ABSTRACT
Ribonuclease, RNase A family, 9 (RNASE9) is a ribonuclease A superfamily member that is expressed only in the epididymis. It is a small, secreted polypeptide, it lacks ribonuclease activity, and its function(s) is unknown. However, epididymis-specific expression suggests a role in sperm maturation. We generated Rnase9−/− mice to study RNASE9 function in vivo. We confirm that RNASE9 expression is restricted to the epididymis. Within the epididymis, RNASE9 is first detected in midcaput, persists through the distal caput and corpus, and wanes in the cauda. Rnase9−/− mice are born at the expected Mendelian ratio, have normal postnatal growth and development, and have no outwardly apparent phenotype. Spermatogenesis is normal, and Rnase9-null sperm are morphologically normal. Rnase9−/− males have normal fertility in unrestricted mating trials, and fertilization rates in in vitro fertilization assays are indistinguishable from wild-type mice. Visual observations coupled with analyses of sperm velocities shortly after swim out from the corpus shows that motility of Rnase9-null sperm is significantly impaired. However, no differences between wild-type and Rnase9-null sperm are detected by computer-assisted sperm analysis 10–90 min after sperm isolation from the corpus or cauda. Assessment of capacitation-dependent signaling pathways in Rnase9-null sperm showed that, while levels of tyrosine phosphorylation of sperm proteins were normal, there was decreased phosphorylation of protein kinase A substrates upon capacitation compared to wild-type mice. In conclusion, RNASE9 is dispensable for fertility, but the absence of RNASE9 during epididymal transit results in impaired sperm maturation.
Keywords: capacitation, epididymis, male reproductive tract, sperm, sperm maturation
INTRODUCTION
The epididymis plays a critical role in male reproduction. Spermatozoa enter the epididymis from the testes as immotile cells that are incapable of fertilization and are converted to motile and fertilization-competent cells in a complex process called maturation [1]. Sperm maturation is associated with a variety of events that include morphologic and structural alterations of sperm, remodeling of the sperm plasma membrane, and addition and/or processing of sperm surface proteins, to name a few [1–3]. These events are to a large extent triggered and influenced by the complex and changing lumenal microenvironment of the epididymis to which sperm are exposed during transit.
A great deal of work has been performed over the years to identify proteins expressed in the epididymis and to study their role(s) in sperm maturation and fertility [4–8]. In addition, proteomic studies have documented a variety of differences between immature and mature sperm [9, 10]. Among the many proteins identified in the epididymis is a subset that appears to be expressed only in the epididymis and, therefore, would likely be important in the primary physiologic function of the epididymis, that is, sperm maturation.
In 2003, Penttinen et al. [11] described five novel epididymis-specific genes that each had unique and highly regionalized expression in the mouse epididymis. Two of the genes they identified were designated Rnase9 (Gene ID: 328401) and Rnase10 (Gene ID: 75019). Both are members of the ribonuclease A superfamily and are expressed only in the epididymis beginning at Postnatal Day 17 [11]. Both genes encode small, secreted polypeptides that lack ribonuclease activity because key catalytic residues conserved in other ribonucleases are absent [12]. The two genes are tightly linked in chromosome region 14C1 with the Rnase9 gene being only 28 kb telomeric to the Rnase10 gene. Rnase9 transcripts were shown to be most highly expressed in the distal caput, whereas Rnase10 transcripts were most highly expressed in the initial segment as assessed by quantitative PCR (qPCR) and in situ hybridization.
Castella et al. [13] independently identified RNASE10 (also known as Train A) as a major constituent in epididymal fluid from the proximal caput in pigs. They purified the protein, cloned the porcine cDNA, and identified the mouse, rat, and human homologs. The physiological importance of Rnase10 in sperm maturation has been subsequently studied in gene-targeted mice [14]. This study showed that Rnase10 transcripts and RNASE10 protein were restricted to the initial segment, consistent with the findings of Penttinen et al. [11]. More importantly, the study of Rnase10−/− males showed that fertility was impaired as a result of the failure of sperm to pass the uterotubal junction that was associated with the loss of ADAM3 and ADAM6 expression on sperm during epididymal transit.
A study of RNASE9 expression in rats by Zhu et al. [15] and the study of Rnase9 transcripts in mice by Penttinen et al. [11] show that RNASE9 expression was epididymis-specific and androgen-dependent. Zhu et al. [15] also presented data indicating that principal cells in the proximal caput are the primary source of RNASE9 synthesis and RNASE9 is expressed on sperm from caput and corpus, but not the cauda. Two studies have been published on human RNASE9 expression but are somewhat inconsistent. In one study, Rnase9 transcripts were detected in many tissues, but RNASE9 protein expression in these tissues was not reported [16]. A second study reported that RNASE9 protein was detected only in epididymis among various tissues examined, but Rnase9 transcript expression in these tissues was not assessed [17]. Be that as it may, nothing is known about the function of RNASE9 in vivo. To study the potential role of RNASE9 in male reproduction, we generated Rnase9−/− mice and performed a detailed analysis of these mice.
MATERIALS AND METHODS
Ethics Statement
All the procedures involving vertebrate animals were reviewed and approved by the Institutional Animal Care and Use Committees at the Oklahoma Medical Research Foundation or the University of Massachusetts at Amherst and were performed in accordance with the eighth edition of the Guide for the Care and Use of Laboratory Animals (National Research Council 2011).
Antibodies
Rabbit polyclonal antibody to mouse RNASE9 was generated and characterized previously [18]. Western blots of a soluble protein fraction of wild-type epididymis using this antiserum detects a single 31-kDa band. Rabbit anti-mouse glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (G9545) and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit immunoglobulin G (IgG) (F0382) were purchased from Sigma-Aldrich. Anti-phosphotyrosine monoclonal antibody (mAb) (clone 4G10, mouse IgG2b-κ) was from Upstate Biotechnology. Phospho-protein kinase A (phospho-PKA) substrate-specific rabbit mAb (clone 100G7E) that detects peptides and proteins containing a phosphorylated serine or threonine residue with arginine at the −3 and −2 positions (corresponding to a PKA consensus phosphorylation sequence) was purchased from Cell Signaling. Affinity-purified polyclonal antibody raised in chicken against the V-ATPase E2 subunit was a kind gift from Dr. Sylvie Breton (Center for Systems Biology, Harvard Medical School) [19]. Horseradish peroxidase (HRP)-conjugated anti-mouse IgG and Cy3-conjugated goat anti-chicken IgG were from Jackson ImmunoResearch Laboratories, HRP-conjugated anti-rabbit IgG (PI-1000) was from Vector Labs or Amersham Biosciences, and rhodamine-conjugated Arachis hypogaea lectin (peanut agglutinin, PNA) (RL-1072) was from Vector Labs.
Gene Targeting
The Rnase9 gene on mouse chromosome 14 contains two exons separated by a 2222 bp intron. Exon 1 is 118 bp and exon 2 is 1071 bp in length. Exon 2 contains the entire Rnase9 open-reading frame and 3′-UTR [11]. Rnase9+/fl mice were generated at Xenogen Biosciences. Briefly, a targeting construct was designed for conditional deletion of a 1.9 kb segment that contains exon 2 and flanking intronic sequence (Fig. 1A). The 5′ homology arm (4.7 kb), conditional knockout region (1.9 kb), and the 3′ homology arm (2.9 kb) were generated by PCR using PrimeSTAR HS DNA Polymerase (Takara Bio) with 129SvJ genomic DNA as the template. All the fragments were confirmed by restriction digestion and end sequencing. These amplicons were cloned into the loxFtNwCD vector (Xenogen Biosciences) that also contains two loxP sites, one immediately 5′ of the conditional knockout region and a second loxP site 3′ to the Neo cassette. In addition, there are flippase recognition target sequences flanking the Neo cassette (for positive selection) and a diphtheria toxin A (DTA) expression cassette (for negative selection). The final vector was confirmed by both restriction mapping and end sequencing.
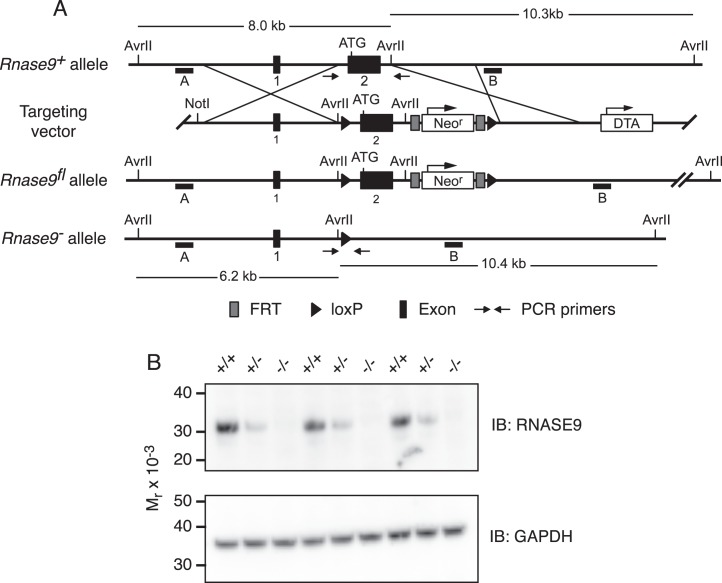
FIG. 1.
Rnase9 gene targeting scheme and confirmation of gene targeted deletion. A) Diagram of the wild-type Rnase9+ allele (top), targeting vector (top middle), Rnase9fl floxed allele (bottom middle), and Rnase9− null allele (bottom). The numbered black boxes indicate exons and the initiating ATG is shown near the 5′ end of exon 2. The 4.7-kb 5′ homology arm and the 2.9-kb 3′ homology arm are indicated. The positive selection neomycin resistance gene (Neor) and the diphtheria toxin A (DTA) negative selection cassette are shown with arrows indicating direction of transcription. Black triangles are loxP recombination sites, and dark gray boxes flanking the Neo cassette are flippase recognition target (FRT) recombination sites. The small horizontal black bars labeled A and B indicate the 5′ and 3′ external probes used for Southern blot analysis, respectively. AvrII restriction sites and predicted AvrII fragment sizes are shown. Small arrows indicate the locations of PCR primers used to detect the Rnase9+ and Rnase9− alleles. B) RNASE9 protein expression. Detergent extracts of mouse epididymis were prepared and analyzed by Western blot analysis using RNASE9 antisera as described in Materials and Methods.
Thirty micrograms of targeting vector was linearized with NotI and electroporated into approximately 107 XSV1 embryonic stem cells that were derived from 129S6/SvEvTac mice (Xenogen Biosciences). After selection in 200 μg/ml G418, 192 embryonic stem cell clones were subjected to primary screening by 3′ PCR. Twenty potential positive clones were identified, and four clones were expanded for Southern blot analysis. The 5′ and 3′ external Southern probes were generated by PCR using 129S6 genomic DNA as the template, cloned into the pCR4.0 vector (Invitrogen), and confirmed by sequencing. Based on the Southern blot analysis using the 5′, 3′, and Neo probes, two clones (A5 and C6) were confirmed for homologous recombination with single neo integration (data not shown). Blastocyst injections were performed using the A5 and C6 clones, and the blastocysts were implanted into pseudopregnant mice to generate chimeras. High-level chimeras were bred with 129S6 females (Taconic Farms) to generate heterozygote Rnase9+/fl mice.
Generation of Rnase9−/− Mice
To generate Rnase9+/− mice, Rnase9+/fl females derived from the A5 clone were mated with 129-Tg (Prm-cre)58Og/J males (Prm-cre) (stock no. 003328; The Jackson Laboratory). Prm-cre mice express Cre recombinase under the control of mouse protamine 1 promoter and mediate efficient recombination only in the male germline [20]. Male offspring from this cross, which have undergone Cre-mediated excision in the germline, were then mated with 129S6 females. Offspring from this cross were genotyped to detect the wild-type and null Rnase9 alleles, as well as the Cre transgene. Male and female Rnase9+/− mice lacking the Cre transgene were mated to generate mice with three Rnase9 genotypes (Rnase9+/+, Rnase9+/−, Rnase9−/−).
Genotyping
All the genotyping was performed by PCR using mouse genomic DNA as a template. Detection of the Rnase9fl allele was performed using the following primers: 5′-GAG GAG GCT GGA TCA TTT CTG GAC A-3′ and 5′-GAA GAG CTT CCT AAT GTT GAC TCT AAA ATA AGT G-3′. Amplification of the wild-type allele yields a 297 bp amplicon, whereas amplification of the floxed allele yields a 388 bp amplicon. Detection of the Rnase9-null allele was performed using the following primers: 5′-GAG GAG GCT GGA TCA TTT CTG GAC A-3′ and 5′-GTA GAA TTT TTG CAG CTC ACT GTC AAG G-3′. Amplification of the wild-type allele yields a 2058 bp amplicon, whereas amplification of the null allele yields a 254 bp amplicon. Detection of the Prm-Cre allele was performed using the following primers: 5′-CAT CTG CCA CCA GCC AGC TAT-3′ and 5′-GGT CCA GCC ACC AGC TTG CAT-3′. Amplification of the Prm-Cre allele yields a 200 bp amplicon.
PCR Analysis
Total RNA was isolated using TRIzol (Invitrogen) and the RNeasy kit (Qiagen) according to the manufacturers' instructions. Complementary DNA was prepared using the iScript cDNA synthesis kit (Bio-Rad) or SuperScript III first-strand synthesis system (Invitrogen).
Rnase9 transcripts in various tissues were detected using semiquantitative RT-PCR. The following PCR primers were used: Rnase9 (5′-GCA AGA GTC TGG TGA AGA GT-3′ and 5′-AGT CCT GAG TTC AGT GTT GC-3′) as described by Penttinen et al. [11] and Gapdh (5′-TCA ACG GCA CAG TCA AGG-3′ and 5′-ACT CCA CGA CAT ACT CAG C-3′). The expected amplicons for Rnase9 and Gapdh are 323 bp and 126 bp, respectively.
Real-time quantitative PCR was performed using SYBR Green PCR master mix (Applied Biosystems) and the CFX96 Real-Time PCR Detection System (Bio-Rad). The relative fold change in transcription was determined using the comparative threshold cycle (CT) method with the Actb (β-actin) and Gapdh housekeeping genes as internal controls. The following qPCR primers were used: Rnase10 (5′-GCC TGC TGT GAG TTG GTG TA-3′ and 5′-AAG GTC CAA CTG TGG CAA AC-3′); Gapdh (5′-TCA ACG GCA CAG TCA AGG-3′ and 5′-ACT CCA CGA CAT ACT CAG C-3′); and Actb (5′-TGT TAC CAA CTG GGA CGA CA-3′ and 5′-GGG GTG TTG AAG GTC TCA AA-3′).
Histology
Testes and epididymides from sexually mature, age-matched Rnase9+/+ and Rnase9−/− mice were fixed in Bouin fixative for 24 h. Fixed tissues were then wax embedded, and 5 μm sections were collected onto slides and stained with hematoxylin-eosin.
Sperm Cytology
Sperm were collected by teasing apart the cauda epididymis into 100 μl phosphate-buffered saline (PBS) maintained at 37°C. The sperm were incubated at 37°C and 5% CO2 for 15 min then washed twice with PBS. Sperm were fixed in 3.8% paraformaldehyde, washed twice with PBS, and mounted in PermaFluor aqueous mounting media (Thermo Scientific). Images were captured using a Nikon E800M microscope equipped with Nikon ACT-1 v.2.62 software.
Immunoblotting
For RNASE9 immunoblotting, tissues from sexually mature mice were homogenized in ice-cold buffer: 100 mM NaCl, 20 mM 3-[N-morpholino] propanesulfonic acid, pH 7.5, 0.25 mM Na3VO4, 1 μg/ml leupeptin, 1 μg/ml antipain, and 1 mM benzamidine. The homogenates were centrifuged (800 × g, 10 min) to obtain a postnuclear supernatant that was then centrifuged (100 000 × g, 1 h, 4°C). The 100 000 × g supernatant (soluble fraction) was collected and stored at −80°C until use. Samples were electrophoresed, electroblotted onto polyvinylidene fluoride membranes, and membranes were blocked as described previously [18]. The membrane was probed with RNASE9 antiserum diluted 1:30 000 in Tris-buffered saline (0.1 M NaCl, 20 mM Tris-HCl) and 0.1% Tween-20 (TBS-T) for 1 h followed by 0.025 μg/ml anti-rabbit HRP in TBS-T for 1 h. After washing, bound secondary antibody was detected using enhanced chemiluminescence (ECL). In some cases, membranes were stripped using OneMinute Stripping Buffer (GM Bioscience) and evaluated by incubating in anti-rabbit HRP with subsequent ECL detection to confirm complete stripping of the initial antibody reagents. The membrane was then stripped again and reprobed using rabbit anti-mouse GAPDH (1:40 000) followed by anti-rabbit HRP as described above.
Phosphotyrosine and phospho-PKA substrate immunoblots of sperm was performed as previously described [21] with some modifications. Briefly, mouse sperm were collected into Toyoda-Yokoyama-Hoshi (TYH) medium [22] and adjusted to 1–2 × 107 cells/ml. The TYH medium used was composed of 119.3 mM NaCl, 4.7 mM KCl, 1.71 mM CaCl2, 1.2 mM KH2PO4, 1.2 mM MgSO4, 5.56 mM glucose, 25.1 mM NaHCO3, 0.51 mM Na pyruvate, 10 μg/ml gentamycin, 0.0006% phenol red, and 5 mg/ml fatty acid- and globulin-free bovine serum albumin (A0281; Sigma-Aldrich) at pH 7.4 when equilibrated with 5% CO2. Sperm samples were incubated in TYH medium at 37°C and 5% CO2 either for 10 min (noncapacitating condition) or 90 min (capacitating condition). Sperm were then washed with PBS, then lysed with Laemmli sample buffer without β-mercaptoethanol, and boiled for 5 min. After centrifugation, β-mercaptoethanol was added to the supernatants to a 5% final concentration and boiled again for 5 min. Protein extracts equivalent to 1–2 × 106 sperm per lane were subjected to SDS-PAGE, and proteins were transferred to polyvinylidene fluoride membranes. Membranes were blocked with 20% fish scale gelatin in TBS-T and then probed with anti-phosphotyrosine mAb (0.1 μg/ml) or anti-phospho-PKA substrate-specific mAb (0.2 μg/ml) followed by HRP-conjugated anti-mouse or anti-rabbit IgG (0.1 μg/ml) and detected using ECL.
Immunofluorescence Microscopy
Sperm from the caput, corpus, or cauda epididymis were collected by mincing epididymal tissue into 250 μl warm PBS. The sperm were incubated at 37°C for 15 min then washed three times with PBS. Sperm were counted, and 50 000 sperm were placed onto slides (Fisherfinest Superfrost), air dried for 90 min, and then washed once with PBS. Slides were fixed with 4% paraformaldehyde in PBS, pH 7.4, for 10 min, permeabilized with PBS and 0.5% Triton X-100 for 5 min, then blocked in PBS, 0.5% bovine serum albumin, and 0.1% Tween-20 overnight. Slides were rinsed with PBS and 0.1% Tween-20 (PBS-T) and then incubated for 1 h with RNASE9 antiserum or preimmune serum diluted 1:1500 in PBS-T. After washing with PBS-T, slides were incubated for 1 h with FITC-goat anti-rabbit IgG (2.75 μg/ml) and rhodamine-PNA (10 μg/ml) in PBS-T. Slides were washed with PBS-T and mounted with Vectashield hard set mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Labs). Sperm was observed in a Nikon E800M microscope.
For immunofluorescence staining of epididymis, the right and left epididymides and vasa deferentia were resected en bloc and placed in freshly prepared 4% paraformaldehyde in PBS, pH 7.4, for 24 h. Tissues were washed in PBS (3 × 15 min) and permeated with 15% sucrose in PBS for 6 h. Tissues were then placed in 30% sucrose in PBS for 18 h, transferred into optimal cutting temperature compound (Sakura Finetek) for 6 h, and then frozen in liquid N2-cooled isopentane. Five micrometer thick frozen sections were placed onto slides (Fisherbrand Colorfrost Plus), air-dried, fixed with 4% paraformaldehyde in PBS, and washed with PBS (3 × 5 min). Sections were quenched with 0.1 M glycine, washed with PBS, and then blocked with DAKO antibody diluent (Dako North America, Inc.) and 0.1% Tween-20 for 1 h. Slides were incubated for 1 h with RNASE9 serum, preimmune serum (1:3000), or chicken anti-mouse V-ATPase (1:300) in DAKO antibody diluent containing 0.1% Tween-20. Slides were washed in PBS-T and then incubated for 1 h with secondary antibodies in DAKO antibody diluent containing 0.1% Tween-20. For RNASE9 staining, FITC-goat anti-rabbit IgG was used at 2.5 μg/ml, while DyLight 594-donkey anti-chicken IgG was used at 5 μg/ml for V-ATPase staining. Sections were washed in PBS-T (3 × 10 min), fixed with 4% paraformaldehyde in PBS for 5 min, washed in PBS-T (3 × 5 min) and then a final time with PBS-T and 0.1% TO-PRO-3 (Invitrogen), and mounted with Vectashield mounting medium containing DAPI. Images were captured using a Nikon E800M microscope equipped with Nikon ACT-1 v.2.62 software. Individual channel images were captured at the same filter and camera settings and later merged in Adobe Photoshop CS5 using the linear dodge blending mode.
Sperm Motility Analysis
Epididymides from either Rnase9+/+ or Rnase9−/− mice were dissected, and then the corpus and cauda regions were placed in separate tubes containing TYH medium. Multiple incisions were made in the tissue, and sperm gently squeezed out into the medium, and incubated in TYH medium at 37°C and 5% CO2 for either 10 min (noncapacitating) or 90 min (capacitating). Sperm suspensions were then loaded into Leja 2 chamber slides (SC 100-01-02-A; Spectrum Technologies), placed on a slide warmer (37°C), and cell motility analyzed using a CEROS Semen Analyzer (Hamilton-Thorne Research). The parameters used for video acquisition were the following: frames acquired, 90; frame rate, 60 Hz; minimum cell size, 4 pixels; static head size, 0.13–2.43; static head intensity, 0.10–1.52; and static head elongation, 5–100. Populations of motile spermatozoa were sorted based on five criteria in the CASAnova software described by Goodson et al. [23].
Corpus Sperm Velocity Analysis
Sperm were collected from the corpus epididymis by cutting the corpus epididymis into 1 mm sections in 100 μl PBS maintained at 37°C. Sperm were allowed to swim out for 5 min. Four microliters of the sperm swim out suspension was placed on a slide, and a 12-mm circular coverslip (12-545-8; Fisher) was placed on top. The slide was then placed in an incubation chamber coupled to a Zeiss 710 inverted microscope maintained at 37°C and 5% CO2. Movies were captured using a Zeiss AxioCam HR camera and exported using the Zeiss Zen Blue v.2011 software as uncompressed AVI files. Sperm straight line velocities (VSL) were determined using the NIS-Element-AR v.3.2 (Nikon) software tracking utility.
In Vitro Fertilization Assays
Six to eight wk-old NSA (CF-1) females (Harlan Laboratories) were superovulated by intraperitoneal injection of 5 units equine chorionic gonadotropin (367222; EMD Millipore) followed by intraperitoneal injection of 5 units of human chorionic gonadotropin (hCG) (C0163; Sigma-Aldrich) 48 h later. Both injections were given in the evening prior to the start of the dark cycle. Thirteen hours after hCG injection, egg clutches were collected from the swollen ampullae and then washed and stored in EmbryoMax human tubal fluid (HTF) (MR-070-D; EMD Millipore) until insemination.
To collect sperm, the cauda epididymis and vas deferens were removed and placed into a 250 μl drop of HTF. The epididymis were gently teased apart and the contents of the vas deferens extruded. The sperm were allowed to swim out into the HTF for 15 min. Sperm were counted, and then 100 μl insemination drops were set up containing 500 000 sperm/ml (50 000 sperm per drop) in HTF under mineral oil. Finally, cumulus-intact eggs were added to each insemination drop. Four hours after insemination, the oocytes were washed once with HTF, transferred to a 50 μl drop of HTF, and then incubated overnight. The numbers of 2-cell embryos were counted 24 h after insemination. All the incubations were in a 37°C incubator with 5% CO2.
RESULTS
Characterization of Rnase9−/− Mice
Male and female Rnase9+/− mice that lack the Cre transgene were generated as described in Materials and Methods and then mated to generate mice with three Rnase9 genotypes (Rnase9+/+, Rnase9+/−, Rnase9−/−). To determine the effect of disruption of the Rnase9 gene on RNASE9 protein levels, immunoblot analysis was performed on epididymal extracts from three sexually mature Rnase9+/+, Rnase9+/−, and Rnase9−/− males (Fig. 1B). This showed that RNASE9 protein was significantly reduced in Rnase9+/− mice compared to wild-type mice and was not detectable in Rnase9−/− mice.
Growth and Development
Rnase9+/− mice appeared normal. The reproductive performance of Rnase9+/− mating pairs was normal. The size of the first two litters from ten Rnase9+/− × Rnase9+/− mating pairs was 5.85 ± 0.32 pups/litter (mean ± SEM, n = 20 litters), compared to that of wild-type mating pairs bred contemporaneously (5.91 ± 0.31 pups/litter [mean ± SEM, n = 11 litters]), consistent with our previous experience for the 129S6 strain [24]. The genotypes of offspring from the first two litters from these Rnase9+/− × Rnase9+/− mating pairs were consistent with Mendelian inheritance (26% wild type, 48% heterozygotes, 25% homozygotes, 117 pups, n = 20 litters), indicating that Rnase9−/− mice develop normally in utero. Like Rnase9+/− mice, Rnase9−/− male and females have no outwardly apparent phenotype.
Spermatogenesis
Histological examination of sexually mature males showed that the testes appear normal as do the caput and cauda epididymides that are filled with spermatozoa (Fig. 2A). In addition, the weights of the testes and epididymides of Rnase9−/− and wild-type males are indistinguishable (Supplemental Table S1; all the Supplemental Data is available online at www.biolreprod.org), and Rnase9-null sperm are cytologically normal (Fig. 2B). Taken together, these data indicate that spermatogenesis is normal in Rnase9−/− males.
FIG. 2.
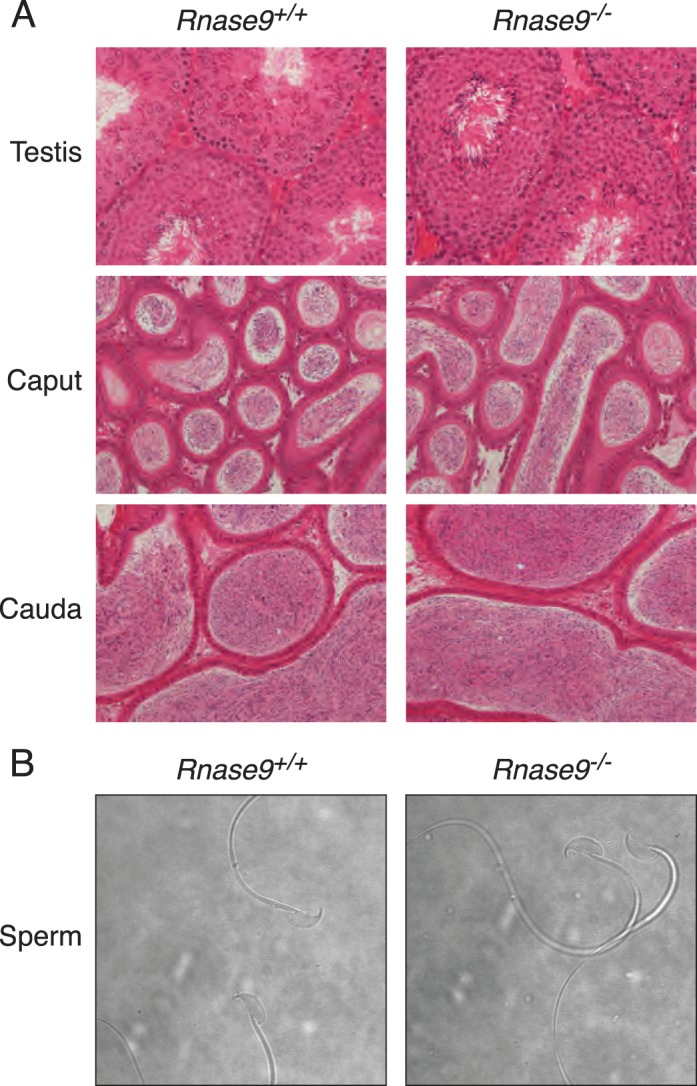
Tissue histology and sperm cytology. A) Hematoxylin-eosin stained sections of tissues. Testes (top panels), caput (middle panels), and cauda epididymides (bottom panels). Images are representative of three different sexually mature, age-matched Rnase9+/+ (left panels) and Rnase9−/− (right panels) mice. Original magnification ×20. B) Phase contrast images of caudal epididymal sperm. Images are representative of three different sexually mature, age-matched Rnase9+/+ (left panel) and Rnase9−/− (right panel) mice. Original magnification ×60.
Reproductive Performance
To examine reproductive performance, sexually mature virgin Rnase9−/− females were mated with Rnase9−/− males. Females were examined for copulatory plugs each morning to determine if and when copulation occurred and the size of the firstborn litter was recorded. We observed no difference in plugging latency between Rnase9−/− mating pairs (3.3 ± 0.8 days, n = 12) and wild-type control mating pairs (2.0 ± 0.3 days, n = 7) bred contemporaneously (mean ± SEM). In addition, we observed no difference in the litter size between Rnase9−/− mating pairs (5.76 ± 0.4, n = 17 litters) and control wild-type mating pairs (5.91 ± 0.3 pups/litter, n = 11 litters) bred contemporaneously (mean ± SEM). These data demonstrate that Rnase9−/− males and females are fertile.
RNASE9 Expression in Wild-Type Mice
Penttinen et al. [11] reported that Rnase9 transcripts were detected only in the epididymis among the various mouse tissues analyzed in their study. To assess RNASE9 protein expression, we performed immunoblot analysis of soluble protein fractions from lung, heart, spleen, liver, kidney, testis, epididymis, uterus, and ovary as described in Materials and Methods. Among these tissues, RNASE9 was detected only in the epididymis (Fig. 3).
FIG. 3.
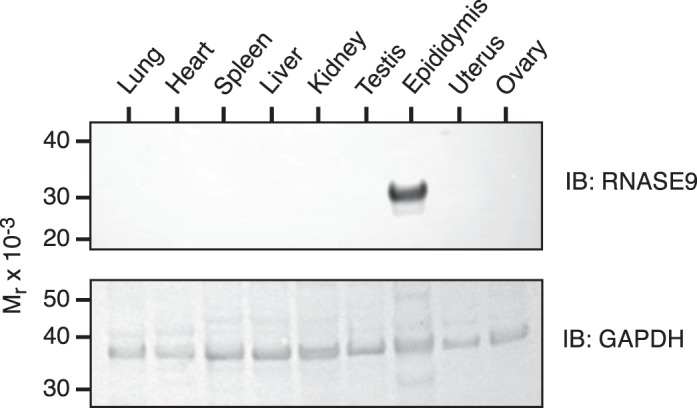
Expression of RNASE9 in mouse tissues. Soluble protein fractions of various tissues were prepared as described in Materials and Methods and subjected to SDS-PAGE followed by immunoblotting with RNASE9 antiserum (top panel). The membrane was then stripped and reprobed using rabbit anti-mouse GAPDH (bottom panel).
Penttinen et al. [11] also reported that within the epididymis Rnase9 transcripts were detected beginning in the initial segment, but Rnase9 transcripts were most abundant in the caput, progressively dropped in the corpus and cauda, and were not detected in the vas deferens. We also examined the relative abundance of RNASE9 protein in soluble protein fractions from various epididymal segments as defined by Cornwall [3] and illustrated in Figure 4A. Note that the overall size and gross anatomy of the epididymis from Rnase9+/+ and Rnase9−/− mice are indistinguishable. Immunoblot analysis of Rnase9+/+ mice detected RNASE9 in the distal caput, corpus, and cauda, but not in the initial segment/proximal caput and midcaput (Fig. 4B).
FIG. 4.
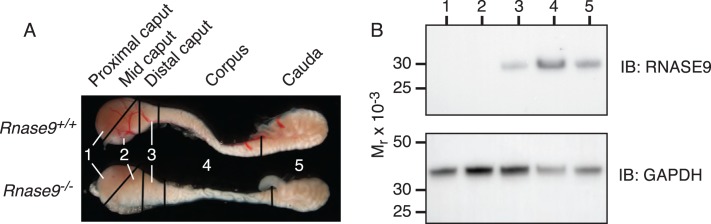
Expression of RNASE9 in mouse epididymis. A) Epididymides were dissected from adult Rnase9+/+ and Rnase9−/− mice and cut into five segments: 1) initial segment/proximal caput; 2) midcaput; 3) distal caput; 4) corpus; and 5) cauda. B) Soluble protein fractions of these segments were prepared as described in Materials and Methods and subjected to SDS-PAGE followed by immunoblotting with RNASE9 antiserum (top panel). The membrane was then stripped and reprobed using rabbit anti-mouse GAPDH (bottom panel).
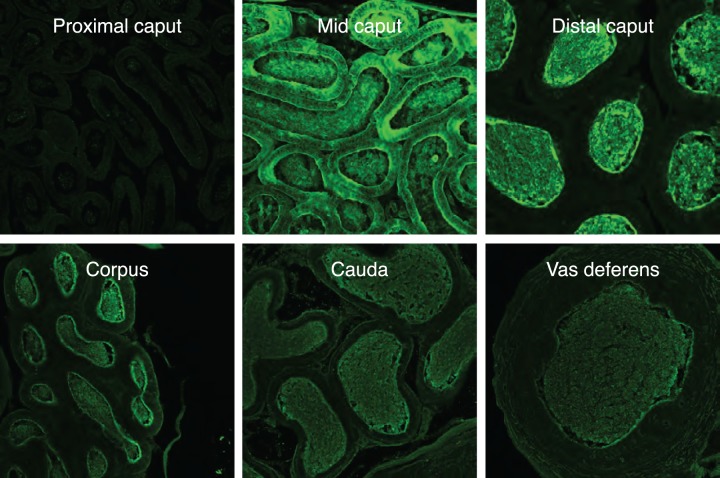
To more precisely examine the localization of RNASE9 in the epididymis, immunofluorescence microscopy was performed. To validate our methodology, we first stained epididymis from Rnase9+/+ and Rnase9−/− mice using RNASE9 antiserum or preimmune serum. This analysis showed strong staining of Rnase9+/+ epididymis using RNASE9 antiserum but no staining using preimmune serum. In addition, there was no staining of Rnase9−/− epididymis with either RNASE9 antiserum or preimmune serum (Supplemental Fig. S1). In the initial segment/proximal caput, there was no RNASE9 staining (Fig. 5). In the midcaput, RNASE9 staining was detected in the epididymal epithelium and lumen, indicating synthesis and secretion of RNASE9 in this segment. In the distal caput, corpus, cauda, and vas deferens, RNASE9 staining was observed only in the lumen.
FIG. 5.
Immunofluorescence microscopy. Epididymal segments as defined in Figure 4A were sectioned and subjected to immunofluorescence microscopy using RNASE9 antiserum (green) as described in Materials and Methods. Original magnification ×20.
The loss of RNASE9 staining in the more distal segments of the epididymis may be due to uptake and degradation by epithelial cells. To address this possibility, we performed dual staining of the cauda for RNASE9 and for V-ATPase, a marker for clear cells. We observed RNASE9 staining in the cytoplasm of some, but not all, V-ATPase-positive clear cells. Interestingly, V-ATPase-negative epithelial cells were also negative for RNASE9 (Fig. 6).
FIG. 6.
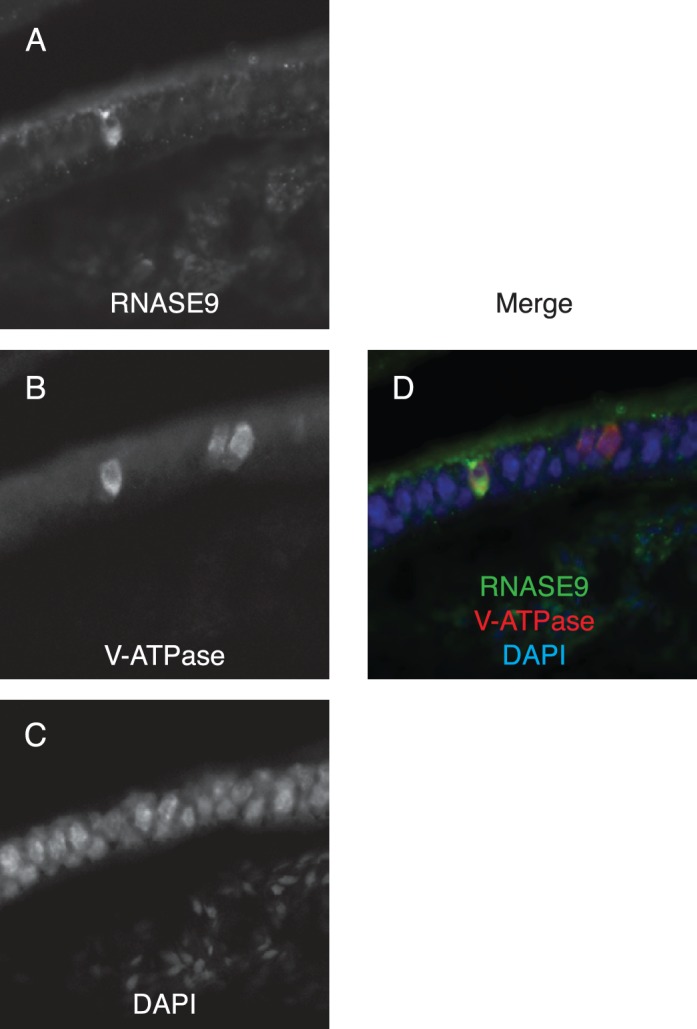
Immunofluorescence microscopy. Cauda epididymis was sectioned and subjected to immunofluorescence staining: (A) RNASE9, (B) V-ATPase, (C) DAPI, and (D) merged. Original magnification ×60.
We also performed immunofluorescence microscopy of sperm. However, we did not detect RNASE9 staining of washed sperm isolated from the caput, corpus, or the cauda (data not shown). During the isolation of sperm for the latter analysis, we consistently observed that media surrounding the Rnase9−/− corpus epididymal segments was less cloudy compared to wild type ≤5 min after incision of the tissue (Supplemental Fig. S2). This difference was not observed when isolating caudal sperm. This suggested that sperm may have defective motility and prompted a detailed assessment of the motility of Rnase9-null sperm.
Computer-Assisted Sperm Analysis
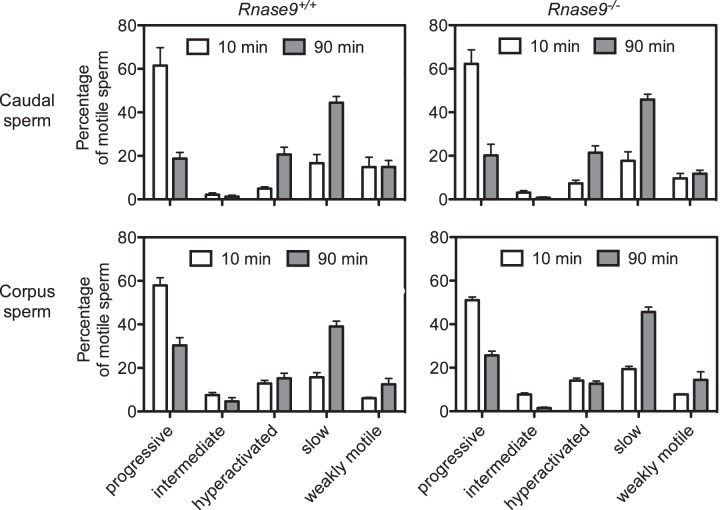
The motility of caudal and corpus sperm from Rnase9+/+ and Rnase9−/− mice was assessed as described in Materials and Methods. Motility parameters were measured, and motile sperm were classified as progressive, intermediate, hyperactivated, slow, or weakly motile using CASAnova software as previously described and validated [23]. For caudal sperm, our analysis showed no significant difference between Rnase9+/+ and Rnase9−/− mice in the percentage of progressive, intermediate, hyperactivated, slow, or weakly motile sperm after 10 or 90 min of capacitation (Fig. 7 and Supplemental Table S2). For both Rnase9+/+ and Rnase9−/− mice, we observed a drop in progressively motile sperm accompanied by a rise in hyperactivated and slowly motile sperm between 10 and 90 min (Fig. 7 and Supplemental Table S3), consistent with the data presented by Goodson et al. [23] for several mouse strains. In addition, all the motility parameters, including average path velocity, VSL, curvilinear velocity, amplitude of lateral head displacement, and beat cross frequency were indistinguishable between the two groups (Supplemental Table S4).
FIG. 7.
Computer-assisted sperm analysis. The motility of caudal (top panels) and corpus sperm (bottom panels) from age-matched Rnase9+/+ (left panels) and Rnase9−/− (right panels) mice was assessed using computer-assisted sperm analysis (CASA) after 10 and 90 min of capacitation. Motile sperm were classified as progressive, intermediate, hyperactivated, slow, or weakly motile using CASAnova software. Results are expressed as mean ± SEM. The number of animals examined were as follows: caudal sperm Rnase9+/+, n = 5; caudal sperm Rnase9−/−, n = 5; corpus sperm Rnase9+/+, n = 6; and corpus sperm Rnase9−/−, n = 6. Statistical significance between groups was determined by using two-tailed Student t-tests with unequal sample variance using PRISM software version 6.03 (GraphPad). Corrections for multiple comparisons were performed using the Holm-Šídák method with an α ≤ 0.05. A summary of the statistical comparisons between Rnase9+/+ versus Rnase9−/− mice and 10 min and 90 min are presented in Supplemental Tables S2 and S3, respectively.
A similar analysis of swim out sperm from the corpus was performed. There was no statistical difference between Rnase9+/+ and Rnase9−/− mice in the percentage of progressive, intermediate, hyperactivated, or slow sperm after 10 or 90 min of capacitation (Fig. 7 and Supplemental Table S2). We observed a statistical difference in the percentage of weakly motile sperm after 10 min (P = 0.005), but not after 90 min. Like caudal sperm, there was a drop in progressively motile sperm accompanied by a rise in slowly motile sperm between 10 and 90 min pattern for both Rnase9+/+ and Rnase9−/− mice (Fig. 7 and Supplemental Table S3). However, in contrast to caudal sperm, there was no significant increase in hyperactivated sperm. We also observed a significant decrease in intermediate motile sperm in Rnase9−/− mice, but not in Rnase9+/+ mice.
We also employed an alternative approach to visualize and quantitate the motility of corpus sperm at times shorter that 10 min. Swim out sperm were collected from groups of age-matched Rnase9+/+ and Rnase9−/− mice (n = 3 per group), video images were recorded as described in Materials and Methods. The raw videos show that Rnase9-null sperm have lower overall sperm motility compared to wild-type sperm (Supplemental Fig. S3 and Supplemental Videos S1 and S2). Straight line velocity was determined for five sperm from three fields of view for each of three mice of each genotype. We observed that the VSL of Rnase9-null sperm (134 ± 215 pixels/sec, mean ± SEM) was significantly lower than wild-type sperm (266 ± 27, pixels/sec) (P = 0.012) (Fig. 8).
FIG. 8.
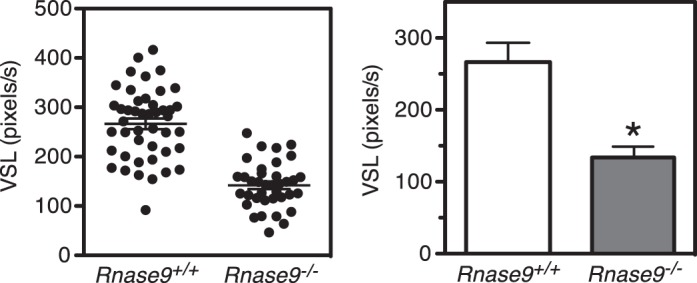
Corpus sperm velocities. The straight line velocity (VSL) of corpus sperm from age-matched Rnase9+/+ and Rnase9−/− mice (n = 3 per group) was quantitated from video images as described in Materials and Methods. Results are expressed as mean ± SEM of five sperm from three fields of view for each of three mice of each genotype. Statistical differences between groups were tested using unpaired, two-tailed t-tests with equal sample variance and an α ≤ 0.05. *P = 0.012.
Capacitation-Associated Protein Phosphorylation
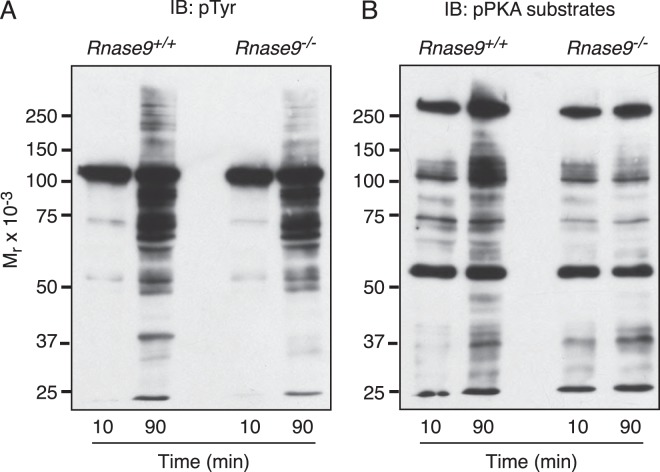
Capacitation of sperm in the female reproductive tract is the key prerequisite for the acquisition of the ability of sperm to fertilize eggs. This process is correlated with PKA activation, phosphorylation of PKA substrates, and subsequent stimulation of protein-tyrosine phosphorylation. To assess the signaling events during capacitation, we performed phosphotyrosine and phospho-PKA substrate immunoblots of capacitated Rnase9-null sperm proteins. We observed no significant differences in the capacitation-associated increase in protein-tyrosine phosphorylation between Rnase9-null and wild-type sperm. However, we observed that Rnase9-null sperm displayed a lack of response in phosphorylation of PKA substrates upon capacitation. (Fig. 9).
FIG. 9.
Capacitation-associated protein phosphorylation. Caudal sperm were collected from Rnase9+/+ and Rnase9−/− mice. Following capacitation, sperm was subjected to SDS-PAGE under reducing conditions followed by immunoblotting with (A) anti-phosphotyrosine (pTyr) mAb or (B) phospho-PKA (pPKA) substrate-specific mAb. The analysis shown is representative of three independent analyses.
In Vitro Fertilization Assays
In vitro fertilization assays were preformed to assess the fertilizing ability of Rnase9-null sperm. We observed no difference in fertilization rates between wild-type (61.3% ± 1.8%) and Rnase9-null sperm (61.7% ± 3.9%) (n = 3, P = 0.89).
Rnase10 qPCR
The mouse Rnase9 gene is located only 28 kb telomeric to the Rnase10 gene in chromosome region 14C1. We therefore performed qPCR analysis to assess if disruption of the Rnase9 gene affected Rnase10 gene expression. We observed no difference in the localization of Rnase10 transcripts or in transcript levels between Rnase9−/− and wild-type mice (Fig. 10).
FIG. 10.
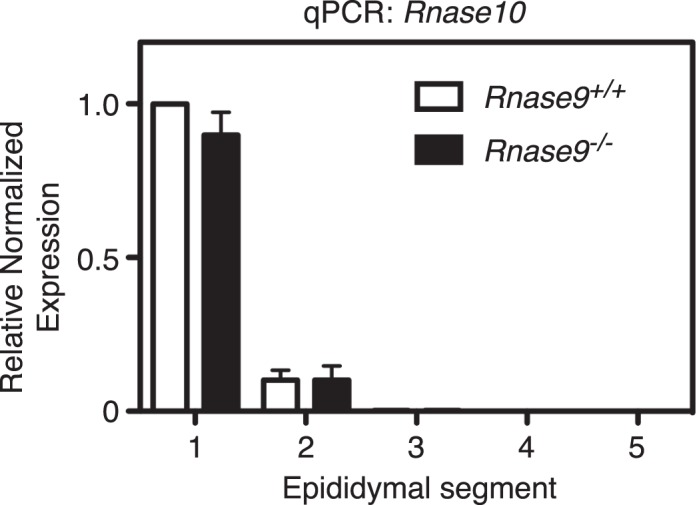
Rnase10 qPCR analysis. Epididymides were dissected from adult Rnase9+/+ and Rnase9−/− mice and cut into five segments as shown in Figure 4A. The relative fold change in transcription was determined using qPCR with Actb and Gapdh genes as internal controls. The results are expressed as mean ± SEM (n = 3 per group). Statistical differences between groups were tested using unpaired, two-tailed t-tests with equal sample variance and an α ≤ 0.05. There was no statistical difference between the groups.
DISCUSSION
We generated Rnase9−/− mice to examine the physiological role of RNASE9 in male reproduction. We first examined RNASE9 expression in wild-type mice using a combination of immunoblotting and immunofluorescence microscopy using a well-characterized rabbit antiserum [18]. The antiserum detects a single 31-kDa protein in the epididymis but not in any other tissue examined. Immunoblot analysis of soluble protein fractions detects RNASE9 in the distal caput, corpus, and cauda. Only soluble protein fractions were analyzed because previous work showed that RNASE9 was only detected in the soluble, and not the membrane, fraction of whole epididymis [18]. Using immunofluorescence microscopy, we observe no RNASE9 staining in the initial segment/proximal caput. In the midcaput, we observe very strong staining of the epithelium and somewhat less intense lumenal staining. RNASE9 staining persists in the distal caput and corpus and wanes in the cauda and vas deferens. However, after the midcaput, RNASE9 staining is restricted to the lumen and is absent from epithelium. The failure to detect RNASE9 in the midcaput by immunoblotting, despite strong immunofluorescence staining, may in part be due to the fact that only the soluble protein fractions were analyzed. We observed RNASE9 staining in the cytoplasm of some, but not all, V-ATPase-positive clear cells but no specific staining of V-ATPase-negative epithelial cells. This suggests that the loss of RNASE9 staining in the distal epididymis may be due to uptake and degradation primarily by clear cells. Our findings are consistent with the findings of Penttinen et al. [11] and Zhu et al. [15] that Rnase9 transcripts are most highly expressed in the distal caput and with the study by Zhu et al. [15] of RNASE9 protein expression in the rat. Zhu et al. detected RNASE9 on rat sperm isolated from caput and corpus, but not in caudal sperm, using immunofluorescence microscopy. However, we did not detect RNASE9 staining of sperm isolated from the caput, corpus, of cauda in our study. This discordance could be due to differences between the rat and mouse or to a technical difference between the two studies. The observation in the rat that RNASE9 staining is lost on caudal sperm coupled with our failure to detect RNASE9 staining of epididymal sperm in the mouse suggests that the interaction of RNASE9 with sperm, if present, is weak and/or rapidly reversible.
Rnase9+/− mice appear normal and when interbred, yield litters of normal size with a Mendelian distribution of the targeted mutation. Both Rnase9−/− males and females have normal postnatal growth and development and have no outwardly apparent phenotype. These observations are not unexpected given that RNASE9 is expressed in the epididymis in males and is not expressed in the female. In Rnase9−/− males, testes and epididymis weights and histology are indistinguishable from wild-type males and Rnase9-null sperm are morphologically normal. We observed that Rnase9−/− males have normal fertility in unrestricted mating trials and the fertilization rates in cumulus-intact in vitro fertilization assays for wild-type and Rnase9-null sperm are indistinguishable.
Spontaneous motility is one of the hallmarks of sperm maturation. Therefore, we also carefully examined the motility of Rnase9-null sperm. Visual observations of swim out drops during isolation of corpus sperm routinely showed obvious difference in the cloudiness of the drops indicating slower or delayed release of sperm from the epididymis. Analysis of the sperm velocities shortly after release (≈1 min) from the corpus showed that the motility of Rnase9-null sperm was significantly impaired. However, using computer-assisted sperm analysis 10 or 90 min after sperm isolation, we observed no difference between Rnase9+/+ and Rnase9−/− mice in the percentage of progressive, intermediate, hyperactivated, slow, or weakly motile sperm. Thus, the impairment in corpus sperm motility of Rnase9-null sperm is transient and normalizes within 10 min after release of sperm from the epididymis. Assessment of capacitation-dependent signaling in Rnase9-null sperm showed that levels of tyrosine phosphorylation of sperm proteins were normal. However, we observed there was decreased phosphorylation of PKA substrates upon capacitation compared to wild-type mice.
In a previous study, we reported that Tpst2−/− males are sterile, while Tpst1−/− have normal fertility suggesting that tyrosine sulfation of unidentified proteins(s) plays a crucial role in male fertility [25]. Subsequently, we reported data that an unidentified 31-kDa protein in sperm/epididymal lysate was undersulfated in Tpst2−/− mice [26]. Later, we identified this protein as RNASE9 [18]. In the latter study, we demonstrated conclusively that RNASE9 was tyrosine-sulfated in wild-type and Tpst1−/−, but not in Tpst2−/−, mice despite comparable expression of RNASE9. Thus, TPST-2 expression is required for sulfation of RNASE9 in vivo, suggesting that lack of sulfation of RNASE9 may contribute mechanistically to the sterility of Tpst2−/− males [25]. Our analysis of Rnase9−/− mice shows that RNASE9 expression is not required for fertility and argues strongly that lack of sulfation of RNASE9 does not contribute significantly to the sterility of Tpst2−/− males.
RNASE9 and RNASE10 are inactive ribonuclease A superfamily members that are expressed only in the epididymis, but are expressed in distinct and highly restricted patterns [11]. A recent study has shown that Rnase10−/− males have impaired fertility [14]. In contrast, disruption of the Rnase9 gene has no apparent impact on fertility. Nevertheless, their homology coupled with the fact that RNASE10 is expressed only in the initial segment while RNASE9 is expressed more distally, suggest that they function cooperatively in vivo. It would be interesting to examine this question in future studies. However, generating a double mutant by interbreeding Rnase9 and Rnase10 knockouts is impractical given that the genes are only 28 kb apart. A different strategy targeting both genes simultaneously would be needed to generate a double knockout.
In summary, we conclude that RNASE9 is not required for fertility either in vitro or in vivo. Nevertheless, lack of RNASE9 has a demonstrable, albeit subtle, role in sperm maturation.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. Sylvie Breton (Center for Systems Biology, Harvard Medical School) for providing the anti-V-ATPase antibody, Dr. Indra Adrianto (Arthritis and Clinical Immunology Program, Oklahoma Medical Research Foundation) for help with the statistical analyses, and Dr. Pablo Visconti (University of Massachusetts-Amherst) for his careful review of this manuscript.
Footnotes
Supported by National Institute of Health Grants RO1HD056022 (to K.L.M.) and RO1HD044044 (to A.M.S.), institutional funds from the Oklahoma Medical Research Foundation (to K.L.M.), and the University of Massachusetts-Amherst Faculty Research Grant/Healey Endowment grant (to A.M.S.). Presented in part at the 38th Annual Meeting of the American Society of Andrology, 13–16 April 2013, San Antonio, Texas.
REFERENCES
- Robaire B, Hinton BT, Orgebin-Crist MC. The Epididymis In Neill JD. (ed.), Knobil and Neill's Physiology of Reproduction, vol. 1, 3rd ed Amsterdam: Elsevier; 2006. 1071 1148 [Google Scholar]
- Yeung C-H, Cooper TG. Acquisition and development of sperm motility upon maturation in the epididymis In Robaire B, Hinton BT. (eds.), The Epididymis: From Molecules to Clinical Practice New York: Kluwer Academic/Plenum Publishers; 2002. 417 434 [Google Scholar]
- Cornwall GA. New insights into epididymal biology and function. Hum Reprod Update. 2009;15:213–227. doi: 10.1093/humupd/dmn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston DS, Wooters J, Kopf GS, Qiu Y, Roberts KP. Analysis of the human sperm proteome. Ann N Y Acad Sci. 2005;1061:190–202. doi: 10.1196/annals.1336.021. [DOI] [PubMed] [Google Scholar]
- Johnston DS, Jelinsky SA, Bang HJ, DiCandeloro P, Wilson E, Kopf GS, Turner TT. The mouse epididymal transcriptome: transcriptional profiling of segmental gene expression in the epididymis. Biol Reprod. 2005;73:404–413. doi: 10.1095/biolreprod.105.039719. [DOI] [PubMed] [Google Scholar]
- Jelinsky SA, Turner TT, Bang HJ, Finger JN, Solarz MK, Wilson E, Brown EL, Kopf GS, Johnston DS. The rat epididymal transcriptome: comparison of segmental gene expression in the rat and mouse epididymides. Biol Reprod. 2007;76:561–570. doi: 10.1095/biolreprod.106.057323. [DOI] [PubMed] [Google Scholar]
- Dacheux JL, Belghazi M, Lanson Y, Dacheux F. Human epididymal secretome and proteome. Mol Cell Endocrinol. 2006;250:36–42. doi: 10.1016/j.mce.2005.12.022. [DOI] [PubMed] [Google Scholar]
- Hsia N, Cornwall GA. DNA microarray analysis of region-specific gene expression in the mouse epididymis. Biol Reprod. 2004;70:448–457. doi: 10.1095/biolreprod.103.021493. [DOI] [PubMed] [Google Scholar]
- Aitken RJ, Nixon B, Lin M, Koppers AJ, Lee YH, Baker MA. Proteomic changes in mammalian spermatozoa during epididymal maturation. Asian J Androl. 2007;9:554–564. doi: 10.1111/j.1745-7262.2007.00280.x. [DOI] [PubMed] [Google Scholar]
- Baker MA, Witherdin R, Hetherington L, Cunningham-Smith K, Aitken RJ. Identification of post-translational modifications that occur during sperm maturation using difference in two-dimensional gel electrophoresis. Proteomics. 2005;5:1003–1012. doi: 10.1002/pmic.200401100. [DOI] [PubMed] [Google Scholar]
- Penttinen J, Pujianto DA, Sipilä P, Huhtaniemi I, Poutanen M. Discovery in silico and characterization in vitro of novel genes exclusively expressed in the mouse epididymis. Mol Endocrinol. 2003;17:2138–2151. doi: 10.1210/me.2003-0008. [DOI] [PubMed] [Google Scholar]
- Cho S, Beintema JJ, Zhang J. The ribonuclease A superfamily of mammals and birds: identifying new members and tracing evolutionary histories. Genomics. 2005;85:208–220. doi: 10.1016/j.ygeno.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Castella S, Fouchécourt S, Teixeira-Gomes AP, Vinh J, Belghazi M, Dacheux F, Dacheux JL. Identification of a member of a new RNase a family specifically secreted by epididymal caput epithelium. Biol Reprod. 2004;70:319–328. doi: 10.1095/biolreprod.103.022459. [DOI] [PubMed] [Google Scholar]
- Krutskikh A, Poliandri A, Cabrera-Sharp V, Dacheux JL, Poutanen M, Huhtaniemi I. Epididymal protein Rnase10 is required for post-testicular sperm maturation and male fertility. FASEB J. 2012;26:4198–4209. doi: 10.1096/fj.12-205211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu CF, Liu Q, Zhang L, Yuan HX, Zhen W, Zhang JS, Chen ZJ, Hall SH, French FS, Zhang YL. RNase9, an androgen-dependent member of the RNase A family, is specifically expressed in the rat epididymis. Biol Reprod. 2007;76:63–73. doi: 10.1095/biolreprod.106.054635. [DOI] [PubMed] [Google Scholar]
- Liu J, Li J, Wang H, Zhang C, Li N, Lin Y. Liu J, Wang W. Cloning, expression and location of RNase9 in human epididymis. BMC Res Notes. 2008;1:111. doi: 10.1186/1756-0500-1-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng GZ, Li JY, Li F, Wang HY, Shi GX. Human ribonuclease 9, a member of ribonuclease A superfamily, specifically expressed in epididymis, is a novel sperm-binding protein. Asian J Androl. 2009;11:240–251. doi: 10.1038/aja.2008.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffhines AJ, Jen CH, Leary JA, Moore KL. Tyrosylprotein sulfotransferase-2 expression is required for sulfation of RNase9 and Mfge8 in vivo. J Biol Chem. 2009;284:3096–3105. doi: 10.1074/jbc.M808434200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietrement C, Sun-Wada GH, Silva ND, McKee M, Marshansky V, Brown D, Futai M, Breton S. Distinct expression patterns of different subunit isoforms of the V-ATPase in the rat epididymis. Biol Reprod. 2006;74:185–194. doi: 10.1095/biolreprod.105.043752. [DOI] [PubMed] [Google Scholar]
- O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc Natl Acad Sci U S A. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapf D, Arcelay E, Wertheimer EV, Sanjay A, Pilder SH, Salicioni AM, Visconti PE. Inhibition of Ser/Thr phosphatases induces capacitation-associated signaling in the presence of Src kinase inhibitors. J Biol Chem. 2010;285:7977–7985. doi: 10.1074/jbc.M109.085845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda Y, Yokoyama M, Hoshi T. Studies on fertilization of mouse eggs in vitro. I. In vitro fertilization of eggs by fresh epididymal sperm. Jpn J Anim Reprod. 1971;16:147–151. [Google Scholar]
- Goodson SG, Zhang Z, Tsuruta JK, Wang W, O'Brien DA. Classification of mouse sperm motility patterns using an automated multiclass support vector machines model. Biol Reprod. 2011;84:1207–1215. doi: 10.1095/biolreprod.110.088989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang YB, Crawley JTB, Aston CE, Moore KL. Reduced body weight and increased postimplantation fetal death in tyrosylprotein sulfotransferase-1-deficient mice. J Biol Chem. 2002;277:23781–23787. doi: 10.1074/jbc.M202420200. [DOI] [PubMed] [Google Scholar]
- Borghei A, Ouyang YB, Westmuckett AD, Marcello MR, Landel CP, Evans JP, Moore KL. Targeted disruption of tyrosylprotein sulfotransferase-2, an enzyme that catalyzes post-translational protein tyrosine O-sulfation, causes male infertility. J Biol Chem. 2006;281:9423–9431. doi: 10.1074/jbc.M513768200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffhines AJ, Damoc E, Bridges KG, Leary JA, Moore KL. Detection and purification of tyrosine-sulfated proteins using a novel anti-sulfotyrosine monoclonal antibody. J Biol Chem. 2006;281:37877–37887. doi: 10.1074/jbc.M609398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.