Background: Rab5a GTPase plays important roles in intracellular transport and cell signaling.
Results: T-cell stimulation through the integrin LFA-1 or the chemokine receptor CXCR4 induces PKCϵ-dependent phosphorylation of Rab5a at Thr-7, which is crucial for cytoskeleton remodeling and cell migration.
Conclusion: PKCϵ-Rab5a-Rac1 axis regulates T-cell motility.
Significance: The study provides novel insights into the role of Rab5a in the adaptive immune response.
Keywords: Cell Migration, Cell Signaling, Immunology, Integrin, Phosphorylation, Rab Proteins, Signal Transduction, T-cell
Abstract
Rab GTPases control membrane traffic and receptor-mediated endocytosis. Within this context, Rab5a plays an important role in the spatial regulation of intracellular transport and signal transduction processes. Here, we report a previously uncharacterized role for Rab5a in the regulation of T-cell motility. We show that Rab5a physically associates with protein kinase Cϵ (PKCϵ) in migrating T-cells. After stimulation of T-cells through the integrin LFA-1 or the chemokine receptor CXCR4, Rab5a is phosphorylated on an N-terminal Thr-7 site by PKCϵ. Both Rab5a and PKCϵ dynamically interact at the centrosomal region of migrating cells, and PKCϵ-mediated phosphorylation on Thr-7 regulates Rab5a trafficking to the cell leading edge. Furthermore, we demonstrate that Rab5a Thr-7 phosphorylation is functionally necessary for Rac1 activation, actin rearrangement, and T-cell motility. We present a novel mechanism by which a PKCϵ-Rab5a-Rac1 axis regulates cytoskeleton remodeling and T-cell migration, both of which are central for the adaptive immune response.
Introduction
Leukocyte trafficking is a key element of an immune response that is critically dependent on a multifunctional molecular array of integrin-mediated adhesions and chemokine signals. Interaction of the αLβ2 integrin leukocyte function associated-antigen-1 (LFA-1), expressed on T-cells, with its ligand intercellular adhesion molecule-1 (ICAM-1), expressed on the surface of high endothelial venules and other cell types, promotes firm adhesion of T-cells to blood vessel walls and their migration through endothelial junctions into secondary lymphoid organs and sites of inflammation (1–3). In addition, chemokines (for example the stromal cell-derived factor-1α (SDF-1α)) and their interactions with specific receptors on T-cells (such as C-X-C chemokine receptor type 4 (CXCR4)) direct T-cells to arrest on post-capillary venules at sites of infection/injury and secondary lymphoid organs (4). This multistep process of T-cell migration is precisely regulated by a plethora of signaling molecules including kinases, adaptors, and motor proteins that ultimately result in the reorganization of cytoskeletal systems and provide mechanical force to propel the cell forward. Emerging evidence suggests an important role for endocytic transport and vesicular traffic in the process of cytoskeletal remodeling and integrin-mediated cell motility (5–8).
Small GTPases of the Rab family are known to function as molecular switches that regulate a variety of cellular processes including proliferation, differentiation, signal transduction, and cytoskeletal reorganization (9–11). In particular, they control intracellular vesicle transport, such as receptor-mediated endocytosis, exocytosis, and receptor recycling (12–14), all of which are important for cell migration. Rab GTPases cycle between a GDP-bound inactive state and a GTP-bound active state. Inactive GDP-bound Rab proteins typically reside in the cytosol, whereas GTP-bound active Rab proteins localize on intracellular membranes where effector targets reside. Thus, the Rab family of proteins typically require turnover of GTP/GDP to function. This reversible control of Rab GTPase activity via GDP/GTP exchange is crucial to their function as regulators of vesicular trafficking (15). One member of the Rab GTPase family, Rab5a, is known to regulate endocytosis, endosomal dynamics, tethering, and fusion of early endosomes. Rab5a contributes to cytoskeletal remodeling in part by regulating the endocytic trafficking of Rac, a member of the Rho GTPase subfamily and an important component of the actin cytoskeletal system (16–18).
The protein kinase C (PKC) family comprises a group of highly related multifunctional serine/threonine kinases that phosphorylate a wide variety of protein targets and play important roles in cellular signaling pathways including those involved in cell migration (19). More than 10 isoforms of PKC have been identified in mammalian tissues, each of which plays a distinct role in the processing and modulation of a variety of physiologic and pathologic responses to external signals. There is substantial evidence that members of the PKC family regulate integrin-mediated cell spreading and migration (20–23). For example, we have established that signaling through the integrin LFA-1 in T-cells results in the association of PKCβ with the centrosome and microtubule cytoskeleton, a process that is essential for cell migration (21). In addition, another PKC family member PKCϵ has been identified as a regulator of β1 integrin-dependent cell migration (23–25).
Although extensive progress has been made to understand T-cell signal transduction pathways, the potential involvement of Rab5a in T-cell migration has not been explored to the similar extent. Moreover, the precise contribution of the PKC isoforms in LFA-1-mediated downstream signaling has remained unclear. In the current study we demonstrate that T-cell stimulation through the integrin LFA-1 or the chemokine receptor CXCR4 triggers PKCϵ-dependent phosphorylation of Rab5a at threonine 7 (Thr-7), which is crucial for Rac1 activation, actin cytoskeleton remodeling, and T-cell migration.
EXPERIMENTAL PROCEDURES
Cells and Reagents
Primary human peripheral blood lymphocyte (PBL) T-cells were isolated from healthy volunteers by standard methods and cultured with phytohemagglutinin and IL-2 as described previously (26). The human T-cell line HuT-78 was obtained from the American Type Culture collection (ATCC) and cultured as described (27–30). Motility-inducing anti-LFA-1 (clone SPV-L7) was from Monosan (Cambridge Biosciences). Human recombinant ICAM-1-Fc was from R&D Systems. Recombinant human IL-2 and SDF-1α were from Peprotech. Phytohemagglutinin, goat anti-human IgG (Fc-specific), goat anti-mouse IgG, mouse anti-α-tubulin, phalloidin-TRITC, and protein A-agarose beads were from Sigma. Mouse anti-PKCϵ and mouse anti-Rac1 antibodies were from BD Biosciences. Mouse anti-Rab5a was from Synaptic Systems GmbH. Rabbit anti-Rab5a and anti-PKCϵ were from Santa Cruz Biotechnology, Inc. HRP-conjugated anti-rabbit and anti-mouse antibodies were from Cell Signaling Technology. The PKC inhibitor bisindolylmaleimide I was from Merck Millipore. Alexa Fluor® 488- and 568-conjugated secondary antibodies, Hoechst 33258, and Alexa Fluor® 568-transferrin conjugate were purchased from Molecular Probes Invitrogen.
Induction of LFA-1-mediated T-cell Migration
We used our well characterized migration-triggering model system where T-cells are stimulated through the LFA-1 receptor via cross-linking with an immobilized anti-LFA-1 antibody or with the physiological ligand ICAM-1 (21, 26–29). These migrating T-cells display dynamic protrusions of the leading edge, flexible oscillatory shape changes of the cell body, and detachment of the trailing edge concomitant with polarized or localized distribution of signaling molecules and cytoskeletal rearrangements. Briefly, 96-well tissue culture plates (flat bottom, NuncTM), 8-well LabTek chamber slides (Nalge Nunc Inc.), or round coverglass, depending on the particular assay type, were coated overnight at 4 °C with 5 μg/ml goat anti-mouse IgG in sterile phosphate-buffered saline (PBS). After washing with PBS, the surface was then incubated with a cross-linking anti-LFA-1 antibody (SPV-L7) for 2 h at 37 °C. ICAM-1-coated surfaces were prepared by adding 5 μg/ml anti-Fc-specific goat anti-human IgG to the culture plates/chamber slides and incubating overnight at 4 °C followed by 1 μg/ml rICAM-1-Fc for 2 h at 37 °C. After washing the surfaces with PBS, HuT-78 or PBL T-cells were loaded into the coated wells and incubated in 5% CO2 at 37 °C for various time points as indicated in “Results” and figure legends of the particular experiment. Migration assays on ICAM-1 also contained 5 mm MgCl2 and 1.5 mm EGTA in the cell culture medium to induce the high affinity form of the LFA-1 receptor on T cells (11).
Plasmid Constructs and Electroporation
Plasmid vector encoding Rab5a fused to green fluorescent protein (Rab5a-GFP), Rab5aQ79L-GFP, and Rab5aS34N-GFP were generous gifts from M. Zerial, Max Planck Institute of Molecular Cell Biology and Genetics. PKCϵ-GFP and kinase-dead (KD) PKCϵ-GFP were kind gifts from D. Romberger, Department of Internal Medicine, University of Nebraska. Lifeact-Ruby (31) was a generous gift from R. Wedlich-Soldner, Max Planck Institute of Biochemistry. Site-directed mutagenesis of Thr-7, Ser-84, and Ser-123 residues of Rab5a to alanine or glutamic acid was performed by Bio S&T Inc., Montreal, Quebec. Plasmid vectors expressing wild-type or Thr-7 mutant Rab5a as a photoactivable GFP fusion (Rab5a-paGFP or Rab5aT7A-paGFP constructs, respectively) were made by replacing the eGFP cDNA with paGFP cDNA (32), a generous gift from J. Lippincott-Schwartz, Cell Biology and Metabolism Branch, National Institutes of Health. The eGFP cDNA of Rab5aT7A-GFP and PKCϵ-GFP constructs were replaced with the mCherry cDNA (a kind gift from R. Tsien, University of California, San Diego) to generate the corresponding cherry-tagged proteins. HuT-78 T-cells were nucleofected with plasmid constructs using the Amaxa NucleofectorTM system according to the manufacturer's recommendations.
RNA Interference
To deplete PKCϵ expression in primary PBL T-cells, a mixture of four siRNA duplexes (Dharmacon ON-TARGETplus SMARTpool® siRNA Reagents, Thermo Fisher Scientific) targeted against PKCϵ or nonspecific control siRNAs was used. Cells (5 × 106) were nucleofected with 1000 nm siRNA using the Amaxa NucleofectorTM system according to the manufacturer's instructions and then harvested after 72 h.
Production of Phospho-Thr-7-specific Rab5a Antibody
A polyclonal antibody against Thr-7-phosphorylated Rab5a was generated by immunizing two rabbits with a synthetic peptide comprising amino acids 2–12 (ASRGApTRPNGP, where pT is phosphorylated Thr) of human Rab5a (NeoMPS PolyPeptide). The total serum was then purified by affinity chromatography on phosphopeptide-coupled columns (Amino-link, Pierce) according to manufacturer's instructions. Briefly, 1 mg of Thr(P)-7 peptide or (OH)Thr-7 peptide was coupled to separate amino-link columns. A total of 0.5 ml of serum was first applied to the (OH)Thr-7-peptide-coupled column to remove nonspecific binding against the (OH)Thr-7 residue of Rab5a. The pre-cleared serum was then purified with a Thr(P)-7-peptide-coupled column and tested by ELISA for specificity and sensitivity.
Cell Lysis, Co-immunoprecipitation, and Western Immunoblotting
Cell lysis was performed as described previously (27) with minor modifications. Briefly, cells were washed with ice-cold PBS and lysed in the lysis buffer containing Triton X-100 (1%) and protease inhibitors phenylmethylsulfonyl fluoride (2 mm), leupeptin (10 μg/ml), and aprotinin (10 μg/ml). The protein content of the cell lysates was determined by the Bradford or BCA assay. The immunoprecipitation was performed as described previously (28, 33). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) of the cellular lysates or immunoprecipitates and subsequent Western immunoblotting using indicated antibodies were performed as described (28). Where indicated, affinity-purified or crude anti-phospho Rab5a(Thr-7) antiserum (1:2000 dilution in blocking buffer) was preincubated with 20 μg/ml non-phosphorylated Thr-7 peptide for 1 h at room temperature before incubation on the membranes. The immunoreactive bands were visualized using the LumiGLO® chemiluminescent detection system (Cell signaling Technology) and subsequent exposure to CL-XPosureTM light-sensitive film (Thermo Scientific). Densitometric analyses of the Western blots were performed by using ImageJ software.
In Vitro Kinase Assay
Rab5a was cloned into the GST expression vector pGEX-6p1, over-expressed in BL21(DE3)pLysS cells, and purified by using glutathione-Sepharose beads (GE Healthcare) according to the manufacturer's instructions. PKCϵ-dependent phosphorylation was measured by incorporation of 32Pi from [γ-32P]ATP. Purified recombinant Rab5a-GST fusion protein, GST alone, or myelin basic protein (MBP; 200 ng) was incubated in 50 μl of kinase assay buffer (40 mm Tris, pH 7.5, 40 mm MgCl2, 0.2 mm HEPES, pH 7.4, 0.2 mm DTT, 0.0002% Triton X-100, 0.3 μg/ml BSA) containing 1 μm ATP, 2 μCi [32P]ATP, 1 μm phorbol 12,13-dibutyrate, and 160 μm phosphatidylserine. After 20 min of incubation at 30 °C, the reaction was stopped by adding 5× SDS-PAGE sample buffer (250 mm Tris, pH 11.5, 10% SDS, 50% glycerol, and 25% β-mercaptoethanol) and analyzed by SDS-PAGE followed by autoradiography. In vitro kinase assays were also carried out as above using nonradioactive ATP (Sigma) in place of [γ-32P]ATP. After allowing the kinase reaction to proceed, SDS-PAGE sample buffer was added, and the samples were resolved on SDS-PAGE gels and probed by Western blotting with the phospho-T7 Rab5a antiserum.
Confocal Imaging and Photoactivation
For confocal imaging and analysis, cells were seeded to rest or migrate on coverslips as described above and then fixed with 3% (w/v) paraformaldehyde in PBS (27). T-cells were permeabilized with 0.3% (v/v) Triton X-100 in PBS and blocked with 5% w/v BSA in PBS for 30 min. After blocking, cells were incubated with primary antibodies for 1 h at room temperature. After washing, cells were incubated with Alexa Fluor® 488- or 568-conjugated secondary antibody for 1 h at room temperature. Cells were also stained with Hoechst to visualize nuclei or phalloidin-TRITC to visualize F-actin. After washing, cells were mounted in fluorescence mounting medium (Dako) and stored at 4 °C. Fluorescence microscopy was performed using a confocal microscope LSM 510 with a Plan-Apochromat differential interference contrast 63× oil objective and 1.4 numerical aperture (Carl Zeiss, Inc.). Images were analyzed using the LSM Imaging software (Carl Zeiss).
For photoactivation and confocal live-cell imaging, cells expressing photoactivable fluorescently labeled proteins were stimulated to migrate on coverslips as described earlier and then placed onto a heated chamber with the internal temperature set at 37 °C (PerkinElmer Life Sciences). Photoactivation was performed with a 405-nm laser using the photobleaching function of LSM Imaging software (Carl Zeiss Inc.) in a time-lapse mode. Generally, 1 pulse of the 405-nm laser was sufficient to activate paGFP so that it produced very bright fluorescence emission that was detected by excitation at 488 nm using a 500–530-nm band pass filter. At least 20 different microscopic fields were observed for each sample.
High Content Analysis
A high content analysis protocol for T-cell morphology analysis has been optimized and established in our laboratory as described (29, 34). Briefly, cells were seeded in triplicate on 96-well flat bottom plates precoated with either poly-l-lysine or anti-LFA-1 for 2 h. After washing, cells were fixed by incubating them for 20 min with 3% (w/v) paraformaldehyde in PBS. Attached cells were then stained for F-actin using phalloidin-TRITC, and the was nucleus stained using Hoechst. Plates were scanned (9 randomly selected fields/well at 20×) using an automated microscope IN Cell Analyzer 1000 (GE Healthcare), and the acquired images were automatically analyzed by IN Cell Investigator software (Version 1.6) using multitarget analysis bio-application module (GE Healthcare).
Transferrin Internalization
Cells were serum-starved for 1 h and stimulated to migrate on anti-LFA-1 as described above before incubating with Alexa Fluor® 568-transferrin conjugate for 30 min on ice. Cells were then rinsed twice in ice-cold PBS and transferred to 37 °C incubator for 10 min to allow internalization before being fixed with 3% (w/v) paraformaldehyde and imaged.
Transwell Migration Assay
Transwell chambers (5-μm pores; Corning Costar) were precoated with 5 μg/ml rICAM-1-Fc at 4 °C overnight and blocked with 5% (w/v) BSA for 1 h at 37 °C. Serum-starved T-cells were loaded in triplicate in the upper chambers and allowed to migrate toward 50 ng/ml SDF-1α-enriched serum-free medium in the lower wells at 37 °C. After 4 h, migrated cells in the lower wells were fixed and stained with Hoechst. IN Cell Analyzer 1000 (GE Healthcare) was employed to perform whole well cell counts. Data from at least three independent experiments were pooled by normalizing the counts with corresponding controls.
Determination of Rac1 Activity in Cells (PBD Binding Assay)
The capacity of Rac1-GTP to bind to GST-PBD (p21-activated kinase binding domain) beads was used to analyze the activity of this GTPase. Serum-starved T-cells (2 × 106) were stimulated with or without immobilized ICAM-1-Fc or SDF-1α for 10 min and lysed in 500 μl of lysis buffer as described (27). Rac1 activity in the cellular lysates was determined using Rac1 activation assay kit according to the manufacturer's instructions (Millipore). Briefly, cleared lysates were incubated overnight at 4 °C with glutathione-Sepharose 4B beads coupled to GST-PBD. After extensive washing, beads were boiled in SDS-PAGE loading buffer, and the amount of bound Rac1-GTP was detected by Western immunoblotting with an anti-Rac1 antibody. In all cases, whole-cell lysates were also analyzed for total Rac1 for normalization purposes. Bands were visualized and quantified as described above, and the Rac1-GTP levels were normalized to the input levels of total Rac1.
Statistical Analysis
The data are expressed as mean ± S.E. For comparison of two groups, p values were calculated by two-tailed unpaired Student's t test. In all cases p values <0.05 were considered to be statistically significant.
RESULTS
PKCϵ Localizes to Endosomes in Migrating T-cells and Associates with Rab5a
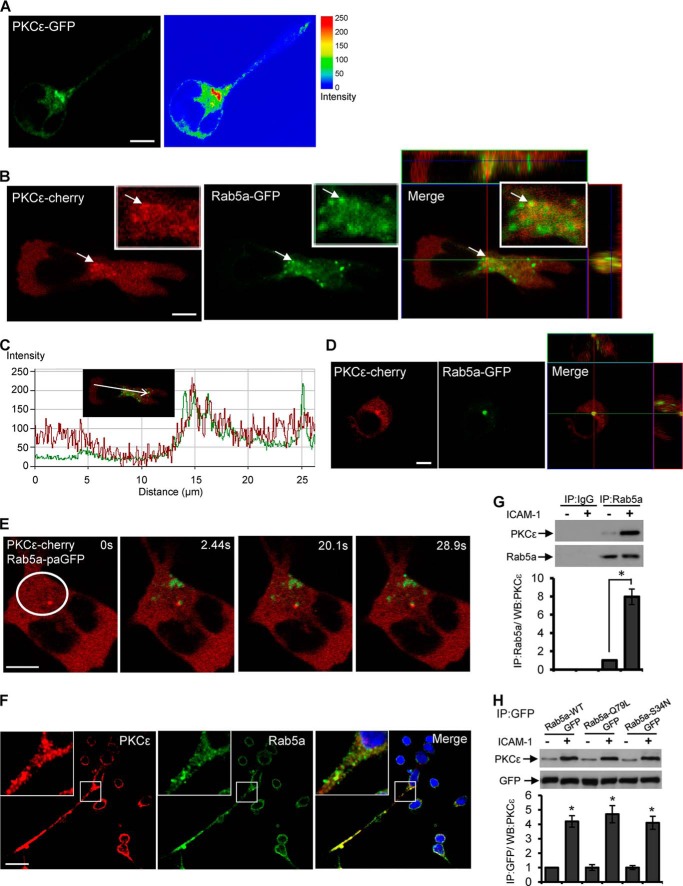
We investigated the subcellular distribution of PKCϵ in resting and migrating T-cells by transfecting the HuT-78 cell line with a plasmid construct encoding PKCϵ-GFP. After allowing the transfected cells to polarize on an anti-LFA-1 cross-linking antibody, we monitored the subcellular distribution of the PKCϵ-GFP expression by confocal microscopy. We noted that PKCϵ-GFP had a distinct localization, as it was distributed around the neck/centrosomal region in migrating cells and also displayed a vesicular pattern that was reminiscent of endosomes (Fig. 1A). To investigate whether PKCϵ was localized to endosomes in T-cells, we transfected HuT-78 cells with a PKCϵ-cherry expression vector in combination with a Rab5a-GFP construct that served as a marker of endosomes. Co-localization of PKCϵ and Rab5a was clearly evident at the neck region and throughout the cell body of migrating HuT-78 T-cells, as demonstrated by confocal microscopy and analysis (Fig. 1, B and C). Notably, both proteins also partially co-localized in resting T-cells (Fig. 1D).
FIGURE 1.
Interaction of PKCϵ and Rab5a in migrating T-cells. A, shown is a representative confocal image of HuT-78 T-cells expressing PKCϵ-GFP (green) stimulated to migrate on an anti-LFA-1 coated surface. The right panel shows an intensity plot generated by using LSM software. B, HuT-78 cells were co-transfected with PKCϵ-cherry (red, left panel) and Rab5a-GFP (green, middle panel) expression vectors, stimulated to migrate on an anti-LFA-1-coated surface, and imaged by confocal microscopy. The merged image (right panel) is an overlay of a series of 0.5-μm spacing Z stack images showing the yz (right) and xz (top) planes around the cell centrosomal region. Magnified images are shown in the corresponding insets, and the arrow indicates co-localized PKCϵ-Rab5a. C, co-localization analysis of the above image was performed along the direction of migration, as indicated, using LSM software. D, shown is a representative confocal fluorescent image of resting HuT-78 cells co-transfected with PKCϵ-cherry (red) and Rab5a-GFP (green) expression vectors. E, HuT-78 cells co-expressing PKCϵ-cherry (red) and Rab5a-paGFP (green) were stimulated to migrate on an anti-LFA-1 coated surface. Shown is a representative cell irradiated within the indicated region at the centrosomal region (white circle) and imaged with time-lapse confocal microscopy at 2.44, 20.1, and 28.9 s. F, primary human PBL T-cells were incubated on ICAM-1-coated plates for 30 min and after fixation immunostained for PKCϵ (red), Rab5a (green), and nuclei (blue). Magnified images are shown in the insets. G, PBL T-cells were left unstimulated or stimulated by rICAM-1 for 30 min and lysed. Cell lysates were immunoprecipitated (IP) with mouse monoclonal anti-Rab5a antibody or control IgG and then subjected to Western immunoblot (WB) analysis using rabbit anti-PKCϵ or anti-Rab5a antibody. H, HuT-78 cells were transfected with plasmid constructs encoding wild-type Rab5a-GFP (Rab5a-WT-GFP), constitutively active GTP-bound Rab5aQ79L mutant (Rab5a-Q79L-GFP), or dominant-negative GDP-bound Rab5aS34N mutant (Rab5a-S34N-GFP) and after 24 h stimulated with rICAM-1 for 30 min or left unstimulated. Cells were lysed, and cellular lysates were immunoprecipitated (IP) with mouse monoclonal anti-GFP and then subjected to Western immunoblot (WB) analysis using rabbit anti-PKCϵ or anti-GFP antibody. Relative densitometry values (mean ± S.E.) of protein bands in PKCϵ blots (G and H) are presented. Data represent three independent experiments and for microscopic imaging at least 20 fields per slide were visualized. Scale bar, 5 μm. *, p < 0.01.
We next examined the interactions between PKCϵ and Rab5a by live cell imaging of migrating T-cells. For this purpose, we expressed Rab5a as a photoactivatable GFP fusion protein (Rab5a-paGFP) in HuT-78 T-cells that also co-expressed PKCϵ-cherry (31). Time-lapse confocal microscopy revealed transient interactions between Rab5a-paGFP-positive endosomes and the PKCϵ-cherry expressing compartments around the neck/centrosomal region of migrating T-cells (supplemental Video S1). A similar localization pattern of Rab5a-containing vesicles has been reported in cultured human U2OS cells (35). Therefore, we focused more closely on the centrosomal region of migrating cells by selecting a population of Rab5a-paGFP positive vesicles and followed their kinetics over time concomitant with PKCϵ-cherry expression (supplemental Video S2). Upon irradiation, Rab5a-paGFP present around the centrosomal area was photo-activated, and in the following 30-s period, Rab5a-positive endosomes fused with a PKCϵ-cherry compartment before separating (Fig. 1E, white circle). Next, we applied irradiation aiming at a “single point” corresponding to a PKCϵ-positive vesicle, and photo-activation of Rab5a-paGFP was visualized within the confines of this structure. The two proteins interacted with each other for a further 60 s and then disengaged (supplemental Video S3). These observations of real-time protein-protein interactions in T-cells after LFA-1 stimulation imply that PKCϵ and Rab5a dynamically interact during the migratory process. Importantly, endogenous PKCϵ co-localization with endogenous Rab5a was also detected in primary human PBL T-cells migrating on ICAM-1, which is the physiological ligand for the LFA-1 integrin (Fig. 1F). To quantify the protein-protein interaction of endogenous PKCϵ with endogenous Rab5a, we performed co-immunoprecipitation studies utilizing primary human PBL T-cells. We observed significantly increased levels (>7-fold) of PKCϵ-associated Rab5a in LFA-1/ICAM-1-stimulated migrating PBL T-cells as compared with that of resting cells (Fig. 1G). Interestingly, the association of Rab5a with PKCϵ was not influenced by the levels of GTP on Rab5a because constitutively active GTP-bound Rab5a (Q79L mutant) and dominant-negative GDP-bound Rab5a (S34N mutant) both associated with the endogenous PKCϵ in migrating T-cells to similar extents (Fig. 1H). Hence, the association of Rab5a with PKCϵ is a nucleotide-independent interaction. Taken together, these studies confirm that PKCϵ localizes to endosomes in migrating T-cells and physically interacts with Rab5a after LFA-1/ICAM-1 stimulation.
PKCϵ Phosphorylates Rab5a on Threonine 7
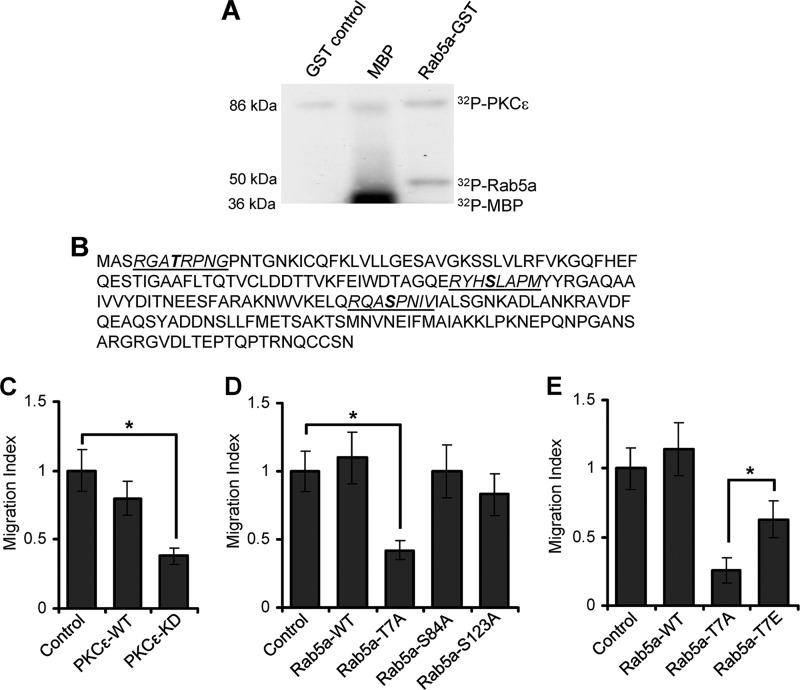
We next investigated whether PKCϵ could phosphorylate Rab5a. An in vitro kinase assay was performed where active recombinant PKCϵ was incubated with a purified Rab5a-GST fusion protein in the presence of radio-labeled ATP ([32P]ATP). Incubation of recombinant PKCϵ with GST alone or with MBP in the presence of [32P]ATP was also used as negative or positive control, respectively. Incorporation of [32P]ATP into Rab5a, GST, or MBP was measured by SDS-PAGE followed by autoradiography. This analysis clearly indicated that PKCϵ was capable of phosphorylating Rab5a in vitro, whereas GST alone was not phosphorylated (Fig. 2A). In contrast, MBP was highly phosphorylated when incubated with PKCϵ in the presence of [32P]ATP (Fig. 2A).
FIGURE 2.
PKCϵ-dependent phosphorylations of Rab5a and its involvement in T-cell motility. A, an in vitro kinase assay was performed with recombinant PKCϵ incubated with purified GST alone (control), MBP, or Rab5a-GST. A representative blot from three independent experiments is shown. B, three potential PKC phosphorylation motifs (underlined) were identified in Rab5a amino acid sequence. The predicted serine/threonine residues (indicated in bold) are Thr-7, Ser-84, and Ser-123. C, HuT-78 T-cells were transfected with the control plasmid, wild-type (PKCϵ-WT), or the kinase-dead mutant of PKCϵ (PKCϵ-KD) expression vectors, and the migratory potential of cells was analyzed using trans-well migration assays. D, HuT-78 cells were transfected with wild-type Rab5a (Rab5a-WT) or mutant Rab5a-T7A, Rab5a-S84A, or Rab5a-S123A plasmid constructs and analyzed in a trans-well migration assay. E, HuT-78 cells were transfected with wild-type Rab5a (Rab5a-WT), mutant Rab5a-T7A, or Rab5a-T7E plasmid constructs and analyzed using trans-well migration assay. Data of at least three independent experiments were normalized to respective control and are presented as the mean ± S.E. *, p < 0.01.
Members of the PKC family generally target serine/threonine residues that lie within (S/T)X(K/R), (K/R)XX(S/T), or (L/R)X(S/T) motifs, where X indicates any amino acid (36). Using an oriented peptide library, Nishikawa et al. (37) also reported that PKCϵ and other novel PKC isozymes prefer hydrophobic amino acids at positions +2, +3, and +4. Based on these criteria, we identified three potential sites on human Rab5a, namely Thr-7, Ser-84, and Ser-123, that may be phosphorylated by PKCϵ (Fig. 2B). These three sites on human Rab5a are evolutionarily conserved, as they are present on Rab5a from all other species (supplemental Fig. S1). Thr-7 lies within the N terminus of Rab5a, a region previously shown to be critical for its functionality, whereas Ser-84 and Ser-123 fall within the highly conserved GTP/GDP binding switch regions of small GTPases (38–40).
We used a number of approaches to determine that Rab5a was phosphorylated on Thr-7 by PKCϵ and that phosphorylation of this site (but not at Ser-84 or Ser-123) was crucial for T-cell migration. First, we demonstrated that transfecting HuT-78 T-cells with a kinase-dead PKCϵ construct perturbed their ability to migrate through ICAM-1-coated filters toward the chemokine SDF-1α in transwell chambers (Fig. 2C). Second, we generated mutant constructs of Rab5a by replacing Thr-7 (Rab5a-T7A), Ser-84 (Rab5a-S84A), or Ser-123 (Rab5a-S123A) residues with alanine, transfected the constructs into HuT-78 T-cells, and analyzed the ability of the cells to migrate toward SDF-1α through transwell filters coated with ICAM-1. Expression of the Rab5a-T7A construct in HuT-78 T-cells perturbed migration in comparison to cells transfected with Rab5a-WT (Fig. 2D). In contrast, mutation of the Ser-84 or Ser-123 sites on Rab5a to alanine did not significantly affect T-cell transwell migration (Fig. 2D). We also addressed the effect of mutating the Thr-7 site with a glutamic acid residue to mimic constitutive phosphorylation of Rab5a (Rab5a-T7E) and analyzed its effect on T-cell migration. We found that the expression of this construct in T-cells also perturbed T-cell migration but not to the same extent as the non-phosphorylated Rab5a-T7A mutant (Fig. 2E). Collectively, these experiments indicate that PKCϵ and phosphorylation/dephosphorylation of Rab5a are independently required for T-cell motility.
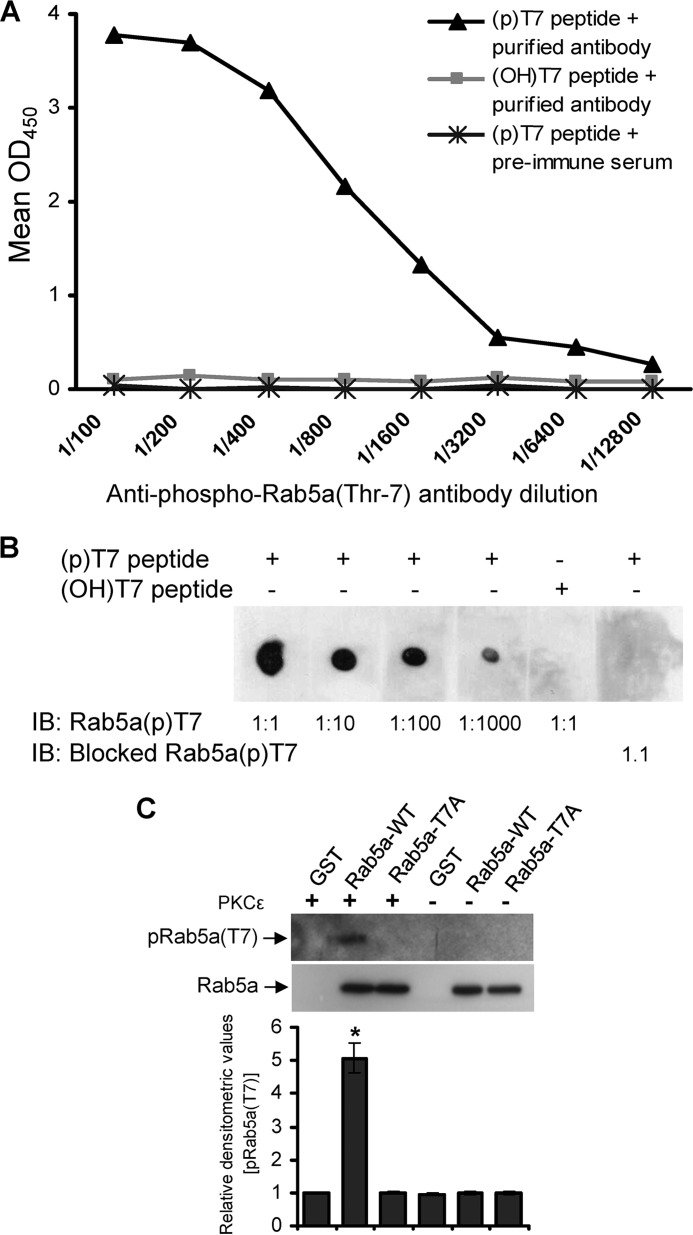
To address whether the Thr-7 residue of Rab5a is indeed phosphorylated, we developed an antibody against the N-terminal phosphorylated Thr-7 site as described under “Experimental Procedures” above. The specificity of the antibody was verified through titration experiments using phospho-T7 containing peptides as analyzed by ELISA (Fig. 3A) and immuno dot-blot assays (Fig. 3B). Furthermore, purified Rab5a-WT-GST, Rab5a-T7A-GST, or GST proteins were incubated in the presence or absence of PKCϵ and analyzed by Western blot using the anti-phospho-Rab5a(Thr-7) antibody. PKCϵ-induced phosphorylation of Rab5a was detected only in the Rab5a-WT-GST sample but not in Rab5a-T7A-GST or GST proteins (Fig. 3C).
FIGURE 3.
Characterization of anti-phospho Rab5a(Thr-7) antibody. A, serial dilutions (1/100 to 1/12,800) of the affinity-purified anti-phospho-Rab5a(Thr-7) antibody was titrated against phospho Thr-7-containing peptide ((p)T7 peptide) and analyzed by ELISA. Preimmune serum and non-phospho (OH)Thr-7-containing peptides ((OH)T7 peptide) were used as controls. B, various dilutions of the purified pRab5a(Thr-7) antibody ranging from 1:1 to 1:1000 were analyzed by immuno dot-blot assay (IB) using Thr(P)-7 peptide. (OH)Thr-7 peptide or pRab5a(Thr-7) antibody preincubated with the Thr(P)-7 peptide was used as the control. C, purified GST, GST-conjugated wild-type Rab5a (Rab5a-WT), or mutant Rab5a-T7A protein was separately incubated in the presence or absence of PKCϵ. The products were analyzed by Western blot using the affinity-purified anti-pRab5a(Thr-7) or anti-Rab5a (loading control). Densitometry analysis of the Western blots was performed and presented (mean ± S.E.). *, p < 0.01. Data represents three independent experiments.
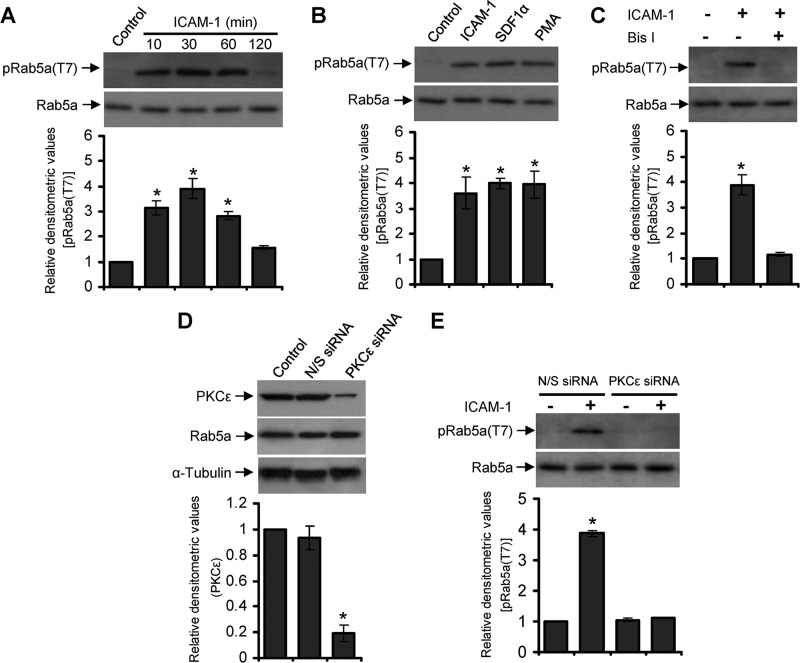
Next, we examined Rab5a phosphorylation at Thr-7 induced via LFA-1 signaling in cells. PBL T-cells were incubated for a range of time points on an immobilized ICAM-1-coated surface, lysed, and analyzed by Western immunoblotting using the anti-phospho-Rab5a(Thr-7) antiserum. Thr-7 phosphorylation of Rab5a was barely detectable in resting T-cells, which was significantly increased after LFA-1/ICAM-1 stimulation, reaching maximum levels within the time frame of 10–30 min, and then declined to basal level at 2 h (Fig. 4A). Similar results were obtained when PBL T-cells were stimulated with either SDF-1α or with the PKC activator phorbol 12-myristate 13-acetate (PMA; Fig. 4B), further suggesting that Rab5a Thr-7 phosphorylation was PKC-dependent.
FIGURE 4.
Rab5a undergoes PKCϵ-dependent phosphorylation on Thr-7 in migrating T-cells. A, human primary PBL T-cells were allowed to migrate via LFA-1 stimulation by incubating on ICAM-1-coated plates for 10, 30, 60, or 120 min and lysed. Cell lysates were analyzed by Western immunoblotting using the crude anti-phospho Rab5a(Thr-7) antiserum (pRab5a(T7)). B, PBL T-cells were left unstimulated or stimulated with ICAM-1 for 30 min, SDF-1α for 2 min, or phorbol 12-myristate 13-acetate (PMA) for 30 min and lysed. Cell lysates were analyzed by Western immunoblotting using the crude pRab5a(Thr-7) antiserum. C, PBL T-cells were untreated or pretreated with PKC inhibitor bisindolylmaleimide I for 30 min, stimulated with ICAM-1 for additional 30 min, and lysed. Cell lysates were analyzed by Western immunoblotting using the crude anti-pRab5a(Thr-7) antiserum. D, PBL T-cells were nucleofected with nonspecific siRNA (N/S siRNA) or siRNA against PKCϵ (PKCϵ siRNA) and then lysed after 72 h. Cell lysates were Western-immunoblotted with anti-PKCϵ antibody. E, PBL T-cells nucleofected with nonspecific siRNA or PKCϵ siRNA were unstimulated or stimulated with ICAM-1 for 30 min and Western-immunoblotted using the crude anti-pRab5a(Thr-7) antiserum. All blots were stripped and re-probed with anti-Rab5a or α-tubulin as indicated. Densitometry analysis of the blots were performed and presented. Results are representative of at least three independent experiments. *, p < 0.01.
To confirm the role of PKC in LFA-1-induced Rab5a Thr-7 phosphorylation, we pretreated PBL T-cells for 30 min with a 10 μm concentration of the PKC inhibitor bisindolylmaleimide I. Western immunoblot analysis of T-cells stimulated with or without LFA-1/ICAM-1 showed that bisindolylmaleimide I pretreatment significantly inhibited Rab5a Thr-7 phosphorylation (Fig. 4C). Furthermore, specific knockdown (>75%) of PKCϵ in T-cells using a siRNA approach (Fig. 4D) significantly inhibited LFA-1-mediated Thr-7 phosphorylation of Rab5a (Fig. 4E). These data clearly indicate that PKCϵ is the specific PKC isoform mediating Thr-7 phosphorylation of Rab5a.
T7A Mutation Partially Inhibits Rab5a Targeting to Endosomal Compartments
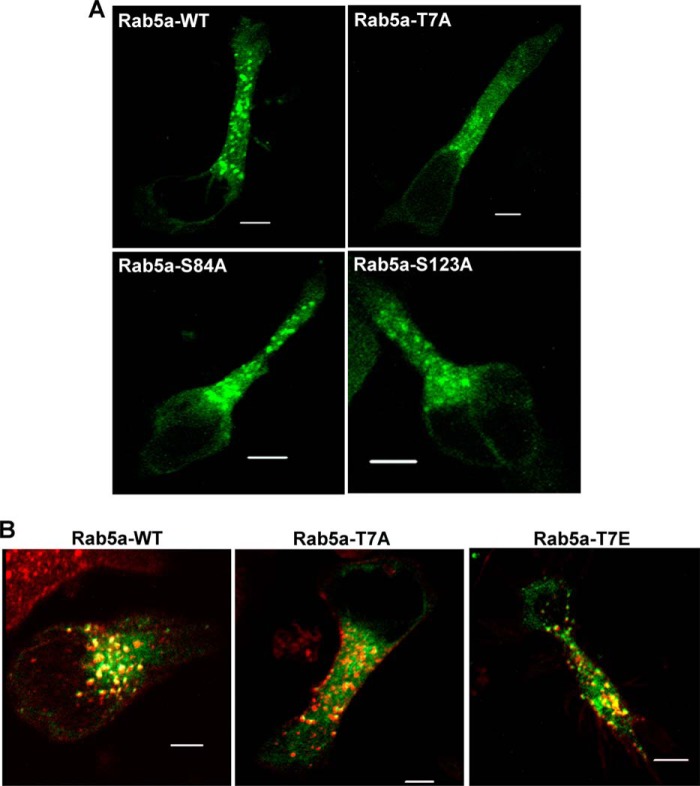
Because mutation of Thr-7 to alanine on Rab5a affected T-cell migration, we investigated the mechanistic basis of this migratory defect. Truncation mutants of Rab5a lacking the first 14–22 N-terminal residues have been shown to disrupt Rab5a functionalities (40–43). However, these mutants remain prenylated, membrane-associated, and functionally interact with guanine nucleotides (40, 41, 44). In addition, functional aspects of protein folding and the crystal structure also remain unaffected (44, 45). Therefore, the T7A mutation would not be predicted to interfere with the GTPase activity of Rab5a and should not affect its three-dimensional conformation. Hence, we investigated whether mutation of Thr-7 on Rab5a affected its subcellular distribution. Expression of wild-type Rab5a-GFP in HuT-78 T-cells demonstrated punctate and globular structures evenly distributed throughout the whole cell body, center, and the periphery (Fig. 5A). Although the expression pattern of Rab5a-S84A-GFP or Rab5a-Ser-123-GFP resembled that of wild type, Rab5a-T7A-GFP appeared to be more cytosolic and less punctate throughout the uropod (Fig. 5A). This distinctive pattern of Rab5a distribution was also verified by co-expressing both wild-type Rab5a-GFP and mutant Rab5aT7A-cherry constructs in T-cells (data not shown). Some punctate Rab5a-T7A-GFP-associated structures were still associated with endocytic vesicles labeled by internalized Alexa Fluor® 568-transferrin-conjugate (Fig. 5B). These studies suggest that phosphorylation of the Thr-7 site partially influences Rab5a subcellular localization and that its mutation does not totally disrupt Rab5a targeting to the transferrin receptor-positive endosomal compartment. Consistent with the finding that the T7E mutant did not affect T-cell migration as much as the T7A mutant (Fig. 2E), the phosphomimetic Rab5a-T7E-GFP transfected cells expressed many small vesicular structures throughout the whole cell body co-localizing with transferrin and resembling the Rab5a-WT-like expression pattern (Fig. 5B).
FIGURE 5.
Subcellular localization of Rab5a and its targeting to T-cell endosomal compartments. A, shown are representative confocal images of HuT-78 T-cells expressing GFP conjugated wild-type Rab5a (Rab5a-WT) or mutant Rab5a-T7A, Rab5a-S84A, or Rab5a-S123A after stimulation by incubating on an anti-LFA-1 coated surface for 1 h. B, HuT-78 cells were transfected with plasmid constructs expressing GFP conjugated wild-type Rab5a (Rab5a-WT), mutant Rab5a-T7A, or Rab5a-T7E and stimulated via LFA-1 as above. Cells were treated with AlexaFluor® 568-labeled transferrin (red) to allow internalization for 10 min and imaged. At least 20 microscopic fields per slide prepared from three independent experiments were visualized, and a representative data is shown. Scale bar, 5 μm.
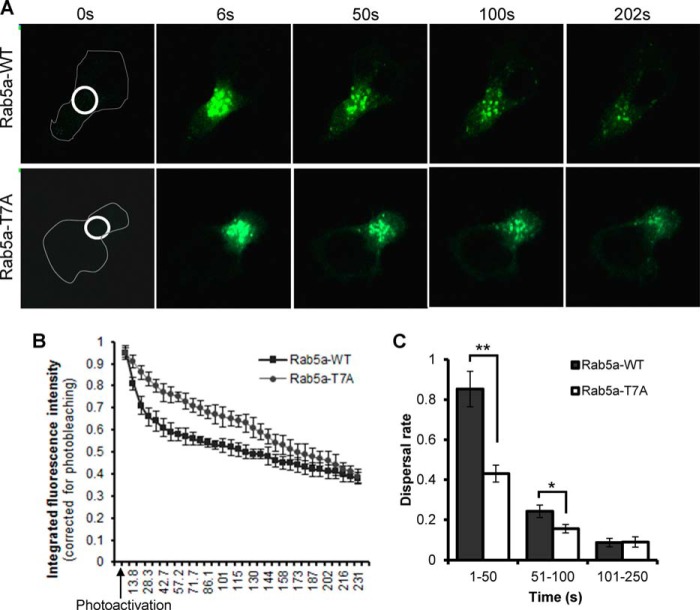
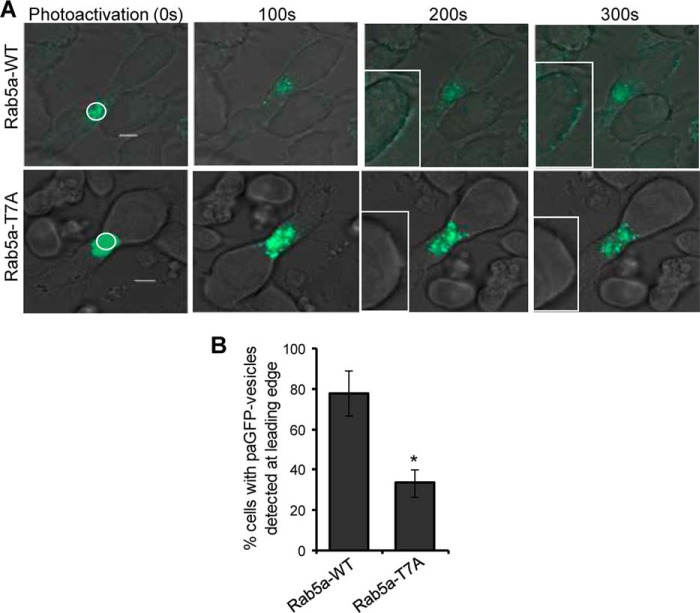
We next employed photoactivation in combination with time-lapse microscopy to directly visualize and compare the dynamics of wild-type Rab5a and Rab5a-T7A at the centrosomal region in transfected cells. After transfection with Rab5a-paGFP or Rab5a-T7A-paGFP constructs, HuT-78 cells were seeded on coverslips coated with a cross-linking anti-LFA-1 antibody for 1 h to allow extended polarization. A pulse of 405-nm laser light aimed around the centrosomal region of the polarized cell resulted in immediate photoactivation of paGFP fusion proteins within the confines of this area. In cells expressing wild-type Rab5a-paGFP, a patch of photoactivated endosomes at the centrosomal region could be seen to rapidly disperse from this area over the following 5 min (Fig. 6A, supplemental Video S4). Conversely, the dispersal rate of photoactivated Rab5a-T7A from the centrosomal region was relatively slow (Fig. 6A, supplemental Video S5). Quantifying a total of 30 Rab5a-paGFP or 40 Rab5a-T7A-paGFP-expressing cells from 3 independent experiments showed that the dispersal rate of Rab5a in phospho-deficient mutant cells was significantly slower than wild-type Rab5a-paGFP-expressing cells in the first 100 s (Fig. 6, B and C). When the dynamics of the photoactivated population of Rab5a-paGFP was tracked for a longer period of time, fluorescence was rapidly dimmed to background levels (Fig. 6C), and this was also accompanied by the appearance of budding vesicles at the leading edge of the polarized cell. This observation was in accordance with a previous study suggesting the half-life of Rab5a-positive early endosomes to be in the range of 2 min (47). In a parallel experiment, we also observed that a majority (∼80%) of Rab5a-paGFP-expressing vesicles continuously redistributed from large structures in the cell centrosomal region, to allow the recycling of Rab5a onto budding endosomes at the leading edge (Fig. 7A, Supplemental Video S6). On the other hand, although the pool of Rab5a-T7A-associated endosomes gradually lost its fluorescence signal (Fig. 7A, supplemental Video S7), the number of cells with emerging and budding vesicles at the leading edge was significantly reduced to 33 ± 7% (Fig. 7B).
FIGURE 6.
Involvement of Rab5a T7A phosphorylation in the diffusion of endosomes from the centrosomal region in migrating T-cells. A, HuT-78 T-cells expressing wild-type Rab5a-paGFP (Rab5a-WT) or mutant Rab5a-T7A-paGFP (Rab5a-T7A) were stimulated to migrate on anti-LFA-1 coated coverglass and imaged by confocal microscopy. Photoactivation was achieved with a pulse of 405-nm laser defined within the indicated white circle at the centrosomal region of the cell. B, the integrated fluorescence intensity of the photoactivated region (white circle shown in A) was quantified for each frame of at least 30 movies, calculated relative to the intensity of the frame immediately after photoactivation and corrected for photo-bleaching and plotted against time (s). C, average dispersal rate of Rab5a calculated for the time intervals 0–50, 51–100, and 101–250 s are shown. Values are the mean integrated fluorescence intensity obtained from three independent experiments ± S.E.; n = 30 for Rab5a-WT and n = 40 for Rab5a-T7A. **, p < 0.001; *, p < 0.01.
FIGURE 7.
Photoactivated Rab5a-paGFP- but not Rab5a-T7A-paGFP-associated vesicles redistribute from the centrosomal region to the leading edge of T-cells during migration. A, HuT-78 T-cells expressing wild-type Rab5a-paGFP (Rab5a-WT) or mutant Rab5a-T7A-paGFP (Rab5a-T7A) were stimulated to migrate on anti-LFA-1-coated coverslips and imaged by confocal microscopy. Photoactivation was performed within the indicated white circle around the centrosomal region of the cell. The leading edges of the migrating cell at 200 and 300 s post-photoactivation are shown enlarged in the insets. B, percentage of cells (±S.E.) with Rab5a-WT- or Rab5a-T7A-associated vesicles detected at the leading edge of migrating cells was plotted. n = 19 for Rab5a-WT and 25 for Rab5a-T7A. *, p < 0.01. Results are representative of three independent experiments.
Rab5aT7A Disrupts Actin Rearrangement during Migration
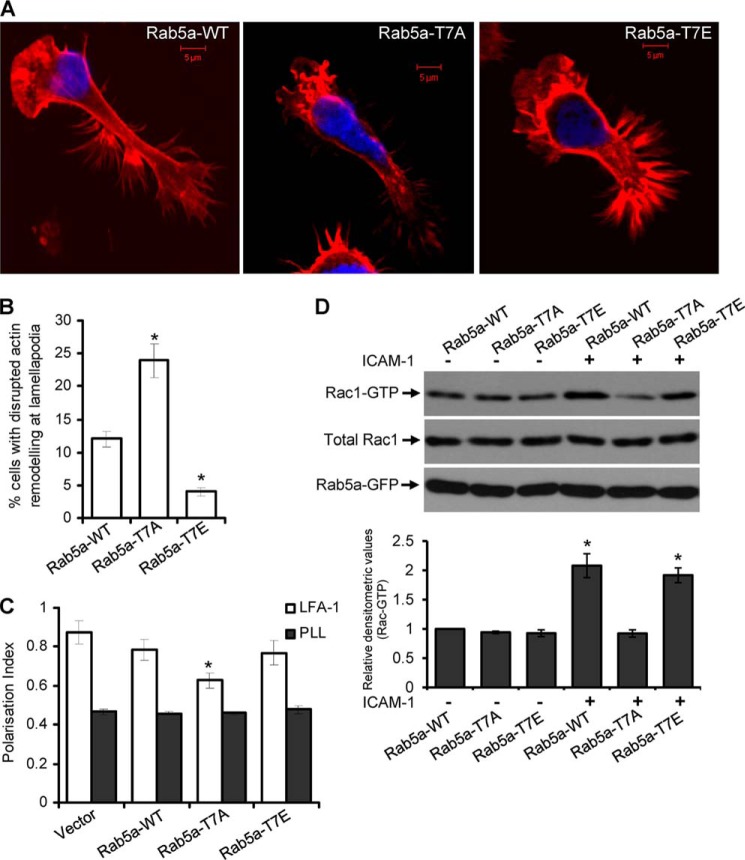
By means of time-lapse microscopy, we observed that T-cells co-expressing Rab5a-GFP and Lifeact-ruby, a 17-amino acid peptide that stains filamentous actin networks (48), displayed highly motile lamellipodia structures extending in a vibrant umbrella-shaped fast protrusion followed almost immediately by active ruffling and retraction in a highly dynamic cycle in the course of moving forward (supplemental Video S8). We investigated if loss of Thr-7 phosphorylation of Rab5a could interfere with actin remodeling at lamellipodia during migration. For this purpose, HuT-78 T-cells expressing wild-type Rab5a, Rab5a-T7A, or Rab5a-T7E were incubated on a cross-linked anti-LFA-1 antibody for 2 h and fixed. These cells were then stained with phalloidin-TRITC, which binds filamentous actin, and Hoechst, which stains the nucleus (Fig. 8A). Looking closely at the leading edge, we observed that in 25% of cells expressing Rab5a-T7A, filamentous actin bundles were disrupted and condensed around the plasma membrane accompanied by asymmetrically shaped lamellipodia and/or irregular ruffling (Fig. 8B, supplemental Video S9). Such disrupted lamellipodia phenotypes were present only in 12% of the wild-type Rab5a or 5% of the Rab5a-T7E-expressing and -migrating T-cells (Fig. 8B). As migrating T-cells rapidly change shape and the lamellipodia are highly dynamic, it was difficult to compare and quantify cell-to-cell differences. Therefore, these measurements reflect gross differences observed at the lamellipodia of only those crawling T-cells that displayed typical amoeboid phenotypes.
FIGURE 8.
Involvement of Rab5a Thr-7 phosphorylation in the actin cytoskeleton rearrangement at the lamellipodia and Rac1 activation in crawling T-cells. A, HuT-78 T-cells expressing wild-type Rab5a (Rab5a-WT), Rab5a-T7A, or Rab5a-T7E were incubated on anti-LFA-1 coated coverslips for 2 h, stained with phalloidin-TRITC (red) and Hoechst (blue), and imaged. Scale bars, 5 μm. B, percentage of cells showing disrupted lamellipodia (as shown in A) were quantified and presented. Phenotypes were scored blindly by three individuals, and data represent the mean ± S.E., n > 100 cells for each samples. C, cells were seeded on poly-l-lysine (PLL)- or anti-LFA-1 (LFA-1)-coated 96-well plates for 2 h and stained for F-actin and nucleus. Images were automatically acquired and analyzed using an InCell AnalyzerTM by selecting only GFP fusion-expressing cells. Results are representative of at least three independent experiments and are presented as the mean ± S.E. D, serum-starved cells expressing Rab5a-WT, Rab5a-T7A, or Rab5T7E were stimulated with or without ICAM-1 for 30 min. Rac1 activation was measured by PAK-1 PBD pulldown assays. The graph shows densitometric analysis of Rac-GTP levels with respect to input levels of total Rac1 for 3 independent experiments. *, p < 0.01.
We employed objective quantification of cytoskeletal change using a previously reported cell-based method of high content analysis that entailed population analysis of the morphology of cells expressing GFP fusion protein (34). Utilizing this approach, we observed that although cells expressing the Rab5a-T7A-GFP mutant were polarized after anti-LFA-1 stimulation, the cell polarization index based on the actin cytoskeleton was significantly reduced (p = 0.003) as compared with cells expressing wild-type Rab5a-GFP or Rab5a-T7E-GFP (Fig. 8C). These results suggest that within the population of Rab5a-T7A-GFP-expressing T-cells, a relatively higher proportion of cells with less elongated or irregularly shaped cytoskeleton is present, implying that they have deficiencies in their ability to reorganize their cytoskeleton in response to a migratory stimulus.
A number of previous studies have established Rac1 as the main driving force in the formation of actin protrusions that are essential for cell migration (47, 48). A crucial role of Rab5a in the activation of Rac1 has also been demonstrated (17, 18). These findings prompted us to investigate if Thr-7 phosphorylation of Rab5a is important for Rac1 activation in migrating T-cells. To address this question, we transfected HuT-78 T-cells with plasmid constructs encoding wild-type, T7A, or T7E forms of Rab5a, and then cells were incubated on ICAM-1 to trigger LFA-1-induced migration. The levels of Rac1 activation in these cells were determined by a PBD binding assay. Consistent with the above observed role of Rab5a in actin remodeling and T-cell polarity, significantly high Rac1 activity (∼2-fold) was detected in T-cells expressing Rab5a-WT or Rab5a-T7E after LFA-1 stimulation in this migration-triggering model system (Fig. 8D). In contrast, expression of Rab5a-T7A perturbed this increase in Rac1 activation after LFA-1/ICAM-1 stimulation (Fig. 8D). Similar results were observed when these cells were stimulated with SDF-1α (data not shown). Taken together, these results clearly demonstrate that PKCϵ-mediated Thr-7 phosphorylation of Rab5a is a necessary step in the regulation of T-cell cytoskeletal reorganization and migration.
DISCUSSION
In this study we establish a novel mechanism involving a PKCϵ-Rab5a-Rac1 axis in the regulation of actin cytoskeletal remodeling and T-cell migration. Specifically, we uncover a key biological function of a previously undescribed phosphorylation site on Rab5a at Thr-7. We show that Rab5a interacts with PKCϵ in migrating T-cells in a nucleotide-independent manner and that Rab5a is phosphorylated on Thr-7 by PKCϵ in response to integrin or chemokine stimulation. We also demonstrate that phosphorylation of this site on Rab5a is required for T-cell migration, as ectopic expression of a phospho-deficient T7A mutant of Rab5a resulted in a significant reduction in the ability of T-cells to migrate across ICAM-1-coated membranes toward SDF-1α. This defect in T-cell motility appears to be due to inefficient trafficking of endosomes toward the leading edge of the cell and correlates with impaired Rac1 activation and actin cytoskeletal remodeling in the lamellipodia, events that are crucial for cell migration.
Rab5 exists as three closely related molecules (Rab5a, Rab5b, and Rab5c) in mammalian cells (40, 49), sharing 88.7% of protein sequence identity (supplemental Fig. S2). Although there is evidence to suggest that Rab5a is involved in chemokine receptor recycling, cell motility, actin organization, Rac distribution, and lamellipodia formation in various cell types (14, 16, 17, 50, 51), its role in the process of T-cell migration has not previously been reported. During migration, the leading edge of the T-cell actively produces filamentous actin-rich lamellipodia that likely facilitate the interaction of LFA-1 with its counter-ligand ICAM-1 on endothelial cells (52). In doing so, T-cells search for a suitable extravasation site on the venular surface that facilitates rapid directional changes that are characteristic of migrating T-cells. The LFA-1 signal in T-cells has been shown to transiently activate Rac1 and induce actin polymerization (53). Here, we found that signaling through LFA-1 or CXCR4 results in Thr-7 phosphorylation of Rab5a, which in turn is critical for Rac1 activation, subsequent actin remodeling, and T-cell motility. This is consistent with previous findings where Rab5 was shown to regulate trafficking of Rac and its guanine nucleotide exchange factor in T-cell lymphoma invasion and metastasis 1 (Tiam1) (16, 17, 54). Whether Tiam1 or other guanine nucleotide exchange factors regulate Rac1 activation downstream of Rab5a or PKCϵ observed in this study requires further investigation.
PKC isoforms have been implicated in T-cell signal transduction including the regulation of migration. For example, our group has demonstrated a role for PKCβ and PKCδ in regulating the microtubule cytoskeleton and active T-cell locomotion induced by LFA-1 integrin or CD44 receptors (21, 55, 56). PKCδ has also been reported to induce integrin-mediated Thr-566 phosphorylation of phospholipase D and direct cell spreading and migration in COS-7 cells (57). In addition, several studies have suggested the involvement of PKCϵ in cell migration in many other model systems. Overexpression of PKCϵ resulted in a highly motile and invasive phenotype in various cancer models (58, 59), and its disruption caused inactivation of Rho family GTPases (60). It has also been shown that PKCϵ can bind directly to both filamentous and globular forms of actin via its actin binding motif (61, 62) or interact indirectly via integrins and scaffolding proteins such as receptors for activated protein kinase C (also known as RACK) (63, 64). In murine embryonic fibroblasts, PKCϵ was shown to phosphorylate vimentin and regulate β1 integrin recycling contributing to cell motility (24, 25). Thus, our identification of Rab5a phosphorylation on the N terminus at Thr-7 represents a novel regulatory mechanism by which PKCϵ integrates extracellular signals via integrins and chemokine receptors and coordinates signal transduction in T-cell migration. Of note, our studies do not exclude the possibility that PKCϵ may also phosphorylate other Ser/Thr residues on Rab5a that could contribute to diverse lymphocyte functions.
The GTP/GDP binding switch regions of Rab proteins are the main features for GTPase activity and are the most conserved structures among family members (38, 39, 65). On the other hand, the N- and C-terminal regions of Rab proteins are variable with the most divergent sequences lying at the extreme N terminus (40, 49). Although the isoprenylated C-terminal motif is required for membrane targeting (66, 67), functions of the extreme N-terminal region are still unknown. Given that deletion of the first 14–22 amino acid residues at the N-terminal domain alone is enough to disrupt Rab5a functionalities (42), the N-terminal region is not only an important element for Rab5a function; it might also act as the domain that specifies isoform functionality. Taking it a step further, we have shown that when phosphorylation of Rab5a on the N-terminal Thr-7 site is disrupted, T-cell migration through ICAM-1-coated membranes toward SDF-1α is significantly repressed. By replacing Thr-7 with glutamic acid (T7E), which mimics constitutive phosphorylation, we were able to show that this mutant also partially inhibits T-cell migration. Although we do not have a clear explanation for this observation, it is possible that Rab5a dynamically cycles between phosphorylated and dephosphorylated states to function in T-cell migration. This is consistent with the observed dynamic interactions between PKCϵ and Rab5a (Fig. 1E, supplemental Video S1). It may be that replacement of Thr-7 with T7E could alter the structure or folding of the Rab5a protein necessary for its function in T-cell migration. Future studies on the kinetics of Rab5a recycling in the context of phosphorylation events in T-cells will further clarify this.
The level of Rab5 dynamically fluctuates on individual early endosomes linked by fusion and fission events into a network in time. Although the bulk of early endosomes are not subjected to rapid turnover, the half-life of Rab5-containing early endosomes has been estimated to be in the range of 2 min (46). Moreover, the amount of Rab5 associated with a given endosome varies considerably over time (46). Utilizing a photoactivation-based imaging approach to track the dynamics of Rab5a after LFA-1 stimulation in motile T-cells, we observed that Rab5a-paGFP-expressing vesicles continuously redistribute from the cell centrosomal region (the site of PKCϵ-Rab5a interaction) to the leading edge. However, the dispersal rate of the phospho-deficient mutant Rab5a-T7A-paGFP was significantly slower than the wild-type Rab5a-paGFP in the first 100 s, and fluorescence dimmed to almost background levels by 250 s. Thus, the dynamic turnover of Rab5a seems to be essential to allow its recycling onto budding endosomes in the cell periphery. In support of this concept, earlier studies suggest that the driving force for cell migration is a polarized endocytic/exocytic cycle that delivers adhesion receptors and proteins from the rear of the cell after focal adhesion disassembly to the leading edge of the migrating cell for extension and substrate attachment. Such events are required for driving the cell forward (68, 69). This could also be due to extraction of Rab5a from the membrane via guanine nucleotide dissociation inhibitors (15, 70, 71). Based on these observations, it can be speculated that Thr-7 phosphorylation may be important in the regulation of GDI-mediated extraction of Rab5a from the endosomal membrane.
The recycling of integrins, which involves their endocytosis from cell surface and recycling back to the plasma membrane, is important for their activities as well as for the regulation of their surface levels (72–74). Previous studies have shown clathrin-dependent or -independent endocytosis of integrins at the leading edge of migrating cells (75). In neutrophils, LFA-1 is associated with detergent-resistant membrane microdomains and is internalized in a filipin-sensitive and dynamin-dependent manner (76). Consistent with these findings, we observed that intracellular vesicles expressing Rab5a-T7A-paGFP were less efficient in redistributing from the centrosomal region to the leading edge. Another Rab5 GTPase family member, Rab5c, has recently been reported to enhance β1-integrin recycling in EGF-induced cancer invasion (74). In addition, LFA-1 recycling has been shown to be independently regulated by Rap2 GTPase as well as Gαq/11 proteins in T-cells (72, 73). Surprisingly, internalization of LFA-1 was not found to be inhibited by a dominant-negative Rab5 construct (N133I) in a Chinese hamster ovary cell model (76). What becomes obvious from all of these studies is that different intracellular pools exist for Rab5a, and its Thr-7 phosphorylation and redistribution may be an important mechanism in the process of cytoskeletal remodeling and lymphocyte migration. It should be noted that although we examined the role of Rab5a mutants in T-cells that still express endogenous Rab5a, it would be interesting to examine these mutants in cells where endogenous Rab5a has been depleted. However, performing such experiments is technically very challenging in T-cells.
In summary, this study provides a new mechanistic insight into the regulation of T-cell migration by a PKCϵ-Rab5a-Rac1 axis. PKCϵ-dependent phosphorylation of Rab5a on Thr-7 regulates its intracellular dynamics and Rac1 activation, which in turn controls actin cytoskeletal remodeling at the lamellipodia and cell migration (Fig. 9). Based on our data, we propose that the Thr-7 phosphorylation of Rab5a might be a missing piece of the structure-function puzzle of this 24-kDa protein and hence may provide new information regarding downstream pathways. Given that Rab5a is central to many fundamental cell processes, this new finding has profound biological implications across cellular organisms. Our results also highlight the concept that LFA-1 not only acts as an adhesion receptor, a role for which it has been known for many years, but also plays an important role in downstream protein-protein interactions and consequent signaling.
FIGURE 9.
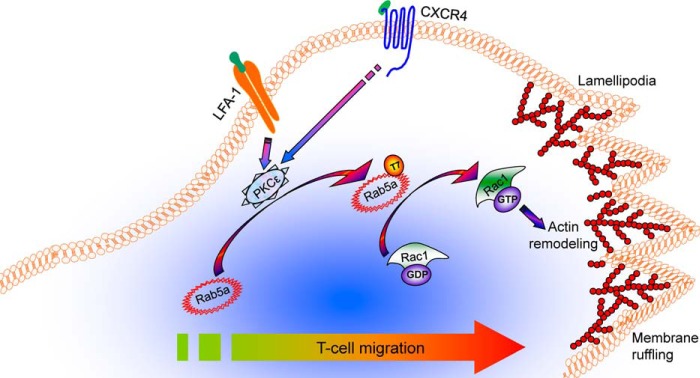
A model for signaling via PKCϵ-Rab5a-Rac1 axis in T-cell migration. T-cell stimulation through LFA-1 or CXCR4 triggers PKCϵ-dependent Rab5a Thr-7 phosphorylation in the centrosomal region, which activates Rac1 GTPase. Activated Rac1 regulates actin remodeling and protrusion necessary for T-cell motility.
Supplementary Material
Acknowledgments
We thank J. Norman and P. Caswell (Beatson Institute, Glasgow) for suggestions and advice on paGFP imaging, A. Davies, C. Edwards, A. M. Byrne, and J. Conroy for technical assistance with the high content analysis and confocal microscopy, and D. Dunican for suggestions and for critically reading the manuscript. We also thank M. Zerial, D. Romberger, R. Wedlich-Soldner, J. Lippincott-Schwartz, and R. Tsien for the generous gifts of plasmid constructs.
This work was supported by grants from Science Foundation Ireland (to A. L.), Higher Education Authority of Ireland under PRTLI cycle 3 (to D. K.), Health Research Board of Ireland (to A. L.), Marie Curie MTKDCT-2005-029798 (to J. S.-Z.), NTU-MOE start-up grant (to N. K. V.), FWF Austrian Science Fund (P19505-B05) (to F. F. and G. B.), and the European Community Program SYBILLA under Grant HEALTH-F4-2008-201106 (to F. F. and G. B.).

This article contains supplemental Figs. 1 and 2 and Movies S1–S9.
- LFA-1
- leukocyte function associated-antigen-1
- ICAM-1
- intercellular adhesion molecule-1
- SDF-1α
- stromal cell-derived factor-1α
- CXCR4
- C-X-C chemokine receptor type 4
- PBL
- peripheral blood lymphocyte
- TRITC
- tetramethylrhodamine isothiocyanate
- rICAM
- recombinant ICAM
- MBP
- myelin basic protein
- paGFP
- photoactivable GFP
- PBD
- p21 binding domain.
REFERENCES
- 1. Hogg N., Patzak I., Willenbrock F. (2011) The insider's guide to leukocyte integrin signalling and function. Nat. Rev. Immunol. 11, 416–426 [DOI] [PubMed] [Google Scholar]
- 2. Hogg N., Laschinger M., Giles K., McDowall A. (2003) T-cell integrins: more than just sticking points. J. Cell Sci. 116, 4695–4705 [DOI] [PubMed] [Google Scholar]
- 3. Ley K., Laudanna C., Cybulsky M. I., Nourshargh S. (2007) Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7, 678–689 [DOI] [PubMed] [Google Scholar]
- 4. Moser B., Wolf M., Walz A., Loetscher P. (2004) Chemokines: multiple levels of leukocyte migration control. Trends Immunol. 25, 75–84 [DOI] [PubMed] [Google Scholar]
- 5. Jones M. C., Caswell P. T., Norman J. C. (2006) Endocytic recycling pathways: emerging regulators of cell migration. Curr. Opin. Cell Biol. 18, 549–557 [DOI] [PubMed] [Google Scholar]
- 6. Pellinen T., Ivaska J. (2006) Integrin traffic. J. Cell Sci. 119, 3723–3731 [DOI] [PubMed] [Google Scholar]
- 7. Caswell P., Norman J. (2008) Endocytic transport of integrins during cell migration and invasion. Trends Cell Biol. 18, 257–263 [DOI] [PubMed] [Google Scholar]
- 8. Caswell P. T., Vadrevu S., Norman J. C. (2009) Integrins: masters and slaves of endocytic transport. Nat. Rev. Mol. Cell Biol. 10, 843–853 [DOI] [PubMed] [Google Scholar]
- 9. Stenmark H., Olkkonen V. M. (2001) The Rab GTPase family. Genome Biol. 2, REVIEWS3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hall A. (2005) Rho GTPases and the control of cell behaviour. Biochem. Soc. Trans. 33, 891–895 [DOI] [PubMed] [Google Scholar]
- 11. Ridley A. J. (2006) Rho GTPases and actin dynamics in membrane protrusions and vesicle trafficking. Trends Cell Biol. 16, 522–529 [DOI] [PubMed] [Google Scholar]
- 12. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 13. Zerial M., McBride H. (2001) Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2, 107–117 [DOI] [PubMed] [Google Scholar]
- 14. Spaargaren M., Bos J. L. (1999) Rab5 induces Rac-independent lamellipodia formation and cell migration. Mol. Biol. Cell 10, 3239–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu S. K., Zeng K., Wilson I. A., Balch W. E. (1996) Structural insights into the function of the Rab GDI superfamily. Trends Biochem. Sci. 21, 472–476 [DOI] [PubMed] [Google Scholar]
- 16. Lanzetti L., Palamidessi A., Areces L., Scita G., Di Fiore P. P. (2004) Rab5 is a signalling GTPase involved in actin remodeling by receptor tyrosine kinases. Nature 429, 309–314 [DOI] [PubMed] [Google Scholar]
- 17. Palamidessi A., Frittoli E., Garré M., Faretta M., Mione M., Testa I., Diaspro A., Lanzetti L., Scita G., Di Fiore P. P. (2008) Endocytic trafficking of Rac is required for the spatial restriction of signaling in cell migration. Cell 134, 135–147 [DOI] [PubMed] [Google Scholar]
- 18. Ramel D., Wang X., Laflamme C., Montell D. J., Emery G. (2013) Rab11 regulates cell-cell communication during collective cell movements. Nat. Cell Biol. 15, 317–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zeng L., Webster S. V., Newton P. M. (2012) The biology of protein kinase C. Adv. Exp. Med. Biol. 740, 639–661 [DOI] [PubMed] [Google Scholar]
- 20. Chun J. S., Ha M. J., Jacobson B. S. (1996) Differential translocation of protein kinase Cϵ during HeLa cell adhesion to a gelatin substratum. J. Biol. Chem. 271, 13008–13012 [DOI] [PubMed] [Google Scholar]
- 21. Volkov Y., Long A., McGrath S., Ni Eidhin D., Kelleher D. (2001) Crucial importance of PKC-β(I) in LFA-1-mediated locomotion of activated T cells. Nat. Immunol. 2, 508–514 [DOI] [PubMed] [Google Scholar]
- 22. Stawowy P., Margeta C., Blaschke F., Lindschau C., Spencer-Hänsch C., Leitges M., Biagini G., Fleck E., Graf K. (2005) Protein kinase Cϵ mediates angiotensin II-induced activation of β1-integrins in cardiac fibroblasts. Cardiovasc. Res. 67, 50–59 [DOI] [PubMed] [Google Scholar]
- 23. Akita Y. (2008) Protein kinase Cϵ: multiple roles in the function of, and signaling mediated by, the cytoskeleton. FEBS J. 275, 3995–4004 [DOI] [PubMed] [Google Scholar]
- 24. Ivaska J., Whelan R. D., Watson R., Parker P. J. (2002) PKCϵ controls the traffic of β1 integrins in motile cells. EMBO J. 21, 3608–3619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ivaska J., Vuoriluoto K., Huovinen T., Izawa I., Inagaki M., Parker P. J. (2005) PKCϵ-mediated phosphorylation of vimentin controls integrin recycling and motility. EMBO J. 24, 3834–3845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Freeley M., O'Dowd F., Paul T., Kashanin D., Davies A., Kelleher D., Long A. (2012) L-plastin regulates polarization and migration in chemokine-stimulated human T lymphocytes. J. Immunol. 188, 6357–6370 [DOI] [PubMed] [Google Scholar]
- 27. Verma N. K., Dempsey E., Conroy J., Olwell P., Mcelligott A. M., Davies A. M., Kelleher D., Butini S., Campiani G., Williams D. C., Zisterer D. M., Lawler M., Volkov Y. (2008) A new microtubule-targeting compound PBOX-15 inhibits T-cell migration via post-translational modifications of tubulin. J. Mol. Med. 86, 457–469 [DOI] [PubMed] [Google Scholar]
- 28. Verma N. K., Dempsey E., Freeley M., Botting C. H., Long A., Kelleher D., Volkov Y. (2011) Analysis of dynamic tyrosine phosphoproteome in LFA-1 triggered migrating T-cells. J. Cell Physiol. 226, 1489–1498 [DOI] [PubMed] [Google Scholar]
- 29. Verma N. K., Dempsey E., Long A., Davies A., Barry S. P., Fallon P. G., Volkov Y., Kelleher D. (2012) Leukocyte function-associated antigen-1/intercellular adhesion molecule-1 interaction induces a novel genetic signature resulting in T-cells refractory to transforming growth factor-β signaling. J. Biol. Chem. 287, 27204–27216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Riedl J., Crevenna A. H., Kessenbrock K., Yu J. H., Neukirchen D., Bista M., Bradke F., Jenne D., Holak T. A., Werb Z., Sixt M., Wedlich-Soldner R. (2008) Lifeact: a versatile marker to visualize F-actin. Nat. Methods 5, 605–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Patterson G. H., Lippincott-Schwartz J. (2002) A photoactivatable GFP for selective photolabeling of proteins and cells. Science 297, 1873–1877 [DOI] [PubMed] [Google Scholar]
- 32. Verma N. K., Davies A. M., Long A., Kelleher D., Volkov Y. (2010) STAT3 knockdown by siRNA induces apoptosis in human cutaneous T-cell lymphoma line Hut78 via downregulation of Bcl-xL. Cell. Mol. Biol. Lett. 15, 342–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Verma N. K., Singh J., Dey C. S. (2004) PPARγ expression modulates insulin sensitivity in C2C12 skeletal muscle cells. Br. J. Pharmacol. 143, 1006–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Freeley M., Bakos G., Davies A., Kelleher D., Long A., Dunican D. J. (2010) A high-content analysis toolbox permits dissection of diverse signaling pathways for T lymphocyte polarization. J. Biomol. Screen 15, 541–555 [DOI] [PubMed] [Google Scholar]
- 35. Serio G., Margaria V., Jensen S., Oldani A., Bartek J., Bussolino F., Lanzetti L. (2011) Small GTPase Rab5 participates in chromosome congression and regulates localization of the centromere-associated protein CENP-F to kinetochores. Proc. Natl. Acad. Sci. U.S.A. 108, 17337–17342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pearson R. B., Kemp B. E. (1991) Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 200, 62–81 [DOI] [PubMed] [Google Scholar]
- 37. Nishikawa K., Toker A., Johannes F. J., Songyang Z., Cantley L. C. (1997) Determination of the specific substrate sequence motifs of protein kinase C isozymes. J. Biol. Chem. 272, 952–960 [DOI] [PubMed] [Google Scholar]
- 38. Bourne H. R., Sanders D. A., McCormick F. (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature 349, 117–127 [DOI] [PubMed] [Google Scholar]
- 39. Valencia A., Chardin P., Wittinghofer A., Sander C. (1991) The ras protein family: evolutionary tree and role of conserved amino acids. Biochemistry 30, 4637–4648 [DOI] [PubMed] [Google Scholar]
- 40. Stenmark H., Valencia A., Martinez O., Ullrich O., Goud B., Zerial M. (1994) Distinct structural elements of rab5 define its functional specificity. EMBO J. 13, 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li G., Stahl P. D. (1993) Structure-function relationship of the small GTPase rab5. J. Biol. Chem. 268, 24475–24480 [PubMed] [Google Scholar]
- 42. Steele-Mortimer O., Clague M. J., Huber L. A., Chavrier P., Gruenberg J., Gorvel J. P. (1994) The N-terminal domain of a rab protein is involved in membrane-membrane recognition and/or fusion. EMBO J. 13, 34–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Barbieri M. A., Roberts R. L., Gumusboga A., Highfield H., Alvarez-Dominguez C., Wells A., Stahl P. D. (2000) Epidermal growth factor and membrane trafficking. EGF receptor activation of endocytosis requires Rab5a. J. Cell Biol. 151, 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sanford J. C., Pan Y., Wessling-Resnick M. (1995) Properties of Rab5 N-terminal domain dictate prenylation of C-terminal cysteines. Mol. Biol. Cell 6, 71–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhu G., Liu J., Terzyan S., Zhai P., Li G., Zhang X. C. (2003) High resolution crystal structures of human Rab5a and five mutants with substitutions in the catalytically important phosphate-binding loop. J. Biol. Chem. 278, 2452–2460 [DOI] [PubMed] [Google Scholar]
- 46. Rink J., Ghigo E., Kalaidzidis Y., Zerial M. (2005) Rab conversion as a mechanism of progression from early to late endosomes. Cell 122, 735–749 [DOI] [PubMed] [Google Scholar]
- 47. Steffen A., Rottner K., Ehinger J., Innocenti M., Scita G., Wehland J., Stradal T. E. (2004) Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 23, 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Radhakrishna H., Al-Awar O., Khachikian Z., Donaldson J. G. (1999) ARF6 requirement for Rac ruffling suggests a role for membrane trafficking in cortical actin rearrangements. J. Cell Sci. 112, 855–866 [DOI] [PubMed] [Google Scholar]
- 49. Bucci C., Lütcke A., Steele-Mortimer O., Olkkonen V. M., Dupree P., Chiariello M., Bruni C. B., Simons K., Zerial M. (1995) Co-operative regulation of endocytosis by three Rab5 isoforms. FEBS Lett. 366, 65–71 [DOI] [PubMed] [Google Scholar]
- 50. Fan G. H., Lapierre L. A., Goldenring J. R., Richmond A. (2003) Differential regulation of CXCR2 trafficking by Rab GTPases. Blood 101, 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Torres V. A., Mielgo A., Barbero S., Hsiao R., Wilkins J. A., Stupack D. G. (2010) Rab5 mediates caspase-8-promoted cell motility and metastasis. Mol. Biol. Cell 21, 369–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Smith A., Carrasco Y. R., Stanley P., Kieffer N., Batista F. D., Hogg N. (2005) A talin-dependent LFA-1 focal zone is formed by rapidly migrating T lymphocytes. J. Cell Biol. 170, 141–151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sánchez-Martín L., Sánchez-Sánchez N., Gutiérrez-López M. D., Rojo A. I., Vicente-Manzanares M., Pérez-Alvarez M. J., Sánchez-Mateos P., Bustelo X. R., Cuadrado A., Sánchez-Madrid F., Rodríguez-Fernández J. L., Cabañas C. (2004) Signaling through the leukocyte integrin LFA-1 in T cells induces a transient activation of Rac-1 that is regulated by Vav and PI3K/Akt-1. J. Biol. Chem. 279, 16194–16205 [DOI] [PubMed] [Google Scholar]
- 54. Stanley P., Smith A., McDowall A., Nicol A., Zicha D., Hogg N. (2008) Intermediate-affinity LFA-1 binds α-actinin-1 to control migration at the leading edge of the T cell. EMBO J. 27, 62–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fanning A., Volkov Y., Freeley M., Kelleher D., Long A. (2005) CD44 cross-linking induces protein kinase C-regulated migration of human T lymphocytes. Int. Immunol. 17, 449–458 [DOI] [PubMed] [Google Scholar]
- 56. Long A., Mitchell S., Kashanin D., Williams V., Prina Mello A., Shvets I., Kelleher D., Volkov Y. (2004) A multidisciplinary approach to the study of T cell migration. Ann. N.Y. Acad. Sci. 1028, 313–319 [DOI] [PubMed] [Google Scholar]
- 57. Chae Y. C., Kim K. L., Ha S. H., Kim J., Suh P. G., Ryu S. H. (2010) Protein kinase Cδ-mediated phosphorylation of phospholipase D controls integrin-mediated cell spreading. Mol. Cell. Biol. 30, 5086–5098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tachado S. D., Mayhew M. W., Wescott G. G., Foreman T. L., Goodwin C. D., McJilton M. A., Terrian D. M. (2002) Regulation of tumor invasion and metastasis in protein kinase Cϵ-transformed NIH3T3 fibroblasts. J. Cell Biochem. 85, 785–797 [DOI] [PubMed] [Google Scholar]
- 59. Pan Q., Bao L. W., Kleer C. G., Sabel M. S., Griffith K. A., Teknos T. N., Merajver S. D. (2005) Protein kinase Cϵ is a predictive biomarker of aggressive breast cancer and a validated target for RNA interference anticancer therapy. Cancer Res. 65, 8366–8371 [DOI] [PubMed] [Google Scholar]
- 60. Pan Q., Bao L. W., Teknos T. N., Merajver S. D. (2006) Targeted disruption of protein kinase Cϵ reduces cell invasion and motility through inactivation of RhoA and RhoC GTPases in head and neck squamous cell carcinoma. Cancer Res. 66, 9379–9384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Prekeris R., Mayhew M. W., Cooper J. B., Terrian D. M. (1996) Identification and localization of an actin-binding motif that is unique to the ϵ isoform of protein kinase C and participates in the regulation of synaptic function. J. Cell Biol. 132, 77–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Hernandez R. M., Wescott G. G., Mayhew M. W., McJilton M. A., Terrian D. M. (2001) Biochemical and morphogenic effects of the interaction between protein kinase Cϵ and actin in vitro and in cultured NIH3T3 cells. J. Cell Biochem. 83, 532–546 [DOI] [PubMed] [Google Scholar]
- 63. Besson A., Davy A., Robbins S. M., Yong V. W. (2001) Differential activation of ERKs to focal adhesions by PKCϵ is required for PMA-induced adhesion and migration of human glioma cells. Oncogene 20, 7398–7407 [DOI] [PubMed] [Google Scholar]
- 64. Besson A., Wilson T. L., Yong V. W. (2002) The anchoring protein RACK1 links protein kinase Cϵ to integrin β chains. Requirements for adhesion and motility. J. Biol. Chem. 277, 22073–22084 [DOI] [PubMed] [Google Scholar]
- 65. Takai Y., Kaibuchi K., Kikuchi A., Kawata M. (1992) Small GTP-binding proteins. Int. Rev. Cytol. 133, 187–230 [DOI] [PubMed] [Google Scholar]
- 66. Magee T., Newman C. (1992) The role of lipid anchors for small G proteins in membrane trafficking. Trends Cell Biol. 2, 318–323 [DOI] [PubMed] [Google Scholar]
- 67. Seabra M. C., Goldstein J. L., Südhof T. C., Brown M. S. (1992) Rab geranylgeranyl transferase. A multisubunit enzyme that prenylates GTP-binding proteins terminating in Cys-X-Cys or Cys-Cys. J. Biol. Chem. 267, 14497–14503 [PubMed] [Google Scholar]
- 68. Bretscher M. S. (1996) Moving membrane up to the front of migrating cells. Cell 85, 465–467 [DOI] [PubMed] [Google Scholar]
- 69. Caswell P. T., Spence H. J., Parsons M., White D. P., Clark K., Cheng K. W., Mills G. B., Humphries M. J., Messent A. J., Anderson K. I., McCaffrey M. W., Ozanne B. W., Norman J. C. (2007) Rab25 associates with α5β1 integrin to promote invasive migration in 3D microenvironments. Dev. Cell 13, 496–510 [DOI] [PubMed] [Google Scholar]
- 70. Soldati T., Riederer M. A., Pfeffer S. R. (1993) Rab GDI: a solubilizing and recycling factor for rab9 protein. Mol. Biol. Cell 4, 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ullrich O., Horiuchi H., Bucci C., Zerial M. (1994) Membrane association of Rab5 mediated by GDP-dissociation inhibitor and accompanied by GDP/GTP exchange. Nature 368, 157–160 [DOI] [PubMed] [Google Scholar]
- 72. Svensson L., Stanley P., Willenbrock F., Hogg N. (2012) The Gαq/11 proteins contribute to T lymphocyte migration by promoting turnover of integrin LFA-1 through recycling. PLoS ONE 7, e38517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Stanley P., Tooze S., Hogg N. (2012) A role for Rap2 in recycling the extended conformation of LFA-1 during T cell migration. Biol. Open. 1, 1161–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Onodera Y., Nam J. M., Hashimoto A., Norman J. C., Shirato H., Hashimoto S., Sabe H. (2012) Rab5c promotes AMAP1-PRKD2 complex formation to enhance β1 integrin recycling in EGF-induced cancer invasion. J. Cell Biol. 197, 983–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rappoport J. Z., Simon S. M. (2003) Real-time analysis of clathrin-mediated endocytosis during cell migration. J. Cell Sci. 116, 847–855 [DOI] [PubMed] [Google Scholar]
- 76. Fabbri M., Di Meglio S., Gagliani M. C., Consonni E., Molteni R., Bender J. R., Tacchetti C., Pardi R. (2005) Dynamic partitioning into lipid rafts controls the endo-exocytic cycle of the αL/β2 integrin, LFA-1, during leukocyte chemotaxis. Mol. Biol. Cell 16, 5793–5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.