ABSTRACT
Retroviral virions initially assemble in an immature form that differs from that of the mature infectious particle. The RNA genomes in both immature and infectious particles are dimers, and interactions between the RNA dimer and the viral Gag protein ensure selective packaging into nascent immature virions. We used high-sensitivity selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) to obtain nucleotide-resolution structural information from scarce, femtomole quantities of Moloney murine leukemia virus (MuLV) RNA inside authentic virions and from viral RNA extracted from immature (protease-minus) virions. Our secondary structure model of the dimerization and packaging domain indicated that a stable intermolecular duplex known as PAL2, previously shown to be present in mature infectious MuLV particles, was sequestered in an alternate stem-loop structure inside immature virions. The intermediate state corresponded closely to a late-folding intermediate that we detected in time-resolved studies of the free MuLV RNA, suggesting that the immature RNA structure reflects trapping of the intermediate folding state by interactions in the immature virion. We propose models for the RNA-protein interactions that trap the RNA in the immature state and for the conformational rearrangement that occurs during maturation of virion particles.
IMPORTANCE The structure of the RNA genome in mature retroviruses has been studied extensively, whereas very little was known about the RNA structure in immature virions. The immature RNA structure is important because it is the form initially selected for packaging in new virions and may have other roles. This lack of information was due to the difficulty of isolating sufficient viral RNA for study. In this work, we apply a high-sensitivity and nucleotide-resolution approach to examine the structure of the dimerization and packaging domain of Moloney murine leukemia virus. We find that the genomic RNA is packaged in a high-energy state, suggesting that interactions within the virion hold or capture the RNA before it reaches its most stable state. This new structural information makes it possible to propose models for the conformational changes in the RNA genome that accompany retroviral maturation.
INTRODUCTION
An early step in the formation of an infectious retroviral particle involves the association of retroviral RNA with the cell membrane via interactions with the viral Gag protein. Budding from the cell yields a particle in which Gag protein domains are arrayed roughly in concentric spheres and the RNA is bound in the interior of the particle. Subsequent cleavage of the Gag structural protein results in large-scale rearrangements to yield a mature particle with a complex structure (1, 2). The genomic RNA also undergoes conformational rearrangements during particle maturation (3–5). In both immature and mature particles, two copies of the genomic RNA are present, and it is known that the structure adopted by the immature form of the viral RNA is less thermodynamically stable than and is structurally distinct from that of the mature form (4, 6). Little is known about the details of the structures adopted by the genomic RNA due to the extraordinary difficulty of producing sufficient retroviral RNA in the immature state for analysis using conventional structure-probing approaches.
Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) allows the RNA structure to be examined at single-nucleotide resolution (7–9). RNA is treated under conditions of interest with 1-methyl-nitroisatoic anhydride (1M7) (10) or other SHAPE reagents (7, 11) that react with flexible (single-stranded) nucleotides to form a bulky RNA adduct. After primer extension, the cDNAs can be separated by capillary electrophoresis (CE), and adduct locations are identified. The reactivity profile provides a measure of flexibility at each nucleotide and, indirectly, the likelihood that a given nucleotide is base paired (Fig. 1A). SHAPE has been used to develop models for many important retroviral RNA structures, including entire HIV-1 (12) and simian immunodeficiency virus (SIVmac) (13) genomes and the mature Moloney murine leukemia virus (MuLV) dimerization and regulatory domain (14). Structural analysis of authentic immature retroviral RNA is challenging because purification of virions from even large volumes of cell cultures from infected or transfected cells typically yields only attomole to femtomole (10−18 to 10−15) quantities of material, and conventional applications of CE-based structural technologies, including SHAPE, require picomole quantities of RNA (8).
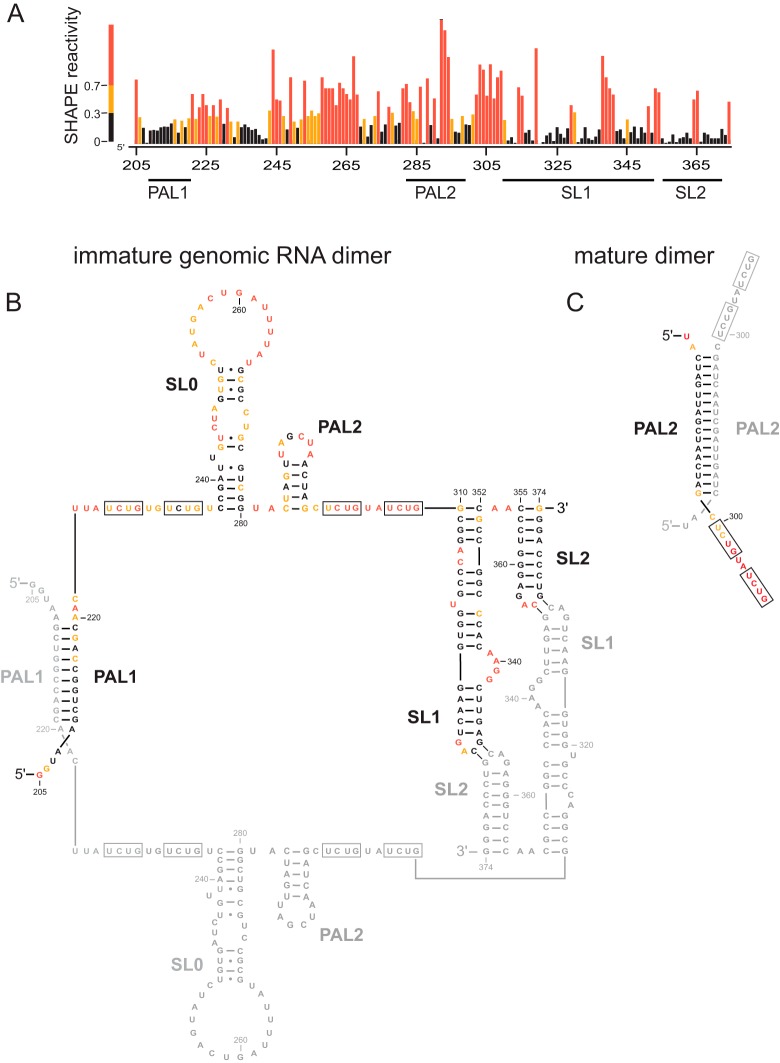
FIG 1.
SHAPE probing of the authentic MuLV immature genomic RNA dimer. (A) Integrated SHAPE data. (B) Secondary-structure model for the immature MuLV RNA genome in the packaging domain region. Nucleotides are colored by their SHAPE reactivities; major structural features are labeled. For clarity, nucleotides in the second strand of the dimer are shown in gray. (C) Secondary-structure model for the mature MuLV genomic RNA dimer in the PAL2 region (39).
We have improved the limits of CE-based detection by 4 orders of magnitude by designing instruments with subfemtomole SHAPE sensitivity (15). Here we describe the application of high-sensitivity SHAPE to an analysis of the secondary structure of the dimerization and packaging domain of MuLV in immature virions. The structure of the regulatory domain inside immature, noninfectious particles differed from that in mature particles. The immature RNA dimer structure was similar to that of a kinetic intermediate observed during the folding of the free RNA. Based on high-resolution structural information for the immature RNA dimer and based on phenotypes of viruses with mutations in the dimerization and packaging domain, we propose a structural model for this domain within packaged, immature genomic RNA.
MATERIALS AND METHODS
Immature MuLV virions and ex virio RNA dimer isolation.
Immature MuLV virion RNA was generated by using a mutant virus in which the active site of the viral protease (PR) contains an aspartate-to-leucine substitution (D32L) (3, 4). Because the PR-negative (PR−) virus cannot cleave Gag, maturation is inhibited, and the virion remains in the immature and noninfectious state. Immature virions were produced by transfection of proviral plasmids into 293T cells and isolated as described previously (12). Briefly, virus was recovered from clarified cell culture supernatants transfected (using TransIT 293 [MirusBio]) with the MuLV-PR− proviral plasmid pRB2719. Virus was collected by ultracentrifugation (Beckman SW-32 rotor at 144,000 × g at 4°C for 1 h). For ex virio RNA isolation, pellets were resuspended in 1 ml lysis buffer (50 mM Tris [pH 7.4], 10 mM EDTA, 1% [wt/vol] SDS, 100 mM NaCl, 100 μg/ml proteinase K, 120 μg/ml glycogen), incubated at 25°C for 45 min, and extracted three times with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and once with an equal volume of chloroform. Samples were precipitated in 70% ethanol with 0.3 M NaCl and stored at −20°C. RNA quantities were determined by real-time reverse transcriptase PCR using an MuLV gag transcript as a standard (nucleotides [nt] 1 to 1560) (GenBank accession number J02255) and quantification of the resulting gag cDNA target (positions 681 to 757) (forward primer 5′-CGG ATC GCT CAC AAC CAG T-3′, reverse primer 5′-AAG GTT GGC CAT TCT GCA GA-3′, and fluorescent probe 5′–6-carboxyfluorescein [FAM]–TAG ATG TCA AGA AGA GAC GTT GGG TTA CCT–6-carboxytetramethylrhodamine [TAMRA]–3′).
High-sensitivity SHAPE structure probing.
Extracted immature MuLV RNA (50 fmol) was incubated in folding buffer (50 mM HEPES [pH 7.5], 200 mM potassium acetate [pH 7.5], and 5 mM MgCl2) at 37°C for 20 min. SHAPE modification was initiated by treating 9 μl of the MuLV RNA sample with 1 μl 1M7 (50 mM in dimethyl sulfoxide [DMSO]), followed by incubation at 37°C for 2 min. RNA was recovered by using a precipitation approach optimized for the quantitative recovery of low-abundance RNAs (15) (2.5 volumes ethanol, 1 volume isopropanol, and 1 μl glycogen [20 mg/ml], with incubation at −20°C for 60 min and centrifugation at 20,000 × g). Pellets were resuspended in 6 μl 1/2× TE (5 mM Tris, 0.5 mM EDTA [pH 8.0]) buffer. For the in virio SHAPE experiments, 0.86 liters of virus in clarified transfected culture supernatants was collected by ultracentrifugation as described above and resuspended in 300 μl of a solution containing 50 mM HEPES, 200 mM NaCl, 0.1 mM EDTA, and 10% (vol/vol) fetal bovine serum (FBS) (pH 8.0). The sample was divided into two parts, and each part was treated with either DMSO or 5 mM 1M7 (final concentration). To isolate viral RNA, DMSO- and 1M7-treated virions were collected by ultracentrifugation, and the pellets were dissolved in 0.2 ml of lysis buffer (50 mM Tris [pH 7.4], 10 mM EDTA, 1% [wt/vol] SDS, 100 mM NaCl, 20 mM dithiothreitol, and 1.0 mg/ml proteinase K), incubated at 25°C for 45 min, and extracted three times with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) and once with an equal volume of chloroform. RNA samples were precipitated in 70% (vol/vol) ethanol with 0.3 M NaCl and stored at −20°C.
Primer extension was performed as described previously (8, 16). DNA primers incorporated locked nucleic acid (LNA) nucleotides to increase binding affinity and stringency for low-copy-number RNAs (17–19). MuLV primers (5′-GGUGC ACCAA AGAGU CCAAA AGC-3′, 5′-end labeled with 5-FAM or 6-JOE [LNA positions are underlined]) annealed to the 3′ end of the MuLV dimerization domain (nucleotides 422 to 445). Primers (0.5 μl; 2 fmol) were annealed to MuLV RNA (6 μl; ∼50 fmol in 1/2× TE buffer) by heating at 65°C and 45°C for 5 min and then snap-cooling on ice. Reverse transcription buffer (3 μl) (200 mM Tris [pH 8.0], 250 mM KCl, 10 mM MgCl2, 2 mM each deoxynucleoside triphosphate [dNTP], 20 mM dithiothreitol [DTT]) was added at 0°C, and primer extension was performed with Superscript III (Invitrogen) reverse transcriptase (0.5 μl; 100 U) at 45°C for 1 min, followed by incubation at 52°C and 65°C for 10 min each. Reaction mixtures were cooled to 4°C and quenched by the addition of 3 M sodium acetate (pH 5.2). A sequencing marker was generated by adding 0.5 μl dideoxy-GTP (10 mM) to the primer extension reaction mixture using unmodified RNA. The 1M7 and DMSO reaction mixes were each combined with equal amounts of dideoxy-GTP-terminated sequencing ladders, precipitated with ethanol, and resuspended in deionized formamide (20 μl).
Capillary electrophoresis.
MuLV cDNA fragments, extended from 50 fmol of input RNA, were resolved by high-sensitivity capillary electrophoresis (15). Samples (10 μl) in deionized formamide were electrokinetically injected (10.2 kV for 30 s) onto a 47-cm (50-μm-inner-diameter) uncoated capillary containing Pop7 (Applied Biosystems) as the internal polymer. The detection window was placed at 36 cm. Voltage was applied by using positive polarity (10.2 kV for 60 min) and immersing the capillary ends in 1× running buffer (Applied Biosystems). The photomultiplier tube voltages were 0.9 kV, and the laser power was 2 mW. Excitation was achieved at 488 and 514 nm by using an argon laser focused within the capillary at the detection window. 5-FAM was used to detect cDNAs corresponding to SHAPE structure probing, and the lower-sensitivity dye 6-JOE was used for sequencing.
Data processing and structure modeling.
Raw sequence traces were corrected for dye variation and signal decay; peak intensities were integrated by using ShapeFinder (20). Traces were analyzed by combining data from two separate capillary runs. Each capillary electrophoresis separation contained (i) a reaction performed in the presence or absence of 1M7 (detected by using a FAM-labeled primer) and (ii) a sequencing reaction (labeled with JOE) performed by using dideoxy-GTP. Data from the two runs were aligned by using identical sequencing lanes and analyzed as described previously (20). SHAPE reactivities were normalized by dividing by the average intensity of the 10% most reactive nucleotides, after excluding outliers identified by box plot analysis (21). SHAPE reactivity information was used to impose a pseudo-free-energy change constraint in conjunction with nearest-neighbor thermodynamic parameters in the RNAstructure prediction program (21–23). For comparison of the free RNA and in-virion RNA experiments, protection factors (24) were calculated as (free RNA − in-virion RNA)/(in-virion RNA), where free RNA and in-virion RNA indicate the SHAPE intensities at each position.
S+/L− viral replication assay.
The S+/L− focus assay enumerates virus particles capable of undergoing several replication cycles in the assay cells over a 4- to 5-day period. The assay was performed as described previously (25). Wild-type and mutant virus particles were produced by transient transfection of 293T cells. The virus-containing culture fluids were serially diluted and inoculated onto the S+/L− assay plates; foci were counted 4 to 5 days later. To quantitate the level of physical virus particles in the fluids, we also measured reverse transcriptase activity (26). Infectivity titers, expressed in focus-inducing units/ml, were normalized to the reverse transcriptase activities so that the results could be expressed as specific infectivity. Results are reported as a percentage of the wild-type specific infectivity with standard errors calculated from duplicate measurements.
RESULTS
SHAPE analysis of MuLV RNA in immature particles.
Immature MuLV virions were produced by transfecting cells with a mutant proviral plasmid expressing a viral protease that is inactivated by an active-site point mutation, D32L (2, 3). As the Gag polyprotein is not cleaved in the resulting virions, viral maturation is halted at the immature stage, and this PR− virus cannot replicate (1, 4, 5). A highly optimized, large-scale preparation from a liter of a transfected-cell culture yielded only a few picomoles of immature viral RNA.
We used high-sensitivity SHAPE, developed precisely for the structural analysis of authentic, low-abundance RNAs, to analyze the MuLV dimerization and packaging regulatory domain (24, 27–29), of approximately 170 nucleotides. RNAs were extracted from virions under mild, nondenaturing conditions designed to preserve preexisting RNA structures. SHAPE was performed on aliquots of 50 fmol of the RNA. Processed SHAPE reactivities were used to develop a model for the secondary structure of the regulatory domain (Fig. 1).
We compared the SHAPE probing data and the resulting data-constrained secondary-structure models obtained for genomic RNA isolated from immature virions with previous data on the mature MuLV RNA genome (14). This comparison supports an overall architecture for the immature RNA that is similar to that of MuLV RNA in mature virions (14, 24). The RNA strands in the mature dimer state are linked by four major interaction motifs (termed PAL1, PAL2, SL1, and SL2). Nucleotides in the apical loops in SL1 and SL2 are generally unreactive toward SHAPE, consistent with the formation of an intermolecular kissing interaction between nucleotides in these loops in both the immature and mature states (30) (Fig. 1B). Modest SHAPE reactivity at C365 in the loop of SL2 may reflect a slight weakening of this structure in the immature state. The self-complementary sequence, termed PAL1, shows relatively low reactivity in both immature and mature structures, consistent with the formation of a 10-bp intermolecular duplex in both types of structures (Fig. 1B). In contrast, many nucleotides in a second self-complementary sequence, PAL2, are reactive by SHAPE in the RNA from immature particles but not in the mature genome, consistent with the presence of an intramolecular stem-loop structure in the RNA from immature particles and an intermolecular duplex in the mature virion (nucleotides 283 through 298) (Fig. 1B). Thus, the RNA conformation is quite different in the PAL2 region for genomes isolated from immature and mature particles (Fig. 1C).
Immature MuLV RNA resembles a dimerization intermediate.
We recently used time-resolved SHAPE to understand the mechanism by which the MuLV nucleocapsid (NC) protein facilitates dimerization of the MuLV RNA genome (31). We also determined the pathway of folding in the absence of protein (Fig. 2A). We compared the SHAPE reactivities for nucleotides in the 170-nt dimerization domain obtained in 16 consecutive 1-s snapshots during dimerization of the RNA in the absence of any protein to the SHAPE reactivities of these nucleotides in the immature MuLV RNA. We quantified the similarity between structures that form during the time-resolved dimerization reaction (Fig. 2B) and those measured for the immature viral RNA (Fig. 1B) by analyses of correlations in SHAPE reactivities.
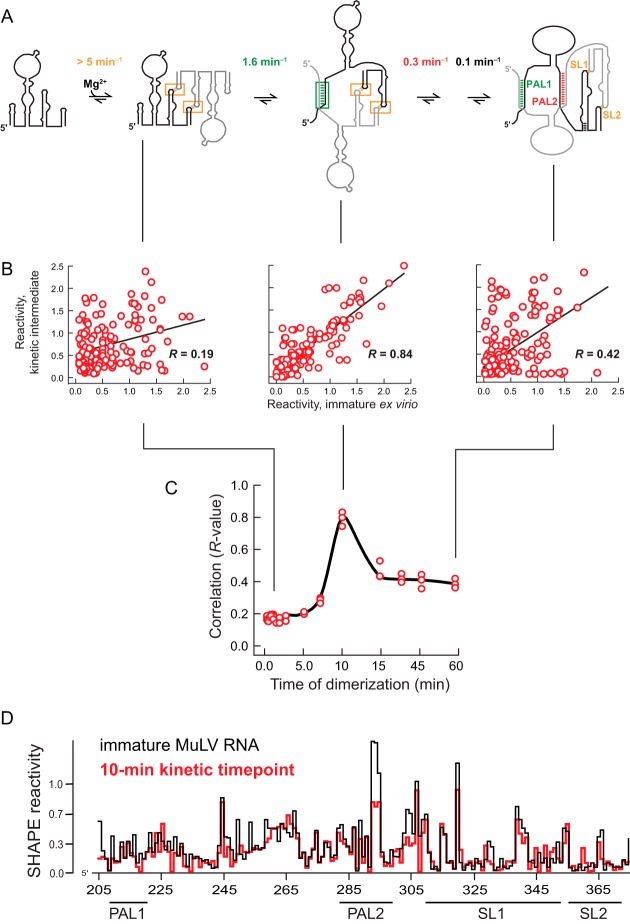
FIG 2.
Correlation between SHAPE reactivities for the immature MuLV RNA and reactivities of intermediates observed during MuLV dimerization. (A) Assembly mechanism for the MuLV packaging domain occurring in solution in the absence of any protein (31). (B) Correlation plots showing the relationship between the structure of the MuLV RNA from immature particles and dimerization states at the 2-, 10-, and 60-min time points based on SHAPE probing. Pearson's R values are shown. (C) Correlations between immature RNA MuLV SHAPE reactivities and reactivities over the course of the dimerization reaction. Correlations from multiple independent in vitro dimerization experiments are shown as separate points. (D) Superposition of SHAPE reactivity profiles corresponding to the ex virio immature MuLV RNA with the 10-min kinetic intermediate.
The profiles obtained during the time-resolved experiment that corresponded to the RNA in the monomer state or in an initial state involving interactions between the SL1 and SL2 stem-loops had low correlations with the immature RNA (Pearson's R, ∼0.2). This low correlation indicates that, as expected, the initial state in the dimerization experiments and the immature state isolated from virions have very different structures (Fig. 2B, left). In contrast, the correlation between the profile for the immature virion and that at the 10-min time point was high, with an R value of 0.84, indicating that these two RNA structures are highly similar. The correlation was low (R, ∼0.4) between the profiles of the RNA from the immature virion and those of the fully folded mature structure (Fig. 2B, compare center and right panels). The immature structure thus correlates most strongly to that observed in a single step in the monomer-to-dimer transition (Fig. 2C). This view is reinforced by directly comparing SHAPE reactivity profiles for protein-free immature RNA and for the 10-min kinetic time point at nucleotide resolution (Fig. 2D). The overall conformation of the immature state is highly similar to that of a kinetic intermediate in the dimerization of MuLV genomic RNA.
SHAPE analysis of MuLV RNA inside immature virions.
To examine the extent to which the SHAPE profile of RNA extracted from PR− immature virions reflected its conformation within these virus particles, we next probed the structure of the MuLV genomic dimer within intact PR− immature viruses, again using femtomole quantities of RNA (15, 24). Intact viral particles were treated with 1M7, the modified RNA was extracted, and primer extension products were quantified. SHAPE profiles from the RNA gently extracted from immature virions and from RNA within virions were used to quantify the effect of the in-virion environment on RNA SHAPE reactivities. We calculated a protection factor, equal to the difference in SHAPE reactivities, normalized by the in-virion reactivity. This analysis revealed that two major regions are protected within the virus particle for both the immature and mature particles (positive protection factors) (Fig. 3). It was previously shown that NC binds to and protects four UCUG sequences in the genomic RNA from chemical modification (24) (Fig. 1B, boxed regions). Although the protected regions are the same in both immature and mature particles, protection patterns were stronger and more distinct in the immature virions (Fig. 3). In PR− immature virions, the NC domain is part of the Gag polyprotein, whereas in mature virions, the polyprotein has been processed. We conclude that the genomic RNA dimer is bound to the NC domain of the Gag protein within immature virions and that these protein-RNA interactions have distinct features in immature and mature virions.
FIG 3.
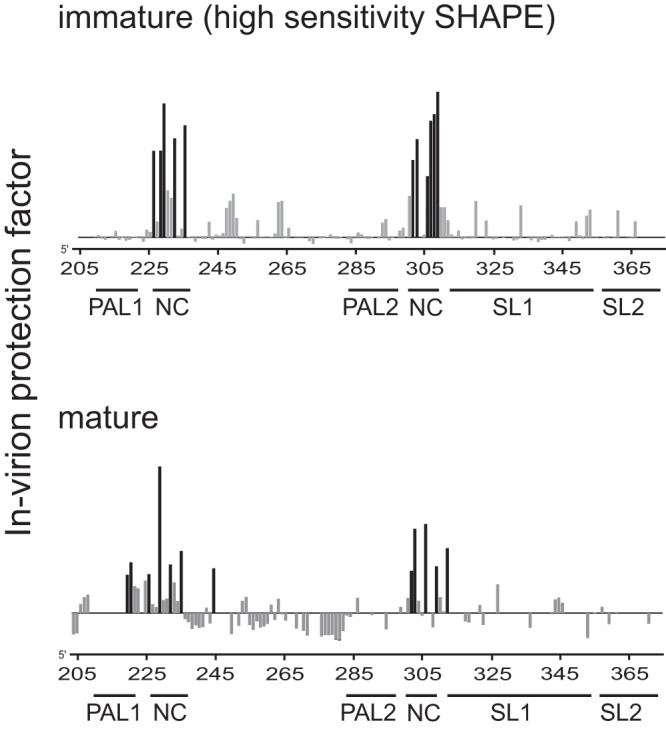
Differences in SHAPE reactivities in MuLV RNAs from immature virions versus mature virions. Shown are protection factors for the immature (top) and mature (bottom) virions. Positive protection factors (see Materials and Methods) report regions constrained inside intact virions relative to genomic RNA gently extracted from virions; negative protection factors (of which there are few) correspond to positions that are more reactive by SHAPE inside virions. Sites of strongest in-virion protection in each case are emphasized in black. Data in the bottom panel were reported previously; experiments probing immature and mature RNAs inside virions were performed under identical conditions (24).
Replication competence of MuLV with mutations in PAL2.
The data presented above show that the immature MuLV dimer differs most strikingly from the mature dimer by the fact that the PAL2 region adopts an intramolecular stem-loop structure rather than an intermolecular base-paired helix (Fig. 1B and C). To examine the biological significance of the differences in PAL2 structure in immature versus mature viruses, we mutated PAL2 sequences and looked for changes in the functional capabilities of the mutant viral genomes. The mutations either completely deleted PAL2 (nt 283 to 297); created a weaker stem in the PAL2 region, which would be unable to form the intermolecular duplex that forms in the mature RNA; or replaced PAL2 (nt 282 to 300) with all uridine residues (Fig. 4, top). The mutations were placed into an infectious MuLV clone, and the clones were transfected into 293T cells. Virus produced by the transfected cells was assayed for infectivity by the S+/L− assay (25). Virus preparations were also assayed for reverse transcriptase activity to normalize for the number of virus particles. We report specific infectivities in each case, corresponding to viral titers, normalized to the reverse transcriptase activity (Fig. 4).
FIG 4.
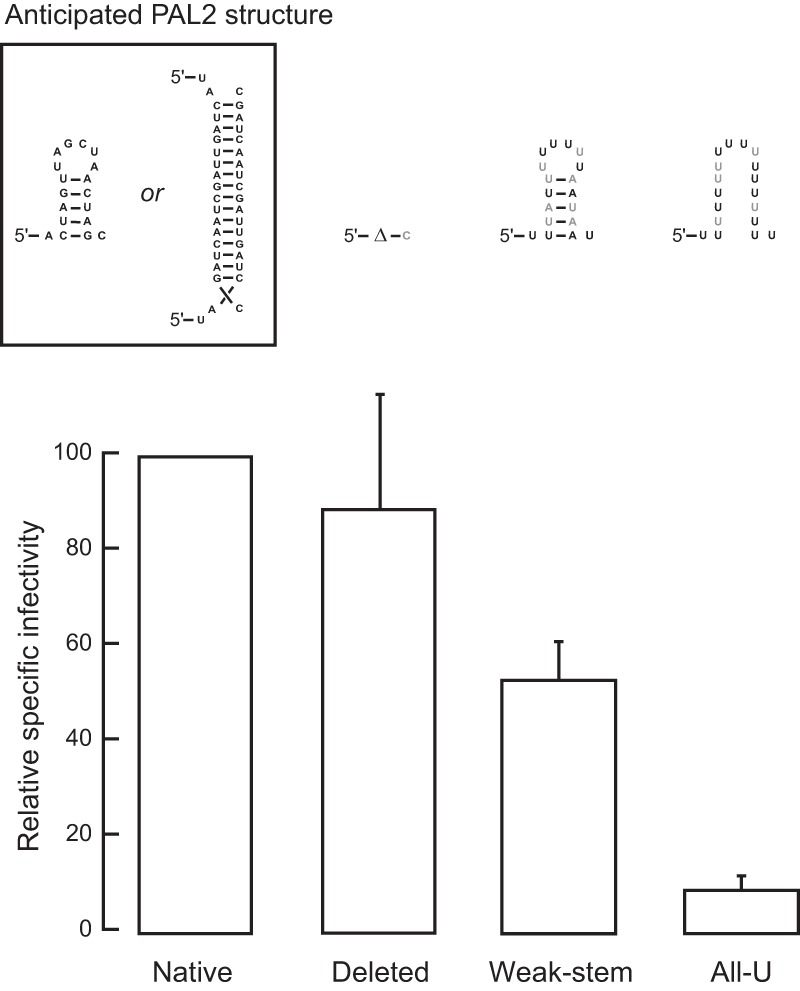
Replication efficiency of MuLV mutants. (Top) Mutations designed to affect the structure in the PAL2 region. Mutated positions are emphasized with black text. (Bottom) Replication efficiencies for each of the four PAL2 mutants were determined by using the S+/L− focus assay (25). Error bars show the standard errors of the means from two trials.
The mutants in which PAL2 was deleted or the ability of PAL2 to dimerize was disrupted had minimal effects on viral infectivity. Replacing all of PAL2 with U residues reduced the infectivity of the virus by roughly 10-fold (Fig. 4). The S+/L− infectivity assay requires an individual virus to undergo multiple rounds of replication during the 4- to 5-day assay interval to produce a focus (25). The ability of the PAL2 mutants to produce any foci implies that they retained the ability to replicate with nearly the efficiency of the native sequence of MuLV. We also introduced the same mutations into pBabeLuc, an MuLV-derived vector expressing the firefly luciferase gene (32). The infectivity data obtained with pBabeLuc also showed that changes in PAL2 had relatively little effect on viral replication under the conditions of this assay (data not shown).
DISCUSSION
Two pieces of physical evidence distinguish the immature and mature states of most retroviral RNA genomes examined to date, including that of MuLV. First, cryo-electron microscopy studies showed that the protein architecture of immature virions contains uncleaved Gag polyprotein subunits arranged such that the matrix, capsid, and nucleocapsid domains form concentric spheres (33, 34). Second, genomic RNA extracted from immature virions has a lower thermal stability than its mature viral counterpart (3, 4, 27, 35), suggesting a significant difference in secondary structure, although the specific locations of the RNA structural differences were not established in previous studies.
In our work, immature and mature virions were isolated under conditions that were vigorous enough to remove viral proteins and membranes but avoided obvious denaturing steps (see Materials and Methods). We note that there is always the possibility that the extraction process affects the final RNA conformation, especially when analyzing a potential kinetic intermediate. However, RNA structures analyzed inside virions and for the protein-free RNA were highly similar, outside the Gag or NC binding sites (Fig. 3). This similarity suggests that structural effects due to RNA genome isolation, while possible, do not strongly affect the conclusions presented here.
Using SHAPE with femtomole (10−15) detection sensitivity (15), we showed that the packaging domain region of MuLV RNA is structurally distinct from that of the RNA in mature particles. Two copies of the genomic RNA are packaged in each virion. In mature particles, the PAL2 sequence is part of an intermolecular duplex, whereas the SHAPE-directed model for the RNA in the immature particle indicates that this region forms a unimolecular stem-loop structure. There are thus fewer intergenome interactions in the immature particle than in the mature particle, consistent with the lower thermodynamic stability of the dimeric structure in the immature genome observed previously (3, 4, 36, 37).
Our data show that the NC domain of Gag and mature NC bind in roughly the same region in immature and mature forms of the genomic RNA (Fig. 3). NC binds to genomic RNA at two sets of tandem UCUG repeats (24). These motifs (boxed regions in Fig. 1 and 5) comprise the primary packaging signal for MuLV; point mutations in these elements reduce genome packaging to background levels (24). The nucleotide-resolution protection patterns in the immature and mature virions differ, suggesting that the protein-RNA interactions change to become less distinct (Fig. 3) as NC is cleaved from the Gag polyprotein and virions undergo maturation.
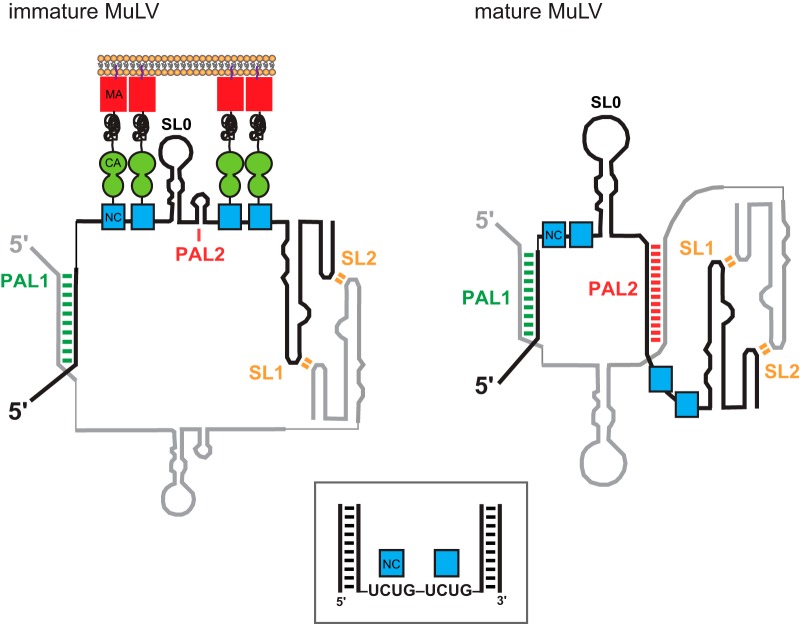
FIG 5.
Model for MuLV retroviral genome packaging in immature and mature states. (Left) Model for the NC domain of the Gag polyprotein bound to the MuLV packaging signal RNA in the immature virion. (Right) Free NC protein bound to the MuLV packaging signal RNA in the mature virion (24). For clarity, Gag and NC interactions are shown for one genomic strand only. (Boxed inset) Model of the high-affinity packaging structure for MuLV based on the recognition of exposed UCUG motifs (40) and the requirement that these sequences occur in a specific three-dimensional structural context involving tandem sequence repeats and flanking double-stranded regions (24).
We evaluated the replication efficiencies of MuLV variants with changes relative to the wild-type sequence in the PAL2 regions in an in vivo replication assay (Fig. 4). The changes that we introduced, which were designed to weaken or destroy the PAL2 stem-loop in the monomer (thus mimicking the structure of the immature dimer) or to completely delete PAL2, had relatively little effect on the ability of the virus to replicate itself. This implies that all of the functions of this region of viral RNA, including packaging into virus particles and expression of viral genes, have only a small dependence on the structure or presence of PAL2 under conditions of replication in cell culture. The data are modestly supportive of a model in which a specific structural spacing between and around the tandem UCUG motifs is important but the sequence making up this structure is not. When PAL2 was deleted, the flanking structured regions around both UCUG repeats, SL0 and SL1, remained intact (Fig. 1 and 5), consistent with previous studies showing that the optimal NC binding site includes flanking duplex structures (Fig. 5, boxed structure) (24). In contrast, replacing the PAL2 stem with an unstructured U sequence had a modest effect on viral replication (Fig. 4, right). This mutation disrupts the structured region-unstructured UCUG repeat region-structured region pattern that mediates optimal NC (or Gag) binding.
Our analysis of MuLV genomic RNA gently extracted from immature viral particles and of RNA inside immature virions revealed that the packaging domain of the immature RNA genome closely resembles an intermediate conformation adopted in a late kinetic step in the dimerization of the MuLV genome (Fig. 2). These data are consistent with two different models for the state formed between Gag and the immature RNA. In one model, the immature Gag-RNA and mature NC-RNA complexes simply have different lowest-free-energy states. In the other model, the immature complex is packaged in a kinetically trapped state due to interactions within the virion.
The RNA is bound by protein components in the intact immature virion, likely the NC domain of Gag, in roughly the same region as in the final mature virion. The differences that we observed in protection patterns when the immature RNA in virion particles was compared to that in mature particles suggest that the protein-RNA interactions are stronger in immature particles due to either direct protein-RNA interactions or cooperative protein interactions (38) (Fig. 5). These interactions appear to be strong enough to arrest RNA folding in what corresponds to a high-energy intermediate folding state in the pathway established by analysis of RNA in the absence of protein (31). Folding into the mature conformation evidently requires processing of the Gag polyprotein.
This work emphasizes the power of SHAPE to identify distinct RNA conformational states in very small quantities of RNA in physiologically relevant complexes. This work supports a model in which MuLV RNA is constrained in the membrane by Gag in a high-energy intermediate state during immature virus assembly. This conformation appears to be trapped due to strong interactions with the nucleocapsid domain of the still-intact Gag polyprotein. Once the Gag polyprotein is cleaved into its four constituent proteins, the genomic RNA folds into its mature infectious form (Fig. 5), potentially facilitated by the chaperone activity of NC.
ACKNOWLEDGMENTS
This work was supported by a grant from the U.S. National Institutes of Health (GM064803 to K.M.W.); in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research (A.R.); and under contract number HHSN261200800001E with Leidos Biomedical Research, Inc. (R.J.G.).
We are indebted to Jane Mirro for superb technical assistance.
Footnotes
Published ahead of print 12 March 2014
REFERENCES
- 1.Coffin JM, Hughes SH, Varmus HE. 1997. Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, NY: [PubMed] [Google Scholar]
- 2.Yeager M, Wilson-Kubalek EM, Weiner SG, Brown PO, Rein A. 1998. Supramolecular organization of immature and mature murine leukemia virus revealed by electron cryo-microscopy: implications for retroviral assembly mechanisms. Proc. Natl. Acad. Sci. U. S. A. 95:7299–7304. 10.1073/pnas.95.13.7299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fu W, Rein A. 1993. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 67:5443–5449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fu W, Gorelick RJ, Rein A. 1994. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 68:5013–5018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berkowitz R, Fisher J, Goff SP. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177–218 [DOI] [PubMed] [Google Scholar]
- 6.Song R, Kafaie J, Yang L, Laughrea M. 2007. HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. J. Mol. Biol. 371:1084–1098. 10.1016/j.jmb.2007.06.010 [DOI] [PubMed] [Google Scholar]
- 7.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. 2005. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 127:4223–4231. 10.1021/ja043822v [DOI] [PubMed] [Google Scholar]
- 8.Wilkinson KA, Merino EJ, Weeks KM. 2006. Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1:1610–1616. 10.1038/nprot.2006.249 [DOI] [PubMed] [Google Scholar]
- 9.Weeks KM, Mauger DM. 2011. Exploring RNA structural codes with SHAPE chemistry. Acc. Chem. Res. 44:1280–1291. 10.1021/ar200051h [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mortimer SA, Weeks KM. 2007. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 129:4144–4145. 10.1021/ja0704028 [DOI] [PubMed] [Google Scholar]
- 11.Mortimer SA, Weeks KM. 2008. Time-resolved RNA SHAPE chemistry. J. Am. Chem. Soc. 130:16178–16180. 10.1021/ja8061216 [DOI] [PubMed] [Google Scholar]
- 12.Watts JM, Dang KK, Gorelick RJ, Leonard CW, Bess JW, Jr, Swanstrom R, Burch CL, Weeks KM. 2009. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 460:711–716. 10.1038/nature08237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pollom E, Dang KK, Potter EL, Gorelick RJ, Burch CL, Weeks KM, Swanstrom R. 2013. Comparison of SIV and HIV-1 genomic RNA structures reveals impact of sequence evolution on conserved and non-conserved structural motifs. PLoS Pathog. 9:e1003294. 10.1371/journal.ppat.1003294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gherghe C, Leonard CW, Gorelick RJ, Weeks KM. 2010. Secondary structure of the mature ex virio Moloney murine leukemia virus genomic RNA dimerization domain. J. Virol. 84:898–906. 10.1128/JVI.01602-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grohman JK, Kottegoda S, Gorelick RJ, Allbritton NL, Weeks KM. 2011. Femtomole SHAPE reveals regulatory structures in the authentic XMRV RNA genome. J. Am. Chem. Soc. 133:20326–20334. 10.1021/ja2070945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gherghe C, Weeks KM. 2006. The SL1-SL2 (stem-loop) domain is the primary determinant for stability of the gamma retroviral genomic RNA dimer. J. Biol. Chem. 281:37952–37961. 10.1074/jbc.M607380200 [DOI] [PubMed] [Google Scholar]
- 17.Latorra D, Arar K, Hurley JM. 2003. Design considerations and effects of LNA in PCR primers. Mol. Cell. Probes 17:253–259. 10.1016/S0890-8508(03)00062-8 [DOI] [PubMed] [Google Scholar]
- 18.Levin JD, Fiala D, Samala MF, Kahn JD, Peterson RJ. 2006. Position-dependent effects of locked nucleic acid (LNA) on DNA sequencing and PCR primers. Nucleic Acids Res. 34:e142. 10.1093/nar/gkl756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fratczak A, Kierzek R, Kierzek E. 2009. LNA-modified primers drastically improve hybridization to target RNA and reverse transcription. Biochemistry 48:514–516. 10.1021/bi8021069 [DOI] [PubMed] [Google Scholar]
- 20.Vasa SM, Guex N, Wilkinson KA, Weeks KM, Giddings MC. 2008. ShapeFinder: a software system for high-throughput quantitative analysis of nucleic acid reactivity information resolved by capillary electrophoresis. RNA 14:1979–1990. 10.1261/rna.1166808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deigan KE, Li TW, Mathews DH, Weeks KM. 2009. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. U. S. A. 106:97–102. 10.1073/pnas.0806929106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Low JT, Weeks KM. 2010. SHAPE-directed RNA secondary structure prediction. Methods 52:150–158. 10.1016/j.ymeth.2010.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. 2004. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl. Acad. Sci. U. S. A. 101:7287–7292. 10.1073/pnas.0401799101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gherghe C, Lombo T, Leonard CW, Datta SA, Bess JW, Jr, Gorelick RJ, Rein A, Weeks KM. 2010. Definition of a high-affinity Gag recognition structure mediating packaging of a retroviral RNA genome. Proc. Natl. Acad. Sci. U. S. A. 107:19248–19253. 10.1073/pnas.1006897107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bassin RH, Tuttle N, Fischinger PJ. 1971. Rapid cell culture assay technique for murine leukaemia viruses. Nature 229:564–566. 10.1038/229564b0 [DOI] [PubMed] [Google Scholar]
- 26.Fu W, Ortiz-Conde BA, Gorelick RJ, Hughes SH, Rein A. 1997. Placement of tRNA primer on the primer-binding site requires pol gene expression in avian but not murine retroviruses. J. Virol. 71:6940–6946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hibbert CS, Mirro J, Rein A. 2004. mRNA molecules containing murine leukemia virus packaging signals are encapsidated as dimers. J. Virol. 78:10927–10938. 10.1128/JVI.78.20.10927-10938.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evans MJ, Bacharach E, Goff SP. 2004. RNA sequences in the Moloney murine leukemia virus genome bound by the Gag precursor protein in the yeast three-hybrid system. J. Virol. 78:7677–7684. 10.1128/JVI.78.14.7677-7684.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Badorrek CS, Weeks KM. 2006. Architecture of a gamma retroviral genomic RNA dimer. Biochemistry 45:12664–12672. 10.1021/bi060521k [DOI] [PubMed] [Google Scholar]
- 30.Badorrek CS, Gherghe CM, Weeks KM. 2006. Structure of an RNA switch that enforces stringent retroviral genomic RNA dimerization. Proc. Natl. Acad. Sci. U. S. A. 103:13640–13645. 10.1073/pnas.0606156103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grohman JK, Gorelick RJ, Lickwar CR, Lieb JD, Bower BD, Znosko BM, Weeks KM. 2013. A guanosine-centric mechanism for RNA chaperone function. Science 340:190–195. 10.1126/science.1230715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rulli SJ, Jr, Muriaux D, Nagashima K, Mirro J, Oshima M, Baumann JG, Rein A. 2006. Mutant murine leukemia virus Gag proteins lacking proline at the N-terminus of the capsid domain block infectivity in virions containing wild-type Gag. Virology 347:364–371. 10.1016/j.virol.2005.12.012 [DOI] [PubMed] [Google Scholar]
- 33.Briggs JA, Wilk T, Welker R, Krausslich HG, Fuller SD. 2003. Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 22:1707–1715. 10.1093/emboj/cdg143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wright ER, Schooler JB, Ding HJ, Kieffer C, Fillmore C, Sundquist WI, Jensen GJ. 2007. Electron cryotomography of immature HIV-1 virions reveals the structure of the CA and SP1 Gag shells. EMBO J. 26:2218–2226. 10.1038/sj.emboj.7601664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stoltzfus CM, Snyder PN. 1975. Structure of B77 sarcoma virus RNA: stabilization of RNA after packaging. J. Virol. 16:1161–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart L, Schatz G, Vogt VM. 1990. Properties of avian retrovirus particles defective in viral protease. J. Virol. 64:5076–5092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oertle S, Spahr PF. 1990. Role of the gag polyprotein precursor in packaging and maturation of Rous sarcoma virus genomic RNA. J. Virol. 64:5757–5763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O'Carroll IP, Crist RM, Mirro J, Harvin D, Soheilian F, Kamata A, Nagashima K, Rein A. 2012. Functional redundancy in HIV-1 viral particle assembly. J. Virol. 86:12991–12996. 10.1128/JVI.06287-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Badorrek CS, Weeks KM. 2005. RNA flexibility in the dimerization domain of a gamma retrovirus. Nat. Chem. Biol. 1:104–111. 10.1038/nchembio712 [DOI] [PubMed] [Google Scholar]
- 40.D'Souza V, Summers MF. 2005. How retroviruses select their genomes. Nat. Rev. Microbiol. 3:643–655. 10.1038/nrmicro1210 [DOI] [PubMed] [Google Scholar]