Background: Tgl3p is the major triacylglycerol lipase showing a dual localization on lipid droplets and the endoplasmic reticulum.
Results: Minimal changes in the C terminus of Tgl3p affect protein stability and functionality.
Conclusion: The membrane environment is crucial for correct protein assembly.
Significance: New aspects of lipase functionality are presented.
Keywords: Lipase, Lipid Droplet, Membrane Protein, Triacylglycerol, Yeast, Membrane Topology
Abstract
Lipid droplets are specific organelles for the storage of triacylglycerols and steryl esters. They are surrounded by a phospholipid monolayer with a small but specific set of proteins embedded. Assembly and insertion of proteins into this surface membrane is an intriguing question of lipid droplet biology. To address this question we studied the topology of Tgl3p, the major triacylglycerol lipase of the yeast Saccharomyces cerevisiae, on lipid droplets. Employing the method of limited proteolysis of lipid droplet surface proteins, we found that the C terminus of Tgl3p faces the inside of the organelle, whereas the N terminus is exposed at the cytosolic side of lipid droplets. Detailed analysis of the C terminus revealed a stretch of seven amino acids that are critical for protein stability and functionality. The negative charge of two aspartate residues within this stretch is crucial for lipase activity of Tgl3p. A portion of Tgl3p, which is located to the endoplasmic reticulum, exhibits a different topology. In the phospholipid bilayer of the endoplasmic reticulum the C terminus faces the cytosol, which results in instability of the protein. Thus, the topology of Tgl3p is important for its function and strongly dependent on the membrane environment.
Introduction
All types of eukaryotic cells such as plant, mammalian, and non-mammalian cells and even some prokaryotes belonging to the Gram-positive genera such as Rhodococcus or Streptomyces contain intracellular lipid droplets (LD)2 (1). These organelles are specialized to store non-polar lipids such as triacylglycerols (TG) and steryl esters (SE). TG and SE are low volume and high energy reserve materials serving as internal depots for sterols and fatty acids, which are important building blocks for membrane biogenesis and a source of cellular energy. Although LD show significant variation in size, shape, and composition in various cell types, they appear to have a general and very special structure. LD of the yeast Saccharomyces cerevisiae are small spherical organelles with an approximate diameter of 400 nm (2). They consist of 95% non-polar lipids with approximately equal amounts of TG and SE. TG and SE seem to be ordered instead of randomly distributed with TG forming the inner core of the LD, which is surrounded by several shells of SE most likely with some TG intercalated. In contrast to other organelles, the surface of LD is covered by a phospholipid monolayer (3).
Several proteome analyzes from S. cerevisiae identified a small set of about 60 proteins on the surface of LD that can adjust to different growth conditions (4, 5). Among the prominent LD proteins, the major TG lipases of the yeast, Tgl3p, Tgl4p, and Tgl5p, were identified (6, 7). All three lipases share the conserved GXSXG lipase motif and a patatin domain instead of a typical α/β hydrolase fold (8). The patatin domain not only differs in the secondary structure from a canonical α/β hydrolase fold but also in the catalytic site of the enzyme (9). The active center of patatin domain containing lipases consists of a serine-aspartate dyad, whereas the active site of a typical lipase with an α/β hydrolase fold comprises a catalytic serine-aspartate-histidine triad.
Tgl3p shows the highest lipolytic activity of the three major TG lipases in the yeast, and a deletion of TGL3 causes accumulation of TG up to 200% compared with wild type cells (7). Interestingly, previous work from our group showed that Tgl3p also harbors an HXXXXD acyltransferase motif and also functions as lysophosphatidylethanolamine acyltransferase (8). A tgl3Δ strain exhibits a reduced amount of total phospholipids, which may be regarded as proof for the role of Tgl3p as acyltransferase in vivo. Multiple enzymatic activities were also shown for Tgl4p and Tgl5p. Tgl5p acts also as a lysophospholipid acyltransferase, and Tgl4p, the functional orthologue of the mammalian adipocyte triacylglycerol lipase, is a multifunctional enzyme exhibiting SE hydrolase, sn2-specific PLA2, and acyl-CoA dependent lysophospholipid acyltransferase activities in vitro (10, 11). Most recently, it was demonstrated that Ayr1p, previously identified as an NADPH-dependent 1-acyl dihydroxyacetone phosphate reductase, also acts as TG lipase (12, 13). These findings clearly illustrate the complex and not yet sufficiently elucidated dynamic network of lipolytic enzymes in the yeast.
During the last decade, our fundamental knowledge about LD constantly increased. However, several important questions remained open, especially those concerning the biogenesis and the assembly of LD. It has been hypothesized that N- or C-terminal hydrophobic stretches of LD proteins are responsible for targeting and anchoring these polypeptides to LD. Removal of hydrophobic C-terminal stretches of the LD proteins Erg1p, Erg6p, and Erg7p disturbed the targeting of these proteins to the LD and led to their accumulation in the ER (14). However, C-terminal stretches of the respective proteins were not sufficient to direct a GFP hybrid to LD. Thus, our knowledge of targeting and anchoring of proteins to LD is still limited. Interestingly, several LD proteins including Tgl3p show a dual localization in LD and in the ER (15–17). This is surprising on one hand, because these proteins have to assemble in two different membrane environments, namely a phospholipid monolayer in LD and a phospholipid bilayer in the ER. How proteins adapt to these two different membrane environments is not known yet. On the other hand, the dual localization of these proteins in LD and the ER can be explained by the close relationship of these two compartments (18).
In the present work topological aspects of Tgl3p, a typical representative of LD proteins, were studied in some detail at the molecular level. The specific roles of N and C termini of the protein were addressed with special emphasis on the stability and functionality of Tgl3p. We show that the C terminus contains small stretches of amino acids whose presence or absence decides about the fate of the protein. The molecular link between topology and functionality of Tgl3p is discussed.
EXPERIMENTAL PROCEDURES
Strains and Culture Conditions
All yeast strains used in this study are listed in Table 1. Cells were grown aerobically to either logarithmic or stationary growth phase at 30 °C in YPD media containing 1% yeast extract, 2% glucose, and 2% peptone. Plasmid containing yeast strains were cultured in minimal media containing 0.67% yeast nitrogen base (U. S. Biochemical Corp), 2% glucose, and the respective amino acid supplements. Growth was monitored by measuring A600.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| Wild type | BY4741 Mat a; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | Euroscarf |
| Δtgl3 | BY4741; tgl3Δ::kanMX4 | Euroscarf |
| QM Δtgl3 | BY4741; dga1 Δ:: kanMX4 lro1Δ:: kanMX4 are1Δ:: kanMX4 are2Δ:: kanMX4 tgl3Δ::hisMX6 | This study |
Genetic Techniques
All primers and plasmids used in this study are listed in Tables 2, 3, and 4. 500 bp of the promoter region and 300 bp of the terminator region of TGL3 were cloned into a pRS315 plasmid. Insertion cassettes were obtained by PCR using genomic DNA from S. cerevisiae. The promotor region of TGL3 was inserted by cleavage with NotI and BamHI, the terminator region by cleavage of PstI and HindIII. TGL3 and all tagged and truncated variants of TGL3 were obtained by PCR and inserted into the pRS315 plasmid containing the promoter and terminator region with BamHI and PstI. All constructs were confirmed by sequencing and then transformed into S. cerevisiae BY4741 tgl3Δ employing the high efficiency lithium acetate transformation protocol (19). After transformation, cells were plated on minimal media lacking the respective amino acids for selection and incubated for 2–3 days at 30 °C.
TABLE 2.
Primers used in this study
The abbreviations used are as follows: Fw, forward; Rev, reverse; underlined small letters, restriction sites; italic letters, tags, start, or stop codons.
| Primer | Sequence (5′– 3′) |
|---|---|
| Promotor Fw | aaaagcggccgcGCTCCCTTGTTTAATAGCTT |
| Promotor Rev | aaaaggatccGCTTAACGGCAACTCAAAG |
| Terminator Fw | aaaactgcagTATCGTTTCCACTTTTTTCTG |
| Terminator Rev | aaaagcttGACCGGTTTTGCAAAGGACG |
| Tgl3 HA (N-term) Fw | aaaaggatccATGtacccatacgatgttcctgactatgcgAAGGAAACGGCGCAGG |
| Tgl3 HA (C-term) Rev | aaaactgcagCTAcgcatagtcaggaacatcgtatgggtaCCTACTCCGTCTTGCTCTTA |
| Tgl3 MYC (N-term) Fw | aaaaggatccATGgaacaaaagctaatctccgaggaagacttgAAGGAAACGGCGCAGG |
| Tgl3 Fw | aaaaggatccATGAAGGAAACGGCGCAGG |
| Tgl3 Rev | aaaactgcagCTACCTACTCCGTCTTGCTCTTA |
| 1-kDa truncation (C-term) | aaaactgcagctaTAATTTAAATTCGACTGCAC |
| 2.4-kDa truncation (C-term) | aaaactgcagctaAATTAAAGCTAGTGCGGGCC |
| 5-kDa truncation (C-term) | aaaactgcagctaACCTTCAATAATCCTTGTTAATG |
| 1-kDa truncation (N-term) | aaaaggatccatgGTGTCTGCTGTAATACCGACC |
| 2-kDa truncation (N-term) | aaaaggatccatgAAAAACTGGATACTGCGTGTAG |
| 5-kDa truncation (N-term) | aaaaggatccatgATCACCGATATTTATTTCTTC |
| Deletion of RARRSR Rev | ttttctgcagctaTATTATGTCGTCTAATTTAAA |
| Deletion of FKLDDII Fw | aagacaagatgtgcagtcgaaAGAGCAAGACGGAGTAG |
| Deletion of FKLDDII Rev | ctactccgtcttgctctTTCGACTGCACATCTTGTC |
| Exchange of F630A Fw | GATGTGCAGTCGAAgcTAAATTAGACGAC |
| Exchange of F630A Rev | GTCGTCTAATTTAgcTTCGACTGCACATC |
| Exchange of L632A Fw | GTGCAGTCGAATTTAAAgcAGACGACATAATAAGAGC |
| Exchange of L632A Rev | GCTCTTATTATGTCGTCTgcTTTAAATTCGACTGCAC |
| Exchange of D633A Fw | TGTGCAGTCGAATTTAAATTAGcCGACATAATAAGAGCAAGACGGAGT |
| Exchange of D633A Rev | ACTCCGTCTTGCTCTTATTATGTCGgCTAATTTAAATTCGACTGCACA |
| Exchange of D634A Fw | TGTGCAGTCGAATTTAAATTAGACGcCATAATAAGAGCAAGACGGAGT |
| Exchange of D634A Rev | ACTCCGTCTTGCTCTTATTATGgCGTCTAATTTAAATTCGACTGCACA |
| Exchange of D633A/D634A Fw | TGTGCAGTCGAATTTAAATTAGcCGcCATAATAAGAGCAAGACGGAGT |
| Exchange of D633A/D634A Fw | ACTCCGTCTTGCTCTTATTATGgCGgCTAATTTAAATTCGACTGCACA |
| Exchange of I635A/I636A Fw | GTCGAATTTAAATTAGACGACgcagcaAGAGCAAGACGGAGTAGGTAG |
| Exchange of I635A/I636A Rev | CTACCTACTCCGTCTTGCTCTtgctgcGTCGTCTAATTTAAATTCGAC |
TABLE 3.
Primers used for RT-PCR
The abbreviations used are as follows: Fw, forward; Rev, reverse.
| Primer | Sequence (5′–3′) |
|---|---|
| RT Act1-Fw | CCAGCCTTCTACGTTTCCATCCAAG |
| RT Act1-Rev | GACGTGAGTAACACCATCACCGGA |
| RT Tgl3-FW | GCCAACAATCCGAGCATAACGGAG |
| RT Tgl3-Rev | TGGTGCCAAGTATGGTCTCGCCA |
TABLE 4.
Plasmids used in this study
aa, amino acids.
| Name | Relevant information | Source |
|---|---|---|
| pRS315 | CEN, LEU2, | 32 |
| pRS315-MYC-TGL3-HA | CEN, LEU2, | This study |
| pRS315-HA-TGL3 | CEN, LEU2 | This study |
| pRS315-HA-tgl3-1kD | CEN, LEU2, TGL3 is N-term HA-tagged and C-term-truncated (last 9 aa missing) | This study |
| pRS315-HA-tgl3-2.4kD | CEN, LEU2, TGL3 is N-term HA-tagged and C-term-truncated (last 20 aa missing) | This study |
| pRS315-HA-tgl3-5kD | CEN, LEU2, TGL3 is N-term HA-tagged and C-term-truncated (last 43 aa missing) | This study |
| pRS315-TGL3-HA | CEN, LEU2 | This study |
| pRS315-1kD-tgl3-HA | CEN, LEU2 TGL3 is C-term HA-tagged and N-term truncated (first 9 aa missing) | This study |
| pRS315-2kD-tgl3-HA | CEN, LEU2, TGL3 is C-term HA-tagged and N-term truncated (first 18 aa missing) | This study |
| pRS315-5kD-tgl3-HA | CEN, LEU2, TGL3 is C-term HA-tagged and N-term truncated (first 43 aa missing) | This study |
| pRS315-HA-tgl3-RARRSR | CEN, LEU2, TGL3 is N-term HA-tagged and the last 6 aa (RARRSR) are missing | This study |
| pSR315-HA-tgl3-FKLDDII | CEN, LEU2, TGL3 is N-term HA-tagged and the aa 630-636 (FKLDDII) are missing | This study |
| pRS315-HA-tgl3-DDAA | CEN, LEU2, TGL3 is N-term HA-tagged and aa Asp-633 Asp-634 are both replaced by A | This study |
| pRS315-HA-tgl3-DDEE | CEN, LEU2, TGL3 is N-term HA-tagged and aa Asp-633 Asp-633 are both replaced by E | This study |
| pRS315-HA-tgl3-D633A | CEN, LEU2, TGL3 is N-term HA-tagged and aa Asp-633 is replaced by Ala | This study |
| pRS315-HA-tgl3-D634A | CEN, LEU2, TGL3 is N-term HA-tagged and aa Asp-634 is replaced by Ala | This study |
| pRS315-HA-tgl3-IIAA | CEN, LEU2, TGL3 is N-term HA-tagged and aa Ile-635 Ile-636 are both replaced by Ala | This study |
| pRS315-HA-tgl3-F630A | CEN, LEU2, TGL3 is N-term HA-tagged and aa Phe-630 is replaced by Ala | This study |
| pRS315-HA-tgl3-L632A | CEN, LEU2, TGL3 is N-term HA-tagged and aa Leu-632 is replaced by Ala | This study |
| pRS315-TGL3-6xHA | CEN, LEU2 TGL3 is C-Term HA6-tagged | This study |
Isolation and Characterization of Subcellular Fractions
Yeast LD and microsomal fractions were obtained at high purity from cells grown to the stationary phase according to published procedures (20, 21). The protein concentration of the isolated fractions was analyzed by the method of Lowry et al. (22) using bovine serum albumin as a standard. Before protein quantification, samples of LD fractions were delipidated with 2–3 volumes of diethyl ether. The organic phase was removed, and samples were dried under a stream of nitrogen. Then proteins were precipitated with trichloroacetic acid at a final concentration of 10% and solubilized in 0.1% SDS, 0.1% NaOH.
SDS-polyacrylamide gel electrophoresis was carried out by the method of Laemmli (23) using 10, 12.5, or 15% separation gels. Western blot analysis was performed as described by Haid and Suissa (24) using the following antibodies: anti-HA-peroxidase conjugated, high affinity (3F10) (Roche Applied Science) (1:1,000); anti-c-Myc antibody monoclonal (9E10) Sigma produced in mouse (1:1,000); anti-Ayr1 (produced in rabbit in house) (1:5,000); anti-Wbp1 (1:5,000) (produced in rabbit and kindly provided by H. Pichler); secondary anti-rabbit horseradish peroxidase-conjugated antibody (Sigma) (1:15,000); secondary anti-mouse horseradish peroxidase-conjugated antibody (Sigma) (1:15,000). Proteins were visualized using an enhanced chemiluminescence detection substrate (Thermo Scientific) following the manufacturer's instructions.
Preparation of Total Cell Extracts for Lipid Analysis and Thin Layer Chromatography (TLC)
Yeast cells were grown to the stationary growth phase, and an equivalent of 30 A600 units was harvested. Cells were disintegrated by vigorous shaking (Vortex) in the presence of glass beads for 10 min at 4 °C, and lipids were extracted as described by Folch et al. (25) using chloroform/methanol (2:1; v/v). For quantification of TG, lipid extracts were applied to Silica Gel 60 plates. Chromatograms were developed in an ascending manner by a two-step developing system. First, light petroleum/diethyl ether/acetic acid (35:15:1, per volume) was used as mobile phase, and plates were developed to two-thirds length of the plate. Then the plates were dried briefly and further developed to the top of the plate using the second mobile phase consisting of light petroleum/diethyl ether (49:1, v/v). Separated bands were visualized by irreversible staining. For this purpose the TLC plates were dipped into a charring solution consisting of 0.63 g of MnCl2·4 H2O, 120 ml of water, 120 ml of methanol, and 8 ml of concentrated sulfuric acid and incubated in a heating chamber at 105 °C for at least 30 min. Bands were quantified by densitometric scanning at 400 nm using a CAMAG TLC Scanner 3.
Proteinase K Protection Assay
LD were isolated as described above, and samples corresponding to 20 μg of LD protein were used for Proteinase K treatment. Proteinase K (Roche Applied Science) was added to a ratio of 1:100, and the incubation was carried out on ice. As control experiments, LD were solubilized in the presence of 1% Triton-X100. The reaction was stopped by adding trichloroacetic acid at a final concentration of 15% and vigorous vortexing. Proteins were precipitated for 1 h and centrifuged in a tabletop centrifuge at 4 °C for 30 min. The protein pellet was washed with ice-cold water and centrifuged again. Aliquots were used for Western blot analysis as described above.
RESULTS
A Proteinase K Protection Assay Reveals the Orientation of the C and N Terminus of Tgl3p
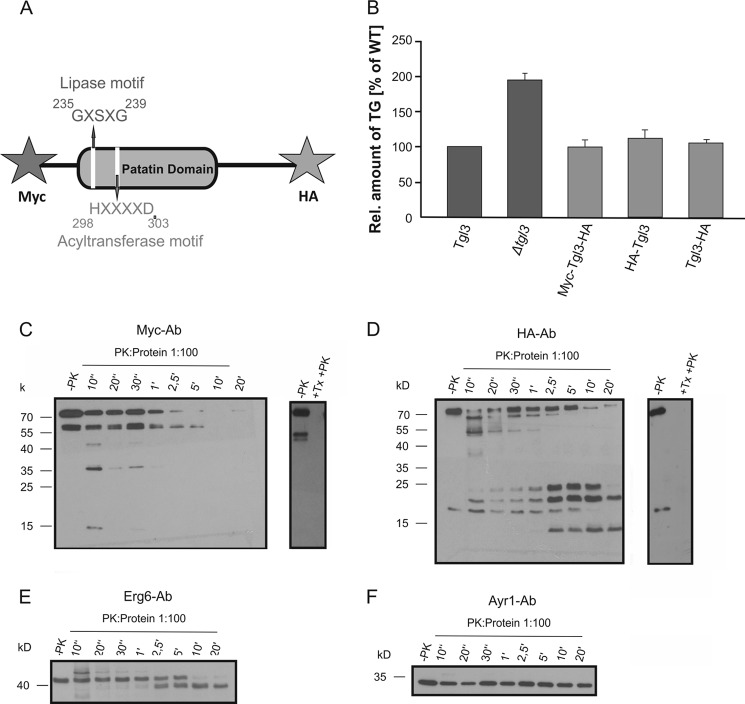
LD are the only organelles of the cell that are covered by a phospholipid monolayer. This fact raises the intriguing question of how proteins are inserted into and/or anchored to the surface membrane of this compartment. To address this question we designed an assay for studying the topology of one prominent LD protein, the TG lipase Tgl3p. This assay is based on limited proteolysis of surface components with proteinase K (PK), which allowed us to test for the orientation of the N terminus and C terminus of Tgl3p in the phospholipid monolayer membrane of LD. For this purpose we constructed a double-tagged variant of Tgl3p carrying an N-terminal Myc tag and an HA tag at the C terminus (Fig. 1A). This construct was cloned into a low copy vector and transformed into a tgl3Δ deletion strain. The functionality of the constructed Myc-Tgl3p-HA hybrid was verified by comparing the TG content of a tgl3Δ strain carrying an untagged version of TGL3 to a tgl3Δ mutant and the strain harboring the double tagged variant of Tgl3p. As Tgl3p is the major TG lipase in S. cerevisiae, deletion of TGL3 resulted in a marked TG accumulation compared with wild type (Fig. 1B). The strain carrying the Myc-Tgl3p-HA protein did not accumulate TG and thus behaved like a wild type strain. Conclusively, the tags did not affect the functionality of Tgl3p, and PK protection assays were carried out with this construct. As an additional control, the functionality of Tgl3p variants tagged only at the N terminus or C terminus were tested. As can be seen from Fig. 1B, both variants were fully functional and exhibited TG levels similar to wild type.
FIGURE 1.
Proteinase K assay to determine the orientation of the N- and C terminus of Tgl3p. A, scheme of Tgl3p showing the patatin domain, the conserved lipase motif, and the acyltransferase motif. For detection of the N terminus and C terminus, an Myc tag and an HA tag were introduced by PCR. B, relative amounts of TG in tgl3Δ and a tgl3Δ strains carrying untagged TGL3, the double-tagged version of TGL3, and N-terminal HA or C-terminal HA-tagged TGL3 are shown. Cells grown to the stationary phase were analyzed. Results are the average from at least three independent experiments with S.D. values (error bars) as indicated. C and D, proteinase K protection assays were performed as described under “Experimental Procedures” with isolated LD from a strain carrying the N-terminal Myc-tagged and C-terminal HA-tagged variant of Tgl3p. Samples were taken at time points from 10 s to 20 min, and 10 μg of LD protein was loaded on each lane. Western blot analysis was performed with antibodies directed against the Myc and HA epitope, respectively. The sample −PK was taken before the addition of PK. The sample +Tx +PK was taken after solubilization of LD with 1% Triton X-100 for 20 min on ice. E and F, the effect of PK treatment on Erg6p and Ayr1p, two LD resident proteins, was investigated with the respective polyclonal antibodies over the time interval from 10 s to 20 min.
The effect of PK treatment on LD surface proteins from isolated LD was monitored at several time points after stopping proteolysis by the addition of TCA. Samples were then analyzed by Western blots using antibodies directed against the N- and C-terminal tags of Tgl3p. Fig. 1C shows a characteristic experiment of this type. As can be clearly seen, the N terminus of Tgl3p was easily accessible to PK. After 2.5 min, hardly any signal of the N terminus of Tgl3p was detectable. This result indicated that the N terminus of Tgl3p faces the cytosolic side of LD protruding into the cytosolic environment.
Western blot analysis with Myc-tagged Tgl3p as described above also detected a truncated version of the protein (see Fig. 1C). Whether this additional band was an unspecific fragment arising during LD purification or the result of specific proteolytic cleavage is currently not known. Interestingly, similar results were obtained with N-terminal HA-tagged Tgl2p, a mitochondrial resident lipase (26). As full-length and truncated Myc-Tgl3p were degraded by PK with similar efficiency (see Fig. 1) and data of these experiments served to demonstrate the instability of the N terminus of Tgl3p, results were not biased.
In contrast to the N terminus, the C terminus of Tgl3p was protected from digestion by PK. During incubation with the proteinase, several smaller fragments of Tgl3p appeared gradually. After 20 min, two stable protein bands were still visible, indicating that PK did not have access to the 20-kDa fragment of the C terminus (Fig. 1D). Thus, we conclude that the C terminus of Tgl3p is protected from degradation by membranes and orientated toward the inside of the organelle. Control experiments with LD samples solubilized with Triton X-100 revealed that N-terminal- as well as C-terminal-tagged variants of Tgl3p were completely digested by PK in the presence of the detergent. This observation indicated that the folding of the two termini per se did not affect the obtained results.
As a control for incubation conditions, we chose to test the effect of PK treatment on Erg6p, a Δ(24)-sterol C-methyltransferase (Fig. 1E), and Ayr1p, an NADPH-dependent 1-acyl dihydroxyacetone phosphate reductase that was recently identified as another TG lipase of S. cerevisiae (12, 13) (Fig. 1F). Both proteins had been identified before as typical LD proteins. A small part of Erg6p seemed to be accessible to PK, whereas Ayr1p was not affected at all after 20 min of PK treatment. Obviously, these two LD proteins are rather deeply embedded in the phospholipid monolayer of LD and, therefore, protected against the PK treatment.
C-terminal Truncations of TGL3 Result in Protein Instability
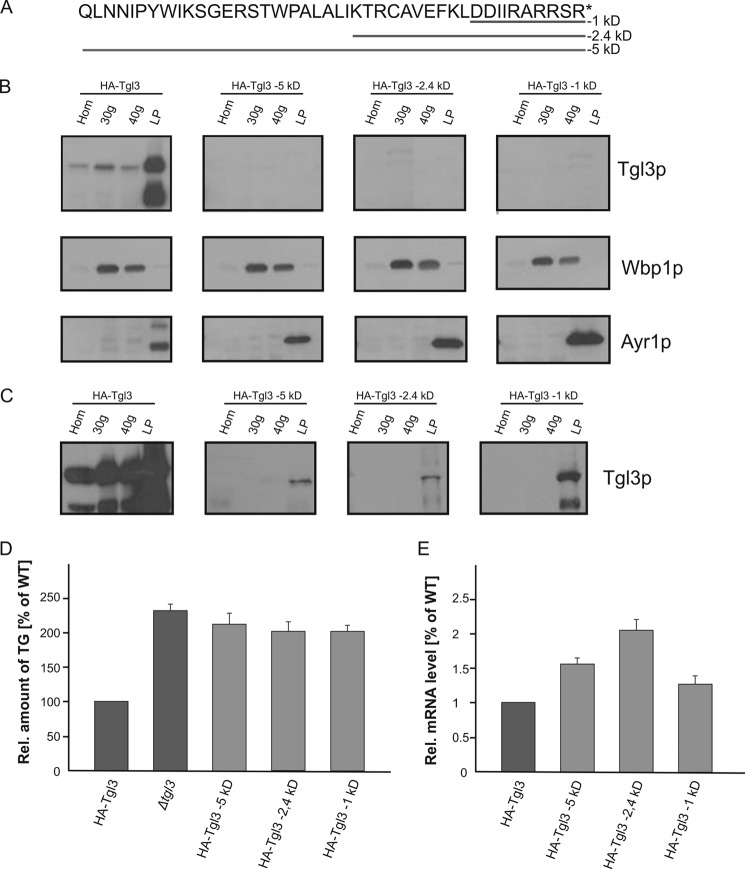
The finding that the C terminus of Tgl3p was inaccessible to proteolysis with PK led us to speculate that this part of the protein might be important for anchoring and/or targeting of the protein to LD. It had been shown before that C-terminal hydrophobic stretches of LD proteins were responsible for their association with the organelle (14). To test this hypothesis for Tgl3p, we constructed three different C-terminal variants of N-terminal HA-tagged Tgl3p that were truncated of 5-kDa (43 amino acids), 2.4-kDa (20 amino acids), and 1-kDa (10 amino acids) (Fig. 2A). LD and microsomal fractions were isolated from the respective strains, and the subcellular distribution of the truncated Tgl3p variants was compared with the full-length protein (Fig. 2B). Full-length Tgl3p and all Tgl3p variants were detected through their N-terminal HA tag. The ER resident protein Wbp1p was used as an ER marker, and Ayr1p was analyzed as a typical LD protein. As already shown before (17), full-length HA-Tgl3p showed a dual localization with the majority of the protein found in LD and a small amount in the 30,000 × g and 40,000 × g microsomal fractions. Surprisingly, all three C-terminal-truncated variants of HA-Tgl3p could not be detected at all under standard Western blot conditions. Only overloading samples 7-fold revealed minimal amounts of truncated Tgl3p proteins present in LD (Fig. 2C). Loss of stable Tgl3p also resulted in a massive loss of Tgl3p function as strains harboring the truncated variants of Tgl3p accumulated TG (Fig. 2D).
FIGURE 2.
C-terminal truncations of Tgl3p result in protein instability. A, amino acid sequence of the C-terminal 5 kDa of Tgl3p and truncations of 1, 2.4, and 5 kDa are shown. The respective amino acids deleted in each construct are underlined. All proteins used were N-terminal HA-tagged. B, homogenates (Hom), 30,000 × g (30g) microsomes, 40,000 × g (40g) microsomes, and LD were isolated from strains carrying full-length Tgl3p and C-terminal truncations of −1, −2.4, and −5 kDa of Tgl3p. Western blot analysis was performed with a primary antibody against the HA tag. Antibodies against Wbp1p (ER marker) and Ayr1p (LD marker) were used for quality control of the subcellular fractions. 10 μg of protein were loaded per lane. C, Western blot analysis of indicated fractions from SDS-PAGE overloaded with ∼70 μg of protein per lane. D, relative TG content of strains grown to the stationary phase was measured to evaluate functionality of Tgl3p in vivo. E, relative mRNA expression levels of full-length TGL3 (set at 1) and truncated variants of TGL3. F, C-terminal-truncated Tgl3p variants in the ER from a dga1Δlro1Δare1Δare2Δ quadruple mutant. Homogenates (Hom), 30,000 × g (30g) microsomes, and 40,000 × g (40g) microsomes were isolated from a QM strain carrying either full-length Tgl3p or C-terminal truncations of −1, −2.4, and −5 kDa of Tgl3p. All proteins were N-terminal HA-tagged. Western blot analysis was performed with a primary antibody against the HA tag. The antibody against Wbp1p (ER marker) was used as quality control of microsomal fractions. 50 μg protein were loaded per lane.
As all truncated variants of Tgl3p were present in a low copy vector with the endogenous promoter and terminator region, we also tested whether the loss of protein was due to insufficient gene expression. Therefore, the respective mRNA levels were quantified by RT-PCR. As can be seen from Fig. 2E, mRNA levels of truncated Tgl3p variants were increased rather than reduced. Thus, decreased transcription efficiency was not the reason for the low protein levels. To further test the possible influence of the lipid/membrane environment on the stability of the protein, we expressed the truncated variants of Tgl3p in the quadruple mutant (QM) strain. In this strain, wild type Tgl3p associated only with the phospholipid bilayer of the ER (Fig. 2F). As truncated variants of Tgl3p were equally unstable in the QM and in wild type, we concluded that the protein degradation occurred immediately after the translation process. Taken together, our results suggested that C-terminal truncations of Tgl3p caused instability of the protein. Even minimal truncations, e.g. 1 kDa, were obviously sufficient to compromise the protein severely. As minor amounts of Tgl3p were still found in the LD, we concluded that the C terminus was involved in protein stability and anchoring rather than targeting of the protein to LD.
Effect of Single and Double Amino Acid Exchanges in the C Terminus on Stability and Functionality of Tgl3p
Experiments described above demonstrated that 10 amino acids of the C terminus are important and critical for the stability of Tgl3p. Therefore, we wished to go into further detail and dissect the role of this part of the protein. First, we removed the last six C-terminal amino acids of Tgl3p (RARRSR) and tested distribution of the protein between LD and microsomal fractions (Fig. 3, A and B). These experiments showed that removing the last six amino acids of the protein led to reduction of the amount but did not change targeting of the protein to LD. Also the TG content of this mutant was similar to wild type with only minor accumulation of TG, which may be due to the reduced amount of Tgl3p (Fig. 3C). As a second step, to elucidate the role of the C terminus, we deleted the amino acids FKLDDII, which overlap the 1- and 2.4-kDa truncation points (Figs. 2A and 3A). Interestingly, deletion of these seven amino acids resulted in a highly unstable protein and, not surprisingly, in TG accumulation (Fig. 3, B and C).
FIGURE 3.
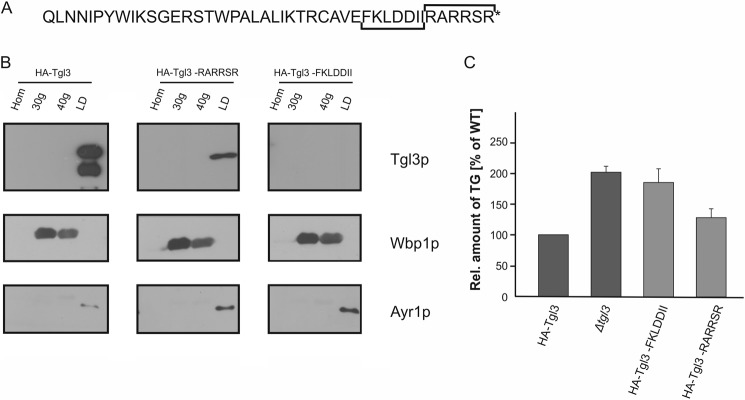
A stretch of seven amino acids within the C terminus of Tgl3p is responsible for stability. A, the C-terminal 5-kDa amino acid sequence of Tgl3p is shown. Truncations are indicated by brackets. B, homogenates (Hom), 30,000 × g (30g) microsomes, 40,000 × g (40g) microsomes, and LD were isolated from strains harboring full-length Tgl3p, Tgl3p minus FKLDDII, and Tgl3p minus RARRSR. All constructs were N-terminal HA-tagged, and Western blot analysis was performed using a primary antibody against the HA tag. Wbp1p (ER marker) and Ayr1p (LD marker) were used for quality control of subcellular fractions. 10 μg of protein were loaded per lane. C, relative amounts of TG from a strain carrying full-length N-terminal HA-tagged Tgl3p, a tgl3Δ deletion mutant, and a tgl3Δ strain harboring Tgl3p minus FKLDDII or RARRSR, respectively, were measured. Cells were grown to the stationary phase. Results are the average from at least three independent experiments with S.D. values (error bars) as indicated.
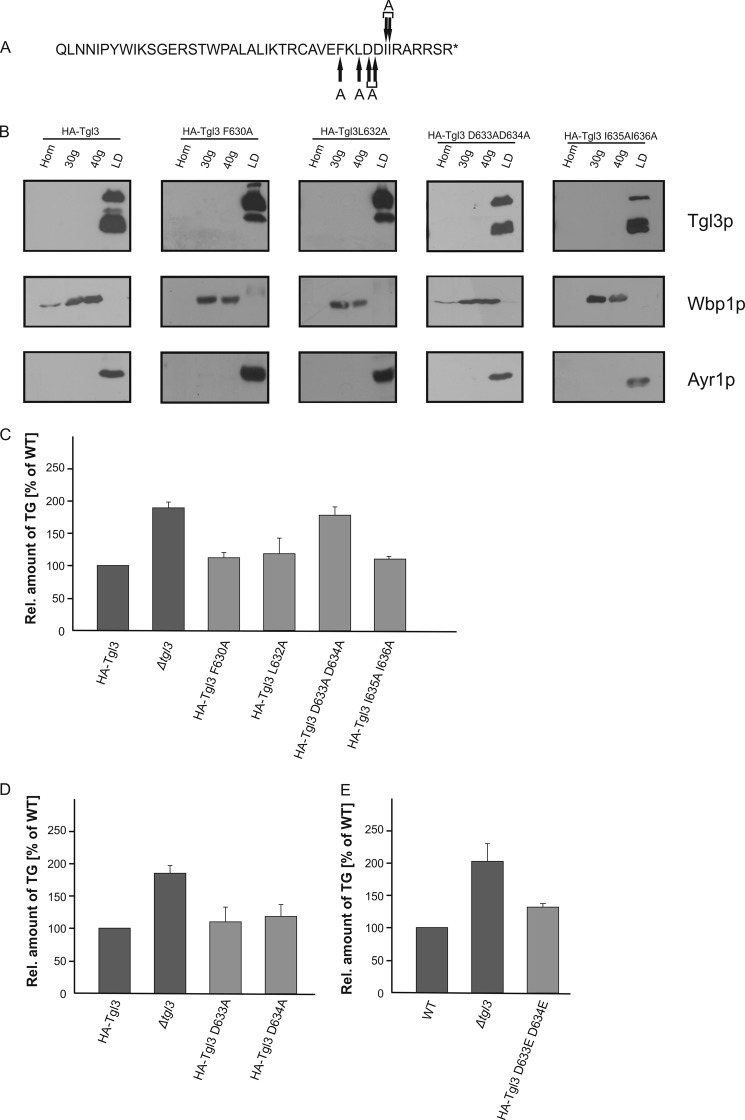
As a last step of molecular investigations we wished to clarify the role of individual amino acids within the FKLDDII region. For this purpose we constructed Tgl3p variants harboring the amino acid exchanges F630A, L632A, D633A/D634A, and I635A/I636A and analyzed microsomal and LD fractions from these strains (Fig. 4, A and B). Interestingly, the protein stability of all Tgl3p constructs bearing single and double amino acid exchanges was the same as full-length HA-Tgl3p. Also the localization of the Tgl3p variants was not altered. Surprisingly, however, HA-Tgl3p D633A/D634A showed TG accumulation comparable to a tgl3Δ deletion strain, whereas all other amino acid exchanges had no effect on Tgl3p functionality (Fig. 4C). Thus, HA-Tgl3p D633A/D634A was the only mutation that compromised activity of Tgl3p but not stability. To test whether both aspartates or only one was responsible for the loss of lipase activity, we also constructed Tgl3p bearing D633A or D634A single amino acid exchanges and compared the TG content with a tgl3Δ strain. Interestingly, lipase activity of Tgl3p D633A and Tgl3p D634A was the same as wild type (Fig. 4D). Consequently, both aspartates in combination are important and essential for the lipase activity of Tgl3p. We further speculated whether the loss of the negative charge caused by the aspartate to alanine exchange was the reason for the loss of lipase activity. Therefore, we exchanged the two aspartates against two glutamates to conserve the negative charge. As can be seen from Fig. 4E, the Tgl3p D633E/D634E variant displayed lipase activity, although at a slightly reduced level. Thus, it appears that the negative charge(s) in the C terminus is crucial for TG lipase activity of Tgl3p.
FIGURE 4.
Single and double amino acid exchanges within the C-terminal region affect stability and activity of Tgl3p. A, amino acid sequence of the C-terminal 5 kDa of Tgl3p is shown, and positions of single and double amino acid exchanges are marked with arrows. All variants were N-terminal HA-tagged. B, homogenates (Hom), 30,000 × g (30g) microsomes, 40,000 × g (40g) microsomes, and LD) from strains harboring full-length Tgl3p, Tgl3p variants with single amino acid exchanges F630A and L632A, and Tgl3p variants bearing double amino acid exchanges D633A/D634A and I635A/I636A were analyzed by Western blotting. Primary antibodies were directed against the HA tag, Wbp1p (ER marker), and Ayr1p (LD marker). 10 μg of protein were loaded per lane. C–E, the relative amounts of TG from the respective strains were analyzed from cells grown to the stationary phase. Results are the average from at least three independent experiments with S.D. values (error bars) as indicated.
The N Terminus of Tgl3p Is Less Sensitive to Truncations Than the C Terminus
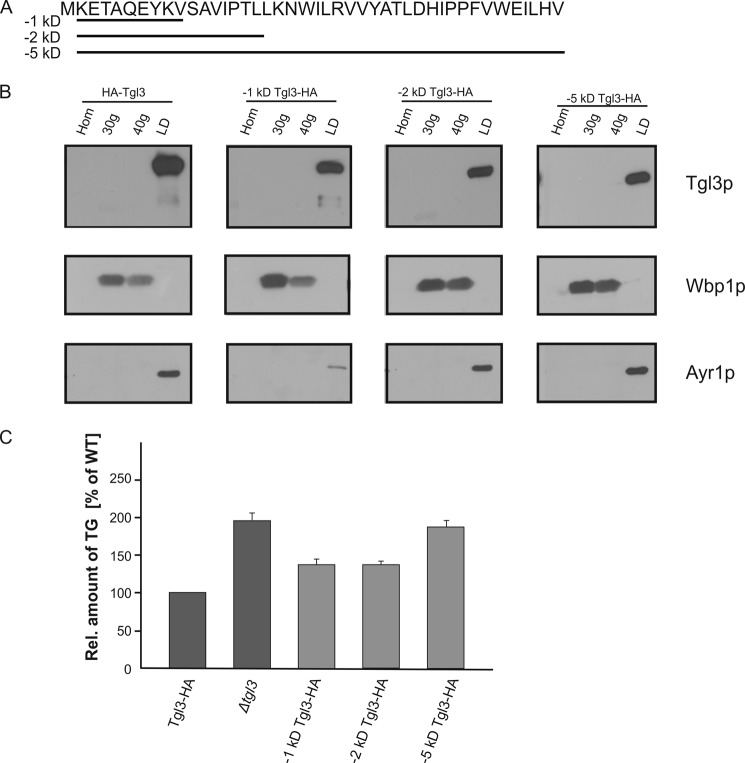
Experiments described above demonstrated that the C terminus of Tgl3p, which is protected by the surface membrane of LD, is critical for stability and activity of the protein. In contrast, the N terminus of Tgl3p was easily digested by PK and thus faces the cytosol (see Fig. 1C). To test the possible effects of N-terminal truncations on targeting and stability of Tgl3p, we constructed strains carrying 1-, 2-, and 5-kDa N-terminal truncations of Tgl3p and performed localization analysis (Fig. 5, A and B). All constructs, including the full-length Tgl3p control, carried C-terminal HA tags. As can been seen from Fig. 5B, targeting of the protein to LD was not affected by all N-terminal truncations. The stability of the 1-, 2-, and 5-kDa N-terminal truncated variants of Tgl3p was only slightly reduced. The TG levels of the strains carrying the 1- and 2-kDa truncations were comparable to wild type, indicating that the lipase activity was not affected (Fig. 5C). The 5-kDa truncation of the N terminus of Tgl3p, however, resulted in significant loss of activity (Fig. 5C).
FIGURE 5.
Truncations of the N terminus of Tgl3p. A, the amino acid sequence of the N-terminal 5 kDa of Tgl3p is shown, and the amino acids deleted in each construct are underlined. B, homogenates (Hom), 30,000 × g (30g) microsomes, 40,000 × g (40g) microsomes, and LD were isolated from strains carrying full-length Tgl3p, and 1, 2, and 5 kDa N-terminal-truncated variants of Tgl3p. All proteins were C-terminal HA-tagged, and Western blot analysis was performed using primary antibodies against the HA tag, Wbp1p (ER marker), and Ayr1p (LD marker). 10 μg of protein were loaded per lane. C, the relative amount of TG from the respective strains grown to the stationary phase was measured. Results are the average from at least three independent experiments with S.D. values (error bars) as indicated.
The C Terminus of Tgl3p Face the Cytosolic Side When Inserted into the Endoplasmic Reticulum
Schmidt et al. (17) showed that a small portion of Tgl3p is found in the ER of wild type cells. In the absence of LD, i.e. in an lro1Δdga1Δare1Δare2Δ QM strain (27), localization of Tgl3p was completely restricted to the ER although in an inactive form. Furthermore, it was shown that the stability of Tgl3p was strongly reduced in the ER of the QM, whereas Tgl3p in LD from a wild type strain was very stable. However, the reason for the short half-life of Tgl3p in the QM remained unclear, and we speculated that inefficient assembly of Tgl3p into the bilayer of the ER in contrast to embedment of the protein in the monolayer membrane of LD might cause this problem. To test this hypothesis we isolated microsomes from the QM and analyzed the topology of Tgl3p in this compartment by PK assays with emphasis on the role of the C terminus of the enzyme. For these experiments we used C-terminal Myc-tagged Tgl3p (Fig. 6). Wpb1p and Kar2p were used as ER markers to ensure the integrity of the microsomal vesicles. Kar2p, a prominent luminal ER protein, was only digested by PK in the presence of detergent. Similarly, Wbp1p, a type I membrane protein with a large luminal domain, was only degraded when Triton X-100 was added during incubation with the protease (28). These control experiments confirmed the integrity of microsomal vesicles in a right-side-out orientation. As can be seen from the Western blot, the C terminus of Tgl3p was easily degraded by PK, indicating its exposure to the cytosol. This orientation of the C terminus of Tgl3p in the ER is in sharp contrast to LD. As the C terminus of Tgl3p is crucial for its stability as demonstrated by the above mentioned experiments, we conclude that exposure of this part of the protein to the environment in ER fractions may be the reason for the short half-life of Tgl3p in this compartment.
FIGURE 6.
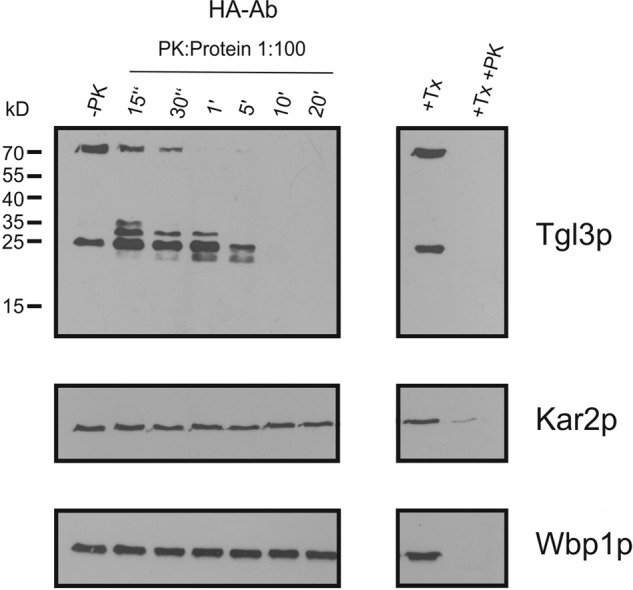
Orientation of the C terminus of Tgl3p in the ER. 30,000 × g microsomes were isolated from a QM lacking lipid droplets. A C-terminal HA-tagged Tgl3p variant was used to perform PK protection assays over a time range from 15 s to 20 min. +Tx, addition of 1% Triton X-100; +Tx +PK, addition of 1% Triton X-100 and PK; −PK, control without PK. Kar2p served as marker protein for luminal ER, and Wbp1p as an integral membrane protein marker. 30 μg of protein were loaded per lane.
DISCUSSION
Tgl3p, the major TG lipase from S. cerevisiae, plays a critical role in TG mobilization and also contributes to phospholipid metabolism through its lysophospholipid acyltransferase activity (8). Recently, Schmidt et al. (17), studying regulatory aspects of this lipase, showed that the activity of Tgl3p was strongly influenced by its localization, and localization depended on substrate availability. Our work presented in this study extends these investigations to the topology of Tgl3p, which led to the identification of protein domains that are critical for the stability and activity of Tgl3p.
PK protection assays using tagged variants of Tgl3p revealed that the N terminus of the protein faces the cytosolic site of the LD, whereas a ∼20-kDa stretch of the C terminus appears to be protected by the surface membrane of the organelle. Surprisingly, in silico analysis demonstrated that the C terminus of Tgl3p is not explicitly hydrophobic and, therefore, not typical for insertion into membranes. The question remains of how such a non-hydrophobic domain can be inserted into a membrane or even protrudes into the highly hydrophobic core of LD. Several mechanisms describing the assembly of proteins in the LD surface have been discussed, but all of them propose direct anchoring of the proteins via hydrophobic helices (29, 30). Thus, the possibility remains that Tgl3p might be associated with a partner protein that allows assembly in the form of a complex into the LD surface. Interestingly, Ayr1p has been identified among some other proteins as a possible partner protein of Tgl3p in a split GFP screening (31). Currently, however, there is no further experimental evidence supporting the view of an interaction of Tgl3p with Ayr1p, and formation of LD surface complexes containing Tgl3p remains pure speculation at present. Our results obtained with Ayr1p (see Fig. 1F) suggest that this protein is deeply embedded within the LD. PK got only access to Ayr1p when LD were incubated at 37 °C and vigorously vortexed during the treatment (data not shown). These results obtained with Ayr1p are in sharp contrast to Tgl3p. Also Erg6p, another typical LD protein seems to be largely inaccessible to PK treatment (Fig. 1E). This result is in line with previous studies from our laboratory (14) which showed that removal of C-terminal hydrophobic domains of Erg6p abolished LD association of the protein. Conclusively, it appears that different types of LD proteins with different hydrophobicity exist that may result in different topology of these proteins at the LD surface.
Results discussed above led us to speculate that the C terminus of Tgl3p may play an important role in targeting of the protein to LD. To test this hypothesis we performed experiments with three different C-terminal-truncated variants of Tgl3p (see Fig. 2, A–C). These experiments, however, indicated that the C terminus of Tgl3p was involved in protein stability rather than in the targeting of the protein to LD. All truncated variants were found to be rather unstable, but residual amounts of the protein were found in LD. We expressed the truncated variants of Tgl3p in the QM strain to explore a possible influence of the membrane/lipid environment on protein stability. In this strain wild type Tgl3p accumulated with the phospholipid bilayer of the ER due to the lack of LD (Fig. 2F). As truncated variants of Tgl3p were equally unstable in the QM and in wild type, we concluded that the protein degradation occurred immediately after the translation process. Further and more detailed molecular analysis of the C terminus of Tgl3p revealed a region of seven amino acids (FKLDDII), which seems to be critical for protein stability (see Fig. 3, A–C). Interesting, single or double amino acid exchanges within this sequence did not affect stability (see Fig. 4, A and B). Most surprisingly we found that a double exchange of D633A/D634A led to an inactive Tgl3p protein and to a strain that accumulated TG like a tgl3Δ strain. This situation is unique insofar as, despite correct targeting, the lipase activity of this Tgl3p variant was severely compromised. Comparable results were obtained previously in our laboratory by Schmidt et al. (17) who showed that Tgl3p mutants inactivated by site-directed mutagenesis in the GXSXG motif targeted normally to lipid droplets. Thus, it appears that activity of Tgl3p is not required for correct targeting.
Previous in silico analysis had revealed that Tgl3p more likely harbors a patatin domain than a canonical α/β hydrolase fold of lipases. The patatin domain differs significantly from a classical hydrolase fold not only in the secondary structure but also in the organization of the active site. Instead of a catalytic triad build from a histidine, serine, and aspartate or glutamate, a patatin domain active site is formed by a serine and aspartate dyad. Thus, it is tempting to speculate that one of the aspartates Asp-633 or Asp-634 may be part of the catalytic dyade. However, there are arguments that speak against this view. First, an amino acid exchange of only one aspartate, either Asp-633 or Asp-634, had no effect on the lipase activity as TG accumulation was not observed in these strains (Fig. 4D). Second, Asp-633 and Asp-634 are not located within the predicted patatin domain where one would usually expect the catalytic active serine-aspartate dyad. Interestingly, however, a double amino exchange of Asp-633 and Asp-634 of Tgl3p to glutamate residues rescued the lipase activity (Fig. 4E). Thus, the negative charge(s) in this position seems to be important for the functionality of Tgl3p. As the three-dimensional structure of Tgl3p is not yet available, the possibility of the above-mentioned interaction of amino acid residues remains hypothetical.
In a previous study from our laboratory Schmidt et al. (17) showed that the half-life of Tgl3p is dramatically reduced when the protein was located to the ER. Experiments described in the present study enabled us to explain this effect at the molecular level. Given the fact that the C terminus of Tgl3p is very important for its stability, exposure of this part of the protein at the cytosolic side of the ER may be the reason for the instability of Tgl3p in this compartment. The membrane bilayer environment of the ER may cause the unfortunate topology of Tgl3p in this organelle, whereas the phospholipid monolayer of the LD surface seems to provide the more appropriate surrounding for the enzyme.
In contrast to the C terminus, the N terminus of Tgl3p is obviously less important for subcellular localization and functionality of the protein. The N terminus of Tgl3p appears to protrude from the LD surface into the cytosol, which makes it susceptible to degradation. However, truncations up to 5 kDa at the N terminus of Tgl3p are not harmful for protein stability. As all N-terminal-truncated variants of Tgl3p were localized to LD, we conclude that at least 5 kDa of the N terminus do not contain targeting sequences to LD.
In summary, our study provides a detailed molecular insight into the membrane topology of Tgl3p, the major TG lipase of the yeast, with emphasis on the specific role of the C terminus of the protein. We were able to narrow critical amino acid stretches important for stability and functionality of the protein down to seven or even two amino acids. Thus, to be or not to be an active lipase in the appropriate subcellular location seems to be a matter of minor changes in the primary sequence of Tgl3p.
Acknowledgment
We thank the Austrian Centre of Industrial Biotechnology Graz for providing the ABI 7500 PCR System.
This work was supported by Austrian Science Fund (FWF) projects P23029 and W901 DK Molecular Enzymology (to G. D.).
- LD
- lipid droplet(s)
- ER
- endoplasmic reticulum
- PK
- proteinase K
- QM
- quadruple mutant
- SE
- steryl ester(s)
- TG
- triacylglycerol(s).
REFERENCES
- 1. Zweytick D., Athenstaedt K., Daum G. (2000) Intracellular lipid particles of eukaryotic cells. Biochim. Biophys. Acta 1469, 101–120 [DOI] [PubMed] [Google Scholar]
- 2. Czabany T., Wagner A., Zweytick D., Lohner K., Leitner E., Ingolic E., Daum G. (2008) Structural and biochemical properties of lipid particles from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 283, 17065–17074 [DOI] [PubMed] [Google Scholar]
- 3. Tauchi-Sato K., Ozeki S., Houjou T., Taguchi R., Fujimoto T. (2002) The surface of lipid droplets is a phospholipid monolayer with a unique fatty acid composition. J. Biol. Chem. 277, 44507–44512 [DOI] [PubMed] [Google Scholar]
- 4. Grillitsch K., Connerth M., Köfeler H., Arrey T. N., Rietschel B., Wagner B., Karas M., Daum G. (2011) Lipid particles/droplets of the yeast Saccharomyces cerevisiae revisited: lipidome meets proteome. Biochim. Biophys. Acta 1811, 1165–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Athenstaedt K., Zweytick D., Jandrositz A., Kohlwein S. D., Daum G. (1999) Identification and characterization of major lipid particle proteins of the yeast Saccharomyces cerevisiae. J. Bacteriol. 181, 6441–6448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Athenstaedt K., Daum G. (2003) YMR313c/TGL3 encodes a novel triacylglycerol lipase located in lipid particles of Saccharomyces cerevisiae. J. Biol. Chem. 278, 23317–23323 [DOI] [PubMed] [Google Scholar]
- 7. Athenstaedt K., Daum G. (2005) Tgl4p and Tgl5p, two triacylglycerol lipases of the yeast Saccharomyces cerevisiae are localized to lipid particles. J. Biol. Chem. 280, 37301–37309 [DOI] [PubMed] [Google Scholar]
- 8. Rajakumari S., Daum G. (2010) Janus-faced enzymes yeast Tgl3p and Tgl5p catalyze lipase and acyltransferase reactions. Mol. Biol. Cell 21, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rydel T. J., Williams J. M., Krieger E., Moshiri F., Stallings W. C., Brown S. M., Pershing J. C., Purcell J. P., Alibhai M. F. (2003) The crystal structure, mutagenesis, and activity studies reveal that patatin is a lipid acyl hydrolase with a Ser-Asp catalytic dyad. Biochemistry 42, 6696–6708 [DOI] [PubMed] [Google Scholar]
- 10. Rajakumari S., Daum G. (2010) Multiple functions as lipase, steryl ester hydrolase, phospholipase, and acyltransferase of Tgl4p from the yeast Saccharomyces cerevisiae. J. Biol. Chem. 285, 15769–15776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kurat C. F., Wolinski H., Petschnigg J., Kaluarachchi S., Andrews B., Natter K., Kohlwein S. D. (2009) Cdk1/Cdc28-dependent activation of the major triacylglycerol lipase Tgl4 in yeast links lipolysis to cell-cycle progression. Mol. Cell 33, 53–63 [DOI] [PubMed] [Google Scholar]
- 12. Ploier B., Scharwey M., Koch B., Schmidt C., Schatte J., Rechberger G., Kollroser M., Hermetter A., Daum G. (2013) Screening for hydrolytic enzymes revealed Ayr1p as a novel triacylglycerol lipase in Saccharomyces cerevisiae. J. Biol. Chem. 288, 36061–36072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Athenstaedt K., Daum G. (2000) 1-Acyldihydroxyacetone-phosphate reductase (Ayr1p) of the yeast Saccharomyces cerevisiae encoded by the open reading frame YIL124w is a major component of lipid particles. J. Biol. Chem. 275, 235–240 [DOI] [PubMed] [Google Scholar]
- 14. Müllner H., Zweytick D., Leber R., Turnowsky F., Daum G. (2004) Targeting of proteins involved in sterol biosynthesis to lipid particles of the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1663, 9–13 [DOI] [PubMed] [Google Scholar]
- 15. Zinser E., Paltauf F., Daum G. (1993) Sterol composition of yeast organelle membranes and subcellular distribution of enzymes involved in sterol metabolism. J. Bacteriol. 175, 2853–2858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Leber R., Landl K., Zinser E., Ahorn H., Spök A., Kohlwein S. D., Turnowsky F., Daum G. (1998) Dual localization of squalene epoxidase, Erg1p, in yeast reflects a relationship between the endoplasmic reticulum and lipid particles. Mol. Biol. Cell 9, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schmidt C., Athenstaedt K., Koch B., Ploier B., Daum G. (2013) Regulation of the yeast triacylglycerol lipase tgl3p by formation of nonpolar lipids. J. Biol. Chem. 288, 19939–19948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Walther T. C., Farese R. V., Jr (2009) The life of lipid droplets. Biochim. Biophys. Acta 1791, 459–466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A. (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11, 355–360 [DOI] [PubMed] [Google Scholar]
- 20. Leber R., Zinser E., Zellnig G., Paltauf F., Daum G. (1994) Characterization of lipid particles of the yeast, Saccharomyces cerevisiae. Yeast 10, 1421–1428 [DOI] [PubMed] [Google Scholar]
- 21. Zinser E., Sperka-Gottlieb C. D., Fasch E. V., Kohlwein S. D., Paltauf F., Daum G. (1991) Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol. 173, 2026–2034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lowry O. H., Rosebrough N. J., Farr A. L., Randall R. J. (1951) Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]
- 23. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 24. Haid A., Suissa M. (1983) Immunochemical identification of membrane proteins after sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Methods Enzymol. 96, 192–205 [DOI] [PubMed] [Google Scholar]
- 25. Folch J., Lees M., Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipids from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 26. Ham H. J., Rho H. J., Shin S. K., Yoon H.-J. (2010) The TGL2 gene of Saccharomyces cerevisiae encodes an active acylglycerol lipase located in the mitochondria. J. Biol. Chem. 285, 3005–3013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sandager L., Gustavsson M. H., Ståhl U., Dahlqvist A., Wiberg E., Banas A., Lenman M., Ronne H., Stymne S. (2002) Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 277, 6478–6482 [DOI] [PubMed] [Google Scholar]
- 28. te Heesen S., Rauhut R., Aebersold R., Abelson J., Aebi M., Clark M. W. (1991) An essential 45-kDa yeast transmembrane protein reacts with anti-nuclear pore antibodies: purification of the protein, immunolocalization and cloning of the gene. Eur. J. Cell Biol. 56, 8–18 [PubMed] [Google Scholar]
- 29. Hinson E. R., Cresswell P. (2009) The antiviral protein, viperin, localizes to lipid droplets via its N-terminal amphipathic α-helix. Proc. Natl. Acad. Sci. U.S.A. 106, 20452–20457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abell B. M., Holbrook L. A., Abenes M., Murphy D. J., Hills M. J., Moloney M. M. (1997) Role of the proline knot motif in oleosin endoplasmic reticulum topology and oil body targeting. Plant Cell 9, 1481–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pu J., Ha C. W., Zhang S., Jung J. P., Huh W.-K., Liu P. (2011) Interactomic study on interaction between lipid droplets and mitochondria. Protein Cell 2, 487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]