Background: Calpain 5 is a ubiquitous, non-classical calpain.
Results: Calpain 5, the second most abundant calpain in the CNS, has two NLS signals, and co-localizes with nuclear PML bodies.
Conclusion: Calpain 5 is a nuclear protease associated with PML bodies.
Significance: The nuclear localization of calpain 5 provides new clues regarding its possible functions.
Keywords: Calpain, Nuclear Transport, Protease, SUMO-interacting Motif (SIM), SUMOylation
Abstract
Calpain 5 (CAPN5) is a non-classical member of the calpain family. It lacks the EF hand motif characteristic of classical calpains but retains catalytic and Ca2+ binding domains, and it contains a unique C-terminal domain. TRA-3, an ortholog of CAPN5, has been shown to be involved in necrotic cell death in Caenorhabditis elegans. CAPN5 is expressed throughout the CNS, but its expression relative to other calpains and subcellular distribution has not been investigated previously. Based on relative mRNA levels, Capn5 is the second most highly expressed calpain in the rat CNS, with Capn2 mRNA being the most abundant. Unlike classical calpains, CAPN5 is a non-cytosolic protein localized to the nucleus and extra-nuclear locations. CAPN5 possesses two nuclear localization signals (NLS): an N-terminal monopartite NLS and a unique bipartite NLS closer to the C terminus. The C-terminal NLS contains a SUMO-interacting motif that contributes to nuclear localization, and mutation or deletion of both NLS renders CAPN5 exclusively cytosolic. Dual NLS motifs are common among transcription factors. Interestingly, CAPN5 is found in punctate domains associated with promyelocytic leukemia (PML) protein within the nucleus. PML nuclear bodies are implicated in transcriptional regulation, cell differentiation, cellular response to stress, viral defense, apoptosis, and cell senescence as well as protein sequestration, modification, and degradation. The roles of nuclear CAPN5 remain to be determined.
Introduction
The calpain (CAPN)2 family of Ca2+-dependent cysteine proteases (clan CA, family C2, EC 3.4.22.52–54) has 15 mammalian members, including the small regulatory subunit CAPNS1 (1). These are subdivided into classical (CAPN1–3, -8, -9, and -11–14) and non-classical (CAPN5–7, -10, -15, and -16) calpains based on primary sequence and into ubiquitous (CAPN1, -2, -5, -7, -10, -13–15, and S1) and tissue-specific (CAPN3, -6, -8, -9, -11, and -12) calpains based on localization (1, 2). CAPN2 is not present in erythrocytes but is present in other cells. The ubiquitous isoforms are presumed to play important roles in all cells, whereas the tissue-specific isoforms are required for more specialized functions. Most investigations have focused on the classical calpains, CAPN1 (also referred to as μ-calpain or μCL) and CAPN2 (also referred to as m-calpain or mCL), which contain a C2-like domain, a penta-EF hand domain, and the cysteine protease domain. They are heterodimers, composed of the large CAPN1 or CAPN2 subunit and the CAPNS1 (CAPN4) small subunit. Their roles include apoptosis cell migration, cytoskeletal remodeling, cell differentiation, necrosis/oncosis, platelet aggregation, and wound healing (3–9). Non-classical calpains, which lack the EF hand motif characteristic of classical calpains but retain catalytic and Ca2+ binding domains, are less well understood. Non-classical calpains are evolutionarily more conserved than classical calpains and are the only calpains present in some organisms such as Caenorhabditis elegans (10, 11).
TRA-3, the C. elegans ortholog of CAPN5, is essential for necrotic neuron death and is also involved in sex determination in nematodes (12, 13). CAPN6 is also an ortholog of TRA-3, but substitution of Cys with Lys at the active site results in a loss of proteolytic activity in eutherians (14). CAPN5, also referred to as hTRA-3, lacks the penta-EF hand domain of classical calpains and has a C2 domain at the C terminus (previously referred to as domain-T) (15). CAPN5 is expressed in all rat and human tissues examined, including various regions of the CNS (16, 17). The subcellular localization of CAPN5 has not been examined previously.
Incubation of SH-SY5Y cell lysates with maitotoxin or the Ca2+ ionophore A23187 results in CAPN5 proteolysis, consistent activation (17). Capn5−/− (Capn5tm1Nde) mice are viable and fertile, although some are severely runted at birth and die by 2 months of age (18). However, another Capn5 null mutant allele (Capn5tm1Dgen) is embryonically lethal (Mouse Genome Informatics ID 3604529). CAPN5 polymorphisms have been associated with autoimmune retinal neurodegeneration (19), polycystic ovary syndrome (20), endometriosis (21), diabetes (22), and Huntington disease (23). Based on the importance of TRA-3 in neuron death in C. elegans and the relatively high expression of Capn5 mRNA in the brain, we sought to further explore CAPN5 in the mammalian CNS.
EXPERIMENTAL PROCEDURES
Experimental Animals
The University of Kentucky Institutional Animal Care and Use Committee approved all procedures involving experimental animals. Animals included Male Sprague-Dawley rats and Capn5 heterozygous mice (C57BL/6J-Capn5tm1Dgen/J; Capn5+/LacZ). These mice, created by DeltaGen (San Mateo, CA) (24), were obtained from The Jackson Laboratory (Bar Harbor, ME) on a B6.129P2 background and then back-crossed onto C57BL/6J for 10 generations. For Western blot and fractionation studies, rats were exsanguinated using CO2 inhalation followed by decapitation. Brains were removed rapidly and homogenized in the appropriate buffer for Western blot, fractionation, or RNA isolation as described below. For immunohistochemical and X-gal staining studies, rats or mice were first perfused with PBS followed by 4% paraformaldehyde in PBS, pH 7.4.
Antibodies and Reagents
Antibodies against CAPN5 (ab28280), adenomatous polyposis coli (APC) tumor suppressor protein (CC-1 clone, ab16749), promyelocytic leukemia protein (PML; ab96051), β-tubulin (ab6046-100), and histone H3 (ab1791) were purchased from Abcam, Cambridge, MA. Anti-CAPN2 (208729, mAb107-82), anti-NeuN (mAb377), and anti-glial fibrillary acidic protein (GFAP; mAb360) were obtained from EMD Millipore, Billerica, MA. An antibody against survival of motor neuron (SMN; 610647) protein was purchased from BD Biosciences, San Jose, CA. Anti-FLAG-M2 antibody (200472) was obtained from Agilent Technologies, Santa Clara, CA. IRDye 800CW anti-rabbit IgG (611-131-132) and IRDye 800CW anti-mouse IgG (610-131-121) were purchased from Rockland, Gilbertsville, PA. Hoechst 33258 (H-3569) and conjugated secondary antibodies Alexa Fluor-680 anti-mouse IgG (A21058), Alexa Fluor-488 anti-rabbit IgG (A11005), and Alexa Fluor-594 anti-mouse IgG (A11034) were purchased from Molecular Probes, Invitrogen, NY. TRI Reagent (T9424), X-Gal (5-bromo-4-chloro-3-indoyl-β-d-galactoside, B4252), pepstatin A (P4265), and β-mecaptoethanol (M6250) were purchased from Sigma. pN1-ZsGreen1 (632448) and pN1-mCherry (632523) vectors were purchased from Clontech Laboratories, Inc. Mountain View, CA. p3X-CMVTM-14-FLAG (E7908) was purchased from Sigma. Human CAPN5 cDNA (MHS1010-58128) was purchased from Thermo Scientific, Open Biosystems, Huntsville, AL. All oligonucleotides were ordered from Integrated DNA Technologies, Coralville, IA. Lipofectamine 2000 CD reagent (12566-014) was obtained from Invitrogen. Pfu DNA polymerase (600135) was ordered from Agilent Technologies, Stratagene Division, La Jolla, CA. EcoRI-HF and BamHI-HF were purchased from New England Biolabs, Ipswich, MA. The Rapid DNA ligation kit (11-635-379-001) was obtained from Roche Applied Science. One Shot® Stbl3TM-competent bacteria (C737303) were purchased from Invitrogen. Additional reagents were purchased from Sigma or Thermo Fisher Scientific.
Quantitative PCR
Brain samples were homogenized in TRI Reagent to extract total RNA. cDNA was prepared using an Applied Biosystems high capacity reverse transcription kit (AB 4368814). Equal amounts of cDNA (100 ng) were used to perform quantitative PCR using TaqMan gene expression master mix (AB 4369016). Reactions were performed in triplicate plus a negative control without cDNA. The quantitative PCR was programmed as an initial denaturation at 50 °C for 2 min followed by 95 °C for 10 min, 40 cycles at 95 °C for 15 s, and 60 °C for 1 min on a StepOne real-time PCR system (Applied Biosystems). The following rat gene transcripts were examined: Capn1 (NCBI reference sequence NM_019152.2, TaqMan gene expression assay Rn00569689_m1); Capn2 (NM_017116.2, Rn00567422_m1); Capn5 (NM_134461.1, Rn00593213_m1); Capn7 (NM_001030037.1, Rn01453530_m1); Capn10 (NM_031673.2, Rn00581535_m1); and Gapdh (NP_058704, Rn99999916-s1). Relative gene expression was determined using comparative CT values. ΔCT of the target gene was obtained as a difference in the CT value from endogenous control GAPDH. The ΔΔCT value of the target gene was calculated by subtracting ΔCT value of the target gene from the ΔCT of a reference gene, Capn1. The relative expression of the target gene was then reported as 2−ΔΔCT.
Cytosol and Crude Nuclear Fractionation
Brain cortices were homogenized in a Dounce homogenizer in isolation buffer containing 215 mm mannitol, 75 mm sucrose, 1 mm EGTA, 20 mm HEPES, and 1 μg/ml pepstatin A. The homogenate was centrifuged at 1300 × g for 3 min to obtain the crude nuclear fraction as a pellet. The supernatant was spun again at 13,000 × g for 10 min to obtain the cytosol as supernatant.
Nuclear Subfractionation
Rat brain cortex was lysed and incubated with buffers provided in a Qproteome nuclear protein kit (Qiagen, catalogue No. 37582). The manufacturer's protocol was followed to obtain cytosol, nucleic acid-binding proteins (NABP), and insoluble nuclear proteins (INP). Briefly, 50 mg of tissue was disrupted in a Dounce homogenizer in 1 ml of lysis buffer supplemented with protease inhibitor solution and 0.1 m DTT followed by incubation on ice for 15 min. 50 μl of detergent solution was added to the solution and vortexed for 10 s. Lysate was then centrifuged at 10,000 × g for 5 min to obtain cytosol as supernatant and pellet for nuclear subfractionation. The pellet was resuspended in 100 μl of extraction buffer NX1 supplemented with protease inhibitor solution and incubated for 30 min on a Rotamix at 4 °C. The suspension was spun down for 10 min at 12,000 × g to separate the NABP fraction as supernatant and INP in the pellet. The pellet was resuspended in extraction buffer NX2 supplemented with protease inhibitor, 0.1 m DTT, and Benzonase® nuclease followed by incubation for 1 h on a Rotamix at 4 °C. The suspension was centrifuged for 10 min at 12,000 × g to obtain INP in the supernatant.
Western Blot
The protein content of samples was assayed using Thermo Scientific Pierce® BCA protein assay reagent A (23228) and reagent B (1859078). 50 μg of each protein was mixed in NuPAGE® sample buffer (NP007) supplemented with 5% β-mercaptoethanol and boiled for 5 min. The boiled samples were separated on 4–12% bis-Tris-HCl gels (NuPAGE NP0335) in MES SDS running buffer (NuPAGE NP0002) and transferred to a 0.2-μm nitrocellulose membrane. After blocking in 5% skim milk in T-TBS (0.05% Tween 20 in Tris-buffered saline, pH 7.6) for 1 h, the membrane was incubated with primary antibody (CAPN5, 1:5000; CAPN2, 1:5000; SMN, 1:5000; histone H3, 1:5000; and β-tubulin, 1:10,000) in 5% skim milk in T-TBS overnight at 4 °C. The membrane was washed in T-TBS three times for 20 min each followed by incubation with an appropriate secondary antibody (anti-rabbit IgG, 1:5000, or anti-mouse IgG, 1:5000, linked to IRDye/Alexa Fluor-680 or IRDye 800) at room temperature for 1 h in dark conditions. The membrane was washed again three times for 20 min each and scanned using an Odyssey infrared imager (LI-COR Biosciences). To measure the intensity of bands on Western blots, Odyssey application software V3.0 (Image Studio) was used. A rectangle was drawn around the band of interest, with the rectangle size being constant for each lane of the blot. The relative densitometric value of the band was determined as compared with the background.
β-Galactosidase Staining
Capn5tm1Dgen/J (Capn5+/−) male mice, ∼3 months of age, were perfused with PBS followed by 4% paraformaldehyde in PBS, pH 7.4. The brains were removed, postfixed overnight, and then cryoprotected in 30% sucrose in PBS. The brains were frozen in powdered dry ice and sectioned at 40 μm in the coronal plane, with sections stored in cryoprotectant (30% ethylene glycol, v/v, and 30% glycerol, v/v, in 1× TBS) at −20 °C until use. Sections were rinsed three times in PBS then incubated with 1 mg/ml X-Gal in 10 mm potassium ferricyanide, 5 mm potassium ferrocyanide, 2 mm MgCl2 at 37 °C overnight or until the dark blue staining appeared. Following dehydration, clearing, and cover-slipping, sections were viewed and photographed using bright-field microscopy.
Immunohistochemistry
Perfusion, fixation, and preparation of male rat brain sections were performed as discussed for β-galactosidase staining. Free-floating brains sections were washed three times in TBS followed by blocking in 5% natural goat serum (NGS) in T-TBS (0.1% Triton X-100) for 30 min at room temperature. Brain sections were incubated with primary antibodies (CAPN5, 1:100; NeuN, 1:200; GFAP, 1:1000; and APC, 1:100) in 5% NGS-T-TBS overnight at 4 °C. Primary antibody was omitted from the negative controls. Sections were washed three times in 1× TBS followed by a 1-h incubation with the appropriate secondary antibodies at 1:1000 dilution (Alexa Fluor-488 anti-rabbit IgG or Alexa Fluor-594 anti-mouse IgG) and then washed three times in TBS. Nuclei were stained with Hoechst 33258 at 10 μg/ml for 1 min. Brain sections were mounted on glass slides with Vectashield (H-1000, Vector Laboratories) florescence mounting medium and examined under a Leica AOBS TCS SP5 inverted laser scanning confocal microscope.
Cell Culture and Immunocytochemistry
SH-SY5Y (ATCC CRL-2266) cells were cultured in Eagle's minimum essential medium (ATCC 30-2003) plus 1% penicillin/streptomycin and 10% FBS at 37 °C in an incubator maintained with 95% air and 5% CO2. The cells were plated on 35-mm glass-bottom culture dishes. The following day, adherent cells were fixed in 4% paraformaldehyde in PBS, pH 7.4, for 15 min at room temperature followed by permeabilization for 10 min with PBS containing 0.25% Triton X-100 (PBS/T). After washing three times with PBS, cells were incubated with 5% NGS-PBS/T for 30 min and then incubated with the primary antibody (PML, 1:100, or CAPN5, 1:100) in 5% NGS-PBS/T overnight at 4 °C. The next day, sections were washed three times in PBS and incubated for 1 h in the dark with 1:1000 Alexa Fluor-488 anti-rabbit IgG or Alexa Fluor-594 anti-mouse IgG followed by three more washes at 5 min each. Nuclei were stained with Hoechst 33258 at 10 μg/ml for 5 min. Cells were viewed under a Nikon Ti-E C2plus confocal microscope.
Plasmid Preparation, Transient Transfection, and Confocal Microscopy
Human CAPN5 cDNA (GenBankTM accession number BC018123.1) and oligonucleotides were purchased as described above under “Antibodies and Reagents.” PCR oligonucleotide primers for CAPN5 were designed to allow cloning into pN1-ZsGreen1, pN1-mCherry, or p3X-CMV-14-FLAG vector, such that ZsGreen1, mCherry, or a FLAG tag was encoded at the C terminus of the fusion protein. PCR was carried out using a Pfu DNA polymerase kit. The resulting DNA product was digested with EcoRI-HF and BamHI-HF. The product was then ligated into the vector using a Rapid DNA ligation kit and transformed into One Shot® Stbl3TM-competent bacteria. Plasmid DNA was isolated using the Qiagen Maxi Prep Kit 12263. Primers for mutagenesis were synthesized by Integrated DNA Technologies, and mutations were introduced using the GeneArt® site-directed mutagenesis system (Invitrogen, catalogue No. 13282).
For transient transfections, SH-SY5Y cells were grown in 35-mm glass-bottom culture dishes. At ∼70% confluency, cells were transfected with 0.2 μg of plasmid vector using Lipofectamine 2000 CD reagent. Transfected cells were imaged at 24 h post-transfection. Prior to imaging, cells were stained for 1 h with 10 μg/ml Hoechst 33258. Images were acquired on a Nikon Ti-E C2plus confocal microscope.
Statistical Analysis
The quantified values of the relative intensity of the CAPN5 immunoreactivity band and quantitative RT-PCR RQ data were analyzed using one-way analysis of variance followed by Tukey's multiple comparison tests.
RESULTS
Calpain 5 Is Highly Expressed in Rat Brain
CAPN5 is present in the CNS (17), but its expression relative to other calpains is unknown. Using the real-time comparative CT method (ΔΔCT) (25), we analyzed the average relative mRNA expression of several ubiquitous calpains (CAPN1, -2, -5, -7, and -10) in the adult rat brain (n = 4). Capn2 had the highest levels of mRNA expression, exceeding that of Capn5 by 2.7-fold. Capn5 was the second most highly expressed calpain in the brain followed by Capn7, -10, and -1, respectively (Fig. 1).
FIGURE 1.

Calpain 5 is highly expressed calpain in rat brain. Relative mRNA expression of calpains 1, 2, 5, 7, and 10 was calculated using the comparative CT (ΔΔCT) method in 3-month-old male Sprague-Dawley rat brain homogenate (n = 4). ΔCT of each of the calpain isoforms was obtained as a difference in the CT value from an endogenous control, Gapdh. ΔΔCT of the target gene was calculated by subtracting ΔCT of the target gene from the ΔCT value of the reference gene, Capn1. Relative mRNA expression of the target gene was then reported as 2−ΔΔCT. The results show that after Capn2, Capn5 is the highest expressing calpain in the brain followed by calpains 7, 10, and 1. The results are expressed as the group means ± S.D., n = 4.
Ab28280 Is Specific to Calpain5
For the immunodetection of CAPN5, we utilized rabbit polyclonal Ab28280 (Abcam), which was raised against the N-terminal 30 amino acids of human CAPN5. On Western blots of rat brain homogenate, the antibody detected a prominent band of ∼75 kDa. The antibody did not detect purified rat CAPN2 (Calbiochem catalogue No. 208718), and preincubation of the antibody with the immunogen peptide abolished the 75-kDa band on Western blots (Fig. 2, A and B). The antibody also detected a band of ∼75 KDa from SH-SY5Y cells expressing full-length human CAPN5 fused with a FLAG tag on the C terminus (Fig. 2C). A second commercial antibody against CAPN5 cross-reacted with CAPN2 (results not shown).
FIGURE 2.
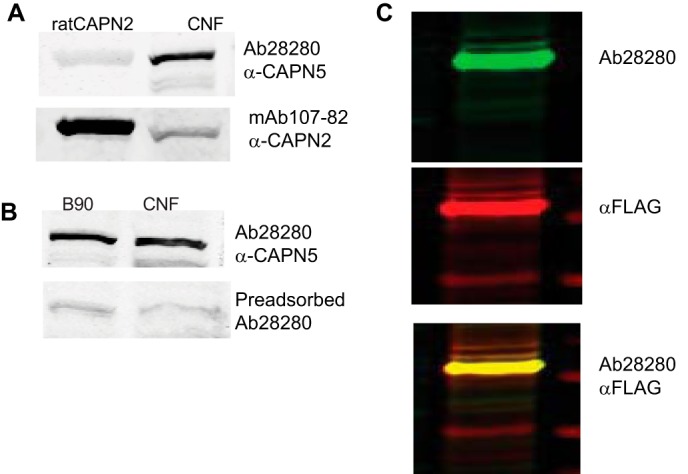
Ab28280 is specific to calpain 5. A, in the crude nuclear fraction (CNF) from rat brain, Ab28280 detects a band of ∼75 kDa but does not detect purified rat calpain 2. B, anti-calpain 2 (mAb 107-82) detected purified calpain 2 and weak calpain 2 immunoreactivity in the crude nuclear fraction. Preincubation of the Ab28280 antibody with the N-terminal peptide used as the immunogen (Abcam Ab41310) abolished the 75-kDa band immunoreactivity on Western blots of brain homogenate from 90-day-old rats. C, as a positive control, we examined extracts of SH-SY5Y cells expressing full-length human CAPN5 fused with a FLAG tag on the C terminus (p3X-CMVTM-14-hCAPN51–640-FLAG). Cell lysate was prepared 24 h post-transfection and probed through Western blotting. Ab28280, with secondary antibody IRDye 800CW anti-rabbit IgG, detected a similar prominent band of ∼75 kDa using the 800 nm channel of the Li-Cor Oydessy imaging system and is presented as a green band in the Li-Cor system. FLAG, detected with an anti-FLAG-M2 antibody and Alexa Fluor-680 anti-mouse IgG, using the 700 nm channel of the Li-Cor Odyssey, is shown in red. The overlap of the bands detected by the anti-FLAG and Ab28280 (yellow) illustrates detection of recombinant human CAPN5 by Ab28280.
Nuclear localization of Calpain 5
To examine the cellular localization of Capn5 mRNA expression, we first utilized Capn5tm1Dgen/J mice in which a LacZ-Neo555G cassette was inserted into the calpain 5 gene. Capn5−/− mice were embryonically lethal, whereas Capn5+/− mice were viable. Despite the lower gene dosage, CAPN5 protein levels were unchanged in the brains of Capn5+/− mice as compared with the wild type (results not shown).
In Capn5+/− mouse brain, X-Gal staining was prevalent in the pyramidal neurons in the hippocampal formation as well as in dentate granule and hilar neurons (Fig. 3A). X-Gal staining was also observed in molecular layers and white matter, indicative of expression in non-neuronal cells including astrocytes and oligodendrocytes. These results are consistent with Capn5 being ubiquitously expressed in all cells.
FIGURE 3.
Calpain 5 is present in the nuclei of neurons and glia. A, X-Gal staining was performed on 40-μm coronal brain sections of ∼3-month-old male Capn5+/LacZ mice (n = 6). In the hippocampal formation, X-Gal staining was prominent in all neurons including CA1 and CA3 pyramidal neurons, granule cells of the dentate gyrus (DG), and hilar neurons (DH). X-Gal staining was also present in molecular layers, suggesting expression in glial cells and consistent with the ubiquitous expression of Capn5 mRNA. B–D, immunohistochemical localization of CAPN5. Confocal images were obtained following double immunolabeling of 40-μm coronal ∼3-month-old male Sprague-Dawley rat brain sections. The co-localization of CAPN5 and NeuN (B), a neuronal nuclear protein (82), indicates that CAPN5 is predominantly localized to neuronal nuclei, although faint extranuclear staining was also observed. CAPN5 immunoreactivity was evident in the nuclei of cells positive for GFAP (C), an intermediate filament protein in astrocytes (83). In cells positive for the APC protein (D), a marker for mature oligodendrocytes (84), CAPN5 immunoreactivity was also localized to the nucleus.
In double-label immunocytochemistry, co-localization of CAPN5 immunoreactivity with anti-NeuN confirmed the neuronal localization of CAPN5, also indicating that expression was predominantly nuclear (Fig. 3B). Double labeling of anti-CAPN5 with anti-GFAP and anti-APC demonstrated nuclear CAPN5 expression in astrocytes and oligodendrocytes (Fig. 3, C and D). However, localization was not exclusively nuclear, as faint immunoreactivity was observed in the neuropil, and a band of CAPN5 immunoreactivity was present in the stratum lacunosum-moleculare in the hippocampal formation (not shown). This is a terminal zone of the perforant path and is rich in mitochondria (26), suggesting a possible presynaptic, postsynaptic, or mitochondrial localization of CAPN5 in addition to the nuclear localization.
To further evaluate the subcellular localization of CAPN5, rat brain cortices were homogenized and separated into cytosolic and crude nuclear fractions. CAPN5 immunoreactivity was abundant in the crude nuclear fraction but was not detected in the cytosolic fraction (Fig. 4A). Additional subfractionation demonstrated that CAPN5 was enriched in the NABP fraction (Fig. 4, B and C). Unlike findings in classical calpains, which are characterized as being mainly cytosolic (2, 27, 28), these results illustrate that CAPN5 is predominantly a non-cytosolic calpain, present in the nucleus in the nucleic acid-binding protein fraction.
FIGURE 4.
Calpain 5 is enriched in the nuclear nucleic acid binding fraction. A, following differential centrifugation, CAPN5 immunoreactivity was prominent in the crude nuclear fraction (CNF) and was not detected in the cytosolic fraction (C). Marker proteins included histone H3 (nuclear) and β-tubulin as a cytosolic marker. B, using the Qiagen nuclear protein kit, rat brain cortex was subfractionated into the cytosolic fraction, the NABP fraction, and the INP fraction followed by probing for CAPN5 and marker proteins by Western blot. CAPN5 was enriched in the NABP fraction but also was detected in the INP fraction. Survival of motor neuron (SMN) protein resides both in the nucleus and the cytosol (85, 86) and was used as a marker for the NABP fraction. Histone H3 is a marker for INP fraction. C, quantitation of the relative intensity of the CAPN5 immunoreactive band in B is shown. The results, reported as group means ± S.D., n = 3, were analyzed as one-way analysis of variance followed by Tukey's multiple comparison test, **, p < 0.001. D, confocal images of 40-μm male Sprague-Dawley rat brain sections co-immunostained with antibodies against CAPN5, NeuN, GFAP, and Hoechst (a nuclear marker) show punctate nuclear localization of CAPN5 in each of the cell types.
Calpain 5 Resides in Punctate Nuclear Domains Associated with PML Bodies
The nuclear staining observed with CAPN5 appeared punctate in each of the cell types examined in rat brain (Fig. 4D). Faint nuclear CAPN5 immunoreactivity was also detected outside of the punctate localization. Perinuclear and extranuclear immunoreactivity was also observed.
Punctate nuclear localization was also observed in SH-SY5Y neuroblastoma cells (Fig. 5), suggesting that CAPN5 is associated with one or more nuclear bodies (29). Non-nuclear, particularly perinuclear, CAPN5 immunoreactivity was also evident (Fig. 5). In SH-SY5Y cells transfected with a plasmid expressing full-length human CAPN5 fused with ZsGreen1 (pN1-CAPN51–640-ZsGreen1), similar punctate nuclear localization was observed with the CAPN5-ZsGreen1 fusion protein (Fig. 5). In several cells, there were also cytoplasmic aggregates of CAPN5 in the perinuclear region (Fig. 5). Dot-like nuclear expression of CAPN5 was also observed following transient transfection of SH-SY5Y cells with a vector encoding human CAPN5 with a C-terminal FLAG tag, as detected with an anti-FLAG antibody (Agilent Technologies, 200472) (result not shown). Endogenous as well as the transiently expressed CAPN5 nuclear dots were co-localized or closely associated with PML bodies, as detected using an antibody against PML protein (Fig. 5). PML-independent CAPN5 dots were also evident.
FIGURE 5.
Calpain 5 is associated with PML bodies. A, co-immunolabeling of endogenous CAPN5 and PML in SH-SY5Y cells indicates that CAPN5 is localized with PML bodies. PML-independent CAPN5 localization was also observed. With every PML nuclear body, a CAPN5 immunoreactive dot or larger cluster was either co-localized with PML or in very close proximity to PML, as indicated by arrows. However, the size of the PML nuclear body did not appear to correlate with the size of the CAPN5 cluster. Extranuclear CAPN5 immunoreactivity was also observed, particularly in the perinuclear region, indicated by an arrowhead. B, PML nuclear bodies were also associated with recombinant hCAPN5. SH-SY5Y cells were transiently transfected with full-length human CAPN5 cDNA with a C-terminal ZsGreen1 tag (pN1-CAPN51–640-ZsGreen1) using Lipofectamine 2000. At 24 h post-transfection, CAPN5-ZsGreen1 expression was detected in intranuclear punctate domains and as extranuclear aggregates. Immunocytochemistry with α-PML antibody reveals co-localization of CAPN5 clusters with PML nuclear bodies.
Calpain 5 Nuclear Localization Signals
Analysis of the primary sequence of human CAPN5 with a motif-scanning tool (30) suggested a bipartite NLS sequence (aa 395–411) in addition to the thiol protease, C2-like, and calpain catalytic domains. PSORTII also revealed the putative bipartite NLS along with several possible monopartite sequences (9). NucPred identified RRRK (aa 21–24) as the most likely monopartite NLS in CAPN5 (31).
To evaluate the putative RRRK NLS, we examined the localization of the first 30 amino acids of CAPN5 fused to ZsGreen1. At 24 h post-transfection, the resultant fusion protein was enriched in the nucleus, where it was diffusely distributed (Fig. 6, A and B, and Table 1), unlike the dot-like domains observed with full-length CAPN5. Using site-directed mutagenesis, the putative NLS residues 21–24 (RRRK) were mutated to alanines (AAAA). The mutated fusion peptide with a C-terminal ZsGreen1 tag was enriched in the cytosol (Fig. 6C).
FIGURE 6.
RRRK (aa 21–24) in human CAPN5 is a nuclear localization signal (NLS1). SH-SY5Y cells were transiently transfected with the first 30 amino acids of CAPN5 fused to ZsGreen1 or CAPN51–30-ZsGreen1 or with full-length CAPN5-ZsGreen1. A, schematic showing the labels of the various constructs and the location of mutated NLS. B, at 24 h post-transfection, CAPN51–30 was enriched in the nucleus. C, mutation of amino acids 21–24, RRRK, to AAAA shifted enrichment to the cytosol. D and E, full-length CAPN5 appears as dot-like domains in the nucleus (D), with large perinuclear aggregates also observed in many cells (E). F, residues 21–24 in the full-length CAPN5 reduced the number of cells with punctate nuclear bodies and the number of nuclear bodies in some cells. G, other cells showed only cytosolic aggregates.
TABLE 1.
Summary of CAPN5 and NLS constructs and their subcellular distribution at 24 h post-transfection
| Constructs | Amino acid sequence | Cellular localization post-transfection |
|---|---|---|
| NLS1 | ||
| pN1-CAPN51–30-ZsGreen1 | MFSCVKPYEDQNYSALRRDCRRRKVLFEDP, aa 1–30 of human CAPN5 | Mainly nuclear |
| pN1-CAPN521–24A1–30-ZsGreen1 | MFSCVKPYEDQNYSALRRDCAAAAVLFEDP, aa 21–24 mutated to Ala in aa 1–30 of human CAPN5 | Mainly cytosolic |
| NLS2 | ||
| pN1-CAPN5393–413-ZsGreen1 | EVKKPEDEVLICIQQRPKRST, PSORTII predicted bipartite NLS extended by 2 amino acids on either sides | Cytosolic |
| pN1-CAPN5395–417-ZsGreen1 | KKPEDEVLICIQQRPKRSTRREG, expanded bipartite NLS to include 2 additional basic residues on C-terminal | Cytosolic |
| pN1-CAPN5391–417-ZsGreen1 | IFEVKKPEDEVLICIQQRPKRSTRREG, addition of upstream sequence to the extended bipartite NLS | Cytosolic/perinuclear |
| pN1-CAPN5390–417-ZsGreen1 | YIFEVKKPEDEVLICIQQRPKRSTRREG, minimal upstream sequence to the bipartite region resulting in nuclear localization | Nuclear |
| pN1-CAPN5390–417-mCherry | ||
| pN1-CAPN5388–417-ZsGreen1 | PQYIFEVKKPEDEVLICIQQRPKRSTRREG, longer version of NLS2 | Nuclear |
| pN1-CAPN5Ala388–417-ZsGreen1 | PQYIFEVAAPEDEVLICIQQAPAASTAAEG, basic residues mutated to Ala (A) | Nuclear |
| pN1-CAPN5A403390–417-ZsGreen1 | YIFEVKKPEDEVLACIQQRPKRSTRREG, Ile403 mutated to Ala | Mainly cytosolic |
| Full-length CAPN5 | ||
| pN1-ZsGreen1 | Empty vector | Cytosolic |
| pN1-CAPN51–640-ZsGreen1 | Full-length human CAPN5, aa 1–640 | Nuclear/perinuclear |
| pN1-CAPN5Asn1–640-ZsGreen1 | Full-length CAPN5 with Lys/Arg of predicted bipartite NLS mutated to Asn (N) (NN[PEDEVLICIQQ]NPNN) | Nuclear/perinuclear |
| pN1-CAPN521–24A1–640-ZsGreen1 | RRRK, aa 21–24 mutated to Ala in full-length CAPN5 | Cytosolic and nuclear |
| pN1-CAPN5Δ390–4171–640-mCherry | YIFEVKKPEDEVLICIQQRPKRSTRREG, aa 390–417 deleted from CAPN5 | Cytosolic and nuclear |
| pN1-CAPN5Δ390–4171–640-ZsGreen1 | ||
| pN1-CAPN521–24A,Δ390–4171–640-mCherry | CAPN5 with aa 21–24 mutated to Ala and aa 390–417 deleted | Cytosolic |
Transient transfection with pN1-CAPN51–640-ZsGreen1 resulted in punctate nuclear and perinuclear CAPN5-ZsGreen1 expression (Fig. 6, D and E). When the AAAA mutation was introduced into full-length CAPN5, nuclear localization persisted in some cells, with fewer punctate nuclear dots observed in some cases (Fig. 6F), whereas other cells had a similar number of punctate nuclear dots as compared with wild-type CAPN5. In other cells, the AAAA mutation abolished the nuclear localization (Fig. 6G). The nuclear localization of CAPN5 in some cells with the 21–24A mutation suggested the possibility of a second NLS.
To determine whether the putative NLS bipartite sequence, KKPEDEVLICIQQRPKR (aa 395–411), represented an NLS, several ZsGreen1 fusion constructs were evaluated (Table 1 and Fig. 7). Extending the bipartite sequence in both directions to form YIFEVKKPEDEVLICIQQRPKRSTRREG (pN1-CAPN5390–417-ZsGreen1) resulted in nuclear localization of the fusion protein in distinct punctate or dot-like domains in transfected cells, with additional perinuclear localization (Fig. 7A). Nuclear localization was also observed with an mCherry tag on the C terminus of the NLS (pN1-CAPN5390–417-mCherry) (Fig. 7B). Shorter constructs exhibited cytosolic and/or perinuclear localization (Fig. 7C and Table 1). The above results suggest that YIFEVKKPEDEVLICIQQRPKRSTRREG represents a second NLS in CAPN5 (NLS2). However, mutation of the basic residues to alanines did not abolish the nuclear localization of the peptide sequence but resulted in a more diffuse nuclear localization (Fig. 7D).
FIGURE 7.

Calpain 5 NLS2 (aa 390–417). A, transient transfection of SH-SY-5Y cells with pN1-CAPN5390–417-ZsGreen1 (YIFEVKKPEDEVLICIQQRPKRSTRREG-ZsGreen1), a peptide sequence enclosing putative bipartite NLS and upstream residues YIFEV, resulted in the punctate nuclear expression of the fusion protein. B, similar results were obtained with a C-terminal mCherry tag. C, reducing this sequence by one amino acid (pN1-CAPN5391–417-ZsGreen1) abolished the nuclear, but not perinuclear, localization. D, a slightly longer sequence, 388–417, exhibited similar punctate nuclear localization to aa 390–417 (not shown). Mutagenesis of the basic amino acid residues to alanines (PQYIFEVAAPEDEVLICIQQAPAASTAAEG) did not prevent the nuclear localization, although the organization appeared less punctate. E, mutating Ile403 to Ala in the predicted SUMO-interacting motif, PEDEVLICI (YIFEVKKPEDEVLACIQQRPKRSTRREG), aborts the nuclear localization. F, deletion of NLS2 (pN1-CAPN5Δ390–417-mCherry) resulted in diffuse cellular localization in many cells, whereas nuclear localization was observed in other cells. Similar results were observed with a ZsGreen1 C-terminal tag (not shown). G, mutation of NLS1 and deletion of NLS2 (pN1-CPAN521–24AΔ390–417-ZsGreen1) resulted in cytosolic localization.
Partner proteins associated with PML bodies are typically SUMOylated or contain a SUMO-interacting motif (SIM) (32, 33). CAPN5 contains a probable SIM (KPEDEVLICI, aa 396–405), predicted using GPS-SBM 1.0 at medium threshold. We sought to determine whether this SIM, which is located within NLS2, might contribute to the punctate CAPN5 nuclear localization. Mutating Ile403 to Ala (pN1-CAPN5I403A388–417-ZsGreen1) abolished the nuclear localization of the ZsGreen1 fusion protein (Fig. 7E and Table 1). Together, the results suggest that NLS2 for CAPN5 contributes to the punctate localization through the SIM domain KPEDEVLICI.
Deletion of the aa 390–417 bipartite NLS2 reduced the extent of nuclear localization, although in some cells the fusion protein went to the nucleus (Fig. 7F). When NLS1 was mutated to AAAA and NLS2 was deleted, the fusion protein was cytosolic (Fig. 7G). For all constructs, similar results were obtained with C-terminal ZsGreen1 and mCherry tags.
NLS2 contains serine and threonine residues adjacent to basic amino acids on the downstream side of the bipartite NLS. Several NLS are regulated by phosphorylation, which can either enhance or impede nuclear import (34). The possible influence of phosphorylation on CAPN5 nuclear import has not been examined in studies to date.
Based on the above results, a schematic of CAPN5 is shown in Fig. 8. NLS1 (aa 21–24) is located within the protease domain, CysPC, whereas NLS2 (aa 390–417) is within the C2-like domain. The conserved CysPC, C2-like, and C2 domains are described in a recent review of calpains by Ono and Sorimachi (1).
FIGURE 8.
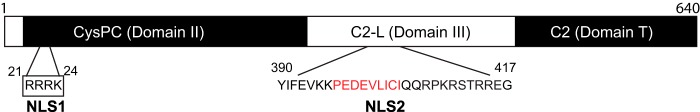
Schematic of calpain 5. NLS1 is located within the CysPC conserved protease domain, also referred to as domain II. NLS2 is within the C2-like domain, also referred to as domain III. Within NLS2, the putative SUMO-interacting motif is indicated by red letters.
DISCUSSION
Most previous investigations of calpains in the CNS have focused on classical calpains 1 and 2, although several other calpains have been detected in the CNS including CAPN3, -5, -10, and -12 (17, 35–37). In this study, we found that Capn5 mRNA levels are second only to Capn2 in relative abundance in the CNS. The much greater expression of Capn2 versus Capn1 mRNA is consistent with previous findings (12). Calpains are largely cytosolic proteases, although CAPN1, -2, and -10 have also been localized to mitochondria, with CAPN2 and -10 additionally detected in the nucleus (2, 27, 28, 36, 38–41). In contrast, CAPN5 localization is predominantly nuclear and is associated with PML nuclear bodies.
PML nuclear bodies are a collection of proteins arranged in spheres of 0.1–1.0 μm in diameter and localized to the nuclear matrix in most tissues and cell lines (42). They are organized by PML protein, which was discovered based on its involvement in acute promyelocytic leukemia (for review see Ref. 43). PML nuclear bodies are implicated in transcriptional regulation as well as the cellular response to stress, viral defense, apoptosis, and cell senescence (29, 33, 44, 45). PML bodies recruit a large number of partner proteins, which are then sequestered, modified, or degraded (33). PML partner proteins are either SUMOylated or contain a SIM (32).
The localization of CAPN5 to PML nuclear bodies suggests that CAPN5 should contain one or more NLS as well as SUMOylation or SIM sites. Classical NLS contain either a single (monopartite) or double (bipartite) group of basic amino acids (46). In bipartite NLS signals, the two clusters of basic amino acids are separated by ∼10–12 amino acids with the prototype being the NLS for nucleoplasmin (47).
We identified two NLS in CAPN5. NLS1 is a monopartite NLS, RRRK, located at residues 21–24 of human CAPN5. NLS2 in CAPN5, residues 390–417, contains two clusters of basic amino acids separated by 11 amino acids, YIFEVKK [PEDEVLICIQQ]RPKRSTRREG (basic amino acids are underlined). In the classical import pathway, basic residues in the NLS bind to either the major or minor binding sites in the importin-α family, with bipartite NLS residues interacting with both sites (46, 48, 49). Monopartite NLS sequences typically have a lysine residue at the second position and bind to the major importin-α binding site (48, 50). In contrast, arginine-rich NLS signals, such as CAPN5 NLS1, bind directly to importin-β (51, 52). The DNA binding transcription factors SOX9 and SRY have similar monopartite NLS sequences, RRRK for SOX 9 and RPRRK for SRY, in which the nuclear import is mediated by importin-β (53–55).
CAPN5 NLS1 is located within the CysPC protease domain of CAPN5, also referred to as domain II, whereas NLS2 is within the C2-like domain, also known as domain III (1, 15). Although these conserved domains are present in both classical and non-classical members of the calpain family, the NLS sequences themselves are not present in other calpains. The NLS1 RRRK sequence is present in gorillas and bats, whereas one arginine is lost (RRK) in the NLS1 in rhesus monkey and other mammals including rats and mice. NLS2 is unique to CAPN5 and highly conserved in mammals, with 100% identity in human and rat and a two-amino acid differences in mouse. With an occasional mismatch, the putative NLS2 aligned with CAPN5 protein in mammals, reptiles, amphibians, and fish.
The CAPN5 NLS2 sequence, YIFEVKKPE DEVLICIQQRPKRSTRREG (aa 390–417), is sufficient for nuclear targeting and consistent with consensus sequences in bipartite NLS, including acidic amino acids in the central linker region, absence of hydrophobic and basic residues in the central region, and proline in the terminal region (48, 56). However, mutagenesis of the basic residues did not abolish nuclear localization, suggesting that NLS2 may function independently of binding to importins. Based on its association with nuclear PML bodies, CAPN5 should also be SUMOylated and/or contain a SIM (32). SIM features include an acidic domain adjacent to a string of three of Val, Ile, and/or Leu amino acids (55–57). A putative SIM (KPEDEVLICI, aa 396–405) in CAPN5 is contained within NLS2. Mutating Ile403 within NLS2 to Ala aborts the nuclear localization, indicating that the SIM motif plays a critical role in both the nuclear import and PML localization of CAPN5. Other PML partner proteins that contain SIMs include Daxx and SP100 in addition to PML itself (57, 58).
SUMOylated PML proteins link to PML nuclear bodies via electrostatic interaction between the PML SUMO moiety and hydrophobic core of partner proteins containing a SIM (45). Analysis of the CAPN5 sequence using SUMOylation prediction algorithms SUMOsp (59) and SUMOplot reveals several high probability SUMOylation sites, including one at Lys395 within the NLS. CAPN5 has been identified previously as a candidate SUMO1-conjugated protein (60). The observation that the mutation of basic residues attenuated the punctate nuclear localization, but not nuclear import, of CAPN5 suggests that SUMOylation may be involved in targeting CAPN5 to the PML nuclear bodies. Daxx, a transcriptional regulator that associates with nuclear PML bodies, also contains two NLS signals and both SIM and SUMO motifs, similar to CAPN5 (61, 62). Daxx influences transcriptional control by acting as a co-repressor to SUMOylated transcription factors that bind to the SIM on Daxx (57, 63).
Consistent with the nuclear localization of CAPN5, a previous genetic association and gene-gene interaction analysis suggests interaction between CAPN5 and the nuclear peroxisome proliferator-activated receptor-δ (64). Calpains have been implicated in the cleavage of nuclear androgen and estrogen receptors, although the specific calpain isoform has not been identified (65–68). CAPN5, along with calpains 1, 7, and 10, was detected in both the nuclear and cytosolic fractions of 293T cells, where the nuclear activity is hypothesized to contribute to the cleavage and toxicity of huntingtin (23). Of related interest is that PML associates with nuclear aggregates in several neurodegenerative disorders, particularly polyglutamine disorders including spinocerebellar ataxia, Huntington disease, and dentatorubral-pallidoluysian atrophy as well as amyotrophic lateral sclerosis (69–76). Several of the aggregated proteins have been demonstrated to be calpain substrates (23, 77–80), with calpain inhibition attenuating the nuclear aggregation (23, 78, 81).
In summary, the results of the present study demonstrate that Capn5 mRNA is expressed at relatively high levels in the CNS, and CAPN5 is a non-cytosolic calpain localized predominantly to the nucleus where it associates with PML nuclear bodies. CAPN5 contains dual NLS: a monopartite NLS1 and bipartite NLS2, which includes a SIM critical to the punctate nuclear localization. Based on the nuclear localization of CAPN5 and its association with nuclear PML bodies, subnuclear localization to the nucleic acid binding fraction, and similarity of the NLS sequences to those found in transcriptional regulators, it is tempting to speculate that CAPN5 is involved in transcriptional regulation, although this remains to be determined experimentally. CAPN5 may also participate in the cleavage of nuclear receptors. Under pathologic conditions, CAPN5 activation may contribute to the nuclear aggregation and accumulation of protein fragments.
This work was supported, in whole or in part, by National Institutes of Health Grants P01NS058484 and P30NS051220. This work was also supported by the Kentucky Spinal Cord and Head Injury Research Trust.
- CAPN
- calpain
- aa
- amino acids
- APC
- adenomatous polyposis coli (tumor suppressor protein)
- GFAP
- glial fibrillary acidic protein
- INP
- integrated nuclear protein
- NABP
- nucleic acid-binding protein
- NeuN
- neuronal nuclei
- NGS
- natural goat serum
- NLS
- nuclear localization signal
- PML
- promyelocytic leukemia protein
- SIM
- SUMO-interacting motif
- bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Ono Y., Sorimachi H. (2012) Calpains: an elaborate proteolytic system. Biochim. Biophys. Acta 1824, 224–236 [DOI] [PubMed] [Google Scholar]
- 2. Goll D. E., Thompson V. F., Li H., Wei W., Cong J. (2003) The calpain system. Physiol. Rev. 83, 731–801 [DOI] [PubMed] [Google Scholar]
- 3. Franco S. J., Huttenlocher A. (2005) Regulating cell migration: calpains make the cut. J. Cell Sci. 118, 3829–3838 [DOI] [PubMed] [Google Scholar]
- 4. Azam M., Andrabi S. S., Sahr K. E., Kamath L., Kuliopulos A., Chishti A. H. (2001) Disruption of the mouse μ-calpain gene reveals an essential role in platelet function. Mol. Cell. Biol. 21, 2213–2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang K. K. (2000) Calpain and caspase: can you tell the difference? Trends Neurosci. 23, 20–26 [DOI] [PubMed] [Google Scholar]
- 6. Liu X., Van Vleet T., Schnellmann R. G. (2004) The role of calpain in oncotic cell death. Annu. Rev. Pharmacol. Toxicol. 44, 349–370 [DOI] [PubMed] [Google Scholar]
- 7. Santos D. M., Xavier J. M., Morgado A. L., Solá S., Rodrigues C. M. (2012) Distinct regulatory functions of calpain 1 and 2 during neural stem cell self-renewal and differentiation. PLoS One 7, e33468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Mellgren R. L., Zhang W., Miyake K., McNeil P. L. (2007) Calpain is required for the rapid, calcium-dependent repair of wounded plasma membrane. J. Biol. Chem. 282, 2567–2575 [DOI] [PubMed] [Google Scholar]
- 9. Horton P., Park K. J., Obayashi T., Fujita N., Harada H., Adams-Collier C. J., Nakai K. (2007) WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Maki M., Maemoto Y., Osako Y., Shibata H. (2012) Evolutionary and physical linkage between calpains and penta-EF-hand Ca2+-binding proteins. FEBS J. 279, 1414–1421 [DOI] [PubMed] [Google Scholar]
- 11. Joyce P. I., Satija R., Chen M., Kuwabara P. E. (2012) The atypical calpains: evolutionary analyses and roles in Caenorhabditis elegans cellular degeneration. PLoS Genet. 8, e1002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Barnes T. M., Hodgkin J. (1996) The tra-3 sex determination gene of Caenorhabditis elegans encodes a member of the calpain regulatory protease family. EMBO J. 15, 4477–4484 [PMC free article] [PubMed] [Google Scholar]
- 13. Syntichaki P., Xu K., Driscoll M., Tavernarakis N. (2002) Specific aspartyl and calpain proteases are required for neurodegeneration in C. elegans. Nature 419, 939–944 [DOI] [PubMed] [Google Scholar]
- 14. Matena K., Boehm T., Dear N. (1998) Genomic organization of mouse Capn5 and Capn6 genes confirms that they are a distinct calpain subfamily. Genomics 48, 117–120 [DOI] [PubMed] [Google Scholar]
- 15. Sorimachi H., Hata S., Ono Y. (2011) Calpain chronicle: an enzyme family under multidisciplinary characterization. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 87, 287–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dear N., Matena K., Vingron M., Boehm T. (1997) A new subfamily of vertebrate calpains lacking a calmodulin-like domain: implications for calpain regulation and evolution. Genomics 45, 175–184 [DOI] [PubMed] [Google Scholar]
- 17. Waghray A., Wang D. S., McKinsey D., Hayes R. L., Wang K. K. (2004) Molecular cloning and characterization of rat and human calpain-5. Biochem. Biophys. Res. Commun. 324, 46–51 [DOI] [PubMed] [Google Scholar]
- 18. Franz T., Winckler L., Boehm T., Dear T. N. (2004) Capn5 is expressed in a subset of T cells and is dispensable for development. Mol. Cell. Biol. 24, 1649–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mahajan V. B., Skeie J. M., Bassuk A. G., Fingert J. H., Braun T. A., Daggett H. T., Folk J. C., Sheffield V. C., Stone E. M. (2012) Calpain-5 mutations cause autoimmune uveitis, retinal neovascularization, and photoreceptor degeneration. PLoS Genet. 8, e1003001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. González A., Sáez M. E., Aragón M. J., Galán J. J., Vettori P., Molina L., Rubio C., Real L. M., Ruiz A., Ramírez-Lorca R. (2006) Specific haplotypes of the CALPAIN-5 gene are associated with polycystic ovary syndrome. Hum. Reprod. 21, 943–951 [DOI] [PubMed] [Google Scholar]
- 21. Penna I., Du H., Ferriani R., Taylor H. S. (2008) Calpain5 expression is decreased in endometriosis and regulated by HOXA10 in human endometrial cells. Mol. Hum. Reprod. 14, 613–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sáez M. E., Martínez-Larrad M. T., Ramírez-Lorca R., González-Sánchez J. L., Zabena C., Martinez-Calatrava M. J., González A., Morón F. J., Ruiz A., Serrano-Ríos M. (2007) Calpain-5 gene variants are associated with diastolic blood pressure and cholesterol levels. BMC Med. Genet. 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gafni J., Hermel E., Young J. E., Wellington C. L., Hayden M. R., Ellerby L. M. (2004) Inhibition of calpain cleavage of huntingtin reduces toxicity: accumulation of calpain/caspase fragments in the nucleus. J. Biol. Chem. 279, 20211–20220 [DOI] [PubMed] [Google Scholar]
- 24. Moore M. W. (2005) High-throughput gene knockouts and phenotyping in mice. Ernst Schering Res. Found. Workshop 50, 27–44 [DOI] [PubMed] [Google Scholar]
- 25. Schmittgen T. D., Livak K. J. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3, 1101–1108 [DOI] [PubMed] [Google Scholar]
- 26. Kageyama G. H., Wong-Riley M. T. (1982) Histochemical localization of cytochrome oxidase in the hippocampus: correlation with specific neuronal types and afferent pathways. Neuroscience 7, 2337–2361 [DOI] [PubMed] [Google Scholar]
- 27. Yoshimura N., Hatanaka M., Kitahara A., Kawaguchi N., Murachi T. (1984) Intracellular localization of two distinct Ca2+-proteases (calpain I and calpain II) as demonstrated by using discriminative antibodies. J. Biol. Chem. 259, 9847–9852 [PubMed] [Google Scholar]
- 28. Suzuki K., Hata S., Kawabata Y., Sorimachi H. (2004) Structure, activation, and biology of calpain. Diabetes 53, S12–S18 [DOI] [PubMed] [Google Scholar]
- 29. Dundr M. (2012) Nuclear bodies: multifunctional companions of the genome. Curr. Opin. Cell Biol. 24, 415–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pagni M., Ioannidis V., Cerutti L., Zahn-Zabal M., Jongeneel C. V., Hau J., Martin O., Kuznetsov D., Falquet L. (2007) MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res. 35, W433–W437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brameier M., Krings A., MacCallum R. M. (2007) NucPred: predicting nuclear localization of proteins. Bioinformatics 23, 1159–1160 [DOI] [PubMed] [Google Scholar]
- 32. Shen T. H., Lin H. K., Scaglioni P. P., Yung T. M., Pandolfi P. P. (2006) The mechanisms of PML-nuclear body formation. Mol. Cell 24, 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lallemand-Breitenbach V., de Thé H. (2010) PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2, a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nardozzi J. D., Lott K., Cingolani G. (2010) Phosphorylation meets nuclear import: a review. Cell Commun. Signal. 8, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. König N., Raynaud F., Feane H., Durand M., Mestre-Francès N., Rossel M., Ouali A., Benyamin Y. (2003) Calpain 3 is expressed in astrocytes of rat and Microcebus brain. J. Chem. Neuroanat. 25, 129–136 [DOI] [PubMed] [Google Scholar]
- 36. Ma H., Fukiage C., Kim Y. H., Duncan M. K., Reed N. A., Shih M., Azuma M., Shearer T. R. (2001) Characterization and expression of calpain 10: a novel ubiquitous calpain with nuclear localization. J. Biol. Chem. 276, 28525–28531 [DOI] [PubMed] [Google Scholar]
- 37. Shin S. J., Lee S. E., Boo J. H., Kim M., Yoon Y. D., Kim S. I., Mook-Jung I. (2004) Profiling proteins related to amyloid deposited brain of Tg2576 mice. Proteomics 4, 3359–3368 [DOI] [PubMed] [Google Scholar]
- 38. Garcia M., Bondada V., Geddes J. W. (2005) Mitochondrial localization of μ-calpain. Biochem. Biophys. Res. Commun. 338, 1241–1247 [DOI] [PubMed] [Google Scholar]
- 39. Raynaud F., Marcilhac A., Chebli K., Benyamin Y., Rossel M. (2008) Calpain 2 expression pattern and subcellular localization during mouse embryogenesis. Int. J. Dev. Biol. 52, 383–388 [DOI] [PubMed] [Google Scholar]
- 40. Arrington D. D., Van Vleet T. R., Schnellmann R. G. (2006) Calpain 10: a mitochondrial calpain and its role in calcium-induced mitochondrial dysfunction. Am. J. Physiol. Cell Physiol. 291, C1159–C1171 [DOI] [PubMed] [Google Scholar]
- 41. Ozaki T., Yamashita T., Ishiguro S. (2009) Mitochondrial m-calpain plays a role in the release of truncated apoptosis-inducing factor from the mitochondria. Biochim. Biophys. Acta 1793, 1848–1859 [DOI] [PubMed] [Google Scholar]
- 42. Stuurman N., de Graaf A., Floore A., Josso A., Humbel B., de Jong L., van Driel R. (1992) A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 101, 773–784 [DOI] [PubMed] [Google Scholar]
- 43. de Thé H., Le Bras M., Lallemand-Breitenbach V. (2012) The cell biology of disease: Acute promyelocytic leukemia, arsenic, and PML bodies. J. Cell Biol. 198, 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Borden K. L. (2002) Pondering the promyelocytic leukemia protein (PML) puzzle: possible functions for PML nuclear bodies. Mol. Cell. Biol. 22, 5259–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bernardi R., Pandolfi P. P. (2007) Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 8, 1006–1016 [DOI] [PubMed] [Google Scholar]
- 46. Lange A., Mills R. E., Lange C. J., Stewart M., Devine S. E., Corbett A. H. (2007) Classical nuclear localization signals: definition, function, and interaction with importin α. J. Biol. Chem. 282, 5101–5105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robbins J., Dilworth S. M., Laskey R. A., Dingwall C. (1991) Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: identification of a class of bipartite nuclear targeting sequence. Cell 64, 615–623 [DOI] [PubMed] [Google Scholar]
- 48. Marfori M., Mynott A., Ellis J. J., Mehdi A. M., Saunders N. F., Curmi P. M., Forwood J. K., Bodén M., Kobe B. (2011) Molecular basis for specificity of nuclear import and prediction of nuclear localization. Biochim. Biophys. Acta 1813, 1562–1577 [DOI] [PubMed] [Google Scholar]
- 49. Görlich D., Vogel F., Mills A. D., Hartmann E., Laskey R. A. (1995) Distinct functions for the two importin subunits in nuclear protein import. Nature 377, 246–248 [DOI] [PubMed] [Google Scholar]
- 50. Pang X., Zhou H. X. (2014) Design rules for selective binding of nuclear localization signals to minor site of importin α. PLoS One 9, e91025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Palmeri D., Malim M. H. (1999) Importin β can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin alpha. Mol. Cell. Biol. 19, 1218–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Truant R., Cullen B. R. (1999) The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin β-dependent nuclear localization signals. Mol. Cell. Biol. 19, 1210–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Forwood J. K., Harley V., Jans D. A. (2001) The C-terminal nuclear localization signal of the sex-determining region Y (SRY) high mobility group domain mediates nuclear import through importin β1. J. Biol. Chem. 276, 46575–46582 [DOI] [PubMed] [Google Scholar]
- 54. Südbeck P., Scherer G. (1997) Two independent nuclear localization signals are present in the DNA-binding high-mobility group domains of SRY and SOX9. J. Biol. Chem. 272, 27848–27852 [DOI] [PubMed] [Google Scholar]
- 55. Kaur G., Jans D. A. (2011) Dual nuclear import mechanisms of sex determining factor SRY: intracellular Ca2+ as a switch. FASEB J. 25, 665–675 [DOI] [PubMed] [Google Scholar]
- 56. Kosugi S., Hasebe M., Matsumura N., Takashima H., Miyamoto-Sato E., Tomita M., Yanagawa H. (2009) Six classes of nuclear localization signals specific to different binding grooves of importin α. J. Biol. Chem. 284, 478–485 [DOI] [PubMed] [Google Scholar]
- 57. Santiago A., Godsey A. C., Hossain J., Zhao L. Y., Liao D. (2009) Identification of two independent SUMO-interacting motifs in Daxx: evolutionary conservation from Drosophila to humans and their biochemical functions. Cell Cycle 8, 76–87 [DOI] [PubMed] [Google Scholar]
- 58. Sternsdorf T., Jensen K., Reich B., Will H. (1999) The nuclear dot protein sp100, characterization of domains necessary for dimerization, subcellular localization, and modification by small ubiquitin-like modifiers. J. Biol. Chem. 274, 12555–12566 [DOI] [PubMed] [Google Scholar]
- 59. Xue Y., Zhou F., Fu C., Xu Y., Yao X. (2006) SUMOsp: a Web server for sumoylation site prediction. Nucleic Acids Res. 34, W254–W257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tirard M., Hsiao H. H., Nikolov M., Urlaub H., Melchior F., Brose N. (2012) In vivo localization and identification of SUMOylated proteins in the brain of His6-HA-SUMO1 knock-in mice. Proc. Natl. Acad. Sci. U.S.A. 109, 21122–21127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yeung P. L., Chen L. Y., Tsai S. C., Zhang A., Chen J. D. (2008) Daxx contains two nuclear localization signals and interacts with importin alpha3. J. Cell. Biochem. 103, 456–470 [DOI] [PubMed] [Google Scholar]
- 62. Wang L., Huang C., Yang M. Q., Yang J. Y. (2010) BindN+ for accurate prediction of DNA and RNA-binding residues from protein sequence features. BMC Syst. Biol. 4, S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lin D. Y., Huang Y. S., Jeng J. C., Kuo H. Y., Chang C. C., Chao T. T., Ho C. C., Chen Y. C., Lin T. P., Fang H. I., Hung C. C., Suen C. S., Hwang M. J., Chang K. S., Maul G. G., Shih H. M. (2006) Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 24, 341–354 [DOI] [PubMed] [Google Scholar]
- 64. Sáez M. E., Grilo A., Morón F. J., Manzano L., Martínez-Larrad M. T., González-Pérez A., Serrano-Hernando J., Ruiz A., Ramírez-Lorca R., Serrano-Ríos M. (2008) Interaction between calpain 5, peroxisome proliferator-activated receptor-γ, and peroxisome proliferator-activated receptor-δ genes: a polygenic approach to obesity. Cardiovasc. Diabetol. 7, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yang H., Murthy S., Sarkar F. H., Sheng S., Reddy G. P., Dou Q. P. (2008) Calpain-mediated androgen receptor breakdown in apoptotic prostate cancer cells. J. Cell Physiol. 217, 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Libertini S. J., Tepper C. G., Rodriguez V., Asmuth D. M., Kung H. J., Mudryj M. (2007) Evidence for calpain-mediated androgen receptor cleavage as a mechanism for androgen independence. Cancer Res. 67, 9001–9005 [DOI] [PubMed] [Google Scholar]
- 67. Pelley R. P., Chinnakannu K., Murthy S., Strickland F. M., Menon M., Dou Q. P., Barrack E. R., Reddy G. P. (2006) Calmodulin-androgen receptor (AR) interaction: calcium-dependent, calpain-mediated breakdown of AR in LNCaP prostate cancer cells. Cancer Res. 66, 11754–11762 [DOI] [PubMed] [Google Scholar]
- 68. Murayama A., Fukai F., Murachi T. (1984) Action of calpain on the basic estrogen receptor molecule of porcine uterus. J. Biochem. 95, 1697–1704 [DOI] [PubMed] [Google Scholar]
- 69. Kaytor M. D., Duvick L. A., Skinner P. J., Koob M. D., Ranum L. P., Orr H. T. (1999) Nuclear localization of the spinocerebellar ataxia type 7 protein, ataxin-7. Hum. Mol. Genet. 8, 1657–1664 [DOI] [PubMed] [Google Scholar]
- 70. Skinner P. J., Koshy B. T., Cummings C. J., Klement I. A., Helin K., Servadio A., Zoghbi H. Y., Orr H. T. (1997) Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature 389, 971–974 [DOI] [PubMed] [Google Scholar]
- 71. Seilhean D., Takahashi J., El Hachimi K. H., Fujigasaki H., Lebre A. S., Biancalana V., Dürr A., Salachas F., Hogenhuis J., de Thé H., Hauw J. J., Meininger V., Brice A., Duyckaerts C. (2004) Amyotrophic lateral sclerosis with neuronal intranuclear protein inclusions. Acta Neuropathol. 108, 81–87 [DOI] [PubMed] [Google Scholar]
- 72. Takahashi J., Fujigasaki H., Iwabuchi K., Bruni A. C., Uchihara T., El Hachimi K. H., Stevanin G., Dürr A., Lebre A. S., Trottier Y., de Thé H., Tanaka J., Hauw J. J., Duyckaerts C., Brice A. (2003) PML nuclear bodies and neuronal intranuclear inclusion in polyglutamine diseases. Neurobiol. Dis. 13, 230–237 [DOI] [PubMed] [Google Scholar]
- 73. Takahashi J., Fujigasaki H., Zander C., El Hachimi K. H., Stevanin G., Dürr A., Lebre A. S., Yvert G., Trottier Y., de Thé H., Hauw J. J., Duyckaerts C., Brice A. (2002) Two populations of neuronal intranuclear inclusions in SCA7 differ in size and promyelocytic leukaemia protein content. Brain 125, 1534–1543 [DOI] [PubMed] [Google Scholar]
- 74. Yamada M., Sato T., Shimohata T., Hayashi S., Igarashi S., Tsuji S., Takahashi H. (2001) Interaction between neuronal intranuclear inclusions and promyelocytic leukemia protein nuclear and coiled bodies in CAG repeat diseases. Am. J. Pathol. 159, 1785–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yamada M., Wood J. D., Shimohata T., Hayashi S., Tsuji S., Ross C. A., Takahashi H. (2001) Widespread occurrence of intranuclear atrophin-1 accumulation in the central nervous system neurons of patients with dentatorubral-pallidoluysian atrophy. Ann. Neurol. 49, 14–23 [DOI] [PubMed] [Google Scholar]
- 76. Fu L., Gao Y. S., Sztul E. (2005) Transcriptional repression and cell death induced by nuclear aggregates of non-polyglutamine protein. Neurobiol. Dis. 20, 656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Simões A. T., Gonçalves N., Koeppen A., Déglon N., Kügler S., Duarte C. B., Pereira de Almeida L. (2012) Calpastatin-mediated inhibition of calpains in the mouse brain prevents mutant ataxin 3 proteolysis, nuclear localization and aggregation, relieving Machado-Joseph disease. Brain 135, 2428–2439 [DOI] [PubMed] [Google Scholar]
- 78. Hübener J., Weber J. J., Richter C., Honold L., Weiss A., Murad F., Breuer P., Wüllner U., Bellstedt P., Paquet-Durand F., Takano J., Saido T. C., Riess O., Nguyen H. P. (2013) Calpain-mediated ataxin-3 cleavage in the molecular pathogenesis of spinocerebellar ataxia type 3 (SCA3). Hum. Mol. Genet. 22, 508–518 [DOI] [PubMed] [Google Scholar]
- 79. Schilling B., Gafni J., Torcassi C., Cong X., Row R. H., LaFevre-Bernt M. A., Cusack M. P., Ratovitski T., Hirschhorn R., Ross C. A., Gibson B. W., Ellerby L. M. (2006) Huntingtin phosphorylation sites mapped by mass spectrometry: modulation of cleavage and toxicity. J. Biol. Chem. 281, 23686–23697 [DOI] [PubMed] [Google Scholar]
- 80. Kim M., Roh J. K., Yoon B. W., Kang L., Kim Y. J., Aronin N., DiFiglia M. (2003) Huntingtin is degraded to small fragments by calpain after ischemic injury. Exp. Neurol. 183, 109–115 [DOI] [PubMed] [Google Scholar]
- 81. Haacke A., Hartl F. U., Breuer P. (2007) Calpain inhibition is sufficient to suppress aggregation of polyglutamine-expanded ataxin-3. J. Biol. Chem. 282, 18851–18856 [DOI] [PubMed] [Google Scholar]
- 82. Mullen R. J., Buck C. R., Smith A. M. (1992) NeuN, a neuronal specific nuclear protein in vertebrates. Development 116, 201–211 [DOI] [PubMed] [Google Scholar]
- 83. Bignami A., Dahl D. (1974) Astrocyte-specific protein and neuroglial differentiation. An immunofluorescence study with antibodies to the glial fibrillary acidic protein. J. Comp. Neurol. 153, 27–38 [DOI] [PubMed] [Google Scholar]
- 84. Bhat R. V., Axt K. J., Fosnaugh J. S., Smith K. J., Johnson K. A., Hill D. E., Kinzler K. W., Baraban J. M. (1996) Expression of the APC tumor suppressor protein in oligodendroglia. Glia 17, 169–174 [DOI] [PubMed] [Google Scholar]
- 85. Fischer U., Liu Q., Dreyfuss G. (1997) The SMN-SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell 90, 1023–1029 [DOI] [PubMed] [Google Scholar]
- 86. Liu Q., Dreyfuss G. (1996) A novel nuclear structure containing the survival of motor neurons protein. EMBO J 15, 3555–3565 [PMC free article] [PubMed] [Google Scholar]






