Background: The hydrogen-evolving membrane-bound hydrogenase (MBH) functions as a simple respiratory system in anaerobic microbes.
Results: Affinity-tagged MBH was solubilized from membranes of a hyperthermophile as an intact 14-subunit complex.
Conclusion: Solubilized MBH was catalytically active, and a structural model based on small angle x-ray scattering (SAXS) was obtained.
Significance: The successful purification of a respiratory hydrogenase has enabled biochemical and structural studies.
Keywords: Bioenergetics, Electron Transfer, Hydrogenase, Membrane Enzyme, Respiration, Affinity Purification, Hydrogen, Membrane Enzyme, Solubilization, Thermophile
Abstract
The archaeon Pyrococcus furiosus grows optimally at 100 °C by converting carbohydrates to acetate, CO2, and H2, obtaining energy from a respiratory membrane-bound hydrogenase (MBH). This conserves energy by coupling H2 production to oxidation of reduced ferredoxin with generation of a sodium ion gradient. MBH is encoded by a 14-gene operon with both hydrogenase and Na+/H+ antiporter modules. Herein a His-tagged MBH was expressed in P. furiosus and the detergent-solubilized complex purified under anaerobic conditions by affinity chromatography. Purified MBH contains all 14 subunits by electrophoretic analysis (13 subunits were also identified by mass spectrometry) and had a measured iron:nickel ratio of 15:1, resembling the predicted value of 13:1. The as-purified enzyme exhibited a rhombic EPR signal characteristic of the ready nickel-boron state. The purified and membrane-bound forms of MBH both preferentially evolved H2 with the physiological donor (reduced ferredoxin) as well as with standard dyes. The O2 sensitivities of the two forms were similar (half-lives of ∼15 h in air), but the purified enzyme was more thermolabile (half-lives at 90 °C of 1 and 25 h, respectively). Structural analysis of purified MBH by small angle x-ray scattering indicated a Z-shaped structure with a mass of 310 kDa, resembling the predicted value (298 kDa). The angle x-ray scattering analyses reinforce and extend the conserved sequence relationships of group 4 enzymes and complex I (NADH quinone oxidoreductase). This is the first report on the properties of a solubilized form of an intact respiratory MBH complex that is proposed to evolve H2 and pump Na+ ions.
Introduction
Over the past decade there has been a major initiative to generate alternative nonfossil fuels to fulfill increasingly critical energy needs. However, such fuels must be energy efficient as well as carbon neutral, and biohydrogen production can meet these criteria (1). Hydrogen is metabolized by microbes from all three domains of life (2). Notably, they all contain the enzyme hydrogenase that functions to catalyze the reversible reduction of protons to molecular hydrogen (H2). Hydrogenases are classified based on the metal content of their active site into the [NiFe]-hydrogenase, [FeFe]-hydrogenase, and the [FeS]-cluster-free hydrogenases. The [NiFe]-hydrogenases are ubiquitous in the microbial world and have been extensively studied from numerous mesophilic bacteria (3–5). The minimum structure is a heterodimer composed of a large and small subunit. The large subunit contains the [NiFe] catalytic site that is coordinated by the sulfur atoms of four cysteine residues organized into two CXXC motifs near the N and C termini. The [NiFe] active site has been extensively studied by EPR spectroscopy (6). The small subunit typically contains three iron-sulfur clusters invariably of the [4Fe-4S] type that shuttle electrons between an acceptor/donor for the enzyme and its active site. [NiFe]-hydrogenases are classified into four groups based on the phylogeny of their catalytic subunits (2). Crystal structures for [NiFe]-hydrogenases are available for group 1 hydrogenases (7, 8), but no structural information is available for the other three hydrogenase groups.
The least studied of the [NiFe]-hydrogenases fall into group 4, and they are defined as the H2-evolving energy-conserving membrane-associated hydrogenases (2). These group 4 enzymes show little sequence similarity to other [NiFe]-hydrogenases, except for the conserved residues that bind the [NiFe] catalytic site and its proximal [4Fe-4S] cluster (9, 10), indicating a distinct evolutionary history (11). Group 4 hydrogenases play an important role in conserving energy by establishing ion gradients across membranes that can be used to generate ATP. These enzymes are much more complex than the characterized dimeric hydrogenases and contain at least six subunits. The simplest members include the seven-subunit hydrogenase 3 from Escherichia coli, which oxidizes formate and evolves H2 (12, 13), the eight-subunit CO-induced hydrogenase of some CO-oxidizing bacteria that conserve energy from coupling the oxidation of CO to H2 production (10, 14), and the six-subunit “energy-conserving” hydrogenase from the archaeon Methanosarcina barkeri, which functions in methanogenesis (15, 16). More complex members of the group 4 enzymes include the 18-subunit formate hydrogen lyase (FHL)3 system from Thermococcus onnurineus (17), which oxidizes formate and evolves hydrogen (80 °C). Six subunits conserved within the group 4 hydrogenases are homologous to six subunits found in the catalytic core of the ubiquitous aerobic respiratory complex NADH quinone oxidoreductase or complex I (NuoBCDIHL (9–11, 18)). This conserved six-subunit homology suggests a close evolutionary history between group 4 enzymes and complex I and shows the importance of the hydrogenases in respiratory processes. However, because of the inherent difficulty of purifying and characterizing large, subunit membrane complexes, little is known about their structure and function (16, 19, 20).
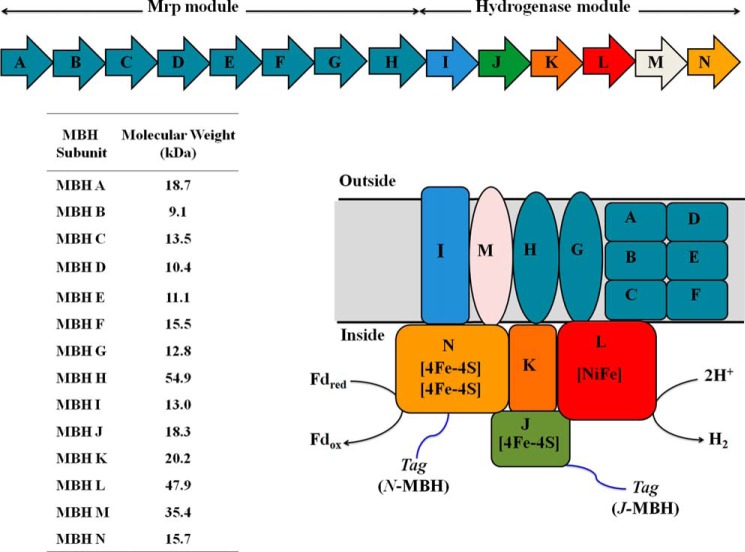
Hyperthermophilic archaea such as Pyrococcus furiosus, which grows optimally at 100 °C (21), contain a complex hydrogenase system (11). P. furiosus grows by fermenting sugars to acetate, CO2, and H2 and its membrane-bound hydrogenase (MBH) catalyzes H2 production using reduced ferredoxin generated from sugar oxidation, as the electron donor (11). Previous studies of P. furiosus MBH showed that it is encoded by a 14-gene operon (mbhA–N: PF1423–PF1436; see Fig. 1; Ref. 22). Six of the last seven genes in the operon are homologous to those encoding the “core” subunits of complex I (mbhH and J–N; see Fig. 1), whereas the other eight subunits (mbhA–H) are homologous to subunits of the Mrp monovalent cation/proton antiporter of some mesophilic bacteria (23). MbhI does not have homology to either complex I subunits or the Mrp subunits, but MbhH has homology to both. Mrp catalyzes the efflux of monovalent cations, such as Na+, K+, and Li+, outward in a coupled reaction that transports protons inwards. Of the 14 subunits of MBH, only mbhJKLN are predicted not to encode transmembrane helices (22, 24). MbhJ and MbhN are proposed to contain one and two [4Fe-4S] clusters, respectively, where MbhJ is the equivalent of the small subunit of the group 1 dimeric [NiFe]-hydrogenases (see Fig. 1). MbhKL are the equivalent to the large subunit and contain the [NiFe] active site, with the four Cys residues provided by MbhL.
FIGURE 1.
Upper panel, operon encoding P. furiosus MBH. The Mrp module is encoded by MbhA–H, and the hydrogenase module is encoded by MbhI–N. Lower panel, schematic representation of P. furiosus MBH. The enzyme is predicted to contain three [4Fe-4S] clusters in addition to the [NiFe]-catalytic site. The His9 affinity tag was attached to either MbhJ or MbhN as indicated. Fdred and Fdox represent the reduced and oxidized forms of P. furiosus ferredoxin. The predicted size (kDa) of each MBH subunit is shown in the table.
The respiratory function of MBH was demonstrated by adding reduced ferredoxin to inverted membrane vesicles of P. furiosus, whereby H2, ATP, and an electrochemical gradient were formed (25). Inhibition studies established that MBH evolved H2, whereas a membrane-bound ATP synthase produced ATP. The much lower reduction potential of ferredoxin (−454 mV (26)) compared with NADH (−320 mV) makes ferredoxin a more thermodynamically favorable electron donor for H2 production and allows energy to be conserved by a respiratory mechanism (25). Because P. furiosus ATP synthase uses Na+ ions rather than protons (27) and the MBH complex encodes a Na+/H+ antiporter (Mrp), it is thought that the hydrogenase module of MBH evolves H2 and generates a proton gradient, whereas the Mrp module transforms it into a Na+ gradient that in turn drives ATP synthesis via ATP synthase (11). Interestingly, MBH in isolated P. furiosus membranes is almost exclusively unidirectional in favor of H2 production in standard in vitro assays (24). This is remarkable because other [NiFe]-hydrogenases preferentially catalyze H2 oxidation, usually by orders of magnitude (2).
This work defines biochemical and structural information on the solubilized and intact 14-subunit MBH complex of P. furiosus. We employed the recent development of genetic tools in this organism (28) that have enabled, for example, its cytoplasmic [NiFe]-hydrogenase to be affinity-labeled, overexpressed, and purified by affinity chromatography (29, 30). Herein we demonstrate that the same strategy can be applied to the MBH complex and show that the purification efficiency depends on the location of the affinity tag on the complex. This is the first report of the affinity purification and characterization of an entire respiratory system from an archaeon as a single intact complex.
EXPERIMENTAL PROCEDURES
Generation of P. furiosus Strains Expressing Affinity-tagged MBH
A competent strain of P. furiosus (COM1) was used to manipulate the MBH operon (28). A one-step marked knock-in genetic protocol was used in which a polyhistidine (His9) affinity tag was inserted at the C terminus of the last gene in the operon (mbhN, PF1436) yielding strain MW0403 (see Fig. 2A) or within the operon at the N terminus of mbhJ (PF1432) yielding strain MW0414 (see Fig. 2B). The knock-in cassette, which contains the selectable marker and the strong constitutive promoter of the gene encoding the S-layer protein (Pslp) with an in frame His9 tag, was generated by using overlapping PCR (31) and Prime Star HS polymerase premix (Clontech) was used to make the knock-in cassette. PyrF was the selectable marker and was placed under the control of the glutamate dehydrogenase promoter (Pgdh). In strain MW0403, expression of the MBH operon was controlled by the native mbh promoter, whereas in strain MW0414, the expression of mbhJ and the subsequent five genes (mbhK-N) was under control of Pslp. Generation of MW0414 required the marker cassette to be placed in front of mbhJ within the MBH operon (see Fig. 2B). Expression of MBH, where mbhN was tagged, was under control of the native mbh promoter, and the tag was placed at the C terminus of the operon (see Fig. 2A). All transformants were PCR screened for correct insertion, and the PCR product was sequenced (Macrogen, MD).
FIGURE 2.
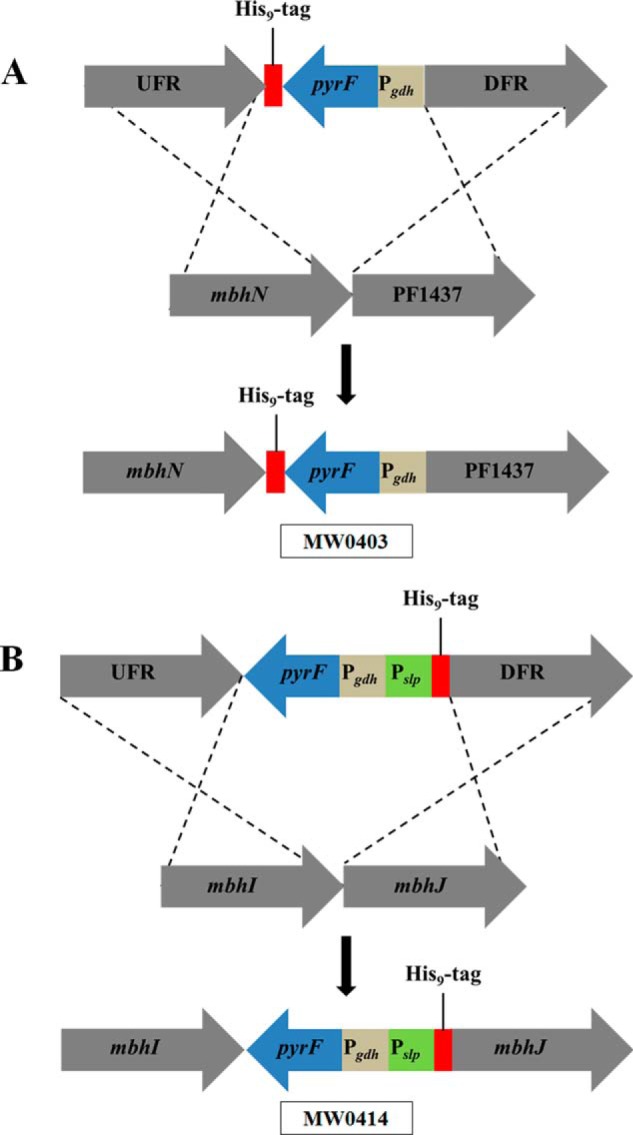
The genetic strategy used to insert the His9 tag into MBH. A, the tag is inserted at the C terminus of mbhN (yielding P. furiosus strain MW0403) where PF1437 is the gene immediately downstream of the MBH operon. B, the tag is inserted at the N terminus of mbhJ (yielding P. furiosus strain MW0414). UFR, upstream flanking region (1 kb) of the MBH operon; DFR, downstream flanking region (1 kb) of the MBH operon; pyrF, selectable marker; Pgdh and Pslp, promoters for the gene encoding glutamate dehydrogenase and the S-layer protein of P. furiosus, respectively. The affinity tag was a His9 tag.
Membrane Preparation
Cells were lysed using 50 mm EPPS (4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid; Sigma-Aldrich), pH 8.0, containing 50 μg/ml DNase I (Sigma-Aldrich), and 2 mm DTT (Inalco, Milano, Italy) in a 5:1 ratio of buffer to cells in an anaerobic chamber (Coy, Grass Lake, MI). After 2 h of incubation at 23 °C, cells were passed twice through a French press at a pressure of 1000 p.s.i. The cell lysate was then centrifuged in a Beckman-Coulter Optima L-90K ultracentrifuge at 100,000 × g for 1 h. The supernatant was removed, and the membrane pellet was resuspended in wash buffer (50 mm EPPS, pH 8.0, containing 5 mm MgCl2, 50 mm NaCl, 10% (v/v) glycerol (all obtained from J. T. Baker), 2 mm DTT, and 0.1 mm PMSF. The membrane pellet was homogenized using 15-ml Pyrex tissue grinders (Pyrex), and the pellet was collected by ultracentrifugation. The washing procedure was repeated twice more, and the pellet was resuspended in 50 mm Tris-HCl (Sigma), pH 8.0, 5 mm MgCl2, 50 mm NaCl, 5% (v/v) glycerol, 2 mm DTT, and 0.1 mm PMSF (resuspension buffer).
Membrane Solubilization by Different Detergents
The detergents tested to solubilize MBH were Cymol, Fos-Choline, sodium deoxycholate, N-dodecyl-β-d-maltoside (Affymetrix), Triton X-114, Triton X-100 (Bio-Rad). For Cymol, Fos-Choline, Triton X-114, Triton X-100, and N-dodecyl-β-d-maltoside, the detergent was added to a final concentration of 2% (w/v) to washed membranes (6 mg of detergent/mg of membrane protein), and the suspension was incubated for 16 h at 40 °C. The same procedure was used for sodium deoxycholate except that the incubation time was 2 h. Concentrations of 5, 10, and 20% (w/v) Triton X-100 were used, and incubations were carried out at both 4 and 40 °C.
Affinity Purification of MBH
All purification steps were carried out anaerobically using a Coy anaerobic chamber. Triton X-100 (10%, w/v) was incubated with washed membranes (30 mg of detergent/mg of membrane protein) for 16 h at 4 °C. The sample was centrifuged in a Beckman-Coulter Optima L-90K ultracentrifuge at 100,000 × g for 1 h. The supernatant was collected, diluted to twice the volume with buffer A (50 mm Tris-HCl, pH 8.0, containing 400 mm NaCl, 0.1% (w/v) Triton X-100, and 4 mm DTT) and applied to a 5-ml FF His-Trap Ni-NTA column (GE Healthcare). The column was washed with buffer A, and the bound protein was eluted with a 20-column volume gradient from 100% buffer A to 100% buffer B (buffer A containing 500 mm imidazole).
Superose 6 Analysis for Determination of MBH Molecular Weight
The molecular weight of the purified MBH sample was analyzed using a calibrated 24-ml Superose 6 30/100 column (GE Healthcare) equilibrated in buffer C (50 mm Tris-HCl, pH 8.0, containing 300 mm NaCl, 0.02% (w/v) Triton X-100 and 2 mm sodium dithionite). MBH purified from the Superose 6 column was used for the SDS-PAGE gel analysis.
SAXS Data
Small angle x-ray scattering was collected at the SIBYLS beamline at the Advanced Light Source (Berkeley, CA) as described (32, 33). The sample was buffer exchanged using an Amicon Ultra centrifugal concentrator (Millipore) in buffer containing 50 mm Tris, pH 8.0, 400 mm NaCl, 0.02% (w/v) Triton X-100, and 4 mm DTT. Thus, Triton X concentration was below the critical micelle concentration. A sample was collected both before and after buffer equilibration. Subtraction of either buffer sample yielded identical results to within experimental error (∼1% of signal). For anaerobic data collection, samples were handled in a positive pressure helium box (containing less than 0.01% O2). MBH was prepared at 5.0, 3.3, and 1.6 mg/ml. The resulting samples were exposed for 0.5, 0.5, 2, and 4 s for data collection. Minor concentration dependence was observed and corrected for by extrapolating to zero concentration. The MBH samples were placed 1.5 m from a MAR165 CCD detector arranged coaxial with the 12 keV monochromatic beam; 1012 photons/second were impingent on the sample. The spot size at the sample was 4 × 1 mm convergent to a 100 μm spot at the detector. Buffer subtraction and raw image data were integrated by beamline software specific for this arrangement (33). Scattering data were plotted on log of x-ray intensity scale versus momentum transfer (q) in inverse Å, where q = (4π sin(θ/2))/λ, θ is the scattering angle relative to the incident beam, and λ is the wavelength. Processing of SAXS data was conducted utilizing the Scatter package. GNOM was utilized to extract the P(r) function (34).
The scattering profile used to calculate the pair distribution function and subsequently in GASBOR (35) was a merged profile combining all four exposures from all concentrations. The two 0.5-s exposures for each concentration were referenced against one another to check for radiation damage. Because none was observed, the two exposures were averaged. The longer exposures were used to reduce noise in the high q region. Once each individual concentration had been merged, the three concentrations were used to apply a concentration-dependent correction. Ten GASBOR calculations generated 10 models from the scattering curve, which were averaged and filtered together using the GASBOR-associated package DAMAVER. The DAMAVER reported normalized spatial discrepancy, which measures agreement between models, was 2.1. The individual models are shown in the supplemental materials, and all support an elongated and asymmetric molecule (supplemental Table S1 and supplemental Figs. S1–S3).
Other Methods
Hydrogenase assays were performed at 80 °C, and H2 was measured using an Agilent Technologies 6850 gas chromatograph. H2 evolution activity was determined using dithionite-reduced methyl viologen (Sigma-Aldrich) as the electron donor (29) or P. furiosus ferredoxin reduced by P. furiosus pyruvate ferredoxin oxidoreductase. The pyruvate ferredoxin oxidoreductase-linked assay contained 100 mm EPPS, pH 8.4, 10 mm sodium pyruvate, 0.2 mm coenzyme A, 0.4 mm TPP, 2 mm MgCl2, 2 mm DTT, pyruvate ferredoxin oxidoreductase (30 μg/ml), and ferredoxin (100 μg/ml). Methyl viologen was used as the electron acceptor in the hydrogen oxidation assays. Oxygen sensitivity assays were carried out by exposing the MBH sample (100 μg/ml) in buffer containing 50 mm Tris, pH 8.0, 400 mm NaCl, and 4 mm DTT to air while shaking (30 rpm), and samples were taken at 0, 2, 4, 8, 16, and 32 h to determine residual hydrogenase activity. Thermal stability at 90 °C was carried out in the same fashion except that the samples were maintained under anaerobic conditions.
Purified MBH complexes were analyzed by electrophoresis using 4–12% Bis-Tris NuPAGE gels (Invitrogen) and 4–20% Tris-glycine NUSEP gels (Bio-Rad). Bands were cut from the SDS-PAGE gel and were analyzed by MALDI-TOF. Purified MBH was also digested in solution with trypsin overnight and identified using two-dimensional LC-MS/MS. Nickel and iron were measured using a octopole-based ICP-MS (7500ce; Agilent Technologies, Tokyo, Japan), equipped with a MicorMist nebulizer (36). X-band (∼9.6 GHz) EPR spectroscopy was carried out using a Bruker ESP-300E EPR spectrometer equipped with an ER-4116 dual-mode cavity and an Oxford Instruments ESR-9 flow cryostat.
RESULTS
Construction of Affinity-tagged MBH
As shown in Fig. 1, the subunits encoded by the first eight genes of the MBH operon, together with mbhI and mbhM, are predicted to be membrane-bound, and therefore they were not considered as targets for affinity tagging. On the other hand, mbhJKLN are not predicted to encode transmembrane helices and are thought to be located on the cytoplasmic side of the membrane complex (22). The His9 affinity tag was inserted either at the C terminus of the subunit encoded by mbhN, which is the last gene in the operon, or at the N terminus of the subunit encoded by mbhJ. The forms of MBH that were generated were designated N-MBH (in P. furiosus strain MW0403) and J-MBH (MW0414), respectively. The His9 tag was chosen because it has been successfully used to purify a form of the cytoplasmic [NiFe]-hydrogenase from P. furiosus (30), suggesting that the tag is unlikely to interfere with synthesis of the [NiFe]-catalytic site of MBH. The recombinant strains generated in this study are summarized in Table 1, and the strategy for constructing N-MBH and J-MBH is shown in Fig. 2.
TABLE 1.
Strains used in this work
COM1 was the competent parent strain. MW0414 and MW0403 were generated by placing the marker cassette at the N terminus of mbhJ and at the C terminus of the operon at mbhN, respectively.
| Strain designation | Genotype | Deleted or inserted ORF/elements | Parent strain | Sources |
|---|---|---|---|---|
| COM1 | ΔpyrF | PF1114 | DSM 3638 | Ref. 30 |
| MW0414 | ΔpyrF::Pgdh pyrF Pslp His9 PF1432 | Pslp His9 inserted in front of PF1432 (mbhJ) | COM1 | This study |
| MW0403 | ΔpyrF::PF1436 His9 Pgdh pyrF | His9 inserted behind PF1436 (mbhN) | COM1 | This study |
Affinity Purification of MBH
Previous attempts to solubilize MBH from the membrane were carried out using the detergents dodecyl-β-d-maltoside and sodium deoxycholate (22, 24). However, dodecyl-β-d-maltoside was not pursued because at higher concentrations it is not compatible with the Ni-NTA affinity purification step and was not successful previously in purifying the intact complex. Sodium deoxycholate (2%, w/v) efficiently solubilized MBH, but incubation for more than 2 h led to deactivation of the enzyme (24). Cymol, Fos-Choline, and Triton X-114 were not as efficient as Triton X-100 at solubilizing MBH as measured by recovery of hydrogenase activity (H2 production from reduced methyl viologen) using 2% (w/v) final detergent concentration (data not shown). Because Triton X-100 is compatible with the Ni-NTA affinity step and was used to successfully purify the ATP synthase from P. furiosus (37), it was investigated over the concentration range of 5–20% (w/v) at both 4 and 40 °C. From this analysis, it was determined that 10% (w/v) Triton X-100 at 4 °C (using 3 mg/ml protein) gave ≥90% recovery of the hydrogenase activity in a solubilized form of both N-MBH and J-MBH. The affinity purification step was carried out under anaerobic conditions, but the placement of the His9 tag clearly affected the efficiency of purification. The J-MBH form yielded ∼3 mg of MBH/50 g of cells (wet weight) with a yield of activity of 27% (Table 2). In contrast, the yield with N-MBH was only 2% with ≤1 mg of protein (data not shown). Hence, placing the affinity tag on the N terminus of the “small” subunit (MbhJ) of this MBH is much more efficient in terms of purification than tagging MbhN. J-MBH was therefore utilized for all of the characterization studies described below, and it will be referred to as purified or solubilized MBH.
TABLE 2.
Purification of J-MBH
| Step | Total units | Total protein | Specific activity | Yield | Fold purification |
|---|---|---|---|---|---|
| μmol min−1 | mg | % | |||
| Wash membrane | 184 | 172 | 1.1 | 100 | 1.0 |
| Triton X-100 extraction | 148 | 147 | 1 | 80 | 0.9 |
| Ni-NTA | 50 | 2.6 | 19 | 27 | 17 |
Characterization of Purified MBH
The purified MBH obtained from affinity purification was analyzed by SDS-PAGE (Fig. 3). Protein bands consistent with the calculated molecular weights from deduced amino acids sequences were observed for all 14 subunits. Note that the proposed catalytic subunit, MbhL (calculated molecular weight of 47,903 Da), undergoes C-terminal proteolysis during the processing of the [NiFe] site, where 47 amino acids are removed, and the calculated size of the mature subunit is 42,899 Da. Thirteen of fourteen MBH subunits were identified both by cutting protein bands from the SDS gel and by using in-solution trypsin digestion followed by LC-MS/MS. MbhJKL was identified by MALDI-TOF analysis from bands cut from the SDS-PAGE gel. The rest of the subunits (including MbhJKL) were identified by LC-MS/MS analysis of in-solution digested samples (supplemental Table S2). MbhD was the only subunit not identified by the LC-MS/MS or MALDI-TOF analyses. This is a small hydrophobic protein (10,412 Da) that is likely resistant to trypsin digestion. A contaminant protein band was observed at approximately the 47-kDa position of the gel and was identified as elongation factor 1α (PF1375; Fig. 3).
FIGURE 3.
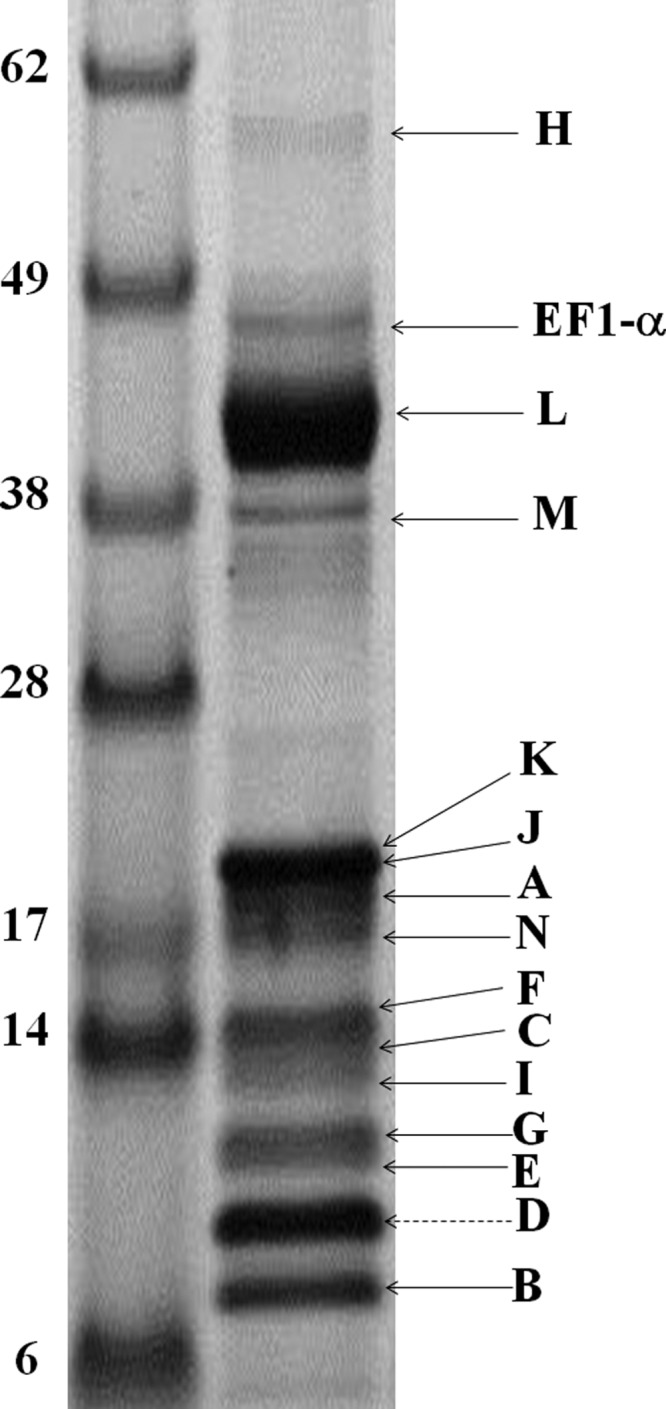
SDS-PAGE of purified J-MBH. MBH subunits were analyzed from bands cut from gel and from samples that were digested in-solution. MBH subunits identified by either MALDI-TOF or LC-MS/MS are labeled with a black arrow. MbhD is the only subunit not identified, and the protein band corresponding to its calculated molecular weight is shown by a dashed arrow.
Purified MBH was also analyzed using a calibrated Superose 6 gel filtration column and gave rise to a single protein peak corresponding to a mass of 310 ± 11 kDa (supplemental Fig. S4). The predicted mass of the entire MBH complex calculated from the deduced amino acid sequences is 298 kDa. These results indicate that the entire complex containing a single copy of each of the 14 subunits has been solubilized and purified.
Purified MBH contained both iron and nickel by analysis using ICP-MS in a ratio of 15.4 ± 0.5:1. This result supports the proposed presence of three [4Fe-4S] clusters in the enzyme (Fig. 1), which together with the [NiFe] active site gives a predicted ratio of 13:1. The protein purified under anaerobic conditions (in the presence of DTT) exhibited a rhombic EPR signal at 50,000 with g values of 2.39, 2.17, and 2.05 (supplemental Fig. S5) indicative of the nickel-boron “ready” state (6). No additional EPR resonances were observed that might be attributable to the iron-sulfur clusters after reduction of the as-purified enzyme with sodium dithionite, and the thionine-oxidized enzyme was also EPR-silent.
MBH uses reduced ferredoxin as an electron donor in vivo, and as shown in Fig. 1, the redox protein is proposed to interact with MbhN. In a previous attempt to characterize MBH, its solubilization led to the loss of the ability to use ferredoxin as an electron donor (22). As shown in Table 3, the solubilized enzyme evolved H2 from reduced ferredoxin, which was reduced using the native pyruvate ferredoxin oxidoreductase system with specific activities of 0.14 and 0.02 unit/mg for the purified and membrane-bound enzymes, respectively, in accordance with increase in purity of the MBH complex. These results suggest that the [Fe-S] clusters and [NiFe] site are intact and functional. The enzyme also retained its catalytic preference upon solubilization. The ratio H2 evolution to H2 oxidation was similar (25:1) for both the membrane-bound and the solubilized forms. The two forms of MBH were similarly insensitive to inactivation by oxygen, with half-lives under air while shaking for ∼14 h. On the other hand, as might be expected, the solubilized enzyme was much less thermostable, with a half-life at 90 °C of 1 h, compared with 25 h for the membrane-bound form.
TABLE 3.
Properties of J-MBH
| Property | J-MBH | Washed membranes |
|---|---|---|
| Pyruvate ferredoxin oxidoreductase-ferredoxin H2 evolution activity (units/mg) | 0.14 | 0.02 |
| H2 evolution:H2 oxidation activity (using methyl viologen as electron carrier) | 25:1 | 26:1 |
| Half-life (t½) at 90 °C under argon (h) | 1 | 25 |
| Half-life (t½) at 25 °C under air (h) | 15 | 13 |
| Metal content (iron:nickel) | 15:1 | 19:1 |
SAXS Data Analysis
The quaternary structure of purified MBH was analyzed by SAXS. Analysis of the SAXS profiles as a function of MBH concentration showed only a minor concentration dependence where the lower concentration (1.5 mg/ml) had a higher radius of gyration than the higher concentration sample (5 mg/ml) providing greater confidence that the sample was not aggregating (Fig. 4A). Also shown in dashed lines is the profile for the six core subunits of complex I (Protein Data Bank code 4HEA). From this, we can further see the similarities between MBH and complex I, because the six core complex I subunits and the MBH calculated profiles agree in much of the low q region, which suggests that there are similar features and dimensions between the two proteins (Fig. 4A). Guineir analysis of the low q (momentum transfer in Å−1) region of the SAXS profile extrapolated to zero concentration yielded a radius of gyration of 63 ± 2 Å. From the radius of gyration and volume of correlation, the mass of the purified MBH was calculated from the SAXS profile (38) to be 310 kDa, which is in good agreement with the predicted value of 298 kDa and the experimental value from the SEC column (310 kDa). In Fourier transform, the real space P(r) function provided an estimated maximum dimension of 288 Å and also suggests a globular shape of the purified MBH (Fig. 4B). The scattering profile decayed as q4 (the value expected for a folded rigid protein (39)). Using 10 GASBOR (34) runs, an average shape was calculated without symmetry (Fig. 5). With a calculated mass in agreement with the mass of the monomer to within error, the Z-shaped structure is expected to characterize the mono-dispersed homogenous species. A comparison with subunits of the atomic resolution structures of NADH quinone oxidoreductase or complex I (Protein Data Bank code 4HEA) is shown within the MBH SAXS model (Fig. 5). The complex I homologous subunits of MBH are drawn in ribbon. These include four cytoplasmic subunits homologous to MbhJKLN (NuoBCDI) and one membrane-bound subunit (MbhM/NuoH). The complex I homolog of MbhH (NuoL) is not continuous with the other homologs within the MBH SAXS model and is shown (green ribbon) with an intervening nonhomologous section of complex I (red space filling). We can distinguish where in the SAXS model the membrane-bound subunits (including the Mrp subunits) and soluble subunits (including the catalytic subunit) are located (Fig. 5). The overall SAXS analysis reveals as a folded assembled complex suitable for the pursuit of other forms of structural analyses for MBH including crystal structures.
FIGURE 4.
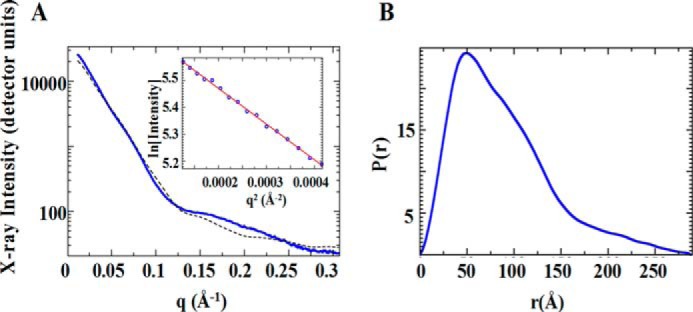
SAXS analysis of purified MBH. A, experimental SAXS profile of MBH (blue) with the Guineir region plotted on the inset with a linear fit (red). The calculated profile from a portion of complex I (NADH quinone oxidoreductase, Protein Data Bank code 4HEA; dashed black line) is shown for comparison. B, the real space pair distribution file extracted from SAXS data.
FIGURE 5.
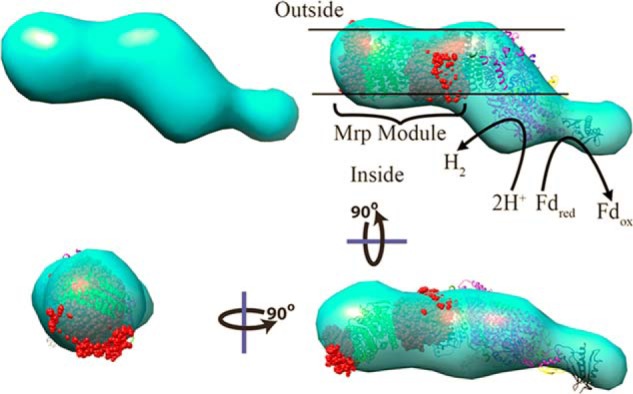
Shape and assembly of MBH from SAXS. Shapes generated from SAXS are elongated and multilobed. The homologous portions of complex I (NADH quinone oxidoreductase, Protein Data Bank code 4HEA) shown as ribbons were augmented by intervening and nonhomologous portions (red spheres) to create an atomic model of equivalent size to MBH. This model was fit into the SAXS generated shape, shown rotated in three orientations. By analogy with the homologous subunits of complex I, assignments were able to the portion of the SAXS model that would likely be associated with the membrane and to the portion that would be responsible for oxidizing reduced ferredoxin.
DISCUSSION
We describe here the first successful affinity purification of a membrane-bound energy-conserving group 4 hydrogenase with an engineered tag. There is only one previous report of affinity purification of any membrane-bound hydrogenase, and this is for the group 1 dimeric enzyme from the mesophilic bacterium Rhizobium japonicum, although in that case affinity purification took advantage of NAD as a substrate and used a reactive red 120-agarose column (40). There is only one previous example of affinity purification of an entire respiratory complex: this is the six-subunit membrane-bound NADH quinone oxidoreductase from the mesophilic bacterium Vibrio cholera, which compares with the 14-subunit MBH. This also used an engineered His9-tagged protein that was solubilized with the detergent dodecyl maltoside (DM) (41). Herein we show that the location of the affinity tag is critical, with a much higher recovery of activity with the tag located on MbhJ compared with MbhN. The former tag location is probably more accessible to the Ni-NTA column and results in the improved purification. The high recovery of enzymatic activity with J-MBH after both solubilization and the affinity step is also noteworthy. For example, the purification of the P. furiosus ATP synthase yielded 2% of the initial activity with ≤1 mg from 50 g of cells (37). This compares with ∼3 mg with a yield of activity of 27% for P. furiosus MBH (Table 2).
Solubilized P. furiosus MBH was purified as an intact complex that evidently contains one copy of each of the 14 subunits encoded by the MBH operon (Fig. 1) with calculated and measured masses of 298 and 310 kDa, respectively. The predicted and measured contents of nickel and iron (13:1 and 15:1, respectively) are also in good agreement with a completely intact complex. Earlier attempts to purify MBH using standard chromatographic techniques resulted in partial purification of complexes containing predominantly MbhLK or MbhJ–N (22, 24). Like the intact MBH complex purified herein, these subcomplexes were catalytically active, and the as-purified enzyme exhibited a rhombic EPR signal with g values very similar to what was observed here with intact MBH (22). However, in that case the signal was assigned to the nickel-carbon state, whereas from the g values it is the same nickel-boron signal that was observed here (6). Interestingly, in the prior study (22), an EPR signal indicative of reduced iron-sulfur clusters signal was observed, but that was not the case here with the complete complex, suggesting perhaps a change in conformation of the cluster-containing subunits when detached from intact MBH. The oxygen tolerance of purified MBH was similar to that in the membrane-bound form, indicating that the infrastructure around metal centers is maintained after detergent extraction. Even though purified MBH was much less thermostable than the membrane-bound enzyme, it was still a very stable complex (half-life at 90 °C of 1 h), illustrating an advantage of solubilizing membrane proteins from hyperthermophilic species.
MBH and other group 4 hydrogenases are proposed to have an evolutionary history in common with the aerobic respiratory complex I (NADH quinone oxidoreductase). This 16-subunit 536-kDa enzyme is the largest complex of the aerobic electron transport chain and couples NADH oxidation to ubiquinone reduction and proton translocation (42, 43). The proton gradient is utilized by ATP synthase to generate ATP. Group 4 hydrogenases have a similar function in generating a proton gradient, whereas P. furiosus MBH has an additional Mrp modules that serves to generate a sodium ion gradient (11). There are six conserved subunits found in the group 4 hydrogenases that are also found in complex I. These subunits comprise the complex I core, and the subunit that binds quinone is the homolog of the [NiFe]-containing catalytic subunit of the group 4 hydrogenases (9, 10). This homology with complex I allows placement of five of these six core subunits from the crystal structure of complex I from Thermus thermophilus (Protein Data Bank code 4HEA) within the SAXS model as cytoplasmic subunits, including the catalytic subunit of MBH. This fit also allows us to postulate where to place the membrane wall in the model, as shown in Fig. 5. Because complex I does not contain any homolog of the Mrp subunits, we would not expect the core subunits to completely fit within the MBH SAXS model. Overall, generation of the SAXS model of MBH shows that our purified protein is sufficiently homogenous for low resolution structural characterization.
P. furiosus MBH represents a large family of membrane-bound respiratory complexes that include those that oxidize formate and carbon monoxide (11). In addition to the Mrp and hydrogenase modules, these enzymes contain additional subunits that oxidize the C-1 substrates. As yet, none of these complexes have been solubilized and purified other than MBH. However, P. furiosus was recently used to heterologously express the membrane-bound 18-subunit FHL of T. onnurineus to give a functional complex that oxidizes formate and evolves H2 (17, 44). FHL is also encoded by a single operon, and in addition to two subunits that show sequence similarity to formate dehydrogenases, the FHL complex contains four homologs of the mbhH subunit of P. furiosus MBH for reasons that are not at all clear. Otherwise, the subunits of the hydrogenase module of FHL are very similar to those of P. furiosus MBH, suggesting that a similar tagging and purification strategy (with the His9 tag attached to the fhlN subunit) might be successful for the intact FHL complex, and this is currently being investigated.
In conclusion, we have devised a one-step affinity purification protocol for the respiratory MBH of P. furiosus that gives a high yield of the entire catalytically active membrane-bound complex. The group 4 hydrogenases show little sequence similarity to the structurally characterized group 1 two-subunit enzymes, except for the residues that bind the [NiFe] active site and the [4Fe-4S] proximal cluster (9, 10). Further characterization of MBH may therefore provide insight into the diversity of hydrogenase structure and function, such as providing a strategy for the characterization of closely related but even more complex membrane-bound respiratory enzymes, as well as providing evolutionary insights into the ubiquitous complex I of aerobic organisms, both prokaryotic and eukaryotic.
Supplementary Material
Acknowledgments
We thank the Mass Spectrometry Facility at John Hopkins University for MS/MS analyses and the Biofermentation and Expression Facility at the University of Georgia for growing P. furiosus.
This work was supported, in whole or in part, by National Institutes of Health Grant GM105404 (to J. A. T.). This work was also supported by Grant DE-FG05-95ER20175 (to M. W. W. A.) from the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the Department of Energy. SAXS analyses were supported by Department of Energy project Integrated Diffraction Analysis Technologies.

This article contains supplemental Tables S1 and S2 and Figs. S1–S5.
- FHL
- formate hydrogen lyase
- MBH
- membrane-bound hydrogenase
- SAXS
- small angle x-ray scattering
- EPPS
- (4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Lee H. S., Vermaas W. F., Rittmann B. E. (2010) Biological hydrogen production: prospects and challenges. Trends Biotechnol. 28, 262–271 [DOI] [PubMed] [Google Scholar]
- 2. Vignais P. M., Billoud B. (2007) Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107, 4206–4272 [DOI] [PubMed] [Google Scholar]
- 3. Fontecilla-Camps J. C. (2009) Structure and function of [NiFe]-hydrogenases. Met. Ions Life Sci. 6, 151–178 [DOI] [PubMed] [Google Scholar]
- 4. Friedrich B., Fritsch J., Lenz O. (2011) Oxygen-tolerant hydrogenases in hydrogen-based technologies. Curr. Opin. Biotechnol. 22, 358–364 [DOI] [PubMed] [Google Scholar]
- 5. Shafaat H. S., Rüdiger O., Ogata H., Lubitz W. (2013) [NiFe] hydrogenases: a common active site for hydrogen metabolism under diverse conditions. Biochim. Biophys. Acta 1827, 986–1002 [DOI] [PubMed] [Google Scholar]
- 6. Foerster S., Stein M., Brecht M., Ogata H., Higuchi Y., Lubitz W. (2003) Single crystal EPR studies of the reduced active site of [NiFe] hydrogenase from Desulfovibrio vulgaris Miyazaki F. J. Am. Chem. Soc. 125, 83–93 [DOI] [PubMed] [Google Scholar]
- 7. Volbeda A., Charon M. H., Piras C., Hatchikian E. C., Frey M., Fontecilla-Camps J. C. (1995) Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373, 580–587 [DOI] [PubMed] [Google Scholar]
- 8. Volbeda A., Darnault C., Parkin A., Sargent F., Armstrong F. A., Fontecilla-Camps J. C. (2013) Crystal structure of the O2-tolerant membrane-bound hydrogenase 1 from Escherichia coli in complex with its cognate cytochrome b. Structure 21, 184–190 [DOI] [PubMed] [Google Scholar]
- 9. Hedderich R. (2004) Energy-converting [NiFe] hydrogenases from archaea and extremophiles: ancestors of complex I. J. Bioenerg. Biomembr. 36, 65–75 [DOI] [PubMed] [Google Scholar]
- 10. Hedderich R., Forzi L. (2005) Energy-converting [NiFe] hydrogenases: more than just H2 activation. J. Mol. Microbiol. Biotechnol. 10, 92–104 [DOI] [PubMed] [Google Scholar]
- 11. Schut G. J., Boyd E. S., Peters J. W., Adams M. W. (2013) The modular respiratory complexes involved in hydrogen and sulfur metabolism by heterotrophic hyperthermophilic archaea and their evolutionary implications. FEMS Microbiol. Rev. 37, 182–203 [DOI] [PubMed] [Google Scholar]
- 12. Böhm R., Sauter M., Böck A. (1990) Nucleotide sequence and expression of an operon in Escherichia coli coding for formate hydrogenlyase components. Mol. Microbiol. 4, 231–243 [DOI] [PubMed] [Google Scholar]
- 13. Sauter M., Böhm R., Böck A. (1992) Mutational analysis of the operon (hyc) determining hydrogenase 3 formation in Escherichia coli. Mol. Microbiol. 6, 1523–1532 [DOI] [PubMed] [Google Scholar]
- 14. Singer S. W., Hirst M. B., Ludden P. W. (2006) CO-dependent H2 evolution by Rhodospirillum rubrum: role of CODH:CooF complex. Biochim. Biophys. Acta 1757, 1582–1591 [DOI] [PubMed] [Google Scholar]
- 15. Kurkin S., Meuer J., Koch J., Hedderich R., Albracht S. P. (2002) The membrane-bound [NiFe]-hydrogenase (Ech) from Methanosarcina barkeri: unusual properties of the iron-sulphur clusters. Eur. J. Biochem. 269, 6101–6111 [DOI] [PubMed] [Google Scholar]
- 16. Meuer J., Bartoschek S., Koch J., Künkel A., Hedderich R. (1999) Purification and catalytic properties of Ech hydrogenase from Methanosarcina barkeri. Eur. J. Biochem. 265, 325–335 [DOI] [PubMed] [Google Scholar]
- 17. Kim M. S., Bae S. S., Kim Y. J., Kim T. W., Lim J. K., Lee S. H., Choi A. R., Jeon J. H., Lee J. H., Lee H. S., Kang S. G. (2013) CO-dependent H2 production by genetically engineered Thermococcus onnurineus NA1. Appl. Environ. Microbiol. 79, 2048–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Marreiros B. C., Batista A. P., Duarte A. M., Pereira M. M. (2013) A missing link between complex I and group 4 membrane-bound [NiFe] hydrogenases. Biochim. Biophys. Acta 1827, 198–209 [DOI] [PubMed] [Google Scholar]
- 19. Soboh B, Linder D., Hedderich R. (2002) Purification and catalytic properties of a CO-oxidizing:H2-evolving enzyme complex from Carboxydothermus hydrogenoformans. Eur. J. Biochem. 269, 5712–5721 [DOI] [PubMed] [Google Scholar]
- 20. Soboh B, Linder D., Hedderich R. (2004) A multisubunit membrane-bound [NiFe] hydrogenase and an NADH-dependent Fe-only hydrogenase in the fermenting bacterium Thermoanaerobacter tengcongensis. Microbiology 150, 2451–2463 [DOI] [PubMed] [Google Scholar]
- 21. Fiala G., Stetter K. (1986) Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100°C. Arch. Microbiol. 145, 56–61 [Google Scholar]
- 22. Sapra R., Verhagen M. F., Adams M. W. (2000) Purification and characterization of a membrane-bound hydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 182, 3423–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Swartz T. H., Ikewada S., Ishikawa O., Ito M., Krulwich T. A. (2005) The Mrp system: a giant among monovalent cation/proton antiporters? Extremophiles 9, 345–354 [DOI] [PubMed] [Google Scholar]
- 24. Silva P. J., van den Ban E. C., Wassink H., Haaker H., de Castro B., Robb F. T, Hagen W. R. (2000) Enzymes of hydrogen metabolism in Pyrococcus furiosus. Eur. J. Biochem. 267, 6541–6551 [DOI] [PubMed] [Google Scholar]
- 25. Sapra R., Bagramyan K., Adams M. W. (2003) A simple energy-conserving system: proton reduction coupled to proton translocation. Proc. Natl. Acad. Sci. U.S.A. 100, 7545–7550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Brereton P. S., Verhagen M. F., Zhou Z. H., Adams M. W. (1998) Effect of iron-sulfur cluster environment in modulating the thermodynamic properties and biological function of ferredoxin from Pyrococcus furiosus. Biochemistry 37, 7351–7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pisa K. Y., Huber H., Thomm M., Müller V. (2007) A sodium ion-dependent A1-AO ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus. FEBS J. 274, 3928–3938 [DOI] [PubMed] [Google Scholar]
- 28. Lipscomb G. L., Stirrett K., Schut G. J., Yang F., Jenney F. E., Jr., Scott R. A., Adams M. W., Westpheling J. (2011) Natural competence in the hyperthermophilic archaeon Pyrococcus furiosus facilitates genetic manipulation: construction of markerless deletions of genes encoding the two cytoplasmic hydrogenases. Appl. Environ. Microbiol. 77, 2232–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chandrayan S. K., McTernan P. M., Hopkins R. C., Sun J., Jenney F. E., Jr., Adams M. W. (2012) Engineering hyperthermophilic archaeon Pyrococcus furiosus to overproduce its cytoplasmic [NiFe]-Hydrogenase. J. Biol. Chem. 287, 3257–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hopkins R. C., Sun J., Jenney F. E., Jr., Chandrayan S. K., McTernan P. M., Adams M. W. (2011) Homologous expression of a subcomplex of Pyrococcus furiosus hydrogenase that interacts with pyruvate ferredoxin oxidoreductase. PLoS One 6, e26569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68 [DOI] [PubMed] [Google Scholar]
- 32. Hura G. L., Menon A. L., Hammel M., Rambo R. P., Poole F. L., 2nd, Tsutakawa S. E., Jenney F. E., Jr., Classen S., Frankel K. A., Hopkins R. C., Yang S.-J., Scott J. W., Dillard B. D., Adams M. W., Tainer J. A. (2009) Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS). Nat. Methods 6, 606–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Classen S., Hura G. L., Holton J. M., Rambo R. P., Rodic I., McGuire P. J., Dyer K., Hammel M., Meigs G., Frankel K. A., Tainer J. A. (2013) Implementation and performance of SIBYLS: a dual endstation small-angle x-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J. Appl. Crystallogr. 46, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Semenyuk A. V., Svergun D. I. (1991) GNOM: a program package for small-angle scattering data-processing. J. Appl. Crystallogr. 24, 537–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Svergun D. I., Petoukhov M. V., Koch M. H. (2001) Determination of domain structure of proteins from x-ray solution scattering. Biophys. J. 80, 2946–2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cvetkovic A., Menon A. L., Thorgersen M. P., Scott J. W., Poole F. L., 2nd, Jenney F. E., Jr., Lancaster W. A., Praissman J. L., Shanmukh S., Vaccaro B. J., Trauger S. A., Kalisiak E., Apon J. V., Siuzdak G., Yannone S. M., Tainer J. A., Adams M. W. (2010) Microbial metalloproteomes are largely uncharacterized. Nature 466, 779–782 [DOI] [PubMed] [Google Scholar]
- 37. Vonck J., Pisa K. Y., Morgner N., Brutschy B., Müller V. (2009) Three-dimensional structure of A1A0 ATP synthase from the hyperthermophilic archaeon Pyrococcus furiosus by electron microscopy. J. Biol. Chem. 284, 10110–10119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rambo R. P., Tainer J. A. (2013) Accurate assessment of mass, models and resolution by small-angle scattering. Nature 496, 477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rambo R. P., Tainer J. A. (2011) Characterizing flexible and intrinsically unstructured biological macromolecules by SAS using the Porod-Debye law. Biopolymers 95, 559–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stults L. W, Moshiri F., Maier R. J. (1986) Aerobic purification of hydrogenase from Rhizobium japonicum by affinity chromatography. J. Bacteriol. 166, 795–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Barquera B., Hellwig P., Zhou W., Morgan J. E., Häse C. C., Gosink K. K., Nilges M., Bruesehoff P. J., Roth A., Lancaster C. R., Gennis R. B. (2002) Purification and characterization of the recombinant Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry 41, 3781–3789 [DOI] [PubMed] [Google Scholar]
- 42. Baradaran R., Berrisford J. M., Minhas G. S., Sazanov L. A. (2013) Crystal structure of the entire respiratory complex I. Nature 494, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Efremov R. G., Baradaran R., Sazanov L. A. (2010) The architecture of respiratory complex I. Nature 465, 441–445 [DOI] [PubMed] [Google Scholar]
- 44. Lipscomb G. L., Schut G. J., Thorgersen M. P., Nixon W. J., Kelly R. M., Adams M. W. (2014) Engineering hydrogen gas production from formate in a hyperthermophile by heterologous production of an 18-subunit membrane-bound complex. J. Biol. Chem. 289, 2873–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.