ABSTRACT
The measles virus (MeV) membrane fusion apparatus consists of a fusion protein trimer and an attachment protein tetramer. To trigger membrane fusion, the heads of the MeV attachment protein, hemagglutinin (H), bind cellular receptors while the 96-residue-long H stalk transmits the triggering signal. Structural and functional studies of the triggering mechanism of other paramyxoviruses suggest that receptor binding to their hemagglutinin-neuraminidase (HN) results in signal transmission through the central segments of their stalks. To gain insight into H-stalk structure and function, we individually replaced its residues with cysteine. We then assessed how stable the mutant proteins are, how efficiently they can be cross-linked by disulfide bonds, whether cross-linking results in loss of function, and, in this case, whether disulfide bond reduction restores function. While many residues in the central segment of the stalk and in the spacer segment above it can be efficiently cross-linked by engineered disulfide bonds, we report here that residues 59 to 79 cannot, suggesting that the 20 membrane-proximal residues are not engaged in a tetrameric structure. Rescue-of-function studies by disulfide bond reduction resulted in the redefinition and extension of the central fusion-activation segment as covering residues 84 to 117. In particular, we identified four residues located between positions 92 and 99, the function of which cannot be restored by disulfide bond reduction after cysteine mutagenesis. These mutant H proteins reached the cell surface as complex oligomers but could not trigger membrane fusion. We discuss these observations in the context of the stalk exposure model of membrane fusion triggering by paramyxoviruses.
IMPORTANCE Measles virus, while being targeted for eradication, still causes significant morbidity and mortality. Here, we seek to understand how it enters cells by membrane fusion. Two viral integral membrane glycoproteins (hemagglutinin tetramers and fusion protein trimers) mediate the concerted receptor recognition and membrane fusion processes. Since previous studies have suggested that the hemagglutinin stalk transmits the triggering signal to the fusion protein trimer, we completed an analysis of its structure and function by systematic Cys mutagenesis. We report that while certain residues of the central stalk segment confer specificity to the interaction with the fusion protein trimer, others are necessary to allow folding of the H-oligomer in a standard conformation conducive to fusion triggering, and still other residues sustain the conformational change that transmits the fusion-triggering signal.
INTRODUCTION
Measles virus (MeV) is an enveloped negative-sense single-stranded RNA virus of the genus Morbillivirus in the family Paramyxoviridae (1). This family includes deadly human and animal pathogens, such as Hendra and Nipah viruses (NiV), and viruses that are a significant cause of morbidity and mortality in humans, like mumps, parainfluenza, and respiratory syncytial viruses (2). Despite a robust vaccination campaign, MeV still affects 10 million people annually and caused an estimated 158,000 deaths in 2011 (3).
Cell entry of MeV and most other Paramyxoviridae occurs at the plasma membrane at neutral pH and is mediated by two glycoproteins, a tetrameric attachment protein and a trimeric fusion protein, F (2, 4). While the F trimer is well conserved, the attachment proteins are divided into three classes: for MeV and the other morbilliviruses, the attachment protein is named hemagglutinin (H) (5), while that of the pneumoviruses and henipaviruses, such as Hendra virus and Nipah virus, is named G (6). The attachment proteins of most other paramyxoviruses have both hemagglutination and neuraminidase activities; thus, they are named HN (2). While the HN proteins bind sialic acid, H and G bind protein receptors (7–10).
All paramyxovirus attachment proteins are type II transmembrane glycoproteins comprised of a large globular head atop a long stalk. In particular, the 617-amino-acid MeV H protein has an N-terminal 33-residue cytoplasmic tail, followed by a transmembrane region, a 96-residue stalk, and a cuboidal head domain (11). The X-ray crystal structures of the H, G, and HN head domains all document a similar six-bladed β-barrel fold (12–14). Attachment protein monomers associate to form dimers stabilized by one or more disulfide bonds in the head-proximal segment of the stalk. A dimer of dimers (tetramer) constitutes the functional unit for efficient fusion triggering (15). While any given attachment protein will only trigger its cognate fusion protein, key aspects of structure are conserved, as documented by the production of functional G-HN chimeric proteins (16–19).
While several structures of paramyxovirus attachment protein heads have been solved, the only stalk structures currently available are those of HN proteins from Newcastle disease virus (NDV) (20) and human parainfluenza virus 5 (PIV5) (21–23). These structures revealed that parts of these stalks adopt a four-helix bundle (4HB) organization with a supercoiled heptad repeat structure that transitions into a straight 11-mer repeat. However, initially the membrane-proximal and head-proximal segments of these HN stalks were not resolved, and only recently was one of the four 16-residue head-proximal segments visualized in a “2-heads-up/2-heads-down” PIV5 HN molecule (22). While the other three head-proximal segments could not be resolved, it is clear that they must assume different conformations. Similarly, the membrane-proximal residues (59 to 89) are not very conserved among the morbilliviruses, and the structure of this region of the stalk is not known for any paramyxovirus.
In the absence of an MeV H-stalk structure, we have been characterizing its function using biochemical and functional assays. Previously we reported on the cysteine scan mutagenesis analysis of the H-stalk top half (residues 103 to 154), which revealed three modules with distinct structure and function (24). Specifically, we identified a head-proximal dimeric linker (residues 139 to 154) followed by a tetrameric spacer (residues 122 to 137) and a conserved central segment (residues 103 to 117). In this study, we extend the Cys scan analysis to the lower half of the stalk (residues 59 to 102). We identify additional residues critical for the fusion trigger function, and our results indicate that the central signal transmission segment extends from residues 84 to 117. Specifically, we identified four conserved residues that are necessary to allow folding of the oligomers in a conformation conducive to fusion triggering. Furthermore, we observed that the bottom of the stalk (residues 59 to 79) adopts an organization different from that of the central segment, and that it does not play a critical role in the fusion triggering function.
MATERIALS AND METHODS
Cells, plasmids, and mutagenesis.
Vero (African green monkey kidney) cells and 293T (human embryonic kidney) cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). All mutations were made in a vaccine-lineage H-protein backbone (H-NSe) (25, 26). Mutations in the H stalk were introduced into the pCG-H expression plasmid by QuikChange site-directed mutagenesis (Agilent Technologies, Santa Clara, CA) according to the manufacturer's instructions. The clones were verified by sequencing the H-protein gene in the vicinity of the mutation. At least two independent clones were tested for each mutation. For expression of tagged human SLAM (SLAM-FLAG), the coding sequence was cloned into the pCG expression plasmid with the FLAG tag (DYKDDDDK) at the carboxy terminus.
Mammalian cell transfection, H-protein expression levels, and determination of oligomeric state.
Vero cells were transfected with plasmid DNA using Lipofectamine 2000 (Life Technologies, Grand Island, NY) according to the manufacturer's instructions. H expression and oligomerization were documented as described previously (24). Briefly, the standard pCG-H (25) or mutated plasmids (2 μg) were transfected into 1.5 × 105 Vero cells in 12-well plates. Thirty-six hours posttransfection, cytoplasmic extracts were made and the proteins separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under nonreducing conditions. The H proteins were visualized on an immunoblot using an H-cytoplasmic tail-specific polyclonal antibody (27) and an ECL Plex Cy3-conjugated secondary antibody on the Typhoon FLA 7000 imager (GE Healthcare, Piscataway, NJ). The image analysis software ImageQuant TL was used for quantification of protein band intensity.
Fusion assays and the partial reduction of disulfide bonds to rescue fusion function.
The semiquantitative cell-to-cell fusion assay was performed as described previously (28). Briefly, 0.5 μg each of three plasmids, encoding the H protein, the F protein, and green florescent protein (GFP), were transfected into 1.0 × 105 Vero cells in 24-well plates. Twenty-four hours posttransfection the extent of fusion was recorded in one field of view (about 2,000 cells) using the following criteria: 0, two or fewer syncytia with 4 to 5 nuclei (background); 1, three or more syncytia with 4 to 5 nuclei; 2, one to three syncytia with more than 10 nuclei; 3, four or more syncytia with more than 10 nuclei.
The fusion function of H-stalk mutants was rescued by limited disulfide bond reduction as described previously (24). Briefly, Vero cells were transfected as described above for the cell-to-cell fusion assay and then incubated with 200 nM fusion inhibitory peptide (FIP) (29, 30). Twenty-four hours posttransfection, cells were washed with phosphate-buffered saline (PBS) and treated with 15 mM dithiothreitol (DTT) or PBS for 30 min. Fusion scores were recorded 3 h after DTT treatment.
Flow cytometry analysis of cell surface H-protein expression.
293T cells (8 × 105 in a 6-well plate) were transfected with H-protein expression plasmids (3 μg) as described above. Thirty-six hours posttransfection, cells were washed with PBS and detached by incubating with Versene (Life Technologies, Grand Island, NY) at 37°C for 10 min. The resuspended cells were washed twice with cold fluorescence-activated cell sorter (FACS) wash buffer (1× PBS, 2% FBS, 0.1% sodium azide) and then incubated with an anti-H monoclonal antibody (1:100 dilution; MAB8905; Millipore, Billerica, MA) for 1 h at 4°C. Cells were washed three times with cold FACS wash buffer and incubated with a phycoerythrin-conjugated secondary antibody (1:100 dilution; 115-116-146; Jackson ImmunoResearch, West Grove, PA) for 1 h at 4°C. After three washes with FACS wash buffer, cells were fixed in 4% paraformaldehyde and analyzed by a FACSCalibur (BD Biosciences, San Jose, CA) cytometer and FlowJo software (Tree Star Inc., Ashland, OR).
Receptor-binding assay.
293T cells (8 × 105 in a 6-well plate) were transfected with 2 μg of the indicated H-protein expression plasmid and 2 μg of the FLAG-tagged SLAM expression plasmid as described above. Thirty-six hours posttransfection, the cells were washed twice with ice-cold PBS and cytoplasmic extracts were generated using coimmunoprecipitation (CoIP) lysis buffer (25 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol, pH 7.4). SLAM-FLAG was immunoprecipitated from this lysate using an anti-FLAG M2 affinity gel (Sigma-Aldrich, St. Louis, MO) according to the manufacturer's instructions. The immunoprecipitated proteins were separated on a 4 to 15% SDS-PAGE under reducing conditions (50 mM β-mercaptoethanol) and transferred to a polyvinylidene difluoride (PVDF) membrane. Coimmunoprecipitated H was revealed by probing with an anti-H cytoplasmic-tail-specific primary antibody (27) and a goat anti-rabbit horseradish peroxidase (HRP)-conjugated secondary antibody. Immunoprecipitation of SLAM was assessed by stripping the immunoblot with Restore Western blot stripping buffer (Thermo Scientific, Waltham, MA) and reprobing with an HRP-conjugated anti-FLAG antibody.
Cell surface protein isolation.
Cell surface proteins were isolated using the Pierce cell surface protein isolation kit according to the manufacturer's instructions. Briefly, Vero cells (8.0 × 105 in a 6-well plate) were transfected with 3 μg of plasmid DNA encoding MeV H constructs as indicated. Twenty-four hours posttransfection, cells were washed in cold PBS and then incubated with 0.25 mg/ml of membrane-impermeable sulfosuccinimidyl-2-(biotinamido)ethyl-1,3-dithio-propionate in PBS for 30 min at 4°C, followed by washing and quenching for 10 min at 4°C. Cells were lysed, and the lysates were cleared by centrifugation for 10 min at 10,000 × g and 4°C. Biotinylated proteins were adsorbed to immobilized streptavidin for 2 h at 4°C and then washed three times. Samples were boiled in Laemmli buffer for 5 min at 95°C and then subjected to SDS-PAGE and immunoblotted using an H-cytoplasmic tail-specific antibody (27).
RESULTS
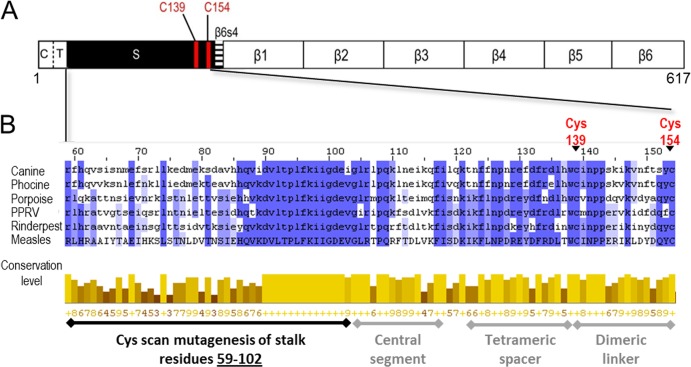
Figure 1A shows a line drawing of the entire MeV H protein, and Fig. 1B shows the sequence of its stalk, including its lower half, which was mutagenized in this study (residues 59 to 102). Multiple-sequence alignment of six morbilliviruses over the stalk region reveals segments with different conservation (Fig. 1B, bottom half). In particular, residues 86 to 109 are very well conserved, whereas the membrane-proximal 27 residues are less conserved.
FIG 1.
Schematic of MeV H protein and sequence alignment of MeV H stalk to other morbilliviruses. (A) H linear structure. From left to right: C, cytoplasmic tail; T, transmembrane region; S, stalk; and β1 to 6, beta sheets 1 through 6. The two Cys residues that cross-link the H dimer are shown as red lines. (B) Sequence alignment of the MeV H stalk (residues 59 to 154) to five other morbilliviruses. The alignment was made using ClustalW2 (42). Canine, canine distemper virus; Phocine, phocine distemper virus; Porpoise, porpoise morbillivirus; PPRV, peste-des-petits-ruminants virus; Rinderpest, rinderpest virus; Measles, measles virus. (Top) Shades of blue represent the degree to which the identity and nature of an amino acid is conserved at a given position. (Bottom) The conservation level of each residue is indicated by the height of the bars below each residue and the associated score below the bar (0 to 9). The black horizontal line indicates the Cys mutants reported in this study. The gray horizontal lines indicate stalk segments, the structure and function of which were defined in a previous study (24).
Expression levels and oligomerization status of H-protein mutants.
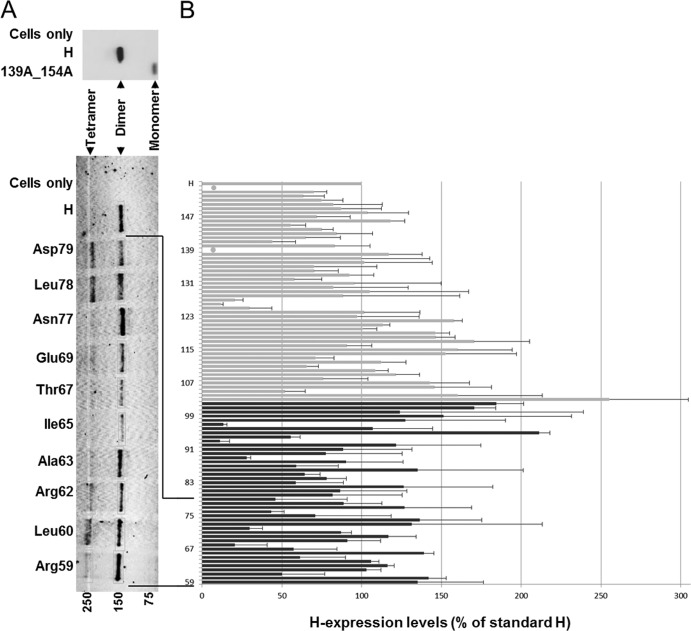
We first assessed the levels of expression of the 44 membrane-proximal H-stalk Cys mutants. The standard H protein runs as a dimer under nonreducing SDS-PAGE conditions, because denaturation disrupts the noncovalent tetramer interactions (Fig. 2A, top). Under the same conditions, a control H-protein where the two stalk Cys residues have been replaced with Ala migrates as a monomer (Fig. 2A, 139A_154A). In contrast, introducing an additional Cys residue in place of any other stalk residue may trap covalent tetramers, which can be visualized under these conditions. In Fig. 2A (bottom), 10 Cys substitution mutants are presented. For most mutants, signals of similar strength were detected at about 150 kDa. In contrast, signals of about 250 kDa had variable strength. The former signal corresponds to H dimers, the latter to H tetramers (15, 24). For each mutant, the total amount of H protein detected was quantified by measuring the respective protein bands and are shown in Fig. 2B (black bars) as the percentage of standard H protein. Data previously obtained for the upper half of the stalk are shown for context (Fig. 2, gray bars) (24, 31). Levels of expression varied, with 37 of 44 mutants having expression levels between 50% and ∼200% of standard H. Remarkably, significantly increased levels of proteins were detected in several mutants, implying that standard H is inherently metastable.
FIG 2.
Expression levels of H-stalk Cys mutants. (A, top gel) Standard H protein (H) and a mutant lacking both stalk Cys residues (139A_154A) separated on a nonreducing SDS-PAGE and immunoblotted. (Bottom gel) Cys mutants (as indicated) and standard H protein was extracted from cells in the presence of 50 mM iodoacetamide, separated on a nonreducing SDS-PAGE, and immunoblotted. Molecular mass markers are indicated at the bottom in kDa. The H monomer, dimer, and tetramer bands are indicated. (B) Total mutant H protein expression as a percentage of standard H. Total protein expression levels were determined by quantifying H dimer and, where present, H tetramer and H monomer protein band intensities using a Typhoon FLA laser scanner and ImageQuant TL software. Results represent the means from three experiments; standard deviations are indicated with error bars. Black bars, expression data for residues 59 to 102; gray bars, expression data for residues 103 to 154 (24). Native Cys at positions 139 and 154 are indicated by filled circles.
About 20 membrane-proximal stalk residues exhibit low tetramer-trapping propensity.
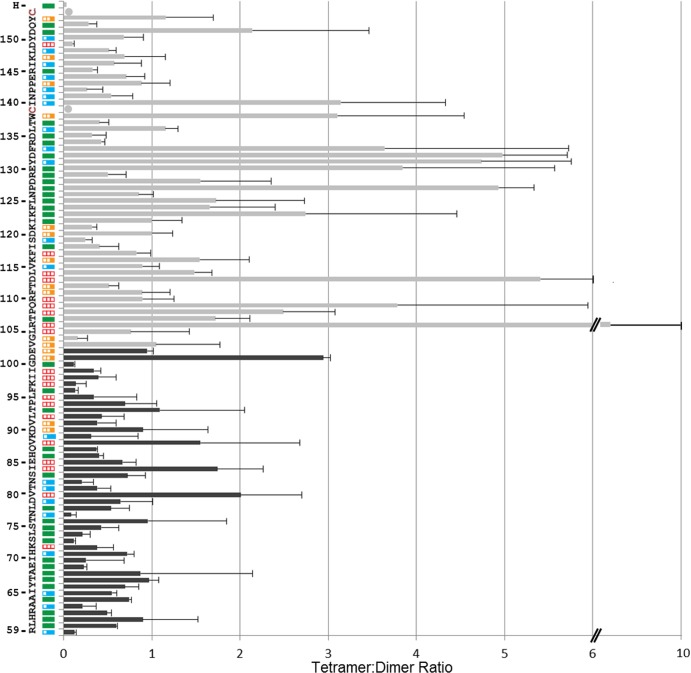
We present the analysis of the tetramer-forming propensity of the lower half of the MeV H stalk (Fig. 3, black bars) in the context of results previously obtained for the upper half (24, 31) (Fig. 3, gray bars). Most residues in the lower half have low tetramer-to-dimer ratios. In particular, none of the 21 membrane-proximal residues and only 5 of the next 24 residues had tetramer-to-dimer ratios above one (Fig. 3, V80, I84, V88, T93, and D101). This is in contrast to residues 106 to 133 in the upper half of the stalk, where 16 of 28 residues had tetramer-to-dimer ratios above one. This suggests that the structures of these stalk segments differ.
FIG 3.
Tetramer trapping propensity and fusion function of H-stalk mutants. The 96 residues (positions 59 to 154, bottom to top) of the H stalk are indicated on the left. Plasmids expressing Cys mutants for each residue were cotransfected in Vero cells with an F-expressing plasmid and a GFP-expressing plasmid. Fusion scores were determined 24 h posttransfection and are represented as colored boxes. Red empty box, fusion score of zero; one-third-filled orange box, fusion score of 1; two-thirds-filled blue box, fusion score of 2; full green box, fusion score of 3. Black or gray bars show tetramer-to-dimer ratios, calculated after loading the cell extracts on a nonreducing SDS-PAGE gel and quantifying the tetramer protein band and dimer protein band. Data for residues 59 to 102 are new (black bars), while data for residues 103 to 154 are from reference 24 (gray bars). Means and standard deviations from three experiments are indicated.
To assess the effect of Cys substitutions on H function, we used a semiquantitative cell-to-cell fusion assay (32). Cotransfection of standard H- and F-protein expression plasmids into Vero cells results in extensive syncytium formation (Fig. 3, green box on the left). The fusion score for each of the stalk mutants is indicated in Fig. 3 on a scale of 0 (no fusion) to 3 (wild-type levels of fusion). Fourteen out of the 21 cysteine mutants in the membrane-proximal segment had no effect on function, 6 resulted in a moderate decrease in fusion efficiency (Fig. 3, blue boxes; fusion score of 2), and one mutation abolished function (Fig. 3, K72; fusion score of 0). Thus, the functional analyses suggest that the membrane-proximal segment is not directly involved in triggering fusion.
Disulfide bond reduction does not restore fusion-triggering function of the most conserved stalk region.
The functional analysis identified additional residues that may be critical for sustaining fusion triggering (Fig. 3, red boxes). Specifically, I84, E85, V88, L92, P94, L95, K97, I98, and I99 completely lost fusion trigger function upon Cys substitution. To assess whether these effects can be reversed by disulfide bond reduction, we subjected Vero cells that were transfected with the corresponding H mutant and the standard F expression plasmid to mild reducing conditions (15 mM DTT). Figure 4A shows that DTT treatment restored fusion function for Cys substitution mutants of I84, E85, and V88. In contrast, Cys mutants of L92, P94, L95, K97, and I99 were not rescued, while I98 showed a very small gain of function. Interestingly, residues 92 to 99 are located in the most conserved stalk segment (Fig. 1B, residues 90 to 102).
FIG 4.
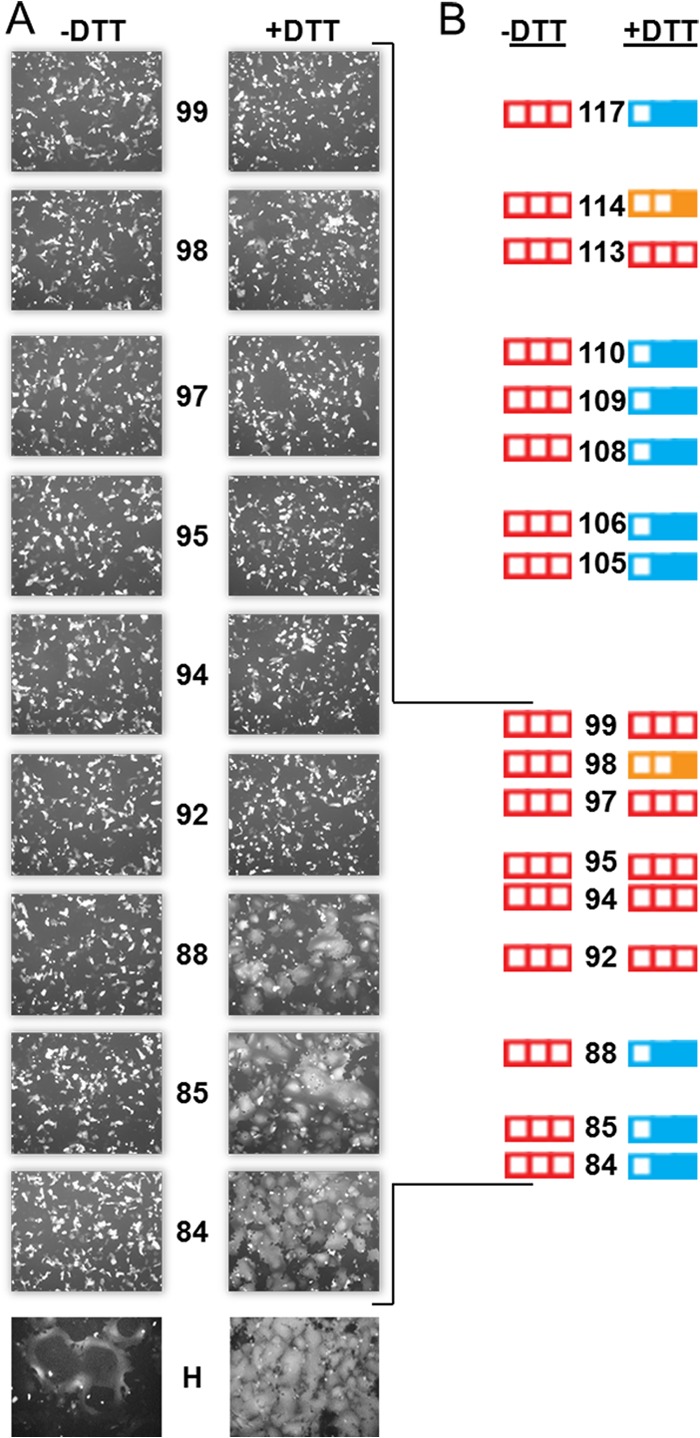
Disulfide bond reduction does not restore fusion trigger function of the most conserved stalk segment. (A) Visual assessment of syncytium formation. Vero cells were cotransfected with plasmids expressing the indicated H construct, standard F, and GFP and incubated with FIP, a fusion inhibitor. Twenty-four hours posttransfection, cells were washed and treated with either PBS (−DTT, left column) or with 15 mM DTT (+DTT, right column) for 30 min. The fusion score was recorded 3 h after DTT treatment. (B) Summary of fusion scores before (−DTT) and after (+DTT) DTT treatment for all of the nonfunctional Cys mutants in the central stalk segment. Fusion scores observed previously (residues 105 to 117) are also indicated (24).
Figure 4B summarizes the DTT-mediated rescue of function for fusion-deficient Cys mutants along the entire central stalk segment (residues 84 to 117), indicating that 17 out of the 34 Cys substitutions completely abolished fusion triggering. The function of 11 out of these 17 Cys mutants was partially restored by limited disulfide bond reduction, suggesting that these residues sustain conformational changes required for the transmission of the fusion trigger. Thus, the central segment of the H stalk involved in signal transmission may extend over more than 30 residues.
Conserved residues in the central stalk segment are required for proper oligomerization and fusion function.
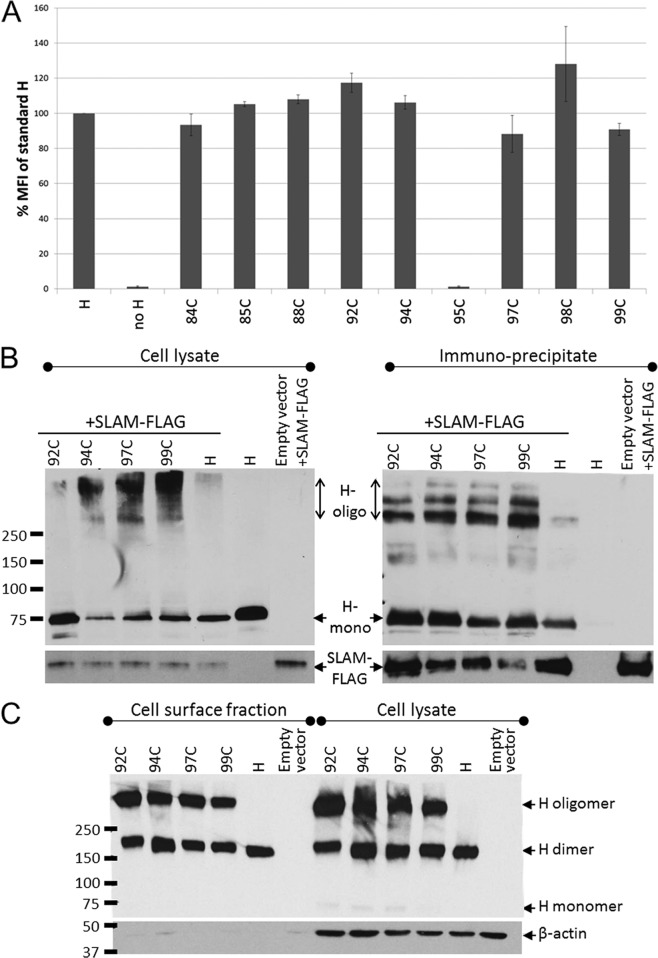
To characterize why the five mutants unable to trigger fusion even after DTT reduction lost function, we measured their surface expression level and receptor binding activity. Figure 5A illustrates the surface expression levels as a percentage of standard H as determined by FACS analysis. The Cys substitution of residue 95 completely blocked surface expression, suggesting protein misfolding, but the remaining four mutants had cell surface expression levels similar to those of the standard H protein.
FIG 5.
Cell surface expression and receptor-binding ability of triggering-deficient H-stalk mutants. (A) H-protein cell surface expression measured by FACS analysis. 293T cells transfected with the indicated H construct were stained with an anti-H ectodomain antibody and phycoerythrin-conjugated secondary antibody. The mean fluorescence intensity (MFI) of each mutant is presented as a percentage of the standard H-protein levels. Error bars indicate standard deviations from three experiments. (B) Receptor-binding ability of H mutants determined by coimmunoprecipitation assay. 293T cells were cotransfected with the indicated H constructs and a SLAM-expressing plasmid (SLAM-FLAG). (Left) Whole-cell lysates of cotransfected 293T cells. (Right) Fraction immunoprecipitated with anti-FLAG affinity gel. (Top) Immunoblot probed with an anti-H cytoplasmic tail-specific antibody. (Bottom) Immunoblot probed with anti-FLAG antibody. H-oligo, H oligomer; H mono, H monomer. (C) Immunoblot of a nonreducing SDS-PAGE of the cell surface fraction (left) and corresponding whole-cell lysate (right) of Vero cells transfected with the indicated H constructs.
We next assessed whether these four mutants retained the ability to interact with the MeV receptor, SLAM. To this end, FLAG-tagged human SLAM was coexpressed with each of the four H mutants, standard H, or with empty vector as a negative control. In Fig. 5B, panels on the left document that the indicated H constructs (top left) and SLAM (bottom left) were expressed in 293T cells 36 h posttransfection. We noted that while the standard H protein ran almost exclusively as a monomer (∼75 kDa) under reducing SDS-PAGE conditions, the Cys mutants showed a substantial amount of a higher oligomeric species (>250 kDa). Coimmunoprecipitation indicated that the Cys stalk mutants bind SLAM at least as well as the standard H protein (Fig. 5B, top right; compare the H+SLAM-FLAG lane to 92C, 94C, 97C, and 99C lanes). H was not coimmunoprecipitated in the absence of SLAM-FLAG, indicating that the H pulldown was SLAM dependent (Fig. 5B, right, second-to-last lane). Again, the H mutants showed species with substantially higher numbers of oligomers, whereas the standard H protein ran as a monomer. Taken together, these data indicate that the conformation of at least one H-head monomer is unaffected based on the ability of tetramers to be coimmunoprecipitated with SLAM. On the other hand, these four Cys mutants exhibit oligomeric H species which are stable under reducing SDS-PAGE conditions under which the standard H protein dissociates into monomers.
To assess which H-protein oligomeric forms reached the cell surface, we biotinylated the cell surface proteins and subsequently pulled them down using streptavidin. Both cell surface proteins and total cell lysates were separated on nonreducing SDS-PAGE and immunoblotted with an H cytoplasmic tail-specific antibody (Fig. 5C, left and right halves, respectively). The standard H protein migrates exclusively as a disulfide-linked dimer (Fig. 5C, H lanes, ∼150 kDa). In contrast, for the four Cys mutants, in addition to the dimer band, we observe a band migrating at >250 kDa (Fig. 5C, lanes marked 92C, 94C, 97C, and 99C). Thus, mutants with Cys substitution of four of the most conserved H-stalk residues can fold into conformations that are efficiently transported to the cell surface. However, these mutants do not generate the triggering-competent H-protein oligomeric forms.
DISCUSSION
Paramyxovirus entry into cells requires the concerted action of receptor-binding protein tetramers (HN, H, or G) and cognate F-protein trimers. Studies of the mechanisms of cell entry of HN-bearing paramyxoviruses, guided by high-resolution structural analyses of the whole HN ectodomain, including the stalk (20, 21), and of the cognate F trimer (33), have begun characterizing how these oligomers interact. These studies also led to the formulation of the “stalk exposure model” of membrane fusion triggering, which postulates that upon receptor binding the attachment protein globular heads undergo a conformational change that disrupts a preexisting head-stalk interaction, exposing an F-activation segment on the stalk (19, 22). Bose et al. (19, 23) noted that ours and other studies of how the central and upper half of the MeV H stalk interacts with the F trimer (24, 31, 34) are generally compatible with the HN-stalk exposure model. The data presented here allow a detailed comparison of H- and HN-based triggering systems, revealing similarities and also interesting differences.
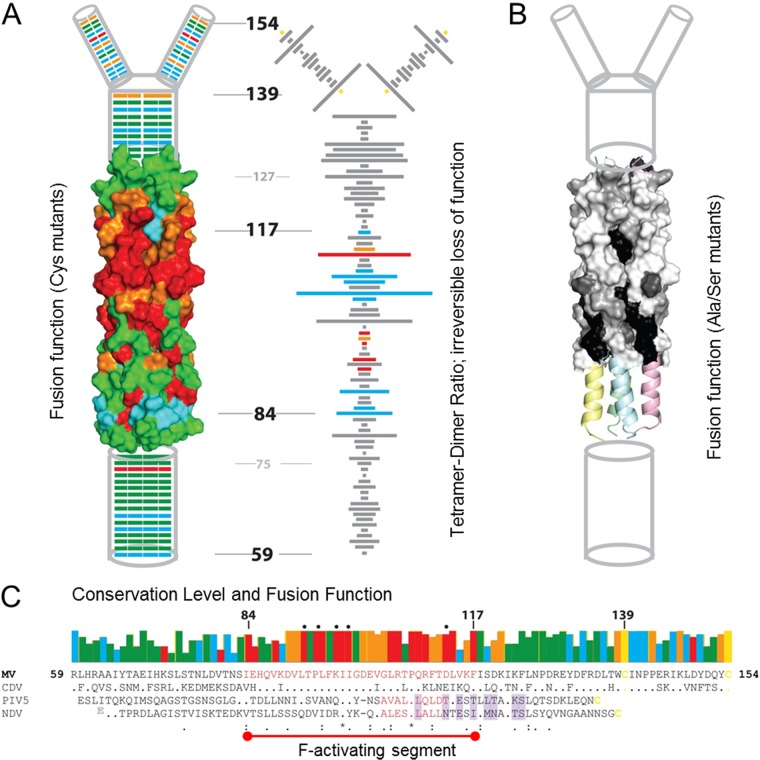
We define here the central fusion-triggering domain of the MeV H stalk as covering approximately 34 residues (84 to 117) (Fig. 6A to C). Most of these residues are highly conserved among the morbilliviruses (Fig. 6C, upper row of boxes and MeV and canine distemper virus [CDV] sequences) but not between H and HN stalks (Fig. 6C, compare MeV and CDV sequences to PIV5 and NDV sequences). Analyses of the tetramer-forming propensity of the central domain of the MeV H stalk reveal a dichotomy: most Cys mutations in the lower segment (residues 84 to 100) cause only limited tetramer trapping (Fig. 6A, right, low tetramer-to-dimer ratios) and preserve fusion function (Fig. 6A, left, green or blue). In contrast, most Cys mutations in the upper segment (residues 101 to 117) efficiently trap tetramers (Fig. 6A, right, high tetramer-to-dimer ratios) and disrupt fusion function (Fig. 6A, left, red).
FIG 6.
Structure and function of the H stalk. (A, left) Fusion-support function of the Cys substitution mutants indicated on a schematic/structure-based model (24) of the H stalk. From the bottom, the membrane-proximal amino acids (59 to 74) are indicated as a cylinder. The 4HB homology model which covers residues 75 to 127 is shown next in surface representation. Residues 128 to 138 are shown again as a cylinder followed by the head-proximal residues 139 to 154, which bifurcate to form two dimeric branches. Different colors denote the fusion score: red, 0; orange, 1; blue, 2; and green, 3. (Right) The tetramer-to-dimer ratio for each Cys mutant along the length of the stalk is indicated by the length of the horizontal bar. The ability to restore fusion by disulfide bond reduction is indicated using the same fusion score color scheme as that for the left panel. Gray bars represent mutants that were not tested for rescue of function by disulfide bond reduction. (B) Fusion support function of Ser/Ala mutants of a part of the central fusion activation segment mapped on the MeV stalk model (31). Shading denotes the fusion score: black, 0; dark gray, 1; gray, 2; and white, 3. (C, top) Sequence conservation of the same selected morbillivirus stalk residues as those depicted in Fig. 1B, with the fusion function color coded as described above for panel A. (Bottom) Multiple-sequence alignment of four paramyxovirus stalks. The MeV F-activation domain is delineated by a red line, and the residues are colored red. Residues comprising the PIV5 and NDV F-activation domains are also colored red. HN-stalk residues that interact with the HN heads based on the PIV5 (22) and NDV (20) heads-down crystal structures are shaded light purple.
On the other hand, reduction of disulfide bonds restores function of six out of eight tetramer-trapping mutants in the upper segment (Fig. 4; data are visualized in Fig. 6A, right, blue bars); function is only partially restored for L114C (Fig. 6A, right, orange bar) and is not restored for D113C (Fig. 6A, right, red bar). This suggests that conformational flexibility of this segment of the F-activation domain is a requirement for efficient fusion triggering. In contrast, disulfide bond reduction does not restore function of six mutants clustered at residues 92 to 99 of the lower segment (Fig. 6A, right, red bars). Interestingly, all four clustered mutants that are expressed at normal levels but cannot be rescued by disulfide bond reduction (L92C, P94C, K97C, and I99C) form oligomers that reach the cell surface but migrate slowly into nonreducing gels (Fig. 5). This suggests that these mutants have a more complex oligomeric structure than the standard H protein, which migrates into this gel almost exclusively as a dimer. Even the semiconservative mutations L92S, F96S, and I98S significantly reduce the fusion support function of the respective mutants (31) (Fig. 6B).
All of these observations are consistent with a model considering that exact conservation of residues in the lower half of the central segment of the MeV H stalk is necessary to support the folding of standard H oligomers that migrate homogeneously as dimers in a denaturing but nonreducing gel. It is possible that only this standard H oligomer can support cognate F-trimer activation. Analogously, it has been postulated that HN has an induced-fit interaction with the cognate F trimer (19). Thus, the conserved central stalk domain of the morbilliviruses may allow the H stalk to fold into a standard or induced-fit conformation that allows functional interactions with the F trimer. As expected for a region highly conserved among morbilliviruses, similar results were also reported for CDV (34).
Bose et al. (19) recently showed that a mainly hydrophobic, 7-amino-acid segment centered around a conserved proline (Fig. 6C, residue 108 in MeV) serves as the F activation region of the HN protein stalk, and although sequence homology of this segment is limited to the central proline and two leucines (Fig. 6C, PIV5 and NDV sequences), the PIV5 HN F-activation region can functionally replace the colinear HN region of NDV (Fig. 6C, PIV5 and NDV sequences, red residues). Even if the inverse substitution did not work, this experiment indicates that components of the fusion activation mechanism are conserved among the HN proteins. The central proline of this HN segment is also conserved in the H protein (Fig. 6C, position 108 in MeV H, asterisk in the lowest line). Similar to MeV H, the conformational flexibility of the HN F-activation domain is critical for efficiently triggering fusion: an engineered disulfide bond that trapped an HN tetramer, constraining the stalk, prevented fusion triggering (19). Finally, MeV H residues R110, L114, F118, and K121, situated above P108 and colinear with HN-stalk residues buried by HN-head dimers in the down position (Fig. 6C, shaded residues), are also implicated in F interactions (31, 35). Thus, the upper half of the central segment of the MeV H stalk could transmit the fusion triggering signal.
We have analyzed here the membrane-proximal ∼20 residues (59 to 79) of the MeV H stalk. No high-resolution structural analysis is available yet for the corresponding stalk region of any paramyxovirus. This H segment does not trap tetramers efficiently, suggesting that the helices are not engaged in a 4HB or another tightly packed structure. Within the morbilliviruses, this segment is not as conserved as the central region, and its mutagenesis had little effect on fusion, with only K72C resulting in a complete loss of function (Fig. 6A and C). Little is known about the structure and function of the HN- and G-stalk membrane-proximal residues, but our results with MeV H suggest that they do not have a direct role in fusion activation.
One interesting difference between the HN and H stalks is the H-stalk 16-residue flexible tetrameric spacer segment (Fig. 1B and 6A and C, residues 122 to 137). This segment can conduct the fusion activation signal even while remaining cross-linked in a tetrameric configuration (24). A comparable segment has not been reported for HN stalks, which are 16 to 19 residues shorter than H stalks (Fig. 6C). Another unique characteristic of the H stalk is the presence of two native Cys residues at positions 139 and 154 (Fig. 6A, right, shown as a yellow dot, and C, MeV and CDV sequences, yellow residues). These Cys residues bracket the H-stalk head-proximal segment that does not trap tetramers efficiently, suggesting bifurcation into dimeric linkers (24) (Fig. 6A, right, residues 139 to 154). Thus, the upper segments of the H and HN stalks are significantly different.
Crystallographic analyses of the whole ectodomains of HN tetramers have revealed that the HN heads can be located atop the stalk, on its sides, or even in an intermediate one-head-dimer-up and one-head-dimer-down arrangement (20–22). When both HN head dimers are down, they cover in part the proline-centered activation segment (Fig. 6C, PIV5 and NDV sequences, compare red residues to shaded residues). This head-stalk arrangement has suggested the stalk-exposure model of fusion triggering: in order for the F-activation segment to first interact with the F trimer and then trigger fusion, at least one head dimer must be in the up position. Even if crystal structures of whole H ectodomains are not yet available, evidence for its broad relevance comes from the observation that headless stalks of HN, H, and G attachment proteins can trigger fusion in a receptor-independent manner (23, 36, 37). On the other hand, since MeV H tetramers and F trimers already interact while being transported to the cell surface (38), it seems likely that at least one H-head dimer is in the up position. This implies that H-stalk exposure alone does not directly trigger membrane fusion. Rather, receptor binding followed by pulling (11) may cause the conformational change of the stalk that triggers membrane fusion. Interestingly, fusion can be triggered by receptor interactions with only one H-head dimer (39).
Finally, we note that an MeV H-head-only construct was crystallized in two distinct tetrameric forms, named I and II (40), even if biochemical data suggest that the stalk and the transmembrane regions are mainly responsible for tetramer formation (41). Form I has been proposed to be a relevant pretriggering structure based on analyses of its dimer-dimer interface by mutagenesis and neutralizing monoclonal antibodies (41). However, whether the H heads adopt the one-head-dimer-up and one-head-dimer-down arrangement observed for PIV5 HN (22) or a different conformation before receptor engagement, such as that in the form I and II H-tetramer structures, remains to be determined.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health grant R01 CA090636 to R.C. and the Edward C. Kendall Fellowship in Biochemistry to C.K.N.
We acknowledge P. Devaux for helpful suggestions.
M.M. is a Merck fellow of the Life Sciences Research Foundation.
Footnotes
Published ahead of print 19 March 2014
REFERENCES
- 1.Griffin D. 2013. Measles, p 1042–1069 In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B. (ed), Fields virology, 6th ed, vol 1 Lippincott, Williams and Wilkins, Philadelphia, PA [Google Scholar]
- 2.Lamb RA, Parks G. 2013. Paramyxoviridae, p 957–995 In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B. Fields virology, 6th ed, vol 1 Lippincott, Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 3.Anonymous. 2011. Vaccines: the case of measles. Nature 473:434–435. 10.1038/473434a [DOI] [PubMed] [Google Scholar]
- 4.Iorio RM, Melanson VR, Mahon PJ. 2009. Glycoprotein interactions in paramyxovirus fusion. Future Virol. 4:335–351. 10.2217/fvl.09.17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navaratnarajah CK, Miest TS, Carfi A, Cattaneo R. 2012. Targeted entry of enveloped viruses: measles and herpes simplex virus I. Curr. Opin. Virol. 2:43–49. 10.1016/j.coviro.2011.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee B, Ataman ZA. 2011. Modes of paramyxovirus fusion: a Henipavirus perspective. Trends Microbiol. 19:389–399. 10.1016/j.tim.2011.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tatsuo H, Ono N, Tanaka K, Yanagi Y. 2000. SLAM (CDw150) is a cellular receptor for measles virus. Nature 406:893–897. 10.1038/35022579 [DOI] [PubMed] [Google Scholar]
- 8.Muhlebach MD, Mateo M, Sinn PL, Prufer S, Uhlig KM, Leonard VH, Navaratnarajah CK, Frenzke M, Wong XX, Sawatsky B, Ramachandran S, McCray PB, Jr, Cichutek K, von Messling V, Lopez M, Cattaneo R. 2011. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature 480:530–533. 10.1038/nature10639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang A, Dutch RE. 2012. Paramyxovirus fusion and entry: multiple paths to a common end. Viruses 4:613–636. 10.3390/v4040613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mateo M, Navaratnarajah CK, Cattaneo R. 2014. Structural basis of efficient contagion: measles variations on a theme by parainfluenza viruses. Curr. Opin. Virol. 5C:16–23. 10.1016/j.coviro.2014.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Navaratnarajah CK, Oezguen N, Rupp L, Kay L, Leonard VH, Braun W, Cattaneo R. 2011. The heads of the measles virus attachment protein move to transmit the fusion-triggering signal. Nat. Struct. Mol. Biol. 18:128–134. 10.1038/nsmb.1967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hashiguchi T, Kajikawa M, Maita N, Takeda M, Kuroki K, Sasaki K, Kohda D, Yanagi Y, Maenaka K. 2007. Crystal structure of measles virus hemagglutinin provides insight into effective vaccines. Proc. Natl. Acad. Sci. U. S. A. 104:19535–19540. 10.1073/pnas.0707830104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crennell S, Takimoto T, Portner A, Taylor G. 2000. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat. Struct. Mol. Biol. 7:1068–1074. 10.1038/81002 [DOI] [PubMed] [Google Scholar]
- 14.Xu K, Rajashankar KR, Chan YP, Himanen JP, Broder CC, Nikolov DB. 2008. Host cell recognition by the henipaviruses: crystal structures of the Nipah G attachment glycoprotein and its complex with ephrin-B3. Proc. Natl. Acad. Sci. U. S. A. 105:9953–9958. 10.1073/pnas.0804797105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brindley M, Plemper R. 2010. Blue native-PAGE and biomolecular complementation reveal tetrameric or higher order oligomer organization of the physiological measles virus attachment (H) protein. J. Virol. 84:12174–12184. 10.1128/JVI.01222-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mirza AM, Aguilar HC, Zhu Q, Mahon PJ, Rota PA, Lee B, Iorio RM. 2011. Triggering of the Newcastle disease virus fusion protein by a chimeric attachment protein that binds to Nipah virus receptors. J. Biol. Chem. 286:17851–17860. 10.1074/jbc.M111.233965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Porotto M, Salah Z, DeVito I, Talekar A, Palmer SG, Xu R, Wilson IA, Moscona A. 2012. The second receptor binding site of the globular head of the Newcastle disease virus hemagglutinin-neuraminidase activates the stalk of multiple paramyxovirus receptor binding proteins to trigger fusion. J. Virol. 86:5730–5741. 10.1128/JVI.06793-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talekar A, Devito I, Salah Z, Palmer SG, Chattopadhyay A, Rose JK, Xu R, Wilson IA, Moscona A, Porotto M. 2013. Identification of a region in the stalk domain of the Nipah virus receptor binding protein that is critical for fusion activation. J. Virol. 87:10980–10996. 10.1128/JVI.01646-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bose S, Song AS, Jardetzky TS, Lamb RA. 22 January 2014. Fusion activation through attachment protein stalk domains indicates a conserved core mechanism of paramyxovirus entry into cells. J. Virol. 10.1128/JVI.03741-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yuan P, Swanson KA, Leser GP, Paterson RG, Lamb RA, Jardetzky TS. 2011. Structure of the Newcastle disease virus hemagglutinin-neuraminidase (HN) ectodomain reveals a four-helix bundle stalk. Proc. Natl. Acad. Sci. U. S. A. 108:14920–14925. 10.1073/pnas.1111691108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bose S, Welch BD, Kors CA, Yuan P, Jardetzky TS, Lamb RA. 2011. Structure and mutagenesis of the parainfluenza virus 5 hemagglutinin-neuraminidase stalk domain reveals a four-helix bundle and the role of the stalk in fusion promotion. J. Virol. 85:12855–12866. 10.1128/JVI.06350-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Welch BD, Yuan P, Bose S, Kors CA, Lamb RA, Jardetzky TS. 2013. Structure of the parainfluenza virus 5 (PIV5) hemagglutinin-neuraminidase (HN) ectodomain. PLoS Pathog. 9:e1003534. 10.1371/journal.ppat.1003534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bose S, Zokarkar A, Welch BD, Leser GP, Jardetzky TS, Lamb RA. 2012. Fusion activation by a headless parainfluenza virus 5 hemagglutinin-neuraminidase stalk suggests a modular mechanism for triggering. Proc. Natl. Acad. Sci. U. S. A. 109:E2625–E2634. 10.1073/pnas.1213813109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Navaratnarajah CK, Negi S, Braun W, Cattaneo R. 2012. Membrane fusion triggering: three modules with different structure and function in the upper half of the measles virus attachment protein stalk. J. Biol. Chem. 287:38543–38551. 10.1074/jbc.M112.410563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cathomen T, Buchholz CJ, Spielhofer P, Cattaneo R. 1995. Preferential initiation at the second AUG of the measles virus F mRNA: a role for the long untranslated region. Virology 214:628–632. 10.1006/viro.1995.0075 [DOI] [PubMed] [Google Scholar]
- 26.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dötsch C, Christiansen G, Billeter MA. 1995. Rescue of measles viruses from cloned DNA. EMBO J. 14:5773–5784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cathomen T, Naim HY, Cattaneo R. 1998. Measles viruses with altered envelope protein cytoplasmic tails gain cell fusion competence. J. Virol. 72:1224–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apte-Sengupta S, Negi S, Leonard VH, Oezguen N, Navaratnarajah CK, Braun W, Cattaneo R. 2012. Base of the measles virus fusion trimer head receives the signal that triggers membrane fusion. J. Biol. Chem. 287:33026–33035. 10.1074/jbc.M112.373308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Norrby E. 1971. The effect of a carbobenzoxy tripeptide on the biological activities of measles virus. Virology 44:599–608. 10.1016/0042-6822(71)90374-6 [DOI] [PubMed] [Google Scholar]
- 30.Richardson CD, Choppin PW. 1983. Oligopeptides that specifically inhibit membrane fusion by paramyxoviruses: studies on the site of action. Virology 131:518–532. 10.1016/0042-6822(83)90517-2 [DOI] [PubMed] [Google Scholar]
- 31.Apte-Sengupta S, Navaratnarajah CK, Cattaneo R. 2013. Hydrophobic and charged residues in the central segment of the measles virus hemagglutinin stalk mediate transmission of the fusion-triggering signal. J. Virol. 87:10401–10404. 10.1128/JVI.01547-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Navaratnarajah CK, Vongpunsawad S, Oezguen N, Stehle T, Braun W, Hashiguchi T, Maenaka K, Yanagi Y, Cattaneo R. 2008. Dynamic interaction of the measles virus hemagglutinin with its receptor signaling lymphocytic activation molecule (SLAM, CD150). J. Biol. Chem. 283:11763–11771. 10.1074/jbc.M800896200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yin H-S, Wen X, Paterson RG, Lamb RA, Jardetzky TS. 2006. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature 439:38–44. 10.1038/nature04322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ader N, Brindley MA, Avila M, Origgi FC, Langedijk JPM, Orvell C, Vandevelde M, Zurbriggen A, Plemper RK, Plattet P. 2012. Structural rearrangements of the central region of the morbillivirus attachment protein stalk domain trigger F protein refolding for membrane fusion. J. Biol. Chem. 287:16324–16334. 10.1074/jbc.M112.342493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paal T, Brindley MA, St Clair C, Prussia A, Gaus D, Krumm SA, Snyder JP, Plemper RK. 2009. Probing the spatial organization of measles virus fusion complexes. J. Virol. 83:10480–10493. 10.1128/JVI.01195-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brindley MA, Suter R, Schestak I, Kiss G, Wright ER, Plemper RK. 2013. A stabilized headless measles virus attachment protein stalk efficiently triggers membrane fusion. J. Virol. 87:11693–11703. 10.1128/JVI.01945-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q, Stone JA, Bradel-Tretheway B, Dabundo J, Benavides Montano JA, Santos-Montanez J, Biering SB, Nicola AV, Iorio RM, Lu X, Aguilar HC. 2013. Unraveling a three-step spatiotemporal mechanism of triggering of receptor-induced Nipah virus fusion and cell entry. PLoS Pathog. 9:e1003770. 10.1371/journal.ppat.1003770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Plemper RK, Hammond AL, Cattaneo R. 2001. Measles virus envelope glycoproteins hetero-oligomerize in the endoplasmic reticulum. J. Biol. Chem. 276:44239–44246. 10.1074/jbc.M105967200 [DOI] [PubMed] [Google Scholar]
- 39.Brindley MA, Takeda M, Plattet P, Plemper RK. 2012. Triggering the measles virus membrane fusion machinery. Proc. Natl. Acad. Sci. U. S. A. 109:E3018–E3027. 10.1073/pnas.1210925109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hashiguchi T, Ose T, Kubota M, Maita N, Kamishikiryo J, Maenaka K, Yanagi Y. 2011. Structure of the measles virus hemagglutinin bound to its cellular receptor SLAM. Nat. Struct. Mol. Biol. 18:135–141. 10.1038/nsmb.1969 [DOI] [PubMed] [Google Scholar]
- 41.Nakashima M, Shirogane Y, Hashiguchi T, Yanagi Y. 2013. Mutations in the putative dimer-dimer interfaces of the measles virus hemagglutinin head domain affect membrane fusion triggering. J. Biol. Chem. 288:8085–8091. 10.1074/jbc.M112.427609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]