Abstract
STUDY QUESTION
Can high-resolution array comparative genomic hybridization (CGH) analysis of DNA samples from women with primary ovarian insufficiency (POI) improve the diagnosis of the condition and identify novel candidate genes for POI?
SUMMARY ANSWER
A mutation affecting the regulatory region of growth differentiation factor 9 (GDF9) was identified for the first time together with several novel candidate genes for POI.
WHAT IS KNOWN ALREADY
Most patients with POI do not receive a molecular diagnosis despite a significant genetic component in the pathogenesis.
STUDY DESIGN, SIZE, DURATION
We performed a case–control study. Twenty-six patients were analyzed by array CGH for identification of copy number variants. Novel changes were investigated in 95 controls and in a separate population of 28 additional patients with POI. The experimental procedures were performed during a 1-year period.
PARTICIPANTS/MATERIALS, SETTING, METHODS
DNA samples from 26 patients with POI were analyzed by a customized 1M array-CGH platform with whole genome coverage and probe enrichment targeting 78 genes in sex development. By PCR amplification and sequencing, the breakpoint of an identified partial GDF9 gene duplication was characterized. A multiplex ligation-dependent probe amplification (MLPA) probe set for specific identification of deletions/duplications affecting GDF9 was developed. An MLPA probe set for the identification of additional cases or controls carrying novel candidate regions identified by array-CGH was developed. Sequencing of three candidate genes was performed.
MAIN RESULTS AND THE ROLE OF CHANCE
Eleven unique copy number changes were identified in a total of 11 patients, including a tandem duplication of 475 bp, containing part of the GDF9 gene promoter region. The duplicated region contains three NOBOX-binding elements and an E-box, important for GDF9 gene regulation. This aberration is likely causative of POI. Fifty-four patients were investigated for copy number changes within GDF9, but no additional cases were found. Ten aberrations constituting novel candidate regions were detected, including a second DNAH6 deletion in a patient with POI. Other identified candidate genes were TSPYL6, SMARCC1, CSPG5 and ZFR2.
LIMITATIONS, REASONS FOR CAUTION
This is a descriptive study and no functional experiments were performed.
WIDER IMPLICATIONS OF THE FINDINGS
The study illustrates the importance of analyzing small copy number changes in addition to sequence alterations in the genetic investigation of patients with POI. Also, promoter regions should be included in the investigation.
STUDY FUNDING/COMPETING INTEREST(S)
The study was supported by grants from the Swedish Research council (project no 12198 to A.W. and project no 20324 to A.L.H.), Stockholm County Council (E.I., A.W. and K.R.W.), Foundation Frimurare Barnhuset (A.N., A.W. and M.B.), Karolinska Institutet (A.N., A.L.H., E.I., A.W. and M.B.), Novo Nordic Foundation (A.W.) and Svenska Läkaresällskapet (M.B.). The funding sources had no involvement in the design or analysis of the study. The authors have no competing interests to declare.
TRIAL REGISTRATION NUMBER
Not applicable.
Keywords: array-CGH, GDF9, DNAH6, TSPYL6, ZFR2
Introduction
A clinical diagnosis of primary ovarian insufficiency (POI) is defined by the presence of primary or secondary amenorrhea (PA or SA) and hypergonadotropic hypogonadism, occurring before the age of 40 (Nelson, 2009; De Vos et al., 2010). This condition has previously also been termed premature ovarian failure in patients with SA and gonadal dysgenesis in PA.
POI pathogenesis can be divided into follicle dysfunction and follicle depletion. Follicle depletion can be caused by a small initial germ cell count or a rapid germ cell loss. External factors, such as surgery, irradiation and infection as well as autoimmune and metabolic disorders also lead to POI. Most cases however remain idiopathic, and there is a significant genetic component (Simpson and Rajkovic, 1999; Persani et al., 2010). The most common are chromosomal aberrations including Turner's syndrome (Baronchelli et al., 2012). Premutation of the FMR1 gene is responsible for 2–5% of sporadic cases with POI presenting with SA (Wittenberger et al., 2007; De Vos et al., 2010).
Other known genetic causes are mutations in genes for oocyte-secreted factors, GDF9 (growth differentiation factor 9) and BMP15 (Dixit et al., 2006; Kovanci et al., 2007), for oocyte nuclear transcription factors NOBOX and FIGLA (Qin et al., 2007; Zhao et al., 2008) and for the hormone receptors FSHR and NR5A1 (Aittomaki et al., 1995; Janse et al., 2012). However, the majority (90%) of patients with POI do not receive a molecular diagnosis (Nelson, 2009) and it is likely that many causative genes still remain unknown.
By genome-wide detection of small genomic rearrangements, it is possible to identify novel genes involved in ovarian development, both on the X chromosome and on autosomes. The aim of this study was to identify novel candidate genes for ovarian development and function by investigating submicroscopic genetic imbalances in patients with POI. A 1 M array-CGH (comparative genomic hybridization) platform with 2.2 kb probe spacing for whole genome coverage and enrichment targeting 78 genes involved in gonadal development was used. At present, all studies reporting copy number variants (CNVs) in patients with POI by array-CGH have used lower resolution platforms (Aboura et al., 2009; Ledig et al., 2010; Liao et al., 2011). Studies using whole genome SNP array (McGuire et al., 2011) and targeted analysis of the X chromosome using array-based techniques have also been reported (Dudding et al., 2010; Quilter et al., 2010; Knauff et al., 2011).
Materials and Methods
Patients
Fifty-four patients with POI (23 with PA, 31 with SA) referred to the Clinical Genetic Laboratory of Karolinska University Hospital Stockholm, Sweden, were included in the study. Samples from 26 patients (17 with PA and 9 with SA) were available for array-CGH experiments.
The diagnosis of POI was set by PA or SA in a girl with female external genitalia as well as internal Müllerian structures (uterus) and hypergonadotropic hypogonadism (FSH > 30 IU/l), determined twice. In the group with SA the overall median age at diagnosis was 22 years (12–37) (Table I). The nine patients with SA included in the array-CGH experiments were diagnosed at a median age of 16 (13–22) years.
Table I.
Clinical overview of investigated patients.
| Clinical data | PA (n = 23) | SA (n = 31) |
|---|---|---|
| Median age at diagnosis, years (range) | 16 (13–18) | 22 (12–37) |
| Median age at menarche, years (range) | – | 13 (10–15) |
| Median FSH level at diagnosis, IU/l (range)a | 89 (39–150) | 80 (34–155) |
| Swedish Caucasian, n (%) | 17 (74) | 26 (84) |
| Known heredity for POI, n (%) | 1 (4) | 6 (19) |
| Spontaneous conception before diagnosis, n (%) | – | 11 (35) |
PA, primary amenorrhea; SA, secondary amenorrhea. aReference value: <30 IU/l.
One patient with PA has several affected sisters with POI who did not want to be included in the study. Among patients with SA, four have mothers with POI, one a sister with POI and one an affected paternal aunt, who is included in the study.
None of the patients had undergone previous ovarian surgery, chemotherapy or radiotherapy or presented with syndromic forms of POI. Five patients presented with hypothyroidism with anti-thyroid autoantibodies, after POI diagnosis in four with SA, and at 11 years of age in one patient with PA. None had autoantibodies against adrenal cortex or 21-hydroxylase protein.
Nine patients with POI underwent laparoscopy with diagnostic ovarian biopsies. In four patients with PA, streak gonads with no visible follicles were found. Three patients with SA also had streak gonads with no visible follicles. One patient with SA at 23 had multiple primordial follicles and one patient with SA at 15 had a few visible primordial and primary follicles.
Ethical approval
All participants gave informed consent and the regional Ethics Committee at Karolinska Institutet, Sweden, approved the study with registration number 2007/263-31/2 and 2011/276-32.
Genetic investigation
A 46, XX karyotype in peripheral blood was confirmed in all but in Patient 24 in whom a balanced Robertsonian translocation identified (45, XX, der (13; 14)) was not considered causative.
Sex chromosome mosaicism was excluded using fluorescent in situ hybridization and DNA probes from chromosomes X and Y on peripheral blood smears, and when available, on touch preparations from gonadal tissue. Genetic investigations included sequencing of the NR5A1, BMP15, FSHR, FIGLA, NOBOX and GDF9 genes. FMR1 mutations were also excluded in patients with SA.
Controls
DNA samples from 95 healthy women were collected. All women were above the age of 40 and had given birth to at least one child. The exclusion criteria were previous egg donation, in vitro fertilization, infertility treatment or menopause before the age of 40.
DNA extraction
DNA was extracted from peripheral blood lymphocytes. Some samples were further purified using the QiAmp DNA minikit (QIAGEN, Sweden) to achieve acceptable quality values for array-CGH analysis.
Array-CGH
Twenty-six unrelated patients with POI were analyzed using a customized 1 M oligomarker array-CGH platform developed at Oxford Gene Technology (OGT, UK). In addition to whole genome coverage, the platform is enriched with probes targeting 78 genes implicated in sex development (Supplementary data, Table SI). The theoretical average probe resolution is 2.2 kb.
Preparation of labeled DNA and subsequent hybridization were performed according to the ‘Agilent oligonucleotide array-based CGH for genomic DNA analysis’ protocol (v6.2) and as previously described (Norling et al., 2013). A commercial DNA sample with pooled human genomic DNA from 10 female controls (Promega, Sweden) was used as reference DNA.
Data was analyzed using the Cytosure interpret software v3.4.3 (OGT). Circular binary segmentation analysis to detect copy number changes was performed using the following parameters: minimum probe count: 5, threshold for gains: 0.35, threshold for losses: 0.65, chromosome average method: median segment.
By comparing data with the online database of genomic variation (DGV) version 10 released Nov 2010 (Iafrate et al., 2004), common CNVs found in healthy control samples were excluded from further investigation. This exclusion was done with caution, as phenotypic effects of genetic imbalances affecting sexual development can be dependent on chromosomal sex. This information is not always available for controls included in the DGV. Aberrations only partially overlapping with rare reported CNVs were not excluded. Small intronic variations and aberrations not affecting genes were excluded after verification that they were not located just upstream or downstream a known gene causing disorders of sex development as a positional effect should then be considered.
Multiplex ligation-dependent probe amplification
Multiplex ligation-dependent probe amplification (MLPA) (Schouten et al., 2002) probes were designed according to the recommendations by Stern et al. (2004) and combined in different probe sets (Supplementary data, Table SII). The reference probes and the pilot probe used have been described earlier (Barbaro et al., 2008).
MLPA reactions were performed according to the EK1 reagent kit (MRC-Holland, the Netherlands) recommendations using 200 ng of DNA and the in-house designed probe set, as previously described (Barbaro et al., 2008). The reference DNA sample used for array-CGH was used as a control in all runs, along with at least another control DNA.
Confirmation of array-CGH findings
Copy number variations left after exclusion criteria were confirmed by MLPA using two probes per aberration. When available, parental samples were analyzed for control of inheritance. Trace data were exported and analyzed in an Excel 2007 spreadsheet. Each sample's peak heights were normalized to the average peak height of the reference probes and subsequently normalized to the average peak height of the control samples. The analysis was accepted if the reference probe ratios were between 0.8 and 1.2. Threshold values for deletions and duplications were set at 0.75 and 1.25, respectively.
MLPA for GDF9
A specific synthetic probe set for MLPA analysis of the GDF9 gene was designed. Two probes were placed in each exon, two upstream GDF9 and one probe downstream (Fig. 1E). The probe set was validated for consistency by analysis of 10 healthy controls, with a standard deviation <0.1 for each probe. Trace data were analyzed using the GeneMarker v1.90 (Soft Genetics, USA) software using internal control probe normalization, quantification by peak height and ratio threshold values for deletion and duplication were 0.75 and 1.3, respectively.
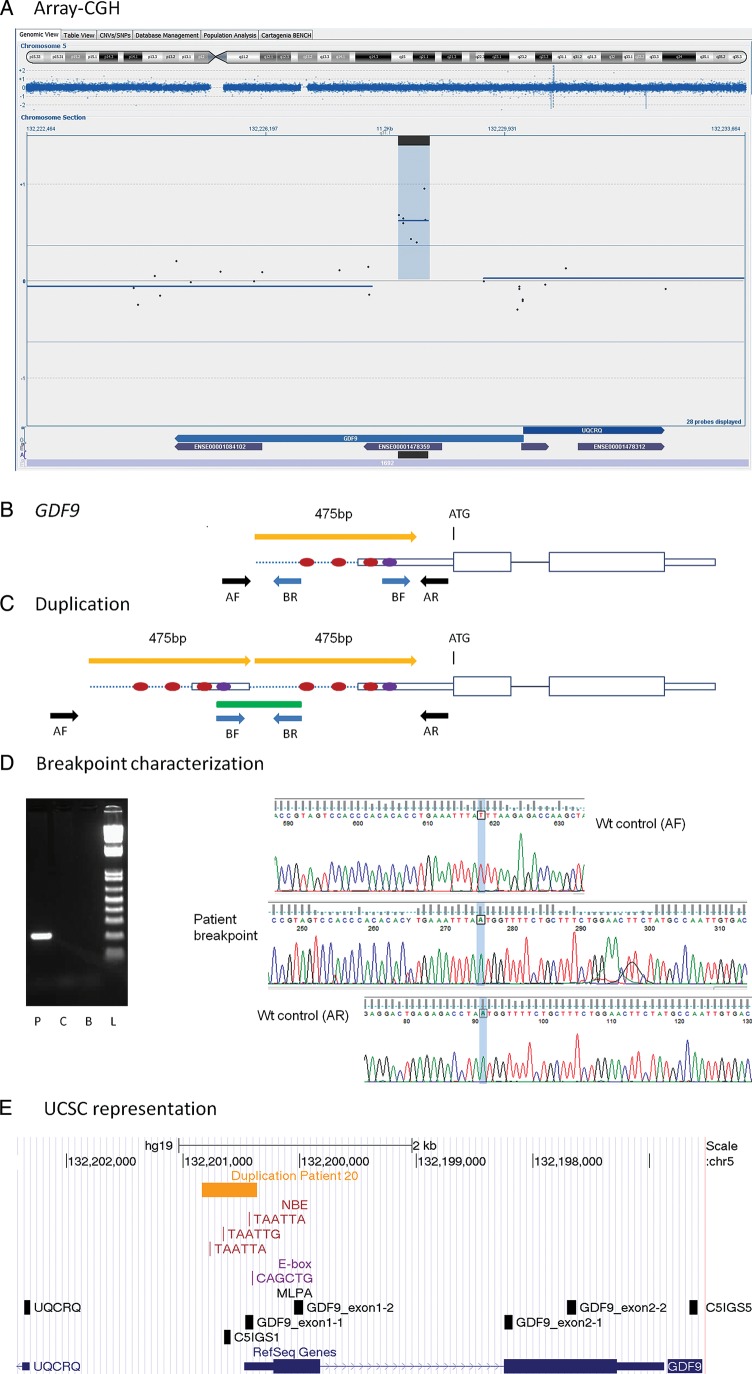
Figure 1.
GDF9 duplication. (A) Representation of an array-CGH result from the Cytosure software. The duplicated segment is indicated by blue background with a positive baseline offset. Blue dispersed dots represent oligomarkers. Blue arrows on bottom indicate gene location and purple smaller arrows represent exons. Black horizontal line on bottom represents location of duplication. Chromosome ideogram shown on top with the current segment magnification indicated by blue background color. (B) Schematic representation of the GDF9 gene. The ATG indicates the initiation of translation. Red dots represent NBE. Violet dot represents the E-box. Yellow arrow indicates the duplicated segment. Black and blue arrows represent PCR primers for wild-type allele (wt) and for duplication breakpoint amplification, respectively. (C) Schematic representation of the duplication. The green line represents the PCR product containing the duplication junction. (D) Breakpoint characterization. Gel image of amplified PCR breakpoint fragment. P, patient; C, control; B, blank; L, ladder. Electropherograms with sequence of the breakpoint (in the middle), partially aligned with the wt allele (upper and lower electropherograms). (E) Representation from the UCSC genome browser, GRCh37/hg19 assembly. Horizontal yellow arrow line indicates the duplicated segment. Locations of NBE (TAATTA, TAATTG) and E-box (CAGCTG) are shown as vertical red and purple lines, respectively. Vertical black boxes represent MLPA probes.
Patient cohort and control screening
A specific synthetic probe set for MLPA analysis of array findings was designed, using one probe per each novel candidate copy number variation identified by array-CGH experiments. In addition, one probe within the keratin-associated protein (KRTAP)2-1 and the KRTAP2-2 gene, respectively, were included. The probe set was validated for consistency. Trace data were analyzed with GeneMarker v1.90.
Sequencing of TSPYL6, KRTAP2-3 and KRTAP2-4
The single exon genes TSPYL6, KRTAP2-3 and KRTAP2-4, including part of the UTRs, were amplified by PCR using DyNAzyme EXT polymerase. Primers were designed using Primer3 software (Koressaar and Remm, 2007; Untergasser et al., 2012) (Supplementary data, Table SIII).
PCR products were cleaned and sequenced using the ABI BigDye Terminator v3.1 kit (Applied Biosystems, Sweden). Electropherograms were analyzed against the reference sequences: NM_001003937.2 for TSPYL6, NM_001165252.1 for KRTAP2-3 and NM_033184.3 for KRTAP2-4, using the SeqScape v2.5 program (Applied Biosystems).
Characterization of the GDF9 duplication
Primers for the amplification and sequencing of the duplication junction were designed using Primer 3 software (Rozen and Skaletsky, 2000) (Supplementary data, Table SIII) with the hypothesis of a tandem duplication.
In silico analysis
For all identified candidate genes the following databases were searched for information. Relevant data are presented in the discussion section.
NCBI (http://www.ncbi.nlm.nih.gov/) including PubMed, UCSC (http://genome.ucsc.edu/) (Dreszer et al., 2012), GeneCards (http://www.genecards.org/), The Human Protein Atlas (http://www.proteinatlas.org/) (Uhlen et al., 2010), Gene expression profiles during sex determination by Dr Serge Nef (http://nef.unige.ch/microarrays.php) (Nef et al., 2005), DECIPHER (Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources) (http://decipher.sanger.ac.uk/), LifeMap Database of Embryonic development, Stem Cell Research and Regenerative Medicine (http://discovery.lifemapsc.com) (Edgar et al., 2013).
Polyphen2 was used to evaluate amino acid substitutions (http://genetics.bwh.harvard.edu/pph2).
Results
Array-CGH
A total of 1720 aberrations were detected by array-CGH analysis of 26 samples with an average of 66 changes per patient (ranging from 29 to 142). After exclusion criteria, 13 copy number changes remained, 6 in the PA group and 7 in the SA (Table II). Two pairs of patients carried the same aberration and one patient carried 3 changes, leaving 11 unique copy number changes identified in a total of 11 patients. One aberration affected the GDF9 gene, already associated with POI, the other 10 changes unraveled novel candidate regions. All aberrations were confirmed by MLPA. When possible, inheritance pattern was investigated.
Table II.
Results from 26 patients, copy numbers shown after application of exclusion criteria.
| Pat. no. | Age at amen. | Chr. band | Start | End | Size (kb) | Probe count | Del/dup | Gene(s) | Inheritance | Other information |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | PA | 2p16.2 | 54296546 | 54398100 | 102 | 39 | Del | ACYP2, TSPYL6 | Not maternal | – |
| 2 | PA | – | – | – | – | – | – | – | – | Ovarian biopsy |
| 3 | PA | 15q26.2 | 92580470 | 92619918 | 39 | 17 | Del | MCTP2 | Maternal | – |
| 4 | PA | 3p21.31 | 47588558 | 47723275 | 135 | 54 | Dup | SMARCC1, CSPG5 | Maternal | Mother breast cancer 41 years |
| 5 | PA | 15q26.2 | 92580470 | 92619918 | 39 | 17 | Del | MCTP2 | Paternal | – |
| 6 | PA | – | – | – | – | – | – | – | – | – |
| 7 | PA | – | – | – | – | – | – | – | – | – |
| 8 | PA | – | – | – | – | – | – | – | – | – |
| 9 | PA | 12p13.33 | 2666671 | 2718905 | 52 | 15 | Dup | CACNA1C | Maternal | – |
| 10 | PA | – | – | – | – | – | – | – | – | – |
| 11 | PA | – | – | – | – | – | – | – | – | – |
| 12 | PA | 15q25.2 | 81917995 | 81997930 | 8 | 25 | Dup | SH3GL3 | Maternal | – |
| 13 | PA | – | – | – | – | – | – | – | – | – |
| 14 | PA | – | – | – | – | – | – | – | – | – |
| 15 | PA | – | – | – | – | – | – | – | – | – |
| 16 | PA | – | – | – | – | – | – | – | – | – |
| 17 | PA | – | – | – | – | – | – | – | – | – |
| 18 | 16 years | – | – | – | – | – | – | – | – | – |
| 19 | 14 years | 19p13.3 | 3688186 | 3816117 | 128 | 51 | Del | RAX2, TJP3, ZFR2, MRPL54, APBA3, MATK | Maternal | Hypothyroidism ovarian biopsy |
| Xp22.33 | 1529221 | 1572694 | 43 | 17 | Dup | P2RY8, ASMTL | Not maternal | – | ||
| 17q21.2 | 36465290 | 36473792 | 9 | 5 | Del | KRTAP2-3, KRTAP2-4 | Not maternal | – | ||
| 20 | 15 years | 5q31.1 | 132228266 | 132228740 | 0.5 | 7 | Dup | GDF9 | NA | – |
| 21 | 22 years | 2p11.2 | 84620455 | 84791661 | 171 | 57 | Del | DNAH6 | NA | Paternal aunt POI 34 |
| 22 | 20 years | 17q21.2 | 36465290 | 36473792 | 9 | 5 | Del | KRTAP2-3, KRTAP2-4a | Mother hemizygous | Hypothyroidism consanguinity mother with bilateral breast cancer |
| 23 | 17 years | 2q34 | 210703481 | 210739467 | 36 | 14 | Del | KANSL1L | Maternal | – |
| 24 | 21 years | – | – | – | – | – | – | – | – | 45,XX,der(13;14) Ovarian biopsy |
| 25 | 15 years | – | – | – | – | – | – | – | – | – |
| 26 | 13 years | – | – | – | – | – | – | – | – | Hypothyroidism with anti-TPO ovarian biopsy |
All coordinates are given using the NCBI36/hg18 build. Pat no., patient number; Age at amen., Age at amenorrhea; Chr. Band., chromosomal band; Del., deletion; Dup., duplication; PA, primary amenorrhea; NA, no available sample.
aHomozygous deletion.
The entire patient cohort of 54 patients with POI and 95 healthy controls were screened by MLPA for additional cases with copy number changes within the novel candidate regions. One patient and two controls were heterozygous for KRTAP2-3 and KRTAP2-4 deletion. One control was hemizygous for the KRTAP2-2 gene. No other patient or control was found to carry any other identified aberration.
GDF9 duplication investigation and GDF9 MLPA analysis
A duplication between 475 and 1729 bp within the GDF9 gene was identified by array-CGH in Patient 20. We amplified the duplication junction (Fig. 1D) and by sequencing, we confirmed a tandem head-to-tail duplication of 475 bp containing the first part of the 5′ UTR and a short upstream DNA sequence (Chr5.hg19:g.132,201,170_132,201,644dup).
Analysis of the entire patient cohort by an MLPA probe set for the identification of deletions/duplications within or encompassing GDF9 did not identify any additional case.
Sequencing
The KRTAP2-3 and KRTAP2-4 genes were sequenced in the two hemizygous patients. No mutations were identified in KRTAP2-4, while the variation c.218G>A (p.Cys73Tyr), that is reported in dbSNP as rs113397060, was identified. KRTAP2-3 sequencing was extended to parental samples and the entire patient–control cohorts. One more patient and one control were homozygous (A/A) for this substitution.
Sequencing of the TSPYL6 gene in the entire patient cohort revealed no mutations.
Discussion
Out of a group of 26 patients with POI, we have identified 11 unique novel aberrations in 11 patients, using a customized high-resolution array-CGH platform. Our detection rate is comparable to Ledig et al. (2010), and higher than studies using platforms with lower resolution (Aboura et al., 2009; Liao et al., 2011; McGuire et al., 2011).
One change affects GDF9 and the other 10 involve new regions. Maternally inherited changes, even though less likely pathogenic if assuming a haploinsufficiency mechanism, have not been excluded by default as autosomal recessive inheritance, and polygenic or environmental factor affecting phenotype penetrance cannot be excluded.
GDF9 on 5q31.1
A duplication affecting GDF9 was detected in Patient 20 with SA at 15 years of age (Fig. 1). This oocyte-secreted factor, expressed from the primary follicle stage, is necessary for normal folliculogenesis and fertility. Mutations affecting the open reading frame of the GDF9 gene have been associated with POI at a low frequency (Dixit et al., 2005; Laissue et al., 2006; Zhao et al., 2007). No such mutations were identified in our patient cohort.
Within the duplicated region there are three NOBOX-binding elements (NBEs) and an E-box sequence. In mouse NBEs regulate Gdf9 expression (Choi and Rajkovic, 2006), and the E-box sequence is a target for transcription factors necessary for the ovarian specific expression of Gdf9 (Yan et al., 2006). Both the NBEs and the E-box are conserved between mice and humans, as well as several other species. Thus, they are likely important also for the regulation of human GDF9 expression (Choi and Rajkovic, 2006; Yan et al., 2006). It is likely that the duplication causes altered GDF9 expression in the developing ovary, leading to POI. This represents the first mutation identified affecting the regulatory region of GDF9.
An enrichment of CNVs has been identified in promoter regions, suggesting that promoter sequences can be mildly unstable. This is a suggested evolutionary mechanism for changes in gene regulation (Conrad et al., 2010).
Candidate genes
TSPY-Like 6, acylphosphatase 2, muscle type 1 and C2ORF73 on 2p16.2
The deletion, affecting TSPY-Like 6 (TSPYL6) and acylphosphatase 2, muscle type 1 (ACYP2), in Patient 1 was not inherited from the mother. The TSPYL6 gene function is unknown, but homozygous mutations of TSPYL1 (TSPY-Like 1) have been found to cause dysgenesis of the testis and sudden infant death syndrome (MIM 608800). We considered TSPYL6 an interesting candidate gene for POI and sequenced the entire patient cohort, but no inactivating mutations were identified.
ACYP2 encodes a muscle type form of acylphosphatase 2 and we consider it a less likely candidate for POI. The deletion occurs just upstream of C2ORF73, an uncharacterized gene expressed in both ovaries and testis. A positional effect of the deletion on this gene expression cannot be excluded as a potential cause of POI.
Multiple C2 domains, transmembrane 2 on 15q26.2
In two unrelated patients with PA (Patients 3 and 5), a 39 kb deletion was detected, removing a large part of intron 1 in one of the several isoforms of the multiple C2 domains, transmembrane 2 (MCTP2) gene (ENST00000543482). MCTP2 is a transmembrane protein that binds calcium in the absence of phospholipids. The deletion could affect splicing or regulation or have no effect at all. It is difficult to find an immediate connection between MCTP2 and POI with the available data.
SWI/SNF-related matrix-associated, actin-dependent regulator of chromatin subfamily c member 1 and chondroitin sulfate proteoglycan 5 on 3p21.31
A 135 kb duplication on chromosome 3p21.31 was found in Patient 4 with PA, spanning the SWI/SNF-related matrix-associated, actin-dependent regulator of chromatin subfamily c member 1 (SMARCC1) gene and at least the first two exons of the CSPG5 gene. SMARCC1 is a member of the SWI/SNF family of proteins that regulate transcription by chromatin remodeling. It is highly expressed in both follicle and ovarian stroma cells. Chondroitin sulfate proteoglycan 5 (CSPG5) is a proteoglycan that may function as a differentiation factor. In mice, there is a sex differential expression profile of Cspg5 in Sf1+ somatic cells in developing gonads at embryonic day 12.5 (E12.5) and E13.5 (Nef et al., 2005) indicating a potential role in gonad differentiation. We consider both SMARCC1 and CSPG5 possible candidate genes for POI.
Calcium channel, voltage-dependent l type alpha 1C subunit on 12p13.33
Patient 9, with PA, has a maternally inherited duplication involving the calcium channel, voltage-dependent l type alpha 1C subunit (CACNA1C) gene, likely not affecting expression as loss of function mutations causes autosomal dominant cardiac conduction abnormalities (Brugada syndrome, MIM 611875; Timothy syndrome, MIM 601005). We do not consider CACNA1C a candidate gene for POI.
SH3-domain GRB2-like 3 on 15q25.2
Patient 12, presenting with PA, carries a maternally inherited duplication of almost the entire sequence of the first intron of the SH3GL3 gene. SH3-domain GRB2-like 3 (SH3GL3) expression is abundant in testis (Ringstad et al., 1997), where it is believed important for spermatogenesis (Li et al., 2009). Moderate expression in follicle cells is reported in the Human Protein Atlas. SH3GL3 interacts with the similar protein SH3GL2 (SH3-domain GRB2-like-2) (Franceschini et al., 2013) that in mice Sh3Gl2 has a sex differential expression profile at E12.5 and E13.5 (Nef et al., 2005). An association between SH3GL3 and POI should be further investigated.
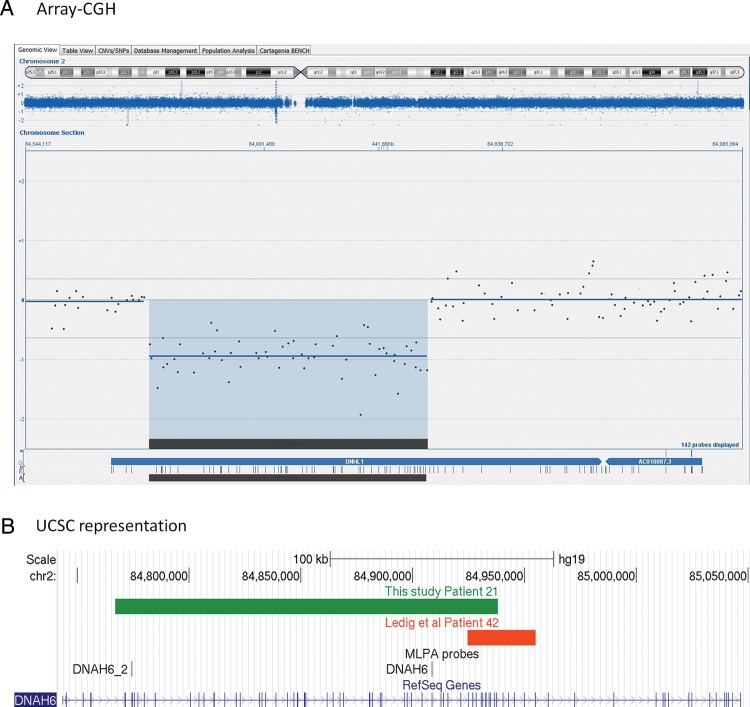
Dynein axonemal heavy chain 6 on 2p11.2
In Patient 21 we have identified a 171 kb deletion that removes at least 52 of the 77 exons of the dynein axonemal heavy chain 6 (DNAH6) gene encoding a heavy axonemal dynein chain. Dyneins are microtubule-associated motor protein complexes, important for ciliar and flagellar motility. There are heavy, light and intermediate chains. The mouse intermediate chain Dnaic2 (dynein, axonemal intermediate chain 2) has been found on the surface of oocytes in secondary and antral stages, but not in primordial or primary follicles suggesting a role for dyneins in ovarian development (Yang and Wu, 2008).
Interestingly, in a previous array-CGH study a smaller but partially overlapping deletion of DNAH6 was detected in a patient with secondary amenorrhea (Fig. 2) (Ledig et al., 2010) strengthening the possible role for DNAH6 in POI. Another study has also identified the related DNAH5 as a candidate gene for POI (Aboura et al., 2009). Mutations in DNAH5 are otherwise known to cause primary ciliary dyskinesia (PCD, MIM244400). We believe that DNAH6 is a very interesting candidate gene for POI.
Figure 2.
DNAH6 deletion. (A) Representation of an array-CGH result from the Cytosure software. The duplicated segment is indicated by blue background with a negative baseline offset. Blue dispersed dots represent oligomarkers. Blue arrows on bottom indicate gene location and purple vertical lines represent exons. The DNAH6 gene is shown with previous name DNHL1. The black horizontal line on bottom represents the location of deletion. Chromosome ideogram shown on top with the current segment magnification indicated by blue background color. (B) Representation from the UCSC genome browser, GRCh37/hg19 assembly. The green line represents the deletion detected in Patient 21. The orange line shows the deletion reported by Ledig et al. Vertical black lines represent MLPA probes.
KRTAP2-3 and KRTAP2-4 on 17q21.2
A 9 kb deletion encompassing the KRTAP2-3 and KRTAP2-4 genes was identified in two unrelated patients with SA and by MLPA screening one additional hemizygous patient was found.
We hypothesized an autosomal recessive causative mechanism, and by sequencing identified a potentially damaging amino acid substitution in the KRTAP2-3 gene in the hemizygous patients and in homozygous form in one more patient with POI. However, also one control was found to be homozygous for the substitution, contradicting KRTAP2-3 mutations as causative for POI. It is possible that the identified KRTAP2-3 and KRTAP2-4 deletion is a normal variant in the Swedish population as also two controls are hemizygous.
Purinergic receptor P2Y G-protein coupled 8 and acetylserotonin O-methyltransferase-like on Xp22.33
In addition to a deletion of KRTAP2-3 and KRTAP2-4, Patient 22 also carries a duplication in the pseudoautosomal region 1 on the X chromosome. The duplication includes the last exon of purinergic receptor P2Y G-protein coupled 8 (P2RY8) and the first exon of the ASMTL gene. P2RY8 is a G-protein coupled receptor expressed in lymphocytes. Acetylserotonin O-methyltransferase-like (ASMTL) is a generally expressed enzyme, with high levels in ovarian follicle cells. We consider both P2RY8 and ASTML less likely candidate genes for POI.
Del 19p13.3
A 128 kb deletion on chromosome 19 was also identified in Patient 22 affecting six genes: TJP3, APBA3, MRPL54, RAX2, MATK and zinc finger RNA-binding protein 2 (ZFR2).
Knock-out mouse models for TJP3 (Xu et al., 2008), APBA3 (Hara et al., 2011) and MATK (Lee et al., 2006) have been described without particular phenotypes and with normal fertility excluding these genes as candidates for POI.
MRPL54 encodes for a mitochondrial ribosome subunit necessary for the synthesis of mitochondrial proteins (Koc et al., 2001). Defects of mitochondrial protein synthesis lead to a more wide and severe phenotype than isolated POI. RAX2 heterozygous missense mutations cause age-related macular degeneration (MIM 610362) and cone-rod dystrophy type 11 (MIM 610381). We exclude MRPL54 and RAX2 as candidate genes for POI.
ZFR2 has no known function, but is expressed in many adult cell types, including ovarian follicle and stroma cells, as well as in female gametocytes and germ cells (Edgar et al., 2013). Ovarian biopsies from the patient show bilateral complete lack of oocytes and follicles and we consider ZFR2 an interesting candidate gene for POI.
KAT8 regulatory NSL complex subunit 1-like on 2q34
In Patient 23, with SA at 17, we detected a 36 kb deletion removing the first coding exon of the KAT8 regulatory NSL complex subunit 1-like (KANSL1L) gene on chromosome 2q34. KANSL1L, together with KANSL1 on chromosome 17, are the human homologs of the Drosophila Waharan gene (Lone et al., 2010). KANSL1 is a regulator of KAT8 influencing gene expression through histone H4 lysine 16 acetylation (Mendjan et al., 2006). Mutations in this gene cause 17q21.31 microdeletion syndrome (MIM 612452). No specific information on KANSL1L function is reported, but as an altered chromatin acetylation could possibly affect developing germ cells and follicle formation, we consider KANSL1L a potential candidate gene for POI.
Conclusion
Using array-CGH, we have identified the first mutation affecting the regulatory region of GDF9 in a patient with SA, most likely pathogenic. Importantly, the small, 475 bp duplication would not have been detected without the customized probe enrichment, making clear that platform resolution and probe targeting are crucial. We have also identified a second DNAH6 deletion in a patient with SA, corroborating DNAH6 as a candidate gene for POI. In addition, we describe several novel candidate genes such as TSPYL6, SMARCC1, CSPG5 and ZFR2.
We consider array-CGH a powerful tool to identify candidate genes for POI and we recommend that patients without a molecular diagnosis should undergo analysis for copy number alterations. Soon next generation sequencing will be available and able to provide reliable data on copy numbers, once this occurs whole genome sequencing should be considered for diagnostics of POI patients, as most known causative genes can either harbor small pathogenic sequence changes or be present in abnormal copy numbers.
Supplementary data
Supplementary data are available at http://humrep.oxfordjournals.org/.
Authors' roles
As first author A.N. performed the experimental work, results interpretation and writing of the first draft of the manuscript. A.L.H., K.R.W. and E.I. performed the clinical characterization and sample acquisition. E.I. also performed sample analysis. A.W. and M.B. were integral in results interpretation. A.L.H., E.I., A.W. and M.B. planned the study and experiments. M.B. performed literature evaluation and manuscript revision. All authors have contributed significantly to critical revision of article for important intellectual content and preparation of the final manuscript submitted for publication.
Funding
The study was supported by grants from the Swedish Research council (project no 12198 to A.W., and project no 20324 to A.L.H.), Stockholm County Council (E.I., A.W., K.R.W.), Foundation Frimurare Barnhuset (A.N., A.W., M.B.), Karolinska Institutet (A.N., A.L.H., E.I., A.W., M.B.), Novo Nordic Foundation (A.W.) and Svenska Läkaresällskapet (M.B.). The funding sources had no involvement in the design or analysis of the study. Funding to pay the Open Access publication charges for this article was provided by the Swedish Research Council.
Conflict of interest
None declared.
Supplementary Material
Acknowledgements
We thank Johan Svensson (Department of Child and Adolescent Medicine, Skåne University Hospital, Malmö, Sweden) for contributing with patient samples and clinical characterization. We thank Oxford Gene Technology, and Duarte Molha, Lee Eastoe, John Shovelton and Brian Woodhouse in particular, for their input and assistance in designing our customized 1 M array-CGH platform, as well as their support throughout experimentation and computerized analysis.
References
- Aboura A, Dupas C, Tachdjian G, Portnoi MF, Bourcigaux N, Dewailly D, Frydman R, Fauser B, Ronci-Chaix N, Donadille B, et al. Array comparative genomic hybridization profiling analysis reveals deoxyribonucleic acid copy number variations associated with premature ovarian failure. J Clin Endocrinol Metab. 2009;94:4540–4546. doi: 10.1210/jc.2009-0186. [DOI] [PubMed] [Google Scholar]
- Aittomaki K, Lucena JL, Pakarinen P, Sistonen P, Tapanainen J, Gromoll J, Kaskikari R, Sankila EM, Lehvaslaiho H, Engel AR, et al. Mutation in the follicle-stimulating hormone receptor gene causes hereditary hypergonadotropic ovarian failure. Cell. 1995;82:959–968. doi: 10.1016/0092-8674(95)90275-9. [DOI] [PubMed] [Google Scholar]
- Barbaro M, Cicognani A, Balsamo A, Löfgren A, Baldazzi L, Wedell A, Oscarson M. Gene dosage imbalances in patients with 46,XY gonadal DSD detected by an in-house-designed synthetic probe set for multiplex ligation-dependent probe amplification analysis. Clin Genet. 2008;73:453–464. doi: 10.1111/j.1399-0004.2008.00980.x. [DOI] [PubMed] [Google Scholar]
- Baronchelli S, Villa N, Redaelli S, Lissoni S, Saccheri F, Panzeri E, Conconi D, Bentivegna A, Crosti F, Sala E, et al. Investigating the role of X chromosome breakpoints in premature ovarian failure. Mol Cytogenet. 2012;5:32. doi: 10.1186/1755-8166-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Rajkovic A. Characterization of NOBOX DNA binding specificity and its regulation of Gdf9 and Pou5f1 promoters. J Biol Chem. 2006;281:35747–35756. doi: 10.1074/jbc.M604008200. [DOI] [PubMed] [Google Scholar]
- Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos M, Devroey P, Fauser BC. Primary ovarian insufficiency. Lancet. 2010;376:911–921. doi: 10.1016/S0140-6736(10)60355-8. [DOI] [PubMed] [Google Scholar]
- Dixit H, Rao LK, Padmalatha V, Kanakavalli M, Deenadayal M, Gupta N, Chakravarty B, Singh L. Mutational screening of the coding region of growth differentiation factor 9 gene in Indian women with ovarian failure. Menopause. 2005;12:749–754. doi: 10.1097/01.gme.0000184424.96437.7a. [DOI] [PubMed] [Google Scholar]
- Dixit H, Rao LK, Padmalatha VV, Kanakavalli M, Deenadayal M, Gupta N, Chakrabarty B, Singh L. Missense mutations in the BMP15 gene are associated with ovarian failure. Hum Genet. 2006;119:408–415. doi: 10.1007/s00439-006-0150-0. [DOI] [PubMed] [Google Scholar]
- Dreszer TR, Karolchik D, Zweig AS, Hinrichs AS, Raney BJ, Kuhn RM, Meyer LR, Wong M, Sloan CA, Rosenbloom KR, et al. The UCSC Genome Browser database: extensions and updates 2011. Nucleic Acids Res. 2012;40:D918–D923. doi: 10.1093/nar/gkr1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudding TE, Lawrence O, Winship I, Froyen G, Vandewalle J, Scott R, Shelling AN. Array comparative genomic hybridization for the detection of submicroscopic copy number variations of the X chromosome in women with premature ovarian failure. Hum Reprod. 2010;25:3159–3160. doi: 10.1093/humrep/deq284. Author reply 3160–3151. [DOI] [PubMed] [Google Scholar]
- Edgar R, Mazor Y, Rinon A, Blumenthal J, Golan Y, Buzhor E, Livnat I, Ben-Ari S, Lieder I, Shitrit A, et al. LifeMap Discovery: the embryonic development, stem cells, and regenerative medicine research portal. PLoS One. 2013;8:e66629. doi: 10.1371/journal.pone.0066629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, et al. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–D815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara T, Mimura K, Abe T, Shioi G, Seiki M, Sakamoto T. Deletion of the Mint3/Apba3 gene in mice abrogates macrophage functions and increases resistance to lipopolysaccharide-induced septic shock. J Biol Chem. 2011;286:32542–32551. doi: 10.1074/jbc.M111.271726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- Janse F, de With LM, Duran KJ, Kloosterman WP, Goverde AJ, Lambalk CB, Laven JS, Fauser BC, Giltay JC. Limited contribution of NR5A1 (SF-1) mutations in women with primary ovarian insufficiency (POI) Fertil Steril. 2012;97:141–146. doi: 10.1016/j.fertnstert.2011.10.032. e142. [DOI] [PubMed] [Google Scholar]
- Knauff EA, Blauw HM, Pearson PL, Kok K, Wijmenga C, Veldink JH, van den Berg LH, Bouchard P, Fauser BC, Franke L. Copy number variants on the X chromosome in women with primary ovarian insufficiency. Fertil Steril. 2011;95:1584–1588. doi: 10.1016/j.fertnstert.2011.01.018. e1581. [DOI] [PubMed] [Google Scholar]
- Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem. 2001;276:43958–43969. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- Koressaar T, Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics. 2007;23:1289–1291. doi: 10.1093/bioinformatics/btm091. [DOI] [PubMed] [Google Scholar]
- Kovanci E, Rohozinski J, Simpson JL, Heard MJ, Bishop CE, Carson SA. Growth differentiating factor-9 mutations may be associated with premature ovarian failure. Fertil Steril. 2007;87:143–146. doi: 10.1016/j.fertnstert.2006.05.079. [DOI] [PubMed] [Google Scholar]
- Laissue P, Christin-Maitre S, Touraine P, Kuttenn F, Ritvos O, Aittomaki K, Bourcigaux N, Jacquesson L, Bouchard P, Frydman R. Mutations and sequence variants in GDF9 and BMP15 in patients with premature ovarian failure. Eur J Endocrinol. 2006;154:739–744. doi: 10.1530/eje.1.02135. [DOI] [PubMed] [Google Scholar]
- Ledig S, Röpke A, Wieacker P. Copy number variants in premature ovarian failure and ovarian dysgenesis. Sex Dev. 2010;4:225–232. doi: 10.1159/000314958. [DOI] [PubMed] [Google Scholar]
- Lee BC, Avraham S, Imamoto A, Avraham HK. Identification of the nonreceptor tyrosine kinase MATK/CHK as an essential regulator of immune cells using Matk/CHK-deficient mice. Blood. 2006;108:904–907. doi: 10.1182/blood-2005-12-4885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Qiao Y, Di Q, Le X, Zhang L, Zhang X, Zhang C, Cheng J, Zong S, Koide SS, et al. Interaction of SH3P13 and DYDC1 protein: a germ cell component that regulates acrosome biogenesis during spermiogenesis. Eur J Cell Biol. 2009;88:509–520. doi: 10.1016/j.ejcb.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Liao C, Fu F, Yang X, Sun YM, Li DZ. Analysis of Chinese women with primary ovarian insufficiency by high resolution array-comparative genomic hybridization. Chin Med J (Engl) 2011;124:1739–1742. [PubMed] [Google Scholar]
- Lone M, Kungl T, Koper A, Bottenberg W, Kammerer R, Klein M, Sweeney ST, Auburn RP, O'Kane CJ, Prokop A. The nuclear protein Waharan is required for endosomal-lysosomal trafficking in Drosophila. J Cell Sci. 2010;123:2369–2374. doi: 10.1242/jcs.060582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire MM, Bowden W, Engel NJ, Ahn HW, Kovanci E, Rajkovic A. Genomic analysis using high-resolution single-nucleotide polymorphism arrays reveals novel microdeletions associated with premature ovarian failure. Fertil Steril. 2011;95:1595–1600. doi: 10.1016/j.fertnstert.2010.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Nef S, Schaad O, Stallings NR, Cederroth CR, Pitetti JL, Schaer G, Malki S, Dubois-Dauphin M, Boizet-Bonhoure B, Descombes P, et al. Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev Biol. 2005;287:361–377. doi: 10.1016/j.ydbio.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Nelson LM. Clinical practice. Primary ovarian insufficiency. N Engl J Med. 2009;360:606–614. doi: 10.1056/NEJMcp0808697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norling A, Hirschberg AL, Iwarsson E, Persson B, Wedell A, Barbaro M. Novel candidate genes for 46,XY gonadal dysgenesis identified by a customized 1 M array-CGH platform. Eur J Med Genet. 2013;12:661–668. doi: 10.1016/j.ejmg.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Persani L, Rossetti R, Cacciatore C. Genes involved in human premature ovarian failure. J Mol Endocrinol. 2010;45:257–279. doi: 10.1677/JME-10-0070. [DOI] [PubMed] [Google Scholar]
- Qin Y, Choi Y, Zhao H, Simpson JL, Chen ZJ, Rajkovic A. NOBOX homeobox mutation causes premature ovarian failure. Am J Hum Genet. 2007;81:576–581. doi: 10.1086/519496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilter CR, Karcanias AC, Bagga MR, Duncan S, Murray A, Conway GS, Sargent CA, Affara NA. Analysis of X chromosome genomic DNA sequence copy number variation associated with premature ovarian failure (POF) Hum Reprod. 2010;25:2139–2150. doi: 10.1093/humrep/deq158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringstad N, Nemoto Y, De Camilli P. The SH3p4/Sh3p8/SH3p13 protein family: binding partners for synaptojanin and dynamin via a Grb2-like Src homology 3 domain. Proc Natl Acad Sci USA. 1997;94:8569–8574. doi: 10.1073/pnas.94.16.8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Schouten JP, McElgunn CJ, Waaijer R, Zwijnenburg D, Diepvens F, Pals G. Relative quantification of 40 nucleic acid sequences by multiplex ligation-dependent probe amplification. Nucleic Acids Res. 2002;30:e57. doi: 10.1093/nar/gnf056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JL, Rajkovic A. Ovarian differentiation and gonadal failure. Am J Med Genet. 1999;89:186–200. doi: 10.1002/(sici)1096-8628(19991229)89:4<186::aid-ajmg3>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- Stern RF, Roberts RG, Mann K, Yau SC, Berg J, Ogilvie CM. Multiplex ligation-dependent probe amplification using a completely synthetic probe set. Biotechniques. 2004;37:399–405. doi: 10.2144/04373ST04. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. Primer3—new capabilities and interfaces. Nucleic Acids Res. 2012;40:e115. doi: 10.1093/nar/gks596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenberger MD, Hagerman RJ, Sherman SL, McConkie-Rosell A, Welt CK, Rebar RW, Corrigan EC, Simpson JL, Nelson LM. The FMR1 premutation and reproduction. Fertil Steril. 2007;87:456–465. doi: 10.1016/j.fertnstert.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Xu J, Kausalya PJ, Phua DC, Ali SM, Hossain Z, Hunziker W. Early embryonic lethality of mice lacking ZO-2, but Not ZO-3, reveals critical and nonredundant roles for individual zonula occludens proteins in mammalian development. Mol Cell Biol. 2008;28:1669–1678. doi: 10.1128/MCB.00891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C, Elvin JA, Lin YN, Hadsell LA, Wang J, DeMayo FJ, Matzuk MM. Regulation of growth differentiation factor 9 expression in oocytes in vivo: a key role of the E-box. Biol Reprod. 2006;74:999–1006. doi: 10.1095/biolreprod.105.050013. [DOI] [PubMed] [Google Scholar]
- Yang Z, Wu J. Mouse dynein axonemal intermediate chain 2: cloning and expression. DNA Cell Biol. 2008;27:479–488. doi: 10.1089/dna.2008.0752. [DOI] [PubMed] [Google Scholar]
- Zhao H, Qin Y, Kovanci E, Simpson JL, Chen ZJ, Rajkovic A. Analyses of GDF9 mutation in 100 Chinese women with premature ovarian failure. Fertil Steril. 2007;88:1474–1476. doi: 10.1016/j.fertnstert.2007.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Chen ZJ, Qin Y, Shi Y, Wang S, Choi Y, Simpson JL, Rajkovic A. Transcription factor FIGLA is mutated in patients with premature ovarian failure. Am J Hum Genet. 2008;82:1342–1348. doi: 10.1016/j.ajhg.2008.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.