Abstract
Cluster of differentiation 4+ (CD4+) effector T-cell subsets [e.g., T-helper (Th) 1 and Th17] are implicated in autoimmune and inflammatory disorders such as multiple sclerosis, psoriasis, and rheumatoid arthritis. Interleukin (IL)-6 is a pleiotropic cytokine that induces Th17 polarization via signaling through the membrane-bound transducer glycoprotein 130 (GP130). Previously, we demonstrated that n–3 (ω-3) polyunsaturated fatty acids (PUFAs) reduce CD4+ T-cell activation and differentiation into pathogenic Th17 cells by 25–30%. Here we report that n–3 PUFAs alter the response of CD4+ T cells to IL-6 in a lipid raft membrane–dependent manner. Naive splenic CD4+ T cells from fat-1 transgenic mice exhibited 30% lower surface expression of the IL-6 receptor. This membrane-bound receptor is known to be shed during cellular activation, but the release of soluble IL-6 receptor after treatment with anti-CD3 and anti-CD28 was not changed in the CD4+ T cells from fat-1 mice, suggesting that the decrease in surface expression was not due to ectodomain release. We observed a significant 20% decrease in the association of GP130 with lipid rafts in activated fat-1 CD4+ T cells and a 35% reduction in GP130 homodimerization, an obligate requirement for downstream signaling. The phosphorylation of signal transducer and activator of transcription 3 (STAT3), a downstream target of IL-6–dependent signaling, was also decreased by 30% in response to exogenous IL-6 in fat-1 CD4+ T cells. Our results suggest that n–3 PUFAs suppress Th17 cell differentiation in part by reducing membrane raft–dependent responsiveness to IL-6, an essential polarizing cytokine.
Introduction
Many chronic diseases are linked to inflammation, including arthritis, multiple sclerosis (1), obesity, diabetes (2), and cancer (3). Cluster of differentiation 4+ (CD4+)10 T-helper (Th) 17 cells play an important role in these inflammatory diseases (4). Specifically, chronic Th17-mediated inflammation in the colon was linked to the onset of inflammatory bowel disease (IBD) and colitis-associated cancer (5). A chronic pronounced elevation in the IL-6/glycoprotein 130 (GP130)/signal transducer and activator of transcription 3 (STAT3) signaling axis is considered a risk factor for colorectal cancer in part due to stimulation of epithelial cell proliferation (6). Additional investigation suggests that inhibition of IL-6 signaling can reduce tumor development in a colitis-induced cancer model (6) and multiple sclerosis (7).
Th17 cells differentiate from naive CD4+ T cells under the influence of IL-6 and TGF-β (8, 9), which activate STAT3. Phosphorylation of STAT3 leads to translocation to the nucleus and subsequent activation of RAR-related orphan receptor-γt (ROR-γt), the master regulator of Th17 transcription. The IL-23 receptor is upregulated on the cell surface and contributes to the maintenance and expansion of mature Th17 cells (10). Th17 cells function primarily to stimulate a neutrophil response and release IL-17A, IL-17F, IL-21, and IL-22 (11).
The IL-6 receptor is an 80 kDa surface protein expressed on naive CD4+ T cells (12). Two molecules of IL-6 bind to 2 membrane-bound IL-6 receptors that form a complex with 2 molecules of GP130, resulting in a hexameric signaling complex (13, 14). Interestingly, many cells, such as smooth muscle and endothelial cells (7), do not express IL-6 receptor, yet they still exhibit trans-signaling in which soluble IL-6 receptor shed from contiguous cells binds to IL-6 and membrane-bound GP130. Activation of T lymphocytes leads to downregulation of IL-6 receptor on the cell surface and an increase in soluble IL-6 receptor (13, 14). Elevated concentrations of soluble IL-6 receptor seen during periods of inflammation are involved in the pathogenesis of rheumatoid arthritis, Crohn’s disease, and colon cancer (14–16).
GP130 is ubiquitously expressed and mediates signal transduction for IL-6 and other cytokines (15). GP130 can be membrane bound or soluble. Although membrane-bound GP130 can activate Janus kinase (JAK) 1, JAK2, and tyrosine kinase 2, soluble GP130 acts as an inhibitor of IL-6 trans-signaling by binding to IL-6/soluble IL-6 receptor without performing subsequent signal transduction (17). In CD4+ T cells, a lack of GP130 leads to a reduced Th17 response and an increased regulatory T-cell response, resulting in protection against experimental autoimmune encephalomyelitis (EAE) (15). The presence of soluble GP130 in the serum also inhibits Th17 development (13), suggesting that GP130 is an important mediator of Th17 differentiation.
n–3 PUFAs modulate the immune response through effects on immune cell activation and abundance (18–20). n–3 PUFAs reduce production of IL-2, a major stimulator of CD4+ T cell proliferation (21), and gene expression of interleukin 2 receptor α (IL-2rα) (22). Furthermore, proinflammatory Th1 and Th17 cell in vivo abundance and differentiation ex vivo are reduced (18–20, 23). Finally, inflammatory cytokines, such as IL-17, and transcription factors, including ROR-γt, are reduced by n–3 PUFAs (19). One immunosuppressive mechanism of n–3 PUFAs contributing to this effect may be linked to the IL-6 signaling pathway.
n–3 PUFAs influence the structure of the plasma membrane (24, 25), a highly heterogeneous lipid bilayer containing liquid ordered and liquid disordered regions (26–28). Liquid ordered regions, also known as lipid rafts, are enriched in cholesterol and sphingolipids and are insoluble in cold, nonionic detergents (28). Expression of IL-6 receptor is modulated by the cholesterol composition of the plasma membrane (29), and reducing membrane glycosphingolipids blocks Th17 polarization without affecting other phenotypes (30), suggesting that Th17 polarization is modulated through a lipid raft–related mechanism. It was shown that n–3 PUFAs suppress the colocalization of T-cell receptor signaling proteins into lipid rafts (24), which decreases the robustness of downstream signaling. With respect to IL-6 signaling, previous reports suggest that GP130 is localized to lipid rafts in kidney (31) and neuroepithelial (32) cells, but this was not investigated in CD4+ T cells. Because IL-6 is a critical initiator of Th17 differentiation, changes in the expression of members of the IL-6 signaling pathway can drastically affect Th17 cell abundance.
This study used the fat-1 mouse, which contains the fat-1 gene from Caenorhabditis elegans and is able to convert n–6 to n–3 PUFAs in vivo (33). fat-1 mice display a similar membrane enrichment of n–3 PUFAs as wild-type (WT) mice given a 4% fish-oil diet (34).These models exhibit a similar lipid raft order, with an ∼1.25-fold increase (relative to control) in their membrane generalized polarization as assessed by Laurdan staining (24, 35). Furthermore, Th17 cell abundance was reduced in fat-1 mice to an amount similar to that observed in WT mice given dietary n–3 PUFAs (20), suggesting that the genetic model is able to duplicate the phenotype of a fish-oil diet. Our hypothesis was that n–3 PUFAs reduce Th17 differentiation through disruption of GP130 localization in lipid rafts, thereby impairing downstream signaling. To test this hypothesis, we examined the effects of n–3 PUFAs on membrane localization of GP130, surface and gene expression of IL-6 receptor and GP130, IL-6–induced GP130 dimerization, and STAT3 phosphorylation in CD4+ T cells from fat-1 mice.
Methods
Experimental mice.
Male and female fat-1 and WT breeder mice were obtained from Dr. Jing Kang, Harvard Medical School. Mice were bred at Texas A&M University facilities, genotyped, and phenotyped as described previously (36) and were maintained on a modified AIN-76A diet containing 10% safflower oil (D03092902; Research Diets) adequate in all required nutrients. All procedures and protocols followed guidelines approved by the U.S. Public Health Service and the Institutional Animal Care and Use Committee at Texas A&M University.
CD4+ T-cell isolation and culture.
WT and fat-1 mice were killed by CO2 asphyxiation. Mouse spleens were extracted aseptically, and CD4+ T cells were isolated using a positive selection magnetic column (Miltenyi Biotec) as described previously (37). CD4+ T-cell purity typically exceeded 90% (20, 34). Viability was assessed by trypan blue exclusion, and cell counts were determined using a Coulter counter (Beckman Coulter) before use in the following experiments.
Colocalization of GP130 in lipid rafts by immunofluorescence.
CD4+ T cells were analyzed by immunofluorescence according to Kim et al. (24) with some modifications. For fluorescence experiments, comparison of fixation with paraformaldehyde vs. paraformaldehyde plus glutaraldehyde (Sigma) was performed. Cells were fixed with 4% paraformaldehyde with or without 0.2% glutaraldehyde for 30 min at room temperature. For analysis of WT and fat-1 cells, freshly isolated CD4+ T cells were plated at 5 × 106 cells per well before fixation with 4% paraformaldehyde (Electron Microscopy Sciences) and 2 quenching steps with warm (37°C) 100 mmol/L glycine. The antibodies used included 10 μg/mL rat anti-mouse GP130 (R&D Systems), 10 μg/mL Alexa Fluor 555–conjugated donkey anti-rat IgG, and 6 μg/mL Alexa Fluor–488 conjugated cholera toxin (Life Technologies). Subsequent visualization was performed using a Zeiss 510 laser scanning microscope in confocal mode. Colocalization was quantitatively assessed by Pearson’s correlation (24), and significance (P < 0.05) was determined by 2-sample Student’s t test (n1 = 15 and n2 = 20 observations for fat-1 and WT, respectively).
Basal GP130 protein expression by Western blot.
Basal concentrations of GP130 were assessed as described previously (38). To determine an optimal protein mass to load for the Western blot, 1, 2, 4, and 8 μg of T-cell protein were electrophoresed in a 4–20% Tris–glycine gel (Expedeon), followed by transfer onto a polyvinylidenedifluoride membrane (Immobilon). Antibodies used included 0.1 μg/mL rabbit anti-GP130 (Santa Cruz Biotechnologies) and 0.1 μg/mL peroxidase-labeled goat anti-rabbit IgG (KPL). Chemiluminescence was quantified using Quantity One software (Bio-Rad) on a Fluor-S Max MultiImager (Bio-Rad) as described previously (38). For analysis of WT and fat-1 protein expression, 4 μg of protein lysate was selected and processed.
Quantification of Gp130 and IL-6 receptor mRNA by qPCR.
CD4+ T cells were isolated for RNA extraction using an RNAqueous kit (AM1914; Life Technologies) according to the instructions of the manufacturer, followed by qPCR analysis as described previously (20). RNA quality was measured using a Bioanalyzer with an RNA integrity number cutoff of ≥8.0. Quantification was assessed on a NanoDrop spectrophotometer before reverse transcription was performed, followed by real-time PCR in which a TaqMan predeveloped assay from Life Technologies was used. To determine the optimal RNA mass for reverse transcription, 0.09, 0.9, 9, 90, and 900 ng of RNA were reverse transcribed and analyzed by qPCR, and the optimal concentration was determined by evaluation of the cycle threshold curves.
GP130 and IL-6 receptor surface expression by flow cytometry.
The antibody concentration was determined by selecting the optimal signal-to-noise ratio. Two hundred thousand freshly isolated splenic CD4+ T cells were stained with either anti-GP130 or anti-IL-6 receptor or the appropriate isotype control (BD Pharmingen). For analysis of WT and fat-1 cells, CD4+ T cells were blocked with 1 μg of CD16/32 (eBioscience) and stained with 0.5 μg of anti-GP130-phycoerythrin (clone AM64; No. 555757; BD Pharmingen) or anti-IL-6 receptor-phycoerythrin (clone D7715A7; No. 554462; BD Pharmingen) before analysis on an Accuri flow cytometer.
Detection of soluble IL-6 receptor by ELISA.
Soluble IL-6 receptor was measured by ELISA (R&D Systems). To generate a threshold concentration of soluble IL-6 receptor that could be detected, 1 × 105, 2 × 105, 5 × 105, and 1 × 106 CD4+ T cells were incubated for 48 h in the presence or absence of stimuli before the supernatant was analyzed by ELISA. The incubation period was also optimized in 2 × 105 cells incubated with or without anti-CD3, anti-CD28, and TNF-α proteinase inhibitor (TAPI) for 24, 48, and 72 h. Forty-eight hours was selected for incubation of 2 × 105 cells, and analysis was conducted according to the instructions of the manufacturer.
IL-6–induced GP130 dimerization by Western blot.
Concentrations of GP130 dimer were also assessed as described previously (38). Briefly, 7 × 106 CD4+ T cells were stimulated with 100 μg/L recombinant IL-6 (No. 575704; Biolegend) for 15 min at 37°C, followed by crosslinking with 3 mmol/L bis(sulfosuccinimidyl)suberate (No. 21585; Thermo Fisher Scientific). Samples were homogenized as described previously (38) and electrophoresed at 125 V for 3 h, followed by an overnight transfer onto a polyvinylidenedifluoride membrane at 400 mA.
IL-6–induced STAT3 phosphorylation by ELISA.
Freshly isolated CD4+ T cells were stimulated with 50 μg/L recombinant IL-6 for 15 or 60 min. A cell population of 1 × 106 cells per well was incubated in a 96-well plate (BD Falcon), followed by release of STAT3 in Cell Lysis Mix provided by the kit. Supernatants were used for STAT3 and phosphorylated STAT3 assessment using an ELISA kit from eBioscience (No. 85-86103) according to the instructions of the manufacturer.
Statistical analyses.
GraphPad Prism (version 6) was used to analyze experimental data using the Student’s t test for direct comparison of 2 populations or 2-factor ANOVA for multiple treatment groups with an upper limit of significance at P < 0.05. Tukey’s test was used for post hoc comparisons. Data not fitting a normal distribution (assessed using GraphPad Prism) was analyzed using the Mann-Whitney U test.
Results
n–3 PUFAs reduce colocalization of GP130 in lipid rafts.
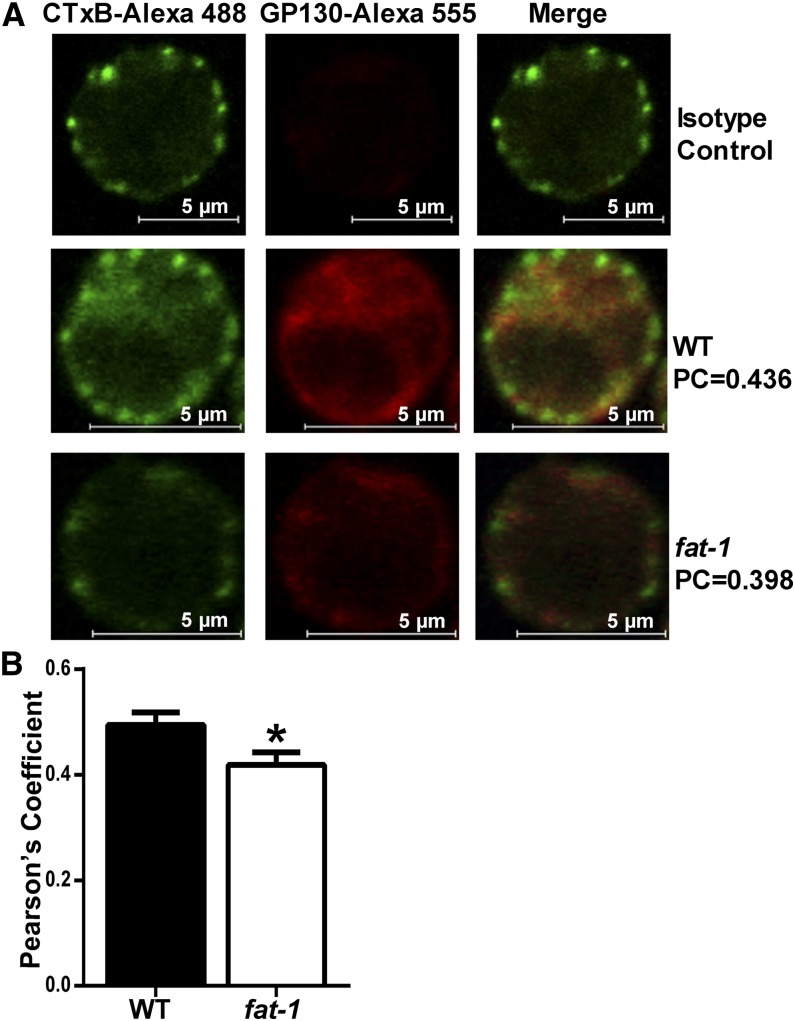
Localization of GP130 in the plasma membrane was assessed by immunofluorescence. To determine the optimal method for fixation, freshly isolated WT CD4+ splenic T cells were fixed with 4% paraformaldehyde or 4% paraformaldehyde plus 0.2% glutaraldehyde for 30 min before analysis by confocal microscopy. Because of high autofluorescence in cells fixed with paraformaldehyde and glutaraldehyde, glutaraldehyde was not included in the fixation process (Supplemental Fig. 1). After fixation with 4% paraformaldehyde, WT and fat-1 splenic CD4+ T cells were visualized using confocal microscopy (Fig. 1). In the left column, lipid rafts were labeled with Alexa Fluor 488–conjugated cholera toxin B subunit, a well-described marker of monosialotetrahexosylganglioside, which is enriched in lipid rafts (39, 40). GP130 was labeled with Alexa Fluor 555 (middle column), and the 2 signals were overlaid (Fig. 1A, right column). The isotype control (top row) had very little signal compared with WT cells (middle row) and fat-1 cells (bottom row) labeled with anti-GP130, suggesting very low nonspecific binding compared with anti-GP130 stained cells. Pearson’s correlation was used to quantitatively determine colocalization between GP130 and lipid rafts using a mathematical equation relating the intensity of each fluorophore in every pixel (41). The fat-1 mice displayed reduced colocalization of GP130 and cholera toxin B subunit compared with WT mice (P = 0.032) (Fig. 1B).
FIGURE 1.
Colocalization of GP130 with lipid rafts in WT and fat-1 mice. Splenic CD4+ T cells were fixed with 4% PFA, followed by incubation with anti-GP130 or IgG2A and Alexa Fluor 555–conjugated anti-rat IgG plus Alexa Fluor 488–conjugated CTxB before images were captured in confocal mode. Representative images of the isotype control, WT, and fat-1 CD4+ T cells with corresponding Pearson’s coefficients (A). Mean Pearson’s coefficient in WT vs. fat-1–derived cells (B). Data represent means ± SEMs. Cells from 3 fat-1 and 4 WT mice were analyzed (n = 15–20 groups of cells). *Different from control, P < 0.05. CD4+, cluster of differentiation 4+; CTxB, cholera toxin B subunit; GP130, glycoprotein 130; PC, Pearson’s coefficient; PFA, paraformaldehyde; WT, wild-type.
n–3 PUFAs do not affect surface or total cell expression of GP130.
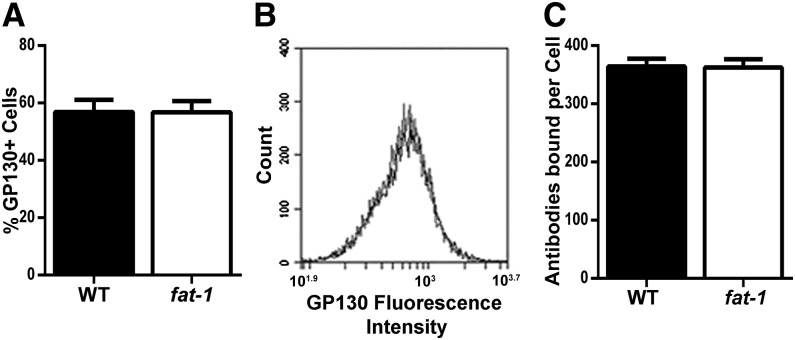
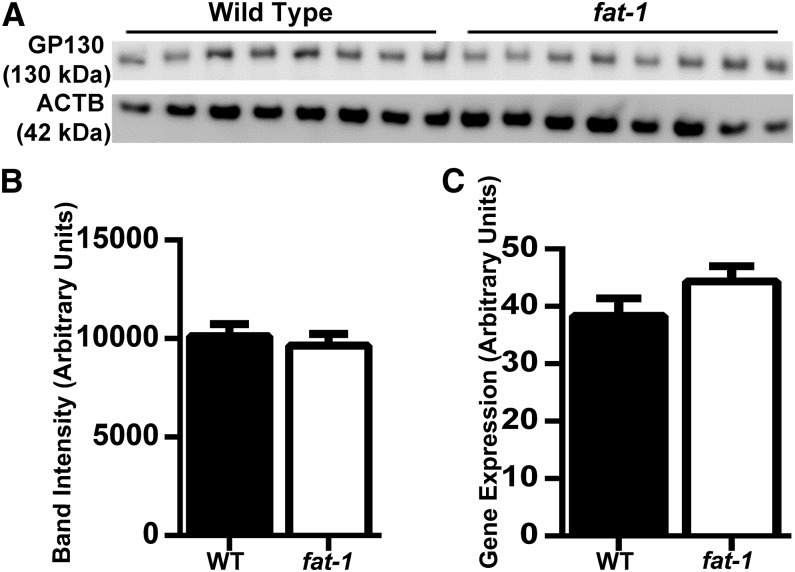
Optimal antibody mass for measuring GP130 expression by flow cytometry was determined by the signal-to-noise ratio. Histograms of fluorescence intensities are shown for each antibody concentration compared with the isotype control along with the signal-to-noise ratio for each concentration (Supplemental Fig. 2). An antibody concentration of 5 μg/mL was selected because of a high signal-to-noise ratio. n–3 PUFAs had no effect on the total number of cells expressing GP130 (Fig. 2A). A representative histogram is also shown to compare GP130 fluorescence intensity between WT (black) and fat-1 (gray) CD4+ T cells (Fig. 2B). Furthermore, n–3 PUFAs had no effect on the number of antibodies bound per cell or the mean fluorescence intensity (Fig. 2C). Subsequently, gene expression was assessed at both the mRNA and protein levels. Before analysis of WT and fat-1 cells, differing amounts of cell protein were analyzed in WT splenic CD4+ T cells by Western blot to determine linearity. Protein masses of 1, 2, 4, and 8 μg were electrophoresed in duplicates (Supplemental Fig. 3A) and quantified by chemiluminescence (Supplemental Fig. 3B), and 4 μg of protein lysate was selected for analysis. Before WT and fat-1 CD4+ T cells were used for gene expression analysis, differing amounts of RNA (0.09, 0.9, 9, 90, and 900 ng) were processed by reverse transcription to determine the linear region of the amplification reaction. An RNA mass of 90 ng was considered optimal (Supplemental Fig. 4). A representative Western blot of GP130 is shown in Figure 3A. No change in GP130 total protein (Fig. 3B) or Gp130 mRNA (Fig. 3C) was detected during the comparison of splenic CD4+ T cells from WT and fat-1 mice.
FIGURE 2.
Surface expression of GP130 in WT and fat-1 mice. Splenic CD4+ T cells were isolated from WT and fat-1 mouse spleens and stained with anti-GP130. Percentage GP130+ cells (A), overlay of WT (black) and fat-1 (gray) fluorescence intensities (B), and antibodies bound per cell (C). Data represent means ± SEMs, n = 4 mice. CD4+, cluster of differentiation 4+; GP130, glycoprotein 130; WT, wild-type.
FIGURE 3.
Basal expression of GP130 and Gp130 gene expression in WT and fat-1 mice. CD4+ T cells were isolated from WT and fat-1 mouse spleens and assessed by Western blot (A), n = 8. GP130 amounts in WT and fat-1 mice (B). Data represent means ± SEMs, n = 8. Gp130 gene expression was measured by qPCR (C), n = 9 WT and 5 fat-1. ACTB, Alexa Fluor–conjugated cholera toxin subunit B; CD4+, cluster of differentiation 4+; GP130, glycoprotein 130; WT, wild-type.
n–3 PUFAs reduce IL-6–induced GP130 dimerization and STAT3 phosphorylation.
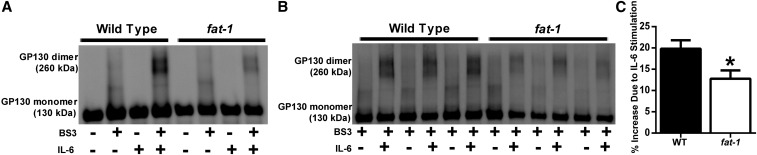
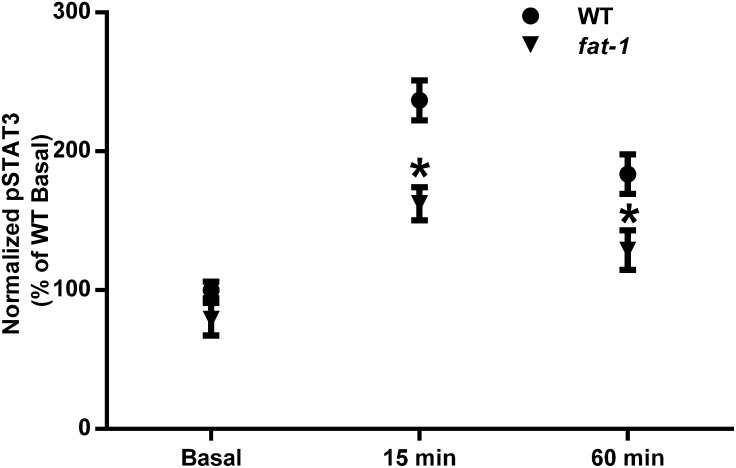
To determine whether n–3 PUFAs affect GP130 function, GP130 dimerization was induced in freshly isolated splenic CD4+ T cells by incubation with 100 μg/L recombinant mouse IL-6. During an optimization step, cells were either stimulated with IL-6 or maintained in resting conditions with or without bis(sulfosuccinimidyl)suberate for crosslinking (Fig. 4A). For analysis of WT and fat-1 cells, stimulated cells were compared with unstimulated cells from the same mouse (Fig. 4B) to determine the percentage increase in dimerization due to stimulation. CD4+ T cells from fat-1 mice exhibited a 35% reduction in dimer formation (P = 0.032) (Fig. 4C). To further ascertain the functional status of the IL-6/GP130/STAT3 axis in the CD4+ T cells from fat-1 mice, STAT3 phosphorylation was assessed after stimulation with 50 μg/L IL-6. CD4+ T cells from fat-1 mice exhibited a 30% reduction in STAT3 phosphorylation in response to IL-6 (Fig. 5) after 15 min (P = 0.001) and 60 min (P = 0.016) of stimulation.
FIGURE 4.
IL-6–induced GP130 dimerization in WT and fat-1 mice. CD4+ T cells were stimulated with IL-6 to induce homodimerization of GP130 and subsequently crosslinked with BS3. Positive (+BS3, +IL-6) and negative (−BS3, −IL-6) controls (A). Representative image of WT and fat-1 immunoblot ± IL-6 (B). IL-6–induced GP130 dimerization was decreased in fat-1 mice (C). Data represent means ± SEMs of 3 separate experiments and were normalized to individual unstimulated values, n = 5. *Different from control, P < 0.05. BS3, bis(sulfosuccinimidyl)suberate; CD4+, cluster of differentiation 4+; GP130, glycoprotein 130; WT, wild-type.
FIGURE 5.
STAT3 phosphorylation in WT and fat-1 mice. CD4+ T cells were incubated with IL-6 before homogenization. Cell lysates were analyzed by ELISA for pSTAT3 and total STAT3. Data represent means ± SEMs, n = 4. *Different from WT at that time, P < 0.05. CD4+, cluster of differentiation 4+; pSTAT3, phosphorylated signal transducer and activator of transcription 3; STAT3, signal transducer and activator of transcription 3; WT, wild-type.
n–3 PUFAs reduce surface IL-6 receptor expression.
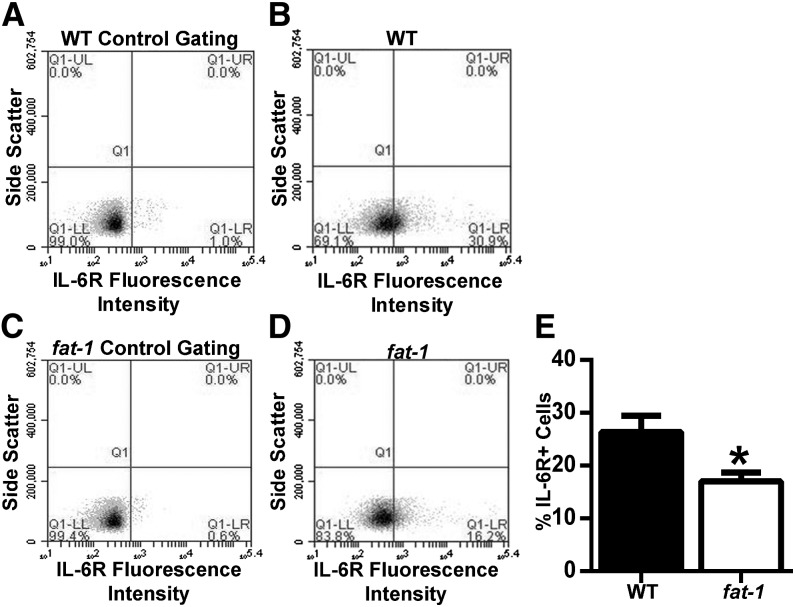
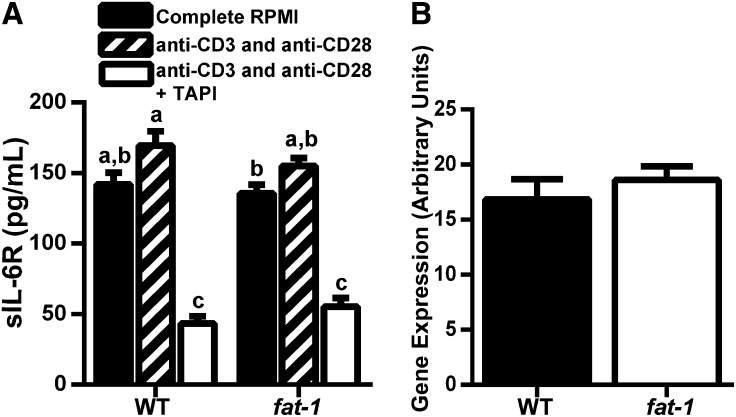
An optimal antibody concentration for IL-6 receptor was determined as above, and 5 μg/mL was selected for analysis. Supplemental Figure 5A–E shows histograms of fluorescence intensity for each antibody concentration compared with the isotype control. Surface expression of membrane-bound IL-6 receptor was assessed by flow cytometry in freshly isolated WT and fat-1 splenic CD4+ T cells. Representative scatter plots of flow cytometric analysis are shown (Fig. 6A–D). Splenic CD4+ T cells from fat-1 mice displayed a 30% reduction in the percentage (P = 0.023) of IL-6 receptor–expressing cells (Fig. 6E). To determine the cause of this decrease, shedding of IL-6 receptor into the media under activating conditions was measured by ELISA. Initially, a range of 1 × 105, 2 × 105, 5 × 105, and 1 × 106 WT CD4+ T cells were incubated with anti-CD3 and anti-CD28 for 48 h, and the supernatants were collected for analysis of soluble IL-6 receptor by ELISA. Supplemental Figure 6A shows the concentration of soluble IL-6 receptor (picograms per milliliter) from each number of incubated CD4+ T cells. TAPI, which inhibits α disintigrin and metalloproteinase (ADAM), was included as a negative control. As expected, stimulation of CD4+ T cells led to an increase in soluble IL-6 receptor detected in the culture media (Supplemental Fig. 6A), and TAPI decreased detectable soluble IL-6 receptor in a cell population of 4 × 106 cells. This cell number was chosen because fewer cells yielded undetectable soluble IL-6 receptor and a higher number of cells resulted in elevated soluble IL-6 receptor despite TAPI treatment, suggesting nonspecific results. Furthermore, an incubation time of 48 h was selected based on the induction of soluble IL-6 receptor shedding compared with unstimulated control (Supplemental Fig. 6B). For direct comparison of WT and fat-1 CD4+ T cells, naive cells were activated with anti-CD3 and anti-CD28, and supernatants were analyzed for soluble IL-6 receptor. n–3 PUFAs had no effect on basal, activated, or TAPI-treated soluble IL-6 receptor concentrations (Fig. 7A). To further probe the effect of n–3 PUFAs on Il-6r expression, mRNA was isolated from naive CD4+ T cells and assessed by qPCR, and optimization was performed as above. Gene expression did not differ (P > 0.05) between WT and fat-1 T cells (Fig. 7B).
FIGURE 6.
Surface expression of IL-6R in WT and fat-1 mice. Splenic CD4+ T cells were isolated from WT and fat-1 mice and stained with anti-IL-6R. Representative gating plots of unstained cells from WT and fat-1 mice (A, C). Representative plots of stained cells of WT and fat-1 (B, D). Percentage IL-6R–expressing cells (E). Data represent means ± SEMs, n = 11. *Different from control, P < 0.05. CD4+, cluster of differentiation 4+; IL-6R, IL-6 receptor; LL, lower left; LR, lower right; Q1, quadrant 1; UL, upper left; UR, upper right; WT, wild-type.
FIGURE 7.
Activation-induced shedding of IL-6R and RNA expression in WT and fat-1 mice. sIL-6R was measured in the supernatant fluids from splenic CD4+ T cells activated with anti-CD3 and anti-CD28 for 48 h (A). TAPI was used to inhibit shedding of IL-6R. Data are combined from 2 separate 48-h incubations, n = 8. Il-6r mRNA concentrations were measured by qPCR (B). Data represent means ± SEMs, n = 5–9. Labeled means without a common letter differ, P < 0.05. CD4+, cluster of differentiation 4+; IL-6R, IL-6 receptor; RPMI, Roswell Park Memorial Institute media; sIL-6R, soluble IL-6 receptor; TAPI, TNF-α protease inhibitor; WT, wild-type.
Discussion
Chronic inflammation can lead to tissue damage, insulin resistance, and disease, including arthritis, diabetes, and IBD (1–3, 5). CD4+ T cells, especially Th1 and Th17 effector cells, are involved in diseases such as arthritis, IBD, and multiple sclerosis (42–44). Modification to cellular differentiation pathways can be protective, e.g., the IL-6–deficient mouse is protected from EAE (45), an animal model of multiple sclerosis. This is relevant because IL-6 is critical for Th17 development, suggesting that EAE is mediated in part by a Th17 response. In addition, IL-17–deficient mice are resistant to collagen-induced arthritis (46).
This study used transgenic fat-1 mice, which generate n–3 PUFAs from n–6 PUFAs de novo due to n–3 FA desaturase from C. elegans (33). These mice were shown to incorporate n–3 PUFAs into the plasma membranes of CD4+ T cells to a similar extent as mice fed a 4% fish-oil diet (34). These data suggest that the fat-1 mouse faithfully recapitulates the phenotype seen in mice fed an n–3 PUFA-enriched diet and is therefore a valid model for the study of the effect of n–3 PUFAs on T-cell biology.
In previous studies, n–3 PUFAs reduced systemic and local Th1 (18, 47) and Th17 abundance and ex vivo differentiation of naive CD4+ T cells into a Th17 phenotype (19, 20, 23), but the mechanism is currently unknown. Based on the essential nature of the IL-6/GP130/STAT3 signaling axis in Th17 differentiation (9), the evidence that membrane modulation disrupts Th17 development (30), and the link between IL-6 signaling and localization in lipid rafts (31, 32), we hypothesized that n–3 PUFAs reduce Th17 differentiation by interfering with IL-6 signaling in a lipid raft–dependent manner. We proposed that n–3 PUFAs might alter the expression, localization, and/or function of IL-6 receptor and/or GP130, the 2 essential membrane proteins involved in the response of CD4+ T cells to the Th17 polarizing effects of IL-6. Therefore, we assessed GP130 expression at the cell surface, total protein and mRNA levels, and the functional capacity of the signaling axis through GP130 dimerization and STAT3 phosphorylation in freshly isolated CD4+ T cells. Furthermore, surface, soluble, and mRNA expression of IL-6 receptor was assessed.
Previous work in our laboratory and others revealed the importance of lipid rafts in the suppressive effect of n–3 PUFAs on CD4+ T-cell activation (24, 25, 38, 48, 49). Therefore, it was essential to measure the lipid raft localization of GP130, a critical coreceptor and signal transducer in the IL-6 signaling pathway. Previous studies suggest that GP130 is localized in lipid rafts in kidney and neuroepithelial cells (31, 32). We showed that n–3 PUFAs did not affect mRNA concentrations of Gp130, cellular ( ) or surface expression of GP130 (Fig. 2). However, n–3 PUFAs reduced localization of GP130 to lipid rafts (Fig. 1). This is noteworthy, because lipid rafts act as signaling platforms for CD4+ T-cell activation and a decrease of GP130 localization in these mesodomains could lead to a reduction in signaling capacity.
GP130 homodimerization is a critical component of the IL-6/GP130/STAT3 axis and can be used as a functional measure of signaling capacity. For this reason, IL-6–induced GP130 dimerization was measured in the splenic CD4+ T cells from WT and fat-1 mice. fat-1 mice exhibited a significant (P = 0.032) 35% reduction in GP130 dimerization (Fig. 4), indicating that the functional ability of the protein is decreased. Furthermore, IL-6–induced STAT3 phosphorylation, a measure of signaling activity through the IL-6 receptor/GP130/STAT3 axis, was also significantly reduced after 15 min (P = 0.001) and 60 min (P = 0.016) in CD4+ T cells from fat-1 mice (Fig. 5).
To the best of our knowledge, this is the first report to document that the ability of n–3 PUFAs to alter the membrane localization of GP130 in lipid rafts is associated with a reduction in IL-6–induced GP130 dimerization and STAT3 phosphorylation. We also observed a significant reduction (P = 0.023) in the surface expression of IL-6 receptor in the splenic CD4+ T cells from fat-1 mice, which suggests a second, independent mechanism by which n–3 PUFAs might interfere with differentiation of Th17 cells (Fig. 6). There are a number of possible mechanisms that might account for this reduction, including increased receptor shedding during cellular activation, reduced production of mRNA or total protein, receptor internalization, or altered trafficking of the receptor to the cell surface. In this study, we determined that the reduction in IL-6 receptor surface expression was not due to increased shedding of IL-6 receptor during activation (Fig. 7A) or a decrease in Il-6r mRNA production (Fig. 7B). Future work will focus on elucidating the mechanism by which n–3 PUFAs reduce IL-6 receptor surface expression.
Together with previous observations, our current findings support the conclusion that raft regions in CD4+ T cells enriched with n–3 PUFAs are modified in structure. GP130 is mislocalized out of lipid rafts, leading to reduced dimerization and downstream activation of STAT3. Finally, membrane-bound IL-6 receptor is decreased without a subsequent increase in soluble IL-6 receptor. These changes lead to reduced Th17 differentiation with n–3 PUFA enrichment. Our results indicate 2 novel mechanisms by which n–3 PUFAs could interfere with Th17 differentiation in vivo and ex vivo. By reducing colocalization of GP130 in lipid rafts in the CD4+ T-cell membrane, n–3 PUFAs attenuate the signaling capacity of the IL-6/GP130/STAT3 axis as evidenced by reduced GP130 dimerization and STAT3 phosphorylation. Furthermore, the ability of n–3 PUFAs to reduce IL-6 receptor surface expression is consistent with a decreased cellular responsiveness to this essential Th17-inducing cytokine.
Our findings using naive splenic CD4+ T cells provide new insights into the mechanism by which n–3 PUFAs suppress Th17 cell differentiation and add to the other mechanisms by which n–3 PUFAs are known to suppress inflammation (50, 51). Unfortunately, the optimal dietary intake of EPA and DHA is currently unknown. The adequate intake for n–3 PUFAs is 1.6 g/d for men and 1.1 g/d for women (52). The AHA recommends 500 mg/d EPA + DHA for healthy individuals and 1 g/d for those with coronary heart disease (53). The FDA currently recognizes 3 g/d EPA + DHA as generally recognized as safe (53), although clinical trials often use higher doses (51). There is some concern that the immune-suppressive effect of n–3 PUFAs could result in increased susceptibility to infections, although this typically occurs at very high doses (54). Furthermore, the degree of immune suppression or even activation by n–3 PUFAs may depend on the location of the insult or total dietary fat (55).
Current anti-inflammatory treatments, such as nonsteroidal anti-inflammatory drugs or glucocorticoids, are expensive and often involve serious side effects (56, 57). Given the high proportion of the population that is afflicted by chronic inflammation, it is important to identify innocuous anti-inflammatory dietary compounds that could ameliorate inflammatory conditions and improve the health of this population. n–3 PUFAs reduce CD4+ T-cell activation and the proinflammatory differentiation and functions of effector Th1 and Th17 subsets (18–20, 23). Understanding the precise mechanisms of these beneficial effects is critical to making informed decisions about recommendations for n–3 PUFA consumption in the diet or as supplements. This study adds new insights to that body of knowledge by furthering the understanding of molecular mechanisms by which n–3 PUFAs act on immune cells.
Supplementary Material
Acknowledgments
The authors thank Dr. Jing Kang for providing fat-1 breeder mice. The authors also thank Dr. Laurie Davidson and Dr. Roger Smith for technical assistance, Evelyn Callaway for technical assistance and mouse breeding, and Jennifer Goldsby for RNA quality analysis and software assistance. M.J.A., T.Y.H., D.N.M., and R.S.C. designed the experiments; M.J.A., J.M.M., and Y.-Y.F. conducted the research; M.J.A. analyzed the data; R.B. performed the confocal imaging; and M.J.A., D.N.M., and R.S.C. wrote the paper. All authors read and approved the final manuscript.
Footnotes
Abbreviations used: ADAM, α disintigrin and metalloproteinase; CD4+, cluster of differentiation 4+; EAE, experimental autoimmune encephalomyelitis; GP130, glycoprotein 130; IBD, inflammatory bowel disease; IL-2rα, interleukin-2 receptor; IL-6r, interleukin 6 receptor; JAK, Janus kinase; ROR-γt, RAR-related orphan receptor-γt; STAT3, signal transducer and activator of transcription 3; TAPI, TNF-α proteinase inhibitor; Th, T-helper; WT, wild-type.
References
- 1.Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell 2010;140:918–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medzhitov R. Inflammation 2010: new adventures of an old flame. Cell 2010;140:771–6. [DOI] [PubMed] [Google Scholar]
- 3.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010;140:883–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sherman E, Barr V, Samelson LE. Super-resolution characterization of TCR-dependent signaling clusters. Immunol Rev 2013;251:21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chalaris A, Garbers C, Rabe B, Rose-John S, Scheller J. The soluble Interleukin 6 receptor: generation and role in inflammation and cancer. Eur J Cell Biol 2011;90:484–94. [DOI] [PubMed] [Google Scholar]
- 6.Neurath MF, Finotto S. IL-6 signaling in autoimmunity, chronic inflammation and inflammation-associated cancer. Cytokine Growth Factor Rev 2011;22:83–9. [DOI] [PubMed] [Google Scholar]
- 7.Rabe B, Chalaris A, May U, Waetzig GH, Seegert D, Williams AS, Jones SA, Rose-John S, Scheller J. Transgenic blockade of interleukin 6 transsignaling abrogates inflammation. Blood 2008;111:1021–8. [DOI] [PubMed] [Google Scholar]
- 8.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 2006;441:235–8. [DOI] [PubMed] [Google Scholar]
- 9.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity 2006;24:179–89. [DOI] [PubMed] [Google Scholar]
- 10.Yang XO, Panopoulos AD, Nurieva R, Chang SH, Wang D, Watowich SS, Dong C. STAT3 regulates cytokine-mediated generation of inflammatory helper T cells. J Biol Chem 2007;282:9358–63. [DOI] [PubMed] [Google Scholar]
- 11.Korn T, Bettelli E, Oukka M, Kuchroo VK. IL-17 and Th17 Cells. Annu Rev Immunol 2009;27:485–517. [DOI] [PubMed] [Google Scholar]
- 12.Betz UA, Müller W. Regulated expression of gp130 and IL-6 receptor alpha chain in T cell maturation and activation. Int Immunol 1998;10:1175–84. [DOI] [PubMed] [Google Scholar]
- 13.Jones GW, McLoughlin RM, Hammond VJ, Parker CR, Williams JD, Malhotra R, Scheller J, Williams AS, Rose-John S, Topley N, et al. Loss of CD4+ T cell IL-6R expression during inflammation underlines a role for IL-6 trans signaling in the local maintenance of Th17 cells. J Immunol 2010;184:2130–9. [DOI] [PubMed] [Google Scholar]
- 14.Briso EM, Dienz O, Rincon M. Cutting edge: soluble IL-6R is produced by IL-6R ectodomain shedding in activated CD4 T cells. J Immunol 2008;180:7102–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Silver JS, Hunter CA. gp130 at the nexus of inflammation, autoimmunity, and cancer. J Leukoc Biol 2010;88:1145–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finotto S, Eigenbrod T, Karwot R, Boross I, Doganci A, Ito H, Nishimoto N, Yoshizaki K, Kishimoto T, Rose-John S, et al. Local blockade of IL-6R signaling induces lung CD4+ T cell apoptosis in a murine model of asthma via regulatory T cells. Int Immunol 2007;19:685–93. [DOI] [PubMed] [Google Scholar]
- 17.Chalaris A, Rabe B, Paliga K, Lange H, Laskay T, Fielding CA, Jones SA, Rose-John S, Scheller J. Apoptosis is a natural stimulus of IL6R shedding and contributes to the proinflammatory trans-signaling function of neutrophils. Blood 2007;110:1748–55. [DOI] [PubMed] [Google Scholar]
- 18.Zhang P, Kim W, Zhou L, Wang N, Ly LH, McMurray DN, Chapkin RS. Dietary fish oil inhibits antigen-specific murine Th1 cell development by suppression of clonal expansion. J Nutr 2006;136:2391–8. [DOI] [PubMed] [Google Scholar]
- 19.Monk JM, Hou TY, Turk HF, Weeks B, Wu C, McMurray DN, Chapkin RS. Dietary n-3 polyunsaturated fatty acids (PUFA) decrease obesity-associated Th17 cell-mediated inflammation during colitis. PLoS ONE 2012;7:e49739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Monk JM, Jia Q, Callaway E, Weeks B, Alaniz RC, McMurray DN, Chapkin RS. Th17 cell accumulation is decreased during chronic experimental colitis by (n-3) PUFA in fat-1 mice. J Nutr 2012;142:117–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arrington JL, McMurray DN, Switzer KC, Fan YY, Chapkin RS. Docosahexaenoic acid suppresses function of the CD28 costimulatory membrane receptor in primary murine and Jurkat T cells. J Nutr 2001;131:1147–53. [DOI] [PubMed] [Google Scholar]
- 22.Jolly CA, McMurray DN, Chapkin RS. Effect of dietary n-3 fatty acids on interleukin-2 and interleukin-2 receptor alpha expression in activated murine lymphocytes. Prostaglandins Leukot Essent Fatty Acids 1998;58:289–93. [DOI] [PubMed] [Google Scholar]
- 23.Monk JM, Hou TY, Turk HF, McMurray DN, Chapkin RS. n3 PUFAs reduce mouse CD4+ T-cell ex vivo polarization into Th17 cells. J Nutr 2013;143:1501–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim W, Fan YY, Barhoumi R, Smith R, McMurray DN, Chapkin RS. n-3 polyunsaturated fatty acids suppress the localization and activation of signaling proteins at the immunological synapse in murine CD4+ T cells by affecting lipid raft formation. J Immunol 2008;181:6236–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim W, Khan NA, McMurray DN, Prior IA, Wang N, Chapkin RS. Regulatory activity of polyunsaturated fatty acids in T-cell signaling. Prog Lipid Res 2010;49:250–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kwiatek JM, Owen DM, Abu-Siniyeh A, Yan P, Loew LM, Gaus K. Characterization of a new series of fluorescent probes for imaging membrane order. PLoS One 2013;8:e52960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Owen DM, Magenau A, Williamson D, Gaus K. The lipid raft hypothesis revisited—new insights on raft composition and function from super-resolution fluorescence microscopy. BioEssays 2012;34:739–47. [DOI] [PubMed] [Google Scholar]
- 28.Lingwood D, Simons K. Lipid rafts as a membrane-organizing principle. Science 2010;327:46–50. [DOI] [PubMed] [Google Scholar]
- 29.Matthews V, Schuster B, Schütze S, Bussmeyer I, Ludwig A, Hundhausen C, Sadowski T, Saftig P, Hartmann D, Kallen KJ, et al. Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by ADAM10 and ADAM17 (TACE). J Biol Chem 2003;278:38829–39. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y, Gumlaw N, Karman J, Zhao H, Zhang J, Jiang JL, Maniatis P, Edling A, Chuang WL, Siegel C, et al. Lowering glycosphingolipid levels in CD4+ T cells attenuates T cell receptor signaling, cytokine production, and differentiation to the Th17 lineage. J Biol Chem 2011;286:14787–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buk DM, Waibel M, Braig C, Martens AS, Heinrich PC, Graeve L. Polarity and lipid raft association of the components of the ciliary neurotrophic factor receptor complex in Madin-Darby canine kidney cells. J Cell Sci 2004;117:2063–75. [DOI] [PubMed] [Google Scholar]
- 32.Yanagisawa M, Nakamura K, Taga T. Roles of lipid rafts in integrin-dependent adhesion and gp130 signalling pathway in mouse embryonic neural precursor cells. Genes Cells 2004;9:801–9. [DOI] [PubMed] [Google Scholar]
- 33.Kang JX. Fat-1 transgenic mice: a new model for omega-3 research. Prostaglandins Leukot Essent Fatty Acids 2007;77:263–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fan YY, Kim W, Callaway E, Smith R, Jia Q, Zhou L, McMurray DN, Chapkin RS. fat-1 transgene expression prevents cell culture-induced loss of membrane n-3 fatty acids in activated CD4+ T-cells. Prostaglandins Leukot Essent Fatty Acids 2008;79:209–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim W, Barhoumi R, McMurray DN, Chapkin RS. Dietary fish oil and DHA down-regulate antigen-activated CD4+ T-cells while promoting the formation of liquid-ordered mesodomains. Br J Nutr 2014;111:254–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia Q, Lupton JR, Smith R, Weeks BR, Callaway E, Davidson LA, Kim W, Fan YY, Yang P, Newman RA, et al. Reduced colitis-associated colon cancer in fat-1 (n-3 fatty acid desaturase) transgenic mice. Cancer Res. 2008;68:3985–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ly LH, Smith R, Chapkin RS, McMurray DN. Dietary n-3 polyunsaturated fatty acids suppress splenic CD4+ T cell function in interleukin (IL)-10−/− mice. Clin Exp Immunol 2005;139:202–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turk HF, Chapkin RS. Membrane lipid raft organization is uniquely modified by n-3 polyunsaturated fatty acids. Prostaglandins Leukot Essent Fatty Acids 2013;88:43–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Day CA, Kenworthy AK. Mechanisms underlying the confined diffusion of cholera toxin B-subunit in intact cell membranes. PLoS One 2012;7:e34923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Margheri G, D’Agostino R, Trigari S, Sottini S, Del Rosso M. The β-subunit of cholera toxin has a high affinity for ganglioside GM1 embedded into solid supported lipid membranes with a lipid raft-like composition. Lipids 2014;49:203–6. [DOI] [PubMed] [Google Scholar]
- 41.Adler J, Parmryd I. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander’s overlap coefficient. Cytometry A. 2010;77:733–42. [DOI] [PubMed] [Google Scholar]
- 42.Jadidi-Niaragh F, Mirshafiey A. Th17 cell, the new player of neuroinflammatory process in multiple sclerosis. Scand J Immunol 2011;74:1–13. [DOI] [PubMed] [Google Scholar]
- 43.Eastaff-Leung N, Mabarrack N, Barbour A, Cummins A, Barry S. Foxp3+ regulatory T cells, Th17 effector cells, and cytokine environment in inflammatory bowel disease. J Clin Immunol 2010;30:80–9. [DOI] [PubMed] [Google Scholar]
- 44.van Hamburg JP, Corneth OBJ, Paulissen SMJ, Davelaar N, Asmawidjaja PS, Mus AMC, Lubberts E. IL-17/Th17 mediated synovial inflammation is IL-22 independent. Ann Rheum Dis 2013;72:1700–7. [DOI] [PubMed] [Google Scholar]
- 45.Samoilova EB, Horton JL, Hilliard B, Liu TST, Chen Y. IL-6-deficient mice are resistant to experimental autoimmune encephalomyelitis: roles of IL-6 in the activation and differentiation of autoreactive T cells. J Immunol 1998;161:6480–6. [PubMed] [Google Scholar]
- 46.Nishihara M, Ogura H, Ueda N, Tsuruoka M, Kitabayashi C, Tsuji F, Aono H, Ishihara K, Huseby E, Betz UA, et al. IL-6–gp130–STAT3 in T cells directs the development of IL-17+ Th with a minimum effect on that of Treg in the steady state. Int Immunol 2007;19:695–702. [DOI] [PubMed] [Google Scholar]
- 47.Zhang P, Smith R, Chapkin RS, McMurray DN. Dietary (n-3) polyunsaturated fatty acids modulate murine Th1/Th2 balance toward the Th2 pole by suppression of Th1 development. J Nutr 2005;135:1745–51. [DOI] [PubMed] [Google Scholar]
- 48.Yog R, Barhoumi R, McMurray DN, Chapkin RS. n-3 polyunsaturated fatty acids suppress mitochondrial translocation to the immunologic synapse and modulate calcium signaling in T cells. J Immunol 2010;184:5865–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan YY, Ly LH, Barhoumi R, McMurray DN, Chapkin RS. Dietary docosahexaenoic acid suppresses T cell protein kinase C theta lipid raft recruitment and IL-2 production. J Immunol 2004;173:6151–60. [DOI] [PubMed] [Google Scholar]
- 50.Miles EA, Calder PC. Influence of marine n-3 polyunsaturated fatty acids on immune function and a systematic review of their effects on clinical outcomes in rheumatoid arthritis. Br J Nutr 2012;107(Suppl 2):S171–84. [DOI] [PubMed] [Google Scholar]
- 51.Calder PC. n−3 Polyunsaturated fatty acids, inflammation, and inflammatory diseases. Am J Clin Nutr 2006;83:1505S–19S. [DOI] [PubMed] [Google Scholar]
- 52.Gebauer SK, Psota TL, Harris WS, Kris-Etherton PM. n−3 Fatty acid dietary recommendations and food sources to achieve essentiality and cardiovascular benefits. Am J Clin Nutr 2006;83:1526S–35S. [DOI] [PubMed] [Google Scholar]
- 53.Kris-Etherton PM, Harris WS, Appel LJ. Fish consumption, fish oil, omega-3 fatty acids, and cardiovascular disease. Arterioscler Thromb Vasc Biol 2003;23:e20–30. [DOI] [PubMed] [Google Scholar]
- 54.Fenton JI, Hord NG, Ghosh S, Gurzell EA. Immunomodulation by dietary long chain omega-3 fatty acids and the potential for adverse health outcomes. Prostaglandins Leukot Essent Fatty Acids 2013;89:379–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hokari R, Matsunaga H, Miura S. Effect of dietary fat on intestinal inflammatory diseases. J Gastroenterol Hepatol 2013;28(Suppl 4):33–6. [DOI] [PubMed] [Google Scholar]
- 56.Bhala N, Emberson J, Merhi A, Abramson S, Arber N, Baron JA, Bombardier C, Cannon C, Farkouh ME, FitzGerald GA; Coxib and Traditional NSAID Trialists’ (CNT) Collaboration. Vascular and upper gastrointestinal effects of non-steroidal anti-inflammatory drugs: meta-analyses of individual participant data from randomised trials. Lancet 2013;382:769–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Everdingen AA, Jacobs JWG, Siewertsz van Reesema DR, Bijlsma JWJ. Low-dose prednisone therapy for patients with early active rheumatoid arthritis: clinical efficacy, disease-modifying properties, and side effects: a randomized, double-blind, placebo-controlled clinical trial. Ann Intern Med 2002;136:1–12. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.