Background: Autoantibodies (anti-Jo-1) to cytoplasmic histidyl-tRNA synthetase (HisRS) are associated with inflammatory myositis.
Results: HisRS and two splice variants (SVs) cross-react with anti-Jo-1 antibodies and are secreted; at least one SV transcript is up-regulated in dermatomyositis.
Conclusion: Secreted HisRS SVs contain major epitopes of anti-Jo-1 autoantibodies.
Significance: Secreted HisRS and its SVs share epitopes for potential extracellular anti-Jo-1 antibody binding.
Keywords: Aminoacyl-tRNA Synthetase, Autoimmune Disease, Epitope Mapping, Immunology, Secretion, Anti-Jo-1 Autoantibody, Dermatomyositis, Myositis
Abstract
Inflammatory and debilitating myositis and interstitial lung disease are commonly associated with autoantibodies (anti-Jo-1 antibodies) to cytoplasmic histidyl-tRNA synthetase (HisRS). Anti-Jo-1 antibodies from different disease-afflicted patients react mostly with spatially separated epitopes in the three-dimensional structure of human HisRS. We noted that two HisRS splice variants (SVs) include these spatially separated regions, but each SV lacks the HisRS catalytic domain. Despite the large deletions, the two SVs cross-react with a substantial population of anti-Jo-l antibodies from myositis patients. Moreover, expression of at least one of the SVs is up-regulated in dermatomyositis patients, and cell-based experiments show that both SVs and HisRS can be secreted. We suggest that, in patients with inflammatory myositis, anti-Jo-1 antibodies may have extracellular activity.
Introduction
Idiopathic inflammatory myositis (IIM)2 is an autoimmune disease that is strongly associated with autoantibodies and is frequently associated with interstitial lung disease (ILD) (1). Myositis-specific antibodies (MSAs) and myositis-associated antibodies define two distinct groups (2). MSAs are directed against histidyl-, threonyl-, alanyl-, glycyl-, isoleucyl-, and asparaginyl-tRNA synthetases. Interestingly, in any single patient, these MSAs are mutually exclusive (1).
Among the myositis-specific anti-aaRS Abs, those directed against cytoplasmic histidyl-tRNA synthetase (HisRS) are the most common (3) and were first described >30 years ago (4). Approximately 25–30% of patients with dermatomyositis (DM) or polymyositis have anti-HisRS Abs (3). In contrast, autoantibodies directed against the other five aaRSs collectively constitute a much smaller percentage (3–5). Anti-HisRS Abs, which were historically designated as anti-Jo-1 Abs, bind to sites that are spread across the entire protein and include both linear and conformational epitopes (6, 7).
Among the various epitopes, the N-terminal portion of HisRS is especially prominent (6–8). In ELISA, recombinant HisRS(1–60) (constituting the first 60 amino acids (aa)) reacted with anti-Jo-1 Abs, whereas a truncated HisRS lacking the first 60 aa failed to react (7). Interestingly, the first 60 aa of HisRS are encoded by the first two exons of the mRNA of HARS and are absent from HisRSs of prokaryotes and lower eukaryotes. As expected, anti-Jo-1 Abs do not react with Escherichia coli HisRS (9). According to our structural analysis, this small domain (designated as a WHEP domain) forms a helical coiled-coil structure (9). Other work showed that HisRS(1–48) induced migration of CD4+ and CD8+ lymphocytes, IL-2-activated monocytes, and immature dendritic cells. In contrast, HisRS(61–509), which lacks the first 60 aa, failed to stimulate these inflammation-related cell migration events (8). Other in vivo studies in mice suggest that HisRS has an etiological relationship to the disease (10).
Despite the wealth of data on the association of HisRS with anti-Jo-1 Ab in IIM/ILD, the cross-reactivity of splice variants (SVs) with anti-Jo-1 Abs is undefined. In this in mind, we previously identified HisRSΔCD, a natural HisRS SV that has an internal deletion that ablates the entire catalytic domain (CD) and joins the N-terminal WHEP domain (1–60 residues) to the C-terminal anticodon-binding domain (ABD) (9). The result is a change in both quaternary and tertiary structures. Thus, HisRSΔCD is a monomer (HisRS is a homodimer) shaped like a dumbbell-like structure, where a flexible linker joins its two domains and the ABD has an altered conformation. Although the epitopes were not mapped, HisRSΔCD reacted with anti-Jo-1 Abs from patient sera (9).
Interestingly, we identified another novel HisRS SV in muscle tissue, which we designated as HisRSWHEP. This SV is composed solely of the first 60 aa of HisRS, which constitute the WHEP domain. It results from a splice event that introduces a stop codon from intron 2. With this discovery, we then set out to investigate whether transcripts for HisRSΔCD and HisRSWHEP are up-regulated in patients with IIM/ILD. In addition, we investigated recombinant forms of these variants and their constituent domains for their reaction with anti-Jo-1 Abs from patients. Our results demonstrate that both the expression and cross-reactivity of HisRSΔCD and of HisRSWHEP are associated with IIM and therefore support the possibility of extracellular anti-Jo-1 antibody binding to HisRS and its SVs.
EXPERIMENTAL PROCEDURES
PCR Identification of HisRSWHEP
A human skeletal muscle cDNA library was used as a template (Clontech, Palo Alto, CA). PCR was performed with a pair of primers (FP1 (AGTGGACAGCCGGGATGGCAGAGC)/RP1 (GCTTGGAGTCTTCCCCATAC)), and the PCR product was validated by direct sequencing. A color-coded trace from sequencing is presented in supplemental Fig. S1.
Sample Preparation for Gene Expression Analysis
All human tissue poly(A)+ RNAs were purchased from Clontech (catalog nos. 636170, 636591, 636128, 636105, 636113, 636119, 636121, 636101, 636118, 636146, 636125, 636162, and 636120). Muscle biopsies from DM patients were kindly provided by the Telethon Network of Genetic Biobanks (Milan, Italy). These samples consisted of 10 muscle biopsies from Caucasian DM patients (including five males and five females). The diagnosis was based on clinical manifestation and histology .Total RNA was isolated from muscle using a PARIS kit (Invitrogen) and was pooled together as the DM group. The control group was pooled total RNA from two healthy Caucasian subjects (including one male and one female; Clontech catalog no. 636534). First-strand cDNAs were synthesized as described previously (9).
Quantitative PCR and Data Analysis
Quantitative PCRs (qPCRs) were performed as described previously (9, 11). The qPCR primer sequences were as follows: qFP1, CACGGTGCAGAAGTCATTGAT; qRP1, TCCCCATACTTTCCCATCAGTG; qFP2, GTGCTCAAAACCCCCAAGTAGAG; qRP2, CACAGTGGCTCACGCCTGT; qFP3, ACCCCCAAGTAGAGACGAG; qRP3, TCTCGCGAACTGCCATCTG; qFPMXA, ACCTGATGGCCTATCACCAG; and qRPMXA, TTCAGGAGCCAGCTGTAGGT.
Detection of HisRS Proteins by Western Blot Analysis
Total cell lysates (TCLs) of monocytic THP-1 cells and human primary skeletal muscle cells (Cell Application, San Diego, CA) were prepared in 50 mm Tris buffer (pH 8.0) containing 1% Triton X-100 and 5 mm EDTA. TCLs (50 μg) were applied to electrophoresis and subsequent Western blot analysis with anti-HisRS mAb (Abnova, Walnut, CA).
Quantification of HisRS Levels in Monocytic THP-1 Cells
The cellular HisRS concentration was determined by standard sandwich ELISA (capture Ab, home-made anti-human HisRS mouse mAb; detection Ab, anti-human HisRS mAb (Abnova), biotinylated in-house). Recombinant human HisRS protein was used as the quantification standard (see below).
Protein Expression and Purification
The cDNAs encoding native human HisRS (aa 1–506), HisRSΔCD (aa 1–60 plus aa 405–506), HisRSWHEP (aa 1–60), CD (aa 54–398), and ABD (aa 406–506) were cloned into the pET21a vector with a C-terminal His6 tag. From our experience, the C-terminal 3 aa (CIC, aa 507–509) reduce protein homogeneity; thus, these residues were removed in all constructs. The constructs were transformed into E. coli BL21(DE3) cells, and expressed proteins were purified by nickel-nitrilotriacetic acid affinity chromatography and further separated by size-exclusion chromatography in 1× PBS buffer with 1 mm DTT. The purity and homogeneity of each protein were checked by analytical size-exclusion chromatography and SDS-PAGE.
Depletion ELISA
Anti-Jo-1 autoantibody-positive patient sera were obtained from RDL Inc. (Los Angeles, CA). A 96-well enzyme immunoassay/radioimmunoassay plate (Corning, Corning, NY) was coated with 50 μl (2 μg/ml) of one of the recombinant proteins (see above) or BSA (as a control) in PBS buffer. After washing and blocking, patient sera containing anti-Jo-1 autoantibodies (in a dilution giving 25% of the maximum effect when applied to a HisRS-coated plate) were added and incubated overnight at 4 °C. After incubation, supernatant was applied to another plate (precoated with the respective recombinant protein) to check the depletion efficiency. The samples with a pre-depletion efficiency of >95% were applied to another plate coated with HisRS for indirect ELISA. The detection Ab was HRP-conjugated goat anti-human IgG (10 ng/well IgG; AbD Serotec, Raleigh, NC). The results were obtained with a FLUOstar OPTIMA instrument (BMG Labtech, Offenburg, Germany).
Secretion Assay
Coding sequences for HisRS, HisRSΔCD, and HisRSWHEP were cloned into the pCI-neo-2×myc vector (Promega, Madison, WI) through the NheI/NotI restriction sites. These constructs were transfected into HEK293T cells or C2C12 myoblasts using Lipofectamine LTX with PlusTM reagents (Invitrogen) following the manufacturer's instructions. To achieve similar overexpression levels, the DNA construct of HisRSΔCD or HisRSWHEP was transfected at 1 μg for 2.8 × 105 cells, whereas that of HisRS was transfected at 0.1 μg. Empty vector was transfected as a control. The transfected cells were split when confluent and plated at 2 × 104 cells/cm2 in a 60-mm dish. Media were refreshed after 3 h, and both media and TCLs were harvested after another 24 h of incubation. The media were precleaned with 5 μl of Dynabeads-protein G (Invitrogen) for 1 h at 4 °C. Anti-Myc polyclonal Ab (1.5 μg; Sigma) was mixed with 5 μl of Dynabeads-protein G in PBS for 1 h at room temperature. The Ab/bead mixture was added to the precleaned media and further incubated for 2 h at 4 °C. The protein-Ab-bead complex was washed with radioimmune precipitation assay buffer (12) and eluted with 0.1 m glycine buffer (pH 2.0). The eluent was neutralized by adding 1 m Tris-HCl (pH 8.0; v/v, 10:1). TCLs were prepared in radioimmune precipitation assay buffer (supplemented with protease inhibitors, Roche Applied Science). Both media and TCLs were subjected to electrophoresis and immunoblotted with anti-Myc mAb (Millipore). Lactate dehydrogenase (LDH) protein was detected with anti-LDHB mAb (Abnova).
Cell injury was assessed by measuring LDH activity in the medium (Roche Applied Science). A LDH activity standard was also conducted, covering 0, 25, 50, 100, 200, 400, and 800 micro-units/100 μl. The detection limit of the assay was defined as the mean absorbance of the blank plus 3 times the standard deviation of the blank.
RESULTS
Identification of HisRSWHEP
Human HisRS is a class II tRNA synthetase composed of a core CD made up of a seven-stranded β-structure with flanking α-helices and a C-terminal ABD. Although absent from prokaryotic and lower eukaryotic HisRSs, an N-terminal coiled-coil WHEP domain was appended at the time of appearance of metazoans. As stated above, this domain is present in our previously identified HisRSΔCD SV. Our goal was to find additional SVs that retained the WHEP domain.
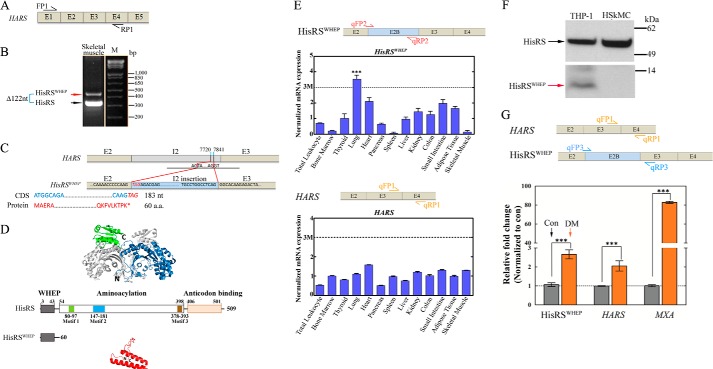
We noted an expressed sequence tag (EST) BP267368 annotation in the University of California Santa Cruz EST database (13). This transcript has a 122-bp insertion of nucleotides from intron 2, located between exons 2 and 3 (supplemental Fig. S1A). Because the intron insertion introduces a stop codon immediately at the end of exon 2, it could, in principle, encode just the WHEP domain of HisRS. To verify this variant, we designed primers that targeted the exon 1 and exon 4 regions of human HARS (Fig. 1A). PCR with a muscle cDNA template and the aforementioned pair of primers yielded a product of 473 nucleotides, which is larger than that expected for the 351-nucleotide transcript that would encode the same region of full-length HARS (Fig. 1B). This product confirmed features of the EST BP267368 annotation. However, in contrast to EST BP267368, our SV had neither a T-to-C substitution in exon 2, which would yield a L56P substitution in HARS, nor a synonymous A-to-G substitution in exon 3 (supplemental Fig. S1, B and C). In addition, our analysis differed in having a synonymous T-to-A substitution in the sequence of the insertion into intron 2. The inserted sequence was flanked by consensus GT-AG splice junctions (Fig. 1C) and created a new exon cassette. We designated this cassette as exon 2B. The transcript that results from this splice event harbors a canonical start codon, so translation would start at the typical initiator ATG and terminate after exon 2 (Fig. 1C). The consequence is a protein composed of solely the first 60 aa of human HisRS. Because this protein is made up of only the WHEP domain, we named it HisRSWHEP (Fig. 1, C and D).
FIGURE 1.
Identification of transcript and protein for HisRSWHEP SV and up-regulation of HisRSWHEP transcript in muscle biopsies of DM patients. A, PCR was used to identify the mRNA encoding HisRSWHEP in human skeletal muscle. Locations of the primers used for PCR are indicated in the schematic. E, exon. B, electrophoretic analysis of the PCR. The upper fragment (red arrow) was amplified from the mRNA for HisRSWHEP, which is 122 nucleotides (nt) longer than the lower fragment amplified from the mRNA for HARS (black arrow). M, mass marker. C, schematic drawing of the intron 2 (I2) insertion in the mRNA for HisRSWHEP and location of the inserted nucleotides in HARS. Notably, the inserted sequence is flanked by canonical splicing signals (preceded by AG and followed by GT) and itself ends with AG. HisRSWHEP is encoded by 183 nucleotides and is translated into 60 aa, which encompass the WHEP domain of HisRS. CDS, coding sequence. D, schematic illustrations of HisRS and HisRSWHEP and their structures (Protein Data Bank code 4G84 for HisRS and code 1X59 for HisRSWHEP). One monomer of HisRS is shown in color (ABD in green and CD in blue), whereas the other monomer is shown in gray. Notably, HisRSWHEP is composed of the N-terminal WHEP domain (shown in red) of human HARS. E, distribution of the transcripts of HisRS and HisRSWHEP in 13 human tissues. Locations of the qPCR primers are indicated in the schematic. The expression levels were normalized to that of the HKG RPL9. The median value was taken as 1.0. Notably, the HisRSWHEP transcript is significantly higher in human lung compare with other tissues and is 3-fold above the median. The expression of the transcript for HARS was normally distributed in the various tissues, with expression levels <3 times that of the median value. F, HisRSWHEP protein was detected in the TCL of monocytic THP-1 cells, but not in that of human skeletal muscle cells (HSkMC). Expected running positions of the proteins are indicated by arrows. G, the transcript for HisRSWHEP is up-regulated in DM muscle biopsies (1.0 ± 0.1 in control (Con) versus 2.7 ± 0.2 in DM). The transcript for HisRS is also up-regulated in the DM samples (1.0 ± 0.01 in control versus 2.1 ± 0.3 in DM). The MXA gene serves as a positive control (1.0 ± 0.06 in control versus 82.8 ± 1.1 in DM). Locations of the qPCR primers are indicated in the schematic. Data are shown as means ± S.D. ***, p < 0.0001.
Expression of Transcripts for HisRSWHEP in 13 Human Tissue Types
We next compared the transcript levels of HisRSWHEP and HARS in 13 human tissue types, which were total leukocytes, bone marrow, spleen, lung, heart, kidney, liver, pancreas, small intestine, colon, thyroid, adipose, and skeletal muscle. The SYBR Green qPCR method was employed. The transcript for HARS was somewhat evenly distributed across all 13 tissue types, deviating no more than 3 times from the median value (Fig. 1E). (Because the transcript for the housekeeping gene (HKG) RPL9 (60 S ribosomal protein L9) is the most evenly distributed among ∼20 HKGs, the levels of the HARS transcripts were normalized to that for RPL9.) In comparison, the transcript level of HisRSWHEP was highest in lung (3.5 times above the median level) (Fig. 1E). The transcript levels of HisRSWHEP were below 0.1% of those of HARS. Interestingly, the expression level of HisRSWHEP was low in normal skeletal muscle tissue in comparison with other tissues.
Detection of HisRSWHEP Protein
We used a standard Western blot method to search for the translation product of the HisRSWHEP transcript. For this purpose, a mAb raised against the N-terminal region (aa 1–97) of human HisRS was used. Considering the relatively small amounts of HisRSWHEP mRNA and the difficulty in obtaining adequate amounts of human tissues, human cell lines cultured in vitro were employed. Although not detected in human skeletal muscle cells, the 6.8-kDa HisRSWHEP protein was readily observed in human monocytic THP-1 cells (Fig. 1F, red arrow). Consistent with the relatively low amount of its mRNA, HisRSWHEP was present at a level estimated close to 1% of that of HisRS, which was detected with the same Ab (Fig. 1F, black arrow). We also determined the cellular HisRS level in monocytic THP-1 cells by standard sandwich ELISA (see “Experimental Procedures”). Our results show that the intracellular HisRS level was 0.94 ± 0.17 μm (mean ± S.E., n = 4).
HisRSWHEP Transcript Is Up-regulated in Pool of Muscle Biopsies from DM Patients
Anti-Jo-1 Abs are present in 15–30% of polymyositis patients and 5–10% of DM patients (14). To evaluate the possibility of HisRSWHEP being an antigen in muscle biopsies of patients with IIM/ILD, we examined its mRNA transcript in pooled muscle biopsies from 10 DM patients. (Because of difficulties in defining primers that were sufficiently specific, the transcript for HisRSΔCD was not measured.) Because MXA (myxovirus resistance gene, a type 1 interferon (α/β)) was reported to be up-regulated in myositis (15), its transcript was included as a positive control. Two HKGs, RPL9 and RPS11 (40 S ribosomal protein S11), were also included in our analysis. As a control, we used total RNA from two healthy Caucasian subjects. Relative to the control, the transcript for HisRSWHEP was significantly up-regulated in RNA samples from DM muscle biopsies (2.7 ± 0.2-fold, p < 0.0001) (Fig. 1G). The transcript for native HARS was also up-regulated (2.1 ± 0.3-fold, p < 0.0001) (Fig. 1G).
Anti-Jo-1 Autoantibodies React Mainly with WHEP and ABD Domains of Human HisRS
The N-terminal portion of HisRS, which includes the WHEP domain, has long been recognized as a major epitope of anti-Jo-1 Abs (6, 7). Considering this point, we focused on the antigenicity of two SVs that harbor the WHEP domain, i.e. HisRSWHEP and HisRSΔCD. We also investigated recombinant forms of HisRS, CD, and ABD. Recombinant versions of each of these five proteins were expressed in E. coli and readily purified. (Thus, all five proteins folded into stable structures.) Depletion ELISAs were used to measure the binding ability of anti-Jo-1 Abs for each of the five proteins (Fig. 2A). Sera from 24 anti-Jo-1 Ab-positive patients were included in our analysis. As expected, full-length HisRS almost completely depleted the anti-Jo-1 Abs (∼100%) in all patient sera (n = 24) (Fig. 2B). Strikingly, the CD and ABD recombinant proteins depleted little of the anti-Jo-1 Ab-positive sera. In contrast, ∼50% of the anti-Jo-1 Abs reacted with HisRSWHEP and WHEP domain-containing HisRSΔCD. Thus, the two SVs are robust targets for anti-Jo-1 Abs. It is of interest that the domains harbored by these two SVs, the WHEP and ABD domains, are well separated on the structure of native HisRS (Fig. 2, C and D).
FIGURE 2.
Anti-Jo-1 patient serum reacts mainly with the N-terminal WHEP domain and C-terminal ABD of human HisRS, and recombinant HisRS SVs are secreted from HEK293T cells and C2C12 myoblasts. A, illustration of depletion ELISAs. Details are provided under “Experimental Procedures.” B, reactivity of anti-Jo-1 Ab-positive patient serum against different HisRS recombinant proteins. Notably, apart from HisRS, anti-Jo-1 Ab-positive patient serum reacted mostly with HisRSWHEP and HisRSΔCD, with a significantly higher reactivity than with the recombinant CD or ABD. C, the two most reactive domains are far apart on the three-dimensional structure of HisRS (Protein Data Bank code 4G84). The C-terminal ABD (shown in green) and N-terminal WHEP domain (shown in red) are highlighted in the structure of HisRS. The N-terminal WHEP domain is not resolved in the structure of HisRS, but is resolved in that of HisRSΔCD (Protein Data Bank code 2LW7). D, the structures of HisRSΔCD and HisRSWHEP show that the two HisRS SVs contain the major anti-Jo-1 epitopes. E and F, recombinant HisRSWHEP, HisRSΔCD, and HisRS proteins (with a C-terminal Myc tag) were transiently expressed in HEK293T cells (E), and recombinant HisRSΔCD and HisRS proteins were transiently expressed in C2C12 myoblasts (F). Expressed proteins were detected in the TCLs with anti-Myc mAb. LDHB in the TCLs served as a loading control. The media were immunoprecipitated by anti-Myc polyclonal Ab and detected with anti-Myc mAb. Notably, all HisRS proteins were detected in the media. The bar graphs show that the LDH activities of all samples were below the detection limit of the assay (indicated by the red line), suggesting little cell damage. The results shown are representative of three separately conducted experiments.
Recombinant HisRS, HisRSΔCD, and HisRSWHEP Are Secreted When Overexpressed in HEK293T Cells
Several examples of secreted human aaRSs have been reported (16–18). Thus, we imagined that, at least as a formal possibility, HisRS and one or both of its SVs could also be secreted under certain conditions, such as in an inflammatory environment. Because our sensitivity of detection was limited by the low abundance of the endogenous splice variants (see above and Ref. 9), as a first step toward investigating secretion, we transiently expressed recombinant HisRS, HisRSWHEP, and HisRSΔCD in HEK293T cells (Fig. 2E) and detected their expression by anti-Myc mAb in the TCLs. (The use of the Myc tag enabled us to distinguish the recombinant proteins from endogenous counterparts.) LDHB Western blotting in TCLs served as a loading control. To check for cell leakiness, we measured medium LDH activity using a cytotoxicity kit (19, 20). As shown in the bar graph in Fig. 2E, all samples had undetectable LDH activity, below the detection limit (indicated by the dashed red line), thus suggesting limited (if any) cell damage. Each of the three HisRS proteins was detected in the cell media fraction (Fig. 2E). These results support the idea that HisRS, HisRSWHEP, and HisRSΔCD can be secreted into the medium.
Recombinant HisRSΔCD and HisRS Are Secreted When Expressed in C2C12 Myoblasts
We also transiently expressed Myc-tagged recombinant HisRS and HisRSΔCD in C2C12 myoblasts. Both HisRS proteins were expressed in C2C12 myoblasts and detected in the cell media (Fig. 2F). LDHB Western blotting in TCLs served as a loading control. (In additional experiments, we could not show that the small WHEP domain alone (HisRSWHEP) could be consistently detected in the media fraction (data not shown), possibly because of variables beyond our immediate control (such as proteases in the media that quickly degrade, like a short polypeptide).) Again, all samples had undetectable LDH activity levels (Fig. 2F). These results suggest the possibility that at least one HisRS SV (HisRSΔCD) can be secreted from a murine muscle cell line.
DISCUSSION
Several previous studies suggest that low-abundant transcripts, which were previously considered as unimportant, are biologically significant in differentiation, metabolism, and phenotypic alternation (21–25). To better understand the expression of HisRSWHEP, we measured the concentration of human HisRS in monocytic THP-1 cells and showed that intracellular HisRS has a concentration of 0.94 ± 0.17 μm (see above), which is roughly comparable with the reported concentration of methionyl-tRNA synthetase in rabbit reticulocytes (∼0.2 μm) (26). On the basis of our estimation that the HisRSWHEP protein is close to 1% of full-length HisRS, we estimate that the cellular content of HisRSWHEP is ∼10 nm. Even if only a fraction is secreted, this concentration is well within the range of known dissociation constants (Kd) for aaRSs in cell signaling events. For example, the aaRS complex-interacting multifunctional protein 1 is reported to bind to CD23 with a Kd of 4.3 nm (27); glycyl-tRNA synthetase binds to CDH6 with a Kd of 3.4 nm (16); and a fragment of tyrosyl-tRNA synthetase, known as mini-tyrosyl-tRNA synthetase, stimulates polymorphonuclear cell migration at 1 nm (28). In addition, these concentrations are higher than the effective concentrations of many cytokines, which are in the picomolar to lower nanomolar range. Thus, in healthy young people (<45 years of age), the serum TNF-α level is estimated to be ∼0.19 pm, IL-6 is estimated to be ∼0.16 pm, and MCP-1 is estimated to be ∼16.4 pm (29). From this perspective, our results harmonize well with what is known about many other systems.
Novel functions for the WHEP domains in tryptophanyl-tRNA synthetase, glutamylprolyl-tRNA synthetase, and glycyl-tRNA synthetase have been reported previously (30–34). Interestingly, the WHEP domain-containing N-terminal 48-aa fragment of HisRS was previously associated with a novel inflammatory function, whereas the residual protein lacking this fragment was inactive (8). Here, we established that two HisRS SVs, unknown at the time of the work of Howard et al. (8) and which each harbor the WHEP domain, are expressed in cultured cells and cross-react with a substantial portion of the anti-Jo-1 Abs from the tested patient population. Both SVs and HisRS can also be secreted. In addition, in a DM patient population pool undiagnosed as to anti-Jo-1 Ab status, expression of HisRS and at least one of these SVs appears to be up-regulated (Fig. 1G).
Non-translational functions for SVs, natural proteolytic fragments, and even a truncated bipartite synthetase (from the recruitment of a novel stop codon) have been reported for various human tRNA synthetases or synthetase-associated proteins (31, 35–43). These non-translational functions reach into many parts of cell biology and homeostatic mechanisms, including angiogenesis, hematopoiesis, and control of tumor growth. In addition, some of these functions are extracellular and are enabled by the capacity of at least some aaRSs to be secreted, as evidenced by their detection in human and mouse sera (16, 17, 44–47). With this in mind, there are suggestions of immunomodulation-related functions, such that aaRSs fragments have activities that can act to resolve inflammation (48, 49). Thus, in light of the many examples of non-catalytic fragments of aaRSs having extracellular functions and given the data presented here showing the reactivity of SVs of HisRS for anti-Jo-1 Abs, the up-regulation of at least one of them in a patient population, and their secretion from cultured cells, we propose that these SVs deserve further investigations related to muscle health and the etiology of inflammatory muscle diseases.
Possibly, HisRS and its two WHEP domain-containing SVs are involved in maintaining immune homeostasis in muscle. When immune surveillance or clearance is needed, HisRS proteins attract immune cells to muscle tissue. In support of this hypothesis, the N-terminal WHEP domain of HisRS may be chemotactic for lymphocytes and activated monocytes (8). Possibly because of their persistent presence, in DM patients, the HisRS proteins are eventually seen as “foreigners,” and autoantibodies against HisRS, especially the WHEP domain, are generated. These autoantibodies may antagonize the immune homeostatic role of HisRS proteins and gradually lead to myositis.
Supplementary Material
Acknowledgment
We thank the Telethon Network of Genetic Biobanks for kindly providing human DM muscle biopsies.
This work was supported, in whole or in part, by National Institutes of Health Grant CA92577 from NCI and Grant GM88278. This work was also supported by Innovation and Technology Fund of Hong Kong Grants UIM181, UIM192, and UIM199; aTyr Pharma; and an NFCR Fellowship (to the Scripps Laboratory for tRNA synthetase research) from the National Foundation for Cancer Research. Professors Mingjie Zhang, Xiang-Lei Yang, and Paul Schimmel are financially benefited by aTyr Pharma in the form of compensation, stock ownership, or both.

This article contains supplemental Fig. S1.
- IIM
- idiopathic inflammatory myositis
- ILD
- interstitial lung disease
- MSA
- myositis-specific antibody
- aaRS
- aminoacyl-tRNA synthetase
- HisRS
- histidyl-tRNA synthetase
- DM
- dermatomyositis
- aa
- amino acid(s)
- SV
- splice variant
- CD
- catalytic domain
- ABD
- anticodon-binding domain
- qPCR
- quantitative PCR
- TCL
- total cell lysate
- LDH
- lactose dehydrogenase
- EST
- expressed sequence tag
- HKG
- house-keeping gene.
REFERENCES
- 1. Love L. A., Leff R. L., Fraser D. D., Targoff I. N., Dalakas M., Plotz P. H., Miller F. W. (1991) A new approach to the classification of idiopathic inflammatory myopathy: myositis-specific autoantibodies define useful homogeneous patient groups. Medicine 70, 360–374 [DOI] [PubMed] [Google Scholar]
- 2. Targoff I. N. (2000) Update on myositis-specific and myositis-associated autoantibodies. Curr. Opin. Rheumatol. 12, 475–481 [DOI] [PubMed] [Google Scholar]
- 3. Mammen A. L. (2011) Autoimmune myopathies: autoantibodies, phenotypes and pathogenesis. Nat. Rev. Neurol. 7, 343–354 [DOI] [PubMed] [Google Scholar]
- 4. Nishikai M., Reichlin M. (1980) Heterogeneity of precipitating antibodies in polymyositis and dermatomyositis. Characterization of the Jo-1 antibody system. Arthritis Rheum. 23, 881–888 [DOI] [PubMed] [Google Scholar]
- 5. Mathews M. B., Bernstein R. M. (1983) Myositis autoantibody inhibits histidyl-tRNA synthetase: a model for autoimmunity. Nature 304, 177–179 [DOI] [PubMed] [Google Scholar]
- 6. Martin A., Shulman M. J., Tsui F. W. (1995) Epitope studies indicate that histidyl-tRNA synthetase is a stimulating antigen in idiopathic myositis. FASEB J. 9, 1226–1233 [DOI] [PubMed] [Google Scholar]
- 7. Raben N., Nichols R., Dohlman J., McPhie P., Sridhar V., Hyde C., Leff R., Plotz P. (1994) A motif in human histidyl-tRNA synthetase which is shared among several aminoacyl-tRNA synthetases is a coiled-coil that is essential for enzymatic activity and contains the major autoantigenic epitope. J. Biol. Chem. 269, 24277–24283 [PubMed] [Google Scholar]
- 8. Howard O. M., Dong H. F., Yang D., Raben N., Nagaraju K., Rosen A., Casciola-Rosen L., Härtlein M., Kron M., Yang D., Yiadom K., Dwivedi S., Plotz P. H., Oppenheim J. J. (2002) Histidyl-tRNA synthetase and asparaginyl-tRNA synthetase, autoantigens in myositis, activate chemokine receptors on T lymphocytes and immature dendritic cells. J. Exp. Med. 196, 781–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Xu Z., Wei Z., Zhou J. J., Ye F., Lo W. S., Wang F., Lau C. F., Wu J., Nangle L. A., Chiang K. P., Yang X. L., Zhang M., Schimmel P. (2012) Internally deleted human tRNA synthetase suggests evolutionary pressure for repurposing. Structure 20, 1470–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Soejima M., Kang E. H., Gu X., Katsumata Y., Clemens P. R., Ascherman D. P. (2011) Role of innate immunity in a murine model of histidyl-transfer RNA synthetase (Jo-1)-mediated myositis. Arthritis Rheum. 63, 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang F., Xu Z., Zhou J., Lo W. S., Lau C. F., Nangle L. A., Yang X. L., Zhang M., Schimmel P. (2013) Regulated capture by exosomes of mRNAs for cytoplasmic tRNA synthetases. J. Biol. Chem. 288, 29223–29228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harlow E., Land David. (1999) Using Antibodies: A Laboratory Manual, p. 449, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 13. Suzuki Y., Yamashita R., Shirota M., Sakakibara Y., Chiba J., Mizushima-Sugano J., Nakai K., Sugano S. (2004) Sequence comparison of human and mouse genes reveals a homologous block structure in the promoter regions. Genome Res. 14, 1711–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jura M., Rychlewski L., Barciszewski J. (2007) Comprehensive insight into human aminoacyl-tRNA synthetases as autoantigens in idiopathic inflammatory myopathies. Crit. Rev. Immunol. 27, 559–572 [DOI] [PubMed] [Google Scholar]
- 15. Greenberg S. A., Pinkus J. L., Pinkus G. S., Burleson T., Sanoudou D., Tawil R., Barohn R. J., Saperstein D. S., Briemberg H. R., Ericsson M., Park P., Amato A. A. (2005) Interferon-α/β-mediated innate immune mechanisms in dermatomyositis. Ann. Neurol. 57, 664–678 [DOI] [PubMed] [Google Scholar]
- 16. Park M. C., Kang T., Jin D., Han J. M., Kim S. B., Park Y. J., Cho K., Park Y. W., Guo M., He W., Yang X. L., Schimmel P., Kim S. (2012) Secreted human glycyl-tRNA synthetase implicated in defense against ERK-activated tumorigenesis. Proc. Natl. Acad. Sci. U.S.A. 109, E640–E647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kapoor M., Zhou Q., Otero F., Myers C. A., Bates A., Belani R., Liu J., Luo J. K., Tzima E., Zhang D. E., Yang X. L., Schimmel P. (2008) Evidence for annexin II-S100A10 complex and plasmin in mobilization of cytokine activity of human TrpRS. J. Biol. Chem. 283, 2070–2077 [DOI] [PubMed] [Google Scholar]
- 18. Wakasugi K., Schimmel P. (1999) Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science 284, 147–151 [DOI] [PubMed] [Google Scholar]
- 19. Rouabhia M., Park H., Meng S., Derbali H., Zhang Z. (2013) Electrical stimulation promotes wound healing by enhancing dermal fibroblast activity and promoting myofibroblast transdifferentiation. PLoS ONE 8, e71660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chan F. K. M., Moriwaki K., De Rosa M. J. (2013) Detection of necrosis by release of lactate dehydrogenase activity. Methods Mol. Biol. 979, 65–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Elowitz M. B. (2002) Stochastic gene expression in a single cell. Science 297, 1183–1186 [DOI] [PubMed] [Google Scholar]
- 22. Kuznetsov V. A., Knott G. D., Bonner R. F. (2002) General statistics of stochastic process of gene expression in eukaryotic cells. Genetics 161, 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ozbudak E. M., Thattai M., Kurtser I., Grossman A. D., van Oudenaarden A. (2002) Regulation of noise in the expression of a single gene. Nat. Genet. 31, 69–73 [DOI] [PubMed] [Google Scholar]
- 24. Blake W. J., KAErn M., Cantor C. R., Collins J. J. (2003) Noise in eukaryotic gene expression. Nature 422, 633–637 [DOI] [PubMed] [Google Scholar]
- 25. Paulsson J. (2004) Summing up the noise in gene networks. Nature 427, 415–418 [DOI] [PubMed] [Google Scholar]
- 26. Kellermann O., Tonetti H., Brevet A., Mirande M., Pailliez J. P., Waller J. P. (1982) Macromolecular complexes from sheep and rabbit containing seven aminoacyl-tRNA synthetases. I. Species specificity of the polypeptide composition. J. Biol. Chem. 257, 11041–11048 [PubMed] [Google Scholar]
- 27. Kwon H. S., Park M. C., Kim D. G., Cho K., Park Y. W., Han J. M., Kim S. (2012) Identification of CD23 as a functional receptor for the proinflammatory cytokine AIMP1/p43. J. Cell Sci. 125, 4620–4629 [DOI] [PubMed] [Google Scholar]
- 28. Vo M. N., Yang X. L., Schimmel P. (2011) Dissociating quaternary structure regulates cell-signaling functions of a secreted human tRNA synthetase. J. Biol. Chem. 286, 11563–11568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kim H., Kim H. S., Youn J. C., Shin E. C., Park S. (2011) Serum cytokine profiles in healthy young and elderly population assessed using multiplexed bead-based immunoassays. J. Transl. Med. 9, 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wakasugi K., Slike B. M., Hood J., Otani A., Ewalt K. L., Friedlander M., Cheresh D. A., Schimmel P. (2002) A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 173–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhou Q., Kapoor M., Guo M., Belani R., Xu X., Kiosses W. B., Hanan M., Park C., Armour E., Do M. H., Nangle L. A., Schimmel P., Yang X. L. (2009) Orthogonal use of a human tRNA synthetase active site to achieve multifunctionality. Nat. Struct. Mol. Biol. 17, 57–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sajish M., Zhou Q., Kishi S., Valdez D. M., Jr., Kapoor M., Guo M., Lee S., Kim S., Yang X. L., Schimmel P. (2012) Trp-tRNA synthetase bridges DNA-PKcs to PARP-1 to link IFN-γ and p53 signaling. Nat. Chem. Biol. 8, 547–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. He W., Zhang H. M., Chong Y. E., Guo M., Marshall A. G., Yang X. L. (2011) Dispersed disease-causing neomorphic mutations on a single protein promote the same localized conformational opening. Proc. Natl. Acad. Sci. U.S.A. 108, 12307–12312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jia J., Arif A., Ray P. S., Fox P. L. (2008) WHEP domains direct non-canonical function of glutamyl-prolyl tRNA synthetase in translational control of gene expression. Mol. Cell 29, 679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wu P. C., Alexander H. R., Huang J., Hwu P., Gnant M., Berger A. C., Turner E., Wilson O., Libutti S. K. (1999) In vivo sensitivity of human melanoma to tumor necrosis factor (TNF)-α is determined by tumor production of the novel cytokine endothelial-monocyte activating polypeptide II (EMAPII). Cancer Res. 59, 205–212 [PubMed] [Google Scholar]
- 36. Shalak V. (2001) The EMAPII cytokine is released from the mammalian multisynthetase complex after cleavage of its p43/proEMAPII component. J. Biol. Chem. 276, 23769–23776 [DOI] [PubMed] [Google Scholar]
- 37. Banin E. (2006) T2-TrpRS inhibits preretinal neovascularization and enhances physiological vascular regrowth in OIR as assessed by a new method of quantification. Invest. Ophthalmol. Vis. Sci. 47, 2125–2134 [DOI] [PubMed] [Google Scholar]
- 38. Tzima E., Reader J. S., Irani-Tehrani M., Ewalt K. L., Schwartz M. A., Schimmel P. (2003) Biologically active fragment of a human tRNA synthetase inhibits fluid shear stress-activated responses of endothelial cells. Proc. Natl. Acad. Sci. U.S.A. 100, 14903–14907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mukhopadhyay R., Jia J., Arif A., Ray P. S., Fox P. L. (2009) The GAIT system: a gatekeeper of inflammatory gene expression. Trends Biochem. Sci. 34, 324–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Han J. M., Park B. J., Park S. G., Oh Y. S., Choi S. J., Lee S. W., Hwang S. K., Chang S. H., Cho M. H., Kim S. (2008) AIMP2/p38, the scaffold for the multi-tRNA synthetase complex, responds to genotoxic stresses via p53. Proc. Natl. Acad. Sci. U.S.A. 105, 11206–11211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Choi J. W., Kim D. G., Park M. C., Um J. Y., Han J. M., Park S. G., Choi E. C., Kim S. (2009) AIMP2 promotes TNF-dependent apoptosis via ubiquitin-mediated degradation of TRAF2. J. Cell Sci. 122, 2710–2715 [DOI] [PubMed] [Google Scholar]
- 42. Choi J. W., Lee J. W., Kim J. K., Jeon H. K., Choi J. J., Kim D. G., Kim B. G., Nam D. H., Kim H. J., Yun S. H., Kim S. (2012) Splicing variant of AIMP2 as an effective target against chemoresistant ovarian cancer. J. Mol. Cell Biol. 4, 164–173 [DOI] [PubMed] [Google Scholar]
- 43. Choi J. W., Kim D. G., Lee A. E., Kim H. R., Lee J. Y., Kwon N. H., Shin Y. K., Hwang S. K., Chang S. H., Cho M. H., Choi Y. L., Kim J., Oh S. H., Kim B., Kim S. Y., Jeon H. S., Park J. Y., Kang H. P., Park B. J., Han J. M., Kim S. (2011) Cancer-associated splicing variant of tumor suppressor AIMP2/p38: pathological implication in tumorigenesis. PLoS Genet. 7, e1001351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Park S. G., Kim H. J., Min Y. H., Choi E. C., Shin Y. K., Park B. J., Lee S. W., Kim S. (2005) Human lysyl-tRNA synthetase is secreted to trigger proinflammatory response. Proc. Natl. Acad. Sci. U.S.A. 102, 6356–6361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Williams T. F., Mirando A. C., Wilkinson B., Francklyn C. S., Lounsbury K. M. (2013) Secreted threonyl-tRNA synthetase stimulates endothelial cell migration and angiogenesis. Sci. Rep. 3, 1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ko Y. G. (2001) A cofactor of tRNA synthetase, p43, is secreted to up-regulate proinflammatory genes. J. Biol. Chem. 276, 23028–23033 [DOI] [PubMed] [Google Scholar]
- 47. Greenberg Y., King M., Kiosses W. B., Ewalt K., Yang X., Schimmel P., Reader J. S., Tzima E. (2008) The novel fragment of tyrosyl-tRNA synthetase, mini-TyrRS, is secreted to induce an angiogenic response in endothelial cells. FASEB J. 22, 1597–1605 [DOI] [PubMed] [Google Scholar]
- 48. Kron M. A., Wang C., Vodanovic-Jankovic S., Howard O. M., Kuhn L. A. (2012) Interleukin-8-like activity in a filarial asparaginyl-tRNA synthetase. Mol. Biochem. Parasitol. 185, 66–69 [DOI] [PubMed] [Google Scholar]
- 49. Kron M. A., Metwali A., Vodanovic-Jankovic S., Elliott D. (2013) Nematode asparaginyl-tRNA synthetase resolves intestinal inflammation in mice with T-cell transfer colitis. Clin. Vaccine Immunol. 20, 276–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.