Abstract
Objective: Several polymorphisms in DNA repair genes have been extensively studied in association with various human cancers, including laryngeal cancer. The present study aimed to investigate the association between polymorphisms of the XRCC1 gene and laryngeal cancer in a Chinese population. Methods: Five polymorphisms of the XRCC1 gene (rs3213403, rs1799778, rs1001581, rs3213282, and rs3810378) were genotyped by TaqMan in 234 patients with larynx cancer and 230 age- and sex-matched controls without cancer. Results: The rs3213403, rs1799778, and rs3213282 polymorphisms of XRCC1 were associated with larynx cancer. Haplotype analysis indicated that CCA (odds ratio [OR], 5.707; 95% confidence interval [CI], 3.277–9.938; p<0.001), TGG (OR, 4.344; 95% CI, 2.804–6.732; p<0.001), ACA (OR, 1.615; 95% CI, 1.159–2.250; p=0.004), and GCG (OR, 1.702; 95% CI, 1.164–2.489; p=0.005) were associated with an increased risk for larynx cancer, respectively. However, TGA (OR, 0.518; 95% CI, 0.398–0.673; p<0.001) and ACC (OR, 0.314; 95% CI, 0.215–0.457; p<0.001) were associated with a decreased risk for larynx cancer. Conclusions: The results indicated that XRCC1 genetic polymorphisms were associated with larynx cancer in a Chinese population.
Introduction
Larynx cancer is a complex disease resulting form interaction among several environmental factors, such as smoking, high alcohol consumption, and occupational and environmental exposure to carcinogens (Parkin et al., 2002; Pawlowska et al., 2009; Kupisz et al., 2010) and genetic polymorphisms (Wang et al., 2013; Ziao et al., 2013). Accumulated evidence suggested that DNA damage, if not repaired or misrepaired, may result in genomic instability, cancer transformation, and/or cell death (Futreal et al., 1994; Khanna and Jackson, 2001; O'Driscoll and Jeggo, 2006).
The x-ray repair cross-complementing group 1 gene (XRCC1) encodes a protein that interacts with nicked DNA and participates with poly-adenosine diphosphate-ribose polymerase, DNA ligase III, and DNA polymerase B to repair single-strand DNA breaks (Thompson and West, 2000). The XRCC1 gene is polymorphic, and dozens of variants have been identified, including several nonsynonymous single-nucleotide polymorphisms (SNPs) in the coding region. In the past decade, a huge amount of studies have investigated the association between XRCC1 polymorphism and cancer, but much of the research has focused on SNPs rather than haplotypes, which better characterize the common patterns of variation in a population (Crawford and Nickerson, 2005).
In the present study, we established haplotypes of the XRCC1 gene, consisting of 5 SNPs (rs3213403, rs1799778, rs1001581, rs3213282, and rs3810378), and assessed the association between these haplotypes and larynx cancer.
Material and Methods
Patients
Blood samples were obtained from 234 patients with laryngeal cancer from the Department of Otolaryngology-Head and Neck Surgery, Jinling Hospital, Nanjing Clinical Medical College, the Second Military Medical University in 2005–2013 and 230 cancer-free age- and sex-matched controls. The patients ranged in age from 40 to 80 years (mean age±standard deviation, 62.2±8.0 years). There were 112 cases of grade 1, 105 cases of grade 2, and 17 cases of grade 3 in total. According to TNM staging, there were 55 cases of stage I, 38 cases of stage II, 88 cases of stage III, 43 cases of stage IVA, and 11 cases of stage IVB. The study was approved by the Bioethics Committee of the Second Military Medical University, and each patient gave written consent.
Genotyping
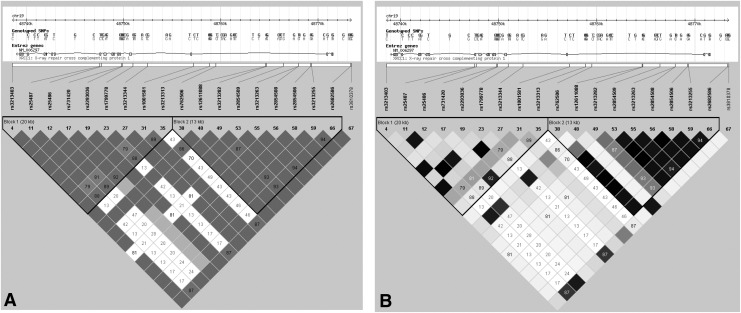
There are 953 SNPs for the human XRCC1 gene listed in the National Center for Biotechnology Information SNP database (http://www.ncbi.nlm.nih.gov/SNP). We also screened the data for the Tag SNPs on the International HapMap Project website (http://www.hapmap.org/). Using the Haploview 4.2 software and the HapMap phrase II database, we obtained five tagging SNPs (rs3213403, rs1799778, rs3810378, rs1001581, and rs3213282) for Han Chinese using minor allele frequency ≥0.10 and linkage disequilibrium patterns with r2 ≥0.8 as a cutoff. As shown in Figure 1, these five SNPs were located in two haplotype blocks.
FIG. 1.
Genetic variation at human XRCC1 gene. By using Haploview 4.2 software and the HapMap phrase II database, 19 genotyped single-nucleotide polymorphisms (SNPs) from Chinese Han were scanned. Linkage disequilibrium (LD) blocks across the locus in Chinese Han were derived by solid spline method in Haploview 4.2. (A) LD value shown: |D′|×100; |D′| color scheme: |D′|=0: white; 0<r2<1: shades of gray, |D′|=1: black; (B) LD value shown: r2×100; r2 color scheme: r2=0: white; 0<r2<1: shades of gray; r2=1: black.
Genomic DNA was extracted from the peripheral blood leukocytes using a DNA extraction kit (Beijing Bioteke Co. Ltd, Beijing, China). Genotyping was confirmed by TaqMan method, as described previously (Xiang et al., 2009).
Statistical analysis
For each polymorphism, departure of the genotype distribution from that expected from Hardy-Weinberg equilibrium was assessed using the standard χ2 test or Fisher exact test. Genotype frequencies in cases and controls were compared by χ2 tests. The genotype-specific risks were estimated as odds ratios (ORs). In all cases, wild-type genotype served as a reference group. On the basis of the genotype data of the genetic variations, we performed linkage disequilibrium (LD) analysis and haplotype-based case–control analysis, using SHEsis software (http://analysis2.bio-x.cn/myAnalysis.php) (Shi and He, 2005; Li et al., 2009). In the haplotype-based case–control analysis, haplotypes with a frequency <0.03 were excluded. Statistical significance was established at p<0.05.
Results
Table 1 shows the distribution of the genotypes and alleles of these five SNPs. The genotype distribution of each SNP did not show significant difference from the Hardy-Weinberg equilibrium values (data not shown). For total participants, the genotype and the allele distribution of rs3213282 differed significantly between the patients with laryngeal cancer and the control participants (p=0.018 and 0.012, respectively). The GG genotype and G allele were more common in the patients with laryngeal cancer than in the control participants. For rs3213403 and rs1799778, although distributions of genotypes did not significantly differ between the two groups in distributions of genotypes, the allele frequencies were significantly different between the groups. However, the genotype and the allele distributions of rs1001581 and rs3810378 did not differ between the patients with laryngeal cancer and the control participants.
Table 1.
Genotype Distribution of XRCC1 Tag Single-Nucleotide Polymorphisms Between Patients with Laryngeal Cancer and Controls
| SNPs | Genotype and allele | Patients with laryngeal cancer (n=234) | Controls (n=230) | p value |
|---|---|---|---|---|
| rs3213403 | AA | 139 (0.594) | 160 (0.696) | 0.073 |
| AG | 86 (0.368) | 63 (0.274) | ||
| GG | 9 (0.038) | 7 (0.030) | ||
| A | 364 (0.778) | 383 (0.833) | 0.035 | |
| G | 104 (0.222) | 77 (0.167) | ||
| rs1799778 | AA | 21 (0.090) | 15 (0.065) | 0.071 |
| AC | 89 (0.380) | 69 (0.300) | ||
| CC | 124 (0.530) | 146 (0.635) | ||
| A | 131 (0.280) | 99 (0.215) | 0.022 | |
| C | 337 (0.720) | 361 (0.785) | ||
| rs1001581 | CC | 16 (0.068) | 11 (0.048) | 0.580 |
| CT | 81 (0.346) | 82 (0.357) | ||
| TT | 137 (0.585) | 137 (0.596) | ||
| C | 113 (0.241) | 104 (0.226) | 0.638 | |
| T | 355 (0.759) | 356 (0.774) | ||
| rs3213282 | CC | 24 (0.103) | 31 (0.135) | 0.018 |
| CG | 105 (0.449) | 125 (0.543) | ||
| GG | 105 (0.449) | 74 (0.322) | ||
| C | 153 (0.327) | 187 (0.407) | 0.012 | |
| G | 315 (0.673) | 273 (0.593) | ||
| rs3810378 | CC | 13 (0.056) | 15 (0.065) | 0.506 |
| CG | 85 (0.363) | 72 (0.313) | ||
| GG | 136 (0.581) | 143 (0.662) | ||
| C | 111 (0.237) | 102 (0.222) | 0.576 | |
| G | 357 (0.763) | 358 (0.778) |
SNP, single-nucleotide polymorphism.
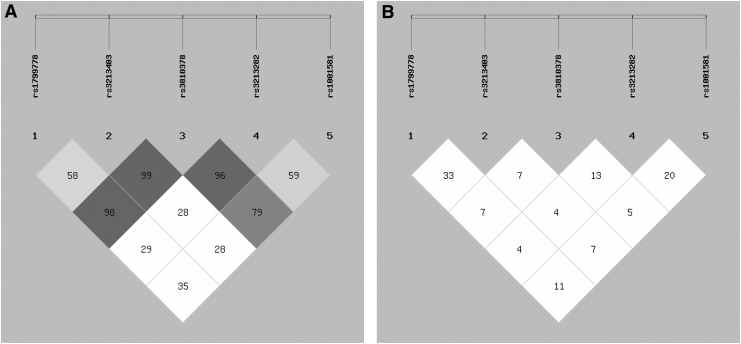
Figure 2 shows patterns of LD in the XRCC1 gene, with their |D′| and r2 values. All 5 SNPs are located in 2 haplotype blocks. In the haplotype-based case–control analysis, for block one, haplotypes were established through the use of rs3213403, rs1799778, and rs3810378; for block 2, haplotypes were established through the use of rs3810378, rs1001581, and rs3213282. As shown in Table 2, CCA (OR, 5.707; 95% CI, 3.277–9.938; p<0.001), TGG (OR, 4.344; 95% CI, 2.804–6.732; p<0.001), ACA (OR, 1.615; 95% CI, 1.159–2.250; p=0.004), and GCG (OR, 1.702; 95% CI, 1.164–2.489; p=0.005) were associated with increased risk for laryngeal cancer. However, TGA (OR, 0.518; 95% CI, 0.398–0.673; p<0.001) and ACC (OR, 0.314; 95% CI, 0.215–0.457; p<0.001) were associated with decreased risk for laryngeal cancer.
FIG. 2.
The patterns of linkage disequilibrium in the XRCC1 gene, with their | D'| (A) and r2 values (B).
Table 2.
Distribution of Haplotype
| Haplotype | Patients with laryngeal cancer | Controls | OR (95% CI) | p value |
|---|---|---|---|---|
| Block 1 | ||||
| C C A* | 79.63 (0.170) | 15.95 (0.035) | 5.707 (3.277–9.938) | <0.001 |
| C G A* | 33.34 (0.071) | 39.65 (0.086) | 0.813 (0.503–1.313) | 0.396 |
| T C A* | 31.37 (0.067) | 37.42 (0.081) | 0.811 (0.495–1.328) | 0.404 |
| T G A* | 219.66 (0.469) | 289.99 (0.630) | 0.518 (0.398–0.673) | <0.001 |
| T G G* | 103.97 (0.222) | 28.37 (0.062) | 4.344 (2.804–6.732) | <0.001 |
| Block 2 | ||||
| A C A* | 106.97 (0.229) | 71.99 (0.157) | 1.615 (1.159–2.250) | 0.004 |
| A C C* | 43.92 (0.094) | 115.01 (0.250) | 0.314 (0.215–0.457) | <0.001 |
| A G C* | 211.19 (0.451) | 195.98 (0.426) | 1.126 (0.868–1.460) | 0.372 |
| G G A* | 22.11 (0.047) | 26.99 (0.059) | 0.803 (0.451–1.430) | 0.455 |
| G G C* | 79.77 (0.170) | 50.01 (0.109) | 1.702 (1.164–2.489) | 0.005 |
OR, odds ratio; CI, confidence interval.
Discussion
In the present study, we found XRCC1 gene haplotypes were significantly associated with laryngeal cancer risk in a Chinese population.
Several studies have reported that the genes involved in DNA repair and in the maintenance of genome integrity play a crucial role in protecting against mutations that lead to cancer. SNPs have been identified in several DNA repair genes, such as XRCC1, XRCC2, and RAD51, and several previous studies indicated that these genes were associated with cancer risk, including laryngeal cancer. Cancer of the larynx is a head and neck cancer. It includes tumors of the nasal cavities, paranasal sinuses, oral cavity, nasopharynx, oropharynx, hypopharynx, and larynx. The contribution of polymorphisms of various DNA repair genes in development of head and neck carcinoma is controversial. Although Krupa et al. did not find an association of XRCC1 polymorphism with laryngeal cancer in a Polish population (Krupa et al., 2011), Mahjabeen and colleagues' finding suggests that XRCC1 is associated with increased risk for head and neck cancer in a Pakistani population (Mahjabeen et al., 2013). In a Chinese population, Yang et al. also found that XRCC1 genetic polymorphism was associated with the increased risk for laryngeal cancer (Yang et al., 2008).
In our study, we genotyped five SNPs in XRCC1 in Chinese participants and assessed the association between XRCC1 and laryngeal cancer using a haplotype-based case–control analysis. The rs3213282 significantly differed between patients with larynx cancer and control participants, indicating that the risk for laryngeal cancer is increased in participants with the G allele of rs3213282. Morris and Kaplan found that for genes with multiple susceptibilities, analysis based on haplotypes has advantages over analysis based on individual SNPs, particularly when LD between SNPs is weak (Morris and Kaplan, 2002). Consequently, in the present study, we successfully established haplotypes for the XRCC1 gene from the different combination of the five SNPs. The frequency of CCA, TGG, ACA, and GCG was associated with increased risk for laryngeal cancer. However, both TGA and ACC were associated with decreased risk for laryngeal cancer.
The present study was limited by the relatively small sample size. This may have led to weak statistical significance and wide CIs when ORs were estimated.
In conclusion, the present results indicate that laryngeal cancer is associated with XRCC1 gene polymorphisms. The CCA, TGG, ACA, and GCG haplotype appears to be a useful genetic marker, and the TGA and ACC haplotypes might be protective factors against laryngeal cancer in Chinese people.
Author Disclosure Statement
No competing financial interests exist.
References
- Crawford DC, Nickerson DA. (2005) Definition and clinical importance of haplotypes. Annu Rev Med 56:303–320 [DOI] [PubMed] [Google Scholar]
- Futreal PA, Liu Q, Shattuck-Eidens D, et al. (1994) BRCA1 mutations in primary breast and ovarian carcinomas. Science 266:120–122 [DOI] [PubMed] [Google Scholar]
- Khanna KK, Jackson SP. (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 27:247–254 [DOI] [PubMed] [Google Scholar]
- Krupa R, Kasznicki J, Gajęcka M, et al. (2011) Polymorphisms of the DNA repair genes XRCC1 and ERCC4 are not associated with smoking- and drinking-dependent larynx cancer in a Polish population. Exp Oncol 33:55–56 [PubMed] [Google Scholar]
- Kupisz K, Stepulak A, Zdunek M, et al. (2010) Preliminary results of prognostic significance of proliferating cell nuclear antigen expression in advanced primary larynx carcinomas and lymph node metastases. Arch Med Sci 6:65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Z, He Z, et al. (2009) A partition-ligation-combination-subdivision EM algorithm for haplotype inference with multiallelic markers: update of the SHEsis (http://analysis.bio-x.cn). Cell Res 19:519–523 [DOI] [PubMed]
- Mahjabeen I, Baig RM, Masood N, et al. (2013) Genetic variations in XRCC1 gene in sporadic head and neck cancer (HNC) patients. Pathol Oncol Res. 19:183–188 [DOI] [PubMed] [Google Scholar]
- Morris RW, Kaplan NL. (2002) On the advantage of haplotype analysis in the presence of multiple disease susceptibility alleles. Genet Epidemiol 23:221–233 [DOI] [PubMed] [Google Scholar]
- O'Driscoll M, Jeggo PA. (2006) The role of double-strand break repair-insights from human genetics. Nat Rev Genet 7:45–54 (2006) [DOI] [PubMed] [Google Scholar]
- Parkin DM, Whelan SL, Ferlay J, et al. (2002) Cancer incidence in five continents. Vol. VIII IARC Scientic Publication No. 155. Lyon, France: International Agency for Research on Cancer [Google Scholar]
- Pawlowska E, Janik-Papis K, Rydzanicz M, et al. (2009) The Cys326 allele of the 8-oxoguanine DNA N-glycosylase 1 gene as a risk factor in smoking- and drinking-associated larynx cancer. Tohoku J Exp Med 219:269–275 [DOI] [PubMed] [Google Scholar]
- Shi YY, He L. (2005) SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res 15:97–98 [DOI] [PubMed] [Google Scholar]
- Thompson LH, West MG. (2000) Xrcc1 keeps DNA from getting stranded. Mutat Res 459:1–18 [DOI] [PubMed] [Google Scholar]
- Wang J, Jin X, Wang H, et al. (2013) The -308G/A polymorphism of the tumor necrosis factor-alpha gene is associated with the risk of upper aerodigestive tract cancer: a meta-analysis. Tohoku J Exp Med 229:245–254 [DOI] [PubMed] [Google Scholar]
- Xie Xiang, Ma YT, Fu ZY, et al. (2009) Haplotype analysis of the CYP8A1 gene associated with myocardial infarction. Clin Appl Thromb Hemost 15:574–580 [DOI] [PubMed] [Google Scholar]
- Xiao H, Li M, Tian L, et al. (2013) Quantitative assessment of the association between GSTM1 null genotype and laryngeal cancer risk. Eur Arch Otorhinolaryngol 270: 615–622 [DOI] [PubMed] [Google Scholar]
- Yang Y, Tian H, Zhang ZJ. (2008) Association of the XRCC1 and hOGG1 polymorphisms with the risk of laryngeal carcinoma. Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 25:211–213 [PubMed] [Google Scholar]