Background: PPARα is an important regulator of hepatic lipid metabolism via target gene regulation.
Results: HILPDA is regulated by PPARα via an upstream PPRE. Targeted overexpression of HILPDA increases hepatic triglyceride storage via reduction of TG secretion.
Conclusion: HILPDA is a novel PPARα target involved in hepatic triglyceride secretion.
Significance: HILPDA might be a potential target in the treatment of non-alcoholic fatty liver disease.
Keywords: Lipid Droplets, Lipoprotein Secretion, Liver Metabolism, Nuclear Receptor, Peroxisome Proliferator-activated Receptor (PPAR), Hig2, Liver Slices, PPRE, Steatosis, VLDL
Abstract
Peroxisome proliferator-activated receptors (PPARs) play major roles in the regulation of hepatic lipid metabolism through the control of numerous genes involved in processes such as lipid uptake and fatty acid oxidation. Here we identify hypoxia-inducible lipid droplet-associated (Hilpda/Hig2) as a novel PPAR target gene and demonstrate its involvement in hepatic lipid metabolism. Microarray analysis revealed that Hilpda is one of the most highly induced genes by the PPARα agonist Wy14643 in mouse precision cut liver slices. Induction of Hilpda mRNA by Wy14643 was confirmed in mouse and human hepatocytes. Oral dosing with Wy14643 similarly induced Hilpda mRNA levels in livers of wild-type mice but not Ppara−/− mice. Transactivation studies and chromatin immunoprecipitation showed that Hilpda is a direct PPARα target gene via a conserved PPAR response element located 1200 base pairs upstream of the transcription start site. Hepatic overexpression of HILPDA in mice via adeno-associated virus led to a 4-fold increase in liver triglyceride storage, without any changes in key genes involved in de novo lipogenesis, β-oxidation, or lipolysis. Moreover, intracellular lipase activity was not affected by HILPDA overexpression. Strikingly, HILPDA overexpression significantly impaired hepatic triglyceride secretion. Taken together, our data uncover HILPDA as a novel PPAR target that raises hepatic triglyceride storage via regulation of triglyceride secretion.
Introduction
The liver is a key organ in the control of lipid metabolism. During the postprandial phase, the liver is responsible for uptake and processing of chylomicron remnants and actively synthesizes cholesterol, bile acids, and fatty acids. In the fasted state, the liver serves as a sink for adipose tissue-derived plasma free fatty acids, which are fully oxidized or converted into ketone bodies. In addition, incoming fatty acids are esterified into triglycerides and stored within lipid droplets or secreted as VLDL (1, 2). Disturbances in these metabolic pathways can impair liver function and predispose to non-alcoholic fatty liver disease. Many genes involved in the above-mentioned processes are regulated by nuclear receptors (3, 4). Nuclear receptors comprise a family of transcription factors that bind to specific response elements across the genome and regulate the expression of a large network of genes (5). As a group, they are activated by a variety of steroid hormones and other lipid-derived compounds, including fat-soluble vitamins, cholesterol, and fatty acids (6). A group of nuclear receptors that has been extensively studied and that plays a central role in the regulation of nutrient homeostasis in liver and other organs are the peroxisome proliferator-activated receptors (PPARs)2 (7). PPARs are ligand-activated transcription factors that bind a variety of (dietary) fatty acids and fatty acid-derived compounds, including various eicosanoids (8). Furthermore, PPARs are the molecular target of the thiazolidinedione and fibrate classes of drugs used in the treatment of diabetes and dyslipidemia, respectively (9). Three different PPARs can be distinguished: PPARα (Nr1c1), PPARβ/δ (Nr1c2), and PPARγ (Nr1c3) (10, 11). PPARγ is highly expressed in adipose tissue, where it regulates genes involved in fat cell development, lipid deposition, insulin signaling, and inflammation (12). PPARβ/δ is found at high levels in many tissues, and compelling evidence indicates that it stimulates fatty acid oxidation, at least in skeletal muscle and adipose tissue (13, 14). Expression of PPARα is highest in tissues that oxidize fatty acids at a rapid rate, including liver, brown adipose tissue, heart, and kidney (15). Activation of PPARα leads to induction of a large set of target genes involved in various pathways of lipid metabolism, including fatty acid transport, activation, storage, and oxidation (16, 17). Indeed, PPARα can be considered as the master regulator of hepatic lipid metabolism. As a consequence, mice lacking PPARα suffer from hepatic steatosis, hypoglycemia, hypoketonemia, and elevated plasma free fatty acids, all of which are aggravated during fasting (18). Inasmuch as almost all PPARα target genes carry out functions related to hepatic lipid homeostasis,3 suggesting that regulation of a gene by PPARα is predictive of a role in lipid metabolism, screening for PPARα targets may be helpful in the identification of novel genes involved in hepatic lipid homeostasis. In this study, we set out to identify novel PPARα targets using mouse liver slices. We identify Hilpda as a PPAR target in mouse and human hepatocytes and show that it plays an important role in hepatic triglyceride storage and secretion.
EXPERIMENTAL PROCEDURES
Recombinant Adeno-associated Viruses (AAVs)
AAVs were made by Vector Biolabs (Philadelphia, PA). In short, mouse Hilpda cDNA was inserted in pAAV-ALBp-3′iALB, a vector containing a modified albumin promoter flanked by two AAV2-derived inverted terminal repeats. To produce viruses, the above construct was packaged into AAV8 capsid in HEK293 cells by transfecting pAAV-ALBp-3′iALB, AAV2/AAV8 rep/cap vector, and Ad helper vector. Viral particles were purified by two sequential CsCl2 gradients and titrated by qPCR to determine viral genomes. The same amount of AAV2/AAV8 hybrid virus expressing GFP was injected to serve as a control.
Animal Studies
Male wild-type Sv129 and Ppara−/− mice on an Sv129 background were obtained from the Jackson Laboratory (Bar Harbor, ME). Five-month-old wild-type and Ppara−/− mice were fed a diet containing Wy14643 at a concentration of 0.1% for 5 days, after which the livers were excised and snap-frozen. Five-month-old wild-type and Ppara−/− mice were fasted for 24 h, and their livers were collected for analysis of Hilpda mRNA and protein levels.
Male 8-week-old C57BL/6J mice were purchased from Charles River Laboratories (Sulzfeld, Germany) and maintained on a 12-h light-dark cycle with ad libitum access to chow and water. Adeno-associated viruses were administrated in a volume of 100 μl via the tail vein. Ten animals were injected with different amounts of AAV ranging from 1 × 1011 to 5 × 1011 genomic copies to determine the dose required for a significant increase in hepatic HILPDA expression. Subsequent experiments were performed after injection of 2.5 × 1011 genomic copies. Body composition analysis of AAV-GFP and AAV-HILPDA animals 4 weeks postinjection was performed using EchoMRI (Echo Medical Systems, Houston, TX).
Male wild-type C57BL/6 animals (9–12 weeks old) were fed a low fat or high fat diet containing 10 or 45 energy percent fat, respectively (D12450B/D12451, Research Diets Services, Wijk bij Duurstede, The Netherlands). After 16 weeks, the livers were analyzed for triglyceride content and HILPDA expression. All animal experiments were approved by the local animal welfare committees of Wageningen University or the German Cancer Research Center (DKFZ, Heidelberg, Germany), respectively.
VLDL Production Test
AAV-GFP and AAV-HILPDA animals were fasted for 16 h and subsequently anesthetized via intraperitoneal injection of a mixture of Dormicum (midazolam, Roche Applied Science) and Hypnorm (fentanyl/fluanisone, VetaPharma, Leeds, UK) prepared in saline at 7 and 14/0.5 mg/kg, respectively. Then a blood sample was drawn, and the animals were injected intravenously with 500 mg/kg body weight tyloxapol (Sigma-Aldrich) diluted in saline to a 10% (w/v) solution. Every 30 min thereafter, blood was collected via the tail vein over the course of 2 h. Serum samples were assayed for triglycerides using a Triglycerides liquicolormono kit (HUMAN, Wiesbaden, Germany).
Hepatic Pparγ1 Overexpression
Ppara−/− mice were injected with adenovirus encoding LacZ or Pparγ1 via the tail vein and sacrificed 6 days later (19).
Liver Slices
Livers from C57BL/6 mice were briefly perfused with saline, excised, and submerged in ice-cold Krebs-Henseleit buffer supplemented with 11 mm glucose, 25 mm sodium bicarbonate, 10 mm HEPES, and penicillin/streptomycin. Next, 5 mm cylindrical liver cores were prepared with a surgical biopsy punch and sectioned to 200 μm slices using a Krumedieck tissue slicer (Alabama Research and Development, Munford, AL) filled with carbonated Krebs-Henseleit buffer. Liver slices were incubated in William's E Medium (Lonza, Verviers, Belgium) in 6-well plates at 37 °C/5% CO2 under continuous agitation. After 1 h, the media were replaced with fresh William's E Medium containing 0.1% DMSO or 20 μm Wy14643, and the slices were incubated for 6 or 24 h.
Hepatocytes and Hepatoma Cell Line
Rat (Wistar) and mouse (Sv129) hepatocytes were isolated by two-step collagenase perfusion. Cells were plated on collagen-coated 6-well plates. Viability was determined by trypan blue exclusion and was at least 75%. Hepatocytes were suspended in William's E medium (Lonza Bioscience, Verviers, Belgium) supplemented with 10% (v/v) fetal calf serum, 20 milliunits/ml insulin, 50 nm dexamethasone, 100 units/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml fungizone, and 50 μg/ml gentamycin. The next day, cells were incubated in fresh medium in the presence or absence of Wy14643 dissolved in DMSO for 24 h, followed by RNA isolation.
Rat hepatoma FAO cells were grown in DMEM containing 10% (v/v) fetal calf serum, 100 units/ml penicillin, and 100 μg/ml streptomycin. Cells were harvested at different time points after the addition of Wy14643 (5 μm) or DMSO.
Transactivation Assays
A fragment of 645 and 519 bp containing the PPRE localized 1200 bp upstream of the transcription start site (TSS) was amplified from human and mouse genomic DNA, respectively, using the following primers: human Fwd1, 5′-GCCTTCACCTGGCAAAGTAG-3′; human Rev1, 5′-AAGAGCCAGGAGGTCACAGA-3′; mouse Fwd1, 5′-GGATGACTCCCAGAAGTGGA-3′; mouse Rev1, 5′-GACCTGCCTTACCAGTCACC-3′. The resulting fragments were gel-purified using a QIAquick gel extraction kit (Qiagen, Venlo, The Netherlands) and employed in a second PCR to introduce KpnI and BglII restriction sites to the resulting 278- and 219-bp fragments containing the human and mouse PPRE, respectively. Primers used for this PCR were as follows: human Fwd2, 5′-aaggtaccAGTTGGCTCCAAATGTCTGC-3′; human Rev2, 5′-cgaaagatctTGGTGGCTTTAGAACTGGCG-3′; mouse Fwd2, 5′-aaggtaccATTTCCACCTCCTGTGCTTG-3′; mouse Rev2, 5′-cgacagatctTGCGTTCTCATGACTTCCAG-3′. To introduce mutations in both the human and mouse PPRE, an overlap PCR strategy was carried out. First, to generate partially overlapping fragments, the human Fwd2 primer was used together with reverse primer 5′-TCCCCACTGCGACCGGGCCCCTCCTTTCCT-3′, and human Rev2 primer was used with forward primer 5′-AGGAAAGGAGGGGCCCGGTCGCAGTGGGGA-3′, using a wild-type insert as template. The resulting fragments were gel-purified and used in a second PCR with human Fwd2 and Rev2 primers. For mice, the Fwd2 primer was combined with reverse primer 5′-CTTCCGCTGTGACCGGGCCCCTACTTTTCA-3′, and Rev2 primer was combined with forward primer 5′-TGAAAAGTAGGGGCCCGGTCACAGCGGAAG-3′. For the final PCR, purified fragments were combined in a PCR to generate a mutant insert using mouse Fwd2 and Rev2 primers. Resulting fragments were cloned in the KpnI and BglII sites of the pGL3-Promoter vector (Promega, Leiden, The Netherlands). The presence of the correct insert was confirmed with sequencing (EZ-Seq, Macrogen, Amsterdam, The Netherlands) using RV3primer (5′-CTAGCAAAATAGGCTGTCCC-3′) (Promega). Reporter vectors were transfected into the human hepatocellular cell line HepG2 (ATCC, Manassas, VA) in the presence or absence of pSG5 vector expressing PPARα, PPARβ/δ, PPARγ, or pSG5 empty vector. pSG5 expression vector encoding RXR was co-transfected with PPAR expression vectors. A β-galactosidase expression vector was co-transfected in all cases for determination of transfection efficiency. All transfections were performed using polyethyleneimine (PEI). Sixteen hours post-transfection, the cells were incubated with Wy14643 (50 μm), GW510516 (5 μm), rosiglitazone (5 μm), or DMSO, respectively. Reporter activity was measured 24 h after adding the corresponding ligands using the Promega luciferase assay kit (Promega) on a Fluoroskan Ascent apparatus (Thermo Scientific). β-Galactosidase activity in cell lysates was determined using 2-nitrophenyl β-d-galactopyranoside as a substrate, and absorbance values were used to normalize the luciferase measurements. Data presented are based on three individual experiments.
ChIP
Primary hepatocytes were isolated from 10–12-week-old wild-type or Ppara−/− animals on a C57BL/6 background according to published methods (20). Hepatocytes were plated on 15-cm dishes coated with rat tail collagen at a density of 1 × 105 cells/cm2. To cross-link proteins to DNA, the cells were incubated with 1% formaldehyde for 10 min, after which glycine was added to a final concentration of 0.125 m to stop the cross-linking reaction. Cells were collected in ice-cold PBS, washed twice, and snap-frozen in liquid nitrogen. Cell pellets were lysed in 500 μl of lysis buffer containing 0.1% SDS, 1% Triton X-100, 0.15 m NaCl, 1 mm EDTA, 20 mm Tris, pH 8, and protease inhibitor mixture (Pierce). Following 30 cycles of sonication (30 s on, 30 s off, high intensity) at 4 °C, the samples were centrifuged at 20,000 × g for 30 min. Next, 100 μl of the supernatant was diluted 4 times with incubation buffer (0.15% SDS, 1% Triton X-100, 0.15 m NaCl, 1 mm EDTA, 20 mm HEPES) and subsequently mixed with antibodies (anti-PPARα (Santa Cruz Biotechnology, Inc., catalog no. H-98) or IgG (Santa Cruz Biotechnology, catalog no. sc2027)) for 2 h at 4 °C on a rotating wheel. To reduce nonspecific binding, Protein A-Sepharose beads (GE Healthcare) were washed twice and preincubated with incubation buffer supplemented with 1 μg/μl BSA for 2 h at 4 °C on a rotating wheel. Then blocked Protein A-Sepharose mixture was combined with chromatin/antibody mixtures and incubated overnight at 4 °C on a rotating wheel. The beads were subsequently washed in a series of buffers: twice in buffer 1 (0.1% SDS, 0.1% NaDOC, 1% Triton X-100, 0.15 m NaCl, 1 mm EDTA, 20 mm HEPES), once in buffer 2 (0.1% SDS, 0.1% NaDOC, 1% Triton X-100, 0.5 m NaCl, 1 mm EDTA, 20 mm HEPES), once in buffer 3 (0.25 m LiCl, 0.5% NaDOC, 0.5% Nonidet P-40, 1 mm EDTA, 20 mm HEPES), and twice in buffer 4 (1 mm EDTA, 20 mm HEPES). Protein-DNA complexes were eluted in 200 μl of elution buffer (1% SDS + 0.1 m NaHCO3). To disrupt the cross-links between protein-DNA, eluted samples were supplemented with 8 μl of 5 m NaCl, 8 μl of RNase A (0.5 mg/ml), and 10 μl of Proteinase K (20 mg/ml; Qiagen) and incubated overnight at 65 °C. DNA was purified using a QiAQuick PCR purification kit (Qiagen) according to the manufacturer's protocol and analyzed by real-time PCR using primers directed against a 125-bp fragment encompassing the PPRE located 1200 base pairs upstream of the TSS of Hilpda. Sequences of the primers that were used were as follows: Hilpda Fwd, 5′-GCACGTCTCTCTTCTTCTAGGT-3′; Hilpda Rev, 5′-TCGCCTCTAAACTAAACGGAA-3′. Primers against an intergenic region 100 kb upstream of Pdk4 served as control: Fwd, 5′-TTGGCTTGCCAAGCTTCTTC-3′; Rev, 5′-AGGGAACGCATTTCCATCAC-3′.
Plasma Analysis
Non-esterified fatty acids were measured using a NEFA-HR(2) kit, and ketone bodies were measured using an Autokit Total Ketone Bodies assay (Wako Chemicals GmbH, Neuss, Germany). Triglycerides were determined with a Triglycerides liquicolormono kit (HUMAN). Glucose, glycerol, and cholesterol were measured using kits from DiaSys (DiaSys Diagnostic Systems, Holzheim, Germany).
Plasma Lipoprotein Analysis
Pooled plasma samples from AAV-GFP and AAV-HILPDA animals were fractionated using gel filtration high-performance liquid chromatography (HPLC) offered by LipoSEARCH (Skylight Biotech, Inc., Akita, Japan). Effluent was subsequently analyzed for triglycerides and cholesterol content.
Histology
For GFP fluorescence microscopy, samples were fixed in 4% formaldehyde (Sigma-Aldrich) overnight, soaked in PBS containing 15% (w/v) sucrose for 12 h, and then incubated in PBS containing 30% (w/v) sucrose overnight. Tissues were subsequently frozen in Tissue-Tek® O.C.T.TM (Sakura Finetek Holland B.V., Alphen aan de Rijn, The Netherlands) and sectioned to 5 μm and mounted on Superfrost microscopy slides. GFP fluorescence was determined using an Olympus CKX31 fluorescent microscope (Olympus Nederland B.V, Zoeterwoude, The Netherlands).
For hematoxylin and eosin staining, liver pieces were fixed in 4% formaldehyde for 24 h and subsequently embedded in paraffin. Samples were sectioned to 5 μm and stained in hematoxylin for 8 min and eosin for 30 s. Following dehydration in 95% ethanol and two xylene changes, the sections were mounted using a xylene-based mounting medium.
To visualize neutral lipids, liver samples were frozen in Tissue-Tek® O.C.T.TM, sectioned to 5 μm using a cryostat, and mounted on Superfrost microscopy slides. After drying at room temperature for 30 min, the tissue sections were fixed in 4% formaldehyde (Sigma-Aldrich) for 10 min. Following three washes with double-distilled H2O, the slides were incubated in Oil Red O (Sigma-Aldrich) solution (30 mg/ml in 60% isopropyl alcohol) for 10 min. Finally, the slides were rinsed with double-distilled H2O twice and mounted using an aqueous mounting medium.
Hepatic Triglycerides
Liver pieces of ∼50 mg were homogenized to a 5% lysate (m/v) using 10 mm Tris, 2 mm EDTA, 0.25 m sucrose, pH 7.5. Lysates were assayed for triglyceride (TGs) using a Triglycerides liquicolormono kit (HUMAN). TG levels were normalized against liver weight.
Hepatic Glycogen
Tissues (∼50 mg) were homogenized in 0.5 ml of 30% KOH, incubated at 95 °C for 30 min, and centrifuged at 15,000 × g to pellet undigested material. Glycogen was precipitated from the supernatants by the addition of 0.75 ml of 95% ethanol and subsequent centrifugation at 3000 × g for 20 min. Glycogen pellets were washed with fresh 95% ethanol, centrifuged, and air-dried. Pellets were dissolved in 0.25 ml of double-distilled H2O and incubated at 37 °C for 30 min. To digest the glycogen, 5 mg of amyloglucosidase (catalog no. A7420, Sigma-Aldrich) was dissolved in 15 ml of 0.2 m sodium acetate, pH 4.8, and 450 μl of the resulting enzyme solution was mixed with 50 μl of glycogen solution. After incubation at 40 °C for 2 h, the samples were neutralized using 10 μl of 30% KOH. Glucose concentrations were determined with a kit from DiaSys, and protein concentrations were measured with a Pierce BCA kit (Thermo Scientific, Landsmeer, The Netherlands).
Western Blot
Liver samples were homogenized in 50 mm Tris, pH 8.0, 150 mm NaCl, 1% (v/v) Nonidet P-40, 0.5% (v/v) sodium deoxycholate, 0.1% SDS, supplemented with Complete EDTA-free protease inhibitor mixture with a Qiagen TissueLyser II. Samples were incubated on a rotating wheel for 30 min at 4 °C and subsequently centrifuged at 15,000 × g for 15 min at 4 °C. Protein concentration was determined using a Pierce BCA kit (Thermo Scientific), and equal amounts of protein were diluted with 2× Laemmli sample buffer. After boiling, the samples were separated on 4–20 or 8–16% gradient Criterion gels. For plasma analysis, 1 μl of plasma from AAV-GFP or AAV-HILPDA animals was loaded on an 8–16% Criterion gel. Proteins were transferred to PVDF membrane using the Transblot turbo system (Bio-Rad). Antibodies against HILPDA (1:1000; Santa Cruz Biotechnology, catalog no. sc-137518), actin (1:2000; Sigma-Aldrich, catalog no. A2066), HSP90 (1:4000; Cell Signaling Technology, Inc., catalog no. 4874), and goat anti-rabbit (1:5000; Jackson ImmunoResearch, catalog no. 111-035-003) were diluted in 5% (w/v) skimmed milk powder in PBS-T. Primary antibodies were applied overnight at 4 °C, and secondary antibody was applied for 1 h at room temperature. Bands were visualized using Pierce ECL Plus (Thermo Scientific) and a ChemiDoc MP (Bio-Rad). The presence of plasma proteins on the PVDF membrane was confirmed using Ponceau S solution (Sigma-Aldrich).
Gene Expression Analysis Including Microarrays
Mouse liver slices and tissues were homogenized in TRIzol® (Invitrogen) using the Qiagen TissueLyser II and stainless steel beads. One microgram of RNA was subsequently employed for cDNA synthesis using the First Strand cDNA synthesis kit (Thermo Scientific). RNA from a selection of slices and tissues was purified with RNeasy Minikit columns (Qiagen) and analyzed for quality with RNA 6000 Nano chips on the Agilent 2100 Bioanalyzer (Agilent Technologies, Amsterdam, The Netherlands). Purified RNA (100 ng) was labeled with the Ambion WT expression kit (Invitrogen) and hybridized to an Affymetrix Mouse Gene 1.1 ST array plate (Affymetrix, Santa Clara, CA). Hybridization, washing, and scanning were carried out on an Affymetrix GeneTitan platform, and readouts were processed and analyzed according to published methods (21). After filtering with interquartile range of 0.25 and expression signal of 20 on a minimum of two arrays, 1575 genes remained, which were used to generate a scatter plot and volcano plot. Microarray data were submitted to the Gene Expression Omnibus (accession number pending).
qPCRs were performed using SensiMix (Bioline, GC Biotech, Alphen aan den Rijn, The Netherlands) and a CFX384 real-time PCR detection system (Bio-Rad). Primer sequences were derived from the Harvard PrimerBank and synthesized by Eurogentec (Seraing, Belgium). Primer sequences are listed in Table 1. Normalization was performed using the expression of 36b4. qPCR analysis of mouse and human hepatocytes exposed to DMSO or Wy14643 for 6 h was performed as described above using RNA from hepatocytes of six different mouse strains and six human hepatocyte sample sets as described (20).
TABLE 1.
Primer sequences of forward and reverse primers used in qPCR analysis
| Gene | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| 36b4 | ATGGGTACAAGCGCGTCCTG | GCCTTGACCTTTTCAGTAAG |
| Hilpda | TGCTGGGCATCATGTTGACC | TGACCCCTCGTGATCCAGG |
| Pparg | CACAATGCCATCAGGTTTGG | GCTGGT CGATATCACTGGAGATC |
| Cpt1a | CTCAGTGGGAGCGACTCTTCA | GGCCTCTGTGGTACACGACAA |
| Cyp4a10 | ACCACAATGTGCATCAAGGAGGCC | AGGAATGAGTGGCTGTGTCGGGGAGAG |
| Pdk4 | TCTACAAACTCTGACAGGGCTTT | CCGCTTAGTGAACACTCCTTC |
| Srebp1c | GGAGCCATGGATTGCACATT | CCTGTCTCACCCCCAGCATA |
| Fasn | TCCTGGGAGGAATGTAAACAGC | CACAAATTCATTCACTGCAGCC |
| Ldlr | GCATCAGCTTGGACAAGGTGT | GGGAACAGCCACCATTGTTG |
| Mttp | ATACAAGCTCACGTACTCCACT | TCCACAGTAACACAACGTCCA |
ATGL and HSL Activity Assay
COS-7 cells were grown in DMEM (Invitrogen) supplemented with 10% fetal calf serum (Sigma-Aldrich) and 100 IU/ml penicillin, 100 μg/ml streptomycin (Invitrogen). cDNAs encoding ATGL/PNPLA2, HSL, HILPDA, and LacZ were cloned in the pcDNA4/His Max vector (Invitrogen) and transfected into COS-7 cells using Metafectene (Biontex GmbH, München, Germany). 50 μg of either ATGL or HSL recombinant lysate was combined with 50 μg of LacZ or HILPDA lysate. Radioactive emulsion was prepared by sonication of 0.3 mm triolein, phosphatidylcholine/phosphatidylinositol, and 9,10-[3H]triolein (PerkinElmer Life Sciences) in 100 mm potassium phosphate buffer (pH 7) supplemented with 2% BSA (essentially fatty acid-free; Sigma). Protein lysates and radioactive emulsion were incubated for 60 min at 37 °C. Next, the reactions were stopped by adding 3.25 ml of methanol/chloroform/heptane (10:9:7) and 1 ml of 0.1 m potassium carbonate/boric acid buffer (pH 10.5). Samples were centrifuged at 800 × g for 20 min, and radioactivity was determined in 1 ml of the supernatant using a scintillation counter (Beckman Coulter, Fullerton, CA).
Total TG Hydrolase Activity Assay
Liver TG hydrolase activity was measured using published methods (22). In short, samples from AAV-GFP and AAV-HILPDA animals were homogenized in lysis buffer (0.25 m sucrose, 1 mm EDTA, protease inhibitor mixture, pH 7.0) and centrifuged at 20,000 g for 30 min at 4 °C to collect the soluble infranatant fraction. Triolein and 9,10-[3H]triolein (PerkinElmer Life Sciences) were sonicated with phosphatidylcholine and phosphatidylinositol in 100 mm potassium phosphate buffer (pH 7.0) to a final concentration of 1.67 mm triolein. 200 μg of infranatant from each liver sample was incubated with 0.1 ml of radioactive substrate at 37 °C for 60 min. The reaction was terminated by adding 3.25 ml of methanol/chloroform/heptane (10:9:7) and 1 ml of 0.1 m potassium carbonate/boric acid buffer (pH 10.5). Following centrifugation at 800 × g for 20 min, radioactivity was determined in 1 ml of the upper phase using an LS 6500 multipurpose scintillation counter (Beckman Coulter).
Statistical Analysis
Student's t tests or two-way analysis of variance with Bonferroni post hoc tests where appropriate were calculated using GraphPad Prism (GraphPad Software, Inc., La Jolla, CA). Results were considered significant at p < 0.05.
RESULTS
Hildpa Is Regulated by PPARα in Liver Slices and Hepatocytes
To identify potential novel target genes of PPARα, mouse liver slices were treated with the PPARα ligand Wy14643. Whole genome microarray analysis using Affymetrix GeneChips revealed increased expression of numerous classical PPARα targets with roles in fatty acid oxidation (e.g. Ehhadh, Slc22a5, Pdk4, Abcd2, and Cpt1b) and other lipid metabolic pathways (e.g. Acot2, Creb3l3, Fabp4, Mgll, and Cidec) (Fig. 1A). However, several genes of the top 40 genes most highly induced by Wy14643 have not been or have only been very poorly characterized. Among those genes, 2310016C08Rik/Hig2, later annotated as Hilpda, was of particular interest because it has recently been identified as a putative lipid droplet-associated protein that is regulated via a hypoxia-dependent mechanism involving HIF1 (23). The Hilpda gene contains one intron and two exons and encodes a highly conserved protein of 63 and 64 amino acids in humans and mice, respectively (Fig. 1B). Induction of Hildpa expression by Wy14643 was confirmed by qPCR analysis, showing a ∼3-fold increase of Hilpda mRNA (Fig. 1C).
FIGURE 1.
Wy14643 increases Hilpda expression in liver slices and hepatocytes. A, the top 40 most highly induced genes in mouse liver slices exposed to 20 μm Wy14643 for 24 h. Values are expressed as -fold changes over DMSO-treated slices from the same animal (n = 4). B, amino acid sequence of mouse and human HILPDA. C, Hilpda mRNA levels determined by qPCR in mouse liver slices exposed to 20 μm Wy14643 for 6 h. Samples include the four mice described in A plus two additional mice (n = 6). Shown are Hilpda mRNA levels in mouse hepatocytes (D, n = 6 animals from different strains) and human hepatocytes (E, n = 6) after exposure to Wy14643 (10 and 50 μm, respectively) for 6 h. F, Hilpda mRNA levels in primary hepatocytes of wild-type and Ppara−/− mice incubated with Wy14643 (10 μm) for 24 h. G, Hilpda mRNA levels in rat hepatocytes incubated with various nuclear receptor ligands for 24 h (RXR agonist LG1069, 1 μm; Wy14643, 5 μm; Fenofibrate, 50 μm; LXR agonist T0901317, 1 μm; FXR agonist chenodeoxycholic acid, 50 μm). H, Hilpda mRNA levels in rat FAO hepatoma cells incubated with Wy14643 (10 μm) for various times. Wy14643 was removed after 4 h of incubation. Data are mean ± S.E. (error bars). Asterisks, significant differences according to Student's t test: *, p < 0.05; **, p < 0.01.
To substantiate our findings in mouse liver slices, we analyzed Hilpda expression in isolated mouse and human hepatocytes exposed to DMSO or Wy14643 for 6 h. Wy14643 treatment significantly increased Hilpda mRNA in both cell types (Fig. 1, D and E). Induction of Hilpda mRNA by Wy14643 was abolished in hepatocytes from Ppara−/− mice (Fig. 1F). In rat hepatocytes Hilpda mRNA was also induced by fenofibrate and by a synthetic ligand for RXR, which serves as a permissive heterodimeric partner for PPARs, but not by ligands for LXR and FXR (Fig. 1G). Finally, Hilpda mRNA was rapidly induced by Wy14643 in rat FAO hepatoma cells, which represent a very suitable cell line to study PPARα-mediated gene regulation, and quickly returned to baseline upon removal of Wy14643 (Fig. 1H). Thus, expression of Hilpda is positively regulated by PPARα in mouse, rat, and human hepatocytes, suggesting that Hilpda may represent a novel putative PPARα target.
Hepatic Hilpda Expression Is Regulated by PPARs in Vivo
We next sought to explore the PPARα-dependent regulation of HILPDA in vivo by employing wild-type and Ppara−/− animals fed either a control diet or the same diet supplemented with Wy14643 for 5 days. Interestingly, Wy14643 significantly up-regulated Hilpda mRNA in wild-type mice, but not Ppara−/− mice, suggesting a PPARα-mediated regulation of Hilpda (Fig. 2A). Wy14643 also markedly increased HILPDA protein levels in a PPARα-dependent manner (Fig. 2B).
FIGURE 2.
PPAR-dependent regulation of Hilpda expression in vivo. Hilpda gene expression (A, n = 4–5/group) and Western blot (B) analysis of livers from wild-type and Ppara−/− mice fed chow containing 0 or 0.1% Wy14643 for 5 days. Hilpda gene expression (C, n = 4–5/group) and Western blot (D) analysis of livers from fed and 24-h fasted wild-type and Ppara−/− mice. E, analysis of Plin4 and Acacb mRNA levels (E) and Pparg mRNA levels (F) in livers from fed and 24-h fasted wild-type and Ppara−/− mice (n = 4–5/group). G, Hilpda mRNA levels in livers from Ppara−/− mice intravenously injected with adenovirus encoding LacZ or Pparg1 (n = 4/group). Data are mean ± S.E. (error bars). Two-way analysis of variance with Bonferroni post hoc test (A, C, E, and F) or Student's t test (G) was performed. *, p < 0.05; **, p < 0.01; ##, p < 0.01. *, difference between WT and Ppara−/− (A, C, E, and F); #, difference between control/Wy14643 (A) or fed/fasted (C, E, and F).
PPARα is an important regulator in the metabolic response to fasting, and the expression of many PPARα regulated genes increases upon fasting (18). However, we found a much less pronounced increase in Hilpda mRNA and protein in livers of fasted wild-type mice compared with Ppara−/− mice. Indeed, Hilpda mRNA was increased 1.4- and 9.2-fold by fasting in wild-type and Ppara−/− mice, respectively (Fig. 2C), which was confirmed by Western blot (Fig. 2D). Other Wy14643-induced genes that show a similar expression pattern during fasting include Plin4 and Acacb (Fig. 2E). PPARγ has been shown to partially compensate for PPARα deletion during high-fat diet feeding or fasting (24, 25). Indeed, Pparg expression was increased in livers of Ppara−/− animals (Fig. 2F). Moreover, adenovirus-mediated hepatic Pparg1 overexpression markedly increased Hilpda mRNA expression (Fig. 2G). Together, these data suggest that Hilpda expression in liver is regulated by both PPARα and PPARγ.
Hilpda Is a Direct PPAR Target
In silico screening using Nubiscan revealed the presence of a potential PPRE around 1200 base pairs (bp) upstream of the TSS of Hilpda, which is highly conserved between mice and humans and part of a highly conserved region (26). To determine whether this PPRE contributes to PPAR-dependent regulation of Hilpda, we generated luciferase reporter vectors containing a ∼200-bp genomic fragment flanking the mouse or human PPRE (Fig. 3A). Notably, all PPARs stimulated luciferase activity driven by mouse wild-type PPRE (Fig. 3, B–D). By contrast, mutating the indicated adenine residues (Fig. 3A) completely abolished the PPAR-dependent increase in luciferase activity (Fig. 3, B–D). Similarly, PPARα induced luciferase reporter activity when co-transfected with human wild-type PPRE but not mutant PPRE (Fig. 3E).
FIGURE 3.
PPAR mediated regulation of Hilpda expression is mediated by a PPRE 1200 bp upstream of the transcription start site. A, sequence of the human and mouse PPRE located 1200 bp upstream of the TSS of Hilpda. Three adenine residues were mutated to cytosine to serve as negative controls. B–D, HepG2 cells were transfected with luciferase reporter constructs containing wild-type or mutant mouse (B–D) or human (E) PPRE together with empty vector or PPAR/RXRα expression vectors as indicated. All conditions contain equal amounts of DNA, and β-galactosidase was co-transfected for normalization. Transfected cells were subsequently exposed to DMSO, 50 μm Wy14643, 5 μm GW501516, or 5 μm rosiglitazone for 24 h. Luciferase readings in cell lysates were normalized against β-galactosidase activity (n = 3). F, chromatin was extracted from wild-type and Ppara−/− primary hepatocytes, and a ChIP assay was performed using antibodies specific for PPARα or IgG. PCR was performed using primers flanking the conserved PPRE 1200 bp upstream of the Hilpda TSS or negative control primers to amplify an intergenic region 100 kb upstream of Pdk4 (n = 3). Data are mean ± S.E. (error bars). *, significant difference according to Student's t test (p < 0.01).
To explore if the PPRE is occupied by PPARα in liver, we performed chromatin immunoprecipitation experiments using primary hepatocytes from wild-type and Ppara−/− mice. Consistent with direct PPAR binding, we efficiently recovered a 125-base pair region surrounding the conserved PPRE from PPARα immunoprecipitates in wild-type hepatocytes compared with Ppara−/− hepatocytes (Fig. 3F). Taken together, these data demonstrate the presence of a functional PPRE 1200 bp upstream of the TSS of Hilpda, suggesting that Hilpda is a novel PPAR target gene.
Hepatic Overexpression of HILPDA Results in a Fatty Liver
To address the functional role of HILPDA in liver, we employed adeno-associated viruses expressing GFP (AAV-GFP) or HILPDA (AAV-HILPDA) under the control of a modified albumin promoter. Intravenous injection of 2.5 × 1011 genomic copies led to the infection of almost all cells and caused a marked increase in HILPDA expression 4 weeks after injection (Fig. 4, A and B). Importantly, HILPDA protein expressed by the AAV had the same molecular weight as the endogenous protein (Fig. 4C). Consistent with the absence of a signal peptide as observed by in silico analysis but in contrast to published data (27), we were unable to detect HILPDA in the circulation, even after injecting higher doses (Fig. 4D). No differences were found in body weight between AAV-GFP and AAV-HILPDA mice 4 weeks postinjection (Fig. 4E). Similarly, the weights of livers, epididymal fat pads, and brown adipose tissue (Fig. 4, F–H) were not different between the two sets of mice. Furthermore, EchoMRI analysis did not show any difference in lean mass or fat mass (Fig. 4, I and J).
FIGURE 4.
Hepatic overexpression of HILPDA does not impact body weight. A, fluorescent microscopy of liver sections from AAV-GFP and AAV-HILPDA mice 4 weeks postinjection of the corresponding AAVs. B, HILPDA protein expression in livers from AAV-GFP and AAV-HILPDA animals. HSP90 served as a loading control. C, Western blot analysis of endogenous and overexpressed HILPDA. For endogenous protein an aliquot of a sample used in Fig. 2B (wild-type animal fed a diet supplemented with Wy14643) was loaded. D, HILPDA Western blot on plasma samples of animals injected with 1–5 × 1011 genomic copies of AAV-HILPDA. An aliquot of diluted liver lysate from an animal injected with 2.5 × 1011 genomic copies of AAV-HILPDA virus was used as a positive control. Ponceau S staining is shown to illustrate the presence of plasma proteins. E, body weight of AAV-GFP and AAV-HILPDA animals 4 weeks after injection of corresponding AAVs (n = 10–13 animals/group). F–H, liver (F), epididymal white adipose tissue (eWAT) (G), and brown adipose tissue (BAT) (H) weight of AAV-GFP and AAV-HILPDA animals (n = 8 animals/group). I and J, fat mass (I) and lean mass (J) in AAV-GFP and AAV-HILPDA animals determined using EchoMRI (n = 10–13 animals/group). Data are mean ± S.E. (error bars).
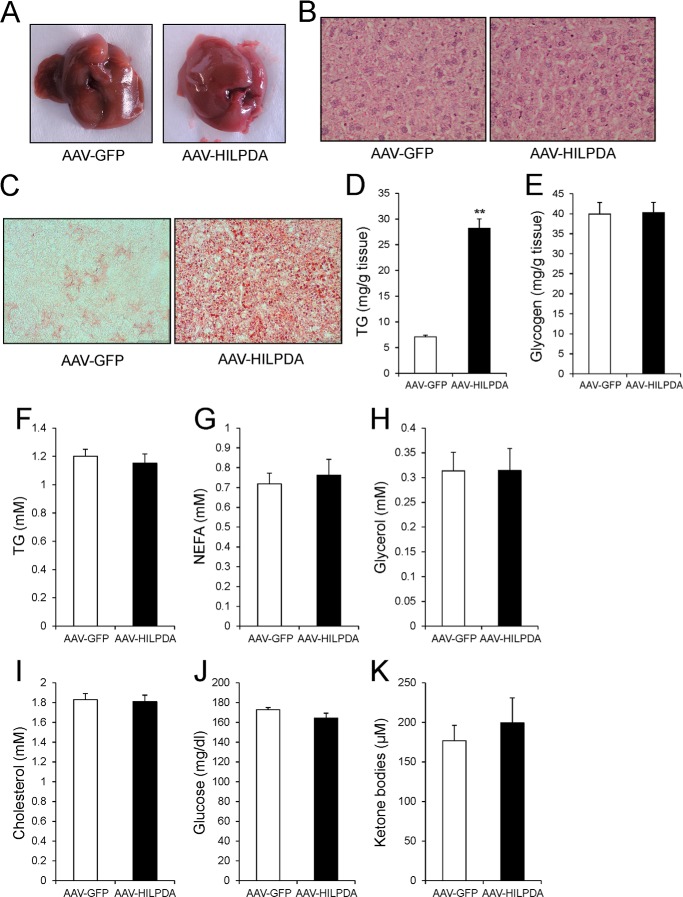
Strikingly, livers from AAV-HILPDA animals showed a different, more yellowish color compared with AAV-GFP animals (Fig. 5A), which often indicates increased fat deposition. Hematoxylin and eosin staining of livers from AAV-GFP and AAV-HILPDA animals indicated no major histological perturbations associated with HILPDA overexpression (Fig. 5B). However, qualitative and quantitative analysis revealed a significant increase in hepatic TG deposition upon HILPDA overexpression (Fig. 5, C and D). In contrast, hepatic hepatic glycogen content was unchanged (Fig. 5E). Plasma TG, NEFA, glycerol, glucose ketone bodies, and cholesterol were not different between AAV-GFP and AAV-HILPDA mice (Fig. 5, F–K). Thus, HILPDA is involved in regulation of hepatic TG metabolism but has no effect on plasma lipid levels.
FIGURE 5.
AAV-mediated HILPDA overexpression induces hepatic steatosis. A, gross morphology of livers from AAV-GFP and AAV-HILPDA animals 4 weeks after injection of viruses. Animals were sacrificed under fed conditions. B, representative H&E staining of livers from AAV-GFP and AAV-HILPDA animals. C, Oil Red O staining of liver sections from AAV-GFP and AAV-HILPDA animals. D, hepatic TG content determined 4 weeks postinjection of AAVs (n = 8 animals/group). E, hepatic glycogen levels in AAV-GFP and AAV-HILPDA animals (n = 8 animals/group). F–K, plasma TG (F), NEFA (G), glycerol (H), cholesterol (I), glucose (J), and ketone body (K) levels in AAV-GFP and AAV-HILPDA animals 4 weeks postinjection of respective viruses (n = 7–8 animals/group). Data are mean ± S.E. (error bars). Asterisks, significant difference according to Student's t test (**, p < 0.01).
HILPDA Does Not Affect Neutral Lipase Activity
Hepatic TG storage is the net result of several processes, including fatty acid β-oxidation, de novo lipogenesis, fatty acid uptake, TG hydrolysis, and TG secretion as VLDL. To identify the mechanism underlying increased lipid deposition in livers of AAV-HILPDA mice, we analyzed the expression of a selection of genes involved in the above mentioned metabolic pathways. However, no differences were found in the expression of relevant genes involved in β-oxidation, lipogenesis, fatty acid uptake, TG hydrolysis, and TG secretion (Fig. 6A). Gene expression profiling by microarray revealed only very minor changes in hepatic gene expression in AAV-HILDPA mice, with only 139 genes significantly altered upon HILPDA overexpression (liberal p value < 0.01), and more than 75% of these genes were changed less than 1.5-fold (Fig. 6, B and C). Gadd45g, a PPARγ target, was the most highly induced gene, showing a 2.5-fold increase in expression (Fig. 6C). Furthermore, Ingenuity Pathway analysis revealed no significantly altered pathways related to lipid metabolism (data not shown). Together, these data suggest that HILPDA overexpression does not induce major changes in expression of genes involved in hepatic lipid metabolism.
FIGURE 6.
Hepatic overexpression of HILPDA does not affect the expression of key genes involved in β-oxidation or de novo lipogenesis. A, qPCR analysis of important genes related to β-oxidation, lipogenesis, hepatic lipid uptake, and VLDL production in livers of AAV-GFP and AAV-HILPDA animals (n = 7–8 animals/group). B, scatter plot of the logarithmic expression signal of 1575 genes left after the set filter criteria (see “Experimental Procedures”). Average expression signal of each gene in the GFP group is plotted on the x axis, and expression in the HILPDA group is plotted on the y axis (n = 2 arrays/group). C, volcano plot displaying the fold induction of 1575 genes (x axis) related to their significance level (y axis) upon HILPDA overexpression. Genes that met the inclusion criteria (see “Experimental Procedures”) were included in the analysis. Arrow, Gadd45g. Data are mean ± S.E. (error bars).
So far, HILPDA shows several similarities to the previously identified G0s2 gene (28): small proteins with no homology to other proteins, PPARα targets, and overexpression causes fatty liver (29, 30). Recently, G0S2 was characterized as a regulator of adipose tissue triglyceride lipase (ATGL/PNPLA2) (31). To explore a possible effect of HILPDA overexpression on intracellular lipase activity, we performed TG hydrolase activity assays. No difference in total TG hydrolase activity was observed between AAV-GFP and AAV-HILPDA livers (Fig. 7A). HILPDA also did not affect the activity of ATGL and HSL in vitro (Fig. 7B). Thus, HILPDA does not seem to directly impact neutral lipase activity.
FIGURE 7.
AAV-HILPDA animals secrete less triglyceride. A, total TG hydrolyase activity in liver samples from AAV-GFP and AAV-HILPDA animals (n = 8 animals/group). B, mouse ATGL or HSL was overexpressed in COS-7 cells, and lysates were assayed for hydrolase activity in the presence of COS-7 lysates from cells transfected with expression vectors for LacZ or HILPDA (n = 2). C, plasma TG levels after intravenous administration of tyloxapol to determine hepatic TG secretion in 16-h fasted AAV-GFP and AAV-HILPDA animals (n = 7 animals/group). D and E, lipoprotein-associated TG (D) and cholesterol (E) levels in HPLC fractions of pooled plasma samples from AAV-GFP and AAV-HILPDA animals. F–H, Hilpda mRNA (F) and protein levels (G) in wild-type animals after a 16-week HFD intervention resulting in the development of hepatic steatosis (H) (n = 8–10 animals/group). Data are mean ± S.E. (error bars). Asterisks, significant differences according to Student's t test (**, p < 0.01).
Hepatic TG Secretion Is Decreased with HILPDA Overexpression
To determine if HILPDA might impact TG secretion, we performed a VLDL production test in AAV-GFP and AAV-HILPDA mice using the lipase inhibitor tyloxapol. Remarkably, HILPDA overexpression markedly reduced hepatic TG secretion (Fig. 7C). To investigate whether HILPDA overexpression is associated with a difference in lipoprotein particle size or composition, we performed lipoprotein profiling using gel filtration high-performance liquid chromatography. However, chylomicron and VLDL particle size and TG content, as well as LDL and HDL size and cholesterol content, were unchanged between the two sets of mice (Table 2 and Fig. 7, D and E).
TABLE 2.
Diameter of chylomicron, VLDL, LDL, and HDL particles in AAV-GFP and AAV-HILPDA animals
| AAV-GFP | AAV-HILPDA | |
|---|---|---|
| nm | nm | |
| Chylomicron | >100 | >100 |
| VLDL | 46.46 | 46.99 |
| LDL | 24.67 | 24.78 |
| HDL | 11.08 | 11.15 |
Finally, we explored the potential role of HILPDA in high fat diet-induced fatty liver. Interestingly, feeding wild-type mice a high-fat diet for 16 weeks did not elicit a change in liver HILPDA expression (Fig. 7, F and G), despite the development of severe hepatic steatosis (Fig. 7H). According to microarray analysis, HILPDA expression in liver was also not significantly changed in healthy obese subjects, in patients with steatosis, and in NASH patients (-fold change ≤1.1, raw p value ≥ 0.2) (32). Similarly, microarray indicated that HILPDA expression was not related to human liver phenotype ranging from healthy to inflamed without steatosis, inflamed with steatosis, and inflamed with steatosis and fibrosis.4
DISCUSSION
PPARα plays important roles in hepatic lipid metabolism by regulating the transcription of numerous genes involved in lipid metabolic pathways. Here we provide evidence that Hilpda is a novel PPARα target gene that promotes hepatic TG deposition by reducing TG secretion.
One of our main findings is the inhibition of VLDL-TG secretion by hepatic HILPDA overexpression. VLDL assembly and secretion have been studied extensively over the last few decades (33). Central to the synthesis of VLDL is the lipidation of apolipoprotein B100 (APOB100) in the endoplasmic reticulum by the microsomal triglyceride transfer protein to generate pre-VLDL (34). Next, large amounts of lipids are transferred to pre-VLDL to yield mature VLDL, although it is not exactly clear whether this lipidation takes place solely in the endoplasmic reticulum or whether the Golgi-apparatus is involved as well (35, 36). Indeed, many details regarding VLDL synthesis, endoplasmic reticulum to Golgi transport, and VLDL exocytosis remain unknown. Recently, additional players have been implicated in the lipidation and secretion of VLDL, including other apolipoproteins (APOC3), lipid droplet-associated proteins (CIDEB), and sorting receptors (SORT1) (37–39). In vitro studies have previously identified HILPDA as a lipid droplet-associated protein (23), and it is therefore of particular interest that the lipid droplet-associated protein PLIN2 has been shown to inhibit VLDL synthesis due to increased shunting of TG to cytosolic lipid droplets (38, 40). Future studies will have to determine the precise molecular mechanism underlying regulation of hepatic TG levels and secretion by HILPDA as well as the molecular details of its association with lipid droplets.
Reduced hepatic VLDL-TG secretion might be expected to lead to a decrease in plasma TG levels. However, we did not observe a change in plasma TG levels when comparing AAV-GFP and AAV-HILPDA animals under either random fed conditions or after an overnight fast. A possible explanation is that VLDL produced in livers of mice overexpressing HILPDA is cleared from the circulation less efficiently, potentially due to an altered apolipoprotein make up decorating the VLDL.
The limited data available on HILPDA suggest that it may represent a lipid droplet-associated protein that is induced by hypoxia (23). Hypoxia is known to elicit numerous metabolic changes driven by the HIF transcription factors, including up-regulation of many glycolytic genes (41, 42). The specific regulatory effect of hypoxia/HIF1 on lipid metabolism is less well characterized, yet it is well known that hypoxia raises intracellular lipids (43). The rationale for hypoxia-induced lipid accumulation is not well characterized, but it may be part of a general mechanism aimed at directing non-oxidized fatty acids toward storage and thus shifting them away from potential lipotoxic effects. Taking into account the ability of HILPDA to promote intracellular lipid storage, it can be hypothesized that HILPDA may play a role in hypoxia-induced lipid accumulation.
The great majority of PPARα target genes identified so far are induced by fasting and by synthetic PPARα agonists specifically in wild-type mice but not Ppara−/− mice, which include Fgf21, Cpt1a, Ehhadh, Hmcgcs2, and many others (44). In contrast, Hilpda expression was induced by fasting specifically in Ppara−/− mice but much less in wild-type mice, resembling the expression pattern of a small set of PPARα targets that includes Lpin2, Plin4, and Acacb. A number of genes with a similar expression pattern were found to be regulated by PPARβ/δ, which may also be true for Hilpda (44). Alternatively, up-regulation of Hilpda in fasted Ppara−/− mice may be mediated by PPARγ, expression of which is elevated in livers of Ppara−/− mice and which potently induces Hilpda expression in liver, as shown by adenoviral overexpression of PPARγ. In this context, it would have been worthwhile to have information on PPARγ protein in Ppara−/− livers. Transactivation experiments indicated that all three PPARs were able to activate a reporter construct driven by a portion of the Hilpda promoter, suggesting that Hilpda probably represents a pan-PPAR target gene. Accordingly, it can be hypothesized that, depending on the specific stimulus, Hilpda can be regulated by either PPARα, PPARβ/δ, or PPARγ.
Induction of HILPDA by PPARα can be predicted to promote liver TG storage. By contrast, administration of synthetic PPARα agonists to rodents decreases fatty liver (45–50). This apparent discrepancy can be reconciled by the notion that Hilpda is one of more than 100 genes involved in lipid metabolism that are induced by PPARα. A number of these PPARα targets, including several genes involved in lipid droplets and TG synthesis, will oppose lowering of liver TG. The disparate effects of different PPARα targets are seemingly counterintuitive but may reflect delicate feedback mechanisms aimed at maintaining lipid homeostasis. It can be hypothesized that induction of genes involved in lipid droplet biology and TG synthesis by PPARα may reflect a broader role of PPARα in metabolizing and neutralizing large amounts of free fatty acids.
Synthetic PPARα agonists in the form of fibrates are used clinically for their ability to raise plasma HDL and reduce plasma TG levels (9). The plasma TG-lowering action of fibrates is primarily achieved via augmentation of plasma TG clearance, although there is in vivo and in vitro evidence that PPARα agonists also lower VLDL-TG production, potentially due to induction of PLIN2 and FABP1 and subsequent storage of TGs in cytosolic lipid droplets (51–54). Conversely, deletion of PPARα leads to elevated VLDL-TG production (55, 56). Conceivably, HILPDA may be involved in mediating the suppressive effect of synthetic PPARα agonists on TG secretion by the liver. Overall, it is evident that the role of PPARs in regulation of hepatic lipid homeostasis goes beyond the induction of genes involved in β-oxidation and ketogenesis. The direct effect of HILPDA on β-oxidation, lipid uptake, and lipogenesis has not been tested; nevertheless, we did not observe any difference in mRNA levels of key genes involved in these processes between AAV-GFP and AAV-HILPDA animals.
In conclusion, we identify Hilpda as a novel PPAR-regulated gene involved in the regulation of hepatic TG metabolism. Increased HILPDA expression decreases the production of VLDL-TG, resulting in the development of a fatty liver. Future experiments aim to elucidate the mechanism underlying the decrease in VLDL production.
Acknowledgments
We thank Yvonne Feuchter and Karin Mudde for technical assistance.
This work was supported by Fondation Leducq Grant 12CVD04 and by Netherlands Organization for Health Research and Development Grant 40-00812-98-08030, NWO ZonMW.
S. Kersten (2014) Mol. Metab., in press.
F. Mattijssen, M. V. Boekschoten and S. Kersten, unpublished data.
- PPAR
- peroxisome proliferator-activated receptor
- HILPDA
- hypoxia-inducible lipid droplet-associated
- PPRE
- PPAR response element
- AAV
- adeno-associated virus
- TG
- triglyceride
- qPCR
- quantitative PCR
- TSS
- transcription start site.
REFERENCES
- 1. McPherson P. A., McEneny J. (2012) The biochemistry of ketogenesis and its role in weight management, neurological disease and oxidative stress. J. Physiol. Biochem. 68, 141–151 [DOI] [PubMed] [Google Scholar]
- 2. Sun Z., Lazar M. A. (2013) Dissociating fatty liver and diabetes. Trends Endocrinol. Metab. 24, 4–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pawlak M., Lefebvre P., Staels B. (2012) General molecular biology and architecture of nuclear receptors. Curr. Top. Med. Chem. 12, 486–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakamura M. T., Cheon Y., Li Y., Nara T. Y. (2004) Mechanisms of regulation of gene expression by fatty acids. Lipids 39, 1077–1083 [DOI] [PubMed] [Google Scholar]
- 5. Chawla A., Repa J. J., Evans R. M., Mangelsdorf D. J. (2001) Nuclear receptors and lipid physiology: opening the X-files. Science 294, 1866–1870 [DOI] [PubMed] [Google Scholar]
- 6. Sonoda J., Pei L., Evans R. M. (2008) Nuclear receptors: decoding metabolic disease. FEBS Lett. 582, 2–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kersten S., Desvergne B., Wahli W. (2000) Roles of PPARs in health and disease. Nature 405, 421–424 [DOI] [PubMed] [Google Scholar]
- 8. Schupp M., Lazar M. A. (2010) Endogenous ligands for nuclear receptors: digging deeper. J. Biol. Chem. 285, 40409–40415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lalloyer F., Staels B. (2010) Fibrates, glitazones, and peroxisome proliferator-activated receptors. Arterioscler. Thromb. Vasc. Biol. 30, 894–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dreyer C., Krey G., Keller H., Givel F., Helftenbein G., Wahli W. (1992) Control of the peroxisomal β-oxidation pathway by a novel family of nuclear hormone receptors. Cell 68, 879–887 [DOI] [PubMed] [Google Scholar]
- 11. Poulsen L., Siersbæk M., Mandrup S. (2012) PPARs: fatty acid sensors controlling metabolism. Semin. Cell Dev. Biol. 23, 631–639 [DOI] [PubMed] [Google Scholar]
- 12. Tontonoz P., Spiegelman B. M. (2008) Fat and beyond: the diverse biology of PPARγ. Annu. Rev. Biochem. 77, 289–312 [DOI] [PubMed] [Google Scholar]
- 13. Kleiner S., Nguyen-Tran V., Baré O., Huang X., Spiegelman B., Wu Z. (2009) PPARδ agonism activates fatty acid oxidation via PGC-1α but does not increase mitochondrial gene expression and function. J. Biol. Chem. 284, 18624–18633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Roberts L. D., Murray A. J., Menassa D., Ashmore T., Nicholls A. W., Griffin J. L. (2011) The contrasting roles of PPARδ and PPARγ in regulating the metabolic switch between oxidation and storage of fats in white adipose tissue. Genome Biol. 12, R75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mandard S., Müller M., Kersten S. (2004) Peroxisome proliferator-activated receptor α target genes. Cell Mol. Life Sci. 61, 393–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Peters J. M., Hennuyer N., Staels B., Fruchart J. C., Fievet C., Gonzalez F. J., Auwerx J. (1997) Alterations in lipoprotein metabolism in peroxisome proliferator-activated receptor α-deficient mice. J. Biol. Chem. 272, 27307–27312 [DOI] [PubMed] [Google Scholar]
- 17. Aoyama T., Peters J. M., Iritani N., Nakajima T., Furihata K., Hashimoto T., Gonzalez F. J. (1998) Altered constitutive expression of fatty acid-metabolizing enzymes in mice lacking the peroxisome proliferator-activated receptor α (PPARα). J. Biol. Chem. 273, 5678–5684 [DOI] [PubMed] [Google Scholar]
- 18. Kersten S., Seydoux J., Peters J. M., Gonzalez F. J., Desvergne B., Wahli W. (1999) Peroxisome proliferator-activated receptor alpha mediates the adaptive response to fasting. J. Clin. Invest. 103, 1489–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yu S., Matsusue K., Kashireddy P., Cao W. Q., Yeldandi V., Yeldandi A. V., Rao M. S., Gonzalez F. J., Reddy J. K. (2003) Adipocyte-specific gene expression and adipogenic steatosis in the mouse liver due to peroxisome proliferator-activated receptor γ1 (PPARγ1) overexpression. J. Biol. Chem. 278, 498–505 [DOI] [PubMed] [Google Scholar]
- 20. Rakhshandehroo M., Hooiveld G., Müller M., Kersten S. (2009) Comparative analysis of gene regulation by the transcription factor PPARα between mouse and human. PLoS One 4, e6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lichtenstein L., Mattijssen F., de Wit N. J., Georgiadi A., Hooiveld G. J., van der Meer R., He Y., Qi L., Köster A., Tamsma J. T., Tan N. S., Müller M., Kersten S. (2010) Angptl4 protects against severe proinflammatory effects of saturated fat by inhibiting fatty acid uptake into mesenteric lymph node macrophages. Cell Metab. 12, 580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Haemmerle G., Lass A., Zimmermann R., Gorkiewicz G., Meyer C., Rozman J., Heldmaier G., Maier R., Theussl C., Eder S., Kratky D., Wagner E. F., Klingenspor M., Hoefler G., Zechner R. (2006) Defective lipolysis and altered energy metabolism in mice lacking adipose triglyceride lipase. Science 312, 734–737 [DOI] [PubMed] [Google Scholar]
- 23. Gimm T., Wiese M., Teschemacher B., Deggerich A., Schödel J., Knaup K. X., Hackenbeck T., Hellerbrand C., Amann K., Wiesener M. S., Höning S., Eckardt K. U., Warnecke C. (2010) Hypoxia-inducible protein 2 is a novel lipid droplet protein and a specific target gene of hypoxia-inducible factor-1. FASEB J. 24, 4443–4458 [DOI] [PubMed] [Google Scholar]
- 24. Patsouris D., Reddy J. K., Müller M., Kersten S. (2006) Peroxisome proliferator-activated receptor α mediates the effects of high-fat diet on hepatic gene expression. Endocrinology 147, 1508–1516 [DOI] [PubMed] [Google Scholar]
- 25. Hashimoto T., Cook W. S., Qi C., Yeldandi A. V., Reddy J. K., Rao M. S. (2000) Defect in peroxisome proliferator-activated receptor α-inducible fatty acid oxidation determines the severity of hepatic steatosis in response to fasting. J. Biol. Chem. 275, 28918–28928 [DOI] [PubMed] [Google Scholar]
- 26. Podvinec M., Kaufmann M. R., Handschin C., Meyer U. A. (2002) NUBIScan, an in silico approach for prediction of nuclear receptor response elements. Mol. Endocrinol. 16, 1269–1279 [DOI] [PubMed] [Google Scholar]
- 27. Kenny P. A., Enver T., Ashworth A. (2005) Receptor and secreted targets of Wnt-1/β-catenin signalling in mouse mammary epithelial cells. BMC Cancer 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zandbergen F., Mandard S., Escher P., Tan N. S., Patsouris D., Jatkoe T., Rojas-Caro S., Madore S., Wahli W., Tafuri S., Müller M., Kersten S. (2005) The G0/G1 switch gene 2 is a novel PPAR target gene. Biochem. J. 392, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y., Zhang Y., Qian H., Lu J., Zhang Z., Min X., Lang M., Yang H., Wang N., Zhang P. (2013) The g0/g1 switch gene 2 is an important regulator of hepatic triglyceride metabolism. PLoS One 8, e72315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang X., Xie X., Heckmann B. L., Saarinen A. M., Czyzyk T. A., Liu J. (2014) Target disruption of G0/G1 switch gene 2 enhances adipose lipolysis, alters hepatic energy balance, and alleviates high fat diet-induced liver steatosis. Diabetes 63, 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yang X., Lu X., Lombès M., Rha G. B., Chi Y. I., Guerin T. M., Smart E. J., Liu J. (2010) The G0/G1 switch gene 2 regulates adipose lipolysis through association with adipose triglyceride lipase. Cell Metab. 11, 194–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ahrens M., Ammerpohl O., von Schönfels W., Kolarova J., Bens S., Itzel T., Teufel A., Herrmann A., Brosch M., Hinrichsen H., Erhart W., Egberts J., Sipos B., Schreiber S., Häsler R., Stickel F., Becker T., Krawczak M., Röcken C., Siebert R., Schafmayer C., Hampe J. (2013) DNA methylation analysis in nonalcoholic fatty liver disease suggests distinct disease-specific and remodeling signatures after bariatric surgery. Cell Metab. 18, 296–302 [DOI] [PubMed] [Google Scholar]
- 33. Tiwari S., Siddiqi S. A. (2012) Intracellular trafficking and secretion of VLDL. Arterioscler. Thromb. Vasc. Biol. 32, 1079–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gordon D. A., Jamil H. (2000) Progress towards understanding the role of microsomal triglyceride transfer protein in apolipoprotein-B lipoprotein assembly. Biochim. Biophys. Acta 1486, 72–83 [DOI] [PubMed] [Google Scholar]
- 35. Gusarova V., Brodsky J. L., Fisher E. A. (2003) Apolipoprotein B100 exit from the endoplasmic reticulum (ER) is COPII-dependent, and its lipidation to very low density lipoprotein occurs post-ER. J. Biol. Chem. 278, 48051–48058 [DOI] [PubMed] [Google Scholar]
- 36. Yamaguchi J., Gamble M. V., Conlon D., Liang J. S., Ginsberg H. N. (2003) The conversion of apoB100 low density lipoprotein/high density lipoprotein particles to apoB100 very low density lipoproteins in response to oleic acid occurs in the endoplasmic reticulum and not in the Golgi in McA RH7777 cells. J. Biol. Chem. 278, 42643–42651 [DOI] [PubMed] [Google Scholar]
- 37. Qin W., Sundaram M., Wang Y., Zhou H., Zhong S., Chang C. C., Manhas S., Yao E. F., Parks R. J., McFie P. J., Stone S. J., Jiang Z. G., Wang C., Figeys D., Jia W., Yao Z. (2011) Missense mutation in APOC3 within the C-terminal lipid binding domain of human ApoC-III results in impaired assembly and secretion of triacylglycerol-rich very low density lipoproteins: evidence that ApoC-III plays a major role in the formation of lipid precursors within the microsomal lumen. J. Biol. Chem. 286, 27769–27780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li X., Ye J., Zhou L., Gu W., Fisher E. A., Li P. (2012) Opposing roles of cell death-inducing DFF45-like effector B and perilipin 2 in controlling hepatic VLDL lipidation. J. Lipid Res. 53, 1877–1889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Strong A., Ding Q., Edmondson A. C., Millar J. S., Sachs K. V., Li X., Kumaravel A., Wang M. Y., Ai D., Guo L., Alexander E. T., Nguyen D., Lund-Katz S., Phillips M. C., Morales C. R., Tall A. R., Kathiresan S., Fisher E. A., Musunuru K., Rader D. J. (2012) Hepatic sortilin regulates both apolipoprotein B secretion and LDL catabolism. J. Clin. Invest. 122, 2807–2816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Magnusson B., Asp L., Boström P., Ruiz M., Stillemark-Billton P., Lindén D., Borén J., Olofsson S. O. (2006) Adipocyte differentiation-related protein promotes fatty acid storage in cytosolic triglycerides and inhibits secretion of very low-density lipoproteins. Arterioscler. Thromb. Vasc. Biol. 26, 1566–1571 [DOI] [PubMed] [Google Scholar]
- 41. Webster K. A. (2003) Evolution of the coordinate regulation of glycolytic enzyme genes by hypoxia. J. Exp. Biol. 206, 2911–2922 [DOI] [PubMed] [Google Scholar]
- 42. Semenza G. L. (2011) Regulation of metabolism by hypoxia-inducible factor 1. Cold Spring Harb. Symp. Quant. Biol. 76, 347–353 [DOI] [PubMed] [Google Scholar]
- 43. Gordon G. B., Barcza M. A., Bush M. E. (1977) Lipid accumulation of hypoxic tissue culture cells. Am. J. Pathol. 88, 663–678 [PMC free article] [PubMed] [Google Scholar]
- 44. Sanderson L. M., Degenhardt T., Koppen A., Kalkhoven E., Desvergne B., Müller M., Kersten S. (2009) Peroxisome proliferator-activated receptor β/δ (PPARβ/δ) but not PPARα serves as a plasma free fatty acid sensor in liver. Mol. Cell. Biol. 29, 6257–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ip E., Farrell G., Hall P., Robertson G., Leclercq I. (2004) Administration of the potent PPARα agonist, Wy-14,643, reverses nutritional fibrosis and steatohepatitis in mice. Hepatology 39, 1286–1296 [DOI] [PubMed] [Google Scholar]
- 46. Ip E., Farrell G. C., Robertson G., Hall P., Kirsch R., Leclercq I. (2003) Central role of PPARα-dependent hepatic lipid turnover in dietary steatohepatitis in mice. Hepatology 38, 123–132 [DOI] [PubMed] [Google Scholar]
- 47. Kashireddy P. V., Rao M. S. (2004) Lack of peroxisome proliferator-activated receptor α in mice enhances methionine and choline deficient diet-induced steatohepatitis. Hepatol. Res. 30, 104–110 [DOI] [PubMed] [Google Scholar]
- 48. Kong L., Ren W., Li W., Zhao S., Mi H., Wang R., Zhang Y., Wu W., Nan Y., Yu J. (2011) Activation of peroxisome proliferator activated receptor α ameliorates ethanol induced steatohepatitis in mice. Lipids Health Dis. 10, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rakhshandehroo M., Sanderson L. M., Matilainen M., Stienstra R., Carlberg C., de Groot P. J., Müller M., Kersten S. (2007) Comprehensive analysis of PPARα-dependent regulation of hepatic lipid metabolism by expression profiling. PPAR Res. 2007, 26839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Shiri-Sverdlov R., Wouters K., van Gorp P. J., Gijbels M. J., Noel B., Buffat L., Staels B., Maeda N., van Bilsen M., Hofker M. H. (2006) Early diet-induced non-alcoholic steatohepatitis in APOE2 knock-in mice and its prevention by fibrates. J. Hepatol. 44, 732–741 [DOI] [PubMed] [Google Scholar]
- 51. Shah A., Rader D. J., Millar J. S. (2010) The effect of PPAR-α agonism on apolipoprotein metabolism in humans. Atherosclerosis 210, 35–40 [DOI] [PubMed] [Google Scholar]
- 52. Kersten S. (2008) Peroxisome proliferator activated receptors and lipoprotein metabolism. PPAR Res. 2008, 132960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Edvardsson U., Ljungberg A., Lindén D., William-Olsson L., Peilot-Sjögren H., Ahnmark A., Oscarsson J. (2006) PPARα activation increases triglyceride mass and adipose differentiation-related protein in hepatocytes. J. Lipid Res. 47, 329–340 [DOI] [PubMed] [Google Scholar]
- 54. Lindén D., Lindberg K., Oscarsson J., Claesson C., Asp L., Li L., Gustafsson M., Borén J., Olofsson S. O. (2002) Influence of peroxisome proliferator-activated receptor α agonists on the intracellular turnover and secretion of apolipoprotein (Apo) B-100 and ApoB-48. J. Biol. Chem. 277, 23044–23053 [DOI] [PubMed] [Google Scholar]
- 55. Lindén D., Alsterholm M., Wennbo H., Oscarsson J. (2001) PPARα deficiency increases secretion and serum levels of apolipoprotein B-containing lipoproteins. J. Lipid Res. 42, 1831–1840 [PubMed] [Google Scholar]
- 56. Tordjman K., Bernal-Mizrachi C., Zemany L., Weng S., Feng C., Zhang F., Leone T. C., Coleman T., Kelly D. P., Semenkovich C. F. (2001) PPARα deficiency reduces insulin resistance and atherosclerosis in apoE-null mice. J. Clin. Invest. 107, 1025–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]