Background: Three catalytically charged residues of F1-ATPase, P-loop lysine, general base, and arginine finger, are thought to be indispensable for catalysis.
Results: Alanine-substituted mutants of the catalytic residues of F1-ATPase drove rotations.
Conclusion: The catalytic residues contribute to efficient catalysis but are not indispensable to chemo-mechanical energy coupling of F1-ATPase.
Significance: The chemo-mechanical coupling mechanism of F1-ATPase is far more robust than previously thought.
Keywords: ATP Synthase, Bioenergetics, F1FO-ATPase, Molecular Motor, Single-molecule Biophysics
Abstract
F1-ATPase (F1) is the rotary motor protein fueled by ATP hydrolysis. Previous studies have suggested that three charged residues are indispensable for catalysis of F1 as follows: the P-loop lysine in the phosphate-binding loop, GXXXXGK(T/S); a glutamic acid that activates water molecules for nucleophilic attack on the γ-phosphate of ATP (general base); and an arginine directly contacting the γ-phosphate (arginine finger). These residues are well conserved among P-loop NTPases. In this study, we investigated the role of these charged residues in catalysis and torque generation by analyzing alanine-substituted mutants in the single-molecule rotation assay. Surprisingly, all mutants continuously drove rotary motion, even though the rotational velocity was at least 100,000 times slower than that of wild type. Thus, although these charged residues contribute to highly efficient catalysis, they are not indispensable to chemo-mechanical energy coupling, and the rotary catalysis mechanism of F1 is far more robust than previously thought.
Introduction
Molecular machines, fueled by nucleotide triphosphate (NTP), play pivotal roles in a wide range of cellular activities, such as gene regulation, organelle transport, membrane transport, protein unfolding, signal transduction, and energy synthesis. Many of these exert mechanical force on their substrates or associated proteins through conformational changes that occur during NTP hydrolysis (1, 2). Although there are some variations in structure, NTP-driven molecular machines share well conserved structural features, especially around the nucleotide binding domain (1). The most highly conserved primary structure among the NTPases is the phosphate-binding loop (P-loop) comprised of the amino acid sequence GXXXXGK(T/S) (where X varies). The P-loop sequence motif was first reported by Walker et al. (3) and is therefore referred to as a Walker A motif. Additionally, the lysine is the most crucial residue for NTP hydrolysis among the P-loop residues (2).
Some subfamilies of NTP-driven molecular machines share other catalytically crucial residues that are also electrically charged. Catalytic glutamic acid is found in the nucleotide-binding pocket of many subfamilies of NTPases, such as the AAA+ proteins, ABC transporters, and RecA-type proteins, although the catalytic glutamic acid of RecA-type proteins occupies a different position in the secondary structure compared with other subfamilies (4). The glutamic acid binds to NTP via a coordinated water molecule at the distal end of the γ-phosphate of bound NTP. Because this residue seems to induce an in-line attack of the water molecule to γ-phosphate and initiate the hydrolysis reaction by activating the water molecule, the carboxylate residue is termed the “general base.” Note that recent theoretical studies have revised the actual working mechanism of the general base in catalysis (5, 6).
A catalytic arginine (Arg) residue is also widely distributed among many NTPases (2). The most studied catalytic arginine is the “arginine finger” of the G-protein-activating protein, which triggers GTP hydrolysis of the G-protein (7). Several studies have reported that the arginine finger stabilizes the transition state of hydrolysis to enhance catalysis. Both AAA+ proteins and RecA-type proteins such as RecA and F1-ATPase also carry the corresponding arginine (or lysine in RecA) in the same region of the arginine finger of G-protein-activating protein (8, 9).
Mutation at these charged residues is fatal for NTPase catalysis and is particularly so when substituted with either a noncharged or an oppositely charged residue, which reduces the catalytic power to undetectable levels in biochemical assays (10–12). Substitution of alanine (Ala) or another noncharged residue for the P-loop lysine (Lys), the most highly conserved of these residues, is the approach used most frequently to knock down the catalytic power of NTPases. However, an explanation as to how the electric charge of these catalytically crucial residues contributes to force generation has remained elusive due to the difficulty of investigating these effects while simultaneously retaining hydrolytic activity. In this study, we evaluated the effects of alanine mutation of the catalytically crucial charged residues on catalytic efficiencies and force generation in a highly sensitive single-molecule assay using the F1-ATPase rotary motor as a model NTP-driven molecular machine.
F1-ATPase (F1),3 the water-soluble portion of the F0F1-ATP synthase, is a member of the RecA-type protein family and possesses all of the catalytically crucial charged residues: P-loop Lys, the general base, and the Arg finger. When isolated from the ATP synthase complex, F1 acts as a rotary motor protein, where it rotates the inner rotary subunit against the stator ring, hydrolyzing ATP (13, 14). The bacterial type F1 is composed of α3β3γδϵ, with the α3β3γ subcomplex representing the minimum complex necessary to function as a rotary motor. The α3β3 subunits form the cylindrical stator ring. The γ subunit is the rotary shaft that penetrates the center of the cylinder (8, 15–17). The catalytic sites for ATP hydrolysis reside on each α-β interface, mainly on the β subunit (8). Therefore, conformational changes of the β subunit are responsible for torque generation. The rotary torque of F1 has repeatedly been reported to be 40 pN·nm for F1 from thermophilic bacteria and the Bacillus PS3 that was investigated in this study (18, 19), although F1 torque from Escherichia coli was reported to be 30–61 pN·nm (14, 15, 20).
Among motor proteins, F1 is unique for its high reversibility in a chemo-mechanical coupling reaction, i.e. F1 catalyzes ATP synthesis when the rotary shaft is forcibly rotated in the reverse direction (21, 22). In cells, F1 binds to Fo, the membrane-embedded part of ATP synthase, to form the whole ATP synthase complex. Under physiological conditions, Fo, powered by the proton-motive force across the membranes, generates a larger torque than F1, thereby reversing F1 to induce the ATP synthesis reaction. This high reversibility means that each reaction step is tightly coupled with a rotary motion of the γ subunit. The tight coupling feature also allows us to elucidate kinetic analysis of individual catalytic reactions from observations of the rotary motion.
To establish a basis for studying the chemo-mechanical coupling mechanism of F1, the reaction scheme of F1 was extensively researched. Although most aspects of the scheme have been resolved, some uncertainties remained (23). Rotations reportedly occur in discrete 120° steps, each coupled to a single turnover of ATP hydrolysis (18). The 120° step is further divided into 80 and 40° substeps (24, 25). The 80° substep is triggered by ATP binding and ADP release, each of which occurs on different β subunits (26). The 40° substep is triggered by ATP hydrolysis and release of inorganic phosphate (Pi), which also occurs on different β subunits (26, 27). The angular positions of the dwell before the 80 and 40° substeps are referred to as the ATP-binding and catalytic angles, respectively.
Establishment of the basic reaction scheme has led to the more fundamental question of determining how the locally occurring chemical reaction induces the large conformational change of the motor protein. Previous molecular genetics and biochemical studies have identified the catalytic residues of F1 based on sequence homology and chemical modifications with ATP analogues (28). The crystal structures of F1 elucidated the atomic details of the catalytic reaction centers (8, 29–31), providing a foundation for structure-based theoretical studies (32–34). All of these studies have identified the abovementioned charged residues as the catalytically critical residues, although some additional residues were also found to be involved in catalysis. Interestingly, when these charged residues are substituted with noncharged or oppositely charged residues, F1 catalytic activity is reduced to undetectable levels in biochemical ATPase assays (10, 11, 35–37), suggesting that these residues are indispensable for catalysis. However, no data have been presented that clearly explain how the electric charges of these residues contribute to force generation of F1. Recently, a single-molecule assay of F1 provided quantitative analysis of very low catalytic activity (i.e. less than 0.005 s−1), which was lower than the detection limit of the biochemical ATPase assay (about 0.3 s−1) (35). Therefore, in this work we re-assessed the catalytic competence of the alanine mutant of the conserved charged residues using this single-molecule rotation assay. All of these mutants exhibited unidirectional rotation, indicating unexpectedly high robustness of catalytic competency. Additionally, we analyzed the impact of the mutation on torque generation.
EXPERIMENTAL PROCEDURES
Wild-type F1 and the F1 mutants F1(αR364A), F1(βK164A), F1(βE190A), and F1(βE190Q) were prepared as reported previously (38). To visualize the rotation of F1, the stator region (α3β3) was fixed onto a glass surface, and magnetic beads (Seradyn, Indianapolis, IN) were attached to the rotor (γ) as a rotation probe, as reported previously (27). The rotating beads were observed under a phase-contrast microscope (IX-70 or IX-71, Olympus, Tokyo, Japan). The rotation assay was performed at 25 °C. The images of rotary motion were recorded at 30–2,000 frames/s (FASTCAM 1024PCI-SE, Photron, Tokyo, Japan; FC300M, Takex, Kyoto, Japan). Images were stored on the HDD of a computer as AVI files and analyzed using custom-made software.
RESULTS
Rotary Motion of Alanine-substituted Mutants αR364A, βK164A, and βE190A
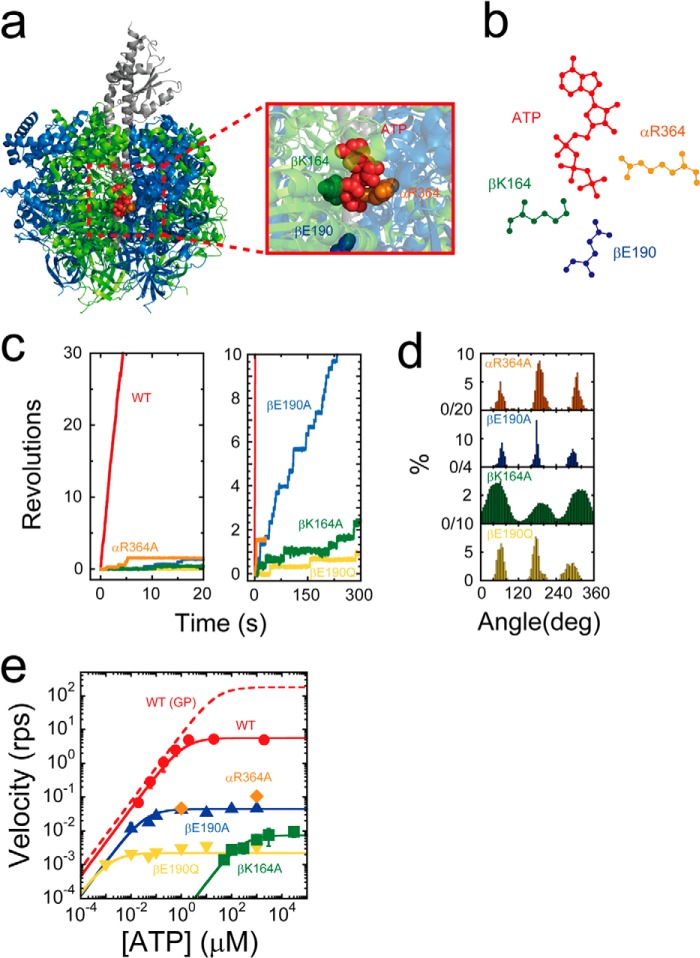
The arginine finger (αR364), the P-loop lysine (βK164), and the general base (βE190) of F1 from thermophilic Bacillus PS3 (TF1) were substituted with alanine to produce the F1 mutants, F1(αR364A), F1(βK164A), and F1(βE190A) (Fig. 1, a and b). βE190A was also substituted with glutamine to produce F1(βE190Q). The ATPase activity of all mutants was undetectable with biochemical assays, which was consistent with the results of previous studies (10, 11, 35–37). The rotation assay was conducted at a saturating ATP concentration (1 mm) by using magnetic beads as rotation markers. Surprisingly, all mutants exhibited continuous rotary motions in an anticlockwise direction (Fig. 1c), although rotation was extremely slow, in some instances taking as long as 5 min to confirm. Nonetheless, the observed rotation indicated that all of the mutants retained the catalytic power of ATP hydrolysis and their chemomechanical coupling nature. The rotational rate of these mutants ranged from 0.002 to 0.1 rps (Fig. 1c). The slowest rotation was observed for F1(βE190Q). As seen in the time courses in Fig. 1, c and d, all the mutants exhibited distinct pauses every 120° throughout the rotation, which limited the overall rotation rate. This observation is significant, as it indicates that one or more catalytic reaction steps are distinctively slowed.
FIGURE 1.
Rotary motion of mutant F1. a, side view of the crystal structure of F1 (Protein Data Bank code 2HLD) and interface between αDP and βDP. The α, β, and γ subunits are shown in blue, green, and gray, respectively. The arginine finger (αArg-364 in TF1; αArg-373 in MF1), P-loop lysine (βLys-164 in TF1; βLys-162 in MF1), general base (βGlu-190 in TF1; βGlu-188 in MF1), and ATP are shown by orange, green, blue, and red space-filling models, respectively. Amino acid residues are numbered according to the TF1 sequence. b, schematic diagrams of the interaction between the catalytic site (βDP) and ATP. c, time courses of rotary motion in the presence of 1 mm ATP; red, orange, green, blue, and yellow represent the time courses of wild-type F1, F1(αR364A), F1(βK164A), F1(βE190A), and F1(βE190Q), respectively. d, histogram of the angular position during rotation calculated from c. e, rotational velocity (v) at various ATP concentrations. The solid curves represent Michaelis-Menten fits with V = Vmax[ATP]/([ATP] + Km), where Vmaxwild type = 5.6 s−1; VmaxαR364A = 1.1 × 10−1 s−1; VmaxβK164A = 7.4 × 10−3 s−1; VmaxβE190A = 4.4 × 10−2 s−1; VmaxβE190Q = 2.2 × 10−3 s−1; Kmwild type = 1.2 μm; KmβK164A = 2.5 × 102 μm; KmβE190A = 3.6 × 10−2 μm; and KmβE190Q = 1.5 × 10−3 μm. From these, the following rate constants for ATP binding were calculated as kon = 3 × Vmax/Km, with konwild type = 1.4 × 107 m−1 s−1; konβK164A = 9.0 × 102 m−1 s−1; konβE190A = 3.7 × 106 m−1 s−1; and konβE190Q = 4.5 × 106 m−1 s−1. The dashed curve represents the rotational velocity of wild-type F1 with a gold colloidal bead, measured in the previous study (44).
The rotational rates of mutants were measured at various ATP concentrations ranging from 1 μm to 30 mm (Fig. 1e). The mutant F1(αR364A) rarely exhibited continuous rotations at low ATP concentrations (less than 1 μm) due to severe inhibition, which did not allow for the assay to detect rotation at an ATP concentration less than 1 μm. The other mutants were observed to follow Michaelis-Menten kinetics, yielding maximum rotation rates (Vmax) and Michaelis constants (Km). Assuming that the chemomechanical coupling ratio of F1 was three ATPs/turn, 3 × Vmax represented the catalytic turnover rate (kcat), and these values were calculated to be 3.3 × 10−1, 2.2 × 10−2, 1.3 × 10−1, and 6.6 × 10−3 s−1 for F1(αR364A), F1(βK164A), F1(βE190A), and F1(βE190Q), respectively (Table 1). The rate constants of ATP binding (konATP) that were determined as 3 × Vmax/Km, were 9.0 × 102, 3.7 × 106, and 4.5 × 106 m−1 s−1 for F1(βK164A), F1(βE190A), and F1(βE190Q), respectively. Compared with kcat and konATP from the wild type (387 s−1 and 2.6 × 107 m−1 s−1, respectively), the impact of the depletion of the conserved charge was significant in the suppression of kcat, whereas the P-loop lysine mutant also resulted in remarkable suppression of konATP. This indicated that the P-loop lysine is involved in both the binding process and catalysis.
TABLE 1.
Kinetic parameters, Stiffness and Torque
Analysis of Stepping Rotation
It is known that F1 has two stable states, pausing at either the binding angle or the catalytic angle, with the latter angle calculated at +80° from the binding angle, in the anticlockwise direction. We therefore attempted to determine the angles where these pauses occurred for the mutants.
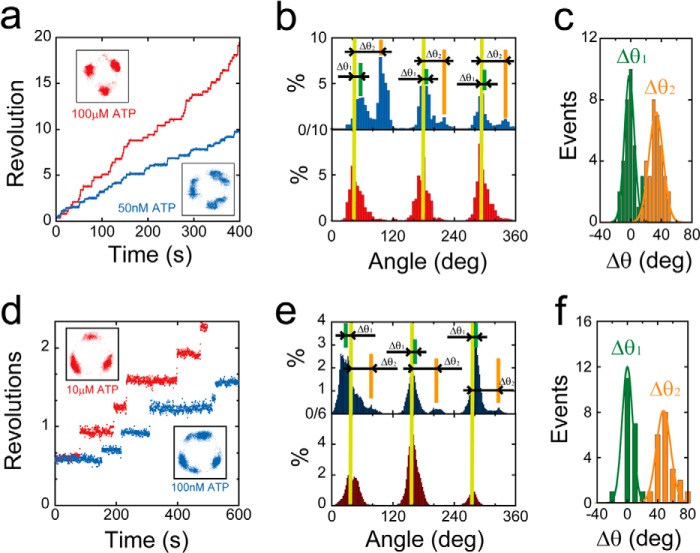
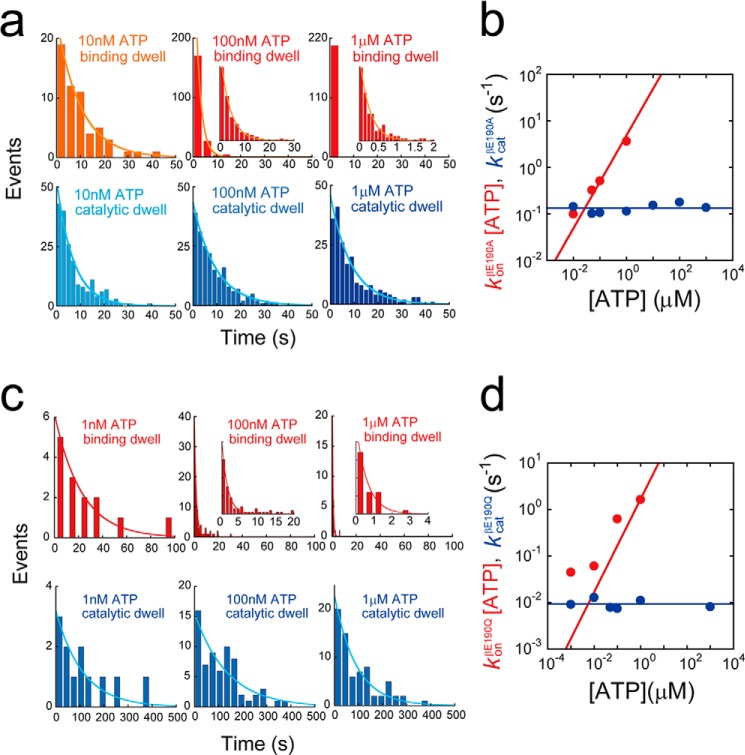
For the analysis of F1(βE190A), we conducted a buffer exchange experiment where the ATP concentration was changed from 1 mm to 50 nm, which was comparable with the Km value of this mutant (Fig. 2a). When the buffer was changed to 50 nm, the mutant exhibited substeps, characterized by pauses at six distinct positions (Fig. 2b). For statistical analysis, we compared the angular position of the pauses appearing at 50 nm ATP from the original pauses at 1 mm ATP for individual rotating molecules (Δθ2 in Fig. 2, b and c). For comparison, the relative angular positions of the original intervening pauses at two ATP concentrations (Δθ1 in Fig. 2, b and c) were also analyzed. The results of this analysis indicated that Δθ1 was −1.7 ± 6.4°, which validated this experimental approach, whereas Δθ2 was 33 ± 9.2°. These results indicated that the intervening pauses of F1(βE190A) occurred at the same catalytic angle as that previously reported for an aspartic acid mutant at βGlu-190 (25). When we conducted the same buffer exchange experiment for F1(βE190Q), we observed that the intervening pause of F1(βE190Q) also occurred at the catalytic pauses (Fig. 2, d–f). Thus, the mutation at the general base caused the distinctly long pause at the catalytic angle. We also analyzed the dwell time of the intervening pause of F1(βE190A) and F1(βE190Q), as well as the dwell time of the ATP-waiting pause (Fig. 3). The histograms of the dwell times were fit to single exponential functions y = C··exp(−kt), allowing us to determine kcat and konATP (Fig. 3, a and c). konATP was proportional to [ATP], but kcat was not (Fig. 3, b and d). konATP was 4.9 × 106 and 1.7 × 106 m−1 s−1, and kcat was 0.13, 9.3 × 10−3 s−1 for F1(βE190A) or F1(βE190Q) (Fig. 5, a and b). These values were consistent with the aforementioned Michaelis-Menten analysis (Fig. 1e).
FIGURE 2.
Stepping rotation of βGlu-190 mutants. a, examples of the time course of rotation by F1(βE190A). Buffer was exchanged during observation, and ATP concentration was reduced from 100 μm (red) to 50 nm (blue). The insets are x-y trajectories during rotation in the presence of 100 μm (left) or 50 nm ATP (right). b, histogram of the angular position of F1(βE190A) in the presence of 100 μm (bottom) or 50 nm ATP (top). Δθ1 and Δθ2 represent the angle distances of the pausing angles at 50 nm ATP compared with those at 100 μm ATP. c, distributions of angle distances of F1(βE190A) (Δθ1 and Δθ2). The average values of Δθ1 and Δθ2 are −1.8 ± 6.4 and 33 ± 9.2° (mean ± S.D., n = 33). d, examples of the time course of rotation by F1(βE190Q). Buffer was exchanged during observation, and ATP concentration was reduced from 10 μm (red) to 10 nm (blue). e, histogram of the angular position of F1(βE190Q) in the presence of 1 mm (bottom) or 1 nm ATP (top). Δθ1 and Δθ2 represent the angle distances of the pausing angles at 1 nm ATP from that at 1 mm ATP. f, distributions of angle distances of F1(βE190Q) (Δθ1 and Δθ2). The average values of Δθ1 and Δθ2 are 9.9 ± 10 and 54 ± 12° (mean ± S.D., n = 21).
FIGURE 3.
Kinetic analysis of βGlu-190 mutants. a, histograms of the dwell time of F1(βE190A) at ATP binding angle (top) or catalytic angle (bottom) in the presence of 1 μm (right), 100 nm (middle), and 10 nm ATP (left). Curves represent the fittings with a single-order reaction scheme, y = C·exp(−kt). b, rate constants of ATP binding and catalysis of F1(βE190A) determined from a. Solid lines represent the linear fittings as follows: konβE190A = 4.9 × 106 m−1 s−1, kcatβE190A = 0.13 s−1. c, histograms of the dwell time of F1(βE190Q) at ATP binding angle (top) or catalytic angle (bottom) in the presence of 1 μm (right), 100 nm (middle), and 1 nm ATP (left). Curves represent the fittings with a single-order reaction scheme, y = C·exp(−kt). d, rate constants of ATP binding and catalysis of F1(βE190Q) determined from c. Solid lines represent the linear fittings as follows: konβE190Q = 1.7 × 106 m−1 s−1, kcatβE190Q = 9.3 × 10−3 s−1.
FIGURE 5.
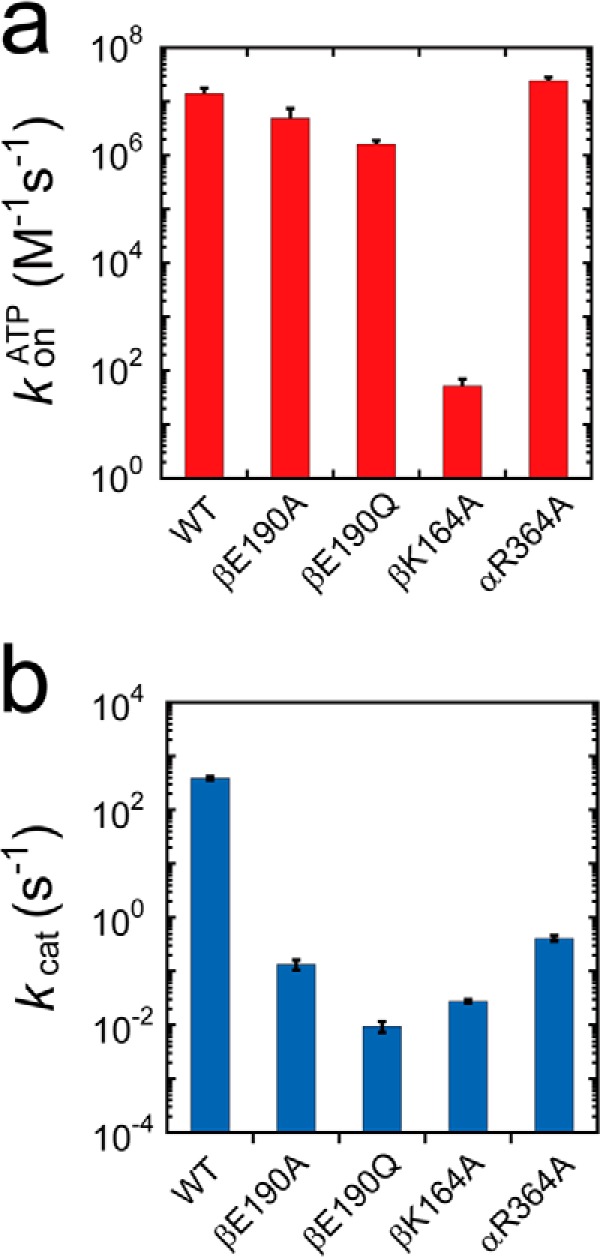
Rate constants of ATP binding and catalysis. a, rate constants of ATP binding for wild type and F1(βK164A) were determined from Fig. 1e, where konATP = 3 × Vmax/Km. Those for F1(βE190A), F1(βE190Q), and F1(αR364A) were determined from the dwell time analyses of Figs. 3, a and c, and 4f. b, rate constants of catalysis (kcat). The rate constants for wild type and F1(βK164A) were determined from Fig. 1e, where kcat = 3 × Vmax. Those for F1(βE190A), F1(βE190Q), and F1(αR364A) were determined from the dwell time analyses of Figs. 3, a and c, and 4f.
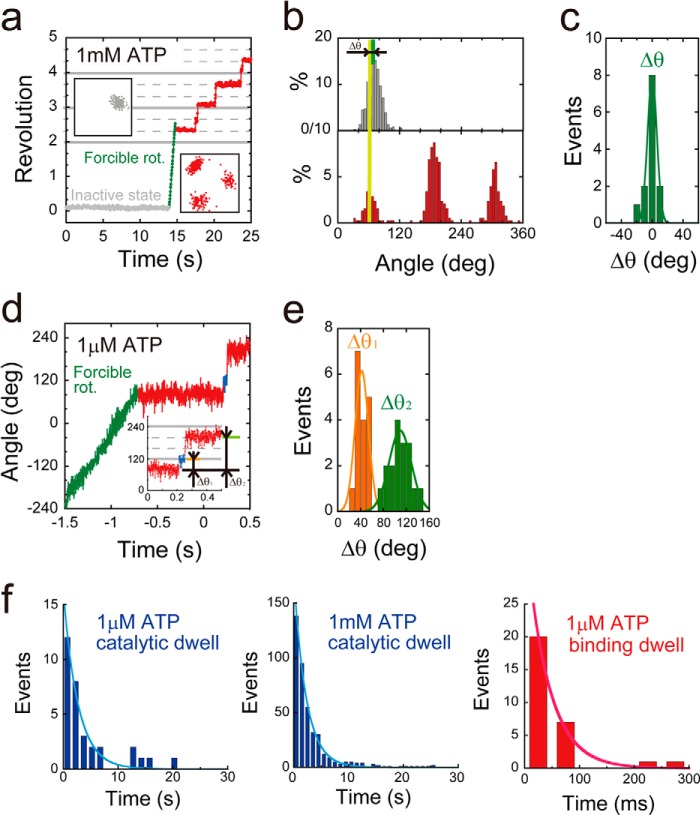
The mutant F1(αR364A) had a tendency to lapse into a long inhibitory state (Fig. 4a). i.e. it paused during the rotation, soon after making turns, and did not spontaneously resume rotation during extensive periods of observation. This inhibitory state was attributed to the ADP-inhibited form based on our previous observations of the lysine-substituted mutant of the arginine finger, F1(αR364K), that also lapsed into a long ADP inhibition state, leading to pauses at the catalytic angle (39). Additionally, as was observed with the ADP inhibition of the wild-type and other mutants F1 (40), F1(αR364A) could also be activated by forcible rotation with magnetic tweezers that allowed us repeated observations of F1(αR364K). We analyzed the relative angular positions of pauses occurring during rotation from ADP inhibition (Δθ in Fig. 4, b and c). Δθ was 0 ± 6.0°, suggesting that F1(αR364A) also paused at the catalytic angles. To confirm this, we compared the intervening pause with the ATP waiting angle by conducting a rotation assay at a low ATP concentration (1 μm). Although the ADP inhibition of F1(αR364A) was more pronounced at low ATP concentrations, F1(αR364A) was observed to rotate only a few steps after forcible activation with magnetic tweezers, allowing the analysis. At low [ATP], F1(αR364A) exhibited substep rotation in each 120° rotation. We analyzed the relative ATP waiting angle from the original intervening pause (Δθ1 in Fig. 4, d and e). The angle measured for Δθ1 was 41 ± 12°, indicating that the original intervening pause occurred at the catalytic angle. We analyzed the dwell time of pauses at both the ATP-binding and catalytic angles. Dwell time histograms were fit with single exponential functions, giving the rate constants of ATP binding or catalysis (Fig. 4f). Values calculated for F1(αR364A) were konATP = 2.4 × 107 m−1 s−1 and kcat = 0.45 s−1 (Fig. 5, a and b). It is interesting that kcat for this mutant was 10-fold smaller than that of the lysine-substituted mutant, F1(αR364K) (39); however, kon was almost the same, which suggests that the positive charge of αArg-364 is necessary for rate enhancement of hydrolysis, but not for efficient binding of ATP.
FIGURE 4.
Stepping rotation of αR364A mutant. a, examples of the time course of rotation by F1(αR364A). This variant lapsed into severe ADP inhibition (gray), and after forcible rotation with magnetic tweezers (green), it exhibited stepwise counterclockwise rotary motion (red). The insets are x-y trajectories during ADP inhibition (left) or stepwise rotation (right). b, histogram of the angular position during ADP inhibition (top) or stepwise rotation (bottom). Δθ represents the angle distance of the pausing angle at 1 mm ATP from that of ADP inhibition. c, distributions of angle distances (Δθ). The average value of Δθ was 0 ± 6.0° (mean ± S.D., n = 13). d, time course of the stepping rotation of F1(αR364A) in the presence of 1 μm ATP. Δθ1 represents the relative angular position of original pause at ATP saturating condition to ATP binding pause. Δθ2 represents the relative angular position of the original pauses. e, distributions of angle distances (Δθ1 and Δθ2). The average values of Δθ1 and Δθ2 are 42 ± 12 and 109 ± 28° (mean ± S.D., n = 36). f, histograms of the dwell time at catalytic angle (top) or ATP binding angle (bottom) in the presence of 1 mm (right) and 1 μm ATP (left). Curves represent the fittings with a single-order reaction scheme, y = C·exp(−kt). The rate constants of ATP binding and catalysis were determined as konαR364A = 2.4 × 107 m−1 s−1, and kcatαR364A = 0.45 s−1.
We also attempted to identify the pausing position of F1(βK164A). This mutant showed large rotary fluctuations during the pauses (Fig. 1d), which implied a significantly looser interaction between the stator and the rotor. When observed at low concentrations of ATP, some molecules exhibited rotation with substeps at 80 and 40°. Although the reduced probability of observing rotation at low [ATP] did not allow for statistical analysis, it is highly probable that the intervening pause of βK164A also occurred at the catalytic angle.
Measurement of Torque
To investigate the roles of the electrical charge of the P-loop lysine, general base, and Arg finger in torque generation, we measured the torque of the alanine mutants by using a recently developed method for torque measurement based on the fluctuation theorem (19). Rotation at 1 mm, where all of the mutants exhibited the 120° stepped rotation, was recorded at 1,000 Hz. For precise torque determination, molecules were selected that displayed symmetric 120° stepping rotation without obvious preferential pausing angles due to surface interactions. From the time trajectory, the portions of rotations were extracted, i.e. the portions of dwells were omitted. It should be noted that F1 showed the backward movements due to thermal fluctuations during rotations, although it showed a net unidirectional movement due to rotary torque. Ratios of the forward and backward movement probabilities (P(Δθ)/P(−Δθ)) over a set time period were then calculated. The ratio of probabilities was then plotted against Δθ, and the torque was determined from the slope (Fig. 6, a–c). Results of this analysis indicated that the torque values for the mutants were evidently smaller than those for wild-type F1 as follows: 28, 10, and 24 pN· nm for F1(αR364A), F1(βK164A), and F1(βE190A), respectively (Fig. 6d). In particular, we observed that the torque of F1(βK164A) was one-fourth that of wild-type F1.
FIGURE 6.
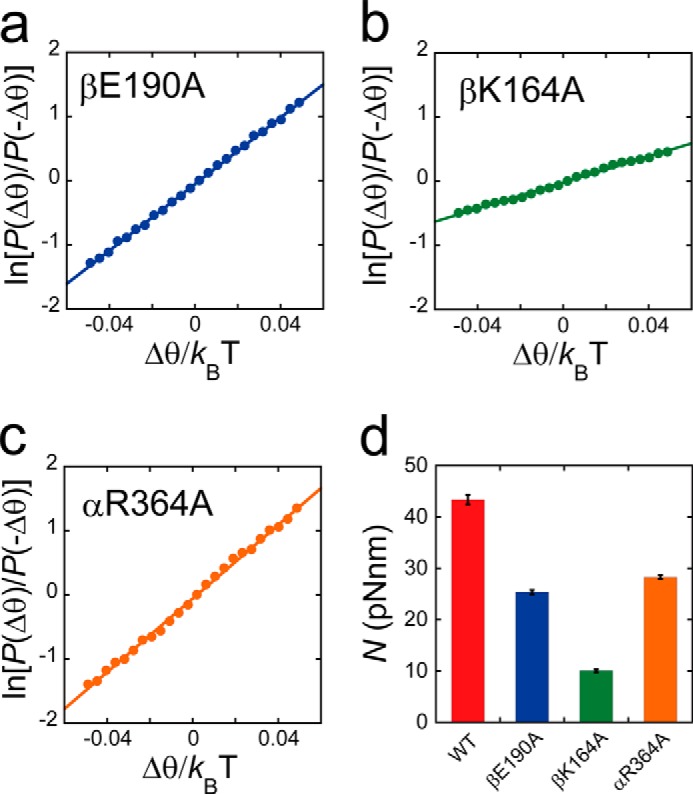
Rotary torque. a–c, fluctuation theorem was employed for the torque measurement of F1(βE190A), F1(βK164A), and F1(αR364A). The ln(P(Δθ)/P(−Δθ)) is plotted against Δθ/kBT. The slope of this plot represents the rotary torque generated by F1. The average torque was determined from a linear approximation of all data points (solid line). d, torque (N) amplitudes generated by wild type F1, F1(βE190A), F1(βK164A), and F1(αR364A) are represented as red, blue, green and orange bars, respectively.
Rotary Potential
We also examined the effect of mutations on the rotary potential during catalytic pauses. The probability densities of γ-orientation during catalytic pauses were measured, and the rotary potentials were determined according to Boltzmann's Law (Fig. 7a). The determined potentials were well fitted with a harmonic function ΔG = 1/2·κ·θ2, where κ is the torsion stiffness. The determined values of stiffness were 79, 42, 22, and 38 pN·nm for wild-type F1, F1(αR364A), F1(βK164A), and F1(βE190A), and these values correlated well with those for the rotary torque (Fig. 7b). The lower stiffness values of the mutants suggested that their rotary potentials became more gradual than those of wild-type F1, which indicated that γ was not tightly held in the cavity of the α3β3 stator ring of the mutants, especially for F1(βK164A). This result concurs with the contention that F1(βK164A) cannot fully induce the open-closed conformational change of the stator and thus weakens the interaction between the stator and rotor.
FIGURE 7.
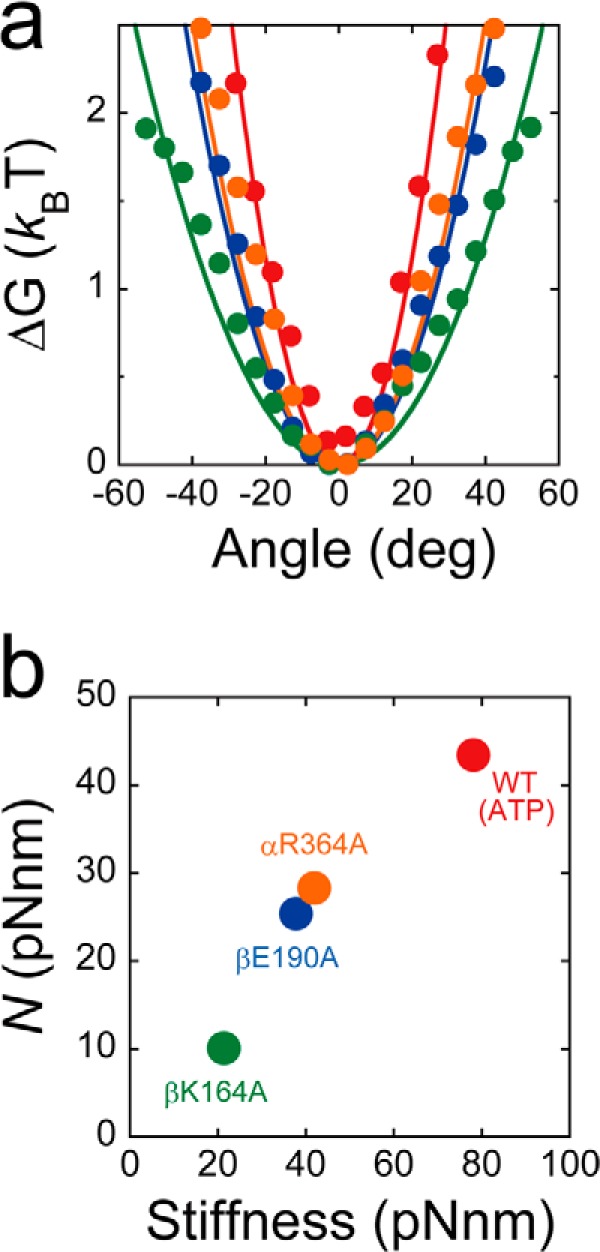
Rotary potential. a, rotary potential of the F1 catalytic waiting state. Probability densities of angular positions during the pause from the five molecules were transformed into rotary potentials according to Boltzmann's law: wild-type F1 (red), F1(βE190A) (blue), F1(βK164A) (green), and F1(αR364A) (orange). Determined potentials were fitted with the harmonic function ΔG = 1/2·κ ·θ2, where κ is the torsion stiffness. Determined stiffness values were 79, 42, 22, and 38 pN·nm for wild-type F1, F1(αR364A), F1(βK164A), and F1(βE190A), respectively. b, rotary torque plotted against rotary potential stiffness.
DISCUSSION
Robustness of Rotary Catalysis Mechanism of F1
All mutants exhibited unidirectional rotation, meaning that all of the charged residues that are highly conserved among P-loop ATPases (P-loop lysine, the general base, and the arginine finger) are dispensable for catalysis. The unidirectionality of the rotation was also comparable with that of wild-type F1, and the frequent backward step was not observed, meaning that the nature of the chemomechanical coupling was fundamentally retained in the absence of the conserved charged residues. This observation suggests that the catalytic power of F1 is much more robust than previously assumed.
The impact of the charge depletion was apparent in the kinetics, as the rotational rate was extremely slow (Figs. 1c and 5). In particular, the βGlu-190 mutant exhibited the slowest rotation (Vmax = 2.2 × 10−3), which was ∼106-fold slower than that of wild-type F1 (Vmax = 1.3 × 103). Analysis on the stepping rotation of the mutants proved that the mutations were responsible for the lengthened pauses at the catalytic angle, suggesting that all of the mutants decelerated the ATP hydrolysis step. In this sense, these findings support the theoretical prediction that these residues remarkably contribute to the rate enhancement of the ATP hydrolysis step (19, 33).
Considering the conserved structural features observed among P-loop ATPases, it is likely that the conserved, charged residues are also dispensable for other ATPases. To confirm the robustness of other ATPases, a highly sensitive assay is required, such as the single-molecule rotation assay of F1. The robustness of catalytic power has not previously been reported for other P-loop motors that have established highly sensitive single-molecule assays, and this is likely due, at least in part, to the relatively low processivity of other molecular motors. Molecular motors translocate their subunit or substrate protein/nucleic acid polymers toward the motor domain with ATPase activity. Because translocation necessarily accompanies the affinity modulation between a motor domain and a substrate, molecular motors dissociate from their substrates at some given probability. If the catalytic turnover rate is largely reduced, the probability of dissociation relatively increases, lowering the processivity of the motors. This could explain why linear motors such as myosin, kinesin, and dynein do not exhibit clear motion when the ATPase is decelerated by a factor of over 100 (41–43).
We also evaluated a set of double mutants: F1(αR364A/βK164A), F1(αR364A/βE190Q), and F1(βK164A/βE190Q). However, we did not observe any rotating particles in the rotation assay. When the roles of the conserved charged residues in rate enhancement are additive, the estimated rotational rate approaches the order of 10−5 revolution/s, and the single-molecule rotation assay might not be sufficiently sensitive to detect such a slow rotation. It should be noted that even if the turnover rate of ATPase is on the order of 10−5 s−1, it is still faster than the rate of spontaneous ATP hydrolysis in aqueous conditions.
Torque Generation
Another impact of the alanine substitution was observed in torque generation. All mutants decreased rotary torque (Fig. 6d). In particular, the rotary torque of F1(βK164A) was significantly reduced to 10 pN·nm. This is evidently lower than that of wild-type F1 and is the lowest among the reported torque values for the F1 mutants. It should be emphasized that although a wide variety of mutations were introduced into F1 in an attempt to identify the residues or structural elements that are crucial for torque generation, several mutations have been reported that evidently reduce the torque of F1. In addition, all of the mutations critical for torque were not found at the catalytic sites but mainly at the β-γ interfaces, such as the DELSEED loop of the β subunit (16, 44). Other nucleotides such as GTP, ITP, and UTP have been previously investigated to determine the effect of base recognition on energy transduction. Although GTP and ITP were found to be slow-binding substrates, GTP and ITP apparently supported torque generation at levels comparable with ATP (45). It should be noted that Martin et al. (46) proposed in a recent paper that ITP-supported rotation decreases angular velocity. However, because the condition was not viscous friction-limiting, it was not possible to determine whether this correlated to a decrease in torque. Thus, the impact of alanine substitution for the charged residues on torque generation is remarkable. Considering that all of these residues are involved in γ- or β-phosphate binding, the interaction between the catalytic site of F1 and the phosphate region of nucleotides is significantly crucial for torque generation.
Interestingly, alanine-substituted mutants did not significantly impact konATP values, except for βK164A, although both βE190A and αR364A reduced konATP by a factor of less than 10, whereas kcat was reduced by a factor between 105 and 106. Thus, it turned out that the apparent binding rate of the substrate was not significantly relevant to torque generation. Our recent work on the mechanical modulation of the kinetic power of F1 indicated that the affinity change process following the first substrate-F1 docking process (substrate recognition process) is responsible for torque generation. The affinity change process would mainly occur at the phosphate-binding region.
The rotary torque of the mutants positively correlated well with the stiffness of rotary potential, as estimated from the catalytic dwell (Fig. 7b). The stiffness of the rotary potential is a good barometer of how strongly the αβ stator ring holds the γ subunit. This correlation was also observed in a previous study of a mutant with extensive alanine substitution at the DELSEED loop of the β subunit (44, 47, 48). Results of this study also indicated that the P-loop lysine, general base, and arginine finger all play a role to tighten the interaction between β and γ, which likely stabilizes the closed conformation of the β subunit.
Role of P-loop Lysine
The alanine mutation on P-loop lysine caused drastic suppression of the rate constants for both ATP binding and hydrolysis as follows: 500,000 and 14,000 times smaller than those of wild type, respectively (Fig. 5 and Table 1). Unlike the general base and the arginine finger, the P-loop lysine is involved in both substrate recognition and the subsequent affinity change process. Thus, it is highly likely that the P-loop lysine is the most catalytically important residue among the ATP-binding residues in F1. In the crystal structures of F1, the P-loop lysine forms hydrogen bonds with β- and γ-phosphate (Fig. 1b). This residue would therefore be responsible for phosphate binding and for maintaining the conformation of the phosphate region of ATP to facilitate the binding of other catalytic residues. An NMR study by Yagi et al. (10) also revealed that this residue is crucial for ATP binding, as well as the conformational transition from the open state to the closed state of the β subunit. Taking these findings into account, it is highly likely that hydrogen bond formation between the P-loop lysine and β- and γ-phosphate of ATP drives the open-to-closed conformational transition that is responsible for torque generation.
Role of General Base
The alanine and glutamine mutations on the general base caused drastic suppression of the ATP hydrolysis rate constants to 2,900 and 41,000 times lower than that of wild type, respectively, although the ATP-binding rate was slightly slower than that of wild type (Fig. 5 and Table 1). Coincidentally, the rotational rate of the glutamine mutant was 2.2 × 10−3 rps, equivalent to an ATPase activity of 6.6 × 10−3 s−1. To our knowledge, this is the lowest ATPase activity value ever measured for F1. Thus, the negative charge at the general base position principally contributes to the acceleration of the ATP hydrolysis step. There are apparent differences in the ATP hydrolysis rates between alanine- and glutamine-substituted mutants, as the alanine mutant hydrolyzes ATP faster (Fig. 5 and Table 1). To retain ATPase activity, it seems likely that similar spatial rearrangements of side chains of other amino acids located at the catalytic sites are required in these mutants, so that the side chain carboxyl groups of other amino acids should occupy spatial positions close to the glutamic acid residues, as indicated in a previous study (35). In view of this, the glutamine, which occupies the aforementioned spatial position with an amide group, prevents spatial rearrangements more readily than the shorter alanine and hence reduces the efficiency of ATP hydrolysis.
The general base has been thought to activate the water molecule between the γ-phosphate and itself, and thereby induce nucleophilic attack of water on the γ-phosphate. However, this prevailing view has been challenged by a recent QM/MM study on the catalysis of F1 (5), which suggested that γ-phosphate dissociation occurs before protonation of the general base and that the general base-facilitated proton transfer is the kinetic bottleneck in the hydrolysis step. Our findings that mutants F1(βE190A or βE190Q) still retain catalytic activity agree with the QM/MM study (5). Therefore, it is likely that proton relay occurs after γ-phosphate dissociation, via the surrounding water molecules, and in the absence of the general base.
Role of Arginine Finger
The alanine mutation on the arginine finger reduced the rate constant of the ATP hydrolysis step by a factor of 950, compared with that of the wild type, whereas the rate of ATP binding was slightly suppressed (Fig. 5 and Table 1). Thus, the positive charge at the arginine finger position principally contributed to rate enhancement of the ATP hydrolysis step. This is consistent with results reported from a structural study on F1 (29). Another prominent impact of the arginine finger mutation is that it caused severe ADP inhibition (Fig. 4a). The enhancement of ADP inhibition was also reported in the lysine-substituted mutant (39). The inhibition of the alanine-substituted mutant was the most severe inhibition, which occurred after only a few turns of rotations, and could not be activated by thermal agitation within 10 min. We also recently learned that irregular Pi release from the catalytic site prior to ADP leads to an ADP-inhibited state (49). Considering that the positive charge of the arginine finger electrostatically interacts with the γ-phosphate of ATP, it is highly probable that the αR364A mutant reduces the binding affinity to the product Pi, which promotes severe ADP inhibition.
Although arginine finger and P-loop lysine possess a positively charged group that interacts with β- and γ-phosphate of bound ATP, the impacts of the Ala substitution are different from each other. The P-loop lysine mutant showed drastic suppression of both nucleotide binding and hydrolytic activity, whereas the arginine finger mutant mainly decreases the hydrolytic activity. This is attributable to the positional difference of the residues in the tertiary structure of F1-ATPase; the P-loop lysine is located in the Walker A motif on the β subunit, whereas arginine finger resides on the α subunit. It is evident from the crystal structure of F1-ATPase (29) that F1-ATPase closes the α-β interface upon the rotation to trigger the hydrolytic reaction. The widely accepted scenario is that after ATP binds to the catalytic site that is mainly formed with the residues on the β subunit, F1-ATPase closes the α-β interface. This conformational change accompanies with approximation of arginine finger toward bound ATP. Several lines of theoretical works (5, 33) strongly suggested that this positional shift of the arginine finger is thought to be crucial for the initiation of the hydrolytic reaction. Conversely, P-loop lysine forms hydrogen bonds with ATP before the closing of the α-β interface. These structural features would be the basis for the distinct difference in the role of these residues.
Acknowledgment
We thank all the members of Noji Laboratory.
This work was supported by Grants-in-aid for Scientific Research 18074005 (to H. N.) and 30540108 (to R. W.) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
- F1
- F1-ATPase
- pN
- piconewton.
REFERENCES
- 1. Vale R. D., Milligan R. A. (2000) The way things move: looking under the hood of molecular motor proteins. Science 288, 88–95 [DOI] [PubMed] [Google Scholar]
- 2. Hanson P. I., Whiteheart S. W. (2005) AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6, 519–529 [DOI] [PubMed] [Google Scholar]
- 3. Walker J. E., Saraste M., Runswick M. J., Gay N. J. (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Muneyuki E., Noji H., Amano T., Masaike T., Yoshida M. (2000) FoF1-ATP synthase: general structural features of ‘ATP-engine’ and a problem on free energy transduction. Biochim. Biophys. Acta 1458, 467–481 [DOI] [PubMed] [Google Scholar]
- 5. Hayashi S., Ueno H., Shaikh A. R., Umemura M., Kamiya M., Ito Y., Ikeguchi M., Komoriya Y., Iino R., Noji H. (2012) Molecular mechanism of ATP hydrolysis in F1-ATPase revealed by molecular simulations and single-molecule observations. J. Am. Chem. Soc. 134, 8447–8454 [DOI] [PubMed] [Google Scholar]
- 6. McGrath M. J., Kuo I. F., Hayashi S., Takada S. (2013) Adenosine triphosphate hydrolysis mechanism in kinesin studied by combined quantum-mechanical/molecular-mechanical metadynamics simulations. J. Am. Chem. Soc. 135, 8908–8919 [DOI] [PubMed] [Google Scholar]
- 7. Scheffzek K., Ahmadian M. R., Kabsch W., Wiesmüller L., Lautwein A., Schmitz F., Wittinghofer A. (1997) The Ras-RasGAP complex: structural basis for GTPase activation and its loss in oncogenic Ras mutants. Science 277, 333–338 [DOI] [PubMed] [Google Scholar]
- 8. Abrahams J. P., Leslie A. G., Lutter R., Walker J. E. (1994) Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature 370, 621–628 [DOI] [PubMed] [Google Scholar]
- 9. Chen Z., Yang H., Pavletich N. P. (2008) Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453, 489–494 [DOI] [PubMed] [Google Scholar]
- 10. Yagi H., Kajiwara N., Iwabuchi T., Izumi K., Yoshida M., Akutsu H. (2009) Stepwise propagation of the ATP-induced conformational change of the F1-ATPase β subunit revealed by NMR. J. Biol. Chem. 284, 2374–2382 [DOI] [PubMed] [Google Scholar]
- 11. Senior A. E., al-Shawi M. K. (1992) Further examination of 17 mutations in Escherichia coli F1-ATPase β-subunit. J. Biol. Chem. 267, 21471–21478 [PubMed] [Google Scholar]
- 12. Li X. D., Rhodes T. E., Ikebe R., Kambara T., White H. D., Ikebe M. (1998) Effects of mutations in the γ-phosphate binding site of myosin on its motor function. J. Biol. Chem. 273, 27404–27411 [DOI] [PubMed] [Google Scholar]
- 13. Yoshida M., Muneyuki E., Hisabori T. (2001) ATP synthase–a marvelous rotary engine of the cell. Nat. Rev. Mol. Cell Biol. 2, 669–677 [DOI] [PubMed] [Google Scholar]
- 14. Junge W., Sielaff H., Engelbrecht S. (2009) Torque generation and elastic power transmission in the rotary FoF1-ATPase. Nature 459, 364–370 [DOI] [PubMed] [Google Scholar]
- 15. Spetzler D., Ishmukhametov R., Hornung T., Day L. J., Martin J., Frasch W. D. (2009) Single molecule measurements of F1-ATPase reveal an interdependence between the power stroke and the dwell duration. Biochemistry 48, 7979–7985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Iko Y., Sambongi Y., Tanabe M., Iwamoto-Kihara A., Saito K., Ueda I., Wada Y., Futai M. (2001) ATP synthase F1 sector rotation. Defective torque generation in the β subunit Ser-174 to Phe mutant and its suppression by second mutations. J. Biol. Chem. 276, 47508–47511 [DOI] [PubMed] [Google Scholar]
- 17. Cherepanov D. A., Junge W. (2001) Viscoelastic dynamics of actin filaments coupled to rotary F-ATPase: curvature as an indicator of the torque. Biophys. J. 81, 1234–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yasuda R., Noji H., Kinosita K., Jr., Yoshida M. (1998) F1-ATPase is a highly efficient molecular motor that rotates with discrete 120 degree steps. Cell 93, 1117–1124 [DOI] [PubMed] [Google Scholar]
- 19. Hayashi K., Ueno H., Iino R., Noji H. (2010) Fluctuation theorem applied to F1-ATPase. Phys. Rev. Lett. 104, 218103. [DOI] [PubMed] [Google Scholar]
- 20. Bilyard T., Nakanishi-Matsui M., Steel B. C., Pilizota T., Nord A. L., Hosokawa H., Futai M., Berry R. M. (2013) High-resolution single-molecule characterization of the enzymatic states in Escherichia coli F1-ATPase. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rondelez Y., Tresset G., Nakashima T., Kato-Yamada Y., Fujita H., Takeuchi S., Noji H. (2005) Highly coupled ATP synthesis by F1-ATPase single molecules. Nature 433, 773–777 [DOI] [PubMed] [Google Scholar]
- 22. Itoh H., Takahashi A., Adachi K., Noji H., Yasuda R., Yoshida M., Kinosita K. (2004) Mechanically driven ATP synthesis by F1-ATPase. Nature 427, 465–468 [DOI] [PubMed] [Google Scholar]
- 23. Weber J. (2010) Structural biology: Toward the ATP synthase mechanism. Nat. Chem. Biol. 6, 794–795 [DOI] [PubMed] [Google Scholar]
- 24. Yasuda R., Noji H., Yoshida M., Kinosita K., Jr., Itoh H. (2001) Resolution of distinct rotational substeps by submillisecond kinetic analysis of F1-ATPase. Nature 410, 898–904 [DOI] [PubMed] [Google Scholar]
- 25. Shimabukuro K., Yasuda R., Muneyuki E., Hara K. Y., Kinosita K., Jr., Yoshida M. (2003) Catalysis and rotation of F1 motor: cleavage of ATP at the catalytic site occurs in 1 ms before 40 degree substep rotation. Proc. Natl. Acad. Sci. U.S.A. 100, 14731–14736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Adachi K., Oiwa K., Nishizaka T., Furuike S., Noji H., Itoh H., Yoshida M., Kinosita K., Jr. (2007) Coupling of rotation and catalysis in F1-ATPase revealed by single-molecule imaging and manipulation. Cell 130, 309–321 [DOI] [PubMed] [Google Scholar]
- 27. Watanabe R., Iino R., Noji H. (2010) Phosphate release in F1-ATPase catalytic cycle follows ADP release. Nat. Chem. Biol. 6, 814–820 [DOI] [PubMed] [Google Scholar]
- 28. Nakamoto R. K., Shin K., Iwamoto A., Omote H., Maeda M., Futai M. (1992) Escherichia coli FoF1-ATPase. Residues involved in catalysis and coupling. Ann. N.Y. Acad. Sci. 671, 335–343 [DOI] [PubMed] [Google Scholar]
- 29. Kagawa R., Montgomery M. G., Braig K., Leslie A. G., Walker J. E. (2004) The structure of bovine F1-ATPase inhibited by ADP and beryllium fluoride. EMBO J. 23, 2734–2744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kabaleeswaran V., Puri N., Walker J. E., Leslie A. G., Mueller D. M. (2006) Novel features of the rotary catalytic mechanism revealed in the structure of yeast F1 ATPase. EMBO J. 25, 5433–5442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cingolani G., Duncan T. M. (2011) Structure of the ATP synthase catalytic complex (F1) from Escherichia coli in an autoinhibited conformation. Nat. Struct. Mol. Biol. 18, 701–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Yang W., Gao Y. Q., Cui Q., Ma J., Karplus M. (2003) The missing link between thermodynamics and structure in F1-ATPase. Proc. Natl. Acad. Sci. U.S.A. 100, 874–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dittrich M., Hayashi S., Schulten K. (2004) ATP hydrolysis in the βTP and βDP catalytic sites of F1-ATPase. Biophys. J. 87, 2954–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mukherjee S., Warshel A. (2011) Electrostatic origin of the mechanochemical rotary mechanism and the catalytic dwell of F1-ATPase. Proc. Natl. Acad. Sci. U.S.A. 108, 20550–20555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Amano T., Tozawa K., Yoshida M., Murakami H. (1994) Spatial precision of a catalytic carboxylate of F1-ATPase β subunit probed by introducing different carboxylate-containing side chains. FEBS Lett. 348, 93–98 [DOI] [PubMed] [Google Scholar]
- 36. Le N. P., Omote H., Wada Y., Al-Shawi M. K., Nakamoto R. K., Futai M. (2000) Escherichia coli ATP synthase α subunit Arg-376: the catalytic site arginine does not participate in the hydrolysis/synthesis reaction but is required for promotion to the steady state. Biochemistry 39, 2778–2783 [DOI] [PubMed] [Google Scholar]
- 37. Löbau S., Weber J., Wilke-Mounts S., Senior A. E. (1997) F1-ATPase, roles of three catalytic site residues. J. Biol. Chem. 272, 3648–3656 [DOI] [PubMed] [Google Scholar]
- 38. Okuno D., Iino R., Noji H. (2010) Stiffness of γ subunit of F1-ATPase. Eur. Biophys. J. 39, 1589–1596 [DOI] [PubMed] [Google Scholar]
- 39. Komoriya Y., Ariga T., Iino R., Imamura H., Okuno D., Noji H. (2012) Principal role of the arginine finger in rotary catalysis of F1-ATPase. J. Biol. Chem. 287, 15134–15142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hirono-Hara Y., Ishizuka K., Kinosita K., Jr., Yoshida M., Noji H. (2005) Activation of pausing F1 motor by external force. Proc. Natl. Acad. Sci. U.S.A. 102, 4288–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yildiz A., Forkey J. N., McKinney S. A., Ha T., Goldman Y. E., Selvin P. R. (2003) Myosin V walks hand-over-hand: single fluorophore imaging with 1.5-nm localization. Science 300, 2061–2065 [DOI] [PubMed] [Google Scholar]
- 42. Sun Y., Sato O., Ruhnow F., Arsenault M. E., Ikebe M., Goldman Y. E. (2010) Single-molecule stepping and structural dynamics of myosin X. Nat. Struct. Mol. Biol. 17, 485–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cho C., Reck-Peterson S. L., Vale R. D. (2008) Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J. Biol. Chem. 283, 25839–25845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tanigawara M., Tabata K. V., Ito Y., Ito J., Watanabe R., Ueno H., Ikeguchi M., Noji H. (2012) Role of the DELSEED loop in torque transmission of F1-ATPase. Biophys. J. 103, 970–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Noji H., Bald D., Yasuda R., Itoh H., Yoshida M., Kinosita K., Jr. (2001) Purine but not pyrimidine nucleotides support rotation of F1-ATPase. J. Biol. Chem. 276, 25480–25486 [DOI] [PubMed] [Google Scholar]
- 46. Martin J. L., Ishmukhametov R., Hornung T., Ahmad Z., Frasch W. D. (2014) Anatomy of F1-ATPase powered rotation. Proc. Natl. Acad. Sci. U.S.A. 111, 3715–3720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Usukura E., Suzuki T., Furuike S., Soga N., Saita E., Hisabori T., Kinosita K., Jr., Yoshida M. (2012) Torque generation and utilization in motor enzyme FoF1-ATP synthase half-torque F1 with short-sized pushrod helix and reduced ATP synthesis by half-torque FoF1. J. Biol. Chem. 287, 1884–1891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kohori A., Chiwata R., Hossain M. D., Furuike S., Shiroguchi K., Adachi K., Yoshida M., Kinosita K., Jr. (2011) Torque generation in F1-ATPase devoid of the entire amino-terminal helix of the rotor that fills half of the stator orifice. Biophys. J. 101, 188–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Watanabe R., Noji H. (2014) Timing of inorganic phosphate release modulates the catalytic activity of ATP-driven rotary motor protein. Nat. Commun. 5, 3486. [DOI] [PMC free article] [PubMed] [Google Scholar]