Background: The role of self-mediated keratin filament cross-linking is addressed in transgenic mice.
Results: A chimeric type II keratin, K8bc, partially rescues Krt5-null mice.
Conclusion: Despite similar assembly properties, K8bc is functionally distinct from its natural counterpart K5 in skin tissue in vivo.
Significance: Type II keratins possess unique properties beyond their potential for filament formation subject to cell type-intrinsic regulation.
Keywords: Epidermis, Intermediate Filament, Mitochondria, Skin, Transgenic Mice, Blistering, Cell Fragility, Epidermolysis Bullosa Simplex, Genodermatosis, Keratin
Abstract
The crucial role of structural support fulfilled by keratin intermediate filaments (IFs) in surface epithelia likely requires that they be organized into cross-linked networks. For IFs comprised of keratins 5 and 14 (K5 and K14), which occur in basal keratinocytes of the epidermis, formation of cross-linked bundles is, in part, self-driven through cis-acting determinants. Here, we targeted the expression of a bundling-competent KRT5/KRT8 chimeric cDNA (KRT8bc) or bundling-deficient wild type KRT8 as a control to the epidermal basal layer of Krt5-null mice to assess the functional importance of keratin IF self-organization in vivo. Such targeted expression of K8bc rescued Krt5-null mice with a 47% frequency, whereas K8 completely failed to do so. This outcome correlated with lower than expected levels of K8bc and especially K8 mRNA and protein in the epidermis of E18.5 replacement embryos. Ex vivo culture of embryonic skin keratinocytes confirmed the ability of K8bc to form IFs in the absence of K5. Additionally, electron microscopy analysis of E18.5 embryonic skin revealed that the striking defects observed in keratin IF bundling, cytoarchitecture, and mitochondria are partially restored by K8bc expression. As young adults, viable KRT8bc replacement mice develop alopecia and chronic skin lesions, indicating that the skin epithelia are not completely normal. These findings are consistent with a contribution of self-mediated organization of keratin IFs to structural support and cytoarchitecture in basal layer keratinocytes of the epidermis and underscore the importance of context-dependent regulation for keratin genes and proteins in vivo.
Introduction
Intermediate filaments (IFs)2 make an essential contribution to the maintenance of cell and tissue integrity in surface epithelia (e.g. skin and oral mucosa), muscle, and other tissues routinely subjected to stress. Loss or disruption of this function accounts for the cell and tissue fragility that underlies a large number of genetically determined diseases that are individually rare but debilitating (1–3). Epidermolysis bullosa simplex, for instance, is a condition in which the basal cell layer of epidermis and related stratified epithelia ruptures in response to trivial frictional trauma and is associated with dominantly acting mutations in either keratin 5 or 14 (K5 or K14), the main keratin pairing that is expressed in this epithelial setting (4–6). Epidermolysis bullosa simplex is the first among more than 80 IF-based disorders (2). There is as yet no effective treatment for epidermolysis bullosa simplex and other disorders rooted in defective IFs (7, 8).
The role of keratin IFs toward the maintenance of cell and tissue integrity is a composite function of their unique micromechanical properties (9, 10) and integration within the cytoskeleton and cell adhesion apparatuses (11–13). This function reflects many key attributes of keratin IFs including their intracellular abundance (14), their organization as cross-linked networks (10, 15), and their attachment at sites of cell-cell and cell-matrix adhesions (11, 12) and at the surface of the nucleus (16). The molecular basis for IF anchorage at sites of integrin-based cell-matrix adhesions and cadherin-based cell-cell adhesions is quite well understood (17, 18). By comparison, the basis for the cross-linked organization of IFs in vivo is poorly understood for most types of cells. In epidermal and hair keratinocytes, for instance, the filament bundling-promoting influence of filaggrin (19) and trichohyalin (20) is well established but restricted to late stages of terminal differentiation, whereas the influence of plectin, epiplakin, and related plakin family members (21–23) or interfilament disulfide bonding (24, 25) remains unclear.
Keratin IFs assembled from the type II K5 (590 residues; ∼58 kDa in humans (26)) and type I K14 (472 residues; ∼50 kDa in humans (27)) exhibit the remarkable property of self-organization into cross-linked networks both as purified entities in vitro (10, 28–30) and when expressed in keratin-free fibroblasts (30). This property of self-organization enhances the mechanical resilience of K5-K14 filament assemblies and depends on interactions involving the distal 25 amino acids of the K14 C-terminal domain, which is exposed at the filament surface, and two separate regions within K5, the so-called head-1A (70 residues) and L2-2B subdomains (129 residues) (30, 31). The property of self-organization appears to be specific to natural keratin pairings (e.g. K5-K14 and K8-K18 (15)) but is transferable upon “chimeragenesis.” For instance, K8 (483 residues; ∼54 kDa in humans (32)), a type II keratin normally restricted to simple epithelial linings, readily co-polymerizes to yield 10-nm filaments when paired with K14, but this pairing does not show self-organization either in vitro or in transfected cells (30). Transferring the head-1A and L2-2B subdomains from K5 into K8 confers the property of self-organization to the resulting chimera (designated K8bc) when artificially paired with K14 in vitro as well as in transfected fibroblasts (30).
Lee and Coulombe (30) predicted that the property of self-organization makes a significant contribution to the structural support role of K5-K14 in the basal layer of the epidermis. A corollary from this prediction is that targeted expression of the K8bc chimera, but not wild type K8, to basal layer keratinocytes should rescue the extensive skin blistering and perinatal lethality phenotype of mice carrying a null mutation at the Krt5 locus (33). Here, we report on our effort to carry out such an experiment using Krt5-null mice.
EXPERIMENTAL PROCEDURES
Cells and Transgenic Mouse Lines
All protocols involving mice were approved by The Johns Hopkins University Animal Care and Use Committee (Baltimore, MD). Mouse lines were maintained under specific pathogen-free conditions and fed chow and water ad libitum. The plasmids pIRES2-GFP hK8bc and pIRES2-GFP hK8 (30) were digested with EcoRI and NheI (New England Biolabs), and the resulting cDNA inserts were treated with calf intestinal phosphatase (New England Biolabs), gel-purified using a QIAquick gel extraction kit (Qiagen), and subcloned into the BamHI site of the modified KRT14 expression vector (34). Clones of the correct orientation were transiently transfected into cell line NIH-3T3 (ATCC CRL-1658) with GeneJuice® transfection reagent (EMD Millipore) for preliminary observations. Plasmids were then linearized with KpnI and AseI (New England Biolabs), and the final linearized ∼5-kb DNA fragment was used to generate transgenic mice by pronuclear injection. Genomic DNA was isolated from potential founders at approximately 6 weeks of age. Relative transgene copy number across lines was determined using PCR with the following primer sets: 5′-CCTGGTCATCATCCTGCC-3′ (rabbit β-globin gene intron), 5′-GGACTTCTGGGTCACCCTGATGG-3′ (KRT8), 5′-AGGTAGGTGGCCAGGCGGTC-3′ (Krt16 forward), and 5′-CCACACCCACAGATCTGGGC-3′ (Krt16 reverse). Band intensity was measured using ImageJ and is reported relative to the Krt16 locus used as a stable internal reference. All subsequent progeny were genotyped using AccustartTM II mouse genotyping kit (Quanta Biosciences) according to the manufacturer's instructions using the primers 5′-TGCATATAAATTCTGGCTGGCG-3′ (forward) and 5′-GCATGAACATGGTTAGCAGAGGG-3′ (reverse), which are directed to the rabbit β-globin intron contained within the expression vector.
Krt5-null mice were described previously (33, 35) and were maintained in a mixed genetic background. These mice were genotyped using a multiplex PCR assay using the universal forward primer 5′-CCCACTAATCATTCACAGCTCG-3′, Krt5 reverse primer 5′-ACCAAAACCAAATCCACTGCCG-3′, and HPRT reverse primer 5′-CGAGTCTGAAGCTCTCGATTTCC-3′. Replacement mice were generated by breeding KRT8bcTg/− or KRT8Tg/− mice with Krt5+/− mice to yield KRT8bcTg/−Krt5+/− or KRT8Tg/−Krt5+/− mice, respectively. These mice were then bred with Krt5+/− mice to yield the following genotypes: Krt5+/+ or Krt5+/− (control), KRT8bcTg/− Krt5+/+ or KRT8Tg/−Krt5+/+ (referred to herein as KRT8bc or KRT8 transgenic mice), Krt5−/− (Krt5-null), and KRT8bcTg/−Krt5−/− or KRT8Tg/−Krt5−/− (referred to herein as KRT8bc or KRT8 replacement mice).
Statistical Analyses
All statistics were performed using GraphPad Prism software. One- or two-way analyses of variance were used to test for significance where appropriate, and adjusted p values are reported.
Protein Purification and Analysis of Transgene Expression
Plasmids pET-K5 and pET-K8 (30) were transformed into the Escherichia coli strain BL21(DE3) to produce the corresponding recombinant proteins as inclusion bodies (30, 36). These recombinant proteins were purified as described previously (10, 15) using HiTrap Q and Mono Q ion exchange chromatography columns (GE Healthcare). Protein concentrations were determined using a Bradford assay kit (Bio-Rad), and serial dilutions were performed to yield standards of known concentration for analyses outlined below.
For analysis of transgene expression in development stage 18.5 (E18.5) embryos, pregnant mice were sacrificed 18 days post-timed mating, and the skins of the embryos were harvested. Skins were chopped repeatedly with a razor blade, and urea-soluble proteins were then extracted as described previously (37). High salt extractions were performed using E18.5 embryonic skin samples as described (38). Protein concentrations were determined with a Bradford assay kit (Bio-Rad). To determine expression levels of endogenous K5 protein and of transgenic proteins, standards of known concentration were run alongside E18.5 samples. Otherwise, equal quantities of protein from genotypes of interest were resolved by 10% SDS-PAGE and transferred to nitrocellulose membranes. Membranes were blocked, and antibody incubations were performed in 5% milk in 1× TBS-Tween 20. Protein epitopes were detected with the following antibodies: rabbit anti-K5 AF-138 (Covance), rabbit anti-K14 AF-64 (Covance), rabbit anti-Hsp70/Hsp72 ADI-SPA-812 (Enzo Life Sciences), chicken anti-K8 ab107115 (Abcam), rabbit anti-K17 (39), and mouse anti-GAPDH antibody sc-365062 (Santa Cruz Biosciences). Secondary antibodies used were horseradish peroxidase-conjugated goat anti-rabbit, goat anti-chicken, and goat anti-mouse. Blots were developed using the SuperSignal West Pico Chemiluminescent substrate kit and were imaged with FluorChem® Q MultiImage III (Alpha Innotech). Band signal intensity was measured from these images with the AlphaView® Q software. The standard curve line of best fit equation and R2 values were calculated using GraphPad Prism. Calculation of K5, K8bc, and K8 quantities was performed using the standard curve equations in Microsoft Excel.
For analysis of transgenic protein expression in adult animals, wild type and transgenic animals were sacrificed at 6 weeks of age, and the back skins were harvested. The tissue was snap frozen, and urea-soluble proteins were extracted as described previously (37). Equal quantities of urea-soluble protein were resolved by 10% SDS-PAGE and transferred onto nitrocellulose membranes. Blots were developed using the SuperSignal West Pico Chemiluminescent substrate kit and were imaged with FluorChem Q MultiImage III (Alpha Innotech). All images were inverted and cropped using ImageJ software.
Quantitative Real Time PCR
Total RNA from either 6-week-old adult animals or E18.5 embryos was isolated using TRIzol® reagent according to the manufacturer's instructions. Complementary DNA was synthesized using an iScriptTM cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. Quantitative real time PCR was carried out using SYBR® Green Real-Time PCR Master Mix from Invitrogen on a Bio-Rad C1000TM thermocycler and CFXTM Real-Time System. Primers used were directed toward KRT8 (5′-CAAGGTGTCCACCTCTGGC-3′ and 5′-ATAGCCGCCGCCCAGGCCA-3′), Krt14 (5′-AGCGGCAAGAGTGAGATTTCT-3′ and 5′-CCTCCAGGTTATTCTCCAGGG-3′), Actb (5′-GGCTGTATTCCCCTCCATCG-3′ and 5′-CCAGTTGGTAACAATGCCATGT-3′), and Gapdh (5′-AAATGGTGAAGGTCGGTGT-3′ and 5′-ACTCCACGACATACTCAGCAC-3′). ΔCq and ΔΔCq for each target were determined by first subtracting the averaged Cq values for Actb and Gapdh from the Cq of the target and then by subtracting the ΔCq of the wild type sample from each of the other samples when applicable. Relative RNA quantity was calculated by taking 2−ΔCq, and relative -fold change was calculated by taking 2−ΔΔCq. Calculations were performed using Microsoft Excel.
Histopathology
Phenotypic adult KRT8bc replacement and control mice were harvested at various ages for morphological study. Prior to sacrifice, images of the mice were taken with a Samsung Digimax S500 digital camera. Tissues analyzed included hairy skin, hairless skin (if present), ulcerative skin, tongue, forepaws, and tail. Tissues were fixed overnight in Bouin's fixative (Bio-Rad), dehydrated, and processed for paraffin embedding by AML Laboratories. Sections were cut 5 μm thick and stained with hematoxylin and eosin. Images were taken on a Zeiss AxioObserver inverted microscope with an AxioCam HRc camera. Images were cropped, and scale bars were added in ImageJ.
Immunofluorescence
For analysis of cultured keratinocytes, E18.5 embryos were again harvested, and the skins were removed. The keratinocytes were isolated using a modified version of the protocol reported by Reichelt and Haase (40). After incubation with Dispase solution, keratinocytes were isolated as reported previously (41). Keratinocytes were cultured on collagen I-coated coverslips in CnT-57 medium (CELLnTEC) supplemented with penicillin/streptomycin at 37 °C and 5% CO2 in the absence of 3T3-J2 feeder cells. Subconfluent coverslips were fixed with ice-cold methanol at −20 °C or with 3.3% paraformaldehyde at room temperature for 10 min. Coverslips fixed with paraformaldehyde were extracted with 1% Triton X-100 in PBS buffer for 5 min at room temperature. Antibodies used to detect K5 and K14 were described above. K8 was detected with chicken anti-human K8 antibody ab14053 (Abcam). Secondary antibodies used were goat anti-chicken Alexa Fluor 488 and goat anti-rabbit Alexa Fluor 594, and nuclei were stained with Hoechst fluorescent dye. Images were taken on a Zeiss AxioObserver inverted microscope with an AxioCam MRm camera.
For analysis of adult transgenic back skin, animals were harvested as described above and a portion of the back skin was embedded in Tissue-Tek® O.C.T. medium (Sakura) and frozen in liquid nitrogen. 5-μm-thick frozen sections were cut and washed once with PBS buffer before probing with the antibodies described above. Images were taken on a Zeiss AxioObserver inverted microscope with an AxioCam MRm camera. All images were cropped, and scale bars were added using ImageJ.
Electron Microscopy
Skins from 6-week-old adult animals or E18.5 embryos were harvested and immersed in 2% paraformaldehyde, 2% glutaraldehyde for 48 h. After primary fixation, tissue samples were postfixed in 1% osmium tetroxide in 0.1 m cacodylate buffer for 1 h on ice. Tissue samples were then rinsed and en bloc stained with 2% aqueous uranyl acetate for 1 h at room temperature. Samples were then dehydrated in a graded series of ethanol (50, 70, 95, and 100%). Tissue was cleared in propylene oxide and then infiltrated in a graded series of Epon 812/propylene oxide (50:50 and 70:30) for 1 h and overnight, respectively, followed by two changes of pure Epon 812 and then polymerized for 48 h at 60 °C. Semithin sections (1 μm) were cut using glass knives on a Sorvall MT2B ultramicrotome. Sections were mounted on glass slides and stained with 1% toluidine blue in 1% sodium borate and coverslipped with Permount. Areas of interest were selected, and thin sections (60–80 nm) were cut using a diamond knife. The sections were mounted on Formvar-carbon-coated 150 mesh copper grids and poststained with 2% aqueous uranyl acetate and Reynold's lead citrate (42). Sections were analyzed using a Hitachi HU12A electron microscope. Eastman Kodak Co. electron microscope film 4489 was developed, and negatives were scanned using an Epson Perfection V500 photo scanner. Images were cropped, and scale bars were added in ImageJ. For cell and nuclear aspect ratio analysis, lines were drawn from the basement membrane to the top of the cells (height) and from side to side (width) at the greatest distance for each cell measured using ImageJ. The lines were measured, and the height was divided by the width. Basal cell desmosomes, hemidesmosomes, and mitochondria were assessed individually from 5000× images.
RESULTS
KRT8 and KRT8bc Transgenes Are Expressed in Transgenic Mouse Epidermis
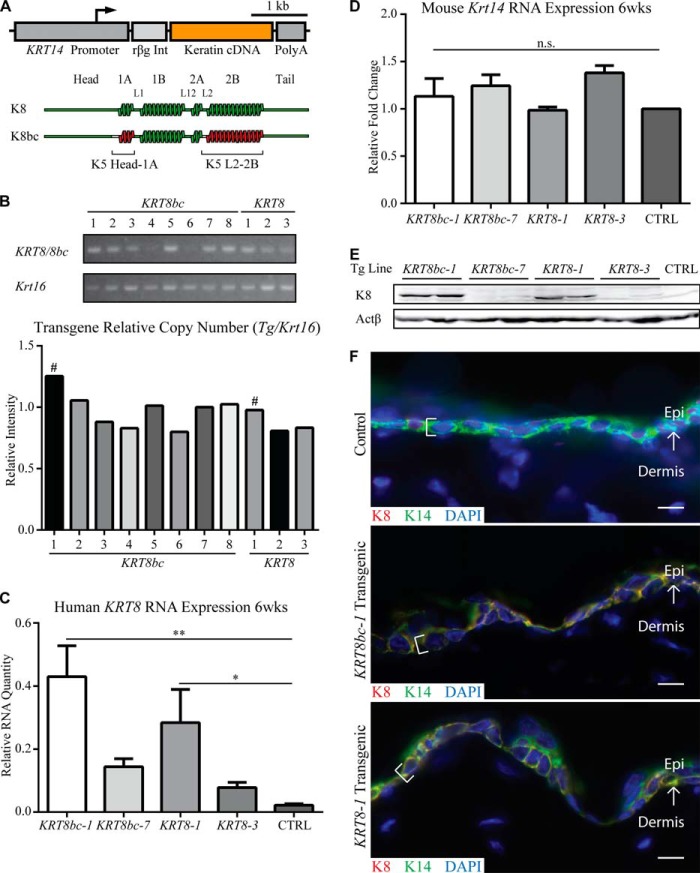
The cDNAs encoding wild type human K8 (15) and the K8bc chimera (30) were subcloned into the KRT14 gene promoter-based expression vector (34) as described (37). Transfection of the final DNA constructs (Fig. 1A) into NIH-3T3 cells along with wild type KRT14 gave rise to keratin IFs (data not shown), confirming that K8 and K8bc can each co-polymerize with K14 as reported (30). The two constructs were isolated and used to generate transgenic founders by pronuclear injection. Transgene-positive founders were then bred to establish transgenic mouse lines. For each construct, the two lines showing the highest transgene copy number in a single insertion site (based on internally controlled PCR and frequency of F1 transgenic offspring; see Fig. 1B and data not shown) were retained for further analysis.
FIGURE 1.
Generation and characterization of transgenes. A, schematic depiction of the transgenes used in this study. The human KRT14 gene promoter was used to drive the tissue-specific expression of keratin cDNAs (KRT8 and KRT8bc), and the rabbit β-globin intron (rβg Int) and human KRT14 3′-UTR sequence (Poly(A)) serve to stabilize the transgene mRNA in mouse cells. B, relative transgene copy number analysis was performed by conventional PCR of genomic DNA with transgene-specific primers. Krt16 was used as a single copy number reference gene. # indicates the highest relative copy number line for each transgene. C and D, quantitative RT-PCR analysis of transgenic human KRT8 mRNA (C) and endogenous mouse Krt14 mRNA (D) in back skin harvested from sex-matched 6-week-old animals. Relative RNA amount is normalized to both actin and Gapdh. CTRL indicates Krt5+/+ skin. Error bars represent S.E. A one-way analysis of variance (Dunnett's test) was used to test for significance, and the adjusted p values are reported. n.s., not significant; *, p < 0.04; **, p < 0.002. E, analysis of total skin protein extracts (10 μg/lane) by Western blotting in 6-week-old sex-matched adult animals. Two mice were analyzed for each of four transgenic lines; CTRL indicates Krt5+/+ skin. F, analysis of transgene expression in frozen skin sections of 6-week-old sex-matched adult animals. K8 epitopes are only present in basal layer keratinocytes (see brackets) of KRT8bc-1 and KRT8-1 transgenic epidermis and co-localizes with endogenous K14 in a normal keratin filament network. Control indicates Krt5+/+ skin. Arrows depict the interface between the epidermis (Epi) and dermis. Bars, 10 μm. wks, weeks.
The resulting progeny from these transgenic lines, designated KRT8bc line 1 (KRT8bc-1), KRT8bc line 7 (KRT8bc-7), KRT8 line 1 (KRT8-1), and KRT8 line 3 (KRT8-3), did not exhibit any visible phenotype. Back skin tissue was harvested from age- and sex-matched animals from these lines for analysis of transgene expression. Of these four lines, two expressed transgene mRNA at appreciable levels by quantitative RT-PCR: lines KRT8bc-1 and KRT8-1 (Fig. 1C). Expression of Krt14 mRNA was unaffected relative to wild type (Fig. 1D). Western blot analyses assessing protein expression revealed similar results with appreciable and comparable levels of transgenic protein detectable for lines KRT8bc-1 and KRT8-1 only (Fig. 1E). Analysis of frozen sections of mouse back skin by immunofluorescence confirmed that K8-specific epitopes occurred in the basal layer of the epidermis as expected (Fig. 1F). The KRT8bc-1 and KRT8-1 lines were selected to test whether KRT14 promoter-driven expression of K8 or K8bc protein can rescue the Krt5-null phenotype.
The KRT8bc, but Not the KRT8, Transgene Is Able to Rescue the Krt5-null Phenotype
Krt5-null mice exhibit severe fragility and blistering of the skin and die with complete penetrance within the 1st h of birth (33), providing a clear readout for the ability of other type II keratins (e.g. K8 and K8bc) to functionally substitute for endogenous K5. The KRT8bc-1 and KRT8-1 transgenic mice (Fig. 1) were each bred to Krt5-hemizygous null (Krt5+/−) mice. Once sexually mature, the resulting KRT8bcTg/−Krt5+/− and KRT8Tg/−Krt5+/− offspring were crossed with Krt5+/− mice. Although this strategy ensures a uniform number of rescue transgene copies within the progeny, only one of every eight offspring is predicted to be a candidate for phenotypic rescue (KRT8bc or KRT8 replacement mice). Replacement events wherein the mouse survived to weaning age (P21) were scored as successful rescues. Of 411 live births analyzed, 16 of 34 mice with the KRT8bc replacement genotype (47%) survived to weaning (Table 1). The surviving KRT8bc replacement animals did not show any detectable blistering at the time of birth and grew normally to adulthood, indicating that the transgene could functionally substitute for endogenous K5 during that period. By contrast, none of the 11 mice born with the KRT8 replacement genotype from 191 live births showed rescue (Table 1); in fact, none lived beyond the first few hours after birth, demonstrating that the KRT8 transgene cannot rescue the K5 deficiency.
TABLE 1.
Summary of type II keratin protein replacement experiment in mouse
Progeny from the replacement breeding strategy (see “Experimental Procedures”) were monitored at and following birth. “Viable replacement” implies that the mouse survived at least until weaning (P21).
| Transgenic line | No. live births | No. animals genotyped | No. replacement animals | No. viable replacements | Viable replacements |
|---|---|---|---|---|---|
| % | |||||
| KRT8bc-1 | 411 | 361 | 34 | 16 | 47 |
| KRT8-1 | 191 | 180 | 11 | 0 | 0 |
Expression of K8bc and K8 Replacement Proteins and mRNAs in the Skin of E18.5 Krt5-null Mice
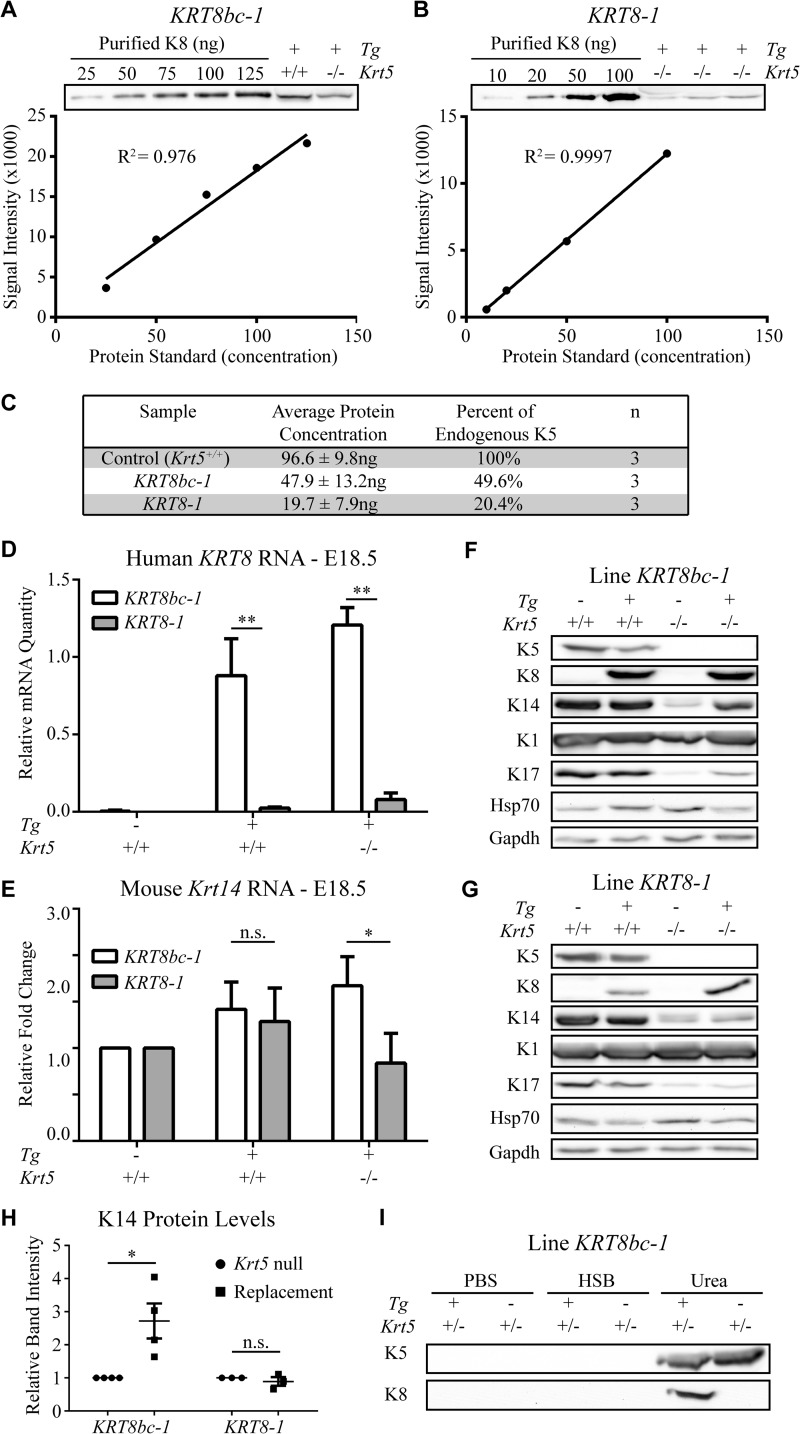
Next, the amount of K8bc and K8 transgenic proteins in the skin of KRT8bc and KRT8 replacement mice were assessed, relating them to the levels of K5 protein in control mice. This analysis was conducted in E18.5 embryos just prior to birth to enable a direct comparison between the two types of rescue proteins and to avoid complications stemming from the extensive skin blistering seen in Krt5-null and KRT8 replacement mice. Purified recombinant proteins were used to generate standard curves for K5 and K8 epitopes (see “Experimental Procedures”), enabling the determination of transgenic protein amount in these lines (Fig. 2, A and B, and data not shown). Using this strategy, we found that endogenous K5 is expressed at 96.6 ng/10 μg of total skin proteins in control E18.5 embryos (n = 3; Fig. 2C). KRT8bc replacement embryos were found to express 47.9 ng of K8bc protein/10 μg of total protein extract (n = 3), corresponding to ∼50% of the concentration of endogenous K5 in control embryos (Fig. 2C). The KRT8 replacement embryos were determined to express 19.7 ng of K8 protein/10 μg of total protein extract (n = 3), accounting for ∼20% of endogenous K5 levels (Fig. 2C). The difference between K8bc and K8 expression was also observed at the mRNA level in E18.5 mouse embryos (Fig. 2D). No difference in the partitioning of K8bc and K5 proteins was observed during fractionation by high salt extraction (Fig. 2I), conveying that K8bc behaves like a bona fide keratin even in the presence of endogenous K5. We infer that the “half-normal” levels of K8bc protein may explain, in part, the partial rescue observed in KRT8bc replacement mice (47% frequency), whereas the even lower levels of K8 protein may have played a role in the failure to obtain successful rescues in KRT8 replacement mice (see “Discussion”).
FIGURE 2.
Analysis of transgene expression in E18.5 mouse embryos. A and B, quantification of transgenic protein expressed in KRT8bc line 1 (KRT8bc-1; A) and KRT8 line 1 (KRT8-1; B) using standard curves of purified recombinant human K8. The line of best fit and R2 values were calculated using GraphPad Prism. C, summary of the transgenic protein quantification from multiple biological (n) and technical replicates. D, transgene RNA expression levels for lines KRT8bc-1 and KRT8-1 in E18.5 skin. E, endogenous Krt14 RNA expression levels for lines KRT8bc-1 and KRT8-1 in E18.5 skin. RNA levels are illustrated relative to actin and Gapdh. Error bars represent S.E. A two-way analysis of variance (Sidak's test) was used to test for significance, and the adjusted p values are reported. n.s., not significant; *, p < 0.005; **, p < 0.0001. F and G, expression of transgenic and endogenous proteins for lines KRT8bc-1 (F) and KRT8-1 (G) in E18.5 skin is shown by Western blot analysis. H, intensity of K14 protein signal relative to loading control signal was quantified, and the increase of K14 in the replacement mouse skin setting is illustrated. I, Western blot analysis of high salt extraction performed on KRT8bc-1 E18.5 skin. Even in the presence of endogenous K5 as a competitor, transgenic K8bc was still found in the insoluble pool. HSB, high salt buffer.
There is substantive evidence indicating interdependence in the steady-state levels of type I and type II keratin proteins in epithelial cells (43, 44). Accordingly, the levels of K5 protein are reduced in newborn Krt14−/− mouse skin (45), whereas the levels of K14 protein are reduced in newborn Krt5−/− mouse skin (33). In the Krt5-null case, in particular, the levels of Krt14 mRNA are also substantially reduced (33), hinting at the complexity of mechanisms regulating the balance between type I and type II keratins in epidermal keratinocytes in vivo. Levels of K14 protein provide a surrogate marker for the “effective dosage” of type II keratin expression in the skin of E18.5 embryos. At the protein level, very weak K14 expression occurred in Krt5-null embryos as well as in KRT8 replacement embryos (Fig. 2, F and G), respectively emulating the complete absence of type II keratins and the low levels of K8 rescue protein observed in these settings. By comparison and as expected, expression of K14 protein was stimulated ∼2.7-fold in KRT8bc replacement embryos over Krt5-null levels (Fig. 2H), consistent with the phenotypic rescue of viable KRT8bc replacement mice (Table 1). Interestingly, the wound-inducible type I keratin K17 followed a similar pattern as K14, further substantiating the status of the type II rescue protein (Fig. 2, F and G). By contrast, the steady state levels of K1, a marker of terminal differentiation, and of Hsp70 (46), a heat shock protein known to interact with keratins, were very similar across genotypes (Fig. 2, F and G).
Examination of Krt14 mRNA levels in E18.5 embryos of relevant genotypes provided additional insight. Similar levels of Krt14 mRNAs occurred in KRT8 and KRT8bc transgenic skin (with a normal Krt5 locus), and these levels were slightly elevated relative to that seen in control embryos (albeit in a non-statistically significant fashion; Fig. 2E). Krt14 mRNA levels were increased nearly 2-fold in the setting of KRT8bc replacement embryos, exhibiting a striking and statistically significant increase compared with KRT8 replacement E18.5 embryos (Fig. 2E). These data clearly point to a mechanism(s) by which basal keratinocytes are able to sense the anomaly stemming from ectopically expressing other type II keratins and to sense the difference between K8bc and K8 at the mRNA and/or protein level.
Analysis of Keratin IFs in Basal Keratinocytes in Primary Culture and in the Epidermis in Situ
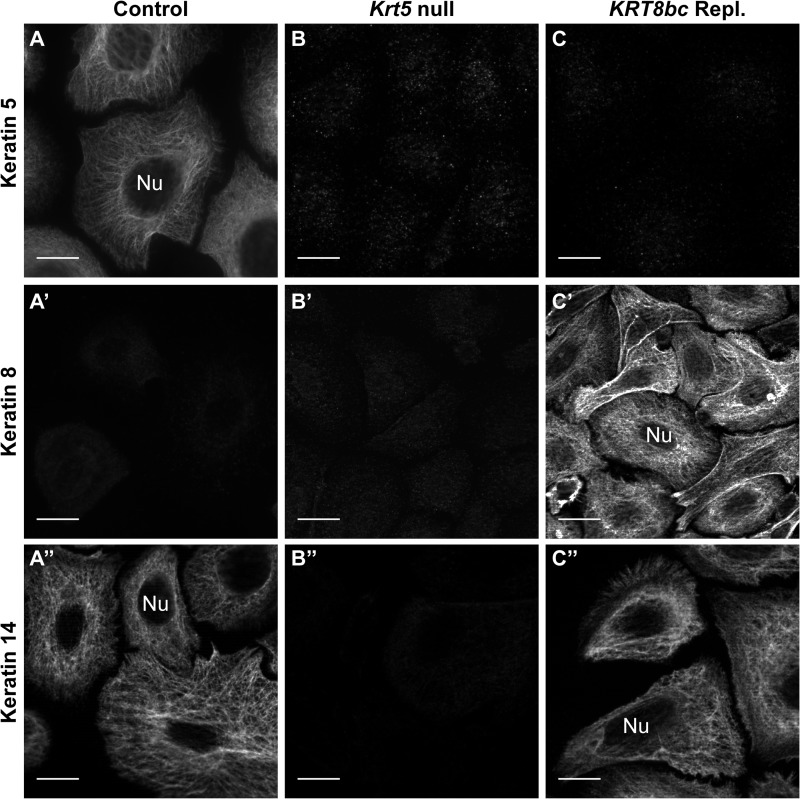
To further assess the behavior of the K8bc protein in vivo, control, Krt5-null, and KRT8bc replacement E18.5 embryos were processed for skin keratinocyte isolation, primary culture, and analysis via indirect immunofluorescence. In control cultures, K5 and K14 were co-localized (data not shown) and formed a classic pan-cytoplasmic array (Fig. 3, A and A″), whereas K8 was typically absent (Fig. 3A′) except in rare cells that may correspond to Merkel cells (data not shown), which express K8 naturally (47). In Krt5-null keratinocyte cultures, no signal was detected for K5 and K8 antigens as expected, whereas the signal for K14 was weak (Fig. 3, B–B″). In KRT8bc replacement cultures, there was no signal for K5 (as expected), and K8bc and K14 were consistently co-localized in the context of normal looking arrays of keratin IFs (Fig. 3, C′ and C″, and data not shown). Images of control and KRT8bc replacement keratinocytes taken under identical exposure conditions did not exhibit reduced levels of K14 protein in contrast to the Western blot data presented in Fig. 2E. This may be due in part to the induced expression of other keratins in the context of primary culture of keratinocytes (48, 49). These findings obtained in mouse skin keratinocytes significantly extend our previous work demonstrating that K8bc is competent for 10-nm filament assembly and IF network formation when paired with type I K14 (30).
FIGURE 3.
Analysis of embryonic skin keratinocytes in primary culture by indirect immunofluorescence. Conventional fluorescence microscopy of keratinocytes harvested from Krt5+/+ (Control; A–A″), Krt5−/− (Krt5 null; B–B″), or KRT8bcTg/−Krt5−/− (KRT8bc Repl.; C–C″) E18.5 embryos (as indicated on top) is shown. Fixed cells were immunostained for keratin 5, keratin 8, and keratin 14 epitopes as indicated at left. Nu, nucleus. Bars, 10 μm.
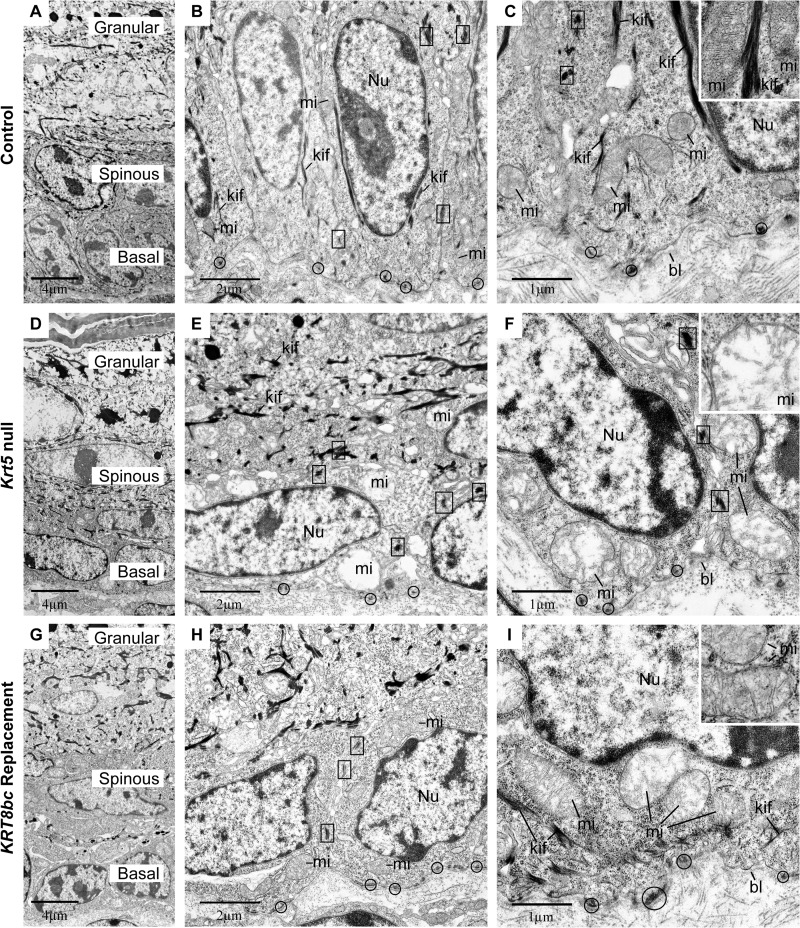
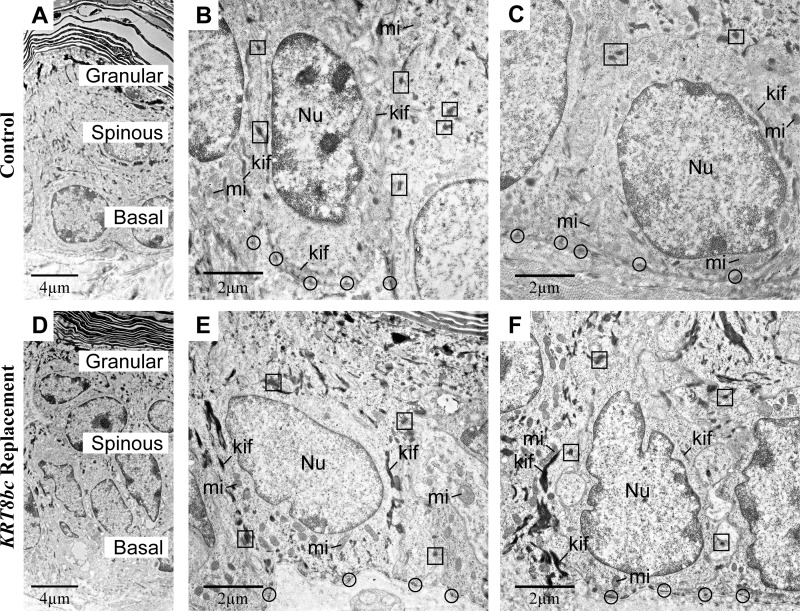
To further examine the innate features of basal keratinocytes including their keratin IF network, we processed E18.5 skin for routine transmission electron microscopy. At this late embryonic stage, the epidermis is mature, and adult-like barrier properties have been acquired (50). In this setting, basal keratinocytes were columnar in shape with an ovoid nucleus oriented along the long axis of the cell (Fig. 4, A–C). Hemidesmosomes and desmosomes were numerous and regularly spaced at the basal pole of basal keratinocytes and at sites of cell-cell contact, respectively, and exhibited association with electron-dense filamentous structures, indicating attachment to keratin IFs (Fig. 4, B and C). In addition, long bundles of keratin IFs oriented along the long axis of the cell could be seen on each side of the nucleus, and mitochondria appeared healthy (Fig. 4, B and C). In areas of Krt5-null embryonic skin for which the epidermis is intact, the thickness, stratification, and differentiated layers of the epidermis appeared normal (Fig. 4D). That said, basal keratinocytes appeared flattened, and their nucleus was oriented parallel, rather than perpendicular, to the surface (Fig. 4, D–F, and Table 2). Although desmosomes and hemidesmosomes persist, they did not appear to associate with electron-dense filaments; in fact, no keratin IF bundles could be detected anywhere in the cytoplasm of basal cells (Fig. 4, E and F). Virtually all mitochondria appeared severely damaged in basal cells as seen through their swelling, loss of cristae, and loss of matrix as reflected by its electron-lucent appearance (Fig. 4, E and F, and Table 2). These alterations did not result from a fixation artifact as mitochondria with normal size and ultrastructure could be found in proximal dermal fibroblasts and in the upper layers of the epidermis (data not shown). Furthermore, the ultrastructural features of nuclei and cytoplasm clearly conveyed that basal keratinocytes are not undergoing apoptosis in intact skin of Krt5-null E18.5 embryos (51). In addition to engendering acute tissue fragility, the genetic loss of Krt5 thus lead to several major cytoarchitectural defects in the epidermis (see Table 2) that were largely confined to basal layer keratinocytes.
FIGURE 4.
Ultrastructural analysis of E18.5 epidermis in situ. Krt5+/+ (Control; A–C), Krt5−/− (Krt5 null; D–F), and KRT8bcTg/−Krt5−/− (KRT8bc Replacement; G–I) mouse epidermis at E18.5 was analyzed by routine transmission electron microscopy of thin sections. A, D, and G provide low magnification surveys of the living layers of epidermis (basal, spinous, and granular), whereas all other panels provide details of basal keratinocytes. bl, basal lamina; kif, keratin intermediate filament bundles; mi, mitochondria; Nu, nucleus. Examples of hemidesmosomes and desmosomes are circled and boxed, respectively, and representative mitochondria are detailed in the insets. Bars, 4 μm in A, D, and G; 2 μm in B, E, and H; and 1 μm in C, F, and I.
TABLE 2.
Summary of ultrastructural observations made in E18.5 (prebirth) epidermis
Control indicates Krt5+/+, Krt5-null indicates Krt5−/−, and KRT8bc replacement indicates KRT8bcTg/−Krt5−/−.
| Features | Control | Krt5-null | KRT8bc replacement |
|---|---|---|---|
| Suprabasal Ultrastructure | Normal | Normal | Normal |
| Hemidesmosomes (per cell) | 4.9 ± 1.7 | 6.7 ± 3.0 | 5.5 ± 1.8 |
| Desmosomes (per cell) | 6.2 ± 1.4 | 4.6 ± 0.8 | 4.1 ± 1.6 |
| Keratin filament bundles | Numerous | Absent | Occasional |
| Cell aspect ratio | 1.58 ± 0.5 | 0.81 ± 0.4 | 1.08 ± 0.4 |
| Nuclear aspect ratio | 2.05 ± 0.9 | 0.66 ± 0.3 | 0.92 ± 0.5 |
| Mitochondria (% normal) | 98.8 | 7.4 | 52.9 |
Several of these ultrastructural attributes were partially restored in basal keratinocytes of KRT8bc replacement E18.5 skin, correlating with a more mechanically sound tissue during harvesting and preparation (data not shown). Basal keratinocytes were cuboidal in shape, and their nuclei were consistently oriented perpendicular to the main axis of epidermis (Fig. 4, G–I, and Table 2). Interestingly, their nuclei still exhibited an aberrant contour rather than the round or ovoid shape observed in control and Krt5-null epidermis. The mitochondrial defects observed in Krt5-null embryonic skin were also partially restored in the KRT8bc replacement skin (Table 2). Ultrastructural features including an electron-dense matrix and intact cristae occurred in nearly half of observed mitochondria (Fig. 4, H and I, and Table 2). Desmosomes and hemidesmosomes appeared normal and were more likely to exhibit association with filament networks (Fig. 4, H and I). Finally, electron-dense bundles of keratin IFs along the cellular periphery and in the general cytoplasm were observed (Fig. 4I). These bundles were smaller, however, and occurred less frequently in KRT8bc replacement mice relative to control epidermis (Table 2). These ultrastructural attributes and their frequency are summarized in Table 2.
Adult KRT8bc Replacement Animals Exhibit Several Skin Abnormalities
KRT8bc replacement mice that survived to adulthood were indistinguishable from their littermates for the first several weeks after birth. Skin tissue was harvested from a successful KRT8bc replacement adult and a control littermate and processed for transmission electron microscopy to examine the epidermis in greater detail. Analysis of these samples revealed a striking similarity between control and KRT8bc replacement epidermis. The shape and orientation of basal keratinocytes and the orientation of their nuclei appeared similar in both samples (Fig. 5). Keratin filament bundles were visible in the cytoplasm and near the nucleus and were observed to be associated with hemidesmosomes and desmosomes in both samples (Fig. 5). Most notably, the mitochondria appeared healthy in KRT8bc replacement epidermis, comparable with control epidermis (Fig. 5, B and D). The most striking difference between the control and rescued skin was the persistence of the aberrant contour of the nuclei (Fig. 5, C and F).
FIGURE 5.
Ultrastructural analysis of adult control and KRT8bc replacement epidermis in situ. Krt5+/+ (Control; A–C), and KRT8bcTg/−Krt5−/− (KRT8bc Replacement; D–F) epidermis in 11-week-old mice was analyzed by routine transmission electron microscopy of thin sections. A and D provide low magnification surveys of the living layers of epidermis (basal, spinous, and granular), whereas B, C, E, and F provide details of basal keratinocytes. kif, keratin intermediate filament bundles; mi, mitochondria; Nu, nucleus. Examples of hemidesmosomes and desmosomes are circled and boxed, respectively. Bars, 4 μm in A and D and 2 μm in B, C, E, and F.
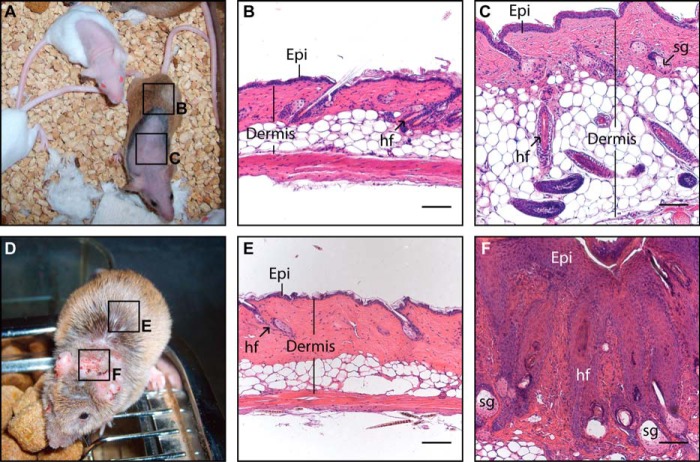
All mice having at least one functional Krt5 allele did not display any skin anomaly at any time regardless of their rescue transgene status. By comparison, adult KRT8bc replacement mice progressively developed various skin anomalies. For instance, most rescued mice developed alopecia starting as early as 6 weeks of age. Hair loss occurred consistently on the nape and often progressed into larger hairless regions along the back, flanks, and occasionally on the face and other body areas (Fig. 6A). Additionally, all KRT8bc replacement animals developed severe ulcerative dermatitis on the ears and nape (Fig. 6D). These areas of ulcerative dermatitis did not respond to topical antibiotic ointment and invariably progressed to become chronic lesions.
FIGURE 6.
Analysis of late onset phenotypes in adult KRT8bc replacement animals. A macroscopic survey (A and D) and histology (B, C, E, and F) illustrating the late onset phenotypes arising in KRT8bc replacement animals are shown. A, 6-month-old adult KRT8bc replacement animals displaying both normal skin and areas of alopecia are shown. B, hairy skin from KRT8bc replacement animal (see box “B” in A) showing normal histology. C, phenotypic skin sample from KRT8bc replacement animal (see box “C” in A) showing misoriented, anagen-staged hair follicles and thickened epidermis. D, a 3-month-old adult KRT8bc replacement animal showing both normal skin and areas of inflamed and hyperkeratotic, scaly skin. E, hairy skin from KRT8bc replacement animal showing normal histology (see box “E” in D). F, phenotypic skin from replacement animals (see box “F” in D) depicting hyperplastic epidermis, pilosebaceous cysts, and a high level of dermal infiltration, suggesting an inflammatory and immune infiltration. Epi, epidermis; hf, hair follicle; sg, sebaceous gland. Bars, 100 μm.
To further assess this phenomenon, routine histology was performed on normal looking, hairy skin and on regions of affected skin. Hairy skin from KRT8bc replacement mice was comparable with wild type skin (data not shown) with a normal epidermis and telogen stage hair follicles (Fig. 6, B and E). Relative to (normal) hairy skin, hairless skin patches in replacement animals showed thickened epidermis and enlarged sebaceous glands (Fig. 6C). Furthermore, hair follicles were aberrantly shaped and oriented and frequently appeared to be at the anagen stage of their cycle (Fig. 6C). In regions of lesional skin, the epidermis was markedly hyperplastic, and the dermis was heavily inflamed (Fig. 6F). As with hairless skin, lesional skin contained hair follicles in anagen stage (when present) that were often improperly oriented. Many follicles were without a visible hair shaft and featured enlarged sebaceous glands (Fig. 6F). In addition, lesional skin contained numerous large cysts of pilosebaceous origin (data not shown).
DISCUSSION
In this study, we set out to test the functional importance of the property of K5/K14 filament self-organization into cross-linked networks by targeting the expression of K8bc or wild type K8 to the epidermis of Krt5-null mice. K8bc is a chimera that is polymerization- and self-organization-competent when paired with K14, whereas K8 readily co-polymerizes with K14 but is unable to form cross-linked networks (30). We found that the KRT14 promoter-driven expression of K8bc rescued Krt5-null mice from massive perinatal blistering and death with a 47% frequency, whereas expression of K8 was unable to afford any rescue in this setting. Although the rescued KRT8bc replacement mice reached adulthood and were reproductively competent, most eventually developed alopecia, and all developed skin erosions; also, their basal keratinocytes exhibited persisting ultrastructural anomalies (see Table 2). These findings imply that, even when expressed at approximately half the levels of endogenous K5 in control skin, the K8bc chimeric protein cannot fully support the function(s) of K5 in mouse epidermis in vivo. Furthermore, the occurrence of rescue from overt cell and tissue fragility in neonatal and adult skin could be correlated with the presence of keratin IF bundles in basal keratinocytes, consistent with (but not proving) the hypothesis put forth in Lee and Coulombe (30).
In the first approximation, the significance of our protein replacement findings and of the differences observed between K8bc and K8, is mitigated by the relatively low and uneven levels at which they were each expressed in Krt5-null mouse epidermis. Partial or lack of rescue cannot be explained by mosaicism either in genotype or expression pattern (Fig. 1 and data not shown). Recent studies suggest that much less than 50% of the normal (control) complement of K5/K14 proteins suffices to rescue key phenotypic traits in keratin-free keratinocytes in culture (13, 52, 53). Our data also suggest that the steady state levels of rescue transgene mRNA differ depending on genotype (e.g. in transgenic mice versus replacement mice) and the rescue construct being expressed (e.g. K8 versus K8bc), extending a previous observation of striking context-dependent post-transcriptional regulation for ectopic keratin mRNAs in basal keratinocytes (54). Although we cannot formally compare the effectiveness of K8 and K8bc proteins in replacing K5 in basal epidermal keratinocytes, it is quite remarkable that subnormal levels of K8bc (Fig. 2C) alongside a partial rescue of endogenous K14 protein levels (Fig. 2F) were sufficient to effectively rescue skin blistering and death in nearly 50% of KRT8bc replacement newborns. Given that ∼30% of its sequence originates from K5, the K8bc protein is hardly a normal keratin protein and in many ways is more aberrant than many of the KRT5 missense alleles known to cause epidermolysis bullosa simplex (8). This element certainly accentuates the significance of the 47% rescue frequency seen in the KRT8bc replacement animals. Otherwise, the outcome of our study conveys that the property of 10-nm filament formation in vitro and in vivo does not suffice to dictate a normal fate and/or function for an IF protein in basal keratinocytes of the epidermis, thereby significantly extending previous rescue efforts of a similar nature (54, 56).
Additionally, our KRT8 transgenic mice did not feature a visible phenotype, whereas Casanova et al. (57) observed severe skin anomalies in transgenic mice that ectopically express wild type K8 in the epidermis. This difference in phenotypic outcome is likely related to differences in K8 expression levels between these two studies. Our findings with KRT8 transgenic mice are in fact similar to those reported by Kirfel et al. (56), who showed that ectopic desmin was able to form 10-nm filaments in basal keratinocytes but could not rescue the Krt5-null mice to any extent, correlating with low transgene expression. For both Kirfel et al. (56) and the current study, the data in hand preclude a formal assignment of the failure to rescue to the low prevailing levels of rescue protein expression achieved versus abnormal protein properties in an ectopic setting. Future studies of this type should exploit a knock-in rather than a transgenic strategy when comparing the properties of allelic variants in vivo.
Our ultrastructural study of embryonic Krt5-null skin as well as adult KRT8bc replacement mice revealed novel and potentially important anomalies associated with K5 protein deficiency in mouse epidermis. Our findings corroborate the previous description of a complete lack of detectable keratin IFs in the cytoplasm of basal keratinocytes and of largely normal appearing suprabasal keratinocytes (33). Beyond these findings, we also observed major cytoarchitectural defects, e.g. aberrancies in cell shape and orientation, the shape and orientation of the nucleus in basal keratinocytes, and major mitochondrial defects. At least in part, the differences between our study and that of Peters et al. (33) may be rooted in the use of different protocols when preparing samples for analysis by transmission electron microscopy. The marked alterations observed in the shape of basal keratinocytes in Krt5-null skin may alter their physical relationship with other key cell types in the epidermis, e.g. melanocytes and Langerhans cells. Such defects, if they exist, may contribute to the aberrations observed in skin pigmentation (58) and Langerhans cell density (59) in mouse models and/or individuals with genetic alterations at the Krt5 locus. The newly described cytoarchitectural alterations may be related to the findings of Lee et al. (24), who proposed that perinuclear K5/K14 filaments impart an oxidation state-dependent influence upon the size and shape of the nucleus in newborn epidermis. On another front, the observation of a difference in the number of desmosomal and hemidesmosomal plaques in Krt5-null basal keratinocytes (which are devoid of keratin IFs) is not surprising in light of similar findings obtained when analyzing type II keratin-free keratinocytes (13) and considering the roles recently assigned to keratins in the localization and maintenance of desmosomes (52, 60).
The mitochondrial defects observed in late stage embryonic Krt5-null mice are particularly interesting. The shape and positioning of mitochondria within cells have been shown to be influenced by keratin IFs in a number of contexts. Similar mitochondrial defects have been reported in Krt14-null mouse neonates (45), although they were shown to be associated with cytolysis and therefore may have been a consequence of overt, postlysis cellular distress. Similar mitochondrial defects were also observed in the myocardium of mice lacking desmin, a type III IF protein found predominantly in muscle (61). Also, we previously reported on the occurrence of intriguing electron-dense inclusions in the mitochondrial matrix of epidermal keratinocytes in KRT16-overexpressing transgenic mice (62) and Krt16-null mice (63). Additionally, smaller mitochondrial size was reported in livers of Krt8-null mice and in mice carrying the KRT18 R89C mutation (64, 65), whereas Stone et al. (55) described an increased mitochondrial size in the skeletal muscle of Krt19-null mice. The significance of the involvement of K5 in mitochondrial physiology and the associated mechanism(s) are worth investigating.
Acknowledgments
We are most grateful to Janet Folmer for expert assistance in tissue preparation and electron microscopy and to Dr. Ryan Hobbs for critical reading of the manuscript. We also thank Drs. Randall Reed, William Wright, and Susan Craig and the members of the Coulombe laboratory for advice and support.
This work was supported, in whole or in part, by National Institutes of Health Grant AR042047.
- IF
- intermediate filament
- K5
- keratin 5
- K8
- keratin 8
- K14
- keratin 14.
REFERENCES
- 1. Omary M. B., Coulombe P. A., McLean W. H. (2004) Intermediate filament proteins and their associated diseases. N. Engl. J. Med. 351, 2087–2100 [DOI] [PubMed] [Google Scholar]
- 2. Szeverenyi I., Cassidy A. J., Chung C. W., Lee B. T., Common J. E., Ogg S. C., Chen H., Sim S. Y., Goh W. L., Ng K. W., Simpson J. A., Chee L. L., Eng G. H., Li B., Lunny D. P., Chuon D., Venkatesh A., Khoo K. H., McLean W. H., Lim Y. P., Lane E. B. (2008) The Human Intermediate Filament Database: comprehensive information on a gene family involved in many human diseases. Hum. Mutat. 29, 351–360 [DOI] [PubMed] [Google Scholar]
- 3. Coulombe P. A., Kerns M. L., Fuchs E. (2009) Epidermolysis bullosa simplex: a paradigm for disorders of tissue fragility. J. Clin. Investig. 119, 1784–1793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bonifas J. M., Rothman A. L., Epstein E. H., Jr. (1991) Epidermolysis bullosa simplex: evidence in two families for keratin gene abnormalities. Science 254, 1202–1205 [DOI] [PubMed] [Google Scholar]
- 5. Coulombe P. A., Hutton M. E., Letai A., Hebert A., Paller A. S., Fuchs E. (1991) Point mutations in human keratin 14 genes of epidermolysis bullosa simplex patients: genetic and functional analyses. Cell 66, 1301–1311 [DOI] [PubMed] [Google Scholar]
- 6. Lane E. B., Rugg E. L., Navsaria H., Leigh I. M., Heagerty A. H., Ishida-Yamamoto A., Eady R. A. (1992) A mutation in the conserved helix termination peptide of keratin 5 in hereditary skin blistering. Nature 356, 244–246 [DOI] [PubMed] [Google Scholar]
- 7. McLean W. H., Moore C. B. (2011) Keratin disorders: from gene to therapy. Hum. Mol. Genet. 20, R189–R197 [DOI] [PubMed] [Google Scholar]
- 8. Coulombe P. A., Lee C. H. (2012) Defining keratin protein function in skin epithelia: epidermolysis bullosa simplex and its aftermath. J. Invest. Dermatol. 132, 763–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Janmey P. A., Euteneuer U., Traub P., Schliwa M. (1991) Viscoelastic properties of vimentin compared with other filamentous biopolymer networks. J. Cell Biol. 113, 155–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ma L., Yamada S., Wirtz D., Coulombe P. A. (2001) A 'hot-spot' mutation alters the mechanical properties of keratin filament networks. Nat. Cell Biol. 3, 503–506 [DOI] [PubMed] [Google Scholar]
- 11. Fuchs E., Cleveland D. W. (1998) A structural scaffolding of intermediate filaments in health and disease. Science 279, 514–519 [DOI] [PubMed] [Google Scholar]
- 12. Coulombe P. A., Wong P. (2004) Cytoplasmic intermediate filaments revealed as dynamic and multipurpose scaffolds. Nat. Cell Biol. 6, 699–706 [DOI] [PubMed] [Google Scholar]
- 13. Kröger C., Loschke F., Schwarz N., Windoffer R., Leube R. E., Magin T. M. (2013) Keratins control intercellular adhesion involving PKC-α-mediated desmoplakin phosphorylation. J. Cell Biol. 201, 681–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Feng X., Zhang H., Margolick J. B., Coulombe P. A. (2013) Keratin intracellular concentration revisited: implications for keratin function in surface epithelia. J. Invest. Dermatol. 133, 850–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamada S., Wirtz D., Coulombe P. A. (2002) Pairwise assembly determines the intrinsic potential for self-organization and mechanical properties of keratin filaments. Mol. Biol. Cell 13, 382–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wilhelmsen K., Litjens S. H., Kuikman I., Tshimbalanga N., Janssen H., van den Bout I., Raymond K., Sonnenberg A. (2005) Nesprin-3, a novel outer nuclear membrane protein, associates with the cytoskeletal linker protein plectin. J. Cell Biol. 171, 799–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Simpson C. L., Patel D. M., Green K. J. (2011) Deconstructing the skin: cytoarchitectural determinants of epidermal morphogenesis. Nat. Rev. Mol. Cell Biol. 12, 565–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Suozzi K. C., Wu X., Fuchs E. (2012) Spectraplakins: master orchestrators of cytoskeletal dynamics. J. Cell Biol. 197, 465–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Steinert P. M., Marekov L. N. (1995) The proteins elafin, filaggrin, keratin intermediate filaments, loricrin, and small proline-rich proteins 1 and 2 are isodipeptide cross-linked components of the human epidermal cornified cell envelope. J. Biol. Chem. 270, 17702–17711 [DOI] [PubMed] [Google Scholar]
- 20. Lee S. C., Kim I. G., Marekov L. N., O'Keefe E. J., Parry D. A., Steinert P. M. (1993) The structure of human trichohyalin. Potential multiple roles as a functional EF-hand-like calcium-binding protein, a cornified cell envelope precursor, and an intermediate filament-associated (cross-linking) protein. J. Biol. Chem. 268, 12164–12176 [PubMed] [Google Scholar]
- 21. Stappenbeck T. S., Bornslaeger E. A., Corcoran C. M., Luu H. H., Virata M. L., Green K. J. (1993) Functional analysis of desmoplakin domains: specification of the interaction with keratin versus vimentin intermediate filament networks. J. Cell Biol. 123, 691–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jang S. I., Kalinin A., Takahashi K., Marekov L. N., Steinert P. M. (2005) Characterization of human epiplakin: RNAi-mediated epiplakin depletion leads to the disruption of keratin and vimentin IF networks. J. Cell Sci. 118, 781–793 [DOI] [PubMed] [Google Scholar]
- 23. Green K. J., Böhringer M., Gocken T., Jones J. C. (2005) Intermediate filament associated proteins. Adv. Protein Chem. 70, 143–202 [DOI] [PubMed] [Google Scholar]
- 24. Lee C. H., Kim M. S., Chung B. M., Leahy D. J., Coulombe P. A. (2012) Structural basis for heteromeric assembly and perinuclear organization of keratin filaments. Nat. Struct. Mol. Biol. 19, 707–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sun T. T., Green H. (1978) Keratin filaments of cultured human epidermal cells. Formation of intermolecular disulfide bonds during terminal differentiation. J. Biol. Chem. 253, 2053–2060 [PubMed] [Google Scholar]
- 26. Lersch R., Fuchs E. (1988) Sequence and expression of a type II keratin, K5, in human epidermal cells. Mol. Cell. Biol. 8, 486–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hanukoglu I., Fuchs E. (1983) The cDNA sequence of a type II cytoskeletal keratin reveals constant and variable structural domains among keratins. Cell 33, 915–924 [DOI] [PubMed] [Google Scholar]
- 28. Wilson A. K., Coulombe P. A., Fuchs E. (1992) The roles of K5 and K14 head, tail, and R/K L L E G E domains in keratin filament assembly in vitro. J. Cell Biol. 119, 401–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bousquet O., Ma L., Yamada S., Gu C., Idei T., Takahashi K., Wirtz D., Coulombe P. A. (2001) The nonhelical tail domain of keratin 14 promotes filament bundling and enhances the mechanical properties of keratin intermediate filaments in vitro. J. Cell Biol. 155, 747–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee C. H., Coulombe P. A. (2009) Self-organization of keratin intermediate filaments into cross-linked networks. J. Cell Biol. 186, 409–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim S., Coulombe P. A. (2010) Emerging role for the cytoskeleton as an organizer and regulator of translation. Nat. Rev. Mol. Cell Biol. 11, 75–81 [DOI] [PubMed] [Google Scholar]
- 32. Leube R. E., Bosch F. X., Romano V., Zimbelmann R., Höfler H., Franke W. W. (1986) Cytokeratin expression in simple epithelia. III. Detection of mRNAs encoding human cytokeratins nos. 8 and 18 in normal and tumor cells by hybridization with cDNA sequences in vitro and in situ. Differentiation 33, 69–85 [DOI] [PubMed] [Google Scholar]
- 33. Peters B., Kirfel J., Büssow H., Vidal M., Magin T. M. (2001) Complete cytolysis and neonatal lethality in keratin 5 knockout mice reveal its fundamental role in skin integrity and in epidermolysis bullosa simplex. Mol. Biol. Cell 12, 1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saitou M., Sugai S., Tanaka T., Shimouchi K., Fuchs E., Narumiya S., Kakizuka A. (1995) Inhibition of skin development by targeted expression of a dominant-negative retinoic acid receptor. Nature 374, 159–162 [DOI] [PubMed] [Google Scholar]
- 35. Kerns M. L., DePianto D., Dinkova-Kostova A. T., Talalay P., Coulombe P. A. (2007) Reprogramming of keratin biosynthesis by sulforaphane restores skin integrity in epidermolysis bullosa simplex. Proc. Natl. Acad. Sci. U.S.A. 104, 14460–14465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Coulombe P. A., Fuchs E. (1990) Elucidating the early stages of keratin filament assembly. J. Cell Biol. 111, 153–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Paladini R. D., Coulombe P. A. (1998) Directed expression of keratin 16 to the progenitor basal cells of transgenic mouse skin delays skin maturation. J. Cell Biol. 142, 1035–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Franke W. W., Schiller D. L., Moll R., Winter S., Schmid E., Engelbrecht I., Denk H., Krepler R., Platzer B. (1981) Diversity of cytokeratins. Differentiation specific expression of cytokeratin polypeptides in epithelial cells and tissues. J. Mol. Biol. 153, 933–959 [DOI] [PubMed] [Google Scholar]
- 39. McGowan K. M., Coulombe P. A. (1998) Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J. Cell Biol. 143, 469–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reichelt J., Haase I. (2010) Establishment of spontaneously immortalized keratinocyte lines from wild-type and mutant mice. Methods Mol. Biol. 585, 59–69 [DOI] [PubMed] [Google Scholar]
- 41. Bernot K. M., Lee C. H., Coulombe P. A. (2005) A small surface hydrophobic stripe in the coiled-coil domain of type I keratins mediates tetramer stability. J. Cell Biol. 168, 965–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Reynolds E. S. (1963) The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17, 208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kulesh D. A., Ceceña G., Darmon Y. M., Vasseur M., Oshima R. G. (1989) Posttranslational regulation of keratins: degradation of mouse and human keratins 18 and 8. Mol. Cell. Biol. 9, 1553–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lersch R., Stellmach V., Stocks C., Giudice G., Fuchs E. (1989) Isolation, sequence, and expression of a human keratin K5 gene: transcriptional regulation of keratins and insights into pairwise control. Mol. Cell. Biol. 9, 3685–3697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lloyd C., Yu Q. C., Cheng J., Turksen K., Degenstein L., Hutton E., Fuchs E. (1995) The basal keratin network of stratified squamous epithelia: defining K15 function in the absence of K14. J. Cell Biol. 129, 1329–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Liao J., Ku N. O., Omary M. B. (1997) Stress, apoptosis, and mitosis induce phosphorylation of human keratin 8 at Ser-73 in tissues and cultured cells. J. Biol. Chem. 272, 17565–17573 [DOI] [PubMed] [Google Scholar]
- 47. Moll R., Moll I., Franke W. W. (1984) Identification of Merkel cells in human skin by specific cytokeratin antibodies: changes of cell density and distribution in fetal and adult plantar epidermis. Differentiation 28, 136–154 [DOI] [PubMed] [Google Scholar]
- 48. Weiss R. A., Eichner R., Sun T. T. (1984) Monoclonal antibody analysis of keratin expression in epidermal diseases: a 48- and 56-kdalton keratin as molecular markers for hyperproliferative keratinocytes. J. Cell Biol. 98, 1397–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McGowan K., Coulombe P. A. (1998) The wound repair-associated keratins 6, 16, and 17. Insights into the role of intermediate filaments in specifying keratinocyte cytoarchitecture. Subcell. Biochem. 31, 173–204 [PubMed] [Google Scholar]
- 50. Hardman M. J., Sisi P., Banbury D. N., Byrne C. (1998) Patterned acquisition of skin barrier function during development. Development 125, 1541–1552 [DOI] [PubMed] [Google Scholar]
- 51. Lu H., Chen J., Planko L., Zigrino P., Klein-Hitpass L., Magin T. M. (2007) Induction of inflammatory cytokines by a keratin mutation and their repression by a small molecule in a mouse model for EBS. J. Invest. Dermatol. 127, 2781–2789 [DOI] [PubMed] [Google Scholar]
- 52. Seltmann K., Roth W., Kröger C., Loschke F., Lederer M., Hüttelmaier S., Magin T. M. (2013) Keratins mediate localization of hemidesmosomes and repress cell motility. J. Invest. Dermatol. 133, 181–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Seltmann K., Fritsch A. W., Käs J. A., Magin T. M. (2013) Keratins significantly contribute to cell stiffness and impact invasive behavior. Proc. Natl. Acad. Sci. U.S.A. 110, 18507–18512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Paladini R. D., Coulombe P. A. (1999) The functional diversity of epidermal keratins revealed by the partial rescue of the keratin 14 null phenotype by keratin 16. J. Cell Biol. 146, 1185–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Stone M. R., O'Neill A., Lovering R. M., Strong J., Resneck W. G., Reed P. W., Toivola D. M., Ursitti J. A., Omary M. B., Bloch R. J. (2007) Absence of keratin 19 in mice causes skeletal myopathy with mitochondrial and sarcolemmal reorganization. J. Cell Sci. 120, 3999–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kirfel J., Peters B., Grund C., Reifenberg K., Magin T. M. (2002) Ectopic expression of desmin in the epidermis of transgenic mice permits development of a normal epidermis. Differentiation 70, 56–68 [DOI] [PubMed] [Google Scholar]
- 57. Casanova M. L., Bravo A., Martínez-Palacio J., Fernández-Aceñero M. J., Villanueva C., Larcher F., Conti C. J., Jorcano J. L. (2004) Epidermal abnormalities and increased malignancy of skin tumors in human epidermal keratin 8-expressing transgenic mice. FASEB J. 18, 1556–1558 [DOI] [PubMed] [Google Scholar]
- 58. Betz R. C., Planko L., Eigelshoven S., Hanneken S., Pasternack S. M., Bussow H., Van Den Bogaert K., Wenzel J., Braun-Falco M., Rutten A., Rogers M. A., Ruzicka T., Nöthen M. M., Magin T. M., Kruse R. (2006) Loss-of-function mutations in the keratin 5 gene lead to Dowling-Degos disease. Am. J. Hum. Genet. 78, 510–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Roth W., Reuter U., Wohlenberg C., Bruckner-Tuderman L., Magin T. M. (2009) Cytokines as genetic modifiers in K5−/− mice and in human epidermolysis bullosa simplex. Hum. Mutat. 30, 832–841 [DOI] [PubMed] [Google Scholar]
- 60. Liovic M., D'Alessandro M., Tomic-Canic M., Bolshakov V. N., Coats S. E., Lane E. B. (2009) Severe keratin 5 and 14 mutations induce down-regulation of junction proteins in keratinocytes. Exp. Cell Res. 315, 2995–3003 [DOI] [PubMed] [Google Scholar]
- 61. Thornell L., Carlsson L., Li Z., Mericskay M., Paulin D. (1997) Null mutation in the desmin gene gives rise to a cardiomyopathy. J. Mol. Cell. Cardiol. 29, 2107–2124 [DOI] [PubMed] [Google Scholar]
- 62. Takahashi K., Folmer J., Coulombe P. A. (1994) Increased expression of keratin 16 causes anomalies in cytoarchitecture and keratinization in transgenic mouse skin. J. Cell Biol. 127, 505–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lessard J. C., Piña-Paz S., Rotty J. D., Hickerson R. P., Kaspar R. L., Balmain A., Coulombe P. A. (2013) Keratin 16 regulates innate immunity in response to epidermal barrier breach. Proc. Natl. Acad. Sci. U.S.A. 110, 19537–19542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tao G. Z., Looi K. S., Toivola D. M., Strnad P., Zhou Q., Liao J., Wei Y., Habtezion A., Omary M. B. (2009) Keratins modulate the shape and function of hepatocyte mitochondria: a mechanism for protection from apoptosis. J. Cell Sci. 122, 3851–3855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kumemura H., Harada M., Yanagimoto C., Koga H., Kawaguchi T., Hanada S., Taniguchi E., Ueno T., Sata M. (2008) Mutation in keratin 18 induces mitochondrial fragmentation in liver-derived epithelial cells. Biochem. Biophys. Res. Commun. 367, 33–40 [DOI] [PubMed] [Google Scholar]