Background: Photolyases are light-driven DNA repair enzymes harboring a catalytic FAD cofactor and usually an antenna chromophore.
Results: 8-Hydroxydeazaflavin is the cognate antenna of the Methanosarcina mazei photolyase, an archaeal representative of the clade of otherwise metazoan class II photolyases.
Conclusion: Phylogenetically, photolyases lost 8-hydroxydeazaflavin as antenna only in higher plants.
Significance: 8-Hydroxydeazaflavin occurs as cofactor within major parts of the metazoan phylome.
Keywords: Algae, Archaea, DNA Repair, Photobiology, Phylogenetics, X-ray Crystallography, Photolyase, Plant Evolution, UV Lesion, Light Harvesting
Abstract
Light-harvesting and resonance energy transfer to the catalytic FAD cofactor are key roles for the antenna chromophores of light-driven DNA photolyases, which remove UV-induced DNA lesions. So far, five chemically diverse chromophores have been described for several photolyases and related cryptochromes, but no correlation between phylogeny and used antenna has been found. Despite a common protein topology, structural analysis of the distantly related class II photolyase from the archaeon Methanosarcina mazei (MmCPDII) as well as plantal orthologues indicated several differences in terms of DNA and FAD binding and electron transfer pathways. For MmCPDII we identify 8-hydroxydeazaflavin (8-HDF) as cognate antenna by in vitro and in vivo reconstitution, whereas the higher plant class II photolyase from Arabidopsis thaliana fails to bind any of the known chromophores. According to the 1.9 Å structure of the MmCPDII·8-HDF complex, its antenna binding site differs from other members of the photolyase-cryptochrome superfamily by an antenna loop that changes its conformation by 12 Å upon 8-HDF binding. Additionally, so-called N- and C-motifs contribute as conserved elements to the binding of deprotonated 8-HDF and allow predicting 8-HDF binding for most of the class II photolyases in the whole phylome. The 8-HDF antenna is used throughout the viridiplantae ranging from green microalgae to bryophyta and pteridophyta, i.e. mosses and ferns, but interestingly not in higher plants. Overall, we suggest that 8-hydroxydeazaflavin is a crucial factor for the survival of most higher eukaryotes which depend on class II photolyases to struggle with the genotoxic effects of solar UV exposure.
Introduction
Besides photosynthetic reaction centers and protochlorophyllide reductases DNA photolyases belong to a few light-driven enzymes, which convert light energy into breakage of UV-B-induced bonds between neighboring pyrimidines (1). As highly efficient DNA repair enzymes, photolyases harvest blue/near-UV light photons for channeling excitation energy into the cleavage of non-oxidative photoproduct lesions in DNA. These lesions are caused by solar exposure of non-shielded DNA to UV-B (λ ∼ 280–315 nm) and UV-C (λ ∼ 100–280 nm) and consist for 10–20% of the pyrimidine (6-4)3 pyrimidone photoproduct, and for 80–90% of the cyclobutane pyrimidine dimer (CPD) lesion in its cis-syn isoform (2). Given the predominance and genotoxicity of the latter, most organisms confronted with sunlight, including archaea, bacteria, and eukaryotes, utilize CPD photolyases to maintain their genomic integrity. Only a few evolutionary later evolved organisms such as the placental mammals afford the loss of photolyase orthologues by relying on alternative DNA repair pathways for the maintenance of their genomic integrity (3, 4).
The catalytic mechanism of photolyases generally employs the photo-induced injection of an electron from a fully reduced and typically U-shaped flavin adenine dinucleotide cofactor (FADH−) onto the DNA lesion. This electron transfer triggers cleavage of CPD or (6-4) photoproducts inside duplex DNA (1, 5) within less than a nanosecond and achieves quantum yields close to one for CPD repair (6, 7). Spectroscopic and structural studies (8–10) showed that class I, class II, and (6-4) photolyases catalyze transient electron transfer to these DNA lesions by binding the lesions next to the FADH− cofactor in the catalytic α-helical C-terminal domain. The N-terminal domain of these photolyases adopts a Rossmann fold, which can accommodate additional prosthetic groups, so called antenna chromophores. Compared with other members of the photolyase-cryptochrome family, class II photolyases are evolutionary distant with pairwise sequence identities of <16%. Nevertheless, structures of class II photolyases from a methanogenic archaeon (8) as well as from a higher plant (11) indicate a similar bilobal architecture as class I and (6-4) photolyases and cryptochromes. Although class II photolyases catalyze like class I CPD photolyases and DASH cryptochromes light-driven DNA-repair and photoreduction of the catalytic FAD cofactor, they show marked differences for their active site structures, the mode of DNA binding, the electron transfer pathways required for photoreduction as well as for the discrimination between intact and UV lesion-containing DNA.
A shortcoming of the catalytic FADH− cofactor is its poor absorption in the blue to near-UV range with extinction coefficients between 2800 m−1cm−1 (400 nm) and 5600 m −1cm−1 (360 nm). For efficient light-driven repair in the blue/near-UV region, photolyases hence rely on a second auxiliary chromophore with large absorption cross-sections to broaden up their limited spectral properties. So far, five different classes of antenna chromophores have been identified from members of the photolyase-cryptochrome superfamily (Fig. 1). These antenna chromophores comprise aromatic moieties absorbing in the range between 380 and 420 nm and include (i) 5,10-methenyltetrahydrofolate (MTHF) as found in many microbial class I photolyases (12) and DASH cryptochromes (13); (ii) 8-hydroxydeazaflavin (8-HDF) from (6-4) photolyases (14, 15) and several class I photolyases (16); (iii and iv) flavin mononucleotide (FMN) and FAD in the class I photolyases from Thermus thermophilus (17) and Sulfolobus tokodaii (18), respectively; and finally (v) 6,7-dimethyl-8-ribityl-lumazin in a novel class of proteo-/cyanobacterial cryptochromes (19). For light harvesting the antenna chromophores absorb a photon and transfer its energy via a Förster mechanism to the catalytic active FADH−. All nucleotide-like chromophores (FAD, FMN, and 8-HDF) are bound within the N-terminal α/β domain of photolyases with its Rossmann-like fold in a distance of 17–18 Å to the catalytic FAD cofactor. In contrast to the deeply buried nucleotide-like antenna, the pterin derivative MTHF is bound close to the protein surface along the interface of the N- and C-terminal domains (20).
FIGURE 1.
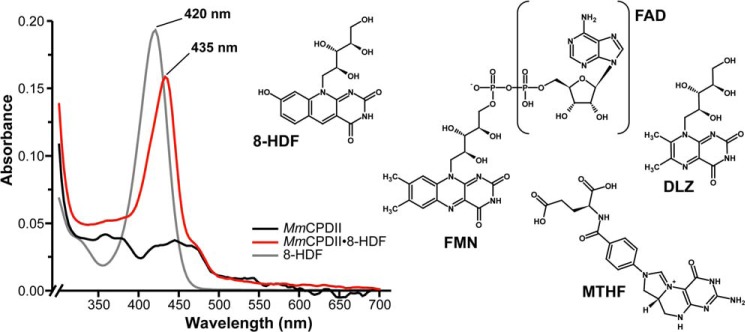
In vitro reconstitution of apoMmCPDII with synthetic 8-HDF and overview of known antenna chromophores of members of the photolyase-cryptochrome family. Binding of 8-HDF to the M. mazei class II photolyase (red curve) causes a red shift by 15 nm compared with free 8-HDF at pH 8 (gray curve), where free 8-HDF is deprotonated at its 8-OH group (pKa 6.3) like MmCPDII-bound deazaflavin.
Like MTHF, the riboflavin derivatives FAD, FMN, as well as the riboflavin biosynthesis intermediate 6,7-dimethyl-8-ribityl-lumazin are commonly found in archaea, bacteria, and eukaryotes, whereas the occurrence of deazaflavin cofactors such as 8-HDF is more limited. For example, deazaflavins are signature molecules of methanogenic archaea, where 8-HDF in its oligoglutamylated and protonated F420 form plays an important role as a low potential hydride carrier (21). Nevertheless, deazaflavins are also found in several other bacterial clades such as the actinobacteria (22) as well as in unicellular green algae. The biological role of 8-HDF in photolyases was demonstrated in the green algae Chlamydomonas reinhardtii. Here, inactivation of the gene for the deazaflavin synthase PHR1 caused a loss of the photorepair of UV lesions as catalyzed in C. reinhardtii by the class II photolyase PHR2 (23). Interestingly, the (6-4) and class II CPD photolyases from the insect Drosophila melanogaster utilize 8-HDF as antenna chromophore as well (15), although this organism misses like other animals a genomic copy of the deazaflavin synthase gene. Instead, the bacterial symbiont Wolbachia was suggested to supply 8-HDF as a vitamin essential for photolyase function (14).
Here, we investigate the antenna chromophore of the class II CPD photolyase from Methanosarcina mazei (MmCPDII) and the spectroscopic and structural characterization of its complex with the 8-HDF antenna. Together with phylogenetic data the structure of the MmCPDII·8-HDF complex and its differences from higher plant photolyases allow us to predict the occurrence of 8-HDF in the whole metazoan branch of life.
EXPERIMENTAL PROCEDURES
Cloning of Streptomyces coelicolor FO Synthase
The gene SCO4429 (fbiC) encoding the FO synthase (7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase) of Streptomyces coelicolor (UniProt entry Q9KZZ7) was amplified from genomic DNA using the Phusion® DNA polymerase (Finnzymes) with primers 5′-GCATTGAATTCGATGACGACTTCCGCGACCTCC-3′ and 5′-GATATTCTCGAGTTAGTCCAGGACCGGCAGCAG-3′ (Metabion). The PCR product was cloned into pCR®2.1-TOPO® vector (Invitrogen) and subcloned via EcoRI and XhoI sites (underlined) into pCDFDuet-1 (Novagen) thus yielding the cofactor plasmid pCDFDuet-His6ScFbiC, which encodes an N-terminally His6-tagged ScFbiC fusion.
Cloning of Photolyases for Co-expression Studies
The gene Synpcc7942_0112 encoding the class I CPD photolyase (UniProt entry Q31S25) of Synechococcus elongatus PCC 7942 (Anacystis nidulans R2) was amplified from isolated genomic DNA with primers 5′-GGTTTGCATATGGCGGCTCCGATTCTGTTTTGG-3′ and 5′-CCAAACTCGAGCTATGAATCGGGCTCAGCCTC-3′, thus introducing restriction sites for NdeI and XhoI (underlined). Accordingly, the PCR product was cloned into expression vector pET-28a (Novagen) to give the plasmid pET28a-His6AnCPDI. The latter encodes N-terminally His6-tagged AnCPDI. Likewise, the gene fragment (bases 1–1560) coding for the (6-4) photolyase domain from D. melanogaster (UniProt entry Q0E8P0) was amplified from the pDEST007 plasmid (9) with primers 5′-GCTTCGCATATGGATTCACAAAGGTCCACG-3′ and 5′-GCCCGTCTCGAGTCAGGTTTCTGATTTCTC-3′ and subcloned via NdeI and XhoI sites into pET-28a to produce pET-28a-His6Dm(6-4). The class I CPD photolyase from T. thermophilus HB8 (UniProt entry P61497) was amplified from genomic DNA with primers 5′-GGAATTCCATATGGGCCCCCTTCTCGTC-3′ and 5′-CGCGGATCCTACCCTCGGGCGAGATCC-3′ (Invitrogen) and subcloned via NdeI and BamHI into pET-28a to yield pET-28aHis6TtCPDI.
Co-expression and Purification of Photolyases (PHR)
Escherichia coli BL21 (DE3) Gold cells (Stratagene) were transformed with either pET28a-His6MmCPDII (8), pET28a-His6AnCPDI, pET28a-His6Dm(6-4), pET28a-His6TtCPDI, or pET28a-His6AtCPDII and the cofactor plasmid pCDFDuet-His6ScFbiC. Autoinduction-triggered co-expression was performed in terrific broth medium at 25 °C (MmCPDII, TtCPDI, AtCPDII), 21 °C (AnCPDI), and 18 °C (Dm(6-4)), respectively. Apart from AtCPDII the putative photolyase·8-HDF complexes were purified according to the protocol of apoMmCPDII apart that cell disruption was performed with a French press. For the purification of AtCPDII buffer ATI (50 mm NaH2PO4, 500 mm NaCl, 20% (v/v) glycerol, pH 7.4) was used for affinity chromatography, and elution was performed by addition of 75 mm imidazole. Size exclusion chromatography was done in buffer ATII (20 mm Tris-HCl, 300 mm NaCl, 10% (v/v) glycerol, pH 7.4). The His6-tagged FO synthase was efficiently removed by the size exclusion chromatography step.
Generation and Preparation of MmCPDII Mutants
Photolyase mutants MmCPDII-S26L and MmCPDII-S26F were obtained from pET28a-MmCPDII by site-directed mutagenesis (QuikChange II; Agilent); resulting plasmids were verified by dideoxy-sequencing (Qiagen). Co-expression and purification of the Ser26 mutants were performed as described for the wild type photolyase.
In Vitro Reconstitution of ApoMmCPDII with Antenna Chromophores
ApoMmCPDII (53 μm) was mixed with the equimolar amount of either FAD, FMN, MTHF, F420, or chemically synthesized 8-HDF. Afterward, all mixtures as well as negative controls were dialyzed at 4 °C against 1.5 ml of buffer I (10 mm Tris-HCl, 100 mm NaCl, pH 8.0) in a pre-greased VDXTM plate (Hampton Research) using 10-μl microdialysis rods (Hampton Research) and a 3.5-kDa cutoff SnakeSkin® dialysis membranes (Pierce). After 24 h, reconstitution was analyzed by UV/visible spectroscopy using a NanoDrop 1000 spectrophotometer (Thermo Scientific). Likewise, the 8-HDF reference spectrum was recorded with the NanoDrop 1000 spectrophotometer and normalized to the MmCPDII concentration used before.
Structure Determination of ApoMmCPDII Crystals Soaked with 8-HDF
MmCPDII crystals grown in 0.5 m lithium sulfate and 7.5% PEG 8000 were soaked for 30 min in crystallization buffer supplemented with 100 μm synthetic 8-HDF. During soaking changes in the integrity of MmCPDII crystals were observed and are most likely reflected by the lower resolution limit of 2.7 Å compared with apoMmCPDII crystals (1.5 Å). Before flash-freezing in liquid nitrogen crystals were soaked in the crystallization solution supplemented with 30% glycerol as cryoprotectant. Diffraction data were collected from a single crystal at 100 K at beamline ID23-2, European Synchrotron Radiation Facility (ESRF). Data processing was carried out using XDS and XSCALE (24) and further refinement using COOT (25) and REFMAC5 (26) at 2.7 Å resolution led to R-factors of Rwork = 20.6% and Rfree = 27.0%. Data processing and refinement statistics are summarized in Table 1.
TABLE 1.
Crystallographic statistics of the MmCPDII·8-HDF complexes
Values in parentheses denote the highest resolution shell.
| MmCPDII·8-HDFsoak (4CDM) | MmCPDII·8-HDF (4CDN) | |
|---|---|---|
| Data collection | ||
| X-ray source | ID23-2 | ID23-1 |
| ESRF, Grenoble, France | ||
| Detector | MarCCD 225 | ADSC Q315r |
| Wavelength (Å) | 0.87260 | 0.97625 |
| Space group | P43212 | P212121 |
| Cell dimensions (a,b,c Å) | 69.89, 69.89, 245.5 | 78.97, 114.7, 141.4 |
| Resolution (Å) | 30.0-2.70 (2.85-2.70) | 35.0-1.90 (2.00-1.90) |
| Total reflections | 64,763 | 407,726 |
| Multiplicity | 4.2 (2.9) | 4.1 (4.1) |
| Unique reflections | 15,494 | 99,823 |
| Completeness (%) | 89.3 (84.9) | 98.5 (98.7) |
| Rmerge (%) | 9.0 (32.0) | 4.5 (43.3) |
| I/σ(I) | 14.7 (4.5) | 23.3 (3.2) |
| Mosaicity (°) | 0.166 | 0.175 |
| Wilson B-factor (Å2) | 38.3 | 25.6 |
| Refinement | ||
| Resolution (Å) | 29.2-2.70 | 34.5-1.90 |
| Rfactor, Rfree | 0.206, 0.270 | 0.149, 0.178 |
| Reflections (working, test set) | 15,081, 216 | 97726, 2,022 |
| Completeness for range (%) | 88.1 | 98.1 |
| r.m.s.d.a from ideal: | ||
| Bond lengths (Å) | 0.010 | 0.009 |
| Bond angles (°) | 1.278 | 1.354 |
| Total number of atoms | 3,826 | 8,267 |
| Mean B-value (Å2) | 24.6 | 30.1 |
a r.m.s.d., root mean square deviation.
Crystallization and Structure Determination of the MmCPDII·8-HDF Complex
An initial 96-well format crystallization screening was performed with a Cartesian robot system and commercially available crystallization screens (Qiagen) using the sitting drop vapor diffusion method at 18 °C. First crystals were obtained in a crystallization condition containing 0.1 m lithium sulfate, 0.1 m trisodium citrate, pH 5.6, and 12% PEG 6000 within 48 h. Using the hanging drop vapor diffusion method and this condition, crystals suitable for data collection were grown in a 24-well plate at a protein concentration of 11.8 mg/ml. For cryoprotection, glycerol was added to a final concentration of 33%, and MmCPDII·8-HDF crystals were flash-frozen in liquid nitrogen. X-ray data were collected from a single crystal at 100 K at beamline ID23-1, ESRF. MmCPDII·8-HDF crystallized in the orthorhombic space group P212121 with unit cell parameters a = 78.97 Å, b = 114.7 Å, c = 141.4 Å. Diffraction data were processed using XDS and XSCALE (24), and initial phases were obtained by molecular replacement using PHASER (27) and the structure of MmCPDII in a tetragonal crystal form (2XRY) as search model. The correct solution with data up to 2.5 Å had two molecules per asymmetric unit with z scores of 25.7 for rotation and 45.9 for translation function for the first molecule and z scores of 26.2 and 79.7 for the second molecule, respectively. After initial automated model building from experimental phases as done by ARP/wARP (28), further refinement using COOT (25) and REFMAC5 (26) at 1.9 Å resolution led to R-factors of Rwork = 14.9% and Rfree = 17.8%. Data processing and refinement statistics are summarized in Table 1. The nomenclature for the secondary structure of MmCPDII is taken from Kiontke et al. (8). Structural analysis and figures were done with PyMOL 1.5 (29).
UV/Visible and Fluorescence Spectroscopy
UV/visible spectra were recorded using a V-660 spectrometer (JASCO) at 8 °C. Fluorescence spectroscopy of MmCPDII in the oxidized state was carried out at 4 °C on a FP-6500 spectrofluorometer (Jasco). Samples of apoMmCPDII and MmCPDII·8-HDF with concentrations of 10 μm or 5.8 μm in buffer I were excited at a wavelength of 420 nm with band widths of 3 nm for excitation and 5 nm for emission, respectively. Reference fluorescence emission spectra were recorded with a high power LED (Roithner Lasertechnik), Maya 2000 Pro spectrometer (Ocean Optics) and SpectraSuite software (Ocean Optics). To generate the reduced state, samples of 16 μm MmCPDII in 10 mm DTT were deoxygenized before being photoreduced on ice by illumination at 450 °nm for 10 min using a 450 °nm high power LED (Roithner Lasertechnik, 0.24 W/cm2). Fluorescence emission/excitation spectra were recorded at 4 °C on a Fluorolog-3 (Horiba) using slit widths of 2 nm.
RESULTS AND DISCUSSION
In Vitro Reconstitution of MmCPDII with Synthetic 8-HDF
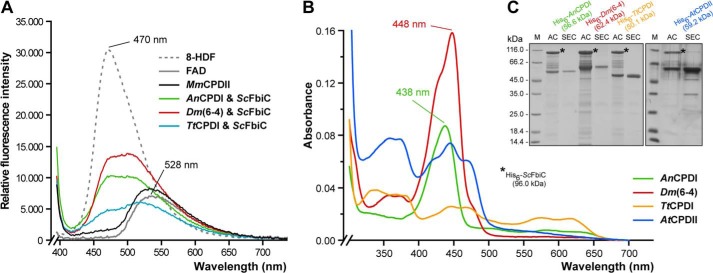
When purified via the E. coli expression system, recombinant class II photolyase from M. mazei (MmCPDII) possesses the catalytic FAD cofactor but lacks any bound and host-derived antenna chromophore as shown before by UV/visible spectroscopy and structural analysis (8). To identify the cognate light-harvesting prosthetic group of MmCPDII, we performed in vitro reconstitution of this so called apoMmCPDII with known antenna chromophores such as FAD, FMN, MTHF, 8-HDF, and F420. Only for synthetic 8-HDF, the UV/visible spectroscopic analysis reveals a characteristically modified absorbance spectrum of the M. mazei class II photolyase. The absorbance spectrum of oxidized apoMmCPDII with peaks at 362, 377, 421, 444, and 469 nm is dominated in the reconstituted MmCPDII·8-HDF complex by a significant peak at 435 nm (Fig. 1). Furthermore, incorporation of deprotonated 8-HDF (λmax 420 nm) into MmCPDII causes a 15-nm red shift of the chromophore absorbance. This red shift of bound 8-HDF complies well with data reported for D. melanogaster (6-4) photolyase (14) and A. nidulans class I photolyase (AnCPDI), the latter either purified directly from A. nidulans cells (16) or in vitro reconstituted with 8-HDF using recombinant enzyme (30, 31).
In Vivo Assembly of the MmCPDII·8-HDF Holoenzyme
To confirm these results and to rule out that incorporation of synthetic 8-HDF during in vitro reconstitution and crystal soaking is enforced by the chosen experimental conditions, an in vivo reconstitution system of MmCPDII with endogenously produced 8-HDF was established. For this purpose, a cofactor plasmid encoding the S. coelicolor FO synthase (ScFbiC), a bifunctional, S-adenosylmethionine-dependent enzyme (26), was constructed that enables the biosynthesis of 8-HDF in E. coli from 4-hydroxypyruvate and 5-amino-6-ribityl-amino-2,4(1H,3H)-pyrimidinedione, an intermediate of the riboflavin synthesis pathway (Fig. 2A). After nickel-nitrilotriacetic acid affinity chromatography, the MmCPDII·8-HDF complex was separated from ScFbiC by size exclusion chromatography (Fig. 2B). Like in vitro reconstituted MmCPDII, the UV/visible spectrum of the MmCPDII·8-HDF holoenzyme in the oxidized state is hallmarked by the distinct absorption maximum at 435 nm (Fig. 2C, black line) indicating that 8-HDF is bound to the photolyase. Moreover, the fluorescence emission spectrum of the complex exhibits a strong fluorescence peak at λem 467 nm (Fig. 2C, thick red line) and differs significantly from apoMmCPDII with the latter exhibiting only broader emission at λem of approximately 528 nm for the oxidized FAD cofactor (Fig. 2C, dotted red line). In accordance with data from AnCPDI (16, 31) and Dm(6-4) (14), the fluorescence maximum of MmCPDII-bound 8-hydroxydeazaflavin resembles the maximum of deprotonated 8-HDF in solution with its λem of 470 nm (Fig. 3A). In contrast, the fully reduced FADH− state exhibits an emission peak for the 8-HDF antenna that is blue-shifted by 12 nm to 455 nm, thus suggesting some interaction between the redox state of the FAD cofactor and the structure of the antenna binding site (Fig. 2D). Furthermore, excitation spectra recorded at λem = 533 nm, the emission maximum of the reduced FADH− chromophore, demonstrate efficient Förster energy transfer between the antenna and the flavin cofactor, as the resulting spectra are clearly dominated by the 8-HDF antenna with its λmax of 435 nm (Fig. 2D).
FIGURE 2.
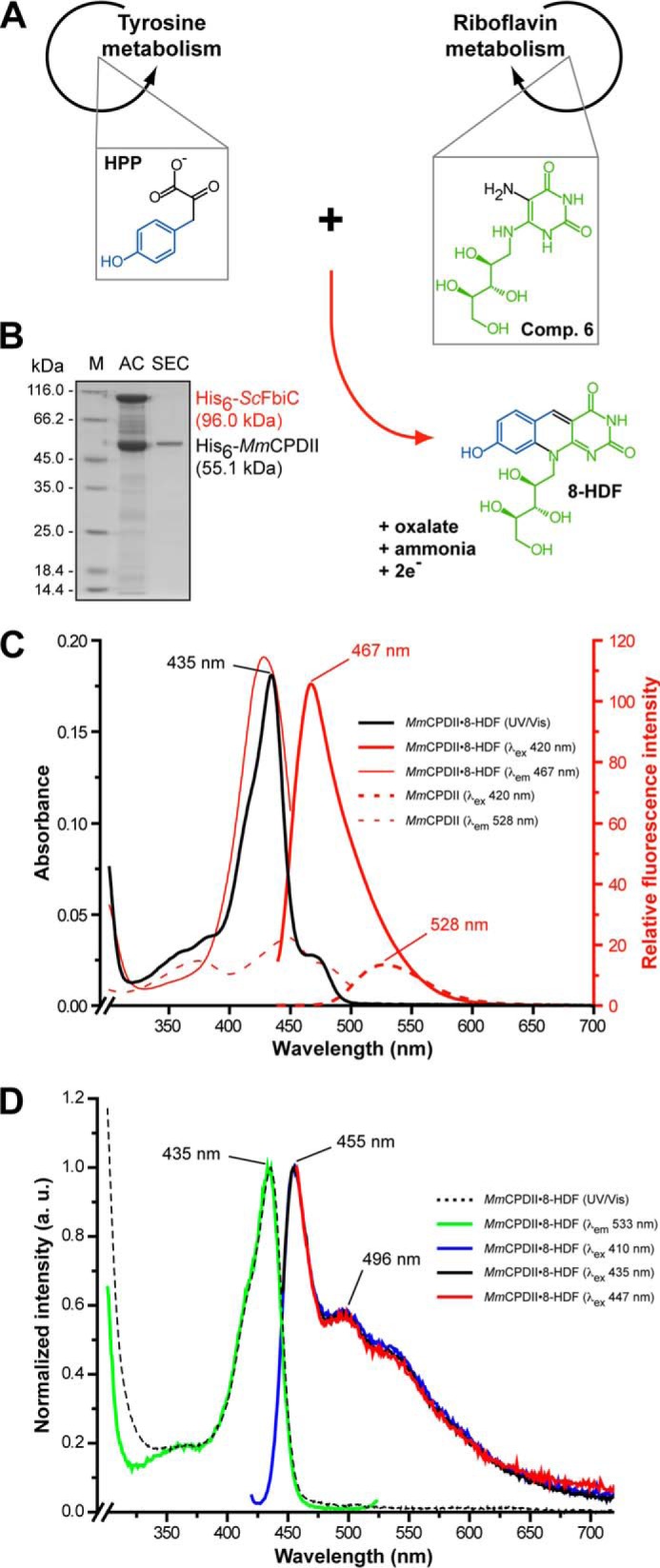
In vivo reconstitution of MmCPDII with an 8-HDF antenna prosthetic group. A, the heterologous synthesis of 8-HDF in E. coli as catalyzed by the S. coelicolor FO synthase (ScFbiC) starting from endogenous metabolites (4-hydroxyphenylpyruvate, compound 6: 5-amino-6-ribityl-amino-2,4(1H,3H)-pyrimidinedione). The reaction mechanism is according to Graham et al. (32). B, SDS-PAGE analysis showing the purification of the MmCPDII·8-HDF complex. C, spectral properties of MmCPDII and its complex with 8-HDF in the oxidized state. The UV/visible spectrum of the MmCPDII·8-HDF complex (black line) features the identical absorbance maximum as the in vitro reconstituted photolyase. Unlike apoMmCPDII (λex 420 nm, dotted red line), the MmCPDII·8-HDF complex features a prominent fluorescence emission maximum at 467 nm (λex 420 nm, solid red line). The λmax of 428 nm for the excitation spectrum of the MmCPDII·8-HDF complex (emission measured at 467 nm, thin red line) indicates a bathochromic shift by about 8 nm for the absorption of the antenna chromophore due to its interactions with the photolyase. D, spectral properties of MmCPDII and its complex with 8-HDF in the fully reduced state. The fluorescence excitation spectrum at 533 nm emission corresponds to 90% of the intensity when recorded at the emission maximum of the 8-HDF antenna of 455 nm.
FIGURE 3.
In vivo reconstitution of different photolyases with 8-HDF. A, fluorescence emission spectra of cofactor extracts from E. coli strains, which express different heterologous photolyases, as obtained by heat denaturation of endogenous proteins (λex 385 nm). For comparison, MmCPDII has been expressed in E. coli without a ScFbiC transgene and shows accordingly only endogenous FAD fluorescence. Reference spectra for FAD (solid) and 8-HDF (dotted) are shown in gray. 3-fold averaged emission spectra were smoothed over three points. B, UV/visible spectra of purified A. nidulans class I CPD photolyase (AnCPDI, green line) and D. melanogaster (6-4) photolyase (Dm(6-4), red line) showing absorbance maxima at 438 nm or 448 nm, respectively, which indicate bound 8-HDF, whereas the class II CPD photolyase from A. thaliana (AtCPDII, blue line) fails to bind 8-HDF as antenna chromophore. Unlike previous results relying solely on crystal soaking (10), the T. thermophilus class I CPD photolyase (TtCPDI, orange line) fails to incorporate 8-HDF. C, SDS-PAGE analysis of photolyase/ScFbiC co-expressions. The asterisks mark the bands for the recombinant, co-expressed His6-ScFbiC. M, protein marker; AC, nickel-nitrilotriacetic acid affinity chromatography; SEC, size exclusion chromatography.
To clear up the question, whether Methanosarcinales are generally capable of synthesizing 8-HDF as cognate antenna of their photolyases, we performed a protein-protein BLAST search with Methanocaldococcus jannaschii CofG and CofH, which have been used before to produce artificial 8-HDF in E. coli (32). Indeed, M. mazei harbors like other methanosarcinal species, e.g. Methanosarcina acetivorans or Methanosarcina barkeri, one CofG and two CofH orthologues (SwissProt entries COFG_METMA, COFH1_METMA, COFH2_METMA).
Validation of the Heterologous in Vivo 8-HDF Reconstitution System
To rule out that our in vivo reconstitution system generates noncognate antenna-photolyase hybrids, class I and (6-4) photolyases were co-expressed with the ScFbiC FO synthase. AnCPDI was chosen as reliable positive control for successful incorporation of the 8-HDF antenna chromophore, because AnCPDI that is directly purified from A. nidulans cells harbors 8-HDF as cognate antenna (16). This cyanobacterium carries genomic copies of the 8-HDF synthase genes cofG and cofH (UniProt entries Q5N4D1, Q5N3T7), on which biosynthesis of the 8-HDF chromophore depends. For comparison, the (6-4) photolyase from D. melanogaster as well as the class I CPD photolyase from T. thermophilus (TtCPDI) were used, as the corresponding organisms lacking CofG/CofH orthologues. Here, 8-HDF has been successfully incorporated by either in vitro reconstitution of Ds(6-4) (14) or crystal soaking of TtCPDI (33).
After purification, all putative recombinant photolyase complexes were analyzed by UV/visible spectroscopy to analyze for in vivo reconstitution with endogenously produced 8-HDF. In case of AnCPDI and Dm(6-4) the UV/visible spectra exhibit prominent absorbance maxima at approximately 440–450 nm (Fig. 3B). Whereas the absorbance maximum of the AnCPDI·8-HDF complex at λmax 438 nm corresponds closely to spectra already published (16, 30, 31), the maximum at λmax 448 nm for the in vivo reconstituted Dm(6-4)·8-HDF complex is red-shifted by 8 nm compared with the in vitro reconstituted photolyase with a λmax of 440 nm (14). Interestingly, in vivo reconstitution with 8-HDF failed for TtCPDI, although the FO synthase was solubly expressed (Fig. 3, A–C). Accordingly, the co-crystal structure of TtCPDI with synthetic 8-HDF, which was generated by soaking TtCPDI crystals with millimolar concentrations of 8-HDF (33), corresponds to a noncognate complex of this photolyase. However, the lack of endogenous 8-HDF biosynthesis pathways is not per se an indicator for 8-HDF playing no role as a cognate antenna chromophore of the respective photolyase. The fruit fly D. melanogaster lacks the genes required for 8-HDF biosynthesis, but its (6-4) and CPD photolyases are nevertheless capable of accommodating 8-HDF as antenna chromophore (15). This insect species may derive 8-HDF from a bacterial, hitherto unknown commensal as suggested before (14).
Structures of the MmCPDII·8-HDF Holoenzyme
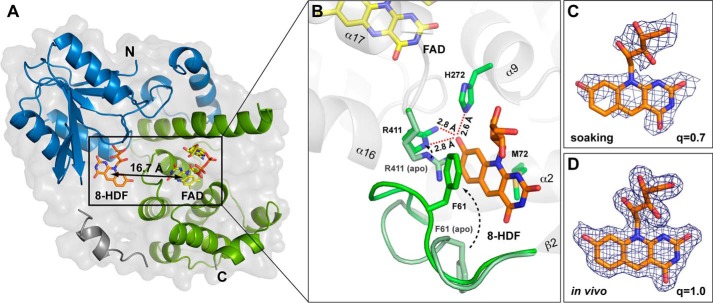
To characterize 8-HDF antenna binding by MmCPDII we derived co-crystal structures either by crystallization of in vivo reconstituted MmCPDII·8-HDF holoenzyme at 1.9 Å resolution or by soaking apoMmCPDII crystals with 8-HDF and subsequent structural analysis at 2.7 Å resolution. Interestingly, the holoenzyme crystallized in a novel, orthorhombic crystal form with two molecules per asymmetric symmetry unit (Table 1). Apart from differences in the linker region between the N- and C-terminal domain (Val186-Glu231) and the antenna binding loop there are almost no deviations between the MmCPDII·8-HDF and the apoMmCPDII structures. Furthermore, the root mean square deviations for complexes A and B of the MmCPDII·8-HDF holoenzyme are low with 0.20 and 0.28 Å (330, 353 Cα atoms), respectively, relative to the apoMmCPDII structure. In both reconstitutions, the 8-HDF chromophore is buried deeply within the N-terminal domain in a distance of 16.7 Å to the catalytic FAD cofactor, which is well in range for efficient Förster energy transfer (Fig. 4, A and B). According to difference electron density the in vivo reconstituted MmCPDII·8-HDF holoenzyme shows quantitative incorporation of the 8-HDF chromophore, whereas soaking achieved under our conditions only 70% occupancy (Fig. 4, C and D). The inhomogeneity of the latter is reflected by the alternative conformations taken up by parts of the antenna loop (Leu57-Ala64) that links β-strand β2 with helix α2. Here, the short helical turn (Glu60-Glu63) that is found in 8-HDF-free MmCPDII structures (PDB codes 2XRY and 2XRZ (8)) undergoes a conformational change by closing the entrance to the 8-HDF binding site. The largest change within the antenna loop is made by Phe61-Leu62, which swivel their packed side chains by a distance of over 12 Å to form π-π-interaction between Phe61 and the middle ring of the deazaflavin (Fig. 4B).
FIGURE 4.
Structural analysis of the MmCPDII·8-HDF holoenzyme. A, cut-away view of the overall structure. The antenna chromophore 8-HDF is bound within the N-terminal α/β domain (blue) in a distance suitable for Förster energy transfer to he catalytic cofactor FAD. B, structural details of the antenna chromophore binding pocket. The binding of 8-HDF causes conformational changes in the chromophore environment between apoMmCPDII (pale green) and MmCPDII·8-HDF holoenzyme (green). Comparison of the 8-HDF occupancies (q) resulting from either crystal soaking (C) or in vivo reconstitution in E. coli (D) is shown. Sigma-A weighted 2Fobs − Fcalc electron densities are contoured at 1σ.
The in vivo reconstituted MmCPDII·8-HDF holoenzyme shows even larger conformational changes compared with the apo-structures of MmCPDII. The rearrangement of the antenna loop involves the region between Asp59 and Ala64 and seals the 8-HDF binding site from bulk solvent access. Furthermore, major parts of the linker between the N- and C-terminal domains of this photolyase (Val186-Glu231) either adopt a conformation different from in 8-HDF lacking MmCPDII structures or are disordered as in complex B (Glu189-Leu217). In complex A, helix α7 (Leu200-Glu214), which is structurally also conserved in the class II photolyase from rice (Oryza sativa, PDB code 3UMV, Asp221-Glu232 (11)), breaks down into three shorter helical segments (α7A, Leu200-Glu205; α7B, Val208-Leu211; α7C, Glu214-Lys219). The newly formed α7C segment hereby stabilizes the closed conformation of the antenna loop by several, newly formed van der Waal interactions. As a result Asp222 of the linker that is exposed before as part of a loop becomes buried and forms a salt bridge with Arg67. Overall, these structural adaptations of the second shell around the antenna chromophore binding site most likely impede reopening and hence release of bound 8-HDF.
The Antenna Binding Site of MmCPDII
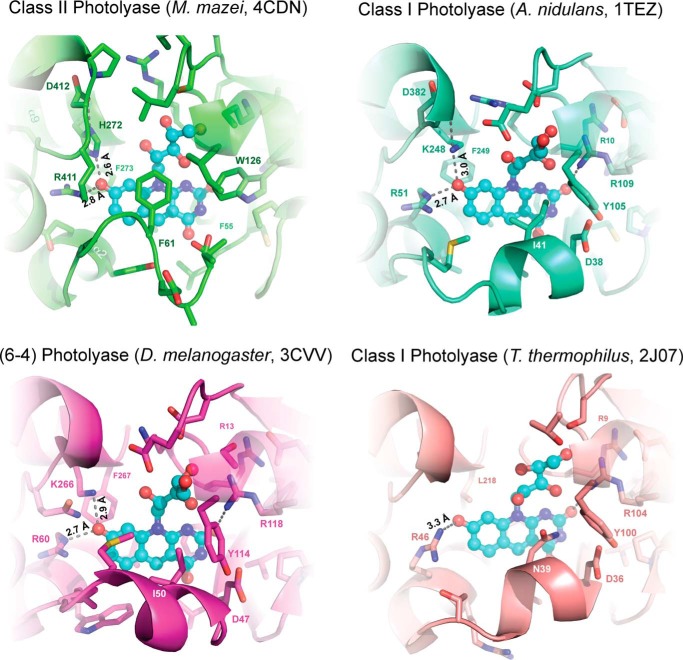
Although the 8-HDF chromophore locates in the same region of the N-terminal domain of MmCPDII as in the cognate 8-HDF complexes of class I and (6-4) photolyases (Fig. 5), there are several peculiar differences for its binding site. First, the re-side of the aromatic ring system of 8-HDF, i.e. in Fig. 5 the backside, makes extensive van der Waal interactions with the bulky side chains of Phe273 and Phe55 as well as with Met72 via its middle ring. In other photolyases, the latter residue is exchanged by a leucine, whereas the walling by phenylalanines is conserved. More important, the si-side of the chromophore aromatic system is covered by Thr58 and Phe61 from the nonregular antenna loop, the indole of Trp126 as well as the side chain of Arg411. In other photolyases, the antenna loop is replaced by a short helical segment, which points only with a leucine toward the middle ring of the 8-HDF chromophore. Second, Trp126 replaces in class II photolyases an otherwise highly conserved arginine residue (AnCPDI, Arg109; Ds(6-4), Arg118), which forms in class I and (6-4) photolyases an H-bond to the C2-carbonyl of the deazaflavin moiety. In MmCPDII, this role is taken by Ser26, which forms hydrogen-bonds to the C2-carbonyl as well as the 3′-hydroxy group of the ribityl moiety (Fig. 5). Finally, two basic residues surround the phenolic ring of the 8-hydroxydeazaflavin to stabilize its deprotonated, anionic state. Only one of these residues, His272, is similar in class I and 6-4 photolyases (AnCPDI, Lys248; Ds(6-4), Lys266), where it forms a salt bridge to the 8-oxy group of 8-HDF. The conformation of this histidine is stabilized by a salt bridge to Asp412 from the C-terminal domain of MmCPDII (Fig. 5). Interestingly, its preceding residue, Arg411, makes the second salt bridge to the 8-oxy group. This residue has no counterpart in class I and (6-4) photolyases and is replaced in the latter by an arginine from the N-terminal domain (AnCPDI, Arg51; Ds(6-4), Arg60).
FIGURE 5.
Cognate and noncognate 8-HDF binding sites in the N-terminal antenna domain of DNA photolyases. For clarity, the 8-HDF chromophore is shown as a ball-and-stick model.
Given other structural differences, which are crucial for function, e.g. the distinct electron-transfer pathways for photoreduction or the binding sites of the catalytic FAD chromophore (8), these observations corroborate the notion of a large evolutionary gap between class II photolyases and other members of the photolyase-cryptochrome superfamily. For example, class I photolyases that lack 8-HDF as cognate antenna such as TtCPDI, miss all of the ascribed motifs of the 8-HDF binding site (Fig. 5).
Conservation of the 8-HDF Antenna Chromophore within Class II Photolyases
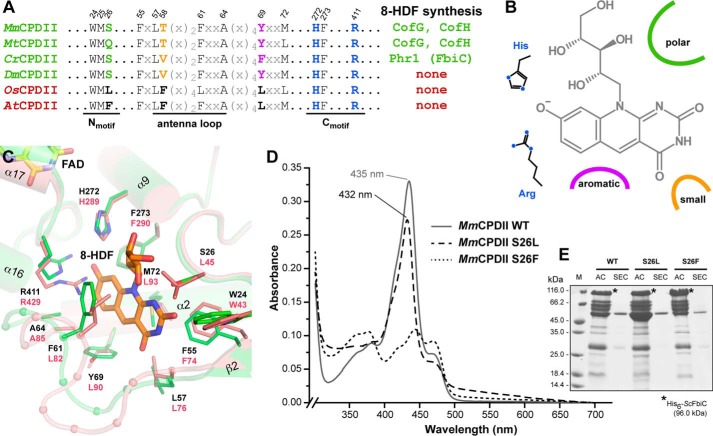
Apart from the structural differences described above, a comparison of the MmCPDII·8-HDF complex and the recently published O. sativa class II CPD photolyase (OsCPDII) shows no other major deviations. Nevertheless, heterologously expressed O. sativa photolyase lacks any kind of an additional antenna chromophore as shown previously by UV/visible spectroscopy (34) as well as by current structural characterization (11). Likewise, the heterologously overexpressed class II photolyases from Arabidopsis thaliana, whose sequence identity to OsCPDII exceeds 65%, fails to incorporate under our in vivo conditions any 8-HDF chromophore (Fig. 3, B and C). A superimposition of the OsCPDII structure with the MmCPDII·8-HDF complex (root mean square deviation 0.60 Å for 338 Cα atoms) shows that although most amino acids lining the antenna binding pocket are either conserved (OsCPDII: Trp43, Leu76, Ala85, His289, Phe290, Arg429) or have at least similar biochemical properties (see Fig. 6C). However, there are several peculiar differences correlating with the loss of 8-HDF binding. First, the “lower” part of the pocket, where the aromatic ring moiety of 8-HDF is accommodated, lacks in OsCPDII the aromatic residues from the antenna loop that are responsible for π-π stacking from the si-side (MmCPDII, Phe61; OsCPDII, Leu82) as well as the edge-to-face interaction with the Cys6 of the deazaflavin (MmCPDII, Tyr69; OsCPDII, Leu90 (35)). Instead, the binding pocket is partly filled in OsCPDII by the bulky side chain of Phe77 (MmCPDII: Thr58) from the antenna loop. Second, in OsCPDII a leucine (Leu45) replaces the polar residue Ser26 of MmCPDII, which is crucial for the recognition of 8-HDF via the H-bonds to the C2-carbonyl and 3′-hydroxy group. In contrast, the basic residues His272 and Arg411 of the C-terminal catalytic domain, which stabilize the deprotonation of the 8-hydroxy group, are preserved in the higher plant photolyases like AtCPDII or OsCPDII and are hence only weak indicators for antenna chromophore binding.
FIGURE 6.
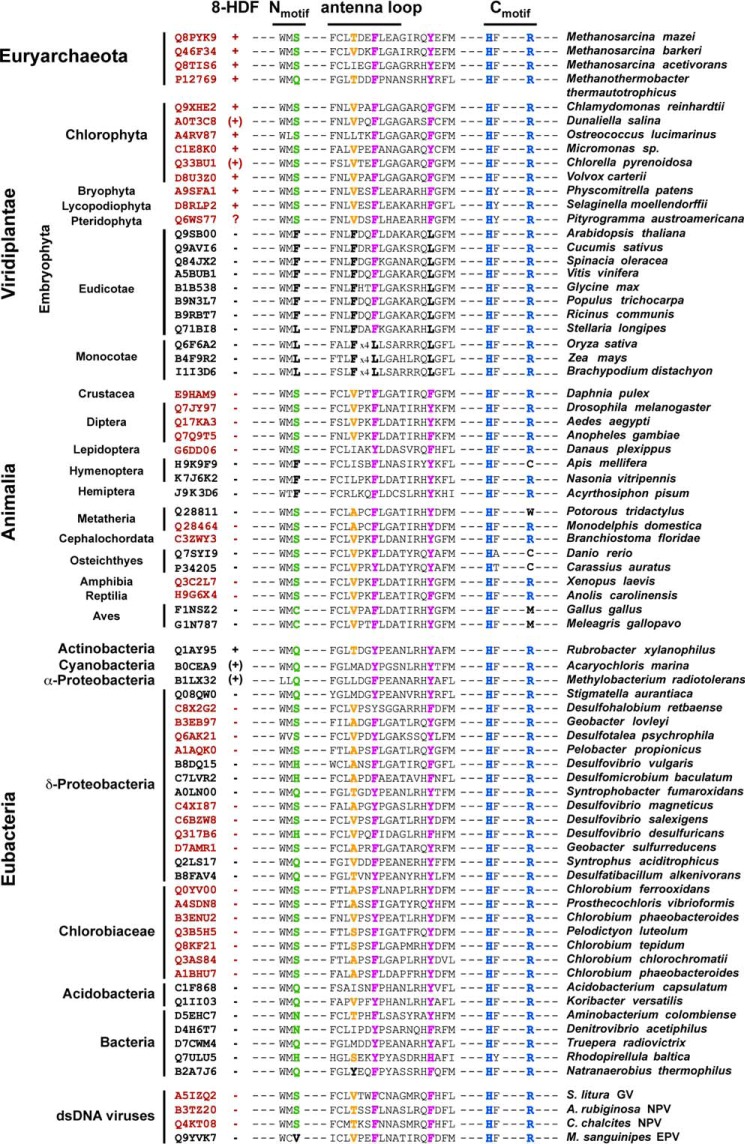
Structural features of the 8-HDF binding pocket within class II photolyases. A, the multiple sequence alignment of selected class II photolyases shows characteristic sequence motifs for 8-HDF binding like the N-motif presenting a polar side chain close to the ribityl moiety and the basic C-motif for salt bridge formation with 8-O− group. B, schematic overview shows structural determinants of the 8-HDF binding motif. C, structural comparison of MmCPDII (green) and OsCPDII (rose) identifies differences crucial for 8-HDF antenna chromophore binding. D, UV/visible spectroscopic analysis shows 8-HDF binding to MmCPDII mutants. Replacement of the polar serine Ser26 within the WMS motif for a nonpolar leucine causes only a slight shift in the absorption maximum (MmCPDII-S26L, dashed line), whereas the bulky, hydrophobic phenylalanine completely impeded 8-HDF incorporation (MmCPDII-S26F, dotted line). E, SDS-PAGE analysis shows MmCPDII mutant/ScFbiC co-expressions. The asterisks mark the bands for the recombinant, co-expressed His6-ScFbiC. M, protein marker; AC, nickel-nitrilotriacetic acid affinity chromatography; SEC, size exclusion chromatography.
Taken together, we can now predict the signature motifs, by which members of the class II photolyase family are capable of utilizing 8-HDF as a cognate antenna (Fig. 6, A and B). First, the C-motif harboring the basic residues His272 and Arg411 has to be intact for interacting with the deprotonated 8-oxy group of 8-HDF. Second, the N-motif (WMS) has to provide a polar residue for H-bonding interactions with 8-HDF as well as formation of the binding site wall. A replacement of this residue by a bulky aromate like in the S26F mutant of MmCPDII and predictably most plant eudicots is incompatible with 8-HDF antenna binding due to steric hindrance, whereas a smaller exchange by leucine is still tolerated for the in vivo incorporation of 8-HDF (Fig. 6, D and E). Third, a small nonbulky residue (MmCPDII: Thr58) as well as the aromatic residues of the antenna loop are required for forming the binding site covering the si-side of the 8-HDF chromophore. Apart from the methanosarcinal photolyases, several bacterial class II photolyases fulfill these criteria such as those from Rubrobacter xylanophilus or members of the classes Chlorobiaceae and Desulfovibrionales. Interestingly, only a few of these microorganisms carry genes for a CofG orthologue for endogenous 8-HDF biosynthesis (Figs. 6A and 7), implying that many microbes may depend on the supply of 8-HDF from the environment for assembling antenna-bearing CPD photolyases. In the plant kingdom, green algae like C. reinhardtii or the Ostreococcus species own class II photolyases with cognate motifs for 8-HDF recognition together with genome-encoded deazaflavin synthases, e.g. PHR1 from C. reinhardtii (23). Interestingly, the ability to utilize 8-HDF antenna comprising class II photolyases for DNA repair is not restricted to these simple, still microbial model systems of higher plants, but is also maintained in land plants like mosses, lycophytes, and ferns. These embryophytes encode class II photolyases with intact 8-HDF binding sites as well as bifunctional CofG/CofH-like synthases for 8-HDF (Physcomitrella patens, A9SZ46; Selaginella moellendorffii, XP_002968233). Seed plants, i.e. angio- and gymnosperms, have apparently lost the ability to equip their class II photolyases with the deazaflavin antenna chromophore due to a loss of the corresponding 8-HDF synthases. Given the structural features at the antenna binding site of the monocot OsCPDII photolyase, where not only the region accommodating the pyrimidine ring is occupied by a leucine replacing Ser26 of MmCPDII, but also affected by a two residue longer antenna loop (Fig. 7), one may argue that the photolyases from monocots and eudicots are capable of recognizing other antenna chromophores than the known ones. For example, colored secondary metabolites such as isoflavonoids, aurones, and anthocyanins have only evolved in gymnosperms and angiosperms (36) and may replace the 8-HDF antenna of class II photolyases, which are predicted to be still present in fern and moss photolyases.
FIGURE 7.
Excerpt of a multiple sequence alignment for class II photolyases highlighting orthologues that participate in the formation of the 8-HDF binding site (shown in red; entry codes are listed in left column). For comparison class II photolyases with binding sites, which are predicted to be incapacitated of 8-HDF binding, are highlighted in black. The column 8-HDF predicts the endogenous occurrence of the 5-deazaflavin chromophore according to the genomic encoding of 8-HDF synthase-like enzymes: +, yes; −, no; ?, unknown; (+) = 8-HDF synthase-like enzymes are present in the genus (Chlorella) and in the order (D. salina: Chlamydomonodales; A. marina: Chroococcales; M. radiotolerans: Rhizobiales), respectively. Further abbreviations: GV, granulovirus; NPV, nucleopolyhedrovirus; EPV, entomopoxvirus.
Interestingly, class II photolyases from animals present a more perplexing view. Clearly, all known animal genomes indicate the absence of any biosynthesis pathway for deazaflavins. Nevertheless, both the (6-4) as well as the CPD photolyases from the insect D. melanogaster can be reconstituted with an 8-HDF antenna (14, 15). Likewise, other diphtheria, e.g. the vectors for malaria and West Nile fever, Anopheles gambiae and Aedes aegyptii, encode class II photolyases compatible with 8-HDF antenna chromophores (Fig. 7). Interestingly, the earlier notion that endosymbiotic Wolbachia or Spiroplasma species may be the source of 8-HDF for D. melanogaster photolyases (14) is not supported by genomic data, which predict a lack of 8-HDF biosynthesis pathways in these bacteria. Accordingly, class II photolyases compatible with 8-HDF antennas are not a general trait in the insect class, because the class II photolyases from hemiptera and hymenoptera like the honey bee (Apis mellifera) show substitutions similar to those found in the higher plant photolyases. The same diversity also holds for the vertebrates. Here, many amphibians, reptilians, and even non-placental mammals encode 8-HDF utilizing class II photolyases (Fig. 7), whereas others like birds and bony fishes show substitutions in their photolyases genes, which ablate 8-HDF binding.
CONCLUSION
8-HDF is an apparently widespread antenna chromophore of photolyases not only among microbial organisms, but also in plants and animals. In the latter, it may fulfill a role as a vitamin as suggested before for the (6-4) photolyases from D. melanogaster (14). Suitable supplies for 8-HDF are many classes of gut microbes, e.g. methanosarcinales, or the environment. Given previous attempts to improve the UV resistance of mammals and plants by introducing photolyase transgenes into these organisms one can postulate that the provision of a suitable pathway for antenna chromophore biosynthesis may be a prerequisite as well.
Acknowledgments
We thank Annegret Wilde (Justus-Liebig University, Giessen) and Verena Helmetag (Philipps-University, Marburg) for genomic DNA, Elvira Happel and Emine Kaya (Ludwig-Maximilians University, Munich) for plasmids, Seigo Shima (Max-Planck Institute for Terrestrial Microbiology, Marburg) for a sample of F420, and Christoph Schwarz, Dennis Walczyk, and Sophie Franz for technical assistance. Synthetic 8-HDF was a gift from Thomas Carell (Ludwig-Maximilians University, Munich). We thank the beamline staff of ID23-1 and ID23-2 at the European Synchrotron Radiation Facility (ESRF Grenoble, France) for excellent support during data collection.
This work was supported by the LOEWE (Landesoffensive zur Entwicklung Wissenschaftlich-Ökonomischer Exzellenz) Center for Synthetic Microbiology (Marburg) and Deutsche Forschungsgemeinschaft Grant BA985/12-1.
The atomic coordinates and structure factors (codes 4CDM and 4CDN) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- (6-4)
- pyrimidine(6-4)pyrimidone dimer
- CPD
- cyclobutane pyrimidine dimer
- FO
- synthase,7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase
- 8-HDF
- 8-hydroxydeazaflavin
- MmCPDII
- class II CPD-photolyase from Methanosarcina mazei
- MTHF
- 5,10-methenyltetrahydrofolate
- PDB
- Protein Data Bank.
REFERENCES
- 1. Essen L. O. (2006) Photolyases and cryptochromes: common mechanisms of DNA repair and light-driven signaling? Curr. Opin. Struct. Biol. 16, 51–59 [DOI] [PubMed] [Google Scholar]
- 2. Sancar A. (2003) Structure and function of DNA photolyase and cryptochrome blue-light photoreceptors. Chem. Rev. 103, 2203–2237 [DOI] [PubMed] [Google Scholar]
- 3. Kato T., Jr., Todo T., Ayaki H., Ishizaki K., Morita T., Mitra S., Ikenaga M. (1994) Cloning of a marsupial DNA photolyase gene and the lack of related nucleotide sequences in placental mammals. Nucleic Acids Res. 22, 4119–4124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lucas-Lledó J. I., Lynch M. (2009) Evolution of mutation rates: phylogenomic analysis of the photolyase/cryptochrome family. Mol. Biol. Evol. 26, 1143–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Müller M., Carell T. (2009) Structural biology of DNA photolyases and cryptochromes. Curr. Opin. Struct. Biol. 19, 277–285 [DOI] [PubMed] [Google Scholar]
- 6. Liu Z., Tan C., Guo X., Kao Y. T., Li J., Wang L., Sancar A., Zhong D. (2011) Dynamics and mechanism of cyclobutane pyrimidine dimer repair by DNA photolyase. Proc. Natl. Acad. Sci. U.S.A. 108, 14831–14836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thiagarajan V., Byrdin M., Eker A. P., Müller P., Brettel K. (2011) Kinetics of cyclobutane thymine dimer splitting by DNA photolyase directly monitored in the UV. Proc. Natl. Acad. Sci. U.S.A. 108, 9402–9407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kiontke S., Geisselbrecht Y., Pokorny R., Carell T., Batschauer A., Essen L. O. (2011) Crystal structures of an archaeal class II DNA photolyase and its complex with UV-damaged duplex DNA. EMBO J. 30, 4437–4449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maul M. J., Barends T. R., Glas A. F., Cryle M. J., Domratcheva T., Schneider S., Schlichting I., Carell T. (2008) Crystal structure and mechanism of a DNA (6-4) photolyase. Angew Chem. Int. Ed. Engl. 47, 10076–10080 [DOI] [PubMed] [Google Scholar]
- 10. Mees A., Klar T., Gnau P., Hennecke U., Eker A. P., Carell T., Essen L. O. (2004) Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science 306, 1789–1793 [DOI] [PubMed] [Google Scholar]
- 11. Hitomi K., Arvai A. S., Yamamoto J., Hitomi C., Teranishi M., Hirouchi T., Yamamoto K., Iwai S., Tainer J. A., Hidema J., Getzoff E. D. (2012) Eukaryotic class II cyclobutane pyrimidine dimer photolyase structure reveals basis for improved ultraviolet tolerance in plants. J. Biol. Chem. 287, 12060–12069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Johnson J. L., Hamm-Alvarez S., Payne G., Sancar G. B., Rajagopalan K. V., Sancar A. (1988) Identification of the second chromophore of Escherichia coli and yeast DNA photolyases as 5,10-methenyltetrahydrofolate. Proc. Natl. Acad. Sci. U.S.A. 85, 2046–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Klar T., Pokorny R., Moldt J., Batschauer A., Essen L. O. (2007) Cryptochrome 3 from Arabidopsis thaliana: structural and functional analysis of its complex with a folate light antenna. J. Mol. Biol. 366, 954–964 [DOI] [PubMed] [Google Scholar]
- 14. Glas A. F., Maul M. J., Cryle M., Barends T. R., Schneider S., Kaya E., Schlichting I., Carell T. (2009) The archaeal cofactor F0 is a light-harvesting antenna chromophore in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 106, 11540–11545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Selby C. P., Sancar A. (2012) The second chromophore in Drosophila photolyase/cryptochrome family photoreceptors. Biochemistry 51, 167–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Eker A. P., Kooiman P., Hessels J. K., Yasui A. (1990) DNA photoreactivating enzyme from the cyanobacterium Anacystis nidulans. J. Biol. Chem. 265, 8009–8015 [PubMed] [Google Scholar]
- 17. Ueda T., Kato A., Kuramitsu S., Terasawa H., Shimada I. (2005) Identification and characterization of a second chromophore of DNA photolyase from Thermus thermophilus HB27. J. Biol. Chem. 280, 36237–36243 [DOI] [PubMed] [Google Scholar]
- 18. Fujihashi M., Numoto N., Kobayashi Y., Mizushima A., Tsujimura M., Nakamura A., Kawarabayasi Y., Miki K. (2007) Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: implication of a novel light-harvesting cofactor. J. Mol. Biol. 365, 903–910 [DOI] [PubMed] [Google Scholar]
- 19. Geisselbrecht Y., Frühwirth S., Schroeder C., Pierik A. J., Klug G., Essen L. O. (2012) CryB from Rhodobacter sphaeroides: a unique class of cryptochromes with new cofactors. EMBO Rep. 13, 223–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chaves I., Pokorny R., Byrdin M., Hoang N., Ritz T., Brettel K., Essen L. O., van der Horst G. T., Batschauer A., Ahmad M. (2011) The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 62, 335–364 [DOI] [PubMed] [Google Scholar]
- 21. Walsh C. (1986) Naturally occurring 5-deazaflavin coenzymes: biological redox roles. Acc. Chem. Res. 19, 216–221 [Google Scholar]
- 22. Isabelle D., Simpson D. R., Daniels L. (2002) Large-scale production of coenzyme F420–5,6 by using Mycobacterium smegmatis. Appl. Environ. Microbiol. 68, 5750–5755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Petersen J. L., Small G. D. (2001) A gene required for the novel activation of a class II DNA photolyase in Chlamydomonas. Nucleic Acids Res. 29, 4472–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 25. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of COOT. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 27. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) PHASER crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Langer G., Cohen S. X., Lamzin V. S., Perrakis A. (2008) Automated macromolecular model building for x-ray crystallography using ARP/wARP version 7. Nat. Protoc. 3, 1171–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 30. Kort R., Komori H., Adachi S., Miki K., Eker A. (2004) DNA apophotolyase from Anacystis nidulans: 1.8 Å structure, 8-HDF reconstitution and x-ray-induced FAD reduction. Acta Crystallogr. D Biol. Crystallogr 60, 1205–1213 [DOI] [PubMed] [Google Scholar]
- 31. Malhotra K., Kim S. T., Walsh C., Sancar A. (1992) Roles of FAD and 8-hydroxy-5-deazaflavin chromophores in photoreactivation by Anacystis nidulans DNA photolyase. J. Biol. Chem. 267, 15406–15411 [PubMed] [Google Scholar]
- 32. Graham D. E., Xu H., White R. H. (2003) Identification of the 7,8-didemethyl-8-hydroxy-5-deazariboflavin synthase required for coenzyme F(420) biosynthesis. Arch. Microbiol. 180, 455–464 [DOI] [PubMed] [Google Scholar]
- 33. Klar T., Kaiser G., Hennecke U., Carell T., Batschauer A., Essen L. O. (2006) Natural and non-natural antenna chromophores in the DNA photolyase from Thermus thermophilus. ChemBioChem 7, 1798–1806 [DOI] [PubMed] [Google Scholar]
- 34. Okafuji A., Biskup T., Hitomi K., Getzoff E. D., Kaiser G., Batschauer A., Bacher A., Hidema J., Teranishi M., Yamamoto K., Schleicher E., Weber S. (2010) Light-induced activation of class II cyclobutane pyrimidine dimer photolyases. DNA Repair 9, 495–505 [DOI] [PubMed] [Google Scholar]
- 35. Chakrabarti P., Bhattacharyya R. (2007) Geometry of nonbonded interactions involving planar groups in proteins. Progr. Biophys. Mol. Biol. 95, 83–137 [DOI] [PubMed] [Google Scholar]
- 36. Rausher M. D. (2006) in The Science of Flavonoids (Grotewold E., ed) pp. 175–211, Springer, New York [Google Scholar]