Abstract
Detergent interaction with extramembranous soluble domains (ESDs) is not commonly considered an important determinant of integral membrane protein (IMP) behavior during purification and crystallization, even though ESDs contribute to the stability of many IMPs. Here we demonstrate that some generally nondenaturing detergents critically destabilize a model ESD, the first nucleotide-binding domain (NBD1) from the human cystic fibrosis transmembrane conductance regulator (CFTR), a model IMP. Notably, the detergents show equivalent trends in their influence on the stability of isolated NBD1 and full-length CFTR. We used differential scanning calorimetry (DSC) and circular dichroism (CD) spectroscopy to monitor changes in NBD1 stability and secondary structure, respectively, during titration with a series of detergents. Their effective harshness in these assays mirrors that widely accepted for their interaction with IMPs, i.e., anionic > zwitterionic > nonionic. It is noteworthy that including lipids or nonionic detergents is shown to mitigate detergent harshness, as will limiting contact time. We infer three thermodynamic mechanisms from the observed thermal destabilization by monomer or micelle: (i) binding to the unfolded state with no change in the native structure (all detergent classes); (ii) native state binding that alters thermodynamic properties and perhaps conformation (nonionic detergents); and (iii) detergent binding that directly leads to denaturation of the native state (anionic and zwitterionic). These results demonstrate that the accepted model for the harshness of detergents applies to their interaction with an ESD. It is concluded that destabilization of extramembranous soluble domains by specific detergents will influence the stability of some IMPs during purification.
Keywords: CFTR, NBD1, DSC, CD, thermal unfolding, detergent interaction, membrane protein, extramembrane domain, soluble domain
Introduction
The options for solubilization and purification of integral membrane proteins (IMP) have been advanced by the continuous development of detergents with diverse chemical structures and physical properties, and by empirically-derived models for how these properties affect IMP stability. Understandably, research has focused on the interactions of detergents with the transmembrane domain (TM), since this region must be liberated from the membrane with an intact native structure.1,2 Some detergents can lead to protein inactivation and denaturation,2,3 while others are less efficient and may result in poor solubility and/or incomplete extraction of the desired protein.4 Detergents are classified into four broad groups based on their ionic properties, i.e., charged (anionic, cationic), zwitterionic, and nonionic. In practice, detergents are often categorized as “harsh” or “mild,” based on their tendency to denature proteins.4–6 As a rule of thumb, charged detergents are harsher than uncharged detergents and detergents with larger head groups and longer hydrophobic tails are milder than their counterparts with smaller head groups and shorter tails.4,6 These general rules are based on empirical evidence, obtained by studying interactions of detergents primarily with membrane proteins,7–17 with a focus on the TM domain. Structural models have been proposed to explain how detergent interactions stabilize the TM domain,8,18 supported in part by X-ray crystal structures.19–21
Detergent effects on soluble proteins have been less well studied. Earlier efforts have focused on establishing the general mechanisms of how charged detergents denature proteins.22–25 It has recently been shown that several proteases used for protein affinity tag removal are significantly inhibited by detergents commonly used for IMP solubilization.26 A recent review by Ozten focuses on the mechanism of soluble protein-detergent interactions, with particular attention to the anionic detergent, SDS.27 It is generally assumed that nonionic and zwitterionic detergents do not bind to soluble proteins except under limited circumstances.28–30 For example, folding intermediates and molten globule states are partially unfolded conformations of soluble proteins that bind detergents at exposed hydrophobic surfaces;30,31 proteins that have explicit binding sites for amphiphiles or hydrophobic molecules, like enzymes with amphiphilic substrates and serum albumins, also bind detergents.29,32–34
Nonetheless, systematic studies of detergent interactions with extramembrane domains1 do not exist, even though IMP structure and function rely on proper communication between the TM and extramembrane domains. Denaturation, or mere destabilization, of the extramembrane domains can lead to the destabilization of the entire protein through loss of contact at the domain interface(s). For example, in vitro assembly/disassembly studies on the Escherichia coli ABC transporter, BtuCD, showed that the disassociation of the extramembrane ATP-binding subunits from the TM subunits is coupled to unfolding of the TM subunits.35,36 Another example is the structural defect arising from the most common cystic fibrosis disease-causing mutation, deletion of the single amino acid F508 within the first nucleotide-binding domain (NBD1) of the cystic fibrosis transmembrane conductance regulator (CFTR). The deletion of F508 (F508del) destabilizes the entire protein as evidenced by its severely impaired folding, trafficking, and channel activity.37–44 Studies on the isolated CFTR NBD1 domain show that F508del results in both kinetic and thermodynamic destabilization, while only causing minor structural changes in the vicinity of the mutation.45–51 On the other hand, incorporation of site-specific mutations in NBD1 that improve NBD1 stability and/or its ability to interact with the intracellular loops, also promote better folding in the full-length CFTR.52–60
Many extramembrane domains of membrane proteins can autonomously fold in solution when isolated from the full-length protein. This observation may be accounted for by a recent study which compared the protein data bank structures of 558 membrane proteins with 43,547 soluble proteins and found that 67% of the extramembrane domains of the membrane proteins share structural similarity with soluble proteins.61 In the present study we focused on the NBD1 of CFTR, whose structure and thermodynamic stability have been well-characterized,45–51,62–64 to dissect detergent effects on the isolated domain and its role in membrane protein stability in the absence of TM domains. We selected members from three detergent classes, anionic, nonionic and zwitterionic, based on their relevance in CFTR purification and/or popularity in membrane protein research. We have adopted a novel quantitative approach by measuring the effects of detergents on the thermostability by differential scanning calorimetry (DSC) and secondary structure by circular dichroism (CD). By modeling the linked equilibria between thermal unfolding and detergent binding, we have identified three distinct mechanisms for thermal destabilization by detergents via global nonlinear least squares fitting of the DSC curves. The effects of these detergents on lysozyme, a soluble protein with a simple two-state unfolding mechanism, were also determined in order to validate our models and suggest our conclusions extend to soluble proteins as well. Recently, Tulumello and Deber showed that the TM segments from several membrane proteins have the same secondary structure in harsh (anionic) or mild (uncharged) detergents and proposed that the denaturing properties of harsh detergents on membrane proteins could be due to their interactions with the non-membrane regions.65 Our results appear to be the first experimental evidence to support this hypothesis. It seems reasonable that the same mechanism may apply to membrane proteins in general. Therefore, our approach could serve as an additional screening tool for detergent selection in membrane protein purification and structural determination.
Results
Overview of the analytical methods
The instrumental techniques used in this study included far-UV circular dichroism (CD), static light scattering (SLS), and differential scanning calorimetry (DSC). With CD, the change in secondary structure was monitored to determine whether or not significant changes in native state structure occurred; with SLS, the formation of large protein/detergent complexes was monitored; together with DSC, we were able to assess whether thermal destabilization and/or unfolding resulted from detergent treatment. DSC measures the difference in heat uptake (heat capacity) between a sample and a reference material during controlled heating or cooling. Typical DSC curves of simple globular proteins show a single endothermic unfolding transition [e.g., Fig. 1(A)]. The mid-point of the unfolding transition peak, Tmax, usually represents the temperature at which 50% of the protein is unfolded and is widely used as a measure of the protein's thermal stability. Also directly measured by DSC is the amount of heat required to unfold the protein, the calorimetric enthalpy, ΔHc, which is obtained from the area under the DSC unfolding transition. As discussed in the present article, conditions that destabilize a protein are defined as those that either lower the Tmax or decrease the ΔHc, or both. If the ΔHc is reduced to zero, then the protein is unfolded or denatured and CD also is expected to report a change in native state structure.
Figure 1.
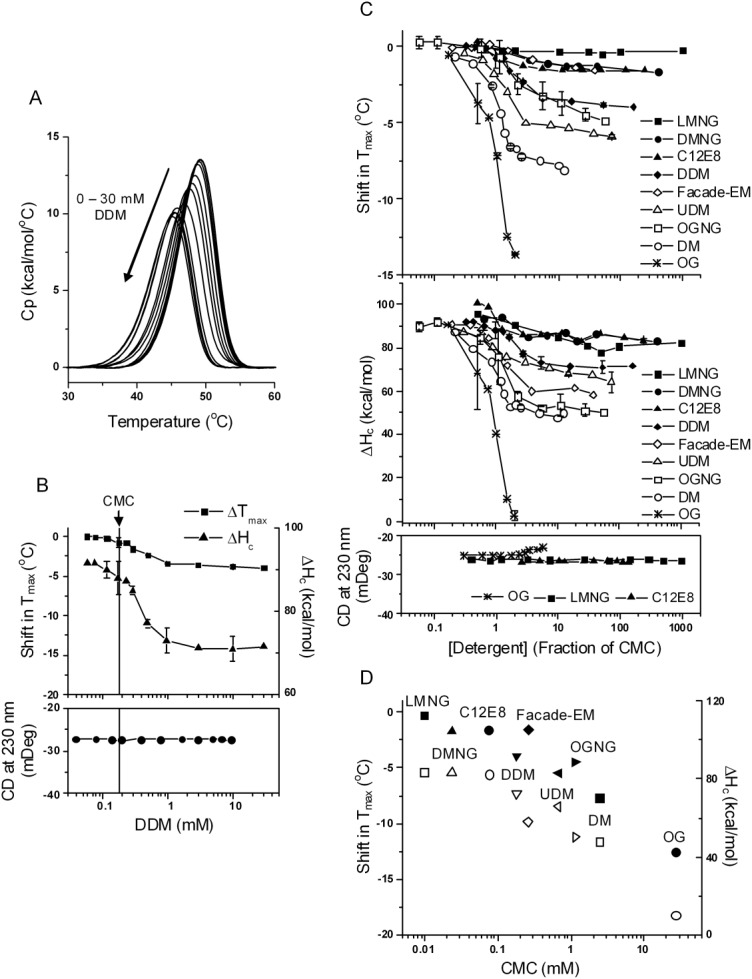
The effect of nonionic detergents. A, DSC curves of NBD1 in the presence of DDM at increasing concentrations. Buffer conditions were 20 mM HEPES pH 7.5, 150 mM NaCl, 10% glycerol, 10% ethylene glycol, 1 mM TCEP, 20 µM ATP, 3 mM MgCl2. Same buffer conditions were used in all DSC and CD experiments. Data represent one set of experiments conducted on the same batch of protein on two separated dates within one week. B, Tmax shift (squares), ΔHc (triangles), and CD (solid circles) as function of DDM concentrations. The DSC data are average of two sets of experiments. The points with error bars represent the average of two or more experiments and the error bars are the standard deviations. The lines connecting the symbols are there to help guide the eye. The vertical line denotes the CMC of DDM. C, Tmax shift (upper panel) and ΔHc (middle panel) in the presence of all the nonionic detergents studied; lower panel, the change in CD signal induced by LMNG, C12E8, and OG. The majority of the data points represent one experiment. The points with error bars represent the average of two or more experiments and the error bars are the standard deviations. D, correlation between CMC and the magnitude of Tmax shift (solid symbols) and ΔHc (open symbols). Data were taken at 31 mM for OG, 10 mM for DMNG, LMNG and Façade-EM, and 20 mM for the rest of the detergents, all corresponding to approximately 1% w/v, a concentration commonly used for membrane protein extraction.
Explicit detergent binding models have previously been used to describe the effects of detergents.30 According to Le Chatelier's Principle, which describes the effects of a concentration change on a dynamic equilibrium, in the presence of an interacting ligand the protein thermal unfolding equilibrium will shift towards either the native or the unfolded state, depending on which state has higher affinity for the ligand. For example, stoichiometric binding of a ligand to the native protein will increase the apparent thermal stability, i.e., Tmax will increase, provided no change (or unfolding) in the native state structure occurs concomitant with binding; conversely, binding to the unfolded state lowers the Tmax.66–68 When binding to both the native and unfolded states occur, the Tmax could shift up or down,68 and only in this case, the Tmax shift may reach a plateau at higher ligand concentrations.67 Therefore, the most important qualitative features of ligand interaction are determined by the observed ligand concentration dependencies of the DSC curves. The quantitative features of the ligand interaction, i.e., binding affinity and enthalpy, can be obtained via thermodynamic modeling (see description of Fig. 4 below). Global nonlinear least squares fitting of DSC curves to thermodynamic models that link the protein unfolding equilibrium and ligand binding equilibria allows for an accurate determination of the ligand binding parameters based on the proposed models, and the goodness of fit supports the chosen model.
Figure 4.
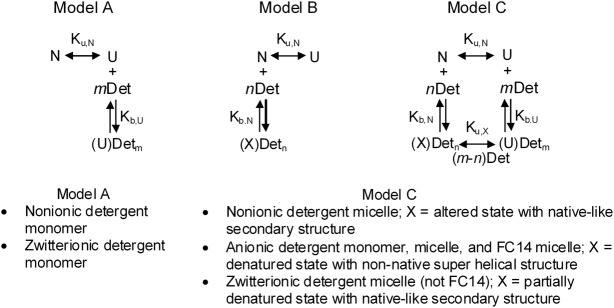
Models for the global curve fitting of the DSC data. The detergent class fit by each model is listed under the model. The double headed arrow indicates an unfolding process that may occur in more than one discrete step. Species are N = native, folded; U = thermally unfolded; Definition of “X” depends on the detergent as shown in the box under Model B and Model C; note that the detergent induced denatured state and thermally unfolded state, U, are not necessarily the same energetic/structural states; Det = detergent; Det may represent monomer or micelle; n, m = number of detergent binding sites on N or U, respectively. See text for how the best-fit model was determined for each detergent class.
In the present studies, it is reasonable to expect that detergents will interact with the unfolded state and unfolding intermediates,30,31 which expose hydrophobic surfaces that can bind multiple detergent molecules. This binding mechanism will lower the Tmax, as expected. On the other hand, binding of multiple detergent molecules to the native state may induce an altered folded state that is less stable than the native state, or such binding may even induce denaturation, and either effect will result in a decrease in both ΔHc and Tmax. Note that this is mechanistically different from native-state stoichiometric ligand binding, which does not produce an altered, partially unfolded, conformation.
A pre-requisite for obtaining accurate thermodynamic parameters via modeling of the DSC curves is the reversibility of the thermal unfolding. Our previously published studies50,51 have shown that the thermal unfolding of NBD1 is irreversible. However, analysis of the DSC data indicates unfolding occurs in two steps, the first of which is equilibrium unfolding to an intermediate that also binds ATP. Further unfolding of the intermediate occurs via a kinetically controlled irreversible step. A total of eight parameters are required to exactly describe the thermal unfolding of NBD1 in the presence of ATP.50 Detergents may affect any or all of these parameters. In the present study, we used low ATP concentration so that the binding of ATP to the intermediate state was negligible, thus reducing the number of parameters by two. However, it was necessary to further reduce the number of fitting parameters, and therefore we fit the DSC curves to a single unfolding transition using three parameters, Tmax, ΔHc and the apparent van't Hoff enthalpy, ΔHv. This is a simplification that nevertheless provides meaningful information on the detergent binding mechanism, since inclusion of the parameter ΔHv allows for ΔHv > ΔHc.2
This type of approximation also has been used in the analyses of ligand binding to proteins that unfold irreversibly, where the apparent ΔHv is used as a true thermodynamic parameter.69–71 This approximation greatly reduced the complexity of the models, but as a consequence, we can not deconvolute the effects of detergent on each unfolding step or on ATP binding affinity (which will affect Tmax and ΔHc). The implications of these approximations will be discussed below. A further consequence is the thermodynamic parameters obtained from curve fitting are necessarily apparent parameters. Nevertheless, as stated above, the purpose of the modeling is to support the general mechanisms of detergent interaction that are established from the observed concentration dependencies of NBD1 stability, and which are not dependent on the quantitative outcome of the modeling.
All classes of detergents destabilize NBD1 but to varying extent
Nonionic detergent micelles destabilize NBD1 primarily via a nondenaturing alteration in native state properties
The nonionic detergent DDM, is often considered as a good starting detergent for membrane protein purification.72 In fact, among all the detergents used for crystallizing α-helical IMPs, DDM has been the most successful.73 For comparison with DDM, we tested DM and UDM (different hydrophobic tail lengths), C12E8 (different head group), OG (smaller head group and shorter tail), new nonionic maltose-neopentyl glycols (MNGs) and Façade-EM (Table I lists the structure and properties of the detergents studied). MNGs and Façade-EM have shown some favorable properties in comparison with previously used nonionic detergents.74,75 The single hydrocarbon tail counterparts of DMNG, LMNG and OGNG are DM, DDM and OG, respectively.
Table I.
List of Detergents Used in this Studya
| Short name | Full name | Structure | CMCb in mM and (% w/v) |
|---|---|---|---|
| DM | n-Decyl−β−D-maltopyranoside | 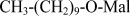 |
2.44 (0.118%) |
| UDM | n-Undecyl−β−D-maltopyranoside | 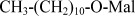 |
0.67 (0.033%) |
| DDM | n-Dodecyl−β−D-maltopyranoside | 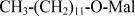 |
0.18 (0.009%) |
| OG | n-Octyl−β−D-glucopyranoside | 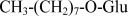 |
20.4 (0.6%) |
| DMNG | Decyl Maltose Neopentyl Glycol | 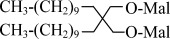 |
0.024 (0.002%) |
| LMNG | Lauryl Maltose Neopentyl Glycol | 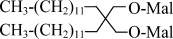 |
0.0099 (0.001%) |
| OGNG | Octyl Glucose Neopentyl Glycol | 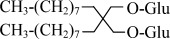 |
1.13 (0.064%) |
| C12E8 | Octaethylene Glycol Monododecyl Ether | 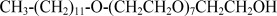 |
0.076 (0.004%) |
| Façade-EM | 3α-hydroxy-7α,12α-di-((O-ß-D-maltosyl)−2-hydroxyethoxy)-cholane |  |
0.26 (0.029%) |
| NaPFO | Sodium Perfluoro-octanoate | 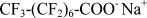 |
10.8 (0.47%) |
| LiPFO | Lithium Perfluoro-octanoate | 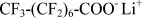 |
7.8 (0.33%) |
| LPG12 | Sodium 1-Lauroyl-2-Hydroxy-sn-Glycero-3-Phospho-(1'-rac-Glycerol) | 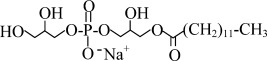 |
1.23 (0.055%) |
| LPG14 | Sodium 1-Myristoyl-2-Hydroxy-sn-Glycero-3-Phospho-(1'-rac-Glycerol) | 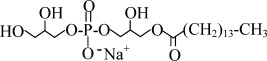 |
0.11 (0.005%) |
| LPG16 | Sodium 1-palmitoyl-2-hydroxy-sn-glycero-3-phospho-(1'-rac-glycerol) | 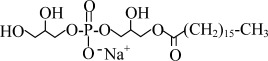 |
0.019 (0.001%) |
| LPC12 | 1-lauroyl-2-hydroxy-sn-glycero-3-phosphocholine | 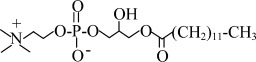 |
0.87 (0.038%) |
| LPC14 | 1-myristoyl-2-hydroxy-sn-glycero-3-phosphocholine | 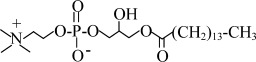 |
0.083 (0.004%) |
| LPC16 | 1-palmitoyl-2-hydroxy-sn-glycero-3-phosphocholine | 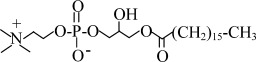 |
0.0089 (0.0004%) |
| FC14 | n-Tetradecyl-phosphocholine | 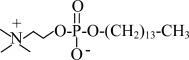 |
0.13 (0.005%) |
| DiC6PC | 1,2-dihexanoyl-sn-glycero-3-phosphocholine | 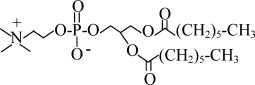 |
10.4 (0.47%) |
| CHAPS | 3-[(3-Cholamidopropyl)-Dimethylammonio]-1-Propane Sulfonate/N,N-Dimethyl-3-Sulfo-N-[3-[[3α,5β,7α,12α)−3,7,12-Trihydroxy-24-Oxocholan-24-yl]Amino]propyl]-1-Propanaminium Hydroxide, Inner Salt |  |
6.83 (0.42%) |
The detergents are grouped based on their ionic property. DDM to Façade-EM are nonionic, NaPFO to LPG16 are anionic, and LPC12 to CHAPS are zwitterionic.
The critical micellar concentrations (CMC) were determined in the DSC buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM TCEP, 10% glycerol and 10% ethylene glycol, 20 µM ATP, 3 mM MgCl2). The standard deviations for the CMC values from duplicate measurements are mostly less than 5% with a few exceptions at 10%.
Mal = maltose; Glu = glucose.
DDM caused a small destabilization in NBD1. Figure 1(A) shows the DSC profiles of NBD1 in the presence of increasing concentrations of DDM. The detergent concentration dependence of Tmax and ΔHc had a similar shape [Fig. 1(B)]. When [DDM] was below the CMC, there was an almost imperceptible decrease in Tmax (<0.5°C) with increasing concentration of detergent monomer and a small change in ΔHc. However, a significant decrease in Tmax occurred above the CMC. This decrease in Tmax was accompanied by a large decrease in ΔHc,3 indicating a reduced structural stability, and suggesting the detergent-bound state was different from the native state but still folded. This native-like state retained native secondary structure based on the lack of change in the CD signal at 230 nm [Fig. 1(B)]. The effect appeared to reach saturation at about 5× CMC, although there was still a small continuous drop in Tmax with further DDM addition. The far-UV CD signal at 230 nm remained constant throughout the concentration range tested, suggesting no change in secondary structure nor denaturation.
The response of NBD1 to other nonionic detergents was similar to DDM. Figure 1(C) shows the change in Tmax, ΔHc, and CD signal at 230 nm induced by all nonionic detergents studied. LMNG and DMNG were the least destabilizing in this series and OG was the most. Nonionic detergents did not induce a change in secondary structure, with the exception of OG, which increased the CD signal slightly. They generally caused little or no change in thermal stability below CMC. Above CMC, the decrease in Tmax and ΔHc usually saturated at 5 to 10× CMC. The magnitude of change at saturation for each detergent correlated with the detergent's CMC [Fig. 1(D)]. A possible mechanism for this correlation will be discussed later. In contrast to all other nonionic detergents studied, OG at 40 mM, approximately 2× its CMC, completely abolished the unfolding transition and this denaturation was also detected by the CD signal above CMC. A similar report in the literature showed that OG induced the formation of a molten globule-like state in glutamate dehydrogenase.76 The small size and high polarity of its head group and the short tail length for OG are consistent with its degree of harshness observed in the present studies.
Anionic detergent monomers and micelles destabilize NBD1 via denaturation of the native state
Members of this class denatured NBD1 either at monomeric concentrations or at concentrations above the CMC often used experimentally. LPGs and PFO are widely used for the purification of full-length CFTR.77–80 Their popularity is due to their high extraction efficiency and the ability to prevent aggregation of the purified CFTR.78 Figure 2(A) shows the DSC detergent concentration dependence of NBD1 in response to LPG14. A steady decrease in both Tmax and ΔHc occurred well below the CMC, and complete loss of the DSC transition was observed at 6.3 mM (60× CMC). LPG14 also induced a change in the secondary structure, as shown by the CD signal at 230 nm. The magnitude of the CD signal increased 77% upon the addition of LPG14, reaching a plateau at concentrations where the DSC peak became very broad [Fig. 2(A), leftmost four traces]. At this point, the protein had lost a cooperative tertiary structure but contained significant amount of non-native helical structure. SDS also induced a similar amount of non-native helical structure at denaturing concentration [compare lower panels of Figs. 2(B,C)], both of which are somewhat larger than the change seen with LPG12 [Fig. 2(C) lower panel].
Figure 2.
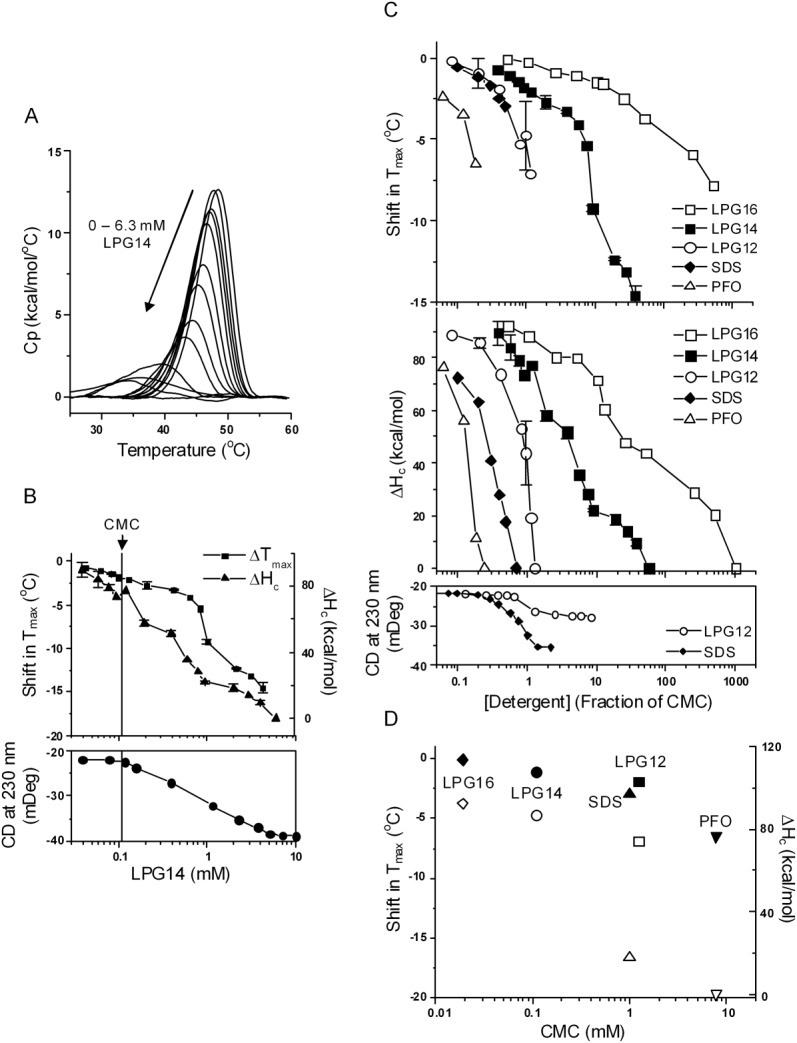
The effect of anionic detergents. A, DSC curves of NBD1 in the presence of LPG14 at increasing concentrations. Data represent one set of experiments conducted on the same day. B, Tmax shift (squares), ΔHc (triangles), and CD (solid circles) as function of LPG14 concentration. The vertical line denotes the CMC of LPG14. The majority of the DSC data represent the average of two sets of experiments. Standard deviation is shown as the error bar. C, Tmax shift (upper panel) and ΔHc (middle panel) in the presence of all the anionic detergents studied; lower panel, the change in CD signal induced by LPG12 and SDS. The lines connecting the symbols are there to help guide the eye. The majority of the data points represent one experiment. The points with error bars represent the average of two or more experiments and the error bars are the standard deviations. D, correlation between CMC and the magnitude of Tmax shift (solid symbols) and ΔHc (open symbols). Data were taken at detergent concentrations that correspond to half of their CMC except PFO. The Tm shift at 0.2× CMC of PFO is shown because it caused complete denaturation at 0.24× CMC.
In addition, there was a noticeable increase in light scattering at LPG14 concentrations below its CMC (data not shown). Previously published studies on SDS/protein interactions27,81,82 show that submicellar SDS binds to protein and clusters at the surface of the protein, and it is possible that LPG14 monomers interact with NBD1 in a similar manner.
The response of NBD1 to LPG12 and LPG16 was similar to LPG14 [Fig. 2(C)]. A continuous drop in ΔHc was observed regardless of whether the detergents are in the monomeric or micellar form. Complete loss of cooperative unfolding occurred in the presence of 1.6 mM LPG12, or 20 mM LPG16. The denaturing power of the LPGs decreased as the alkyl tail length increases [Fig. 2(D)]. This observation agrees with the general trend that detergents with a longer tail length are milder.
Data for PFO are shown in Figure 2(C). Like SDS, PFO was much harsher than LPG14, causing complete loss of the DSC transition at concentrations as low as 2 mM, which corresponds to 1/4 CMC and 1/100 of the typical concentration used in initial protein extractions from membrane77,80 [Fig. 2(C)].
Zwitterionic detergent micelles destabilize NBD1 by partial denaturation of the native state
This class of detergents have the advantage of being more “lipid-like,” and are believed to better mimic natural membranes through matching head group chemistry and tail length.83 For example, lysophosphatidylcholines (LPCs) and short-chain phosphatidylcholine lipids (DiC6–9PC) have been shown to maintain the native structure of membrane proteins.83,84 Nevertheless, they caused complete loss of the DSC transition at concentrations above the CMC, although usually at concentrations higher than used for membrane protein extraction. Compared to the anionic LPG14, LPC14 was milder [Fig. 3(A)]. As a monomer, LPC14 caused almost no change in Tmax and ΔHc, and no change in CD signal [Fig. 3(B)], similar to the nonionic detergents. Above the CMC, a decrease in both Tmax and ΔHc was observed. The Tmax shift saturated at >10× CMC, but the decrease in ΔHc did not. Extrapolation to higher detergent concentration suggested that complete loss of ΔHc might occur, but only above a detergent concentration that is relevant to membrane protein extraction or purification. Unlike the anionic detergents, LPC14 did not change the secondary structure of NBD1, suggesting the change induced by this detergent was mainly in the tertiary structure.
Figure 3.
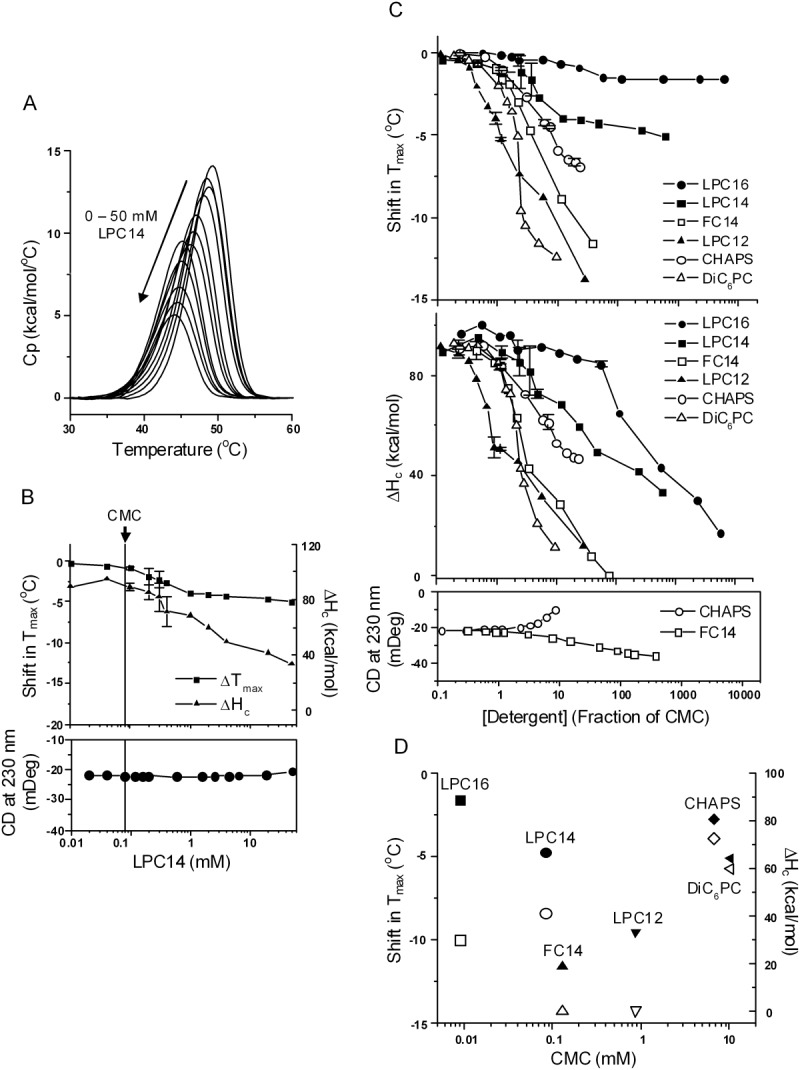
The effect of zwitterionic detergents. A, DSC curves of NBD1 in the presence of LPC14 at increasing concentrations. Data represent one set of experiments conducted on three separate dates. B, Tmax shift (squares), ΔHc (triangles), and CD (solid circles) as function of LPC14 concentration. The vertical line denotes the CMC of LPC14. The DSC data are average of two sets of experiments. The points with error bars represent the average of two experiments and the error bars are the standard deviations. C, Tmax shift (upper panel) and ΔHc (middle panel) in the presence of all the zwitterionic detergents studied; lower panel, the change in CD signal induced by FC14 and CHAPS. The majority of the data points represent one experiment. The points with error bars represent the average of two or more experiments and the error bars are the standard deviations. The lines connecting the symbols are there to help guide the eye. D, correlation between CMC and the magnitude of Tmax shift (solid symbols) and ΔHc (open symbols). Data were taken at 20 mM detergent concentration except FC14. The Tm shift at 5 mM FC14 is shown because it caused complete denaturation at 10 mM.
Figure 3(C) shows the effects of several other commonly used zwitterionic detergents, including DiC6PC, CHAPS and FC14. These lowered the Tmax and ΔHc to varying degrees, and LPC12 and FC14 completely abolished the DSC transition at 10 to 100× CMC. They also caused different changes in the secondary structure: FC14 increased helicity, while CHAPS decreased it. The formation of non-native helical structure (inferred from a decrease in ellipticity at 230 nm) characterized the denaturation of NBD1 by both the anionic and the harsher zwitterionic detergents (e.g. FC14). Among the zwitterionic detergents, the thermal unfolding parameters did not correlate with detergent CMC [Fig. 3(D)], perhaps due to structural diversity, although there appeared to be a correlation between the Tmax and CMC within the LPC series.
Detergent binding mechanisms from global fitting of DSC curves
Global curve fitting of the respective concentration-dependent DSC data [e.g., as shown in Figs. 1(A), 2(A), and 3(A)] was performed using the thermodynamic models shown in Figure 4. These models comprise the simplest combination of the unfolding process and the detergent binding mechanisms: Model A, destabilization with no change in native structure, via binding to the unfolded state only; Model B, destabilization via alteration or denaturation of the native state only; Model C, destabilization via binding to both the native and the unfolded states. These are reasonable molecular models and can also account for all the observed changes in Tmax and ΔHc. A thermodynamic cycle similar to that shown in Model C has long been used to model the effects of ligand binding on protein stability.67,85 Simulations using the models showed that only models A and C correctly predicted the lowering of Tmax and ΔHc in the presence of the detergents. However, only Model C also fits the saturation of the destabilization effect at higher detergent concentrations, such as observed for nonionic detergents (see below). The mathematical derivations of all the models are shown in the Supporting Information Section SD1.
In order to select the best and simplest model that adequately described the data, detergent concentration-dependent datasets from each detergent class were globally fit to all three models. In addition, because the aggregation state of the detergents is different below and above the CMC, the primary interacting detergent species may be different, and therefore the DSC data in these two regions were fit separately. In other words, three different fits were performed for each selected detergent below and above their CMC. The model with the lowest reduced χ24 value was considered to be the best-fit model. Supporting Information Table ST1 lists the three reduced χ2 values obtained for the detergents that were fitted with a micelle species. In Supporting Information Figure SF1, the DDM dataset above CMC is used to illustrate the different fit curves obtained by the various models. The resulting fits for the best model for a representative from each detergent class is shown superimposed on the actual DSC data in Supporting Information Figure SF2.
With very few exceptions, detergents belonging to the same class were best fit by the same model. The Tmax and ΔHc values were extracted from the fits and compared with the experimental values obtained for each detergent as shown in Figure 5, illustrating the predicted effects of the best model on these two experimentally-determined parameters.
Figure 5.
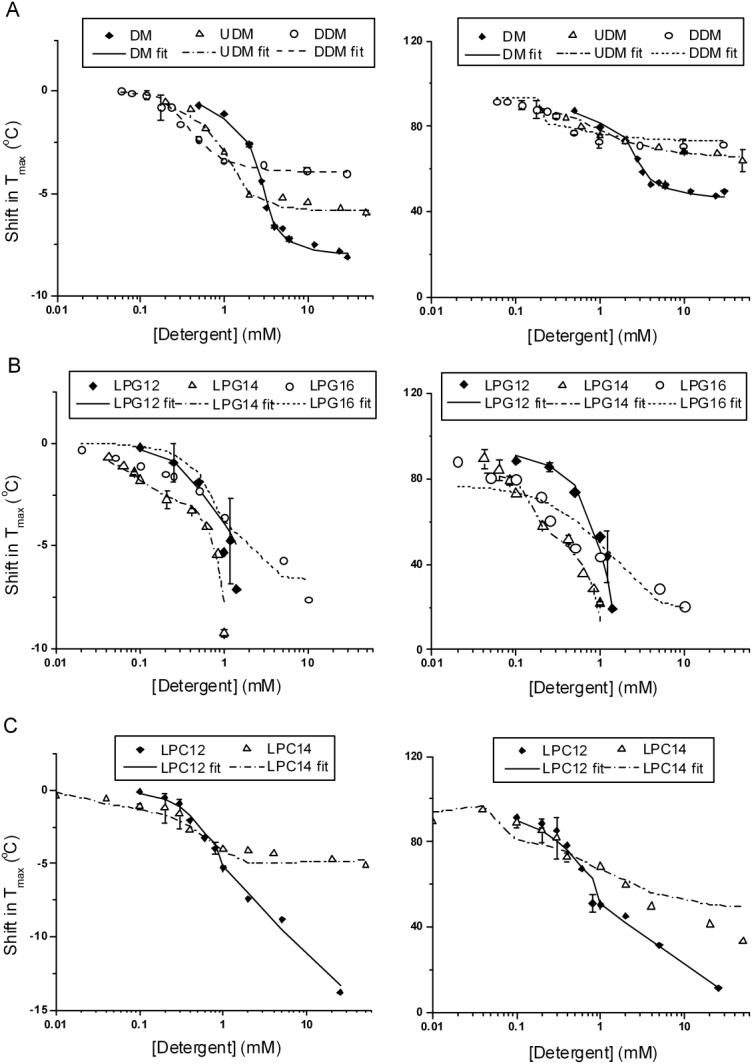
Comparison of experimental and fitted Tmax and ΔHc values for selected representatives from each detergent class. A, maltoside; B, LPGs; C, LPCs. Left panels are for shift in Tmax, and right panels are for ΔHc. The fitted Tmax and ΔHc values were extracted from the fitted DSC curves. The symbols represent the experimental values with error bars representing the standard deviations of two experiments, and the lines represent the fitted values; see Supporting Information for further discussion of the models and the actual experimental and fitted DSC curves (Supporting Information Fig. SF2). Global curve fitting was performed on all DSC curves collected for each detergent, except for DDM and LPG14, in which case one complete set of DSC curves collect on the same day or two separate dates within a week were used.
Nonionic detergents
We focused curve fitting efforts on the three alkyl maltosides with increasing tail lengths (DM, UDM, and DDM). Below CMC, Model A was the best-fit model with the detergent monomers as the interacting species, which describes detergent binding exclusively to the unfolded state. Intuitively, one may expect detergent monomers to interact with the native state as well and, in fact, Model C fit the DSC curves below CMC equally well as Model A. However, the fit parameters suggested the binding to the native state is extremely weak, which in effect reduces Model C to Model A. Among the three maltosides, DDM had the lowest CMC, i.e., monomer concentration, and possibly as a consequence, DDM also caused the least destabilization in this region. Above CMC, Model C better described the binding processes with the detergent micelles as the interacting species. The destabilizing effect decreased in the order of DM > UDM > DDM, as the micelle size of the detergent increased.
Anionic detergents
The denaturing effects of the anionic detergents resembled those of chemical denaturants, such as urea and guanidine-HCl, in that a linear relationship exists between the free energy of unfolding and the denaturant concentration (Supporting Information Fig. SF5 for LPG12).86,87 Denaturation by urea and guanidine is generally thought to occur by both preferential stabilization of the unfolded state88 and binding to the native state (changes in the water activity and dynamics in the native state also likely apply89,90).
Model C (Fig. 4) was used to fit the anionic detergent data both below and above CMC. For anionic detergents that completely denatured NBD1 in the submicellar region, such as SDS, PFO, and LPG12, (recall that CD data shown in Fig. 2 were consistent with denaturation), it is reasonable to conclude that the monomers are the denaturing species, although there is evidence in the literature that premicellar clusters of SDS formed on the protein surface are more potent denaturants than the monomers.27 For LPG14 and LPG16, because of their low CMC (i.e. monomer concentration), there was no significant destabilization below CMC, and complete denaturation occurred at 60× CMC for LPG14 and 1000× CMC for LPG16. Curve fitting using either the monomer or micelle as the interacting species above CMC yielded fits with similar reduced χ2 values and consequently did not distinguish which species was the denaturing species above CMC.
Zwitterionic detergents
Below CMC, this class was similar to nonionic detergents. Model C and Model A fit equally well in this region, but fit parameters suggested that binding to N was weak and therefore reducing Model C to Model A. Above CMC, with the exception of LPC165 the detergents were best fit by Model C, with micelles as the interacting species. Curve fitting on CHAPS data above CMC by Model A and Model C resulted in similar reduced χ2 values (see Supporting Information Table ST1). The parameters obtained from fitting to Model C suggested binding to N was weak but with large uncertainty in the estimated binding affinity. Because CD showed loss of secondary structure in the presence of high concentration of CHAPS, the destabilization by CHAPS seems more likely to follow Model C. However, it was impossible to estimate the apparent binding affinity to N from curve fitting.
Egg white lysozyme as a model protein for validation of binding mechanisms
Lysozyme was chosen for study to shed light on the applicability of the observed detergent effects for soluble proteins. The same modeling approach was applied to egg white lysozyme, which has been well-studied and known to unfold by a reversible, two-state mechanism.67,91 This also allowed us to validate the approach of using three parameters, Tmax, ΔHc, and ΔHv, to describe the unfolding process. Curve fitting results showed that for lysozyme, the fitted ΔHc and ΔHv were within 10% of each other, an indication of a bona fide two-state reversible process. Even though the lysozyme and NBD1 unfolding mechanisms are different, we found the effects of the detergents on lysozyme were similar to NBD1. Furthermore, the same respective models best described these effects (Supporting Information Section SD2). Notable differences between lysozyme and NBD1 were the greater destabilization of NBD1 seen with nonionic detergents, and the response to OG; while OG eventually denatures NBD1 (recall the significant changes in ellipticity as well as ΔHc), OG did not denature lysozyme. The validation of the models using the lysozyme DSC datasets is shown in Supporting Information Section SD3.
Doping anionic detergents with nonionic detergents or phospholipids reduces their denaturing effects
The ability of anionic detergents to prevent aggregation is likely due to the increased electrostatic repulsion between CFTR/detergent complexes.78 If charge is a requirement for solubilization, it may be possible to include nonionic detergents, which do not denature, with the anionic detergents, in order to mitigate denaturation by anionic detergents, while maintaining an overall net negative charge for the mixed micelle. The effectiveness of using a mixed micelle system to reduce the denaturing effects of the LPGs was demonstrated by the increase in both Tmax and ΔHc upon the addition of DDM into the NBD1/LPG14 complex, shown in Figure 6(A). Inclusion of more DDM resulted in less destabilization. In addition, the CD signal of NBD1 remained the same in the presence of the mixed detergents (data not shown) in contrast to the large increase seen with LPG14 alone [Fig. 2(B)], suggesting the native secondary structure is preserved. The effect of added lipids was also studied. Many membrane proteins have conserved lipid binding sites,92 and lipids are often required in order to maintain the stability of IMP during purification and structural determination.3,93 The denaturing effect of LPG14 was also reduced by the inclusion of lipids [Fig. 6(B)]. The rescuing effects of the lipids were dependent on the detergent concentration and the time the protein spent in the presence of LPG14 without the lipids.
Figure 6.
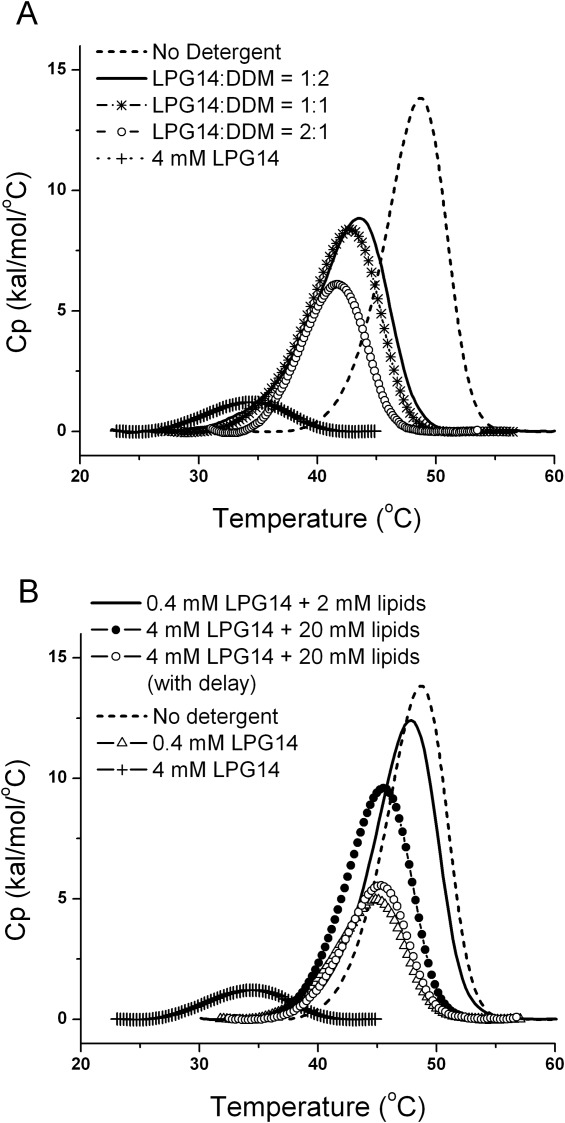
Increase in both Tmax and ΔHc due to the addition of DDM or phospholipids into the NBD1/LPG14 complex. A, the effect of added DDM. The total detergent concentrations were held constant at 10 mM, while the DDM/LPG14 ratio was increased. DSC curve in the presence of 4 mM LPG14 is shown as the control because LPG14 caused complete denaturation at 10 mM. B, the effect of added phospholipids. LPG14 and lipid concentrations are shown in the data labels. When lipids were added with delay, the delay time was 4 hrs. The composition of the lipids was POPC: POPE = 4:1 (w/w).
Reversibility of detergent denaturation
There is precedent in the literature showing complete recovery of enzymatic activity upon SDS-removal from membrane proteins that were purified with SDS.94,95 Most functional data on CFTR have been collected after the protein was reconstituted into lipids, which usually is accompanied by complete removal of the detergent used for purification. Since all detergents affected the stability of NBD1 more or less depending on the concentrations used, it was of interest to test whether the effects were reversible, i.e. could the native state be regained upon detergent removal.
Table II shows the recovery of native NBD1 after incubation with various detergents for a certain length of time and subsequent removal of detergents by hydrophobic-interaction spin columns. The recovery of total protein (column 2) and the fraction of folded protein (column 3) were monitored. Recovery from nonionic detergents was comparable with the detergent-free control, while recovery from the anionic detergents or FC14 was much lower. Addition of DDM improved the recovery from LPG14. For FC14 and LPG14, the percentage of irreversibly denatured NBD1 increased with both incubation time and detergent concentration.
Table II.
Recovery of the Native hNBD1 After Detergent Removala
| Detergent and incubation time | Detergent concentration | Concentration recovery (%)b | Calorimetric enthalpy (%)c | Combined recovery (%) |
|---|---|---|---|---|
| no detergent | 93 ± 7 | 100 | 93 ± 7 | |
| DDM, 4 hrs | 20 mM (1% w/v, 111× CMC) | 98.5 ± 0.7 | 101 ± 1 | 99 ± 1 |
| DDM, 20 hrs | 20 mM (1% w/v, 111× CMC) | 81 ± 23 | 95 ± 4 | 77 ± 22 |
| DMNG, 4 hrs | 20 mM (1.9% w/v, 833× CMC) | 95 ± 4 | 101 ± 6 | 96 ± 7 |
| DMNG, 20 hrs | 20 mM (1.9% w/v, 833× CMC) | 95 ± 8 | 100 ± 8 | 95 ± 11 |
| LMNG, 4 hrs | 20 mM (2% w/v, 2020× CMC) | 99 ± 4 | 104 ± 1 | 103 ± 4 |
| LMNG, 20 hrs | 20 mM (2% w/v, 2020× CMC) | 96 ± 3 | 100 ± 4 | 96 ± 5 |
| LPG14, 4 hrs | 4 mM (0.19% w/v, 36× CMC) | 47 ± 6 | 99 ± 4 | 47 ± 6 |
| LPG14, 4 hrs | 10 mM (0.48% w/v, 90× CMC) | 55 ± 10 | 48 ± 4 | 26 ± 5 |
| LPG14, 20 hrs | 4 mM (0.19% w/v, 36× CMC) | 16 ± 4 | 74 ± 6 | 12 ± 3 |
| PFO, 4 hrs | 2 mM (0.09% w/v, 0.2× CMC) | 13 ± 2 | 57 ± 5 | 7 ± 1 |
| PFO, 4 hrs | 20 mM (0.9% w/v, 2× CMC) | 1 ± 1 | 0 | 0 |
| DDM/LPG14 1(1:1), 4 hrs | 10 mM (0.5% w/v, 69× CMC) | 94 ± 12 | 102 ± 1 | 96 ± 12 |
| DDM/LPG14 (1:1), 20 hrs | 10 mM (0.5% w/v, 69× CMC) | 91 ± 13 | 88 ± 16 | 81 ± 18 |
| FC14, 2 hrs | 5 mM (0.19% w/v, 38× CMC) | 63.5 ± 0.7 | 70 ± 14 | 44 ± 9 |
| FC14, 2 hrs | 25 mM (0.95% w/v, 192× CMC) | 31 ± 6 | 35 ± 1 | 11 ± 2 |
| FC14, 20 hrs | 5 mM (0.19% w/v, 38× CMC) | 33 ± 4 | 27 ± 9 | 9 ± 3 |
| LPC14, 4 hrs | 25 mM (1.2% w/v, 301× CMC) | 70.5 ± 0.7 | 83 ± 2 | 59 ± 2 |
| LPC14, 20 hrs | 25 mM (1.2% w/v, 301× CMC) | 83 ± 3 | 81 ± 4 | 67 ± 4 |
| CHAPS, 4 hrs | 10 mM (0.6% w/v, 1.5× CMC) | 96 ± 6 | 99 ± 3 | 95 ± 7 |
| CHAPS, 20 hrs | 10 mM (0.6% w/v, 1.5× CMC) | 91 ± 3 | 97 ± 2 | 88 ± 3 |
| CHAPS, 4 hrs | 68 mM (4.2% w/v, 10× CMC) | 85 ± 16 | 96 ± 11 | 82 ± 18 |
| CHAPS, 20 hrs | 68 mM (4.2% w/v, 10× CMC) | 86 ± 18 | 92 ± 6 | 79 ± 17 |
Tmax of all the samples after detergent removal matches the no-detergent control sample, indicating the spin column effectively removes all detergents. Results were averages of two sets of experiments conducted with different batches of protein on different dates.
The loss of protein in the spin column is likely due to the irreversible association of the protein with some of the detergents. When the detergents were retained by the spin column, the protein molecules were retained with the detergents.
A lowered apparent molar enthalpy indicates the existence of some denatured NBD1 in the recovered samples from the spin column.
Milder detergents improve the purification yield of full-length CFTR and help maintain its ATP binding ability
We have carried out small-scale purification trials of full-length CFTR expressed in mammalian cells96,97 with the three different classes of detergents to determine whether recovery results obtained for NBD1 are transferrable to the full-length protein. The extraction efficiency and overall protein recovery after immobilized metal-affinity chromatography (IMAC) are summarized in Table III. Although CFTR is quite soluble in many detergents, recovery after a single IMAC chromatography step (overnight exposure to the detergent) varied significantly. Nonionic detergents resulted in much better recovery after IMAC than the anionic and zwitterionic detergents which likely reflects the better (short-term) stability of full-length CFTR in non-ionic detergents as seen for the isolated NBD1 (Fig. 1).
Table III.
Extraction Efficiency and IMAC Recovery of Full-Length Human CFTR
| Detergent used for extraction and purification | Detergent concentration during extraction (mM) | Extraction efficiencya (%) | IMAC recoveryb (%) | Overall recovery (%) |
|---|---|---|---|---|
| UDM | 10 | 84 | 54 | 45 |
| DDM | 9.8 | 79 | 51 | 40 |
| DMNG | 5.3 | 74 | 55 | 41 |
| LMNG | 5.0 | 85 | 43 | 37 |
| C12E8 | 9.3 | 73 | 60 | 44 |
| LPC14 | 10.7 | 117 | 19 | 22 |
| LPC16 | 10.1 | 100 | 16 | 16 |
| CHAPS | 8.1 | 52 | 26 | 14 |
| LPG14 | 10.4 | 115 | 28 | 32 |
| LPG16 | 10.1 | 100 | 13 | 13 |
Extraction efficiency = total CFTR extracted/total CFTR found in the membrane preparation.
IMAC recovery = total CFTR eluted/total CFTR applied.
To determine if the influence of detergents on the extramembrane NBD was similar in the native full-length CFTR as with the isolated domain, the ability of the full-length CFTR to bind ATP was monitored as a function of detergent and incubation time. ATP binding was monitored by incubating membranes containing CFTR with 8-azido-[γ-32P]-ATP followed by exposure to the detergent for the times and concentrations indicated prior to photo-labeling (Fig. 7). The amount of ATP that remained bound through the detergent treatment was quantitated by autoradiography after immunoprecipitation of CFTR with the monoclonal antibody L12B4 and SDS/PAGE. Previous studies98,99 have shown that the NBD1 site has a higher affinity to ATP and a low hydrolytic turnover rate, whereas the binding of ATP at the NBD2 site is followed by rapid hydrolysis and product release. In the present experiments, the occlusion of the unhydrolyzed ATP in the NBD1 site was monitored. A loss of ATP occlusion suggests a loss or significant reduction in the ATP binding affinity to NBD1. Figure 7(A) shows a comparison among DDM, FC14, and LPG14. Overall, there was a positive correlation between the extent of nucleotide occluded in these three detergents and the recovery of native NBD1 after detergent removal from the isolated domain (Table II). Furthermore, the loss of the ability of CFTR to retain the bound nucleotide in 20 mM LPG14 or 26 mM FC14 is entirely consistent with the complete denaturation of NBD1 by 6 mM LPG14 or 10 mM of FC14 as observed by DSC. Longer incubation with detergents was more detrimental to the ATP binding. After 24 hrs, CFTR retained ATP only in 2 mM DDM or LPG14, but not in FC14. In the absence of added lipids, some ATP occlusion persisted in DMNG and LMNG [Fig. 7(B)] and these detergents also caused less destabilization to NBD1 in comparison with DDM. In the presence of lipids, the level of ATP occlusion increased significantly in all three nonionic detergents. This result is also similar to the result with isolated NBD1 which showed that inclusion of lipids improved its stability in the presence of detergent.
Figure 7.
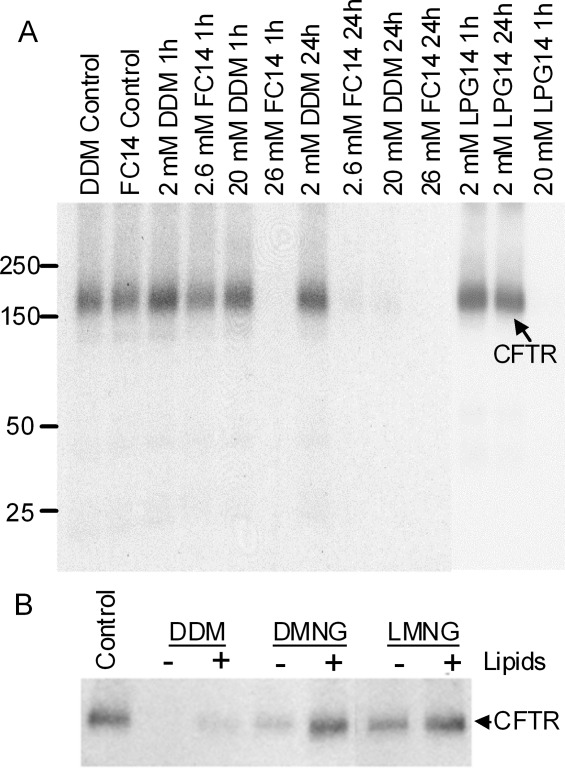
Photolabeling of full-length CFTR with 8-azido-[γ-32P]ATP. Membranes (10 µg protein) from BHK-21 cells expressing CFTR were incubated with 8-azido-[γ-32P]ATP (25 µM for 5 min), and then crosslinked by UV irradiation after exposure to different detergents for the indicated times (controls indicating 0 time). After crosslinking, CFTR was immunoprecipitated and subjected to SDS/PAGE and autoradiography. 32P radioactivity associated with the CFTR band was determined by electronic autoradiography (Packard Instant Imager). A, A comparison among DDM, FC14, and LPG14, in the absence of lipids. B, Membranes were incubated with 1% (w/v) of DDM, DMNG or LMNG for 2 hrs, followed by 0.2% (w/v) for 24 hrs, in the presence (+) or absence (−) of liver polar lipids (0.1% w/v final concentration). The control membranes in both panels were crosslinked immediately after solubilization.
Discussion
In this work, we systematically studied the interactions between NBD1 and three classes of detergents. DSC and CD were used to monitor the effect of detergent on NBD1, revealing two general thermodynamic models for thermal destabilization (Models A and C in Fig. 4). Global fitting of the detergent concentration-dependent DSC data using these models that link unfolding and detergent binding processes allowed us to analyze the detergent interaction mechanisms. In all cases where the DSC data suggested partial or complete unfolding by detergent, changes in CD signal at 230 nm also pointed to changes in secondary structure consistent with a global loss of native structure. To summarize, anionic detergents denature NBD1 mainly through binding to the native state, a process that is complete at or below the CMC for the harshest detergents. NBD1 is also susceptible to denaturation by zwitterionic detergents, with complete denaturation occurring via micelle binding at very high concentrations for most zwitterionic detergents. Nonionic detergents destabilize NBD1 at monomeric concentrations by binding primarily to the unfolded state, and above their CMC through binding to the native state. OG was the only member of this class to denature NBD1. The extent of structural change induced by the bound detergents determines the detergent harshness, which follows the general trend observed for membrane proteins, i.e., anionic > zwitterionic > nonionic. There is an apparent correlation between destabilization and CMC. This correlation is most convincing for the nonionic detergents. Within a given structural group, the correlation is also apparent for the anionic and zwitterionic detergents.
The anionic detergents LPG and PFO are generally considered to be milder than SDS and have been used for CFTR purification.77–80 They have been shown to support the intramolecular helix-helix interactions in a so-called helical hairpin consisting of just the third and fourth adjacent TM segments of the 12 TM helices in CFTR.9 LPG16 is effective in yielding high quality NMR spectra for five membrane proteins,84 although all five proteins are small helical membrane proteins that do not possess extramembrane domains. Those results may not be indicative of the detergent's effect on complex membrane proteins, such as the ABC transporters, which contain significant extramembrane domains. In our study, LPG and PFO were found to denature NBD1 at moderate concentrations and short exposure times. They appear to denature NBD1 via the same mechanism as SDS, i.e., the detergents bind to the native state and apparently induce a highly helical conformation (inferred from ellipticity changes at 230 nm) that lacks organized tertiary structure. For those that denature via monomer binding, the harshness also correlates with their CMC, which in turn is a measure of the monomer concentration. The formation of non-native helical structure in NBD1 may be the cause of the irreversible denaturation observed for these detergents. Overnight incubation with LPG14 at a concentration typically used during purification, or incubation for 4 hrs at a concentration typically used during initial extraction results in more than 75% permanent loss of the native NBD1. Recovery from PFO is ever lower.
Zwitterionic detergents, such as those studied here, are used extensively in membrane protein purification and structure determination. These detergents cause significant thermal destabilization by apparently disrupting primarily tertiary structure, since there is no change in CD signal, while there is a significant decrease in ΔHc. Complete denaturation of NBD1, as evidenced by a complete loss of ΔHc, requires high micelle concentrations and does not involve alteration in secondary structure. The exception is FC14, which induces non-native helical structure in a similar manner to the anionic detergents. Like the anionic detergents, denaturation by FC14 is not reversible. Incubation with FC14 for only 2 hrs, at a concentration typically used during initial extraction, results in approximately 90% loss of the native NBD1. In contrast to FC14, more than 60% of the native NBD1 is recoverable from LPC14 and CHAPS, possibly because their micelles destabilize NBD1 without affecting its secondary structure.
Destabilization caused by nonionic detergents is almost completely reversible, which is encouraging for the prospect of reconstitution of full-length membrane proteins from these detergents. The degree of destabilization correlates well with CMC. A similar correlation has been observed for the few soluble proteins that bind uncharged detergents.30,34 To explain this correlation, Otzen proposed that detergent monomers bind to discrete sites on the protein and prime it for interaction with micelles. Consequently, the destabilizing effects depend on monomer concentration, i.e., the CMC.27,34 In contrast, our current models suggest nonionic and zwitterionic detergent monomers do not alter the native conformation, but lower the apparent thermal stability through binding to the unfolded state. It appears that nonionic detergent micelles bind to the native state and induce a less stable, folded conformation. The apparent correlation between destabilization and CMC may be due to the influence of the micelle size. Larger micelles can accommodate more proteins, resulting in less detergent molecule bound per protein, and less destabilization. A dependence on micelle size would manifest itself as a correlation with CMC, because micelle size and CMC are highly correlated within the same class of detergents.
One area that needs further investigation is whether minor reduction in Tmax and ΔHc is the result of a protein conformational change or loss of ATP binding or both. Destabilization caused by most nonionic and zwitterionic detergent monomers and several nonionic detergent micelles is less than one would expect from the complete loss of ATP binding and, therefore, either or both changes in ATP affinity and conformation could account for the destabilization. However, it would be difficult to tweeze out these different effects because they may be linked; i.e., even small changes in tertiary structure could result in a weaker affinity for ATP. Loss of ATP binding may also account for the more significant destabilization of NBD1 by the nonionic detergents in comparison to lysozyme. However, it's equally possible that NBD1 is more susceptible to detergent destabilization because thermal unfolding is irreversible. We have conducted preliminary isothermal titration calorimetry experiments and found UDM or DDM monomers do not alter the ATP binding parameters. This lends support to the interpretation that these detergent monomers destabilize NBD1 by binding to the unfolded state. Similar experiments will enable us to further differentiate the mechanisms.
Relevance to full length CFTR and other IMP
Our preliminary results on full-length CFTR recovery after detergent extraction suggest that detergent effects on NBD1 stability and recovery can predict to some degree their effects on CFTR. In fact, it has recently been communicated that harsh detergents such as LPG14 were shown to eliminate the thermal unfolding transition of full-length CFTR, as shown here for isolated NBD1.100 Moreover, as shown here, the ATP binding affinity of CFTR is reduced or abolished by select detergents, which could also be anticipated from the destabilization or denaturation of the isolated NBD1 domain by the same detergents. While we interpret the reduction in NBD1 unfolding Tmax and ΔHc by nonionic detergents as a result of an induced conformational change, we can not rule out the possibility that these parameters are reduced as a result of the lower ATP affinity of the altered conformation. Collectively, we hope these observations will prove useful for future work on CFTR purification. We anticipate that conditions that improve recovery of isolated NBD1, such as the inclusion of excipients that are known to stabilize the native state and/or phospholipids during purification, will promote better recovery of the native full-length CFTR because of the known coupling between the stability of NBD1 and the stability of CFTR.52–60 Beyond CFTR, the destabilization by detergents is not unique to NBD1. As we showed for lysozyme, the molecular details of detergent destabilization appear to be generally applicable to soluble proteins. We have learned that the widely used empirical relationship between detergent harshness and detergent structure describes the effects of detergents on NBD1, lysozyme, and by extrapolation should also be applicable to the extramembranous soluble domains of other IMP, many of which are structurally homologous to soluble proteins.
Materials and Methods
Materials
The full name and structure of the detergents are listed in Table I. Detergents were from Anatrace (Maumee, OH), except DiC6PC was from Avanti Polar Lipids (Alabaster, AL), and CHAPS was from Pierce (Rockford, IL). POPC, POPE and liver polar lipids were from Avanti Polar Lipids (Alabaster, AL). Bisbenzimide H 33342 (aka Hoechst 33342) was from Sigma (St. Louise, MO). The purity of all chemicals was 99%. Chicken egg white lysozyme (6× crystallized) was from Seikagaku Corp (Tokyo, Japan), and was used without further purification. Stable BHK-21 (baby-hamster kidney) cell line expressing full-length CFTR was generated and maintained as described previously;98 8-azido-[γ-32P]ATP was obtained from Affinity Labeling Technologies.
Protein purification
NBD1 purification was conducted as previously described,50,51 yielding protein in 150 mM NaCl, 20 mM HEPES pH 7.5, 10% glycerol, 10% ethylene glycol, 1 mM tris(2-carboxymethyl) phosphine (TCEP), 2 mM ATP, 3 mM magnesium chloride. Proteins were >98% pure as judged by Coomaisse Blue staining of SDS-PAGE gels, showed no evidence of aggregation and ran as monomers during gel filtration. Protein concentration was determined with the Pierce 660 nm assay in microtiter plate format, calibrated using Bacillus subtilis NAD synthetase. Protein was stored at −80°C.
Critical micellar concentration (CMC) determination
CMC was determined via a hydrophobic dye partitioning method.101 Detergents were prepared as 10% w/v stock solutions in pure water, and filtered through a 0.45 µm filter. The stock solutions were diluted to various concentrations in 150 mM NaCl, 20 mM HEPES pH 7.5, 10% glycerol, 10% ethylene glycol, 1 mM TCEP, 20 μM ATP, and 3 mM magnesium chloride. All detergent solutions contained 7 μM of the fluorescent dye, Bisbenzimide H 33342. Fluorescence of the solutions were measured in 96-well microtiter plates in a PolarStar Optima fluorimeter (BMG Labtech) using an excitation wavelength of 355 ± 5 nm and an emission wavelength of 460 ± 5 nm. CMC was determined by non-linear curve fitting to the fluorescence data. There were at least 24 points on each curve.
Differential scanning calorimetry (DSC)
Calorimetry was carried out on the VP-Capillary DSC System (MicroCal Inc., GE HealthCare), in 0.130 mL cells, at a heating rate of 2°C/min. Since the Tmax and ΔHc for NBD1 thermal unfolding varies with scan rate, for a selected detergent, UDM, we conducted the concentration-dependent DSC experiment at three different scan rates and performed global curve fitting analysis for all three data sets. We confirmed that the best fit model remained the same regardless of scan rate and the apparent detergent binding parameters obtained were similar (see Supporting Information Fig. SF4 and Supporting Information Table ST2), suggesting the approximation to a single unfolding transition for NBD1 did not bias the outcome or conclusions. An external pressure of 2.0 atm was maintained during all DSC runs to prevent possible degassing of the solutions upon heating. Unless otherwise indicated, the buffer for NBD1 DSC experiments was 20 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM TCEP, 10% glycerol and 10% ethylene glycol, 20 μM ATP, 3 mM MgCl2 with various amount of detergents added before experiments. Stock protein solution in 2 mM ATP was buffer-exchanged three times into the DSC buffer using the Amicon ultrafiltration devices with a MWCO of 10 kDa. The buffer for lysozyme DSC experiments was 50 mM sodium acetate, pH 3.9. Lysozyme was first dissolved in this buffer, and then buffer-exchanged two times into the same buffer using an Amicon ultrafiltration device. Lysozyme concentration was determined by UV absorbance at 280 nm with an extinction coefficient of 2.64 (mg/mL)−1cm−1.
DSC data were analyzed with the MicroCal Origin 7.0 software, from which the unfolding temperature (Tmax), and the calorimetric (ΔHc) and van't Hoff (ΔHv) unfolding enthalpies were obtained. A detergent-free control was included in each set of experiments conducted on the same day. The average Tmax and ΔHc of the controls was 49.0 ± 0.4°C and 91 ± 4 kcal/mol, respectively, from total of 44 DSC runs. The shift in Tmax (ΔTmax) was calculated based on the difference between the curves in the presence of detergents and the detergent-free control on the same day. For data points with duplicate experiments conducted on different days, the ΔTmax was averaged. There was less variation in ΔTmax than Tmax because Tmax is highly sensitive to the ATP concentration which varied slightly in each different preparation of the DSC samples.
Circular dichroism (CD) and static light scattering (SLS)
CD and SLS measurements were conducted using a J-815 spectropolarimeter (Jasco, Easton, MD) equipped with a PFD-425 Peltier temperature-controlled cell, an FMO-427 fluorescence detector. The monochromator of the FMO-427 detector was set to 230 nm for SLS with sensitivity of 850 Volts. SLS were acquired simultaneously with CD data, which were collected at 230 nm.
Prism 5 (GraphPad, San Diego, CA) was used for plotting and least-squares curve fitting. Background subtraction used the standard buffer in the absence of detergent for CD and 90° SLS measurements. An additional linear normalization factor was applied to produce equivalent CD signals at detergent-free condition, to correct for the dilution caused by addition of detergents.
Detergent removal
NBD1 (0.5 mg/mL) was incubated with detergents at 4°C for 2, 4, or 20 hrs. Detergents were removed using the Pierce detergent removal spin column (Thermo Fisher Scientific Inc.) per manufacture's instruction. The recovered protein samples were immediately subjected to DSC. Protein concentration after detergent removal was determined by the Pierce 660 nm assay. Protein-free detergent control samples were tested in parallel to ensure the absence of interference by possible residual detergent. The total native protein recovery rate is the product of the % protein concentration recovery and the % molar enthalpy recovery (see Table II for detail).
Full-length CFTR extraction and IMAC recovery
Full-length CFTR containing a C-terminal GFP fusion was expressed in HEK293 cells under doxycycline-inducible transcriptional control elements, including the reverse tet-transactivator (M2) and the TRE promoter (Tet-on system) as described.96 CFTR extraction and purification was carried out as described.97 Briefly, Microsomal membranes (2 mg/mL) prepared from these cells were incubated with 0.5% detergent for 30 min on ice, then insoluble material pelleted at 100,000g. Extracts were diluted fivefold and incubated 16 hrs with NiNTA. Resin was washed with like detergent and eluted with 0.35M imidazole. CFTR was quantitated in detergent extracts and NiNTA eluates by in-gel GFP fluorescence and densitometry, in comparison to a Sumo-GFP standard (LifeSensors).102 100% is the amount of CFTR found in microsomes dissolved in Laemmli sample buffer.
Photoaffinity labeling of full-length CFTR
Photoaffinity labeling of full-length CFTR with 8-azido-[γ-32P]ATP was carried out as previously described.98,99 Membranes (10 µg protein) from BHK-21 cells expressing CFTR were incubated with 25 µM 8-azido-[γ-32P]ATP for 5 min, and then exposed to different detergents at the concentrations and times as indicted in the figure legend. Following the detergent incubation, the suspension was irradiated at 254 nm in a Stratalinker UV cross-linker for 2 min. CFTR was then immunoprecipitated and subjected to SDS/PAGE and autoradiography. 32P radioactivity associated with the CFTR band was determined by electronic autoradiography (Packard Instant Imager).
Acknowledgments
The authors thank members of the Cystic Fibrosis Foundation Therapeutics CFTR 3D Structure Consortium for insightful discussions.
Glossary
- CD
circular dichroism
- CFTR
cystic fibrosis transmembrane conductance regulator
- DSC
differential scanning calorimetry
- ESD
extramembranous soluble domain
- IMAC
immobilized metal-affinity chromatography
- IMP
integral membrane protein
- NBD1
first nucleotide binding domain
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- POPE
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine
- SLS
static light scattering
- TM
transmembrane. Detergent abbreviations (i.e. short names) are listed in Table I.
Footnotes
The terms extramembrane domain and extramembranous soluble domain are used interchangeably.
For a true two-state unfolding equilibrium, ΔHv is equal to ΔHc. In the case of NBD1 unfolding in the presence of ATP, ΔHc is mainly comprised of the enthalpy of unfolding and the enthalpy of ATP dissociation; the apparent ΔHv is larger than ΔHc because the DSC peak is artificially sharpened by irreversibility.50
A small positive heat capacity change (ΔCp) usually accompanies protein unfolding, The ΔCp of NBD1 unfolding has been previously determined to be 1.4 kcal/mol/°C.50 On the basis of the relationship δ(ΔH) = (ΔCp)δ(ΔT) alone, the ΔHc is expected to decrease 5.6 kcal/mol with a 4°C decrease in Tmax. The actual decrease in ΔHc seen with DDM was much greater than that.
Reduced χ2 = sum of squared errors/(# of data points − # of fitting parameters).
Curve fitting on the LPC16 dataset by all three models resulted in large χ2 values indicating no convergence. Therefore, the best-fit model for LPC16 was not determined.
Supporting Information
Additional Supporting Information may be found in the online version of this article.
Supplementary Information
References
- 1.Wiener MC. A pedestrian guide to membrane protein crystallization. Methods. 2004;34:364–372. doi: 10.1016/j.ymeth.2004.03.025. [DOI] [PubMed] [Google Scholar]
- 2.Seddon AM, Curnow P, Booth PJ. Membrane proteins, lipids and detergents: not just a soap opera. Biochim Biophys Acta. 2004;1666:105–117. doi: 10.1016/j.bbamem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 3.Garavito RM, Ferguson-Miller S. Detergents as tools in membrane biochemistry. J Biol Chem. 2001;276:32403–32406. doi: 10.1074/jbc.R100031200. [DOI] [PubMed] [Google Scholar]
- 4.Prive GG. Detergents for the stabilization and crystallization of membrane proteins. Methods. 2007;41:388–397. doi: 10.1016/j.ymeth.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 5.Linke D. Detergents: an overview. Methods Enzymol. 2009;463:63034–63042. doi: 10.1016/S0076-6879(09)63034-2. [DOI] [PubMed] [Google Scholar]
- 6.Tate CG. Practical considerations of membrane protein instability during purification and crystallisation. Methods Mol Biol. 2010;601:187–203. doi: 10.1007/978-1-60761-344-2_12. [DOI] [PubMed] [Google Scholar]
- 7.Lund S, Orlowski S, de Foresta B, Champeil P, le Maire M, Møller JV. Detergent structure and associated lipids determinants in the stabilization of solubilized Ca2+-ATPase from sarcoplasmic reticulum. J Biol Chem. 1989;264:4907–4915. [PubMed] [Google Scholar]
- 8.Le Maire M. Interaction of membrane proteins and lipids with solubilizing detergents. Biochim Biophys Acta. 2000;1508:86–111. doi: 10.1016/s0304-4157(00)00010-1. [DOI] [PubMed] [Google Scholar]
- 9.Therien AG, Deber CM. Interhelical packing in detergent micelles. Folding of a cystic fibrosis transmembrane conductance regulator construct. J Biol Chem. 2002;277:6067–6072. doi: 10.1074/jbc.M110264200. [DOI] [PubMed] [Google Scholar]
- 10.Fernández C, Hilty C, Wider G, Wüthrich K. Lipid-protein interactions in DHPC micelles containing the integral membrane protein OmpX investigated by NMR spectroscopy. Proc Natl Acad Sci USA. 2002;99:13533–13537. doi: 10.1073/pnas.212515099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fisher LE, Engelman DM, Sturgis JN. Effect of detergents on the association of the Glycophorin A transmembrane helix. Biophys J. 2003;85:3097–3105. doi: 10.1016/S0006-3495(03)74728-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kiefer H. In vitro folding of alpha-helical membrane proteins. Biochim Biophys Acta. 2003;1610:57–62. doi: 10.1016/s0005-2736(02)00717-4. [DOI] [PubMed] [Google Scholar]
- 13.Borths EL, Poolman B, Hvorup RN, Locher KP, Rees DC. In vitro functional characterization of BtuCD-F, the Escherichia coli ABC transporter for vitamin B12 uptake. Biochemistry. 2005;44:16301–16309. doi: 10.1021/bi0513103. [DOI] [PubMed] [Google Scholar]
- 14.Poget SF, Girvin ME. Solution NMR of membrane proteins in bilayer mimics: small is beautiful, but sometimes bigger is better. Biochim Biophys Acta. 2007;1768:3098–3106. doi: 10.1016/j.bbamem.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sonoda Y, Newstead S, Hu NJ, Alguel Y, Nji E, Beis K, Yashiro S, Lee C, Leung J, Cameron AD, Byrne B, Iwata S, Drew D. Benchmarking membrane protein detergent stability for improving throughput of high-resolution X-ray structures. Structure. 2011;19:17–25. doi: 10.1016/j.str.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cross TA, Mukesh S, Myunggi Y, Zhou HX. Influence of solubilizing environments on membrane protein structures. Trends Biochem Sci. 2011;36:117–125. doi: 10.1016/j.tibs.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lai JY, Poon YS, Kaiser JT, Rees DC. Open and shut: crystal structures of the dodecylmaltoside solubilized mechanosensitive channel of small conductance from Escherichia coli and Helicobacter pylori at 4.4 Å and 4.1 Å resolutions. Protein Sci. 2013;22:502–509. doi: 10.1002/pro.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popot JL. Amphipols, nanodiscs, and fluorinated surfactants: three nonconventional approaches to studying membrane proteins in aqueous solutions. Annu Rev Biochem. 2010;79:737–775. doi: 10.1146/annurev.biochem.052208.114057. [DOI] [PubMed] [Google Scholar]
- 19.Luecke H, Schobert B, Richter HT, Cartailler JP, Lanyi JK. Structure of bacteriorhodopsin at 1.55 Å resolution. J Mol Biol. 1999;291:899–911. doi: 10.1006/jmbi.1999.3027. [DOI] [PubMed] [Google Scholar]
- 20.Qin L, Hiser C, Mulichak A, Garavito RM, Ferguson-Miller S. Identification of conserved lipid/detergent-binding sites in a high-resolution structure of the membrane protein cytochrome c oxidase. Proc Natl Acad Sci USA. 2006;103:16117–16122. doi: 10.1073/pnas.0606149103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cogdell RJ, Gardiner AT, Roszak AW, Stončius S, Kočovský P, Isaacs NW. Mapping lipid and detergent molecules at the surface of membrane proteins. Biochem Soc Trans. 2011;39:775–779. doi: 10.1042/BST0390775. [DOI] [PubMed] [Google Scholar]
- 22.Reynolds JA, Tanford C. The gross conformation of protein-sodium dodecyl sulfate complexes. J Biol Chem. 1970;245:5161–5165. [PubMed] [Google Scholar]
- 23.Nozaki Y, Reynolds JA, Tanford C. The interaction of a cationic detergent with bovine serum albumin and other proteins. J Biol Chem. 1974;249:4452–4459. [PubMed] [Google Scholar]
- 24.Jones MN. Surfactant interactions with biomembranes and proteins. Chem Soc Rev. 1992;21:127–136. [Google Scholar]
- 25.Otzen DE. Protein unfolding in detergents: effect of micelle structure, ionic strength, pH, and temperature. Biophys J. 2002;83:2219–2230. doi: 10.1016/S0006-3495(02)73982-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vergis JM, Wiener MC. The variable detergent sensitivity of proteases that are utilized for recombinant protein affinity tag removal. Protein Expr Purif. 2011;78:139–142. doi: 10.1016/j.pep.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otzen D. Protein–surfactant interactions: a tale of many states. Biochim Biophys Acta. 2011;1814:562–591. doi: 10.1016/j.bbapap.2011.03.003. [DOI] [PubMed] [Google Scholar]
- 28.Lundahl P, Mascher E, Kameyama K, Takagi T. Water-soluble proteins do not bind octyl glucoside as judged by molecular sieve chromatographic techniques. J Chromatogr. 1990;518:111–121. doi: 10.1016/s0021-9673(01)93166-8. [DOI] [PubMed] [Google Scholar]
- 29.Kragh-Hansen U, Hellec F, de Foresta B, le Maire M, Møller JV. Detergents as probes of hydrophobic binding cavities in serum albumin and other water-soluble proteins. Biophys J. 2001;80:2898–2911. doi: 10.1016/S0006-3495(01)76255-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Otzen DE, Sehgal P, Westh P. Alpha-lactalbumin is unfolded by all classes of surfactants but by different mechanisms. J Colloid Interface Sci. 2009;329:273–283. doi: 10.1016/j.jcis.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 31.Nath D, Rao M. Artificial chaperone mediated refolding of xylanase from an alkalophilic thermophilic Bacillus sp. Implications for in vitro protein renaturation via a folding intermediate. Eur J Biochem. 2001;268:5471–5478. doi: 10.1046/j.0014-2956.2001.02485.x. [DOI] [PubMed] [Google Scholar]
- 32.Nielsen AD, Arleth L, Westh P. Analysis of protein–surfactant interactions—a titration calorimetric and fluorescence spectroscopic investigation of interactions between Humicola insolens cutinase and an anionic surfactant. Biochim Biophys Acta. 2005;1752:124–132. doi: 10.1016/j.bbapap.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 33.Mogensen JE, Sehgal P, Otzen DE. Activation, inhibition, and destabilization of Thermomyces lanuginosus lipase by detergents. Biochemistry. 2005;44:1719–1730. doi: 10.1021/bi0479757. [DOI] [PubMed] [Google Scholar]
- 34.Sehgal P, Bang Nielsen S, Pedersen S, Wimmer R, Otzen DE. Modulation of cutinase stability and structure by phospholipid detergents. Biochim Biophys Acta. 2007;1774:1544–1554. doi: 10.1016/j.bbapap.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 35.Di Bartolo ND, Hvorup RN, Locher KP, Booth PJ. In vitro folding and assembly of the Escherichia coli ATP-binding cassette transporter, BtuCD. J Biol Chem. 2011;86:18807–18815. doi: 10.1074/jbc.M110.176891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Di Bartolo N, Booth PJ. Unravelling the folding and stability of an ABC (ATP-binding cassette) transporter. Biochem Soc Trans. 2011;39:751–760. doi: 10.1042/BST0390751. [DOI] [PubMed] [Google Scholar]
- 37.Cheng SH, Gregory RJ, Marshall J, Paul S, Souza DW, White GA, O'Riordan CR, Smith AE. Defective intracellular transport and processing of CFTR is the molecular basis of most cystic fibrosis. Cell. 1990;63:827–834. doi: 10.1016/0092-8674(90)90148-8. [DOI] [PubMed] [Google Scholar]
- 38.Dalemans W, Barbry P, Champigny G, Jallat S, Dott K, Dreyer D, Crystal RG, Pavirani A, Lecocq JP, Lazdunski M. Altered chloride ion channel kinetics associated with the delta F508 cystic fibrosis mutation. Nature. 1991;354:526–528. doi: 10.1038/354526a0. [DOI] [PubMed] [Google Scholar]
- 39.Lukacs GL, Chang XB, Bear C, Kartner N, Mohamed A, Riordan JR, Grinstein S. The delta F508 mutation decreases the stability of cystic fibrosis transmembrane conductance regulator in the plasma membrane. Determination of functional half-lives on transfected cells. J Biol Chem. 1993;268:21592–21598. [PubMed] [Google Scholar]
- 40.Qu BH, Strickland EH, Thomas PJ. Localization and suppression of a kinetic defect in cystic fibrosis transmembrane conductance regulator folding. J Biol Chem. 1997;272:15739–15744. doi: 10.1074/jbc.272.25.15739. [DOI] [PubMed] [Google Scholar]
- 41.Skach WR. Defects in processing and trafficking of the cystic fibrosis transmembrane conductance regulator. Kidney Int. 2000;57:825–831. doi: 10.1046/j.1523-1755.2000.00921.x. [DOI] [PubMed] [Google Scholar]
- 42.Riordan JR. Assembly of functional CFTR chloride channels. Annu Rev Physiol. 2005;67:701–718. doi: 10.1146/annurev.physiol.67.032003.154107. [DOI] [PubMed] [Google Scholar]
- 43.Du K, Sharma M, Lukacs GL. The DeltaF508 cystic fibrosis mutation impairs domain-domain interactions and arrests post-translational folding of CFTR. Nat Struct Mol Biol. 2005;12:17–25. doi: 10.1038/nsmb882. [DOI] [PubMed] [Google Scholar]
- 44.Cheung JC, Deber CM. Misfolding of the cystic fibrosis transmembrane conductance regulator and disease. Biochemistry. 2008;47:1465–1473. doi: 10.1021/bi702209s. [DOI] [PubMed] [Google Scholar]
- 45.Lewis HA, Buchanan SG, Burley SK, Conners K, Dickey M, Dorwart M, Fowler R, Gao X, Guggino WB, Hendrickson WA, Hunt JF, Kearins MC, Lorimer D, Maloney PC, Post KW, Rajashankar KR, Rutter ME, Sauder JM, Shriver S, Thibodeau PH, Thomas PJ, Zhang M, Zhao X, Emtage S. Structure of nucleotide-binding domain 1 of the cystic fibrosis transmembrane conductance regulator. EMBO J. 2004;23:282–293. doi: 10.1038/sj.emboj.7600040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thibodeau PH, Brautigam CA, Machius M, Thomas PJ. Side chain and backbone contributions of Phe508 to CFTR folding. Nat Struct Mol Biol. 2005;12:10–16. doi: 10.1038/nsmb881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lewis HA, Zhao X, Wang C, Sauder JM, Rooney I, Noland BW, Lorimer D, Kearins MC, Conners K, Condon B, Maloney PC, Guggino WB, Hunt JF, Emtage S. Impact of the deltaF508 mutation in first nucleotide-binding domain of cystic fibrosis transmembrane conductance regulator on domain folding and structure. J Biol Chem. 2005;280:1346–1353. doi: 10.1074/jbc.M410968200. [DOI] [PubMed] [Google Scholar]
- 48.Atwell S, Brouillette CG, Conners K, Emtage S, Gheyi T, Guggino WB, Hendle J, Hunt JF, Lewis HA, Lu F, Protasevich II, Rodgers LA, Romero R, Wasserman SR, Weber PC, Wetmore D, Zhang FF, Zhao X. Structures of a minimal CFTR first nucleotide-binding domain as a monomer, head-to-tail homodimer, and pathogenic mutant. Protein Eng Des Sel. 2010;23:375–384. doi: 10.1093/protein/gzq004. [DOI] [PubMed] [Google Scholar]
- 49.Lewis HA, Wang C, Zhao X, Hamuro Y, Conners K, Kearins MC, Lu F, Sauder JM, Molnar KS, Coales SJ, Maloney PC, Guggino WB, Wetmore DR, Weber PC, Hunt JF. Structure and dynamics of NBD1 from CFTR characterized using crystallography and hydrogen/deuterium exchange mass spectrometry. J Mol Biol. 2010;396:406–430. doi: 10.1016/j.jmb.2009.11.051. [DOI] [PubMed] [Google Scholar]
- 50.Protasevich I, Yang Z, Wang C, Atwell S, Zhao X, Emtage S, Wetmore D, Hunt JF, Brouillette CG. Thermal unfolding studies show the disease causing F508del mutation in CFTR thermodynamically destabilizes nucleotide-binding domain 1. Protein Sci. 2010;19:1917–1931. doi: 10.1002/pro.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang C, Protasevich I, Yang Z, Seehausen D, Skalak T, Zhao X, Atwell S, Spencer Emtage J, Wetmore DR, Brouillette CG, Hunt JF. Integrated biophysical studies implicate partial unfolding of NBD1 of CFTR in the molecular pathogenesis of F508del cystic fibrosis. Protein Sci. 2010;19:1932–1947. doi: 10.1002/pro.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Teem JL, Berger HA, Ostedgaard LS, Rich DP, Tsui LC, Welsh MJ. Identification of revertants for the cystic fibrosis delta F508 mutation using STE6-CFTR chimeras in yeast. Cell. 1993;73:335–346. doi: 10.1016/0092-8674(93)90233-g. [DOI] [PubMed] [Google Scholar]
- 53.DeCarvalho AC, Gansheroff LJ, Teem JL. Mutations in the nucleotide binding domain 1 signature motif region rescue processing and functional defects of cystic fibrosis transmembrane conductance regulator delta F508. J Biol Chem. 2002;277:35896–35905. doi: 10.1074/jbc.M205644200. [DOI] [PubMed] [Google Scholar]
- 54.Loo TW, Bartlett MC, Clarke DM. The V510D suppressor mutation stabilizes DeltaF508-CFTR at the cell surface. Biochemistry. 2010;49:6352–6357. doi: 10.1021/bi100807h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thibodeau PH, Richardson JM, III, Wang W, Millen L, Watson J, Mendoza JL, Du K, Fischman S, Senderowitz H, Lukacs GL, Kirk K, Thomas PJ. The cystic fibrosis-causing mutation deltaF508 affects multiple steps in cystic fibrosis transmembrane conductance regulator biogenesis. J Biol Chem. 2010;285:35825–35835. doi: 10.1074/jbc.M110.131623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.He L, Aleksandrov LA, Cui L, Jensen TJ, Nesbitt KL, Riordan JR. Restoration of domain folding and interdomain assembly by second-site suppressors of the DeltaF508 mutation in CFTR. FASEB J. 2010;24:3103–3112. doi: 10.1096/fj.09-141788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Aleksandrov A, Kota P, Aleksandrov L, He L, Jensen T, Cui L, Gentzsch M, Dokholyan N, Riordan J. Regulatory insertion removal restores maturation, stability and function of ΔF508 CFTR. J Mol Biol. 2010;401:194–210. doi: 10.1016/j.jmb.2010.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Aleksandrov AA, Kota P, Cui L, Jensen T, Alekseev AE, Reyes S, He L, Gentzsch M, Aleksandrov LA, Dokholyan NV, Riordan JR. Allosteric modulation balances thermodynamic stability and restores function of ΔF508 CFTR. J Mol Biol. 2012;419:41–60. doi: 10.1016/j.jmb.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mendoza JL, Schmidt A, Li Q, Nuvaga E, Barrett T, Bridges RJ, Feranchak AP, Brautigam CA, Thomas PJ. Requirements for efficient correction of ΔF508 CFTR revealed by analyses of evolved sequences. Cell. 2012;148:164–174. doi: 10.1016/j.cell.2011.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong Q, Ostedgaard LS, Rogers C, Vermeer DW, Zhang Y, Welsh MJ. Mouse cystic fibrosis transmembrane conductance regulator (CFTR) chimeras identify regions that partially rescue CFTR-ΔF508 processing and alter its gating defect. Proc Natl Acad Sci USA. 2012;109:917–922. doi: 10.1073/pnas.1120065109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nam H-J, Han SK, Bowie JU, Kim S. Rampant exchange of the structure and function of extramembrane domains between membrane and water soluble proteins. PLoS Comput Biol. 2013;9:e1002997. doi: 10.1371/journal.pcbi.1002997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanelis V, Hudson RP, Thibodeau PH, Thomas PJ, Forman-Kay JD. NMR evidence for differential phosphorylation-dependent interactions in WT and DeltaF508 CFTR. EMBO J. 2010;29:263–277. doi: 10.1038/emboj.2009.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmidt A, Mendoza JL, Thomas PJ. Biochemical and biophysical approaches to probe CFTR structure. Methods Mol Biol. 2011;741:365–376. doi: 10.1007/978-1-61779-117-8_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hudson RP, Chong PA, Protasevich II, Vernon R, Noy E, Bihler H, An JL, Kalid O, Sela-Culang I, Mense M, Senderowitz H, Brouillette CG, Forman-Kay JD. Conformational changes relevant to channel activity and folding within the first nucleotide binding domain of the cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2012;287:28480–28494. doi: 10.1074/jbc.M112.371138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tulumello DV, Deber CM. Effciency of detergents at maintaining membrane protein structures in their biologically relevant forms. Biochim Biophys Acta. 2012;1818:1351–1358. doi: 10.1016/j.bbamem.2012.01.013. [DOI] [PubMed] [Google Scholar]
- 66.Brandts JF, Lin LN. Study of strong to ultratight protein interactions using differential scanning calorimetry. Biochemistry. 1990;29:6927–6940. doi: 10.1021/bi00481a024. [DOI] [PubMed] [Google Scholar]
- 67.Cooper A, Nutley MA, Wadood A. Differential scanning microcalorimetry. In: Harding SE, Chowdhry BZ, editors. Protein-ligand interactions: hydrodynamics and calorimetry. Oxford, New York: Oxford University Press; 2000. pp. 287–318. [Google Scholar]
- 68.Waldron TT, Murphy KP. Stabilization of proteins by ligand binding: application to drug screening and determination of unfolding energetics. Biochemistry. 2003;42:5058–5064. doi: 10.1021/bi034212v. [DOI] [PubMed] [Google Scholar]
- 69.Hu CQ, Sturtevant JM. Thermodynamic study of yeast phosphoglycerate kinase. Biochemistry. 1987;26:178–182. doi: 10.1021/bi00375a025. [DOI] [PubMed] [Google Scholar]
- 70.Sturtevant JM. Biochemical applications of differential scanning calorimetry. Ann Rev Phys Chem. 1987;38:463–488. [Google Scholar]
- 71.Edge V, Allewell NM, Sturtevant JM. Differential scanning calorimetric study of the thermal denaturation of aspartate transcarbamoylase of Escherichia coli. Biochemistry. 1988;27:8081–8087. doi: 10.1021/bi00421a017. [DOI] [PubMed] [Google Scholar]
- 72.Sonoda Y, Cameron A, Newstead S, Omote H, Moriyama Y, Kasahara M, Iwata S, Drew D. Tricks of the trade used to accelerate high-resolution structure determination of membrane proteins. FEBS Lett. 2010;584:2539–2547. doi: 10.1016/j.febslet.2010.04.015. [DOI] [PubMed] [Google Scholar]
- 73.Newstead S, Ferrandon S, Iwata S. Rationalizing alpha-helical membrane protein crystallization. Protein Sci. 2008;17:466–472. doi: 10.1110/ps.073263108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chae PS, Rasmussen SG, Rana RR, Gotfryd K, Chandra R, Goren MA, Kruse AC, Nurva S, Loland CJ, Pierre Y, Drew D, Popot JL, Picot D, Fox BG, Guan L, Gether U, Byrne B, Kobilka B, Gellman SH. Maltose-neopentyl glycol (MNG) amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nat Methods. 2010;7:1003–1008. doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee SC, Bennett BC, Hong WX, Fu Y, Baker KA, Marcoux J, Robinson CV, Ward AB, Halpert JR, Stevens RC, Stout CD, Yeager MJ, Zhang Q. Steroid-based facial amphiphiles for stabilization and crystallization of membrane proteins. Proc Natl Acad Sci USA. 2013;110:E1203–E1211. doi: 10.1073/pnas.1221442110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ghobadi S, Safarian S, Moosavi-Movahedi AA, Ranjbar B. Octyl glucoside induced formation of the molten globule-like state of glutamate dehydrogenase. J Biochem. 2001;130:671–677. doi: 10.1093/oxfordjournals.jbchem.a003033. [DOI] [PubMed] [Google Scholar]
- 77.Ramjeesingh M, Li C, Garami E, Huan LJ, Hewryk M, Wang Y, Galley K, Bear CE. A novel procedure for the efficient purification of the cystic fibrosis transmembrane conductance regulator (CFTR) Biochem J. 1997;327:17–21. doi: 10.1042/bj3270017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang P, Liu Q, Scarborough GA. Lysophosphatidylglycerol: a novel effective detergent for solubilizing and purifying the cystic fibrosis transmembrane conductance regulator. Anal Biochem. 1998;259:89–97. doi: 10.1006/abio.1998.2633. [DOI] [PubMed] [Google Scholar]
- 79.Ketchum CJ, Rajendrakumar GV, Maloney PC. Characterization of the adenosinetriphosphatase and transport activities of purified cystic fibrosis transmembrane conductance regulator. Biochemistry. 2004;43:1045–1053. doi: 10.1021/bi035382a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ramjeesingh M, Ugwu F, Stratford FL, Huan LJ, Li C, Bear CE. The intact CFTR protein mediates ATPase rather than adenylate kinase activity. Biochem J. 2008;412:315–321. doi: 10.1042/BJ20071719. [DOI] [PubMed] [Google Scholar]
- 81.Giehm L, Oliveira CL, Christiansen G, Pedersen JS, Otzen DE. SDS-induced fibrillation of alpha-synuclein: an alternative fibrillation pathway. J Mol Biol. 2010;401:115–133. doi: 10.1016/j.jmb.2010.05.060. [DOI] [PubMed] [Google Scholar]
- 82.Nielsen MM, Andersen KK, Westh P, Otzen DE. Unfolding of beta-sheet proteins in SDS. Biophys J. 2007;92:3674–3685. doi: 10.1529/biophysj.106.101238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Koehler J, Sulistijo ES, Sakakura M, Kim HJ, Ellis CD, Sanders CR. Lysophospholipid micelles sustain the stability and catalytic activity of diacylglycerol kinase in the absence of lipids. Biochemistry. 2010;49:7089–7099. doi: 10.1021/bi100575s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Krueger-Koplin RD, Sorgen PL, Krueger-Koplin ST, Rivera-Torres IO, Cahill SM, Hicks DB, Grinius L, Krulwich TA, Girvin ME. An evaluation of detergents for NMR structural studies of membrane proteins. J Biomol NMR. 2004;17:43–57. doi: 10.1023/B:JNMR.0000012875.80898.8f. [DOI] [PubMed] [Google Scholar]
- 85.Becktel WJ, Schellman JA. Protein stability curves. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- 86.Greene RF, Jr, Pace CN. Urea and guanidine hydrochloride denaturation of ribonuclease, lysozyme, alpha-chymotrypsin, and beta-lactoglobulin. J Biol Chem. 1974;249:5388–5393. [PubMed] [Google Scholar]
- 87.Schellman JA. Fifty years of solvent denaturation. Biophys Chem. 2002;96:91–101. doi: 10.1016/s0301-4622(02)00009-1. [DOI] [PubMed] [Google Scholar]
- 88.Makhatadze GI. Thermodynamics of protein interactions with urea and guanidinium hydrochloride. J Phys Chem. 1999;103:4781–4785. [Google Scholar]
- 89.Timasheff SN, Xie G. Preferential interactions of urea with lysozyme and their linkage to protein denaturation. Biophys Chem. 2003;105:421–448. doi: 10.1016/s0301-4622(03)00106-6. [DOI] [PubMed] [Google Scholar]
- 90.Schellman JA. Protein stability in mixed solvents: a balance of contact interaction and excluded volume. Biophys J. 2003;85:108–125. doi: 10.1016/S0006-3495(03)74459-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Privalov PL. Stability of proteins: small globular proteins. Adv Prot Chem. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- 92.Qin L, Sharpe MA, Garavito RM, Ferguson-Miller S. Conserved lipid-binding sites in membrane proteins: a focus on cytochrome c oxidase. Curr Opin Struct Biol. 2007;17:444–450. doi: 10.1016/j.sbi.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Callaghan R, Berridge G, Ferry DR, Higgins CF. The functional purification of P-glycoprotein is dependent on maintenance of a lipid-protein interface. Biochim Biophys Acta. 1997;1328:109–124. doi: 10.1016/s0005-2736(97)00079-5. [DOI] [PubMed] [Google Scholar]
- 94.Dong M, Penin F, Baggetto LG. Efficient purification and reconstitution of P-glycoprotein for functional and structural studies. J Biol Chem. 1996;271:28875–28883. doi: 10.1074/jbc.271.46.28875. [DOI] [PubMed] [Google Scholar]
- 95.Dong M, Baggetto LG, Falson P, Le Maire M, Penin F. Complete removal and exchange of sodium dodecyl sulfate bound to soluble and membrane proteins and restoration of their activities, using ceramic hydroxyapatite chromatography. Anal Biochem. 1997;247:333–341. doi: 10.1006/abio.1997.2103. [DOI] [PubMed] [Google Scholar]
- 96.DeLucas LJ, Dai Q, Ray M, Brackin W, McCombs D, Johnson D, Ding H, Hildebrandt E, Purna B, Riordan JR, Urbatsch IL, Kappes JC. Progress toward elucidating the 3D structure of full-length CFTR. Pediatric Pulmonology. 2012;47:248. [Google Scholar]
- 97.Hildebrandt EI, Kappes JC, DeLucas LJ, Cant N, Ford R, Zhang Q, Urbatsch IL. Evaluation of detergents for CFTR purification, concentration, and reconstitution of ATPase activity. Pediatric Pulmonol. 2013;48:231. [Google Scholar]
- 98.Aleksandrov L, Aleksandrov AA, Chang XB, Riordan JR. The first nucleotide binding domain of cystic fibrosis transmembrane conductance regulator is a site of stable nucleotide interaction, whereas the second is a site of rapid turnover. J Biol Chem. 2002;277:15419–15425. doi: 10.1074/jbc.M111713200. [DOI] [PubMed] [Google Scholar]
- 99.Aleksandrov L, Aleksandrov A, Riordan JR. Mg2+-dependent ATP occlusion at the first nucleotide-binding domain (NBD1) of CFTR does not require the second (NBD2) Biochem J. 2008;416:129–136. doi: 10.1042/BJ20081068. [DOI] [PubMed] [Google Scholar]
- 100.Cant N, Rimington T, Meng X, Pollock N, Ford B. Biophysical measures of full-length CFTR thermostability: comparison of wild-type and F508del CFTR. Pediatric Pulmonol. 2013;48:209–210. [Google Scholar]
- 101.Jumpertz T, Tschapek B, Infed N, Smits SH, Ernst R, Schmitt L. High-throughput evaluation of the critical micelle concentration of detergents. Anal Biochem. 2011;408:64–70. doi: 10.1016/j.ab.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 102.Drew D, Lerch M, Kunji E, Slotboom DJ, de Gier JW. Optimization of membrane protein overexpression and purification using GFP fusions. Nat Methods. 2006;3:303–313. doi: 10.1038/nmeth0406-303. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information


