Background: The biological activities of 1,25(OH)2D3 in osteoblasts are dependent upon their differentiation state.
Results: Genome-wide analyses reveal transcriptomic responses to 1,25(OH)2D3, and VDR, RUNX2, and C/EBPβ cistromes are modified during differentiation.
Conclusion: Differentiation-induced changes in expression and transcription factor genome occupancy underlie the response to 1,25(OH)2D3.
Significance: Transcription factor occupancy at genome sites is dynamic and significantly influenced by the state of cellular differentiation.
Keywords: CCAAT/Enhancer-binding Protein (C/EBP), ChIP-sequencing (ChIP-seq), Chromatin Modification, Osteoblast, Transcription Enhancer, Vitamin D
Abstract
The biological effects of 1α,25-dihydroxyvitamin D3 (1,25 (OH)2D3) on osteoblast differentiation and function differ significantly depending upon the cellular state of maturation. To explore this phenomenon mechanistically, we examined the impact of 1,25(OH)2D3 on the transcriptomes of both pre-osteoblastic (POBs) and differentiated osteoblastic (OBs) MC3T3-E1 cells, and assessed localization of the vitamin D receptor (VDR) at sites of action on a genome-scale using ChIP sequence analysis. We observed that the 1,25(OH)2D3-induced transcriptomes of POBs and OBs were quantitatively and qualitatively different, supporting not only the altered biology observed but the potential for a change in VDR interaction at the genome as well. This idea was confirmed through discovery that VDR cistromes in POBs and OBs were also strikingly different. Depletion of VDR-binding sites in OBs, due in part to reduced VDR expression, was the likely cause of the loss of VDR-target gene interaction. Continued novel regulation by 1,25(OH)2D3, however, suggested that factors in addition to the VDR might also be involved. Accordingly, we show that transcriptomic modifications are also accompanied by changes in genome binding of the master osteoblast regulator RUNX2 and the chromatin remodeler CCAAT/enhancer-binding protein β. Importantly, genome occupancy was also highlighted by the presence of epigenetic enhancer signatures that were selectively changed in response to both differentiation and 1,25(OH)2D3. The impact of VDR, RUNX2, and C/EBPβ on osteoblast differentiation is exemplified by their actions at the Runx2 and Sp7 gene loci. We conclude that each of these mechanisms may contribute to the diverse actions of 1,25(OH)2D3 on differentiating osteoblasts.
Introduction
The ability of 1α,25-dihydroxyvitamin D3 (1,25(OH)2D3)2 to regulate calcium and phosphorus homeostasis in higher vertebrates is orchestrated through its actions on intestine, kidney, and bone (1). In the skeleton, 1,25(OH)2D3 functions both to preserve bone integrity and strength and to create a viable mineral reservoir accessible during times of negative calcium and/or phosphate balance (2, 3). These activities are both direct and indirect; accordingly, 1,25(OH)2D3 acts indirectly via the intestine and kidney to maintain the blood levels of calcium and phosphorus necessary for mineralization and directly to modulate bone cell differentiation and function (4). The direct actions of 1,25(OH)2D3 on bone cells are complex and include both cell autonomous and paracrine mechanisms, the latter arising from the ability of the hormone to stimulate or repress the expression of growth factors and cytokines from chondrocytes, osteoblasts, and osteocytes that affect cells located nearby (2). One such paracrine factor is receptor activator of nuclear factor-κB ligand that is produced in a variety of bone cell types and plays a paramount role in regulating the formation, activation, function, and survival of hematopoietic cell-derived osteoclasts (5). 1,25(OH)2D3 also plays an endocrine role in regulating the expression of fibroblast growth factor 23 (FGF23) from osteocytes; this factor acts on the distal kidney to regulate phosphate metabolism and may have independent functions in other tissues as well (6–8). Evidence for these homeostatic actions of 1,25(OH)2D3 derives from studies conducted both in normal rodents in vivo and in VDR- or cytochrome p450 27B1 (CYP27B1)-null mice (9–12).
Early in vitro studies revealed that 1,25(OH)2D3 both stimulated and inhibited osteoblast-lineage cell growth and function depending upon the source of the cells and their initial state of differentiation (2). Additional studies have confirmed this pleiotropic effect of the hormone in primary osteoblast preparations, cells of distinct skeletal origin, and in osteoblastic cell lines representative of different stages of maturation. Interestingly, mixed regulatory effects of 1,25(OH)2D3 have been recapitulated both in vivo and ex vivo using osteoblasts derived from the VDR- and/or CYP27B1-null mouse models referred to above (13, 14) and in a mouse model in which the VDR is overexpressed in late osteoblasts (15). These studies are more difficult to interpret, however, as the biological effects of the hormone are much more complex in vivo (16). In all of these cases, the mechanisms that underlie these dynamic and stage-specific effects of the vitamin D hormone on cells are not well understood. However, these findings have significant implications for the effects of 1,25(OH)2D3 in the skeleton during unique physiological states or in various disease conditions.
Regardless of whether 1,25(OH)2D3 functions in an anabolic or catabolic fashion on the skeleton or via direct actions to regulate additional biological activities associated with vitamin D, its actions involve VDR-dependent changes in gene expression. Like many members of this class of DNA binding factors, the VDR functions in a largely 1,25(OH)2D3-controlled manner by interacting directly with vitamin D response elements (VDREs) located within regulatory regions that are linked functionally to specific target genes (17, 18). Recent studies suggest that in addition to their positions adjacent to gene promoters, enhancers are also frequently located distal to promoter regions (19, 20) and may consist of multiple component sites. Whether located near or far in linear terms from transcriptional start sites (TSS), however, most of these elements appear to function through direct molecular interaction with the promoter regions of their target genes via looping mechanisms (21). Regardless of this detail, VDR binding enables the recruitment of co-regulatory enzyme complexes that are frequently chromatin-active (22, 23); they covalently modify histone tails that precipitate decondensation, move or displace nucleosomes thereby improving factor access, and/or facilitate direct enhancer linkage to transcription initiation complexes that form at promoters and that contribute to altered gene output (17). The VDR also participates in the down-regulation of gene expression as well either directly through DNA binding or via protein-protein interaction mechanisms with DNA-bound transcriptional activators that suppress gene expression (24). Neither of these mechanisms are well understood.
The mouse MC3T3-E1 pre-osteoblast has been utilized for several decades as an inducible in vitro model for studying osteoblast differentiation to the mature cell phenotype (25–27). Recently, we showed that differentiation leads to a significant alteration in the transcriptome of these cells and that this change correlates directly with both a partial redistribution of RUNX2 and C/EBPβ at binding sites across the genome and modifications to the epigenome (28). In this study, we explore how differentiation affects the transcriptional response of these cells to 1,25(OH)2D3. The results suggest that differentiation causes a pervasive and selective change in the cistromic patterns for VDR/RXR, RUNX2, and C/EBPβ following treatment with 1,25(OH)2D3 that likely underlies transcriptional output.
EXPERIMENTAL PROCEDURES
Reagents
1,25(OH)2D3 was obtained from SAFC Global (Madison, WI). ChIP antibodies to VDR (C-20, sc-1008, lot A0912), RXR (ΔN-197, sc-774, lot G2310), C/EBPβ (C-19, sc-150, lot F3011), and RUNX2 (M-70, sc-10758, lot D1411) were purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Antibodies to H4K5Ac (07-327, lot DAM1794310) and H3K9Ac (06-942, lot 31636) were purchased from Millipore (Billerica, MA). Antibodies for H3K4me1 (ab8895, lot GR43807-1), H3K4me3 (ab1012, lot GR10853-1), and H3K36me3 (ab9050, lot GR52625-1) were purchased from Abcam (Cambridge, MA). All quantitative real time PCR (qPCR) reagents (Fast Start SYBR Green Master Mix (with ROX)) were obtained from Roche Applied Science, and TaqMan gene expression assays were from Life Technologies (Applied Biosystems (ABI) Foster City, CA). All qPCR was conducted on the StepOnePlus from Applied Biosystems. Primers were obtained from IDT (Coralville, IA). All Primers and TaqMan expression assays are listed in supplemental Table 1.
Cell Culture and Differentiation
MC3T3-E1 cells (pre-osteoblast, POB) (early passage line from Sudo et al. (25)) were cultured in minimum Eagle's medium α-modification supplemented with 10% heat-inactivated fetal bovine serum from Hyclone (Logan, UT), and 1% penicillin/streptomycin from Invitrogen. For differentiation, cells were grown to confluency and then changed into differentiation media (10 mm β-glycerophosphate and 50 μg/ml ascorbic acid) for 15 days (osteoblast, OB), replenishing the media every 2–3 days until assay. Cells were stained for alkaline phosphatase, alizarin red, and von Kossa as reported previously (28).
Gene Expression Analysis
POB or OBs were first prepared from MC3T3-E1 cells as described above. They were then treated with vehicle or 10−7 m 1,25(OH)2D3 for 0 and 24 h prior to RNA isolation. RNA was isolated using the TRI Reagent protocol (MRC, Cincinnati, OH) and amplification methods according Roche NimbleGen, Inc., as reported recently (28). All data passing 95% confidence limits greater than 2-fold were included in up- or down-regulated data analysis. Data are displayed as log2 values (log2) as well as fold change values (1,25(OH)2D3/vehicle) in supplemental Table 1. For all other RT-qPCR and validation, POB or OB cells were treated with 10−7 m 1,25(OH)2D3 for 0, 3, 6, 12, or 24 h; 1 μg of isolated total RNA was DNase-treated, reverse-transcribed using the High Capacity cDNA kit (Applied Biosystems), and then diluted to 100 μl with RNase/DNase I free water. qPCR was performed using primers specific to a select set of differentially expressed genes by TaqMan analyses available in supplemental Table 1 and validation data are found in supplemental Dataset 1.
Western Blot Analysis
Cells were differentiated in 10-cm plates for 0 (POB) through 15 (OB) days as described above. Prior to collection, the cells were treated with vehicle or 10−7 m 1,25(OH)2D3 for 24 h. Cells were collected in PBS and centrifuged for 4 min at 4,000 rpm (4 °C). Pellet was dissolved in 1× Laemmli buffer (Bio-Rad) diluted from 4× with buffer A (10 mm Hepes-KOH, pH 7.8, 400 mm KCl, 0.1 mm EDTA, 5 mm MgCl2, 0.1% Nonidet P-40, 1 mm DTT, and 0.5 mm PMSF) and incubated on ice for 10 min. The differentiated samples contained substantial mineralized matrix and were subjected to three 15-s pulses with a Polytron Homogenizer (Power Gen 125, Fisher). The resulting sample was sonicated with a microtip sonic dismembrator (Fisher) for three 10-s pulses. 20 μg of sample was heated to 95 °C for 5 min and loaded for samples on Tris-glycine 4–20% gels (Invitrogen). Blots were transferred to PVDF membrane and blocked with 5% milk in PBS with 1% Tween 20 (PBST) overnight at 4 °C. Primary antibodies for VDR (9A7, 1:2,000), RUNX2 (D130-3, MBL International, 1:1,000), C/EBPβ (C-19, Santa Cruz Biotechnology, 1:5,000), or β-tubulin (H-235, Santa Cruz Biotechnology, 1:5,000) were applied in 1% milk PBST at room temperature for 1 h. Blots were washed three times for 15 min with PBST. Secondary antibodies were goat α-rat IgG-HRP (Santa Cruz Biotechnology, 1:2,500), goat α-mouse IgG2b-HRP (Santa Cruz Biotechnology, 1:10,000), and goat α-rabbit IgG-HRP (Santa Cruz Biotechnology, 1:5,000) for both C/EBPβ and β-tubulin in 1% milk PBST at room temperature for 1 h. PVDF membrane was stripped three times (50 mm glycine, pH 2.5, 1% SDS) at room temperature and washed three times with PBST prior to β-tubulin western analysis. Detection was completed with SuperSignal West Dura Extended Duration Substrate (Thermo Scientific) on an ImageQuant LAS-4000 (GE Healthcare).
siRNA
Gene knockdown was performed using siRNA duplexes as reported previously (29) that were obtained from Dharmacon RNA Technologies (Thermo Fisher). siControl, siLAMIN, siVDR, and siRUNX2 siRNA duplex pools were transfected at 50 nm using DharmaFECT reagent 1 according to the manufacturer's protocols. In some experiments, siRNA duplexes were co-transfected with 50 ng of pCH110-βgal and 250 ng of luciferase reporters for Runx2 and Sp7 peak reporter regions. Cells were cultured for 48 or 72 h, at which point the media were changed, and the cells were treated for an additional 24 h with 10−7 m 1,25(OH)2D3 or ethanol vehicle and processed as indicated above for RNA and RT-qPCR TaqMan analyses. For 24-well reporter assays, cells were harvested, and the lysates were assayed for luciferase and β-galactosidase activities as described below. Luciferase activity was normalized in all cases using β-galactosidase activity.
Molecular Cloning
The pCH110-β galactosidase reporter plasmid was previously described (30). All gene-specific pTK plasmids were constructed by cloning the appropriate mouse DNA fragments obtained through DNA amplification of genomic MC3T3-E1 DNA into the pTK-luc vector using BamHI, SalI, and/or HindIII restriction sites. Mutations were introduced using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) as per the manufacturer's protocol and recommendations.
Analysis of Enhancer Activity Using Reporter Genes
MC3T3-E1 cells were seeded into 24-well plates in α-minimal essential medium containing 10% FBS at a concentration of 5.0 × 104 cells/well and transfected 24 h later with Lipofectamine PLUS (Invitrogen) in serum and antibiotic-free medium. Individual wells were co-transfected with 250 ng of a luciferase reporter vector and 50 ng of pCH110-βgal. After transfection, the cells were cultured in medium supplemented with 20% FBS with or without 1,25(OH)2D3. Cells were harvested 18 h after treatment, and the lysates were assayed for luciferase and β-galactosidase activities as described previously (31). Luciferase activity was normalized to β-galactosidase activity in all cases.
Chromatin Immunoprecipitation (ChIP) Followed by Sequencing (ChIP-seq)
Chromatin immunoprecipitation was performed as described previously (32). Briefly, MC3T3-E1 cells were treated for 3 h with vehicle or 10−7 m 1,25(OH)2D3 and optimized as reported previously (29). Samples were subjected to immunoprecipitation using either a control IgG antibody or the indicated experimental antibody listed under “Reagents” (VDR, RXR, C/EBPβ, RUNX2, H3K5Ac, H3K9Ac, H3K4me1, H3K4me3, or H3K36me3). All ChIP and ChIP-seq methodologies, including statistical information and data processing, were performed as reported recently (28). All ChIP-seq statistics, ChIP-seq peaks, and validation qPCR primers are listed in supplemental Table 1.
All current data are deposited in the Gene Expression Omnibus (accession number GSE51515). RUNX2 and C/EBPβ ChIP-seq data tracks from POBs and OBs in the vehicle condition are deposited under accession number GSE41955 (28).
RESULTS
Effects of 1,25(OH)2D3 on Gene Expression during MC3T3-E1 Osteoblast Differentiation
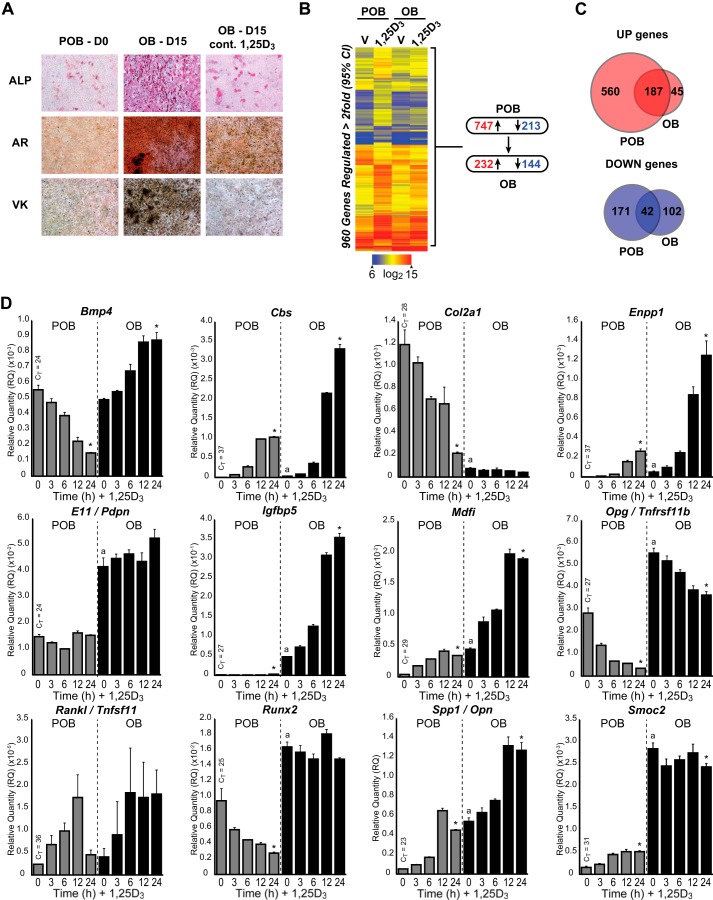
As documented in Fig. 1A, ascorbic acid and β-glycerophosphate induce bone formation in MC3T3-E1 pre-osteoblasts (POB) during a 15-day culture period, a well described outcome that is a manifestation of the mature osteoblast phenotype in vitro. The inclusion of 1,25(OH)2D3 during this time period blocks bone formation, and numerous studies have suggested that this involves an inhibition of differentiation. Surprisingly, however, the impact of osteoblast differentiation on the response of bone cells to 1,25(OH)2D3 has not been systematically explored, despite the suggestion that the actions of 1,25(OH)2D3 on osteoblast-lineage cells are stage-specific both in vitro and in vivo (2). To address this issue directly, we treated undifferentiated POB and 15-day differentiated OB MC3T3-E1 cells with either vehicle or 1,25(OH)2D3 for 24 h and then examined the effects of both differentiation and 1,25(OH)2D3 on the two cell types via gene expression profiling. As reported recently, POB differentiation to OBs altered the expression of 721 genes (>2-fold, 95% confidence limit), including the genes for Runx2, Col1a1, Spp1, osteoprotegerin (Tnfrsf11b), parathyroid hormone receptor (Pth1r), podoplanin (E11, Pdpn), and MyoD family inhibitor (Mdfi) (28). Interestingly, treatment of POBs and OBs with 1,25(OH)2D3 in this context also resulted in differential gene regulation profiles as well. As summarized in Fig. 1B, the expression of 960 genes was altered (>2-fold, 95% confidence limit) (747 increased and 213 decreased) by 1,25(OH)2D3 in POBs, whereas only 376 were modulated by the hormone (232 increased and 144 decreased) in differentiated OBs. The Venn diagrams (Fig. 1C) demonstrate that although 187 of these genes were up-regulated in common (>2-fold, 95% confidence limit), 560 and 45 genes were uniquely up-regulated in POBs and OBs, respectively. A similar qualitative feature was noted with down-regulated genes. These findings suggest that although a subset of genes are similarly regulated by 1,25(OH)2D3 in both POBs and OBs, cells also respond uniquely to the hormone at each stage of differentiation as well.
FIGURE 1.
Effects of 1,25(OH)2D3 on gene expression during MC3T3-E1 osteoblast differentiation. A, POB cells differentiated for 15 days to OB cells were treated with 10−7 m 1,25(OH)2D3 (1,25D3) or vehicle every 3 days during culture. Plates were stained with alkaline phosphatase (ALP), von Kossa (VK), and Alizarin Red (AR). B, gene expression heatmap of vehicle (V) or 1,25(OH)2D3 (1,25D3)-treated POB and OB cells. Blue, low expression; red, high expression. Genes shown in each row refer to POB differentially regulated genes. Summary of gene changes is shown to the right, with down-regulated <2-fold shown in blue and up-regulated >2-fold shown in red. C, Venn diagram representation of up-regulated (top, red) and down-regulated (bottom, blue) genes that overlap in POB and OB cells. D, gene expression time course (0, 3, 6, 12, and 24 h) of 10−7 m 1,25(OH)2D3 treatment in POB and OB cells by TaqMan RT-qPCR. Data are displayed as relative quantitation (RQ) compared with β-actin ± S.E. *, p < 0.05, 24 h versus 0 h within cell type by one-way analysis of variance. a, p < 0.05, POB 0 h versus OB 0 h by two-way analysis of variance with Dunnett's post test.
GO term analysis of the gene subsets revealed that 1,25(OH)2D3 induced gene categories in both POBs and OBs that were largely related to skeletal system development (biological process (BP), GO:0001501), bone development (BP, GO:0060348), and ossification (BP, GO:0001503). Genes within these categories included those involved in development of the osteoblast lineage (Runx2, Sp7, and Vdr), matrix secretion (Col1a1 and Col2a1), hormones, growth factors, signaling (bone morphogenic protein 4 (Bmp4), Tnfrsf11b, Pth1r, and insulin-like growth factor-binding protein 5 (Igfbp5)), bone resorption mediators (Tnfsf11, matrix metallopeptidase 13 (Mmp13), and cystathionine-β-synthase (Cbs)), and regulators of mineralization (Enpp1, Enpp3, dentin matrix acidic phosphoprotein 1 (Dmp1), and Spp1). Genes that were down-regulated by 1,25(OH)2D3 in POBs included those involved in cell cycle; those uniquely down-regulated in OBs were related to cell adhesion (BP, GO:0007155), including cadherin 1 (Cdh1), Col2a1, fibulin 5 (Fbln5), integrin binding sialoprotein (Ibsp), and osteomodulin (Omd). Although some regulated genes were novel, others are well recognized targets of vitamin D hormone action both in vitro and in vivo (33–35).
A cohort of these genes was examined further by direct RT-qPCR analysis both to validate the microarray data and to explore additional comparative details of the changes in gene expression that occur in response to 1,25(OH)2D3 in the context of differentiation (supplemental Dataset 1). Aside from confirming the microarray analyses, the data substantiate significant changes in baseline expression for many genes as a result of differentiation. Most interesting, however, is the observation that the regulation of this subset of genes by 1,25(OH)2D3 is both qualitatively and quantitatively different, and this finding extends to many others not documented here (data not shown). Thus, as seen in Fig. 1D and supplemental Dataset 1, many genes are up- or down-regulated from different baselines (e.g. Spp1, Cbs, Enpp1, Enpp3, Igfbp5, and Mdfi), whereas a few are regulated only in POBs or OBs but not both (e.g. Runx2, Col2a1, Dmp1, and Pth1r). Three genes are not regulated directly by 1,25(OH)2D3 (Mepe, Col1a1, and E11), but one gene, although low in expression, is equivalently regulated in both cell types (Tnfsf11); others are differentially regulated in POBs and OBs (Bmp4 and Smoc2). The time course of response suggests that the vast majority of these genes are direct responders to 1,25(OH)2D3; however, several may be regulated indirectly, perhaps via differentiation. Many genes that respond to 1,25(OH)2D3 in both cell states frequently show striking differences in the degree of response. Perhaps most interesting is Bmp4, which is suppressed by 1,25(OH)2D3 in POBs and stimulated in OBs. As defined by this set of target genes, and consistent with the microarray analysis overall, these data suggest that although transcriptomic response to 1,25(OH)2D3 declined significantly, numerous qualitative changes were also observed that likely resulted in the two biological states of osteoblast differentiation.
Delineation of the VDR Cistrome in POBs and OBs
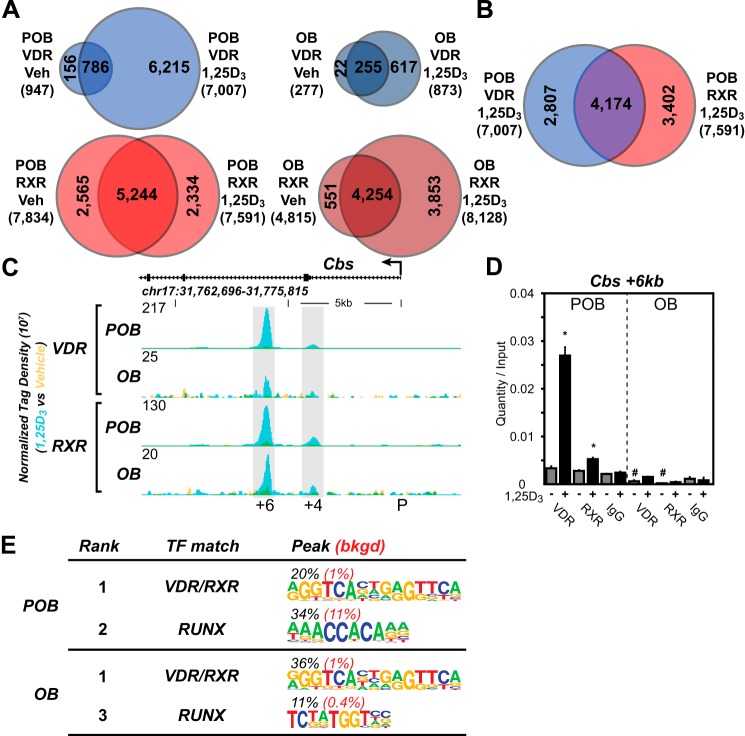
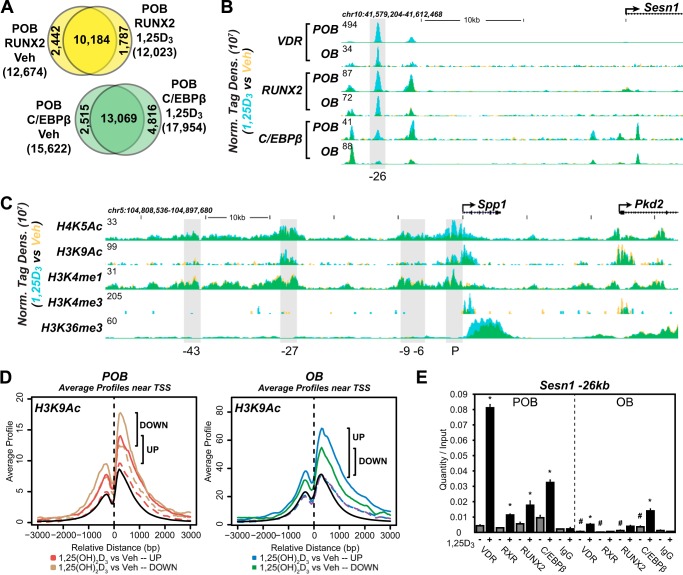
Based upon the above findings, we explored the mechanisms that were responsible for these altered responses to 1,25(OH)2D3 by first conducting a ChIP-seq analysis of the VDR and RXR cistromes in both POBs and OBs. RXR is an active heterodimer partner of the VDR and co-occupies a high percentage of VDR-binding sites when examined on a genome-wide scale (18, 29, 36). Fig. 2A summarizes the VDR cistromes quantitatively in both POBs and OBs following an optimized 3-h treatment with either vehicle or 1,25(OH)2D3. Although 947 genomic binding sites for the VDR (false discovery rate of <0.01) were evident in POBs in the absence of 1,25(OH)2D3, this number increased to 7,007 following hormone treatment. The finding that many hormone-independent VDR-binding sites (786) overlapped those detected in the presence of 1,25(OH)2D3 suggests a novel class of VDR-binding sites with potentially unusual activity. In the absence of 1,25(OH)2D3, the RXR cistrome in POBs was much larger than that of VDR, and although only modestly changed numerically via hormone treatment, it appeared to be composed of both a pre-bound and a new hormone-dependent class (2,334) of binding sites for RXR. Importantly, the results of a co-occupancy analysis shown in Fig. 2B revealed that almost 60% of VDR-binding sites also contained RXR and furthermore that RXR was frequently pre-bound to many sites prior to hormone-induced VDR binding. Most surprisingly, ChIP-seq analysis of VDR and RXR cistromes in OBs seen in Fig. 2A (right side) revealed a dramatic and unique quantitative decrease in the number of VDR-binding sites; 277 sites were detected before and only 873 were detected after hormonal activation. The RXR cistrome, however, was only slightly reduced; a redistribution effect of 1,25(OH)2D3 on RXR in OBs was also observed, although the basis for this change is not understood. Fig. 2C depicts the overlaid input normalized sequence density tracks for the Cbs gene that documents not only a representative effect of 1,25(OH)2D3 on VDR and RXR binding in POBs but the dramatic restriction or loss of VDR binding that occurs following OB differentiation. These data are confirmed using direct ChIP-qPCR analysis as seen in Fig. 2D. These data as well as VDR- and RXR-binding sites at each of the gene targets depicted in supplemental Dataset 1 provide additional evidence for both of these concepts.
FIGURE 2.
Delineation of the VDR cistromes in POBs and OBs. A, Venn diagram depiction of replicate normalized VDR- (blue) and RXR (red)-binding sites in vehicle (Veh) and 1,25(OH)2D3 (1,25D3)-treated undifferentiated (POB) cells (left) and differentiated (OB) cells (right). B, overlapping Venn diagrams for VDR and RXR in the POB-treated cells from the 1,25(OH)2D3 condition. C, ChIP-seq tag density tracks for the Cbs gene locus for VDR and RXR binding (Veh, yellow; 1,25(OH)2D3, blue; overlap, green) in the POB and OB cells. Genomic location and scale are indicated, and maximum height of tag sequence density for the data track is indicated on the y axis (normalized to input and 107 tags). Gene transcriptional direction is indicated by an arrow and exons by boxes. Peak regions of interest are highlighted by gray boxes, and their distance from TSS (P, promoter) is indicated below. D, ChIP-qPCR analysis of Cbs +6-kb peak region. Data displayed as quantitation normalized to ChIP input ± S.E. *, p < 0.05, 10−7 m 1,25(OH)2D3 (+) versus vehicle (−) within ChIP antibody by Student's t test. #, p < 0.05, POB versus OB by Student's t test. E, de novo over-representation analysis of VDR 1,25(OH)2D3 peak sequences and matching transcription factor-binding sites found through HOMER. Abundance shown as percentage (black) compared with 50,000 GC content matched sequences (red).
ChIP-seq analysis of the VDR and RXR cistromes in both POBs and OBs also suggests additional properties of these cistromes as well. First, VDR/RXR binding occurs within introns and at distal intergenic sites greater than 95% of the time (5% promoter (±1 kb of the TSS), 60% intergenic, 30% introns, and 5% exon), an observation consistent with our earlier findings in cancer cells and with those for other nuclear receptors such as estrogen receptor and peroxisome proliferator-activated receptor γ as well (32, 37–39). Second, a de novo search for the most highly represented sequences found at sites bound by VDR or by both VDR and RXR in the two cistromes revealed the presence of a classic VDRE motif (Fig. 2E). This analysis suggests that although the predominant DNA-binding site for the VDR is represented by two repeated half-sites separated by 3 bp, other motifs, including those that mediate suppression, may also be present. A position weight matrix-based assessment using Genomatix (40), however, revealed the presence of this classic VDRE(s) in a much higher percentage of ChIP-seq identified VDR-binding sites (95%), many located directly under ChIP peak maxima. Finally, sequences frequently over-represented in our de novo analysis also included the motif for the osteoblast master regulator RUNX2 (Fig. 2E), and the binding motif for the chromatin remodeler C/EBPβ.
Specific VDR Binding to Genome Correlates with the 1,25(OH)2D3-regulated Expression of Genes
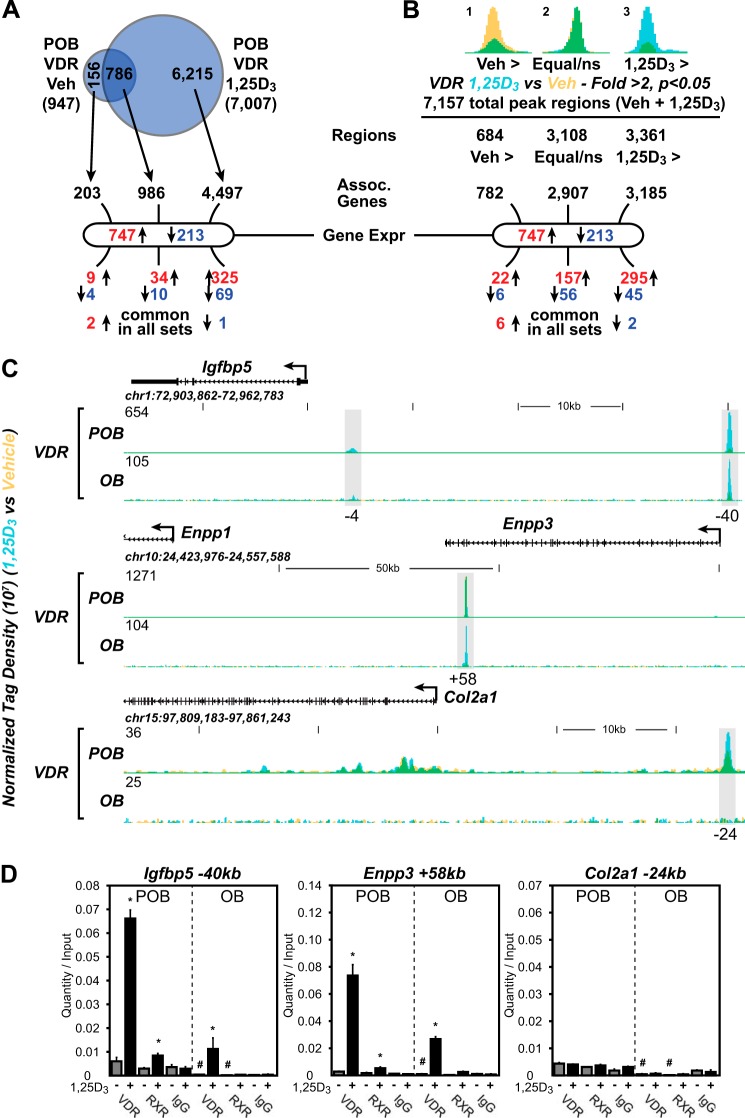
Given acquisition of both the VDR cistrome and 1,25(OH)2D3 response profiles for genes in both POBs and OBs, we used the nearest neighbor algorithm genomic regions enrichment of annotations tool (GREAT (41)) to determine the nature of the relationship between these two parameters on a genome-wide level focusing specifically on POBs. This analysis, as seen in Fig. 3A, revealed that the 7,007 1,25(OH)2D3-induced VDR-binding sites found in POBs were located within the proximity of 5,483 genes, 4,497 of which were associated with VDR sites present only following treatment with 1,25(OH)2D3. Interestingly, however, only 438 of these 5,483 genes were actually regulated by the hormone (46% of the 960 gene regulated, 359 increased and 79 decreased). Thus, the majority of VDR-binding sites identified by cistrome analysis was not linked to genes that were regulated by 1,25(OH)2D3 under the culture conditions utilized in this study. Interestingly, a third category was also evident; none of the 525 genes that comprise the remainder of the 1,25(OH)2D3-regulated transcriptome in POBs contained demonstrable VDR-binding sites near their surrounding genetic loci. This indicates that the presence of a detectable VDR-binding site in the vicinity of a gene by ChIP analysis is not a prerequisite for regulation by 1,25(OH)2D3. Although reduced numerically, the results in OBs were similar (data not shown).
FIGURE 3.
Specific VDR binding to the genome correlates with the 1,25(OH)2D3-regulated expression of genes. A, Venn diagram depiction of replicate normalized VDR-binding sites assessed using HOMER in undifferentiated (POB) cells treated with vehicle (Veh) or 1,25(OH)2D3 (1,25D3). Arrows link the VDR-binding site subsets (Veh-specific, 1,25(OH)2D3 (1,25D3)-specific, and Veh/1,25(OH)2D3 overlap) to annotated neighboring genes using GREAT. B, statistical enrichment of VDR ChIP-seq tag density. Peaks were divided into Veh > 2-fold (Veh >, yellow), equivalent (not statistically different, Equal/ns), and 1,25(OH)2D3 > 2-fold (1,25D3 >, blue) and then correlated to genes using GREAT nearest neighbor analysis. Overlapping regions appear as green. Below A and B, genes found to be differentially regulated by 1,25(OH)2D3 for POB cells (747, up-regulated; 213, down-regulated) are shown. The GREAT-associated genes are cross-referenced to these gene sets and listed below. Up-regulated genes, red; down-regulated genes, blue. C, ChIP-seq tag density tracks for the Igfbp5, Enpp1/3, and Col2a1 gene loci for VDR binding (Veh, yellow; 1,25(OH)2D3, blue; overlap, green) in POB and OB cells. Additional details are the same as in Fig. 2. D, ChIP-qPCR analysis of Igfbp5 −40-kb, Enpp3 +58-kb, and Col2a1 −24-kb peak regions. Data displayed as quantitation normalized to ChIP input ± S.E. *, p < 0.05, 10−7 m 1,25(OH)2D3 (+) versus vehicle (−) within ChIP antibody by Student's t test. , p < 0.05, POB versus OB by Student's t test.
Perhaps a more accurate representation of these data from the Venn diagram can be interpreted in Fig. 3B. Here, the sequencing tag density is taken into account after each peak region is determined (total 7,157 regions), and statistics can be utilized from the replicate samples to understand fold changes in protein occupancy. Fig. 3, A and B, show that the regions enriched in response to vehicle contain a very small portion of the VDR-binding events and that these regions correlate poorly with genes that are differentially expressed, whereas those that contain 1,25(OH)2D3-mediated VDR binding correlate highly to genes that are regulated (157 up/56 down and 295 up/45 down). In this analysis, VDR occupies a higher percentage of sites in both the absence and presence of 1,25(OH)2D3 demonstrating the increased sensitivity of this approach. This quantitative approach leads to the same overall conclusion, however.
Closer examination of those gene cohorts that were regulated by 1,25(OH)2D3 in both POBs and OBs using GO term analysis (sets in common and unique) revealed similar gene categories highlighting skeletal development and/or function. One of the highest enriched GO categories for BP of those genes that contained VDR and were up-regulated by 1,25(OH)2D3 was related to phosphate metabolic process (GO:0006796). This group contained genes such as AP2-associated kinase 1 (Aak1), eph receptor B2 (Ephb2), Janus kinase 2 (Jak2), Mdfi, protein kinase C (Prkca), as well as several other protein kinase and phosphatases. Full ChIP-seq profiles for these and other genes are found in supplemental Dataset 1.
Regardless of the above uncertainties, additional correlative information can be obtained by focusing on specific gene candidates and exploring the unique relationship between the differential binding of the VDR to these individual gene loci in POBs and OBs (supplemental Dataset 1) and the differential effects of 1,25(OH)2D3 on their expression (Fig. 1D and supplemental Dataset 1). Noteworthy are the genes for Igfbp5, Enpp1 and Enpp3, and Col2a1 (Fig. 3C and expanded view in supplemental Dataset 1). As seen in the ChIP-seq tracks across the locus for Igfbp5, for example, despite the striking increase in this gene's response to 1,25(OH)2D3 in OBs as compared with POBs, the amount of VDR binding associated with this robust up-regulation is decreased by 85%. Similar observations can be seen at many of the additional gene loci that are documented. These results were confirmed using direct ChIP-qPCR, as documented in Fig. 3D. Interestingly, although VDR is present in low amounts at enhancers found at and associated with a down-regulation of Col2a1 gene expression by 1,25(OH)2D3 in POBs, both VDR binding and regulation is completely lost at these sites in differentiated OBs. These and other examples suggest that although the VDR cistrome declines substantially on a genome-wide basis as a function of differentiation, the loss of VDR at numerous sites is not associated with a reduction but rather with the maintenance of and/or an increase in response to 1,25(OH)2D3.
VDR Expression Is Suppressed in OBs
Selective desensitization of the OB transcriptome to 1,25(OH)2D3 on a genome-wide scale together with an apparent decline in the VDR cistrome and/or the amount of VDR bound at many sites on the genome suggest the possibility that VDR protein levels may have declined during OB differentiation. To explore this possibility (reinforced by the reduced VDR transcript levels seen in OBs in supplemental Dataset 1), we assessed the level of VDR transcripts using RT-qPCR analysis and VDR protein levels directly by Western blot analysis. As can be seen in Fig. 4A, VDR transcript levels were reduced by 80% in OBs relative to POBs; the presence of 1,25(OH)2D3 over this 15-day period prevented this reduction, however, highlighting an auto-regulatory effect of 1,25(OH)2D3 on Vdr gene expression in bone cells. Importantly, as seen in Fig. 4B, transcript down-regulation during differentiation was mirrored by a substantial decrease in VDR protein levels as well, a decrease that is similarly reversed by 1,25(OH)2D3. This result illustrates the dynamic nature of the regulation of Vdr gene expression. Nevertheless, it is clear that the reduction in response to 1,25 (OH)2D3 in OBs overall is due, at least in part, to a decline in the level of the VDR.
FIGURE 4.
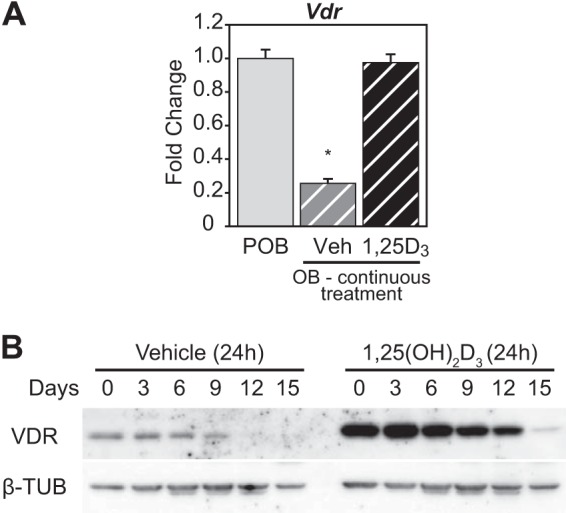
VDR expression is suppressed in OBs. A, POB cells differentiated for 15 days to OB cells were treated with 10−7 m 1,25(OH)2D3 (1,25D3) or Veh every 3 days continuously during culture. RT-qPCR was performed on RNA isolated from the POB and OB cells for Vdr levels. Values are relative quantitation (RQ) normalized to β-actin levels with fold change set to 1 for the POB sample (POB, gray; OB Veh, gray striped; OB 1,25(OH)2D3, black striped). Samples completed in biological triplicate ± S.E. *, p < 0.05 compared with POB sample. B, Western blot of VDR and β-tubulin (β-TUB) proteins from whole cell lysates collected every 3 days during differentiation from POB to OB (15 days). The cells were treated with vehicle or 10−7 m 1,25(OH)2D3 for 24 h prior to isolation.
VDR-binding Sites Contain RUNX2 and/or C/EBPβ Cofactors
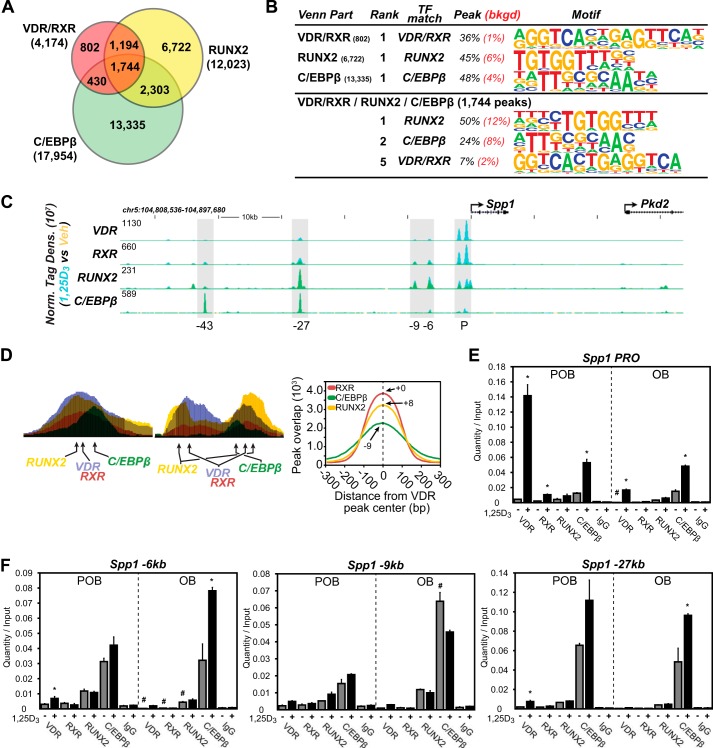
Although VDR protein levels are reduced, the altered response to 1,25(OH)2D3 exhibited by a large subset of genes in OBs indicates that factors other than receptor concentration are likely involved. RUNX2 and C/EBPβ represent two lineage maturation factors that function similarly to regulate chromatin structure. Both are induced by ascorbic acid (42, 43) and are central to osteoblast differentiation. VDR and RUNX2 are known to physically interact (44); therefore, we examined whether these factors might be co-localized to sites of VDR occupancy on a genome-wide scale. This analysis was supported by the considerable frequency with which RUNX2- and/or C/EBPβ-binding site motifs were predicted at active VDR binding regions, as documented in Fig. 2D, and more directly by our recent determination of the cistromes for these two factors in this cellular model of differentiation (28). As can be seen in Fig. 5A, a comparative bioinformatic analysis of the cistromes for VDR/RXR, RUNX2, and C/EBPβ did indeed indicate the presence of a significant overlap between the VDR/RXR cistrome and these two additional factors in POBs. Accordingly, 2,938 VDR/RXR-binding sites contained RUNX2 (70%), 2,174 sites contained C/EBPβ (52%), and 1,744 sites contained both factors (42%). Fig. 5B documents the results of a de novo motif analysis at these 1,744 sites which revealed in turn the frequent presence of consensus DNA sequences preferential for each of these factors. Further analysis, as documented in Fig. 5C, revealed several of the complexities of this relationship using the Spp1 gene locus, a known target of 1,25(OH)2D3, as an example. As can be seen in the ChIP-seq tracks for VDR, RXR, RUNX2, and C/EBPβ (Fig. 5C), at least four binding regions for one or more of these factors are present upstream of the Spp1 TSS. Although all contain variable levels of 1,25(OH)2D3-induced VDR and RXR, three contain significant levels of RUNX2, and two contain significant levels of C/EBPβ. It is important to note here that both RUNX2 and C/EBPβ are pre-bound to DNA, whereas VDR binding represents a consequence of activation by 1,25(OH)2D3. Thus, RUNX2 and C/EBPβ binding to DNA precedes that of the VDR.
FIGURE 5.
VDR-binding sites contain RUNX2 and/or C/EBPβ cofactors. A, Venn diagram depiction of replicate-normalized VDR/RXR-, RUNX2-, and C/EBPβ-binding sites in POB cells. B, de novo over-representation analysis of VDR/RXR, RUNX2, and C/EBPβ peak sequences and matching transcription factor-binding sites found through HOMER. Abundance shown as percentage (black) compared with 50,000 GC content-matched sequences (red). C, ChIP-seq tag density tracks for the Spp1 gene locus for VDR, RXR, RUNX2, and C/EBPβ binding (Veh, yellow; 1,25(OH)2D3, blue; overlap, green). Additional details are the same as in Fig. 2. D, schematic depiction of VDR-, RUNX2-, and C/EBPβ-binding elements in relationship to peak maxima as found in ChIP-seq data sets, left. Right, ChIP-seq peaks for RXR, RUNX2, and C/EBPβ were plotted with their relationship to VDR-binding peak centers. Peak maxima for each factor indicated by number upstream (−) or downstream (+) from VDR peak center. E and F, ChIP-qPCR analysis of Spp1 peak regions. Data displayed as quantitation normalized to ChIP input ± S.E. *, p < 0.05, 10−7 m 1,25(OH)2D3 (+) versus vehicle (−) within ChIP antibody by Student's t test. #, p < 0.05, POB versus OB by Student's t test.
The overall organization of two of the regulatory sites and the putative binding sites in Spp1 that interact with each of these binding factors is documented in Fig. 5D, left panel, placing RUNX2 and C/EBPβ on either side of the VDR/RXR heterodimer. The results in Fig. 5D, right panel, summarize the spatial relationship that can be quantified for RUNX2 and C/EBPβ binding, respectively, on a global scale at +8 and −9 bp versus VDR/RXR. The results in Fig. 5, E and F, using ChIP-qPCR analysis confirm the binding of these factors to several of the regions of the Spp1 gene seen in Fig. 5C. It is therefore possible that these factors define the organization of at least one “osteoblast-specific” regulatory complex whose overall activity integrates inputs from multiple signaling pathways.
1,25(OH)2D3 Induces a Partial Genomic Redistribution of RUNX2 and C/EBPβ
Interestingly, in addition to the above effects on VDR activity, the reverse effect of the hormone on the genomic distribution of RUNX2 and C/EBPβ may also occur as well. Thus, as documented in Fig. 6A, a 3-h treatment with 1,25(OH)2D3 results in a significant partial redistribution of RUNX2 and C/EBPβ binding across the POB genome, accompanied by minor changes in the number of binding sites for both factors. These results suggest that in addition to the impact of RUNX2 and C/EBPβ on vitamin D action, the hormone also has an impact on RUNX2 and C/EBPβ binding, both of which could affect transcriptomic output in POBs. The sestrin-1 (Sesn1) gene tracks seen in Fig. 6B depict the induction of novel RUNX2 and C/EBPβ sites by 1,25(OH)2D3 at the −26-kb site, changes that are confirmed by direct ChIP-qPCR analysis as seen in Fig. 6E. Although the RUNX2 and C/EBPβ cistromes are reduced, a similar overall effect of 1,25(OH)2D3 on OBs was also noted. Thus, the actions of 1,25(OH)2D3 are not only strongly linked to the presence and activity of RUNX2 and C/EBPβ in bone cells, but perhaps also to the hormone's actions to selectively influence their binding and genomic activities with the VDR.
FIGURE 6.
RUNX2 and C/EBPβ binding are influenced by 1,25(OH)2D3 as are histone modification markers. A, Venn diagram depiction of replicate normalized RUNX2- and C/EBPβ-binding sites assessed using HOMER in undifferentiated (POB) cells treated with Veh or 1,25(OH)2D3 (1,25D3). B, ChIP-seq tag density tracks for the Sesn1 gene locus for VDR, RUNX2, and C/EBPβ in the POB and OB cells (Veh, yellow; 1,25D3, blue; overlap, green). Additional details are the same as in Fig. 2. C, ChIP-seq tag density tracks for the Spp1 gene locus for H4K5Ac, H3K9Ac, H3K4me1, H3K4me3, and H3K36me3 binding (Veh, yellow; 1,25(OH)2D3, blue; overlap, green). Further details as in B. D, ChIP-seq tag density compared near the TSS (±3 kb) of genes up-regulated (red and blue) versus down-regulated (yellow and green) in POB (left) or OB (right) cells by cis-regulatory element annotation system. Solid lines represent the H3K9Ac tag density in the 1,25(OH)2D3 condition, and dashed lines represent the vehicle condition. All TSS of genes in genome are indicated by a black solid line. Specific changes in density are highlighted by vertical black bars. E, ChIP-qPCR analysis of Sesn1 −26-kb peak region. Data displayed as quantitation normalized to ChIP input ± S.E. *, p < 0.05, 10−7 m 1,25(OH)2D3 (+) versus vehicle (−) within ChIP antibody by Student's t test. #, p < 0.05, POB versus OB by Student's t test.
Regulatory Sites Are Enriched for Epigenetic Histone Enhancer Signatures
Our previous studies suggest that key enhancer signature marks at H3 and H4 (28) are associated with binding sites for RUNX2 and C/EBPβ and that modest changes in the levels of these marks occur in a site-specific manner as a function of POB differentiation. We therefore explored whether these marks were also associated with VDR binding on the POB and OB genomes. Enhancer histone modifications were indeed enriched at sites of VDR/RXR binding as well as at sites of RUNX2 and C/EBPβ binding, as exemplified by the data for H3K9ac, H4K5ac, H3K4me1, and H3K4me3 documented at the Spp1 gene locus in Fig. 6C and at other genes documented in supplemental Dataset 1. Activation by 1,25(OH)2D3 also led to an up-regulation of several of these marks at the Spp1 locus, including H3K9ac, H4K5ac, and H3K4me3, but only at regions that become occupied by VDR and RXR. Importantly, cis-regulatory element annotation system analysis (CEAS) (45), as seen in Fig. 6D, showed that 1,25(OH)2D3 induced an increase in H3K9ac near the TSS of many genes that are up-regulated by 1,25(OH)2D3; a similar relationship was seen with other histone marks as well (data not shown). H3K9ac was also increased at genes that are down-regulated by the hormone, indicating that repressive mechanisms can also be highlighted by marks such as these. These results suggest that epigenetic modifications at histones identify enhancers in osteoblast lineage cells that perhaps contribute to the associated gene's regulation during both differentiation and as a function of 1,25(OH)2D3.
1,25(OH)2D3 Blocks POB Differentiation and Mineralization via Direct Actions to Suppress Both Runx2 and Sp7 Gene Expression
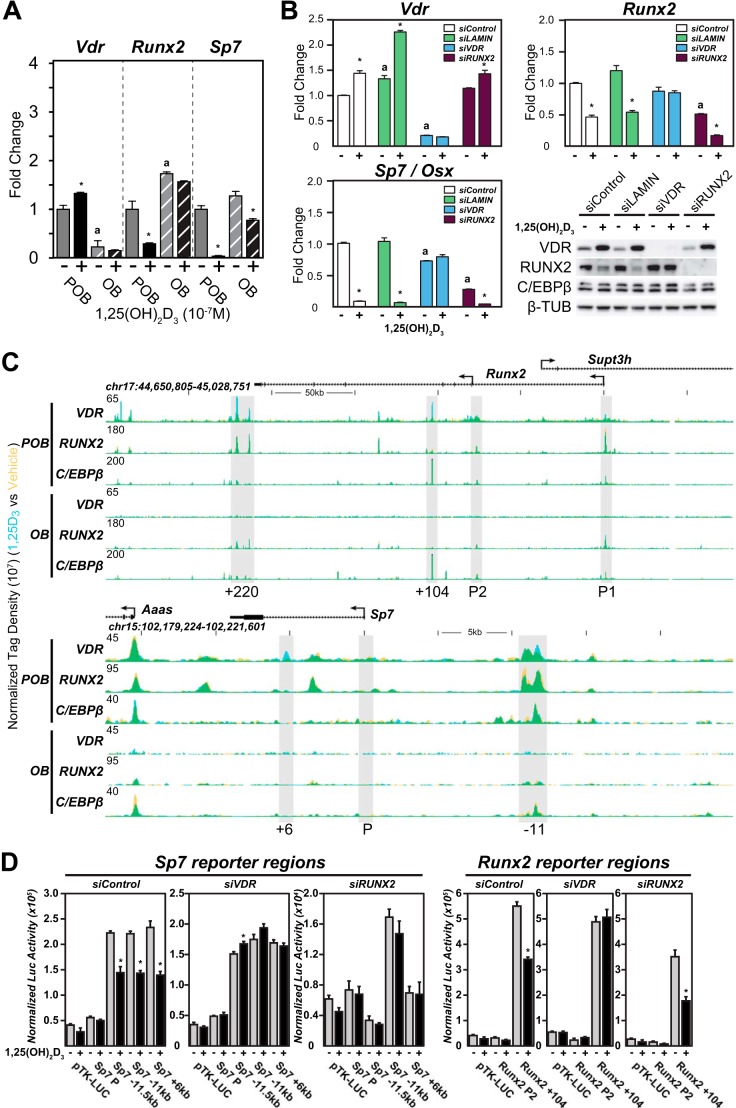
Earlier studies suggest that continuous administration of 1,25(OH)2D3 suppresses both Runx2 and Sp7 gene expression in early osteoblast-lineage cells in culture and affects the expression of other genes that are involved in mineralization such as Spp1. Indeed, it was recently reported that 1,25(OH)2D3 also regulates the expression of mineralization inhibitors such as Ank, Enpp1, and Enpp3 both in vitro and in vivo, activities that involve enhancers to which these genes are linked (16). Interestingly, these activities of 1,25(OH)2D3 can change for some genes in mature osteoblasts. Thus, as seen in Fig. 7A and supplemental Dataset 1, although 1,25(OH)2D3 down-regulates both RUNX2 and Osterix, the gene product of Sp7, in POBs, this regulation is reduced or lost in OBs. Importantly, RUNX2 protein levels were similarly down-regulated following a 24-h treatment with 1,25(OH)2D3, consistent with transcript levels (Fig. 7B) (46). To explore these actions in more detail at the Runx2 and Sp7 loci, we first assessed the impact of siRNA down-regulation of Vdr and Runx2 and on both basal and 1,25(OH)2D3-induced transcript expression in POBs. VDR and RUNX2 mRNA and protein were both reduced significantly with these siRNAs, as seen in Fig. 7B. The results in this figure also show that although VDR knockdown eliminated the expression of the Vdr gene, it also prevented the down-regulation of both Runx2 and Sp7 as well. siRNA knockdown of Runx2, however, had no effect on Vdr expression but strikingly down-regulated Sp7 without eliminating the ability of 1,25(OH)2D3 to further suppress its expression. These results establish that 1,25(OH)2D3 directly suppresses both Runx2 and Sp7 expression but that RUNX2 is a primary regulator of basal Sp7 expression.
FIGURE 7.
1,25(OH)2D3 blocks POB differentiation and mineralization via direct actions to suppress both Runx2 and Sp7 gene expression. A, RT-qPCR was performed on RNA isolated from the POB and OB cells treated with 24 h of vehicle (−) or 10−7 m 1,25(OH)2D3 (+) for Vdr, Runx2, and Sp7 genes. Values are relative quantitation normalized to β-actin levels with fold change set to 1 for the POB vehicle sample (POB, solid; OB, striped). Samples completed in biological triplicate ± S.E. *, p < 0.05 compared with vehicle (−) sample; a, p < 0.05, vehicle OB compared with vehicle POB. B, POB cells were transfected with siRNA for siControl (white), siLAMIN (green), siVDR (blue), and siRUNX2 (purple). RT-qPCR was performed on RNA isolated from the siRNA cells treated for 24 h with vehicle (−) or 10−7 m 1,25(OH)2D3 (+) for Vdr, Runx2, and Sp7 genes. Values are relative quantitation normalized to β-actin levels with fold change set to 1 for the siControl vehicle sample. Samples completed in biological triplicate ± S.E. *, p < 0.05 compared with vehicle (−) sample; a, p < 0.05 vehicle compared with vehicle siControl. Western blot was performed on whole cell lysates from cells treated with siRNA as indicated and vehicle (−) or 10−7 m 1,25(OH)2D3 (+) for 24 h. C, ChIP-seq tag density tracks for the Runx2 and Sp7 gene loci for VDR, RUNX2, and C/EBPβ binding (Veh, yellow; 1,25(OH)2D3, blue; overlap, green) in the POB and OB cells. Additional details are the same as in Fig. 2. D, cloned pTK-luciferase enhancer reporter constructs transfected into POB cells were treated with siControl (left), siVDR (middle), and siRUNX2 (right) for VDR binding regions in Sp7 and Runx2 genes. Post-transfection, samples were treated with vehicle (−) or 10−7 m 1,25(OH)2D3 (+) for 12–16 h. Results displayed as relative light units normalized with β-galactosidase co-transfection levels. Triplicate set of assays ± S.E. *, p < 0.05, 1,25(OH)2D3 compared with vehicle within each construct.
Given these results, we then further explored the underlying mechanism that might account for the differential sensitivity of the Runx2 and Sp7 genes to 1,25(OH)2D3 in POBs and OBs by examining the ChIP-seq data tracts for VDR, RUNX2, and C/EBPβ across these two gene loci in both cell types. As can be seen in Fig. 7C, upper track, VDR binding is induced by 1,25(OH)2D3 across the Runx2 locus at both an intronic site at +104 kb, several intergenic sites at the 3′ end of the gene, and very modestly at the P1 and P2 promoters. Several of these sites also contain pre-bound RUNX2 and one (+104 kb) contains C/EBPβ as well. As also seen in Fig. 7C (lower track), VDR binding is similarly evident at several locations across the Sp7 locus, including one at a distal intergenic site −11 kb upstream and several more modest binding sites at the promoter and within an intron at +6 kb. Both RUNX2 and C/EBPβ are also bound to the Sp7 distal site at −11 kb and at several additional sites across the locus independent of the VDR. To confirm the validity of these sites, we cloned these regions into a reporter vector, introduced them into POBs via transfection, and examined whether they were indeed capable of mediating response to 1,25(OH)2D3. As can be seen in Fig. 7D, 1,25(OH)2D3 prompted a down-regulation of luciferase activity in the presence of the +104-kb enhancer but not with the Runx2 P2 promoter region; this activity of the hormone at the +104-kb enhancer was lost upon VDR knockdown. The high basal activity of the enhancer at +104 kb, but not its response to 1,25(OH)2D3, was also abrogated following knockdown of RUNX2, supporting the role of RUNX2 binding at this enhancer. In contrast, the activities of virtually all of the enhancers derived from the Sp7 locus were suppressed by 1,25(OH)2D3 in a VDR-dependent fashion. It is clear, therefore, that VDR-binding sites in addition to the site previously reported at the Runx2 P2 promoter (47) may be critical for Runx2 gene regulation, and similar conclusions can also be drawn for Sp7. Perhaps most interestingly, the binding of both the VDR and RUNX2 seen in POBs is lost following ChIP-seq analysis in OBs; C/EBPβ, however, is maintained. This finding suggests that the failure of 1,25(OH)2D3 to down-regulate both Runx2 and Sp7 expression in mature osteoblasts is likely due to the loss of both VDR and RUNX2 binding at these two genes.
DISCUSSION
Our previous studies suggest that both quantitative and qualitative transcriptomic changes are evident upon the differentiation of POBs to bone-forming OBs in vitro (28). These changes are accompanied by a striking decrease in the cistromes for both RUNX2 and C/EBPβ, the former a master regulator of osteoblast lineage development and the latter a chromatin regulatory factor active in numerous mesenchyme-derived cells (48). In this study, we examined whether these changes associated with differentiation were capable of altering a response to the vitamin D hormone 1,25(OH)2D3, a question sparked by previous studies, which suggested that the actions of this hormone are cell maturation-dependent both in vitro and in vivo (2). Using genome-scale studies, we show that transcriptomic responses to the hormone are indeed altered in OBs following differentiation. These changes in mature osteoblasts include both a striking overall reduction in transcriptional responses to 1,25(OH)2D3 as well as an alteration in response profiles for a large cohort of genes. They are also accompanied by a considerable reduction in the cistrome for the VDR/RXR heterodimer. Although this overall reduction in the VDR cistrome is likely a result of a concomitant down-regulation of VDR protein levels in OBs, the continued or increased responsiveness of a substantial subset of genes to 1,25(OH)2D3 together with a significant reduction in VDR binding at these genes was surprising. We show that these differences in sensitivity to 1,25(OH)2D3 at many genes may be due to contributory changes in the level of occupancy and activity of RUNX2 and/or C/EBPβ both at independent sites of action at target genes as well as at sites that specifically contain the VDR. The effects of 1,25(OH)2D3 are potentially magnified by the ability of the hormone to promote a rapid and significant redistribution of RUNX2 and C/EBPβ binding at genomic sites and to down-regulate the expression of not only RUNX2 but an osteoblast differentiation factor Osterix that is dependent upon RUNX2 for expression. We conclude that although the activity of 1,25(OH)2D3 in osteoblast lineage cells is differentiation state-sensitive, responsiveness to 1,25(OH)2D3 is dependent on not only the levels of the VDR but additional transcription factors as well.
Although significant changes were noted in both the transcriptomes and the cistromes in POBs and OBs, the general features of the VDR cistromes in these two cell types were similar and typical of those observed previously for the VDR (19, 20). They include the receptor's general dependence upon 1,25(OH)2D3 for DNA binding, the involvement of RXR as a heterodimer partner, the presence of classic VDREs within sites of VDR binding (95% of peaks as determined by position weight matrix analyses), and the frequently distal location of VDR binding loci. Interestingly, although VDR-binding sites were proximally linked to several thousand annotated protein-coding genes via the nearest neighbor algorithm GREAT, only 46% of the 960 genes actually regulated by 1,25(OH)2D3 exhibited one or more adjacent VDR-binding sites; the remaining regulated genes did not contain detectable sites of VDR binding. There are many possible explanations for this latter finding, although the simplest is that the location of the regulatory region lies outside the regions inspected. ChIA-PET analysis, for example, suggests that some genes are regulated not only by highly remote enhancers but by enhancers on other chromosomes or via genes that share their enhancers (49, 50). Surprisingly, the vast majority of VDR-binding sites identified were located near genes that were not regulated by 1,25(OH)2D3. Although the explanation for this finding is likely complex, our results suggest that our understanding of the relationship between detectable VDR binding activity and the regulation of specific target genes is relatively incomplete.
As indicated earlier, the VDR protein undergoes a dynamic down-regulation during osteoblast differentiation in the absence of 1,25(OH)2D3. We have previously reported that auto-regulation of VDR gene expression occurs in bone cells both in vitro and in vivo (51). Mechanistically, this regulation is mediated directly through the activity of at least two enhancers, one located within a VDR gene intron and the other upstream of the TSS (52–54). Thus, it could be argued that 1,25(OH)2D3 is trophic for VDR expression in bone and that the basal concentration of 1,25(OH)2D3 in this tissue represents an indirect determinant of net transcriptional output in response to the hormone. Interestingly, a recent report in mice using immunocytochemistry has suggested that the level of the VDR in osteoblast lineage cells may decrease as a function of differentiation, supporting a potential change in the differentiated osteoblast genome due to VDR down-regulation (55). However, whether basal VDR levels are dependent upon circulating concentrations of 1,25(OH)2D3 in vivo is unknown. Importantly, however, the VDR is also regulated in bone and other cell types by a number of additional signaling pathways and factors (53). Regardless of the individual components or their contributions, the importance of regulating and maintaining adequate levels of VDR may provide an explanation for why the VDR gene is auto-regulated by 1,25(OH)2D3.
The striking overlap between 1,25(OH)2D3-induced VDR occupancy at target gene enhancers and pre-bound RUNX2 and C/EBPβ suggests that the pre-bound factors are likely determinants of a receptive regulatory region whose activities can be modulated by secondary factors such as the VDR. Although both RUNX2 and C/EBPβ occupy independent sites at many regulated genes, the simultaneous presence of the RUNX2, C/EBPβ, and VDR/RXR at many enhancers suggests the possibility that these DNA-binding proteins and additional regulatory factors known to associate with these factors may include components of an important “osteoblast-specific” enhancer complex. We have not shown a direct interaction of these factors with each other. Nevertheless, the idea that they may interact directly is supported by the close association of the DNA sequence motifs that are found in these osteoblast-specific enhancer regions. The concept that active protein complexes might regulate the activities of 1,25(OH)2D3 on bone cell gene expression was advanced earlier through the seminal work of Stein and co-workers (43, 56, 57). This study, however, provides an additional, broader genome-wide perspective to these original insights. The identity of additional non-DNA binding co-regulatory factors that may participate is unknown, although VDR, RUNX2, and C/EBPβ can recruit numerous classes of chromatin regulators such as histone acetyltransferase, histone deacetylase, and others as well (58–61). Why should pre-bound RUNX2 and/or C/EBPβ contribute to 1,25(OH)2D3 action? RUNX2 is both an inducer and suppressor of gene regulation (62). Its activity is highly regulated in osteoblast lineage cells through inputs from multiple signaling pathways that can impact RUNX2 concentration, DNA binding, post-translational modification, and direct interaction with numerous other DNA binding and non-DNA binding factors (48, 62–64). C/EBPβ exhibits a similar complexity in its actions (65). Thus, enhancers that represent targets of VDR activity and contain these additional factors are not only regulatory in complex ways but are capable of integrating inputs from multiple signaling pathways. Progressive mineralization, for example, may selectively impact RUNX2 activity, as has been shown during osteocyte differentiation (66). The presence of RUNX2 at potential VDR-binding sites is therefore likely to exert a significant influence on both VDR binding and transcriptional activity, and thus it is likely to impose a considerable influence on both selectivity and sensitivity to the 1,25(OH)2D3 hormone. Perhaps most importantly, these studies point to the highly dynamic nature and rapidly changing configuration of the regulatory cistromes for several transcription factors during even modest changes in the differentiation state of cells.
As indicated earlier, our studies demonstrate that in addition to the effects of 1,25(OH)2D3 on RUNX2 distribution across the genome, 1,25(OH)2D3 also directly regulates the expression of both Runx2 and Sp7 genes. Previous work using traditional methods suggested that the Runx2 gene is auto-regulated via RUNX2 and suppressed by 1,25(OH)2D3 through action near the Runx2 P2 promoter (47, 67). In contrast, our studies reveal multiple binding sites for RUNX2 within the Runx2 gene locus, sites near P1 and P2 as well as a site located within an intron and a cluster at the 3′ end of the gene. These binding regions contain consensus DNA sequences for RUNX2. We also note several potential VDR binding regions at both P1 and P2 and additional intronic and downstream sites as well; several of these overlapping sites bind RUNX2. Interestingly, although the Runx2 P1 promoter was relatively inactive and unresponsive, RUNX2 siRNA reduced basal expression in a site that contained RUNX2 alone, and VDR siRNA led to a loss of 1,25(OH)2D3 regulated activity at an intronic site that contained only the VDR. Sp7, however, although known to be strongly induced by RUNX2 (68), is also suppressed by 1,25(OH)2D3. Accordingly, several enhancers capable of binding both RUNX2 and/or VDR are seen spanning the gene locus. Reporter studies of several of these segments in the presence of control, RUNX2, or VDR siRNA support the idea that the loss of RUNX2 at two of the three RUNX2-bound sites results in reduced basal expression of the reporter and that each of the enhancers mediates VDR-dependent down-regulation by 1,25(OH)2D3. Thus, these studies support the direct actions of RUNX2 and 1,25(OH)2D3 on both Runx2 and Sp7 gene expression in POBs.
In conclusion, our studies demonstrate that differentiation of POBs to OBs leads to a qualitative and quantitative change in transcriptional response to 1,25(OH)2D3. These changes are due to a reduction in VDR binding at genomic sites due, in part, to a striking down-regulation of VDR expression and protein. A large subset of genes show increased or novel responsiveness to 1,25(OH)2D3, however, supporting the idea that additional factors such as RUNX2 and C/EBPβ may also contribute. With respect to these two factors, 1,25(OH)2D3 also prompts a minor redistribution of their binding across the genome and regulates the expression of RUNX2. Thus, our studies highlight the critical role of transcription factor interaction in altered transcriptomic response to 1,25(OH)2D3 and the effect of these changes on selective gene regulation.
Supplementary Material
Acknowledgments
We thank members of the Pike Lab for their helpful discussions and contributions to this manuscript and the technical contributions of Amber Mael. We also acknowledge the University of Wisconsin DNA Sequencing Facility supported by Marie Adams, John Alliet, and Josh Hyman.
This work was supported, in whole or in part, by National Institutes of Health Grants DK-072281 from NIDDK and AR-045173 from NIAMS (to J. W. P.).
This article was selected as a Paper of the Week.

This article contains supplemental Table S1 and Dataset 1.
- 1,25(OH)2D3
- 1α,25-dihydroxyvitamin D3
- RT-qPCR
- reverse transcription–quantitative polymerase chain reaction
- GREAT
- genomic regions enrichment of annotations tool
- GO
- gene ontology
- TSS
- transcriptional start site
- C/EBP
- CCAAT/enhancer-binding proteins
- RUNX2
- runt-related transcription factor 2
- VDR
- vitamin D receptor
- RXR
- retinoid X receptor
- OB
- osteoblast
- POB
- pre-osteoblast
- VDRE
- vitamin D response element
- RXR
- retinoid X receptor
- Veh
- vehicle
- BP
- biological process.
REFERENCES
- 1. Lieben L., Carmeliet G., Masuyama R. (2011) Calcemic actions of vitamin D: effects on the intestine, kidney and bone. Best Pract. Res. Clin. Endocrinol. Metab. 25, 561–572 [DOI] [PubMed] [Google Scholar]
- 2. Bikle D. D. (2012) Vitamin D and bone. Curr. Osteoporos Rep. 10, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eisman J. A., Bouillon R. (2014) Vitamin D: direct effects of vitamin D metabolites on bone: lessons from genetically modified mice. Bonekey Rep. 3, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Plum L. A., DeLuca H. F. (2010) Vitamin D, disease and therapeutic opportunities. Nat. Rev. Drug Discov. 9, 941–955 [DOI] [PubMed] [Google Scholar]
- 5. Lacey D. L., Timms E., Tan H. L., Kelley M. J., Dunstan C. R., Burgess T., Elliott R., Colombero A., Elliott G., Scully S., Hsu H., Sullivan J., Hawkins N., Davy E., Capparelli C., Eli A., Qian Y. X., Kaufman S., Sarosi I., Shalhoub V., Senaldi G., Guo J., Delaney J., Boyle W. J. (1998) Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell 93, 165–176 [DOI] [PubMed] [Google Scholar]
- 6. Shimada T., Mizutani S., Muto T., Yoneya T., Hino R., Takeda S., Takeuchi Y., Fujita T., Fukumoto S., Yamashita T. (2001) Cloning and characterization of FGF23 as a causative factor of tumor-induced osteomalacia. Proc. Natl. Acad. Sci. U.S.A. 98, 6500–6505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shimada T., Kakitani M., Yamazaki Y., Hasegawa H., Takeuchi Y., Fujita T., Fukumoto S., Tomizuka K., Yamashita T. (2004) Targeted ablation of Fgf23 demonstrates an essential physiological role of FGF23 in phosphate and vitamin D metabolism. J. Clin. Invest. 113, 561–568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Inoue Y., Segawa H., Kaneko I., Yamanaka S., Kusano K., Kawakami E., Furutani J., Ito M., Kuwahata M., Saito H., Fukushima N., Kato S., Kanayama H. O., Miyamoto K. (2005) Role of the vitamin D receptor in FGF23 action on phosphate metabolism. Biochem. J. 390, 325–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bouillon R., Bischoff-Ferrari H., Willett W. (2008) Vitamin D and health: perspectives from mice and man. J. Bone Miner. Res. 23, 974–979 [DOI] [PubMed] [Google Scholar]
- 10. Bouillon R., Carmeliet G., Verlinden L., van Etten E., Verstuyf A., Luderer H. F., Lieben L., Mathieu C., Demay M. (2008) Vitamin D and human health: lessons from vitamin D receptor null mice. Endocr. Rev. 29, 726–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lieben L., Carmeliet G. (2013) The delicate balance between vitamin D, calcium and bone homeostasis: lessons learned from intestinal- and osteocyte-specific VDR null mice. J. Steroid Biochem. Mol. Biol. 136, 102–106 [DOI] [PubMed] [Google Scholar]
- 12. Dardenne O., Prud'homme J., Arabian A., Glorieux F. H., St-Arnaud R. (2001) Targeted inactivation of the 25-hydroxyvitamin D(3)-1(α)-hydroxylase gene (CYP27B1) creates an animal model of pseudovitamin D-deficiency rickets. Endocrinology 142, 3135–3141 [DOI] [PubMed] [Google Scholar]
- 13. Amling M., Priemel M., Holzmann T., Chapin K., Rueger J. M., Baron R., Demay M. B. (1999) Rescue of the skeletal phenotype of vitamin D receptor-ablated mice in the setting of normal mineral ion homeostasis: formal histomorphometric and biomechanical analyses. Endocrinology 140, 4982–4987 [DOI] [PubMed] [Google Scholar]
- 14. Panda D. K., Miao D., Bolivar I., Li J., Huo R., Hendy G. N., Goltzman D. (2004) Inactivation of the 25-hydroxyvitamin D 1α-hydroxylase and vitamin D receptor demonstrates independent and interdependent effects of calcium and vitamin D on skeletal and mineral homeostasis. J. Biol. Chem. 279, 16754–16766 [DOI] [PubMed] [Google Scholar]
- 15. Gardiner E. M., Baldock P. A., Thomas G. P., Sims N. A., Henderson N. K., Hollis B., White C. P., Sunn K. L., Morrison N. A., Walsh W. R., Eisman J. A. (2000) Increased formation and decreased resorption of bone in mice with elevated vitamin D receptor in mature cells of the osteoblastic lineage. FASEB J. 14, 1908–1916 [DOI] [PubMed] [Google Scholar]
- 16. Lieben L., Masuyama R., Torrekens S., Van Looveren R., Schrooten J., Baatsen P., Lafage-Proust M. H., Dresselaers T., Feng J. Q., Bonewald L. F., Meyer M. B., Pike J. W., Bouillon R., Carmeliet G. (2012) Normocalcemia is maintained in mice under conditions of calcium malabsorption by vitamin D-induced inhibition of bone mineralization. J. Clin. Invest. 122, 1803–1815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pike J. W., Meyer M. B. (2013) Fundamentals of vitamin D hormone-regulated gene expression. J. Steroid Biochem. Mol. Biol. 10.1016/j.jsbmb.2013.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pike J. W., Lee S. M., Meyer M. B. (2014) Regulation of gene expression by 1,25-dihydroxyvitamin D3 in bone cells: exploiting new approaches and defining new mechanisms. Bonekey Rep. 3, 482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meyer M. B., Goetsch P. D., Pike J. W. (2010) A downstream intergenic cluster of regulatory enhancers contributes to the induction of CYP24A1 expression by 1α,25-dihydroxyvitamin D3. J. Biol. Chem. 285, 15599–15610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Meyer M. B., Goetsch P. D., Pike J. W. (2010) Genome-wide analysis of the VDR/RXR cistrome in osteoblast cells provides new mechanistic insight into the actions of the vitamin D hormone. J. Steroid Biochem. Mol. Biol. 121, 136–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. de Wit E., de Laat W. (2012) A decade of 3C technologies: insights into nuclear organization. Genes Dev. 26, 11–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dowd D. R., Sutton A. L., Zhang C., MacDonald P. N. (2005) in Vitamin D (Feldman D., Pike J. W., Glorieux F. H., eds) 2nd Ed., pp. 291–304, Elsevier/Academic Press, New York [Google Scholar]
- 23. Meyer M. B., Pike J. W. (2013) Corepressors (NCoR and SMRT) as well as coactivators are recruited to positively regulated 1α,25-dihydroxyvitamin D3-responsive genes. J. Steroid Biochem. Mol. Biol. 136, 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murayama A., Kim M. S., Yanagisawa J., Takeyama K., Kato S. (2004) Transrepression by a liganded nuclear receptor via a bHLH activator through co-regulator switching. EMBO J. 23, 1598–1608 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25. Sudo H., Kodama H. A., Amagai Y., Yamamoto S., Kasai S. (1983) In vitro differentiation and calcification in a new clonal osteogenic cell line derived from newborn mouse calvaria. J. Cell Biol. 96, 191–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Elford P. R., Felix R., Cecchini M., Trechsel U., Fleisch H. (1987) Murine osteoblastlike cells and the osteogenic cell MC3T3-E1 release a macrophage colony-stimulating activity in culture. Calcif. Tissue Int. 41, 151–156 [DOI] [PubMed] [Google Scholar]
- 27. Udagawa N., Takahashi N., Akatsu T., Sasaki T., Yamaguchi A., Kodama H., Martin T. J., Suda T. (1989) The bone marrow-derived stromal cell lines MC3T3-G2/PA6 and ST2 support osteoclast-like cell differentiation in cocultures with mouse spleen cells. Endocrinology 125, 1805–1813 [DOI] [PubMed] [Google Scholar]
- 28. Meyer M. B., Benkusky N. A., Pike J. W. (2014) The RUNX2 cistrome in osteoblasts: characterization, down-regulation following differentiation and relationship to gene expression. J. Biol. Chem. 289, 16016–16031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meyer M. B., Watanuki M., Kim S., Shevde N. K., Pike J. W. (2006) The human transient receptor potential vanilloid type 6 distal promoter contains multiple vitamin D receptor binding sites that mediate activation by 1,25-dihydroxyvitamin D3 in intestinal cells. Mol. Endocrinol. 20, 1447–1461 [DOI] [PubMed] [Google Scholar]
- 30. Kim S., Yamazaki M., Zella L. A., Shevde N. K., Pike J. W. (2006) Activation of receptor activator of NF-κB ligand gene expression by 1,25-dihydroxyvitamin D3 is mediated through multiple long-range enhancers. Mol. Cell. Biol. 26, 6469–6486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yamamoto H., Shevde N. K., Warrier A., Plum L. A., DeLuca H. F., Pike J. W. (2003) 2-Methylene-19-nor-(20S)-1,25-dihydroxyvitamin D3 potently stimulates gene-specific DNA binding of the vitamin D receptor in osteoblasts. J. Biol. Chem. 278, 31756–31765 [DOI] [PubMed] [Google Scholar]
- 32. Meyer M. B., Goetsch P. D., Pike J. W. (2012) VDR/RXR and TCF4/β-catenin cistromes in colonic cells of colorectal tumor origin: impact on c-FOS and c-MYC gene expression. Mol. Endocrinol. 26, 37–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kriebitzsch C., Verlinden L., Eelen G., van Schoor N. M., Swart K., Lips P., Meyer M. B., Pike J. W., Boonen S., Carlberg C., Vitvitsky V., Bouillon R., Banerjee R., Verstuyf A. (2011) 1,25-Dihydroxyvitamin D3 influences cellular homocysteine levels in murine preosteoblastic MC3T3-E1 cells by direct regulation of cystathionine β-synthase. J. Bone Miner. Res. 26, 2991–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carlberg C., Dunlop T. W., Saramäki A., Sinkkonen L., Matilainen M., Väisänen S. (2007) Controlling the chromatin organization of vitamin D target genes by multiple vitamin D receptor binding sites. J. Steroid Biochem. Mol. Biol. 103, 338–343 [DOI] [PubMed] [Google Scholar]
- 35. Meyer M. B., Benkusky N. A., Pike J. W. (2013) 1,25-Dihydroxyvitamin D3 induced histone profiles guide discovery of VDR action sites. J. Steroid Biochem. Mol. Biol. 10.1016/j.jsbmb.2013.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Meyer M. B., Zella L. A., Nerenz R. D., Pike J. W. (2007) Characterizing early events associated with the activation of target genes by 1,25-dihydroxyvitamin D3 in mouse kidney and intestine in vivo. J. Biol. Chem. 282, 22344–22352 [DOI] [PubMed] [Google Scholar]
- 37. Carroll J. S., Meyer C. A., Song J., Li W., Geistlinger T. R., Eeckhoute J., Brodsky A. S., Keeton E. K., Fertuck K. C., Hall G. F., Wang Q., Bekiranov S., Sementchenko V., Fox E. A., Silver P. A., Gingeras T. R., Liu X. S., Brown M. (2006) Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 38, 1289–1297 [DOI] [PubMed] [Google Scholar]
- 38. Nielsen R., Pedersen T. A., Hagenbeek D., Moulos P., Siersbaek R., Megens E., Denissov S., Børgesen M., Francoijs K. J., Mandrup S., Stunnenberg H. G. (2008) Genome-wide profiling of PPARγ:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 22, 2953–2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Welboren W. J., Stunnenberg H. G., Sweep F. C., Span P. N. (2007) Identifying estrogen receptor target genes. Mol. Oncol. 1, 138–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cartharius K., Frech K., Grote K., Klocke B., Haltmeier M., Klingenhoff A., Frisch M., Bayerlein M., Werner T. (2005) MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942 [DOI] [PubMed] [Google Scholar]
- 41. McLean C. Y., Bristor D., Hiller M., Clarke S. L., Schaar B. T., Lowe C. B., Wenger A. M., Bejerano G. (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol. 28, 495–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lian J. B., Stein J. L., Stein G. S., Montecino M., van Wijnen A. J., Javed A., Gutierrez S. (2001) Contributions of nuclear architecture and chromatin to vitamin D-dependent transcriptional control of the rat osteocalcin gene. Steroids 66, 159–170 [DOI] [PubMed] [Google Scholar]
- 43. Gutierrez S., Javed A., Tennant D. K., van Rees M., Montecino M., Stein G. S., Stein J. L., Lian J. B. (2002) CCAAT/enhancer-binding proteins (C/EBP) β and δ activate osteocalcin gene transcription and synergize with Runx2 at the C/EBP element to regulate bone-specific expression. J. Biol. Chem. 277, 1316–1323 [DOI] [PubMed] [Google Scholar]
- 44. Marcellini S., Bruna C., Henríquez J. P., Albistur M., Reyes A. E., Barriga E. H., Henríquez B., Montecino M. (2010) Evolution of the interaction between Runx2 and VDR, two transcription factors involved in osteoblastogenesis. BMC Evol. Biol. 10.1186/1471-2148-10-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ji X., Li W., Song J., Wei L., Liu X. (2006) CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 34, W551–W554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Peng Y., Shi K., Wang L., Lu J., Li H., Pan S., Ma C. (2013) Characterization of Osterix protein stability and physiological role in osteoblast differentiation. PLoS One 8, e56451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Drissi H., Pouliot A., Koolloos C., Stein J. L., Lian J. B., Stein G. S., van Wijnen A. J. (2002) 1,25-(OH)2-vitamin D3 suppresses the bone-related Runx2/Cbfa1 gene promoter. Exp. Cell Res. 274, 323–333 [DOI] [PubMed] [Google Scholar]
- 48. Long F. (2012) Building strong bones: molecular regulation of the osteoblast lineage. Nat. Rev. Mol. Cell Biol. 13, 27–38 [DOI] [PubMed] [Google Scholar]
- 49. Li G., Ruan X., Auerbach R. K., Sandhu K. S., Zheng M., Wang P., Poh H. M., Goh Y., Lim J., Zhang J., Sim H. S., Peh S. Q., Mulawadi F. H., Ong C. T., Orlov Y. L., Hong S., Zhang Z., Landt S., Raha D., Euskirchen G., Wei C. L., Ge W., Wang H., Davis C., Fisher-Aylor K. I., Mortazavi A., Gerstein M., Gingeras T., Wold B., Sun Y., Fullwood M. J., Cheung E., Liu E., Sung W. K., Snyder M., Ruan Y. (2012) Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148, 84–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Handoko L., Xu H., Li G., Ngan C. Y., Chew E., Schnapp M., Lee C. W., Ye C., Ping J. L., Mulawadi F., Wong E., Sheng J., Zhang Y., Poh T., Chan C. S., Kunarso G., Shahab A., Bourque G., Cacheux-Rataboul V., Sung W. K., Ruan Y., Wei C. L. (2011) CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pike J. W., Meyer M. B., Watanuki M., Kim S., Zella L. A., Fretz J. A., Yamazaki M., Shevde N. K. (2007) Perspectives on mechanisms of gene regulation by 1,25-dihydroxyvitamin D3 and its receptor. J. Steroid Biochem. Mol. Biol. 103, 389–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zella L. A., Kim S., Shevde N. K., Pike J. W. (2006) Enhancers located within two introns of the vitamin D receptor gene mediate transcriptional autoregulation by 1,25-dihydroxyvitamin D3. Mol. Endocrinol. 20, 1231–1247 [DOI] [PubMed] [Google Scholar]
- 53. Zella L. A., Meyer M. B., Nerenz R. D., Lee S. M., Martowicz M. L., Pike J. W. (2010) Multifunctional enhancers regulate mouse and human vitamin D receptor gene transcription. Mol. Endocrinol. 24, 128–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lee S. M., Bishop K. A., Goellner J. J., O'Brien C. A., Pike J. W. (2014) Mouse and human BAC transgenes recapitulate tissue-specific expression of the vitamin D receptor in mice and rescue the VDR-null phenotype. Endocrinology 155, 10.1210/en.2014-1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang Y., Zhu J., DeLuca H. F. (2014) Identification of the vitamin D receptor in osteoblasts and chondrocytes but not osteoclasts in mouse bone. J. Bone Miner. Res. 29, 685–692 [DOI] [PubMed] [Google Scholar]
- 56. Sierra J., Villagra A., Paredes R., Cruzat F., Gutierrez S., Javed A., Arriagada G., Olate J., Imschenetzky M., Van Wijnen A. J., Lian J. B., Stein G. S., Stein J. L., Montecino M. (2003) Regulation of the bone-specific osteocalcin gene by p300 requires Runx2/Cbfa1 and the vitamin D3 receptor but not p300 intrinsic histone acetyltransferase activity. Mol. Cell. Biol. 23, 3339–3351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lian J. B., Javed A., Zaidi S. K., Lengner C., Montecino M., van Wijnen A. J., Stein J. L., Stein G. S. (2004) Regulatory controls for osteoblast growth and differentiation: role of Runx/Cbfa/AML factors. Crit. Rev. Eukaryot. Gene Expr. 14, 1–41 [PubMed] [Google Scholar]
- 58. Jensen E. D., Nair A. K., Westendorf J. J. (2007) Histone deacetylase co-repressor complex control of Runx2 and bone formation. Crit. Rev. Eukaryot. Gene Expr. 17, 187–196 [DOI] [PubMed] [Google Scholar]
- 59. Jensen E. D., Schroeder T. M., Bailey J., Gopalakrishnan R., Westendorf J. J. (2008) Histone deacetylase 7 associates with Runx2 and represses its activity during osteoblast maturation in a deacetylation-independent manner. J. Bone Miner. Res. 23, 361–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schroeder T. M., Kahler R. A., Li X., Westendorf J. J. (2004) Histone deacetylase 3 interacts with runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J. Biol. Chem. 279, 41998–42007 [DOI] [PubMed] [Google Scholar]
- 61. Schroeder T. M., Jensen E. D., Westendorf J. J. (2005) Runx2: a master organizer of gene transcription in developing and maturing osteoblasts. Birth Defects Res. C Embryo Today 75, 213–225 [DOI] [PubMed] [Google Scholar]
- 62. Jonason J. H., Xiao G., Zhang M., Xing L., Chen D. (2009) Post-translational regulation of Runx2 in bone and cartilage. J. Dent. Res. 88, 693–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Franceschi R. T., Xiao G., Jiang D., Gopalakrishnan R., Yang S., Reith E. (2003) Multiple signaling pathways converge on the Cbfa1/Runx2 transcription factor to regulate osteoblast differentiation. Connect. Tissue Res. 44, 109–116 [PMC free article] [PubMed] [Google Scholar]
- 64. Franceschi R. T., Xiao G. (2003) Regulation of the osteoblast-specific transcription factor, Runx2: responsiveness to multiple signal transduction pathways. J. Cell. Biochem. 88, 446–454 [DOI] [PubMed] [Google Scholar]
- 65. Steinberg X. P., Hepp M. I., Fernández García Y., Suganuma T., Swanson S. K., Washburn M., Workman J. L., Gutiérrez J. L. (2012) Human CCAAT/enhancer-binding protein β interacts with chromatin remodeling complexes of the imitation switch subfamily. Biochemistry 51, 952–962 [DOI] [PubMed] [Google Scholar]
- 66. Dallas S. L., Bonewald L. F. (2010) Dynamics of the transition from osteoblast to osteocyte. Ann. N.Y. Acad. Sci. 1192, 437–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Liu J. C., Lengner C. J., Gaur T., Lou Y., Hussain S., Jones M. D., Borodic B., Colby J. L., Steinman H. A., van Wijnen A. J., Stein J. L., Jones S. N., Stein G. S., Lian J. B. (2011) Runx2 protein expression utilizes the Runx2 P1 promoter to establish osteoprogenitor cell number for normal bone formation. J. Biol. Chem. 286, 30057–30070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nishio Y., Dong Y., Paris M., O'Keefe R. J., Schwarz E. M., Drissi H. (2006) Runx2-mediated regulation of the zinc finger Osterix/Sp7 gene. Gene 372, 62–70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.