The Middle East respiratory syndrome coronavirus (MERS-CoV) has killed ∼50% of individuals known to be infected, making understanding its mechanisms of transmission and pathogenesis and identifying candidate treatments a high priority. Heinz Feldmann, Vincent J. Munster and Michael G. Katze and their colleagues now report that treating MERS-CoV-infected rhesus macaques with interferon-α2b and ribavirin reduced virus replication and infection severity, suggesting the potential use of this combination for the clinical treatment of infected humans.
Supplementary information
The online version of this article (doi:10.1038/nm.3362) contains supplementary material, which is available to authorized users.
Subject terms: Viral infection, Drug therapy
Abstract
The emergence of Middle East respiratory syndrome coronavirus (MERS-CoV) is of global concern: the virus has caused severe respiratory illness, with 111 confirmed cases and 52 deaths1 at the time of this article's publication. Therapeutic interventions have not been evaluated in vivo; thus, patient management relies exclusively on supportive care, which, given the high case-fatality rate, is not highly effective. The rhesus macaque is the only known model organism for MERS-CoV infection, developing an acute localized to widespread pneumonia with transient clinical disease2,3 that recapitulates mild to moderate human MERS-CoV cases4,5. The combination of interferon-α2b and ribavirin was effective in reducing MERS-CoV replication in vitro6; therefore, we initiated this treatment 8 h after inoculation of rhesus macaques. In contrast to untreated, infected macaques, treated animals did not develop breathing abnormalities and showed no or very mild radiographic evidence of pneumonia. Moreover, treated animals showed lower levels of systemic (serum) and local (lung) proinflammatory markers, in addition to fewer viral genome copies, distinct gene expression and less severe histopathological changes in the lungs. Taken together, these data suggest that treatment of MERS-CoV infected rhesus macaques with IFN-α2b and ribavirin reduces virus replication, moderates the host response and improves clinical outcome. As these two drugs are already used in combination in the clinic for other infections, IFN-α2b and ribavirin should be considered for the management of MERS-CoV cases.
Supplementary information
The online version of this article (doi:10.1038/nm.3362) contains supplementary material, which is available to authorized users.
Main
MERS-CoV is the first lineage-C betacoronavirus known to infect humans7. Similar to SARS (severe acute respiratory syndrome) coronavirus infection, MERS-CoV infection can result in an acute respiratory distress syndrome with multiorgan dysfunction8. The rapid identification of therapeutics for MERS-CoV is a high priority as there is currently no specific therapy or vaccine. A meta-analysis of data from the SARS outbreak in 20039 found that the benefit of different treatment strategies10,11,12,13,14,15,16,17,18 was inconclusive at best9. We recently demonstrated in vitro that the antiviral effect of interferon-α2b (IFN-α2b) against MERS-CoV is augmented by the concomitant use of ribavirin, and when we combined the treatments, the effective concentrations were such that they could be used in the clinic6. Here we evaluate the effectiveness in vivo of combination IFN-α2b and ribavirin treatment, in an attempt to identify a therapeutic approach that can be immediately implemented in the clinic for MERS-CoV cases.
Two groups of three rhesus macaques were infected with 7 × 106, the TCID50 (50% tissue culture infectious dose), of MERS-CoV (hCoV-EMC/2012, a clinical isolate8), as previously described2. We initiated treatment 8 h after infection, with subcutaneous delivery of 5 mega international units (MIU)/kg of IFN-α2b and an intravenous loading dose of ribavirin (30 mg per kg body weight). We subsequently treated the rhesus macaques with ribavirin intramuscularly (10 mg per kg body weight) every 8 h and IFN-α2b (5 MIU/kg) subcutaneously every 16 h until 72 h after infection, when all animals were euthanized (Fig. 1a) at the peak of clinical signs and viral loads in this model2,3.
Figure 1. Schedule of treatment and selected clinical laboratory parameters.
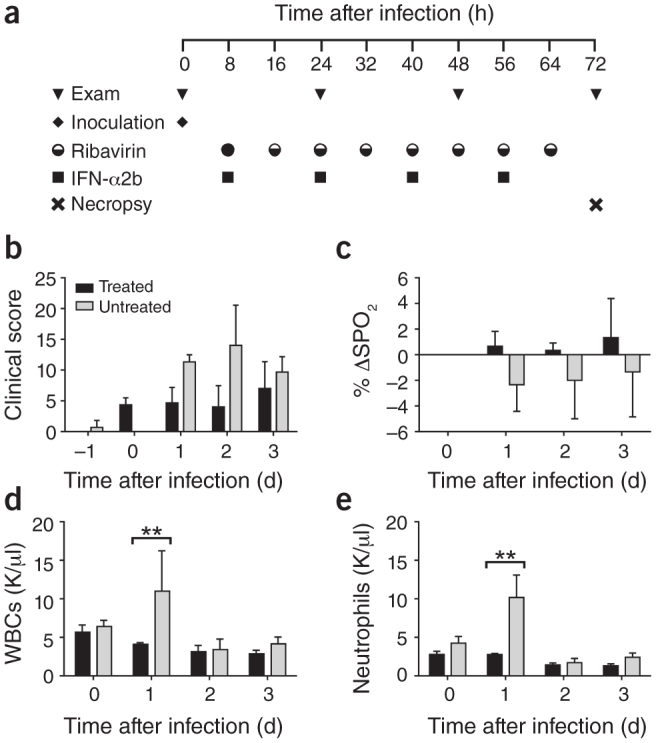
(a) Schematic showing the experiment schedule. IFN-α2b was delivered subcutaneously at 5 MIU/kg. Ribavirin was delivered as an initial loading dose intravenously at 30 mg/kg (full circle) with subsequent doses delivered intramuscularly at 10 mg/kg (half-full circle). Rhesus macaques were given either IFN-α2b and ribavirin (treated, RM1–RM3, n = 3) or sham treatment (untreated, RM4–RM6, n = 3) on the same schedule. (b) Clinical score (see also Supplementary Table 1) determined from an established scoring sheet34. (c) Changes in oxygen saturation from preinoculation values (% δSPO2). (d,e) The total number of white blood cells (WBCs) (d) and neutrophils (e) in blood samples obtained from animals 0, 24, 48 and 72 h after infection. All values are mean ± s.d. (two-way analysis of variance (ANOVA), Bonferroni's post hoc test, **P < 0.01). A single sample per time point per animal was used for the analysis. K, cell count × 103.
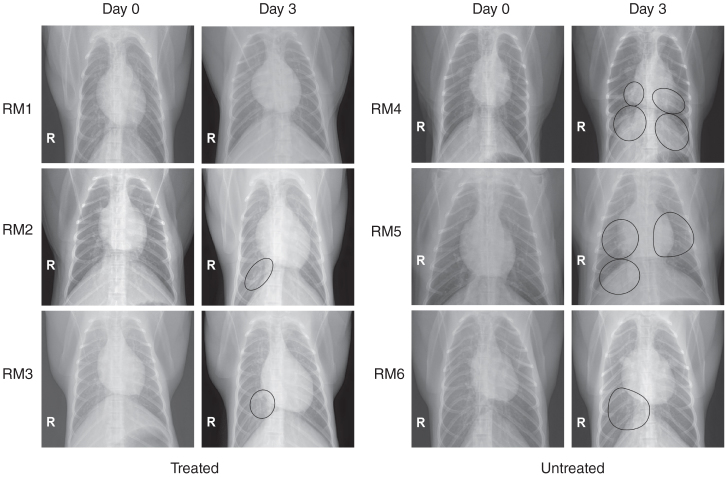
Infection with MERS-CoV in the absence of treatment led to similar clinical signs as previously reported, including increased respiration rates, abdominal breathing and piloerection2,3 (Fig. 1b and Supplementary Table 1). The animals' observed breathing difficulties were supported by evidence of decreased oxygen saturation in untreated animals (2–5% decrease from baseline) (Fig. 1c). We observed significantly increased white blood cell counts in untreated animals 1 d after infection, but they returned to normal levels on subsequent days (Fig. 1d). We attributed the increase in white blood cells mainly to a significant increase in neutrophils (Fig. 1e). In contrast, treated animals did not show evidence of increased respiration, piloerection or decreased oxygen saturation levels, nor did they show significant changes in hematology (Fig. 1d,e). To monitor signs of pneumonia, we X-rayed the animals daily. Radiographs from treated animals remained normal 1 d after infection and showed, at most, light infiltration in a single lobe 2 or 3 d after infection in one or two of three animals, respectively (Supplementary Table 2). In contrast, untreated animals showed evidence of increasing amounts of interstitial infiltration between 1 and 3 d after infection, ranging from localized (single lobe) to diffuse severe pulmonary interstitial infiltration throughout the lungs (Fig. 2 and Supplementary Table 2).
Figure 2. Radiographic alterations.
Ventrodorsal thoracic X-rays from IFN-α2b– and ribavirin–treated (RM1–RM3) and untreated (RM4–RM6) rhesus macaques imaged before MERS-CoV infection (day 0) and on day 3 after infection. Areas of interstitial infiltration, indicative of pneumonia, are circled. R indicates the right side of the animal.
Upon necropsy, gross pathology revealed lungs that were normal in appearance in all treated animals (Fig. 3a). In contrast, lungs from untreated animals were firm and edematous, failed to collapse (Fig. 3a) and contained gross lesions with a mean affected area of 14.3% (Fig. 3a and Supplementary Table 3). Multifocal consolidation and dark red discoloration, consistent with interstitial pneumonia, was most prevalent on the dorsal surface of the lungs of untreated animals. We did not observe any other pathological changes at necropsy.
Figure 3. Pathology and viral loads in selected tissue samples.
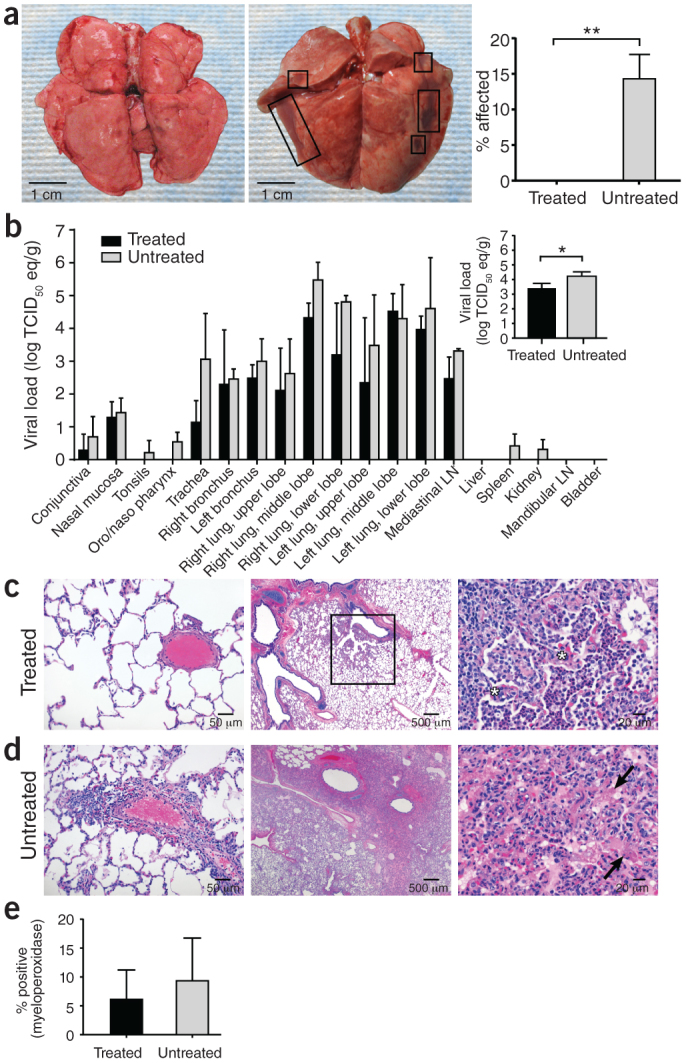
(a) Gross pathology of lungs from IFN-α2b– and ribavirin–treated (left) and untreated (middle) MERS-CoV–infected rhesus macaques 72 h after inoculation. The percentage area showing gross pathology is indicated on the right. Six lung lobes per animal (dorsal and ventral) from three animals per group were scored. (b) The mean viral load determined by qRT-PCR from individual tissues collected at necropsy. The mean viral load of all lung lobes combined is indicated in the inset. Log TCID50 eq/g, log TCID50 equivalents per gram tissue; one sample per tissue per animal from three animals per group were analyzed. (c,d) Lung tissue collected at necropsy stained with H&E. Pulmonary arterioles appear normal in treated animals (RM2) (c, left). Treated RM2 shows mild acute bronchointerstitial pneumonia demonstrated by multifocal accumulations of inflammatory cells centered on terminal bronchioles (c, middle). Higher magnification of the boxed region showing thickening of the alveolar septae by small numbers of neutrophils, macrophages, fibrin and edema (asterisk) (c, right). Pulmonary arterioles are bounded by moderate numbers of lymphocytes indicative of perivasculitis in untreated animals (d, left). Larger numbers of inflammatory cells and fibrin also extend into alveolar spaces. Untreated RM4 shows marked coalescing bronchointerstitial pneumonia with abundant alveolar fibrin and edema (d, middle). Higher magnification of untreated RM4 showing airways filled with organizing fibrin (arrow) and moderate numbers of viable and degenerate neutrophils and macrophages (d, right). (e) Percentage of neutrophils in lung sections stained with myeloperoxidase. Two sections per lobe and six lobes per animal from three animals per group were analyzed. All values are mean ± s.d. (t-test, *P < 0.05, **P < 0.01).
We used quantitative real-time RT-PCR (qRT-PCR) to determine viral loads as TCID50 equivalents in tissues, fluids and nasal and throat swabs19. There was no detectable viral RNA in the blood at any time point or in urine taken at necropsy. Daily bronchial alveolar lavage samples showed similar viral loads between treated and untreated groups (Supplementary Fig. 1). With the exception of the oronasopharynx in treated animals, all respiratory tract tissues collected at necropsy were positive for MERS-CoV by qRT-PCR (Fig. 3b). Viral RNA was also absent from the tonsils, spleen and kidney of treated animals, but it was present in these tissues in two of three untreated animals. In all of the tissues analyzed, with the exception of left middle lobe lung specimens, the viral loads were lower in specimens from treated animals as compared to untreated animals (Fig. 3b). When we combined the values from all lung specimens, the mean viral load in treated animals was 0.81 log lower than in untreated animals (P = 0.0428, unpaired t-test, one tail), demonstrating a statistically significant reduction in virus replication throughout the lung (Fig. 3b, inset).
All animals developed some degree of pulmonary pathology with mild to marked multifocal bronchointerstitial pneumonia, predominantly centered on the terminal bronchioles (Fig. 3c,d). However, untreated rhesus macaques 4 and 5 showed moderate to marked lesions with abundant alveolar edema, fibrin with formation of hyaline membranes and low to moderate numbers of macrophages (Fig. 3d), a lesion severity not observed in the treated animals (Fig. 3c and Supplementary Table 3). Only untreated animals showed significant scoring for perivascular infiltrates within and adjacent to affected areas of the lung (Fig. 3d) (P < 0.001, Mann-Whitney). We observed higher numbers of neutrophils in lungs of untreated animals compared to treated ones (9.3% compared to 6.1%, respectively) (Fig. 3e).
We monitored serum IFN-α levels to assess the delivery of IFN-α2b. Treated animals had at least 2.3 times the level of IFN-α measured in untreated animals (1 and 3 d after infection), whereas 2 d after infection, treated animals had 37 times the level of untreated animals (Supplementary Fig. 2), which probably reflects the timing of IFN-α2b administration in relation to blood collection. Untreated animals showed evidence of an IFN response 1 and 2 d after infection, with means of 190 and 103 pg/ml, respectively; however, IFN-α was undetectable by 3 d after infection. Treated animals did not show alterations in systemic (serum) interleukin-2 (IL-2), monocyte chemotactic protein-1 (MCP-1), IL-1 receptor antagonist (IL-1RA), IL-6, IL-15 and IFN-γ levels compared to those observed in untreated animals (Supplementary Fig. 3). We observed elevated local levels of IL-6, IFN-γ and MCP-1 in homogenate from lung tissue collected 3 d after infection of untreated animals compared to treated animals (Supplementary Fig. 3), which indicates a tissue-specific host response to infection that is moderated by treatment.
We analyzed the effect of treatment on the lung transcriptome by microarray of RNA extracted from the right lower lobe (RLL). We performed a singular value decomposition–coupled multidimensional scaling (SVD-MDS) analysis, providing an overall view of the transcriptome, on samples from infected and uninfected animals in addition to control lung tissue samples from naive rhesus macaques (Fig. 4a). Comparison of group means revealed a clear separation between the treated and untreated lung RLL samples; treated and untreated samples were also separated from naive control samples, indicating that treatment induces unique transcriptional changes. Using a relaxed P value cutoff (P < 0.05), we observed 966 differentially expressed genes (DEGs) when treated, infected and untreated, infected animals were compared to naive animals (Supplementary Table 4), many associated with innate antiviral processes induced by IFN-α2b and ribavirin20,21. These included genes encoding type 1 and 3 IFN, pattern recognition receptors and IFN-induced with helicase C domain 1 (IFIH1, or MDA5), effector molecules and associated signal transducers. Comparison of uninfected, untreated animals to infected, treated and infected, untreated animals revealed lower expression of inflammatory genes in treated animals suggestive of milder disease (Supplementary Fig. 5).
Figure 4. Transcriptional signatures.
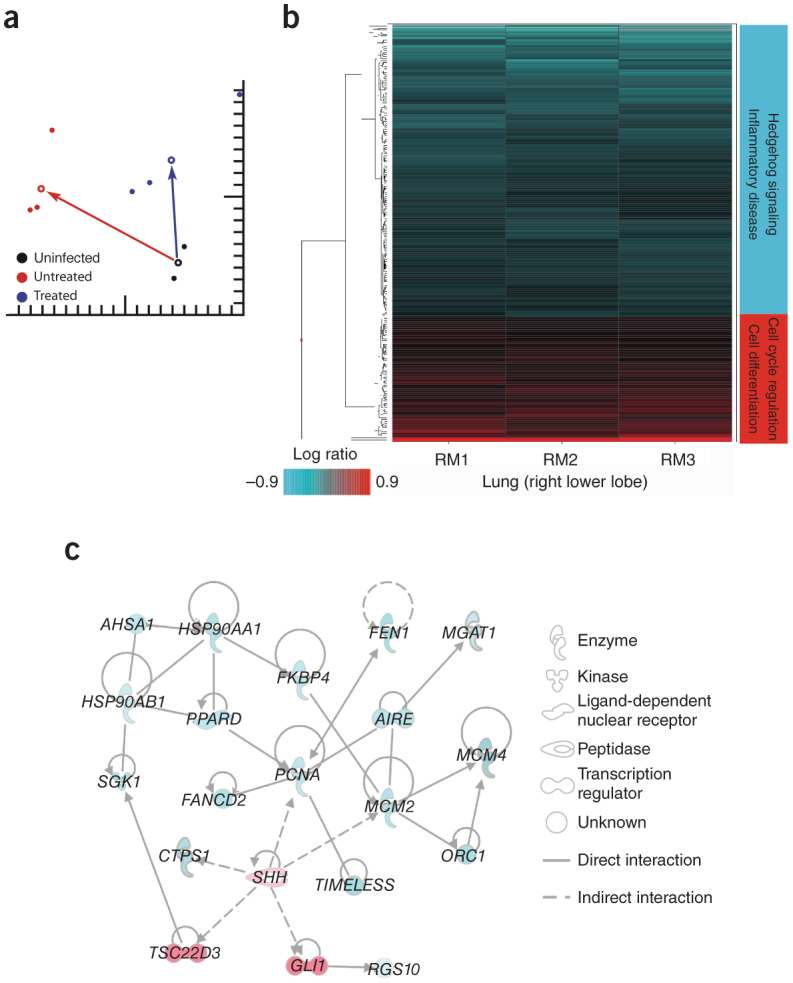
(a) Singular value decomposition–coupled multidimensional scaling (SVD-MDS) representation showing the distribution of individual samples following dimensionality reduction. This represents the similarity of the gene expression patterns of the following groups: uninfected, untreated control lung (black), MERS-CoV–infected, untreated lung (red, RM4–RM6) and MERS-CoV–infected, treated lung (blue, RM1–RM3). All lung samples were from the lower right lobe. Group means are shown as open circles, and distances from uninfected, untreated controls for MERS-CoV–infected, treated and MERS-CoV–infected, untreated macaques are shown as arrows between group means. The distance between points and groups is indicative of the differences in gene expression between the different groups. (b) Heatmap showing log10 expression ratios to untreated, MERS-CoV–infected group mean of 205 DEGs associated with IFN-α2b and ribavirin treatment, as determined by Welch's t-test (P < 0.01, fold change ≥ 1.5) and grouped by hierarchical clustering. (c) Molecular interaction network built using DEGs shown in b. Solid lines show direct molecular interactions; dashed lines show indirect molecular interactions. Red molecules are those upregulated in treated animals relative to untreated, whereas turquoise molecules are downregulated.
To elucidate the mechanism by which treatment reduces inflammation and disease during MERS-CoV infection, we sought to identify DEGs that are not associated with known IFN-stimulated gene induction programs. By examining the 205 DEGs identified using a more stringent P value cutoff (P < 0.01) (Fig. 4b and Supplementary Table 5) and Ingenuity Pathway Analysis, we identified many genes that were downregulated in untreated animals. Many of these were associated with hedgehog signaling (Fig. 4c), a pathway involved in respiratory morphogenesis and vascularization22,23,24,25,26,27 and potentially involved in inflammatory injury28. In treated animals, we observed upregulation of sonic hedgehog (SHH) and subsequent downregulation of genes associated with cell proliferation (Supplementary Table 5). SHH, the priming protein of the hedgehog pathway, and the associated molecule GLI family zinc finger 1 have been shown to block injurious responses in the pulmonary vasculature induced by multiple types of inflammatory and stress stimuli29,30,31,32. Consistent with the reduced inflammation, reduction in cellular infiltrates and lack of perivasculitis in the lungs of treated animals, the upregulation of SHH suggests that the hedgehog pathway may regulate the host response to protect the lung from potentially injurious inflammation.
Rhesus macaques treated with IFN-α2b and ribavirin 8 h after MERS-CoV infection showed improved clinical parameters with no breathing abnormalities and little evidence of pneumonia by X-ray. In addition, treated animals showed reduced systemic and local levels of proinflammatory markers. Lung tissues collected from treated animals 3 d after infection showed a significant reduction in viral genome copies, an absence of gross lesions and less severe histopathological changes compared to untreated animals. Microarray analysis of the lung supported the success of treatment and identified the hedgehog signaling pathway as a putative contributor to decreased lung damage. These data suggest that treatment of MERS-CoV–infected rhesus macaques with IFN-α2b and ribavirin improves outcome of infection.
IFN-α2b and ribavirin treatment would be expected to provide the greatest benefit early in infection; however, the prolonged disease course in humans8,33 suggests that the treatment window may be considerably wider in the clinic. The disease severity in the rhesus macaque model, which seems most similar to mild to moderate human MERS-CoV cases, makes it difficult to extrapolate the outcome of this treatment in severe cases in humans. Thus, it is currently unknown whether initiation of treatment upon progression to severe respiratory illness would have any benefit, a situation faced in many acute infectious diseases. Additional therapeutics should be evaluated; however, given the severity and poor outcome of MERS-CoV infection, combined IFN-α2b and ribavirin therapy should be considered as an early intervention therapy for MERS-CoV.
Methods
Biosafety statement.
All infectious work with MERS-CoV was approved by the Rocky Mountain Laboratories (RML) Institutional Biosafety Committee (IBC) and performed in a high-containment facility at the RML, Division of Intramural Research (DIR), National Institute of Allergy and Infectious Diseases (NIAID), National Institutes of Health (NIH). IBC-approved standard operating procedures were applied for all infectious work.
Virus and cells.
Vero cells (African green monkey kidney) were maintained at 37 °C in 5% CO2 in Dulbecco's modified Eagle's media (DMEM) supplemented with 10% FBS (FBS), 50 U/ml penicillin and 50 μg/ml of streptomycin. MERS-CoV (isolate hCoV-EMC/2012)8 was subsequently propagated on Vero cells using DMEM as above with 2% FBS (complete DMEM).
Animals.
All animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of the Rocky Mountain Laboratories and performed following the NIH Guide for the Care and Use of Laboratory Animals and the guidelines of the Association for Assessment and Accreditation of Laboratory Animal Care, International (AAALAC) by certified staff in an AAALAC-approved facility. We inoculated six healthy rhesus macaques (Macaca mulatta), aged 4–6 years, with a total of 7 × 106 TCID50 of MERS-CoV by combined intratracheal, intranasal, oral and ocular routes as previously established2,3,35. Animals were randomly assigned to either the treated (two males, one female) or untreated (two females, one male) group in a nonblinded manner.
Treatment.
Treatment was initiated at 8 h after infection. The initial ribavirin (MP Bioscience) loading dose (30 mg per kg body weight) was delivered intravenously; subsequent doses (10 mg per kg body weight) were delivered every 8 h intramuscularly. Human interferon-α2b, as IntronA (Merck) (5 MIU/kg) was delivered subcutaneously every 16 h. Three animals received IFN-α2b and ribavirin (treated), whereas three animals received sham treatment (untreated) by the same routes with a comparable volume per kg body weight of sterile saline. The animals were monitored three times daily through clinical scoring and/or examinations as described previously34. Clinical examinations included daily ventral-dorsal and lateral X-rays, evaluations of body temperature, blood pressure, heart rate and respiration rate, pulse oximetry, venous bleeding, bronchoalveolar lavage and collection of swabs from nasal and oral mucosa. A board-certified veterinarian interpreted the radiographic data. On day 3 after infection, all animals were necropsied, and conjunctiva, nasal mucosa, tonsil, oronasopharynx, mandibular lymph node, salivary gland, trachea, right bronchus, left bronchus, all six lung lobes, mediastinal lymph node, inguinal lymph node, axillary lymph node, mesenteric lymph node heart, liver, spleen, kidney and urinary bladder were collected for virological and histopathological assays.
Hematology assays.
Hematological analysis was carried out following standard protocols. The total white blood cell, lymphocyte, platelet, reticulocyte and red blood cell counts, hemoglobin and hematocrit values, mean cell volume, mean corpuscular volume, and mean corpuscular hemoglobin concentrations were determined from EDTA blood with the HemaVet 950FS+ laser-based hematology analyzer (Drew Scientific).
Serum and lung homogenate cytokine and chemokine analysis.
Samples from all lung lobes were weighed and subsequently homogenized in 400 μl of Bio-Plex cell lysis buffer (BioRad) with a stainless steel bead on the Qiashredder (Qiagen). Debris were pelleted and the supernatant removed. Both serum and tissue samples were inactivated with 5 MRads γ-radiation according to standard operating procedures. The total protein concentration in the tissue homogenate was determined with the DC Protein Assay (BioRad) and used to normalize the input. Concentrations of granulocyte colony–stimulating factor, granulocyte-macrophage colony–stimulating factor, IFN-γ, IL-1β, IL-1RA, IL-2, IL-4, IL-5, IL-6, IL-8, IL-10, IL-12/23 (p40), IL-13, IL-15, IL-17, MCP-1 and macrophage inflammatory protein-1α (MIP-1α), MIP-1β, soluble CD40 ligand (sCD40L), transforming growth factor-α (TGF-α), tumor necrosis factor-α (TNF-α), vascular endothelial growth factor (VEGF) and IL-18 were measured on a Bio-Plex 200 instrument (Bio-Rad) using the Non-Human Primate Cytokine MILLIPLEX map 23-plex kit (Millipore) according to the manufacturer's instructions. Levels of IFN-α in the serum were determined with the VeriKine human IFN-α ELISA kit (PBL, Piscataway, NJ) according to the manufacturer's instructions.
Histopathology.
Tissues were placed in cassettes and fixed in 10% neutral buffered formalin for 7 d. Subsequently, tissues were processed with a Sakura VIP-5 Tissue Tek, on a 12-h automated schedule, using a graded series of ethanol, xylene and ParaPlast Extra. Embedded tissues were sectioned at 5 μm and dried overnight at 42 °C before staining. Tissue sections were stained with H&E. Specific anti-CoV immunoreactivity was detected using a polyclonal rabbit antibody generated against hCoV-EMC/2012 at a 1:1,000 dilution. Tissues were also stained with a myeloperoxidase-specific antibody (1:450 dilution; 760-159, R&D Systems, Minneapolis, MN). The tissues were then processed for immunohistochemistry using the Discovery XT automated processor (Ventana Medical Systems) with a DAPMap (Ventana Medical Systems) kit, scanned with the Aperio ScanScope XT (Aperio Technologies, Inc.) and the entire section analyzed with the ImageScope Positive Pixel Count algorithm (version 9.1).
Genome quantification and microarray sample preparation.
RNA from AVL-treated blood and RLT-treated tissues were extracted with the QiaAmp Viral RNA and the RNeasy kits, respectively (Qiagen). qRT-PCR using primers and probe previously described19 were performed on the RotorGene Q (Qiagen). A tenfold dilution series of viral RNA based on TCID50 equivalents was used as a standard. For microarray analysis, lung homogenates were stored at −80 °C in equal volumes RLT buffer (Qiagen) and 70% ethanol until preparation. Samples were thawed, and two additional volumes of RLT buffer with 0.01 volumes of 2-mercaptoethanol were added, followed by an additional two volumes of 70% ethanol. RNA was then extracted using RNeasy spin columns per the manufacturer's protocol. Low-yield samples were concentrated using the RNA Clean and Concentrator (Zymo Research). Probe labeling was carried out using the Agilent Low Input processing protocol, and probes were hybridized to Agilent Rhesus macaque 4×44K microarrays (Agilent Technologies) using the manufacturer's one-color analysis protocol.
Microarray data and functional analysis.
For comparisons of differentially expressed genes (DEGs) in infected lungs of both IFN-α2b– and ribavirin-treated and untreated animals, raw array data were uploaded to Genedata Analyst 7.6 (Genedata). Data were normalized using the quantile normalization method, and the log10 ratio expression was calculated relative to the mean probe values of the three untreated animals. Statistically significant DEGs were determined using Welch's t-test (P < 0.01, fold change ≥ 1.5). Hierarchical clustering of DEGs was performed by the unweighted average method (unweighted pair group with arithmetic mean; UPGMA) using Spotfire DecisionSite 9.1.1 (Tibco). Analysis of functional enrichment was performed using Ingenuity Pathway Analysis software (Ingenuity Systems).
For measures of gene expression dynamics, the array data were normalized using NeONORM36 and subjected to SVD-MDS analysis37. In parallel, the data for each individual were merged using weighted averages to generate combined profiles. These were visualized using SVD-MDS.
Statistical analyses.
With the exception of the data from the microarray experiment, data were analyzed in Prism (GraphPad Software) using the unpaired t-test, two-way ANOVA with Bonferroni's post hoc test and the Mann-Whitney test as indicated in the figure legends.
Accession codes.
Raw microarray data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus database under accession number GSE47957.
Supplementary information
Supplementary Figures 1–4 and Supplementary Tables 1–5 (PDF 3002 kb)
Acknowledgements
We would like to thank B. Haagmans and R. Fouchier, Erasmus Medical Center, for providing MERS-CoV (isolate hCoV-EMC/2012). We also thank all the members of the Rocky Mountain Veterinary Branch (Division of Intramural Research, NIAID, US National Institutes of Health (NIH)) for their assistance, especially K. Cordova, J. Faris, K. Hardcastle, R. LaCasse, D. Long, K. Meade-White, R. Rosenke, T. Thomas and A. Weidow. A. Mora and A. Athman assisted with editing the figures.
Author Contributions
D.F., E.d.W., V.J.M. and H.F. conceived of and designed the study. D.F., E.d.W., A.L.R., F.F., A.O., D.P.S., T.B., C.M. and D.B. performed the experiments. D.F., E.d.W., A.L.R., A.O., D.P.S., L.B., A.G.B., V.J.M., M.G.K. and H.F. analyzed the data. D.F., A.L.R., M.G.K. and H.F. wrote the manuscript. This work was supported in part by the Intramural Research Program, NIAID, NIH, in addition to the NIAID Regional Centers of Excellence (U54 AI081680), Systems Virology (NIH/NIAID contract number HHSN272200800060C) and Washington National Primate Research Center (P51OD010425) to M.G.K.
Accession codes
Primary accessions
Gene Expression Omnibus
Competing interests
The authors declare no competing financial interests.
References
- 1.Anonymous. MERS-COV - Eastern Mediterranean (65) Saudi Arabia, RFI, Qatar. ProMED 20130904.1919328 (2013).
- 2.Munster VJ, de Wit E, Feldmann H. Pneumonia from human coronavirus in a macaque model. N. Engl. J. Med. 2013;368:1560–1562. doi: 10.1056/NEJMc1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Wit, E. et al. Novel human betacoronavirus causes a transient lower respiratory tract infection in a rhesus macaque model. Proc. Natl. Acad. Sci. USA (in the press).
- 4.Centers for Disease Control and Prevention (CDC). Update: Severe respiratory illness associated with Middle East Respiratory Syndrome Coronavirus (MERS-CoV)–worldwide, 2012–2013. MMWR Morb. Mortal. Wkly. Rep.62, 480–483 (2013). [PMC free article] [PubMed]
- 5.Omrani AS, et al. A family cluster of Middle East Respiratory Syndrome Coronavirus infections related to a likely unrecognized asymptomatic or mild case. Int. J. Infect. Dis. 2013;17:e668–e672. doi: 10.1016/j.ijid.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Falzarano D, et al. Inhibition of novel beta coronavirus replication by a combination of interferon-alpha2b and ribavirin. Sci. Rep. 2013;3:1686. doi: 10.1038/srep01686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Boheemen S, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. mBio. 2012;3:e00473–12. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 9.Stockman LJ, Bellamy R, Garner P. SARS: systematic review of treatment effects. PLoS Med. 2006;3:e343. doi: 10.1371/journal.pmed.0030343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Soo YO, et al. Retrospective comparison of convalescent plasma with continuing high-dose methylprednisolone treatment in SARS patients. Clin. Microbiol. Infect. 2004;10:676–678. doi: 10.1111/j.1469-0691.2004.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ho JC, et al. Pentaglobin in steroid-resistant severe acute respiratory syndrome. Int. J. Tuberc. Lung Dis. 2004;8:1173–1179. [PubMed] [Google Scholar]
- 12.Cheng Y, et al. Use of convalescent plasma therapy in SARS patients in Hong Kong. Eur. J. Clin. Microbiol. Infect. Dis. 2005;24:44–46. doi: 10.1007/s10096-004-1271-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peiris JS, et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sung JJ, et al. Severe acute respiratory syndrome: report of treatment and outcome after a major outbreak. Thorax. 2004;59:414–420. doi: 10.1136/thx.2003.014076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Booth CM, et al. Clinical features and short-term outcomes of 144 patients with SARS in the greater Toronto area. J. Am. Med. Assoc. 2003;289:2801–2809. doi: 10.1001/jama.289.21.JOC30885. [DOI] [PubMed] [Google Scholar]
- 16.Chan KS, et al. Treatment of severe acute respiratory syndrome with lopinavir/ritonavir: a multicentre retrospective matched cohort study. Hong Kong Med. J. 2003;9:399–406. [PubMed] [Google Scholar]
- 17.Chu CM, et al. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loutfy MR, et al. Interferon alfacon-1 plus corticosteroids in severe acute respiratory syndrome: a preliminary study. J. Am. Med. Assoc. 2003;290:3222–3228. doi: 10.1001/jama.290.24.3222. [DOI] [PubMed] [Google Scholar]
- 19.Corman VM, et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17:20285. doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 20.Thomas E, et al. Ribavirin potentiates interferon action by augmenting interferon-stimulated gene induction in hepatitis C virus cell culture models. Hepatology. 2011;53:32–41. doi: 10.1002/hep.23985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarasin-Filipowicz M, et al. Interferon signaling and treatment outcome in chronic hepatitis C. Proc. Natl. Acad. Sci. USA. 2013;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Menshykau D, Kraemer C, Iber D. Branch mode selection during early lung development. PLOS Comput. Biol. 2012;8:e1002377. doi: 10.1371/journal.pcbi.1002377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang M, Wang H, Teng H, Shi J, Zhang Y. Expression of SHH signaling pathway components in the developing human lung. Histochem. Cell Biol. 2010;134:327–335. doi: 10.1007/s00418-010-0738-2. [DOI] [PubMed] [Google Scholar]
- 24.Li M, et al. Mesodermal deletion of transforming growth factor-β receptor II disrupts lung epithelial morphogenesis: cross-talk between TGF-β and Sonic hedgehog pathways. J. Biol. Chem. 2008;283:36257–36264. doi: 10.1074/jbc.M806786200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, et al. Hedgehog signaling in neonatal and adult lung. Am. J. Respir. Cell Mol. Biol. 2013;48:703–710. doi: 10.1165/rcmb.2012-0347OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.White AC, Lavine KJ, Ornitz DM. FGF9 and SHH regulate mesenchymal Vegfa expression and development of the pulmonary capillary network. Development. 2007;134:3743–3752. doi: 10.1242/dev.004879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.White AC, et al. FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development. 2006;133:1507–1517. doi: 10.1242/dev.02313. [DOI] [PubMed] [Google Scholar]
- 28.Cigna N, et al. The hedgehog system machinery controls transforming growth factor-beta-dependent myofibroblastic differentiation in humans: involvement in idiopathic pulmonary fibrosis. Am. J. Pathol. 2012;181:2126–2137. doi: 10.1016/j.ajpath.2012.08.019. [DOI] [PubMed] [Google Scholar]
- 29.Fitch PM, Howie SE, Wallace WA. Oxidative damage and TGF-β differentially induce lung epithelial cell sonic hedgehog and tenascin-C expression: implications for the regulation of lung remodelling in idiopathic interstitial lung disease. Int. J. Exp. Pathol. 2011;92:8–17. doi: 10.1111/j.1365-2613.2010.00743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang G, et al. Activation of the sonic hedgehog signaling controls human pulmonary arterial smooth muscle cell proliferation in response to hypoxia. Biochim. Biophys. Acta. 2010;1803:1359–1367. doi: 10.1016/j.bbamcr.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang Y, et al. Protection from lipopolysaccharide-induced pulmonary microvascular endothelial cell injury by activation of hedgehog signaling pathway. Mol. Biol. Rep. 2011;38:3615–3622. doi: 10.1007/s11033-010-0473-8. [DOI] [PubMed] [Google Scholar]
- 32.Pogach MS, Cao Y, Millien G, Ramirez MI, Williams MC. Key developmental regulators change during hyperoxia-induced injury and recovery in adult mouse lung. J. Cell. Biochem. 2007;100:1415–1429. doi: 10.1002/jcb.21142. [DOI] [PubMed] [Google Scholar]
- 33.Health Protection Agency (HPA) UK Novel Coronavirus Investigation team. Evidence of person-to-person transmission within a family cluster of novel coronavirus infections, United Kingdom, February 2013. Euro Surveill.18, 20427 (2013). [DOI] [PubMed]
- 34.Brining DL, et al. Thoracic radiography as a refinement methodology for the study of H1N1 influenza in cynomologus macaques (Macaca fascicularis) Comp. Med. 2010;60:389–395. [PMC free article] [PubMed] [Google Scholar]
- 35.Kobasa D, et al. Aberrant innate immune response in lethal infection of macaques with the 1918 influenza virus. Nature. 2007;445:319–323. doi: 10.1038/nature05495. [DOI] [PubMed] [Google Scholar]
- 36.Noth S, Brysbaert G, Benecke A. Normalization using weighted negative second order exponential error functions (NeONORM) provides robustness against asymmetries in comparative transcriptome profiles and avoids false calls. Genomics Proteomics Bioinformatics. 2006;4:90–109. doi: 10.1016/S1672-0229(06)60021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bécavin C, Tchitchek N, Mintsa-Eya C, Lesne A, Benecke A. Improving the efficiency of multidimensional scaling in the analysis of high-dimensional data using singular value decomposition. Bioinformatics. 2011;27:1413–1421. doi: 10.1093/bioinformatics/btr143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1–4 and Supplementary Tables 1–5 (PDF 3002 kb)