethylmaleimide-sensitive factor attachment protein receptors) Syp21 (Syntaxin of plants 21) and Syp52 (37) proteins (kindly provided by M. Sato [Kyoto University]); pSAT2/Cherry-PTS1, encoding the monomeric cherry fluorescent protein fused to the C-terminal 10 amino acid residues of pumpkin hydroxypyruvate reductase, including its type 1 peroxisomal targeting signal (38); pRTL2/TIC40-RFP, encoding the Arabidopsis 40-kDa component of the translocon at the inner membrane of chloroplasts fused to the N terminus of RFP (39); and pSAT4A/AtPAP26-mCherry, encoding the Arabidopsis purple acid phosphatase isoform 26 fused to the N terminus of the monomeric cherry fluorescent protein (40). Yeast two-hybrid expression vectors, pGADT7 and pGBKT7 (Clontech) encoding Arabidopsis Vps23, Vps25, Vps37, and Vps28 were also previously described (35).
Plasmids used for bimolecular fluorescence complementation (BiFC) assays were based on pSAT4/nEYFP and pSAT4/cEFYP, which encode the N-terminal and C-terminal halves of the enhanced yellow fluorescent protein (nEYFP and cEYFP), respectively (kindly provided by S. Gelvin [Purdue University]) (41). Briefly, pSAT4/p36-cYFP, p3691-330-cYFP, and pSAT4/nYFP-Vps23 were constructed by PCR amplifying the p36 or Vps23 open reading frames (ORFs), or portions thereof, and cloned into pSAT4/nEFYP or pSAT4/cEFYP. Complete details on the oligonucleotide primers used for generating these and other plasmids are available upon request.
Plant expression vectors encoding modified (truncated) versions of p36 (i.e., p3691-330, p3668-330, and p3623-330), Vps23 (GFP-UEV, Myc-CC+StBox [CC stands for coiled-coil, and StBox stands for steadiness box], Myc-CC, Myc-StBox, and Myc-UEV+CC) were generated by PCR amplification of the sequence corresponding to the indicated proteins or protein domains and ligated into either pRTL2/MCS (containing a multiple cloning site [MCS]) (42), pRTL2/Myc-MCS (encoding an initiation methionine, followed by the Myc epitope tag and then an MCS), pRTL2/GFP-MCS (encoding GFP followed by an MCS) (14), or pUC18/NheI-GFP (encoding GFP with a 5′ unique NheI restriction site) (43). Similarly, yeast two-hybrid expression vectors encoding p36 or p361-90 or modified (truncated) versions of Vps23 (UEV, CC+StBox, CC, StBox, and UEV+CC) were generated by PCR amplification and subcloned into pGBKT7 (encoding the GAL4 DNA-binding domain [BD] and a Myc epitope tag, followed by an MCS) or pGADT7 (encoding the GAL4 activation domain [AD] and an HA epitope tag, followed by an MCS), respectively.
For construction of pRTL2/Myc-NtVps23, the 3′ sequence of VPS23 from Nicotiana tabacum was obtained by 3′ rapid amplification of cDNA ends (RACE)-PCR using cDNA synthesized from N. tabacum suspension cell mRNA, and a gene-specific forward primer based on the available N. tabacum VPS23 expressed sequence tag (EST) (NCBI accession number EB680173). Subsequently, full-length NtVPS23 was amplified from BY-2 cDNA using gene-specific primers and ligated into pRTL2/Myc-MCS (via pCR2.1TOPO serving as a shuttle vector), yielding pRTL2/Myc-NtVps23. pRTL2/Myc-NtVps23 was then used as the template DNA for constructing pRTL2-Myc/NtUEV, which includes the entire UEV domain of N. tabacum Vps23.
pRTL2/p36Δ7-22 was constructed using PCR-based site-directed mutagenesis, whereby sequences encoding amino acid residues 7 to 22 in the p36 ORF were deleted using the appropriate forward and reverse mutagenic primers and pRTL2/p36 as the template DNA. pRTL2/p331-74-p3691-330 and pRTL2/p361-28-p3313-74-p3691-330 were constructed in the following manner. First, sequences encoding the N-terminal 74 amino acid residues of p33 were amplified from pRTL2/p33 (4), and the resulting PCR products were ligated into pRTL2/p3691-330, yielding pRTL2/p331-74-p3691-330. Next, sequences encoding the N-terminal 12 amino acid residues of p33 were replaced (via successive site-directed mutagenesis reactions and pRTL2/p331-74-p3691-330 as the initial template DNA) with sequences encoding the N-terminal 28 residues of p36, yielding pRTL2/p361-28-p3313-74-p3691-330. pRTL2/Myc-p361-90-TraB and pRTL2/Myc-p361-28-TraB, encoding an N-terminal Myc epitope tag fused to the N-terminal 90 or 28 amino acids of p36 followed by the C-terminal 34 or 75 amino acid residues of Arabidopsis TraB (At1g05270), respectively, were constructed by first introducing (using PCR-based site-directed mutagenesis and pRTL2/Myc-TraB as the template DNA) an NheI site immediately upstream of either codon 338 or codon 297 (of 371) in the TraB ORF. The modified plasmids were then digested with NheI (removing sequences coding for residues 1 to 337 or 1 to 296 of TraB and the N-terminal Myc epitope tag) followed by ligation with NheI-digested PCR products encoding the N-terminal 90 or 28 residues of p36, along with an appended Myc epitope tag from pRTL2/Myc-p36 (see above). pRTL2/Myc-TraB was constructed by ligating the full-length TraB ORF (cDNA provided by the ABRC) into pRTL2/Myc-MCS.
Agrobacterium infiltration and rub inoculation of N. benthamiana and RNA gel blot analysis.
N. benthamiana plants were grown in chambers at 21°C with a 16-h/8-h light/dark cycle. The leaves of approximately 3-week-old plants were infiltrated or, for experiments involving TRV, triple infiltrated with cultures of Agrobacterium tumefaciens (LBA4404) carrying the appropriate binary vectors. Procedures involving Agrobacterium have been described previously (4). Rub inoculations were performed 2 days after infiltration with 10 μg of plasmid DNA encoding full-length infectious TBSV or CIRV cDNA diluted in 30 μl of RNA inoculation buffer (11, 44). Approximately 4 to 6 days after inoculation or, for experiments involving TRV, 2 or 4 days after infiltration, the leaves were flash frozen in liquid nitrogen followed by total RNA extraction (45). Aliquots of isolated RNA were separated in nondenaturing 1.2% (wt/vol) agarose gels, and viral RNAs were detected by electrophoretic transfer to nylon (Hybond-N; Amersham Biosciences) followed by incubation with 32P-end-labeled oligonucleotide probes complementary to the CIRV and TBSV genome or TRV RNA1 genome (45, 46). Complete details of oligonucleotides used for RNA gel blot analysis are available upon request. Labeled RNAs were visualized using a phosphorimager. Results presented are representative of at least two separate experiments.
Biolistic bombardment and fluorescence microscopy of BY-2 cells.
N. tabacum Bright Yellow-2 suspension cell cultures were maintained and prepared for biolistic bombardment with a Biolistic PDS-1000/He particle delivery system (Bio-Rad Laboratories) as described previously (47). Bombarded cells were incubated for ∼4 to 8 h to allow for expression and sorting of the introduced gene product(s). Cells were fixed in 4% (wt/vol) formaldehyde, followed by permeabilization with 0.01% (wt/vol) pectolyase Y-23 (Kyowa Chemical Products) and either 0.3% (vol/vol) Triton X-100, which permeabilizes the plasma membrane and all organellar membranes, or 25 μg/ml digitonin, which permeabilizes only the plasma membrane. Details on the differential detergent permeabilization of BY-2 cells have been previously described (48).
Primary and dye-conjugated secondary antibodies used for immunofluorescence staining of cells and their sources were as follows: rabbit anti-Myc IgGs and mouse anti-hemagglutinin (anti-HA) IgGs (Bethyl Laboratories); mouse anti-Myc antibodies in hybridoma medium (Princeton University, Monoclonal Antibody Facility); mouse anti-α-tubulin (Sigma-Aldrich Ltd.); mouse anti-CAT antibodies in hybridoma medium (provided by S. Subramani); mouse anti-maize β-ATPase antibodies in hybridoma medium (49); rabbit anti-cytochrome c oxidase subunit II (CoxII) IgGs (50); rabbit anti-p36 IgGs raised against a synthetic peptide corresponding to an amino acid sequence either in the C terminus of p36 (residues 218 to 237 [4] or the intervening loop [residues 147 to 160]) of p36 (Cedarlane Laboratories); goat anti-mouse and goat anti-rabbit Alexa Fluor 488 IgGs (Molecular Probes); and goat anti-rabbit rhodamine red-X IgGs (Jackson ImmunoResearch Laboratories).
Microscopic images of cells were acquired using an Axioscope 2 MOT epifluorescence microscope (Carl Zeiss Inc.) or a Leica DM RBE microscope. Figure compositions and merged images were generated using Openlab (Improvision) or Northern Eclipse (Empix Imaging Inc.) software and Adobe Photoshop CS (Adobe Systems). Images presented in all figures are representative of the results obtained from analyzing ≥25 independently transformed cells from at least three separate experiments. Colocalization of proteins was quantified using the ImageJ plugin “Co-localization Finder” and methods as described previously (51). Pearson's correlation coefficient r values of −1.0 to 1.0 are considered to be equivalent to all of the pixels from the regions of interest within the individual red and green channels of the images being 100% noncolocalized to 100% colocalized, respectively.
BiFC assays.
Bimolecular fluorescence complementation assays were performed as previously described (11). BY-2 cells were transformed via biolistic bombardment with plasmid DNA encoding RFP serving as a transformation control and an internal reference for assessing any cell-to-cell variability in RFP/YFP fluorescence values due to differences in protein expression, together with nYFP-Vps23, and p36-cYFP or p3691-330-cYFP. Transformed cells were visualized (via epifluorescence microscopy) based on RFP fluorescence, and both RFP and reconstituted YFP fluorescence intensities were collected with identical image acquisition settings (e.g., gain, offset, and exposure). Acquisition settings, amounts of plasmid DNA bombarded, and postbombardment cell incubation times employed in BiFC assays were chosen based on preliminary optimization experiments aimed at minimizing the possibility of nonspecific interactions. Likewise, p3691-330-cYFP, rather than empty cYFP vector, was chosen as a potential negative control based on guidelines for assessing membrane-bound protein interactions using the BiFC assay (52). The mean intensity of RFP and YFP fluorescence in transformed cells was calculated by defining the boundary of each cell followed by quantification of the mean pixel intensity using ImageJ software. The raw data for at least 25 cells were then expressed as a mean YFP-to-RFP ratio and a Student two-tailed t test assuming unequal variance between samples was used to determine statistical significance. Results shown are representative of three independent experiments.
Yeast two-hybrid analysis.
Yeast two-hybrid assays were carried out as described previously (53) with some modifications (35). Yeast cells (PJ69-4A) containing pGADT7 (activation domain fusions) and pGBKT7 (DNA-binding domain fusions) plasmids were cultured in synthetic dextrose medium (2% [wt/vol] dextrose, 0.67% [wt/vol] yeast nitrogen base without amino acids, 2 g/liter synthetic mix of amino acid supplements [SD-Leu,Trp; Bufferad]), diluted in a 1:5 dilution series, and then replica plated on agar plates containing SD-Leu,Trp or SD-Leu,Trp,His,Ade. Results of growth assays presented in figures are representative of the results obtained from analyzing three isolated yeast colonies from at least two separate cotransformations. In addition, all fusion proteins described in this study were confirmed to be properly expressed based on Western blot analysis of protein lysates obtained from yeast (co)transformed with two-hybrid plasmids, as described elsewhere (35).
In vitro coimmunoprecipitations.
Myc-tagged versions of p361-90, p36, and Vps28 were synthesized in vitro using the TNT T7 coupled reticulocyte lysate system (Promega), with the corresponding pGBKT7-based plasmids serving as the template DNA. S-epitope- tagged Vps23 (S-Vps23) was expressed in Escherichia coli BL21 Codon Plus (Stratagene). Cultures were grown to an optical density at 600 nm (OD600) of ∼0.6 to 0.8, and protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma-Aldrich Ltd.) at 30°C for 3 h. For experiments involving “mock” lysate, E. coli cells were transformed with empty vector (pET29a). Total soluble proteins were isolated in extraction buffer using a French press, and lysate was cleared by centrifugation as described elsewhere (54). Total soluble protein from cleared S-Vps23-containing or mock lysates were then separated using SDS-PAGE and stained with Coomassie blue R250 to ensure approximately equal total protein. Coimmunoprecipitations were carried out as described elsewhere (54), and proteins were resolved by SDS-PAGE, subjected to Western blotting using rabbit anti-Myc or mouse anti-S-tag (Novagen) antibodies, and detected using chemiluminescence.
RESULTS
CIRV replication in N. benthamiana is disrupted by a mutant of Vps4.
To begin to test whether ESCRT plays a role in CIRV replication, a dominant-negative version of the Arabidopsis ESCRT protein Vps4 (Vps4E232Q) was expressed in CIRV-infected N. benthamiana leaves. Vps4 is an AAA-ATPase, and Vps4E232Q contains a Glu-to-Gln mutation in the ATPase domain that blocks ATP hydrolysis and causes defects in endosomal protein sorting (33, 55), presumably by preventing disassembly of the endogenous ESCRT-III machinery at the late endosomal surface, as it does in yeast (56). Leaves of N. benthamiana, which is a host of tombusviruses, including CIRV (57), were infiltrated with Agrobacterium harboring a plasmid expressing Vps4E232Q (33) (agro-infiltrated). The same leaves were then rub inoculated 2 days later with a CaMV 35S promoter-containing plasmid encoding the full-length infectious CIRV cDNA (11), and 4 or 6 days thereafter, viral genomic and subgenomic RNAs were analyzed by Northern blotting.
As shown in Fig. 2A, compared to CIRV-infected N. benthamiana leaves that were agro-infiltrated with an empty vector (35S), expression of Vps4E232Q dramatically reduced the amounts of CIRV genomic RNA and both subgenomic mRNAs (sg1 and sg2 mRNA). Consistent with the results reported previously on the inhibition of TBSV replication in N. benthamiana by overexpression of another dominant-negative, ATPase-deficient version of Arabidopsis Vps4, i.e., Vps4K178A (23), we found that overexpression of Vps4E232Q also inhibited the accumulation of TBSV genomic and subgenomic mRNAs (Fig. 2A). Overexpression of Vps4E232Q did not, however, inhibit the replication of the Tobacco rattle virus (TRV) in N. benthamiana leaves (Fig. 2B), which is a (+)RNA tobravirus (46) and does not rely on ESCRT for its replication (23). This confirms that the inhibition of CIRV (and TBSV) replication by Vps4E232Q is not simply a consequence of an indirect, inhibitory effect(s) due to the overexpression of this ESCRT mutant in plant cells. As expected, no viral RNAs were detectable in mock-infected N. benthamiana leaves expressing Vps4E232Q (results are presented for TRV only [Fig. 2B]).
FIG 2.
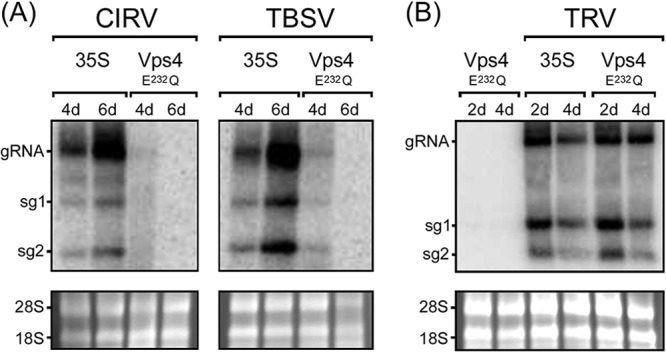
Inhibition of CIRV and TBSV replication in N. benthamiana by Vps4E232Q. (A) RNA blot analysis of N. benthamiana leaves infiltrated with Agrobacterium harboring either an empty vector (35S) or a vector encoding Arabidopsis Vps4E232Q, followed by rub inoculation with full-length infectious CIRV or TBSV cDNA. The relative positions of the CIRV and TBSV genomic (gRNA) and the two subgenomic mRNAs (sg1 and sg2) are shown to the left of the gel. The numbers of days after infiltration or inoculation are indicated above the lanes. (B) RNA blot analysis of N. benthamiana leaves agro-infiltrated with either Vps4E232Q alone, empty vector (35S), and two other vectors (TRV1 and TRV2) encoding the full-length infectious TRV genome (TRV), or Vps4E232Q, TRV1, and TRV2. In panels A and B, the 28S and 18S rRNAs in the corresponding ethidium bromide-stained agarose gels are shown as a loading control.
CIRV p36 recruits Vps23 to mitochondria in plant cells.
Given the well-documented role of Tsg101 (the mammalian homolog of Vps23) in HIV budding from the plasma membrane in mammalian cells (21, 22) and that yeast Vps23p is redirected to peroxisomes in yeast cells coexpressing TBSV p33 (23), we tested whether, in an analogous manner, CIRV causes the relocalization of Vps23 to mitochondria in plant cells. Toward this end, N. tabacum BY-2 suspension-cultured cells—which serve as a well-characterized system for studying protein targeting in plant cells (57), including viral proteins (58, 59)—were transiently (co)transformed (via biolistic bombardment) with plasmids encoding full-length CIRV and/or an N-terminal Myc epitope-tagged version of Arabidopsis Vps23 (Myc-Vps23) and then processed for immunoepifluorescence microscopy.
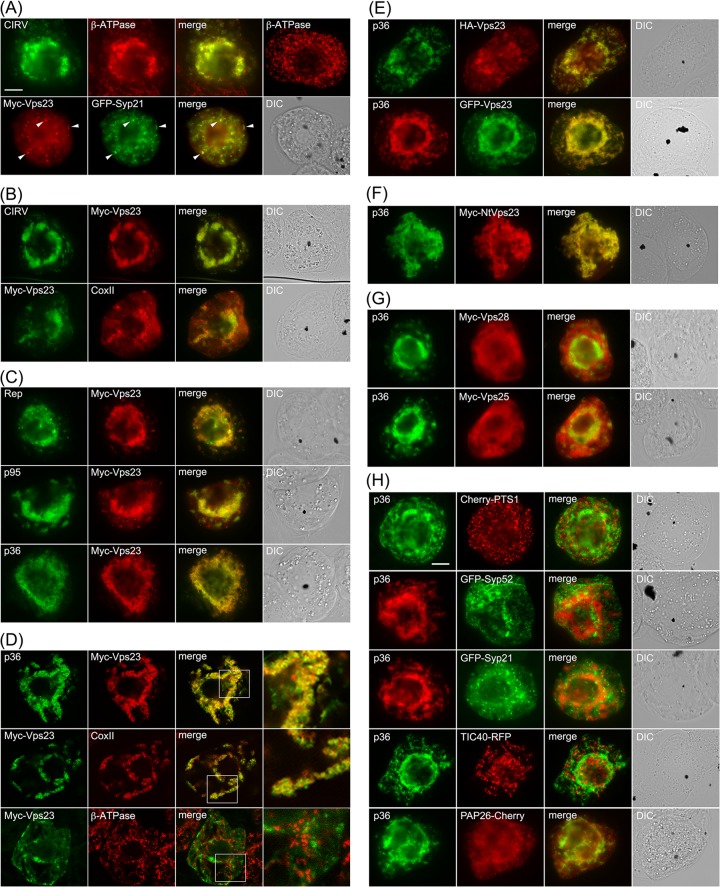
As shown in Fig. 3A and consistent with previously published results (11), both the p36 and p95 replicase proteins in CIRV-transformed BY-2 cells localized to endogenous β-ATPase-containing mitochondria, which as a consequence of the expression of the viral proteins, were conspicuously altered (i.e., aggregated) in terms of their intracellular distribution. For comparison purposes, refer to the normal appearance of the β-ATPase-containing mitochondria that are distributed throughout the cytosol in a nontransformed BY-2 cell (Fig. 3A). Also consistent with previously published results (35), Myc-Vps23 localized in BY-2 cells to the cytosol and late endosomes, shown by its partial colocalization with GFP-Syp21 (Fig. 3A), which is a well-known late endosomal marker protein (38). In contrast, coexpression of CIRV and Myc-Vps23 in the same cell resulted in relocalization of Myc-Vps23 to CIRV-induced aggregated mitochondria (Fig. 3B), which was confirmed as such based on immunostaining of both Myc-Vps23 and endogenous mitochondrial CoxII in the same batch of CIRV- and Myc-Vps23-cotransformed cells (Fig. 3B). These results support the premise that, similar to HIV and TBSV, CIRV causes a change in the intracellular localization of Vps23.
FIG 3.
Relocalization of Vps23 from the cytosol and late endosomes to mitochondria in BY-2 cells coexpressing CIRV p95 and/or p36. (A to C) Representative epifluorescence micrographs of BY-2 cells (co)transformed (as indicated by panel labels) with either full-length CIRV, Rep (both p95 and p36 together), p95, p36, Myc-Vps23, or GFP-Syp21, or mock transformed with empty plasmid DNA and then immunostained for the endogenous mitochondrial protein β-ATPase (top right-hand photo in panel A). All cells shown in Fig. 3 were formaldehyde fixed and processed for immunofluorescence microscopy as described in Materials and Methods. Note that p95 and p36 were immunodetected using primary antibodies raised against a synthetic peptide that corresponds to an amino acid sequence in both proteins. p95 is produced by the translational read-through of the p36 amber stop codon (8). Selected transformed cells were also immunostained (as indicated) for endogenous mitochondrial β-ATPase or CoxII. Also shown are the corresponding merge and differential interference contrast (DIC) images. The yellow color in the merged images indicates colocalization, and the white arrowheads in panel A indicate obvious examples of Myc-Vp23 and GFP-Syp21 colocalization at late endosomes. Bar = 10 μm. (D) Representative confocal micrographs of BY-2 cells cotransformed with either p36 and Myc-Vps23 (top and middle rows), or GFP-Syp21 and Myc-Vps23 (bottom row), which were immunostained for Myc-Vps23 and endogenous CoxII (GFP fluorescence is not shown). All images are medial (midcell) optical sections of cells, and boxes represent the portions of the cells shown at higher magnification in the panels on the right. The Pearson's correlation coefficient r values were as follows: r = 0.81 for p36 and Myc-Vps23, r = 0.71 for Myc-Vps23 and CoxII, and r = 0.21 for Myc-Vps23 and CoxII. (E to H) Representative epifluorescence micrographs of BY-2 cells cotransformed (as indicated) with either p36 and either N-terminal HA epitope- or GFP-tagged Vps23 (E); p36 and a Myc-tagged N. tabacum Vps23 (F), p36 and Myc-tagged Vps28 or Vps25 (G), or p36 and various organelle marker fusion proteins, including Cherry-PTS1 (peroxisome), GFP-Syp52 (early endosome/trans-Golgi network), GFP-Syp21 (late endosome), TIC40-RFP (plastid), PAP26-Cherry (lytic vacuole) (H). Bar = 10 μm.
We determined next whether the replicase proteins p36 and/or p95 expressed outside of the context of full-length CIRV were capable of recruiting Vps23 to mitochondria in plant cells. As shown in Fig. 3C, coexpression of Myc-Vps23 with either or both replicase proteins resulted in the relocalization of Myc-Vps23 to aggregated mitochondria (Fig. 3C), similar to when Myc-Vps23 was coexpressed with full-length CIRV (Fig. 3B). Colocalization between p36 and Myc-Vps23 is also shown in Fig. 3D using confocal microscopy, and quantification of colocalization in images obtained from medial (midcell) optical sections using the mean Pearson's correlation coefficient, r, confirmed a high degree of fluorescence signal overlap for both sets of proteins (see the legend to Fig. 3D for r values). In contrast, confocal microscopy and quantification of colocalization confirmed that Myc-Vps23 was not associated with mitochondria in BY-2 cells in the absence of coexpressed p36, as expected (see Fig. 3D and the legend for r value).
We also performed a number of additional control experiments to confirm that the recruitment of Vps23 to mitochondria by p36 was not dependent on the appended peptide sequence or fluorescent reporter protein (Fig. 3E) or the heterologous expression of Arabidopsis Vps23 in tobacco cells (Fig. 3F) and that it was specific for Vps23, since at least two other ESCRT proteins, Vps28 (ESCRT-I) and Vps25 (ESCRT-II), were not recruited by p36 (Fig. 3G). Moreover, we demonstrated that the appearance (i.e., distribution) of organelles other than mitochondria was unaffected in p36 (co)expressing cells (Fig. 3H), indicating that colocalization of Vps23 and p36 at mitochondria is not simply due to a general aggregation of organelles.
Taken together, the results presented in Fig. 3 indicate that CIRV specifically recruits the ESCRT-I component Vps23 to mitochondria in plant cells and that the p36 replicase protein is the minimal viral component necessary for this recruitment.
The cytosol-facing N-terminal region of p36 recruits Vps23 to mitochondria.
On the basis of previously published models (11, 26) and as illustrated in Fig. 4A, p36 is an integral outer mitochondrial membrane protein that is orientated with both the N- and C-terminal regions facing the cytosol. p36 also possesses two TMDs and an intervening loop region that contains the mitochondrial targeting signal and is predicted to face the intermembrane space (11). Keeping in mind this predicted topology, we began to explore regions within p36 that could be responsible for recruitment of Vps23 to mitochondria by examining mutant versions of p36 lacking the cytosol-facing N- and/or C-terminal portions of the protein, since these should be available to interact with cytosolic Vps23.
FIG 4.
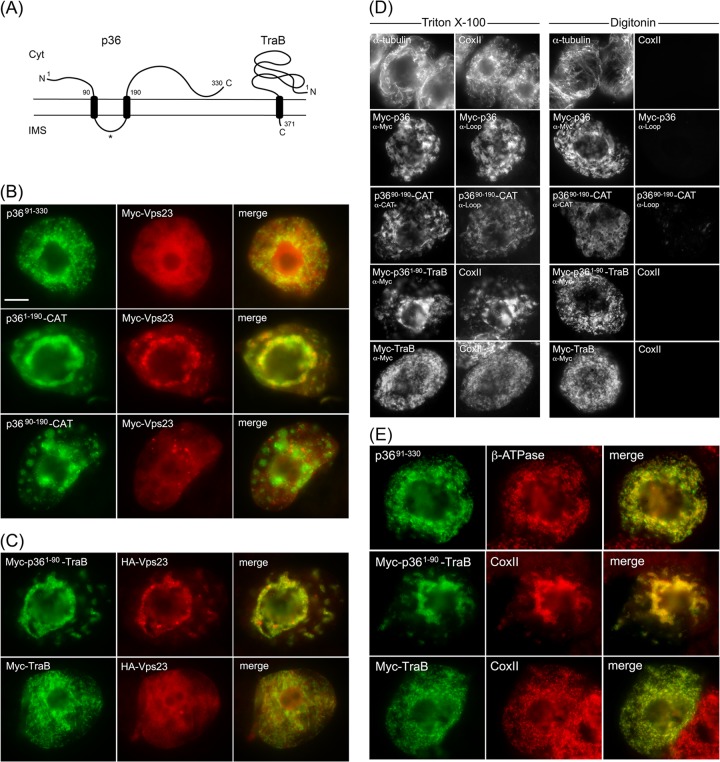
The cytosol-facing N-terminal region of p36 is both necessary and sufficient for relocalizing Vps23 to mitochondria in BY-2 cells. (A) Topology models of p36 and TraB in the mitochondrial outer membrane based on previously published results (11, 26, 50) and those presented in panel D. The numbers show the numbers of specific amino acid residues in full-length p36 (330 residues) and TraB (371 residues), including those in the names of the modified versions of p36 and p36-CAT (and p36-TraB) shown in panels B to E. The asterisk represents the relative position of the peptide sequence in the intervening loop of p36 used to generate the antiloop antibodies used in panel D. The N and C termini of the proteins are shown. Cyt, cytosol; IMS, intermembrane space. (B and C) Representative epifluorescence micrographs of BY-2 cells cotransformed with proteins as indicated by panel labels. Cells were formaldehyde fixed and processed for immunofluorescence epimicroscopy as described in the legend to Fig. 3. The Pearson's correlation coefficient r values based on confocal microscopy of cells coexpressing p3691-330 and Myc-Vps23 or Myc-TraB and HA-Vps23 were r = 0.31 and r = 0.25, respectively. Bar = 10 μm. (D) Representative epifluorescence micrographs of BY-2 cells either nontransformed (top row) or cotransformed with the indicated proteins, fixed with formaldehyde, and then permeabilized with either Triton X-100 (which permeabilizes both the plasma membrane and all organellar membranes) or digitonin (which permeabilizes only the plasma membrane) (16). The cells were then processed for immunoepifluorescence microscopy using primary antibodies raised against endogenous cytosolic α-tubulin, endogenous mitochondrial matrix protein CoxII (anti-CoxII [α-CoxII]), the Myc epitope tag (anti-Myc [α-Myc]), an amino acid sequence in the intervening loop of p36 (antiloop [α-Loop]), or the bacterial passenger protein CAT (anti-CAT [α-CAT]), as indicated by panel labels. Note that the presence of immunostaining in digitonin-permeabilized cells indicates that endogenous α-tubulin and the expressed protein's appended Myc epitope tag(s) or CAT moiety are exposed to the cytosol. Conversely, endogenous CoxII or the intervening loop sequence of p36 is not immunodetectable in the same corresponding digitonin-permeabilized cells. (E) Representative epifluorescence micrographs of BY-2 cells transformed with p3691-330, Myc-p361-90-TraB, or Myc-TraB and immunostained for endogenous mitochondrial β-ATPase or CoxII. Note that the mitochondria in the cells expressing p361-90-TraB, but not Myc-TraB, were aggregated, similar to the mitochondrial aggregation observed in cells expressing full-length p36 (Fig. 3).
As shown in Fig. 4B, Myc-Vps23 localized to the cytosol and not to mitochondria when coexpressed with p3691-330, a mutant of p36 in which the protein's cytosol-facing, N-terminal 90 amino acid residues were deleted (see the legend to Fig. 4B for the r value of coexpressed Myc-Vps23 and p3691-330, based on confocal microscopy). In contrast, p361-190-CAT, which is a previously characterized p36 mutant wherein the cytosol-facing C-terminal region of the protein (i.e., residues 191 to 330) was replaced with the passenger protein CAT (11), retained the ability to recruit Myc-Vps23 to mitochondria (Fig. 4B). Taken together, these data indicate that the N terminus, but not the C terminus, of p36 is necessary for the recruitment of Vps23 to mitochondria.
Consistent with this conclusion, Myc-Vps23 was not recruited to mitochondria when coexpressed with p3690-190-CAT (Fig. 4B), which lacks the N-terminal 90 residues of p36, and also has its cytosol-facing C terminus replaced with CAT (11). These results further indicate that the intervening loop region of p36 alone is not capable of recruiting Vps23 to mitochondria, as expected from the protein's predicted topology (Fig. 4A), wherein the intervening loop faces the mitochondrial intermembrane space and, thus, is inaccessible to interact with Vps23 in the cytosol. Confirmation that the intervening loop in both full-length p36 and p3690-190-CAT is oriented toward the mitochondrial intermembrane space was obtained using BY-2 cell differential detergent permeabilization assays (Fig. 4D). For instance, applied antibodies specific for a peptide sequence within the intervening loop of p36 were able to immunodetect the protein (or p3690-190-CAT) in cells only when both the plasma membrane and mitochondrial membranes (and all other organelle membranes) were permeabilized with Triton X-100, but not when only the plasma membrane was permeabilized with digitonin. We also confirmed that all of the p36 mutants mentioned above were properly targeted to mitochondria based on their colocalization with an endogenous mitochondrial marker protein (results are presented for p3691-330 only [Fig. 4E]).
To further demonstrate the importance of the N-terminal region (i.e., residues 1 to 90) of p36 in the recruitment of Vps23 to mitochondria, we replaced the N terminus of a mitochondrial outer membrane protein (TraB) with that of p36 and tested the fusion protein's ability to recruit coexpressed Vps23 to mitochondria. TraB is a “tail”-anchored membrane protein (Fig. 4A) (60), consisting of (i) an N-terminal region that faces the cytosol and represents the majority of the protein, (ii) a single TMD located near its C terminus, and (iii) a short C-terminal tail region orientated toward the intermembrane space that, along with the TMD, constitutes the protein's mitochondrial outer membrane targeting information (11). As shown in Fig. 4C, Myc-p361-90-TraB, consisting of a Myc epitope tag fused to the N-terminal 90 amino acids of p36 and the C-terminal TMD and tail of TraB (i.e., residues 348 to 371), colocalized with coexpressed HA epitope-tagged Vps23 (HA-Vps23; refer also to Fig. 3E) at mitochondria in BY-2 cells; mitochondrial recruitment was confirmed by colocalization of Myc-p361-90-TraB and endogenous CoxII in the same batch of Myc-p361-90-TraB- and HA-Vps23-cotransformed cells (Fig. 4E). In contrast, HA-Vps23 did not colocalize with full-length Myc-TraB at mitochondria (Fig. 4C and E) (refer also to the Fig. 4C legend for the r value for coexpressed Myc-TraB and HA-Vps23, based on confocal microscopy), a result that was not due to an aberrant topology of Myc-TraB, which like Myc-p361-90-TraB is orientated with its N terminus facing the cytosol (Fig. 4D).
Overall, the data presented in Fig. 4 indicate that the cytosol-facing N-terminal portion of p36, which represents the first 90 amino acids of the protein, is both necessary and sufficient for recruitment of Vps23 to mitochondria in plant cells.
The N-terminal region of p36 interacts directly with Vps23.
We investigated next whether the N-terminal region of p36 interacts directly with Vps23 by employing several different approaches. For example, yeast two-hybrid assays revealed that coexpression of p361-90 and Vps23 resulted in significant yeast growth on high selection media (Fig. 5A), indicating that the two fusion proteins interact. Similarly, coexpression of Vps23 and ESCRT-I protein Vps28 resulted in growth on high selection media, which has been reported previously using the yeast two-hybrid assay (36, 61, 62). As a negative control and consistent with our in vivo data (Fig. 3G), no interaction was observed between p361-90 and the ESCRT-II component Vps25. Likewise, coexpression of p361-90, Vps23, Vps28, or Vps25 with the corresponding empty vectors yielded no yeast growth on high selection media (Fig. 5A).
FIG 5.
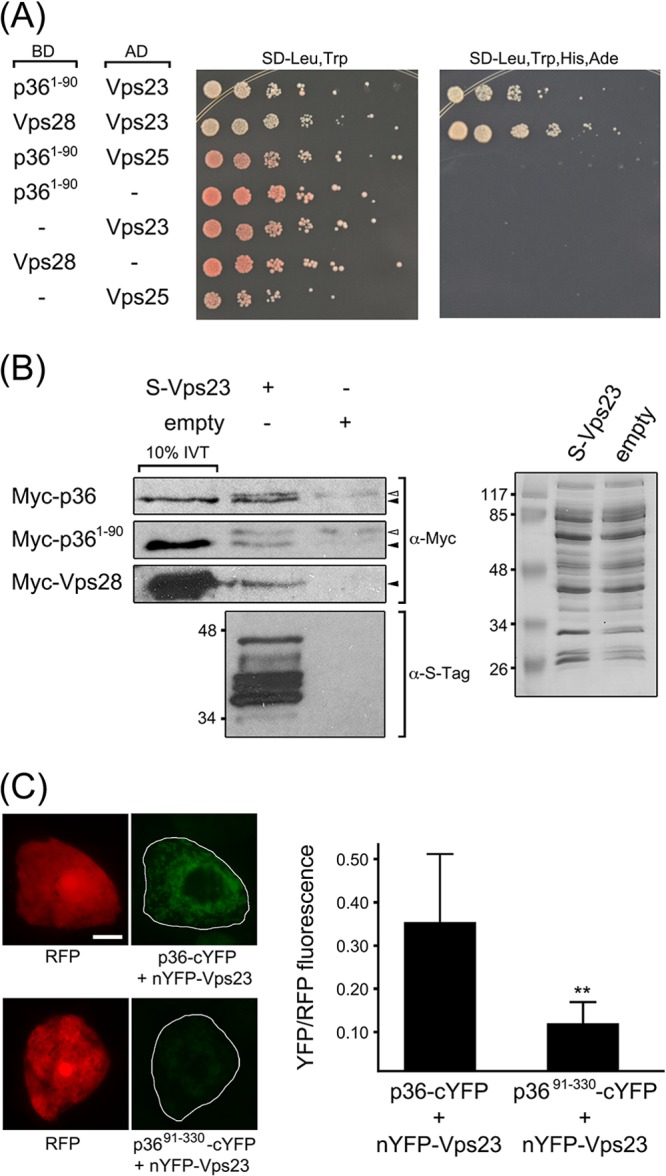
The N-terminal region of p36 interacts with Vps23. (A) p361-90 interacts with Vps23 in the yeast two-hybrid assay. Yeast strains were cotransformed with the indicated pairs of GAL4-binding domain (BD) and GAL4-activiating domain (AD) fusion proteins or the corresponding empty BD or AD control plasmids, denoted by a hyphen in the BD or AD column. Serial (1:5) dilutions of cells were spotted onto agar plates containing either low-stringency media (SD-Leu,Trp) or high-stringency media (SD-Leu,Trp,His,Ade), where cell growth is dependent on two-hybrid protein interactions. (B) In vitro coimmunoprecipitation of p36 or p361-90 and Vps23. Whole-cell lysates of E. coli transformed with S-Vps23 or the corresponding empty vector were incubated with S-protein agarose and in vitro-translated (IVT) full-length Myc-p36, Myc-p361-90, or Myc-Vps28, and washed. Eluted proteins (and 10% of the total IVT reaction mixture) were subjected to SDS-PAGE and protein blotting and then probed with either anti-Myc or anti-S-tag antibodies. The solid (black) arrowheads indicate the relative positions of specific immunodetected proteins; the open (white) arrowheads indicate nonspecific, immunodetected proteins. Note that the immunodetected proteins indicated by the solid arrowheads are not present in the coimmunoprecipitation reactions with lysates of E. coli containing empty vector. Shown to the right is a Coomassie blue-stained gel of total soluble protein from lysates containing S-Vps23 or empty vector prior to incubation with S-protein agarose, confirming equivalent input. The numbers to the left of the gels are the molecular masses (in kilodaltons) of protein standards. (C) Interaction of p36 and Vps23 in the BiFC assay. Representative epifluorescence micrographs of BY-2 cells triple transformed (as indicated) with RFP, nYFP-Vps23, and either p36-cYFP or p3691-330-cYFP. Relative RFP and YFP fluorescence values of transformed cells, which are delineated by the solid lines in the panels on the right were quantified using ImageJ (refer to Materials and Methods for details). Note the relatively low YFP fluorescence in the representative cell coexpressing the negative control, p3691-330-cYFP. At least 25 transformed cells were analyzed from at least three independent experiments, and the mean YFP-to-RFP ratios (plus standard deviation [SD]) are plotted in the bar graph on the right. The two asterisks denote a statistically significant difference between the two samples (P ≤ 0.001).
The interaction between p36 and Vps23 was confirmed using in vitro coimmunoprecipitation. Briefly, recombinant S-epitope-tagged Vps23 (S-Vps23) from cleared E. coli cell lysate was immobilized on S-protein agarose and then incubated with in vitro-translated, Myc-tagged versions of either full-length p36 (Myc-p36), the N-terminal 90 amino acids of p36 (Myc-p361-90), or Vps28 (Myc-Vps28); Myc-Vps28 served as a positive control based on its previously published interaction with Vps23 via coimmunoprecipitation (35, 63). For a negative control, the same three in vitro-translated proteins were incubated with S-protein agarose preincubated with cleared cell lysate prepared from E. coli transformed with empty vector, but containing an approximately equal amount of total protein (Fig. 5B, right panel). As shown also in Fig. 5B, both Myc-p36 and Myc-p361-90, similar to Myc-Vps28, were coimmunoprecipitated with immobilized S-Vps23, but not by the corresponding empty vector negative controls (marked by solid arrowheads in Fig. 5B, left panel), indicating that both full-length p36 and the N terminus of p36 interact directly with Vps23.
We further demonstrated the interaction between p36 and Vps23 using the in vivo BiFC assay. In this experiment, p36 or p36 lacking its N terminus (p3691-330) fused to the C-terminal half of YFP (p36-cYFP and p3691-330-cYFP, respectively) were each coexpressed with Vps23 fused to the N-terminal half of YFP (nYFP-Vps23) in BY-2 cells. In addition, cells were transformed with cytosolic RFP, which was used to identify transformed cells and also served as an internal normalization for reconstituted YFP fluorescence, minimizing differences due to cell-to-cell variability in expression levels. p3691-330, which does not recruit Vps23 to mitochondria in plant cells (Fig. 4B), served as a negative control. As shown in Fig. 5C, BY-2 cells cotransformed with p36-cYFP and nYFP-Vps23 displayed significantly more YFP/RFP fluorescence than cells cotransformed with p3691-330-cYFP and nYFP-Vps23. While we cannot rule out the possibility that the relatively small amount of YFP/RFP fluorescence still observed upon coexpression of p3691-330-cYFP and nYFP-Vps23 may be a result of weaker or, as discussed below, “secondary” interactions between the C terminus of p36 and Vps23, these results indicate that, just as in yeast two-hybrid and in vitro coimmunoprecipitation assays (Fig. 5A and B), p36 and Vps23 interact in plant cells in a manner that is dependent upon the N terminus of p36.
A unique polypeptide sequence near the N terminus of p36 is both necessary and sufficient for recruiting Vps23 to mitochondria.
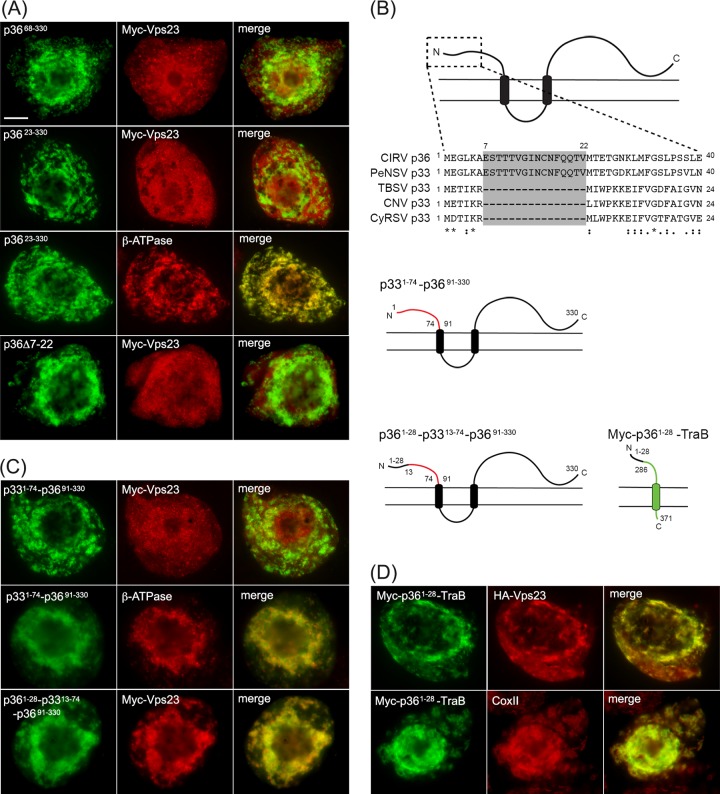
Since the N-terminal 90-amino-acid sequence of p36 contains no sequence motifs that match the peptide late-domain motifs (e.g., P[S/T]AP) responsible for the recruitment of ESCRT by enveloped retroviral proteins (20) or by TBSV p33 (23, 25), we carried out a mutational analysis of this region to define any potentially novel Vps23 recruitment motifs. The first mutants examined were those in which either the N-terminal 67 (p3668-330) or 22 (p3623-330) amino acid residues were deleted from the protein. As shown in Fig. 6A, both p3668-330 and p3623-330, which were properly targeted to mitochondria (results are presented for p3623-330 only [Fig. 6A), did not recruit Myc-Vps23. Instead, Myc-Vps23 localized in these cells primarily to the cytosol and/or late endosomes, just as it does when it is expressed on its own (Fig. 3A) or coexpressed with the p36 mutant lacking the entire 90-amino-acid-long N-terminal region (p3691-330) (Fig. 4B). These data suggest that at least the first 22 amino acids of the p36 N terminus are essential for recruiting Vps23 to mitochondria.
FIG 6.
A unique 16-amino-acid-long sequence at the N terminus of p36 is both necessary and sufficient for relocalizing Vps23 to mitochondria in BY-2 cells. (A) Representative epifluorescence micrographs of BY-2 cells cotransformed with proteins as indicated by panel labels and immunostained (as indicated) for endogenous mitochondrial β-ATPase. Numbers in the name of the construct denote the specific amino acid residues derived from full-length p36 (330 residues) or specific residues deleted from p36. Bar = 10 μm. (B) Amino acid sequence alignment of the N termini of various tombusvirus p36 and p33 proteins and cartoon illustrations of p33-36 and p36-TraB hybrid proteins. Sequences were obtained from GenBank and aligned using ClustalW. Identical and similar amino acids in each protein are indicated with asterisks and colons or periods, respectively. The unique amino acid sequence present in CIRV p36 and PeNSV p33 (residues 7 to 22) but absent in TBSV, CNV, and CyRSV p33 are shaded gray. Cartoons depict the structure and topology of p33–p36 and p36-TraB hybrid proteins in the mitochondrial outer membrane. Lines representing amino acid sequences from p33, p36, and TraB are colored red, black, and green, respectively. Numbers represent specific amino acid residues derived from either full-length p33 (296 residues), p36 (330 residues), or TraB (371 residues) and correspond to the numbers in the names of the p33–p36 and p36-TraB hybrid proteins described in panels C and D. (C and D) Representative epifluorescence micrographs of BY-2 cells cotransformed with proteins as indicated by panel labels and immunostained (as indicated) for endogenous mitochondrial β-ATPase. Numbers in the name of the construct denote the specific amino acid residues derived from full-length p36 or p33 and are as illustrated in panel B.
Comparison of the N-terminal sequences of p36 with its counterparts from various other tombusviruses (Fig. 6B) revealed that only p36 and the replicase protein of Pelargonium necrotic spot virus (PeNSV), which like CIRV, replicates at host cell membranes derived specifically from mitochondria (64), possess a unique and identical stretch of 16 amino acids (residues 7 to 22) that is not present in the replicase proteins of tombusviruses that replicate at peroxisome-derived membranes (i.e., TBSV p33, Cucumber necrosis virus [CNV] p33, Cymbidium ringspot virus [CyRSV] p33). Notably, this unique sequence in CIRV p36 is due to the initiation codon of the p36 open reading frame being positioned further upstream than the corresponding codon in TBSV or CyRSV (5, 8) and while it contains a negative determinant for satellite RNA replication (65), it is not suspected to play a significant role in virus symptom development (66). Nonetheless, we chose to focus on whether this unique region in p36 (and PeNSV p33) is involved in the recruitment of Vps23 based on the inability of p3623-330 to recruit Vps23 to mitochondria. As shown in Fig. 6A, Myc-Vps23 did not localize to mitochondria when coexpressed with a p36 mutant lacking residues 7 through 22 (p36Δ7-22), supporting the premise that this region in p36 is specifically involved in Vps23 recruitment. As a complementary approach, we also replaced the N-terminal 90 residues of p36 with the equivalent N-terminal soluble region of TBSV p33 (residues 1 to 74), which is similarly orientated toward the cytosol (4, 26), but does not contain the unique sequence found within the N terminus of p36 (Fig. 6B). Moreover, this N-terminal region of p33 is not considered on its own to be sufficient for interacting with Vps23 (23). As shown in Fig. 6C, the resulting fusion protein, p331-74-p3691-330, did not colocalize with Myc-Vps23 at mitochondria (refer to Fig. 6C, which shows colocalization between p331-74-p3691-330 and endogenous β-ATPase for evidence that this fusion protein was properly targeted to mitochondria). In contrast, when the first 28 residues of p36 were reintroduced into p331-74-p3691-330, yielding p361-28-p3313-74-p3691-330 (Fig. 6B), the recruitment of Myc-Vps23 to mitochondria was restored (Fig. 6C).
We also showed that the N-terminal 28 amino acid residues of p36 were capable of recruiting Vps23 to mitochondria when this region was fused to the N terminus of TraB. That is, similar to Myc-p361-90-TraB (Fig. 4C), Myc-p361-28-TraB, which as illustrated in Fig. 6B, consists of the N-terminal 28 residues of p36 fused to a portion of the cytosol-facing domain of TraB (in order to preserve the overall length of the soluble N-terminal region), as well as its C terminus, also recruits HA-Vps23 to mitochondria in coexpressing cells (Fig. 6D).
Taken together, the data presented in Fig. 6 indicate that the unique N-terminal 16-amino-acid-long sequence present in CIRV p36, but absent in the replicase proteins from tombusviruses that rely on peroxisomal membranes for their replication, contains a novel Vps23 recruitment motif.
Recruitment of Vps23 to mitochondria by p36 requires its StBox domain rather than the UEV domain.
As illustrated in Fig. 7A, Arabidopsis Vps23, similar to Vps23 in all species examined, consists of three unique structural/functional domains, including (i) an N-terminal UEV domain, (ii) a central coiled-coil (CC) region, which is involved in ESCRT-I protein-protein interactions, and (iii) a C-terminal steadiness box (StBox), which is also involved in protein interactions with other ESCRT-I components, as well as regulating Vps23 protein turnover (63). Given the importance of the UEV domain in facilitating the interaction between Vps23 and p33 during recruitment to peroxisomes (23) and the interactions between Tsg101 and HIV Gag p6 at the plasma membrane (20) and between certain ESCRT-0 proteins and Vps23/Tsg101 during MVB biogenesis in yeasts and mammals (13), we postulated that the UEV domain might also be involved in the recruitment of Vps23 to mitochondria by p36.
FIG 7.
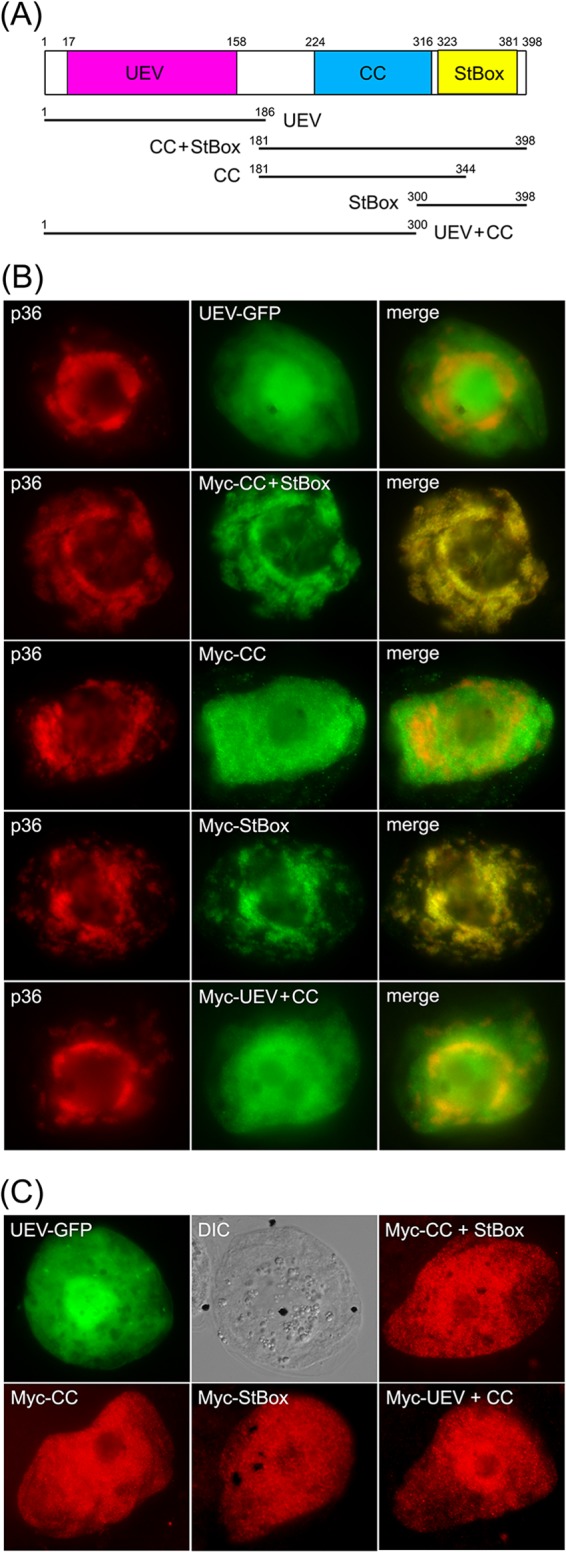
The C-terminal StBox of Vps23 is necessary and sufficient for its recruitment to mitochondria by p36 in BY-2 cells. (A) Schematic diagram illustrating the domain organization of Arabidopsis Vps23 based on Spitzer et al. (63). Numbers denote the specific amino acid residues derived from full-length Vps23 (398 residues) that delineate the protein's UEV, CC, and StBox domains, which are colored red, blue, and yellow, respectively, and which are depicted below as lines representing the various Vps23 truncation mutants described in panels B and C. Numbers denote the specific amino acid residues at the N and C termini of each Vps23 mutant. (B and C) Representative epifluorescence micrographs of BY-2 cells (co)transformed with proteins as indicated by panel labels. Names of mutants represent the specific domain(s) derived from Vps23, as illustrated in panel A. The corresponding merged images (B) and the corresponding DIC images of the UEV-GFP-transformed BY-2 cells (C) are also shown.
To investigate this possibility, the UEV domain of Vps23 fused to green fluorescent protein (UEV-GFP) was coexpressed with full-length p36 in BY-2 cells. As shown in Fig. 7B, UEV-GFP was not localized to mitochondria in cells coexpressing p36, but instead localized throughout the cytosol, just as it does when expressed on its own, as do all other Vps23 mutants described in this study [see below]) (Fig. 7C). The lack of recruitment of the UEV domain by p36 was somewhat unexpected and led us to investigate next whether the CC and/or StBox domains play a role in its recruitment to mitochondria by p36. As shown in Fig. 7B, a Vps23 mutant consisting of both the CC and StBox domains fused to an N-terminal Myc epitope tag (i.e., Myc-CC+StBox; refer to schematic diagram in Fig. 7A), colocalized with coexpressed p36 at mitochondria in BY-2 cells, reinforcing the notion that the recruitment of Vps23 by p36 is mediated by a region(s) other than the UEV domain. As shown also in Fig. 7B, a Vps23 mutant consisting of the CC domain alone (Myc-CC) localized to the cytosol in cells coexpressing p36, while the StBox domain (Myc-StBox) alone was recruited to p36-containing mitochondria. These results suggest that Vps23 recruitment is mediated by the StBox domain. Indeed, the Vps23 mutant consisting of the UEV and CC domains (Myc-UEV+CC) did not colocalize with coexpressed p36 but instead remained in the cytosol (Fig. 7B), just as it does when coexpressed without full-length p36 (Fig. 7C). Together, these results indicate that the C-terminal StBox domain is critical for the recruitment of Vps23 to mitochondria by p36 and that the UEV domain either does not play a role or plays a minor role that is undetectable using this method.
Consistent with this conclusion, yeast two-hybrid assays performed with p361-90 and the equivalent set of Vps23 mutants as those described above revealed that only those proteins containing the StBox (i.e., CC+StBox and StBox) conferred yeast growth on high selection media (Fig. 8A). A notable exception was the UEV+CC mutant, which when coexpressed with p361-90, also resulted in growth on high selection media, albeit relatively less than that observed for yeast coexpressing CC+StBox, StBox, or full-length Vps23 (Fig. 8A). However, as mentioned above, the equivalent UEV-CC mutant is not recruited to mitochondria by full-length p36 in plant cells (Fig. 7B), suggesting that if the CC domain does indeed participate in the recruitment of Vps23 by p36, it does so to a lesser extent than the StBox domain does. While we confirmed that UEV+CC (or any of the other Vps23 mutants) does not autoactivate the yeast two-hybrid reporter genes (Fig. 8B), we cannot rule out the possibility that the observed p361-90/UEV+CC interaction may be the result of a “bridge” by an endogenous yeast ESCRT component(s). Moreover, and as discussed in more detail below, we also confirmed that all of the Vps23 mutants (and full-length Vps23) interacted in yeast two-hybrid assays with the Arabidopsis ESCRT-I proteins Vps28 and Vps37 (Fig. 8C and D) in ways that are consistent with how these domains are involved in the corresponding ESCRT protein-protein interactions in yeasts and mammals (27–30). For instance, like their yeast and mammalian counterparts, the interaction of Arabidopsis Vps28 and Vps23 is mediated by the Vps23 StBox domain (Fig. 8C), while the interaction of Arabidopsis Vps37 and Vps23 is mediated by the Vps23 CC domain, although the Vps23 UEV domain also appears to be involved in Vps23-Vps37 binding (Fig. 8D).
FIG 8.
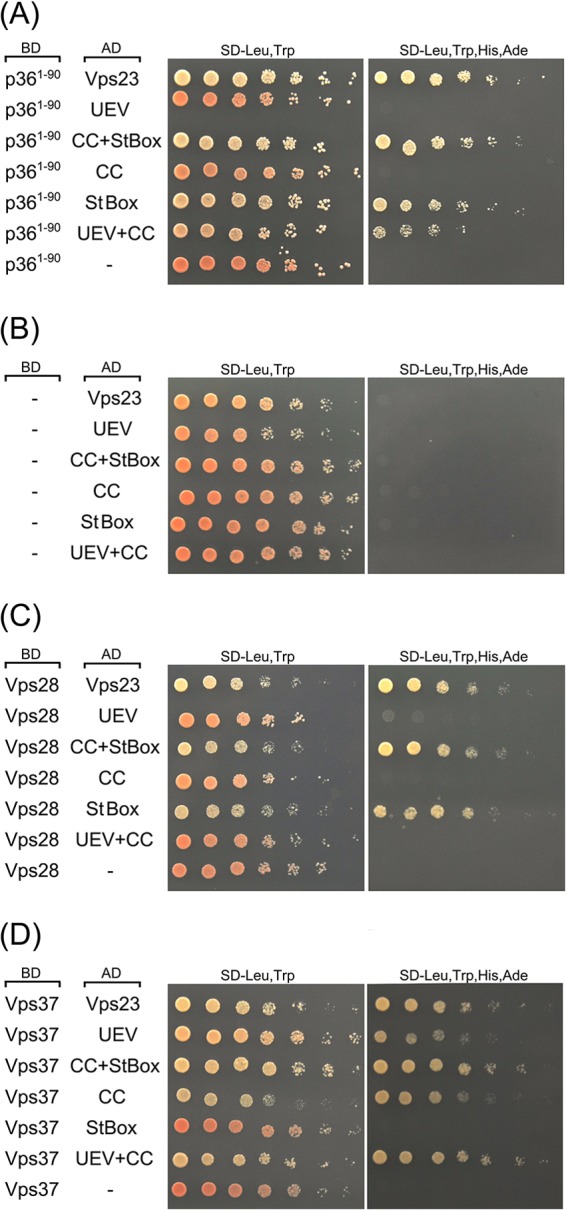
The N-terminal region of p36, like Vps28, interacts with the C-terminal StBox of Vps23 in the yeast two-hybrid assay. (A to D) Yeast strains were cotransformed with the indicated pairs of GAL4-binding domain (BD) and GAL4-activating domain (AD) fusion proteins or the corresponding empty BD or AD control plasmids (indicated by a hyphen in the BD or AD column). Serial (1:5) dilutions of cells were spotted onto agar plates containing either low-stringency medium (SD-Leu,Trp) or high-stringency medium (SD-Leu,Trp,His,Ade), where cell growth is dependent on two-hybrid protein interactions. Note that neither full-length Vps23, nor any of the truncation mutants of Vps23, Vps28, and Vps37 autoactivated the two-hybrid reporter gene system on their own.
DISCUSSION
Understanding how viruses appropriate host cell organelles in order to carry out their replication is not only an important step in the development of strategies to prevent infection, it often provides novel insights into the molecular mechanisms underlying organelle biogenesis and dynamics in uninfected cells. For example, recent studies examining how the Bunyavirus Tomato spotted wilt virus buds from the endoplasmic reticulum (ER) in plant cells or how Turnip mosaic virus infection leads to the fusion of ER-derived vesicles with chloroplasts has shed new light on the formation of ER exit sites (67) and the role(s) of the previously uncharacterized SNARE protein Syp71 (68). Likewise, the characterization of the late-domain motifs in certain retroviral structural proteins and their interaction with the host cell ESCRT-I protein Tsg101 (69) was a key step in the subsequent discovery that similar peptide motifs exist in certain ESCRT-0 proteins. As such, it is now known that, in uninfected cells, the sequential recruitment of the ESCRT subcomplexes (i.e., ESCRT-0, -I, -II, and -III) during MVB biogenesis begins with ESCRT-0 binding to ESCRT-I and that retroviral proteins such as the HIV Gag p6 mimic ESCRT-0 in order to hijack ESCRT-I (Tsg101) to the sites of viral budding at the plasma membrane (20).
Presently, our understanding of ESCRT in plants is relatively poor, and thus, working models based on studies of ESCRT in yeasts and mammals are often employed to infer the mechanisms of ESCRT assembly and function in plants. While this approach has generally proven to be valid because of the overall conserved nature of ESCRT function among evolutionarily diverse organisms, particularly during MVB biogenesis (35, 61, 62), there are also several features of ESCRT in plants that are distinct, including the ability of TOL proteins to compensate for the lack of ESCRT-0 proteins in terms of ubiquitinated cargo recognition and sorting (16, 17). Thus, understanding how different tombusviruses appropriate the plant ESCRT machinery, apparently through different forms of molecular mimicry, as discussed below, not only gives important insight into how tombusviruses manipulate membranes to facilitate their replication but may also provide valuable clues to previously unexplored aspects of normal ESCRT assembly and function in plants.
The recruitment of ESCRT to mitochondria by CIRV is mediated by a unique sequence located near the N terminus of p36.
Plant tombusviruses are a well-established model for studying (+)RNA virus replication and virus-host interactions (70) and are also emerging as an especially useful tool for understanding how (+)RNA viruses recruit and manipulate specific host cell organelle membranes in order to create unique compartments that house the viral replication machinery. For instance, based on studies that focused on TBSV and employed yeast as a model organism for tombusvirus replication (71), Nagy and coworkers recently extended an earlier working model (72) whereby ESCRT is hijacked by TBSV in order to facilitate the progressive invagination of the peroxisomal boundary membrane, which according to the model, leads to the concentration and assembly of viral replicase complexes within nascent spherules (23). While several aspects of this model remain to be confirmed experimentally, one of the most notable tenets is that the TBSV replicase protein p33 binds to the ESCRT-I protein Vps23 via a peptide sequence (i.e., PSVP) in p33 that resembles the proline-rich late-domain motif found in HIV Gag p6. Consequently, p33, analogous to Gag p6, is considered to “mimic” ESCRT-0 in terms of its ability to bind and recruit Vps23 (ESCRT-I) (21, 22). However, as mentioned above, the lack of plant homologs of ESCRT-0 proteins implies that the precise nature by which p33 interacts with Vps23 is not completely analogous to the interaction between HIV Gag p6 and Tsg101 and, instead, may reflect a unique mechanism for ESCRT recruitment in plants, which may or may not involve the TOL proteins, and which is currently an open question.
Here we investigated whether an ESCRT recruitment process is utilized by CIRV, another member of the tombusvirus family that also causes pronounced changes to a specific organellar membrane in infected plant cells, but unlike TBSV, does so at the mitochondrial outer membrane (73). Our initial interest in investigating CIRV stemmed from the observation that the late-domain-like motif (i.e., PSVP) identified in TBSV p33 (23) is not conserved in CIRV p36 (Fig. 1), suggesting that, if CIRV does rely on the ESCRT machinery, it does so in a distinct manner compared to TBSV. To begin to test this hypothesis, we showed that the replication efficiency of CIRV does indeed depend on ESCRT. That is, coexpression of a mutant version of Vps4 (Vps4E232Q), which is one of the best-characterized ESCRT components in plants (33, 35, 55, 63), inhibited CIRV replication in N. benthamiana plants (Fig. 2), presumably by disrupting the ESCRT-dependent assembly of the replicase complex, a conclusion that was reinforced by the observation that TBSV, but not TRV, replication was also inhibited by Vps4E232Q, as expected (23). We also showed that the ESCRT-I component Vps23 is redirected to mitochondria in plant cells expressing either full-length CIRV or the p36 replicase-associated protein alone (Fig. 3), indicating that p36, like TBSV p33 (23), is the minimal viral component necessary for Vps23 recruitment. Moreover, the recruitment of Vps23 to mitochondria by p36 was shown to be mediated by the viral protein's N-terminal 90 amino acids (Fig. 4 and 5), a region that precedes the first of the protein's two TMDs and is orientated toward the cytosol (Fig. 4A), which we considered a prerequisite for interaction with cytosolic Vps23. In contrast, neither the cytosol-facing C-terminal portion, nor the intervening loop sequence of p36, which contains the mitochondrial targeting signal and is orientated toward the mitochondrial intermembrane space (11) (Fig. 4), were directly involved in Vps23 recruitment (Fig. 4). The latter observation is particularly notable because the intervening loop region of TBSV p33, which contains the PSVP late-domain-like motif implicated in recruitment of Vps23 to peroxisomes (23), is also considered to be orientated toward the peroxisomal matrix (11, 26). Thus, it remains to be determined how the PSVP motif in p33 is accessible to bind to Vps23 in the cytosol, although one possibility is that the association of nascent p33 with cytosolic chaperones prior to its targeting to and insertion into the peroxisomal membranes (74) may render this motif accessible to Vps23. While we did not detect in vivo recruitment activity for the intervening loop region and C-terminal region of p36, we also cannot rule out the possibility that additional motifs within these regions of p36 interact, albeit weakly, with Vps23 and that these secondary interactions might help CIRV to more efficiently compete with the cellular ESCRT machinery, for example, Vps28 (as discussed below) for Vps23. Indeed, while the BiFC results presented in Fig. 5C support the premise that p36 and Vps23 interact in plant cells and do so in a manner that is dependent upon the N terminus of p36, the observation that coexpression of the p3691-330 mutant and Vps23 yielded a BiFC signal, although much weaker than that from full-length p36 and Vps23, implies that such secondary interactions might exist.
To gain an understanding of the mechanism by which the N terminus of p36 recruits Vps23 to mitochondria, we carried out a mutational analysis of this region, the results of which highlighted a 16-amino-acid-long sequence (i.e., residues 7 to 22) that is present in tombusviruses that replicate at mitochondria (i.e., CIRV and PeNSV) and absent in tombusviruses that replicate at peroxisomes (i.e., TBSV and Cucumber necrosis virus [CNV]) (Fig. 6B). We showed that when this unique polypeptide sequence in p36 was specifically deleted, Vps23 recruitment to mitochondria was abolished (Fig. 6A). Conversely, the inclusion of this sequence from p36 within the context of various hybrid fusion proteins consisting of other portions of p36 and either p33 or TraB conferred the ability to recruit Vps23 to mitochondria (Fig. 6C and D). This implies that this region in p36 can function as a Vsp23-binding domain outside of the context of the full-length viral protein. Despite its involvement in Vps23 recruitment, results from previous studies indicate that this unique N-terminal sequence in p36 is not essential for virus replication, since a mutant version of CIRV with this region deleted induced the same necrotic symptoms in infected N. benthamiana plants as that observed for the wild-type virus, although the appearance of these symptoms was conspicuously delayed (66). These results suggest that the N-terminal motif may mediate a more robust infection. Indeed, the retention of this motif in CIRV indicates that it confers a competitive advantage to the virus, which we suggest is related to its demonstrated role in Vps23 recruitment. Nonetheless, the virus viability observed in the absence of the motif implies that CIRV might rely on other secondary interactions between p36 and Vps23, as mentioned above, and/or additional ESCRT factors (e.g., Bro1), analogous to how other viruses, such as HIV and TBSV, interact with multiple ESCRT proteins (20, 23). This study, in combination with the rapidly growing interest in the plant ESCRT machinery in general, should now facilitate a more detailed analysis of the other ESCRT factors involved in CIRV replication.
It is also worth mentioning that while the N terminus of p36, including the sequence corresponding to amino acid residues 7 through 22, is devoid of any obvious functional domains that resemble those used by other viruses to recruit Vps23/Tsg101 (e.g., proline- or leucine-rich late-domain motifs) (75), a portion of this region is predicted to be relatively disordered (i.e., based on IUPred [76]). Disordered (unstructured) regions are commonly found in viral proteins, where they are known to mediate interactions with different host cell components in dynamic ways (77). Indeed, intrinsically disordered regions in TBSV p33 have been implicated in its RNA chaperone activity (78), as well as potentially mediating interactions with host proteins and viral factors (79). Currently, we are assessing the structural attributes (or lack thereof) of the p36 N terminus, as well as employing peptide “mimetics” (80) to determine the possible structure-function relationship of this region of p36 and its binding to Vps23.
p36, similar to the ESCRT-I protein Vps28, interacts with the C-terminal StBox domain of Vps23.
Additional evidence that CIRV utilizes a distinct mechanism in order to hijack ESCRT was our finding that the binding and recruitment of Vps23 by p36 do not depend on the Vps23 N-terminal UEV domain but instead are mediated by an interaction between p36 and the C-terminal StBox domain of Vps23 (Fig. 7 and 8). In mammalian cells, the StBox in Tsg101 is involved in coordinating the regulation of the protein's stability and its interaction with Vps28 during ESCRT-I assembly (81). Similarly, we showed that Arabidopsis Vps28 and Vps23 interact via the Vps23 StBox (Fig. 7 and 8). As such, it is possible that p36 “mimics” Vps28 by binding to the StBox in Vps23, thereby facilitating its recruitment to mitochondria. It now remains to be determined whether p36 and Vps28 compete for an interaction with Vps23 in plant cells, based on our assumption that the molecular mechanisms underlying ESCRT-I assembly in plants and mammals, including the interaction of Vps28 with the Vps23 StBox, are similar. This appears to be plausible, since the core components of Arabidopsis ESCRT-I (i.e., Vps23, Vps28, and Vps37) are already known to interact in a manner similar to their mammalian counterparts (33, 35, 55, 63), and as we showed in this study (Fig. 8), Arabidopsis Vps37 and Vps28 interact with the UEV/CC domains and StBox of Arabidopsis Vps23, respectively, which is consistent with models of ESCRT-I assembly in mammals and yeasts (13). Presently, we are comparing the nature of the interaction between p36 or Vps28 and the Vps23 StBox in plant cells and comparing these with the well-characterized interactions between Vps28 and Vps23 in yeasts and mammals (27–30).
Our model that the binding of p36 and Vps28 to Vps23 is mutually exclusive also prompts the question of how additional downstream ESCRT components would be recruited and assembled at the mitochondrial membranes in CIRV-infected cells, since Vps28 is considered to act as a bridge between ESCRT-I and ESCRT-II during normal MVB biogenesis (82). However, there is also a growing appreciation that not all ESCRT components participate in the cellular processes involving ESCRT-dependent membrane deformation. For instance, during HIV infection in mammalian cells, the binding of Tsg101 by HIV Gag p6 does not lead to the recruitment of Vps28 and subsequently ESCRT-II. Instead, Tsg101 and Gag p6 interact with the ESCRT accessory protein Alix, which recruits ESCRT-III, which ultimately facilitates the viral budding process (20). Interestingly, a mutant of Bro1, the Alix ortholog in plants, was shown to inhibit TBSV replication and TBSV p33 interacts with yeast Bro1p (23). This suggests a role for Bro1 in ESCRT assembly at peroxisomes in TBSV-infected plant cells that may be analogous to the role for Alix during HIV budding. Likewise, it is possible that while CIRV and TBSV seem to rely on distinct mechanisms for the initial recruitment of Vps23, the recruitment of additional ESCRT components, either in parallel with the initial interaction(s) of p36 and Vps23 or further downstream, may be mechanistically similar, including the recruitment of Bro1. Future experiments aimed at dissecting the recruitment of additional ESCRT components, including Bro1 and ESCRT-III, during CIRV replication, will be important for obtaining a clear picture of the molecular events that underlie spherule formation, which is critical for understanding the spatial dynamics of tombusvirus replication at host cell membranes.
ACKNOWLEDGMENTS
We thank M. Otegui, S. Gelvin, T. Elthon, and S. Subramani for their generous gifts of plasmids or antibodies used in this study and T. Nguyen for maintaining tobacco plants and BY-2 cell cultures. We are also indebted to all the members of our laboratories and J. Dyer for their helpful discussions during the course of this work. We also thank the anonymous referees for their constructive comments that led to an improved version of the article.
This study was supported by grants from the Natural Sciences and Engineering Research Council of Canada (NSERC) to R. T. Mullen and K. A. White, as well as an NSERC CGS-D Postgraduate Scholarship to L. G. L. Richardson. R. T. Mullen holds a University of Guelph Research Chair, and K. A. White holds a Canada Research Chair in plant biotechnology and structural biology.
Footnotes
Published ahead of print 26 March 2014
REFERENCES
- 1.Russo M, Burgyan J, Martelli GP. 1994. Molecular biology of Tombusviridae. Adv. Virus Res. 44:381–428. 10.1016/S0065-3527(08)60334-6 [DOI] [PubMed] [Google Scholar]
- 2.den Boon JA, Ahlquist P. 2010. Organelle-like membrane compartmentalization of positive-strand RNA virus replication factories. Annu. Rev. Microbiol. 64:241–256. 10.1146/annurev.micro.112408.134012 [DOI] [PubMed] [Google Scholar]
- 3.Laliberté JF, Sanfaçon H. 2010. Cellular remodeling during plant virus infection. Annu. Rev. Phytopathol. 48:69–91. 10.1146/annurev-phyto-073009-114239 [DOI] [PubMed] [Google Scholar]
- 4.McCartney AW, Greenwood JS, Fabian MR, White KA, Mullen RT. 2005. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell 17:3513–3531. 10.1105/tpc.105.036350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgyan J, Rubino L, Russo M. 1996. The 5′-terminal region of a tombusvirus genome determines the origin of multivesicular bodies. J. Gen. Virol. 77:1967–1974. 10.1099/0022-1317-77-8-1967 [DOI] [PubMed] [Google Scholar]
- 6.Scholthof KB, Scholthof HB, Jackson AO. 1995. The tomato bushy stunt virus replicase proteins are coordinately expressed and membrane associated. Virology 208:365–369. 10.1006/viro.1995.1162 [DOI] [PubMed] [Google Scholar]
- 7.Oster SK, Wu B, White KA. 1998. Uncoupled expression of p33 and p92 permits amplification of tomato bushy stunt virus RNAs. J. Virol. 72:5845–5851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rubino L, Burgyan J, Russo M. 1995. Molecular cloning and complete nucleotide sequence of carnation Italian ringspot tombusvirus genomic and defective interfering RNAs. Arch. Virol. 140:2027–2039. 10.1007/BF01322690 [DOI] [PubMed] [Google Scholar]
- 9.Cimino PA, Nicholson BL, Wu B, Xu W, White KA. 2011. Multifaceted regulation of translational readthrough by RNA replication elements in a tombusvirus. PLoS Pathog. 7:e1002423. 10.1371/journal.ppat.1002423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Navarro B, Rubino L, Russo M. 2004. Expression of the Cymbidium ringspot virus 33-kilodalton protein in Saccharomyces cerevisiae and molecular dissection of the peroxisomal targeting signal. J. Virol. 78:4744–4752. 10.1128/JVI.78.9.4744-4752.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hwang YT, McCartney AW, Gidda SK, Mullen RT. 2008. Localization of the Carnation Italian ringspot virus replication protein p36 to the mitochondrial outer membrane is mediated by an internal targeting signal and the TOM complex. BMC Cell Biol. 9:54. 10.1186/1471-2121-9-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagy PD, Barajas D, Pogany J. 2012. Host factors with regulatory roles in tombusvirus replication. Curr. Opin. Virol. 2:691–698. 10.1016/j.coviro.2012.10.004 [DOI] [PubMed] [Google Scholar]
- 13.Hanson PI, Cashikar A. 2012. Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 28:337–362. 10.1146/annurev-cellbio-092910-154152 [DOI] [PubMed] [Google Scholar]
- 14.Otegui MS, Spitzer C. 2008. Endosomal functions in plants. Traffic 9:1589–1598. 10.1111/j.1600-0854.2008.00787.x [DOI] [PubMed] [Google Scholar]
- 15.Schellmann S, Pimpl P. 2009. Coats of endosomal protein sorting: retromer and ESCRT. Curr. Opin. Plant Biol. 12:670–676. 10.1016/j.pbi.2009.09.005 [DOI] [PubMed] [Google Scholar]
- 16.Herman EK, Walker G, van der Giezen M, Dacks JB. 2011. Multivesicular bodies in the enigmatic amoeboflagellate Breviata anathema and the evolution of ESCRT 0. J. Cell Sci. 124:613–621. 10.1242/jcs.078436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Korbei B, Moulinier-Anzola J, De-Araujo L, Lucyshyn D, Retzer K, Khan MA, Luschnig C. 2013. Arabidopsis TOL proteins acts as gatekeepers for vacuolar sorting of PIN2 plasma membrane protein. Curr. Biol. 16:2500–2505 [DOI] [PubMed] [Google Scholar]
- 18.Morita E. 2012. Differential requirements of mammalian ESCRTs in multivesicular body formation, virus budding and cell division. FEBS J. 279:1399–1406. 10.1111/j.1742-4658.2012.08534.x [DOI] [PubMed] [Google Scholar]
- 19.Jouvenet N, Zhadina M, Bieniasz PD, Simon SM. 2011. Dynamics of ESCRT protein recruitment during retroviral assembly. Nat. Cell Biol. 13:394–401. 10.1038/ncb2207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin-Serrano J, Neil SJ. 2011. Host factors involved in retroviral budding and release. Nat. Rev. Microbiol. 9:519–531. 10.1038/nrmicro2596 [DOI] [PubMed] [Google Scholar]
- 21.Pornillos O, Higginson DS, Stray KM, Fisher RD, Garrus JE, Payne M, He GP, Wang HE, Morham SG, Sundquist WI. 2003. HIV Gag mimics the Tsg101-recruiting activity of the human Hrs protein. J. Cell Biol. 162:425–434. 10.1083/jcb.200302138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Im YJ, Kuo L, Ren X, Burgos PV, Zhao XZ, Liu F, Burke TR, Jr, Bonifacino JS, Freed EO, Hurley JH. 2010. Crystallographic and functional analysis of the ESCRT-I /HIV-1 Gag PTAP interaction. Structure 18:1536–1547. 10.1016/j.str.2010.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barajas D, Jiang Y, Nagy PD. 2009. A unique role for the host ESCRT proteins in replication of Tomato bushy stunt virus. PLoS Pathog. 5:e1000705. 10.1371/journal.ppat.1000705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Serviene E, Shapka N, Cheng CP, Panavas T, Phuangrat B, Baker J, Nagy PD. 2005. Genome-wide screen identifies host genes affecting viral RNA recombination. Proc. Natl. Acad. Sci. U. S. A. 102:10545–10550. 10.1073/pnas.0504844102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barajas D, Nagy PD. 2010. Ubiquitination of tombusvirus p33 replication protein plays a role in virus replication and binding to the host Vps23p ESCRT protein. Virology 397:358–368. 10.1016/j.virol.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rubino L, Russo M. 1998. Membrane targeting sequences in tombusvirus infections. Virology 252:431–437. 10.1006/viro.1998.9490 [DOI] [PubMed] [Google Scholar]
- 27.Stuchell MD, Garrus JE, Müller B, Stray KM, Ghaffarian S, McKinnon R, Kräusslich HG, Morham SG, Sundquist WI. 2004. The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J. Biol. Chem. 279:36059–36071. 10.1074/jbc.M405226200 [DOI] [PubMed] [Google Scholar]
- 28.Eastman SW, Martin-Serrano J, Chung W, Zhang T, Bieniasz PD. 2005. Identification of human VPS37C, a component of endosomal sorting complex required for transport-I important for viral budding. J. Biol. Chem. 280:628–636 [DOI] [PubMed] [Google Scholar]
- 29.Kostelansky MS, Sun J, Lee S, Kim J, Ghirlando R, Hierro A, Emr SD, Hurley JH. 2006. Structural and functional organization of the ESCRT-I trafficking complex. Cell 125:113–126. 10.1016/j.cell.2006.01.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teo H, Gill DJ, Sun J, Perisic O, Veprintsev DB, Vallis Y, Emr SD, Williams RL. 2006. ESCRT-I core and ESCRT-II GLUE domain structures reveal role for GLUE in linking to ESCRT-I and membranes. Cell 125:99–111. 10.1016/j.cell.2006.01.047 [DOI] [PubMed] [Google Scholar]
- 31.Hearne PQ, Knorr DA, Hillman BI, Morris TJ. 1990. The complete genome structure and synthesis of infectious RNA from clones of tomato bushy stunt virus. Virology 177:141–151. 10.1016/0042-6822(90)90468-7 [DOI] [PubMed] [Google Scholar]
- 32.Liu Y, Schiff M, Dinesh-Kumar SP. 2002. Virus-induced gene silencing in tomato. Plant J. 31:777–786. 10.1046/j.1365-313X.2002.01394.x [DOI] [PubMed] [Google Scholar]
- 33.Haas TJ, Sliwinski MK, Martinez DE, Preuss M, Ebine K, Ueda T, Nielsen E, Odorizzi G, Otegui MS. 2007. The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 19:1295–1312. 10.1105/tpc.106.049346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung SM, Frankman EL, Tzfira T. 2005. A versatile vector system for multiple gene expression in plants. Trends Plant Sci. 10:357–361. 10.1016/j.tplants.2005.06.001 [DOI] [PubMed] [Google Scholar]
- 35.Richardson LG, Howard AS, Khuu N, Gidda SK, McCartney A, Morphy BJ, Mullen RT. 2011. Protein-protein interaction network and subcellular localization of the Arabidopsis thaliana ESCRT machinery. Front. Plant Sci. 2:20. 10.3389/fpls.2011.00020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shockey JM, Gidda SK, Chapital DC, Kuan JC, Dhanoa PK, Bland JM, Rothstein SJ, Mullen RT, Dyer JM. 2006. Tung tree DGAT1 and DGAT2 have nonredundant functions in triacylglycerol biosynthesis and are localized to different subdomains of the endoplasmic reticulum. Plant Cell 18:2294–2313. 10.1105/tpc.106.043695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uemura T, Ueda T, Ohniwa RL, Nakano A, Takeyasu K, Sato MH. 2004. Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29:49–65. 10.1247/csf.29.49 [DOI] [PubMed] [Google Scholar]
- 38.Ching SL, Gidda SK, Rochon A, van Cauwenberghe OR, Shelp BJ, Mullen RT. 2012. Glyoxylate reductase isoform 1 is localized in the cytosol and not peroxisomes in plant cells. J. Integr. Plant Biol. 54:152–168. 10.1111/j.1744-7909.2012.01103.x [DOI] [PubMed] [Google Scholar]
- 39.Dhanoa PK, Richardson LG, Smith MD, Gidda SK, Henderson MP, Andrews DW, Mullen RT. 2010. Distinct pathways mediate the sorting of tail-anchored proteins to the plastid outer envelope. PLoS One 5:e10098. 10.1371/journal.pone.0010098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hurley BA, Tran HT, Marty NJ, Park J, Snedden WA, Mullen RT, Plaxton WC. 2010. The dual-targeted purple acid phosphatase isozyme AtPAP26 is essential for efficient acclimation of Arabidopsis to nutritional phosphate deprivation. Plant Physiol. 153:1112–1122. 10.1104/pp.110.153270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Citovsky V, Lee LY, Vyas S, Glick E, Chen MH, Vainstein A, Gafni Y, Gelvin SB, Tzfira T. 2006. Subcellular localization of interacting proteins by bimolecular fluorescence complementation in planta. J. Mol. Biol. 362:1120–1131. 10.1016/j.jmb.2006.08.017 [DOI] [PubMed] [Google Scholar]
- 42.Restrepo MA, Freed DD, Carrington JC. 1990. Nuclear transport of plant potyviral proteins. Plant Cell 2:987–998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clark SM, Di Leo R, Dhanoa PK, Van Cauwenberghe OR, Mullen RT, Shelp BJ. 2009. Biochemical characterization, mitochondrial localization, expression, and potential functions for an Arabidopsis gamma-aminobutyrate transaminase that utilizes both pyruvate and glyoxylate. J. Exp. Bot. 60:1743–1757. 10.1093/jxb/erp044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scholthof HB. 1999. Rapid delivery of foreign genes into plants by direct rub-inoculation with intact plasmid DNA of a tomato bushy stunt virus gene vector. J. Virol. 73:7823–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White KA, Morris TJ. 1994. Nonhomologous RNA recombination in tombusviruses: generation and evolution of defective interfering RNAs by stepwise deletions. J. Virol. 68:14–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.MacFarlane SA, Vassilakos N, Brown DJ. 1999. Similarities in the genome organization of Tobacco rattle virus and pea early-browning virus isolates that are transmitted by the same vector nematode. J. Gen. Virol. 80:273–276 [DOI] [PubMed] [Google Scholar]
- 47.Lingard MJ, Gidda SK, Bingham S, Rothstein SJ, Mullen RT, Trelease RN. 2008. Arabidopsis PEROXIN11c-e, FISSION1b, and DYNAMIN-RELATED PROTEIN3A cooperate in cell cycle-associated replication of peroxisomes. Plant Cell 20:1567–1585. 10.1105/tpc.107.057679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee MS, Mullen RT, Trelease RN. 1997. Oilseed isocitrate lyases lacking their essential type 1 peroxisomal targeting signal are piggybacked to glyoxysomes. Plant Cell 9:185–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luethy MH, Horak A, Elthon TE. 1993. Monoclonal antibodies to the α- and β-subunits of the plant mitochondrial F1-ATPase. Plant Physiol. 101:931–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frelin O, Agrimi G, Laera VL, Castegna A, Richardson LG, Mullen RT, Lerma-Ortiz C, Palmieri F, Hanson AD. 2012. Identification of mitochondrial thiamin diphosphate carriers from Arabidopsis and maize. Funct. Integr. Genomics 12:317–326. 10.1007/s10142-012-0273-4 [DOI] [PubMed] [Google Scholar]
- 51.Gidda SK, Shockey JM, Falcone M, Kim PK, Rothstein SJ, Andrews DW, Dyer JM, Mullen RT. 2011. Hydrophobic-domain-dependent protein-protein interactions mediate the localization of GPAT enzymes to ER subdomains. Traffic 12:452–472. 10.1111/j.1600-0854.2011.01160.x [DOI] [PubMed] [Google Scholar]
- 52.Lee LY, Wu FH, Hsu CT, Shen SC, Yeh HY, Liao DC, Fang MJ, Liu NT, Yen YC, Dokladal L, Sykorova E, Gelvin SB, Lin CS. 2012. Screening a cDNA library for protein-protein interactions directly in planta. Plant Cell 24:1746–1759. 10.1105/tpc.112.097998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Clontech Laboratories 2007. Matchmaker™ GAL4 two-hybrid system 3 & libraries user manual. PT3247-1 (PR742219). Clontech Laboratories, Inc., Mountain View, CA [Google Scholar]
- 54.Park S, Gidda SK, James CN, Horn PJ, Khuu N, Seay DC, Keereetaweep J, Chapman KD, Mullen RT, Dyer JM. 2013. The α/β hydrolase CGI-58 and peroxisomal transport protein PXA1 coregulate lipid homeostasis and signaling in Arabidopsis. Plant Cell 25:1726–1739. 10.1105/tpc.113.111898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shahriari M, Keshavaiah C, Scheuring D, Sabovljevic A, Pimpl P, Hausler RE, Hulskamp M, Schellmann S. 2010. The AAA-type ATPase AtSKD1 contributes to vacuolar maintenance of Arabidopsis thaliana. Plant J. 64:71–85 [DOI] [PubMed] [Google Scholar]
- 56.Babst M, Wendland B, Estepa EJ, Emr SD. 1998. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 17:2982–2993. 10.1093/emboj/17.11.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brandizzi F, Irons S, Kearns A, Hawes C. 2003. BY-2 cells: culture and transformation for live cell imaging. Curr. Protoc. Cell Biol. Chapter 1:Unit 1.7. 10.1002/0471143030.cb0107s19 [DOI] [PubMed] [Google Scholar]
- 58.Laporte C, Vetter G, Loudes AM, Robinson DG, Hillmer S, Stussi-Garaud C, Ritzenthaler C. 2003. Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of Grapevine fanleaf virus movement protein in tobacco BY-2 cells. Plant Cell 15:2058–2075. 10.1105/tpc.013896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wei T, Zhang C, Hong J, Xiong R, Kasschau KD, Zhou X, Carrington JC, Wang A. 2010. Formation of complexes at plasmodesmata for potyvirus intercellular movement is mediated by the viral protein P3N-PIPO. PLoS Pathog. 6:e1000962. 10.1371/journal.ppat.1000962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Duncan O, Taylor NL, Carrie C, Eubel H, Kubiszewski-Jakubiak S, Zhang B, Narsai R, Millar AH, Whelan J. 2011. Multiple lines of evidence localize signaling, morphology, and lipid biosynthesis machinery to the mitochondrial outer membrane of Arabidopsis. Plant Physiol. 157:1093–1113. 10.1104/pp.111.183160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shahriari M, Richter K, Keshavaiah C, Sabovljevic A, Huelskamp M, Schellmann S. 2011. The Arabidopsis ESCRT protein-protein interaction network. Plant Mol. Biol. 76:85–96. 10.1007/s11103-011-9770-4 [DOI] [PubMed] [Google Scholar]
- 62.Ibl V, Csaszar E, Schlager N, Neubert S, Spitzer C, Hauser MT. 2012. Interactome of the plant-specific ESCRT-III component AtVPS2.2 in Arabidopsis thaliana. J. Proteome Res. 11:397–411. 10.1021/pr200845n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Spitzer C, Schellmann S, Sabovljevic A, Shahriari M, Keshavaiah C, Bechtold N, Herzog M, Muller S, Hanisch FG, Hulskamp M. 2006. The Arabidopsis elch mutant reveals functions of an ESCRT component in cytokinesis. Development 133:4679–4689. 10.1242/dev.02654 [DOI] [PubMed] [Google Scholar]
- 64.Heinze C, Wobbe V, Lesemann DE, Zhang DY, Willingmann P, Adam G. 2004. Pelargonium necrotic spot virus: a new member of the genus Tombusvirus. Arch. Virol. 149:1527–1539 [DOI] [PubMed] [Google Scholar]
- 65.Célix A, Burgyan J, Rodriguez-Cerezo E. 1999. Interactions between tombusviruses and satellite RNAs of tomato bushy stunt virus: a defect in sat RNA B1 replication maps to ORF1 of a helper virus. Virology 262:129–138. 10.1006/viro.1999.9865 [DOI] [PubMed] [Google Scholar]
- 66.Burgyán J, Hornyik C, Szittya G, Silhavy D, Bisztray G. 2000. The ORF1 products of tombusviruses play a crucial role in lethal necrosis of virus-infected plants. J. Virol. 74:10873–10881. 10.1128/JVI.74.23.10873-10881.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ribeiro D, Jung M, Moling S, Borst JW, Goldbach R, Kormelink R. 2013. The cytosolic nucleoprotein of the plant-infecting bunyavirus tomato spotted wilt recruits endoplasmic reticulum-resident proteins to endoplasmic reticulum export sites. Plant Cell 25:3602–3614. 10.1105/tpc.113.114298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wei T, Zhang C, Hou X, Sanfacon H, Wang A. 2013. The SNARE protein Syp71 is essential for turnip mosaic virus infection by mediating fusion of virus-induced vesicles with chloroplasts. PLoS Pathog. 9:e1003378. 10.1371/journal.ppat.1003378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pornillos OP, Garrus JE, Sundquist WI. 2002. Mechanisms of enveloped RNA virus budding. Trends Cell Biol. 12:569–579. 10.1016/S0962-8924(02)02402-9 [DOI] [PubMed] [Google Scholar]
- 70.White KA, Nagy PD. 2004. Advances in the molecular biology of tombusviruses: gene expression, genome replication, and recombination. Prog. Nucleic Acid Res. Mol. Biol. 78:187–226. 10.1016/S0079-6603(04)78005-8 [DOI] [PubMed] [Google Scholar]
- 71.Nagy PD, Pogany J. 2006. Yeast as a model host to dissect functions of viral and host factors in tombusvirus replication. Virology 344:211–220. 10.1016/j.virol.2005.09.017 [DOI] [PubMed] [Google Scholar]
- 72.Mullen RT, McCartney AW, Flynn CR, Smith GST. 2006. Peroxisome biogenesis and the formation of multivesicular peroxisomes during tombusvirus infection: a role for ESCRT? Can. J. Bot. 84:551–564. 10.1139/b06-005 [DOI] [Google Scholar]
- 73.Di Franco A, Russo M, Martelli GP. 1984. Ultrastructure and origin of cytoplasmic multivesicular bodies induced by Carnation Italian ringspot virus. J. Gen. Virol. 65:1233–1237. 10.1099/0022-1317-65-7-1233 [DOI] [Google Scholar]
- 74.Serva S, Nagy PD. 2006. Proteomics analysis of the tombusvirus replicase: Hsp70 molecular chaperone is associated with the replicase and enhances viral RNA replication. J. Virol. 80:2162–2169. 10.1128/JVI.80.5.2162-2169.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ren X, Hurley JH. 2011. Proline-rich regions and motifs in trafficking: from ESCRT interaction to viral exploitation. Traffic 12:1282–1290. 10.1111/j.1600-0854.2011.01208.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dosztányi Z, Csizmok V, Tompa P, Simon I. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics 21:3433–3434. 10.1093/bioinformatics/bti541 [DOI] [PubMed] [Google Scholar]
- 77.Xue B, Williams RW, Oldfield CJ, Goh GK, Dunker AK, Uversky VN. 2010. Viral disorder or disordered viruses: do viral proteins possess unique features? Protein Pept. Lett. 17:932–951. 10.2174/092986610791498984 [DOI] [PubMed] [Google Scholar]
- 78.Stork J, Kovalev N, Sasvari Z, Nagy PD. 2011. RNA chaperone activity of the tombusviral p33 replication protein facilitates initiation of RNA synthesis by the viral RdRp in vitro. Virology 409:338–347. 10.1016/j.virol.2010.10.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nagy PD, Richardson CD. 2012. Viral replication–in search of the perfect host. Curr. Opin. Virol. 2:663–668. 10.1016/j.coviro.2012.11.001 [DOI] [PubMed] [Google Scholar]
- 80.Ross NT, Katt WP, Hamilton AD. 2010. Synthetic mimetics of protein secondary structure domains. Philos. Trans. A Math Phys. Eng. Sci. 368:989–1008. 10.1098/rsta.2009.0210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.McDonald B, Martin-Serrano J. 2008. Regulation of Tsg101 expression by the steadiness box: a role of Tsg101-associated ligase. Mol. Biol. Cell 19:754–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gill DJ, Teo H, Sun J, Perisic O, Veprintsev DB, Emr SD, Williams RL. 2007. Structural insight into the ESCRT-I/-II link and its role in MVB trafficking. EMBO J. 26:600–612. 10.1038/sj.emboj.7601501 [DOI] [PMC free article] [PubMed] [Google Scholar]