Background: Ephrin A2 (EphA2) Sterile α Motif (SAM) domains undergo phosphorylation at Tyr921, Tyr930, and Tyr960.
Results: Recruitment of the Grb7 SH2 domain by EphA2 SAM is phosphorylation site-specific.
Conclusion: Tyrosine phosphorylation of the EphA2 SAM domain has wide implications for the differential recruitment of binding partners.
Significance: SAM tyrosine phosphorylation imparts specificity to its adaptor protein interactions and network formation, easily studied in vitro.
Keywords: Adaptor Protein, Biophysics, Endocytosis, Phosphorylation, Protein-tyrosine Kinase (Tyrosine Kinase), Ephrin Receptors, SAM Domains, SHIP2, Receptor Clustering and Signaling
Abstract
The sterile α motif (SAM) domain of the ephrin receptor tyrosine kinase, EphA2, undergoes tyrosine phosphorylation, but the effect of phosphorylation on the structure and interactions of the receptor is unknown. Studies to address these questions have been hindered by the difficulty of obtaining site-specifically phosphorylated proteins in adequate amounts. Here, we describe the use of chemically synthesized and specifically modified domain-length peptides to study the behavior of phosphorylated EphA2 SAM domains. We show that tyrosine phosphorylation of any of the three tyrosines, Tyr921, Tyr930, and Tyr960, has a surprisingly small effect on the EphA2 SAM structure and stability. However, phosphorylation at Tyr921 and Tyr930 enables differential binding to the Src homology 2 domain of the adaptor protein Grb7, which we propose will lead to distinct functional outcomes. Setting up different signaling platforms defined by selective interactions with adaptor proteins thus adds another level of regulation to EphA2 signaling.
Introduction
Phosphorylation plays a major role in the regulation of protein function (1, 2). Although there are many cellular studies using phosphorylation-deficient proteins, there are relatively few systems where the effects of phosphorylation on the structure and the interactions of a protein has been tested in vitro (3, 4). Biophysical studies of phosphorylated proteins have been hampered by low yields, difficulties in obtaining site-specific phosphorylation, or the lack of a good phosphomimetic. Recent progress in peptide synthesis has made it possible to generate sizeable protein domains with the incorporation of phosphotyrosines at specific positions (peptides up to 100 residues can now be synthesized by several companies). Here, we report a biophysical study of synthesized and specifically phosphorylated protein domains. To our knowledge, this is the first report of a biophysical study utilizing full-length tyrosine-phosphorylated domains that have been generated by chemical synthesis.
Eph proteins belong to the family of transmembrane protein receptor tyrosine kinases (5–7). Signaling through Eph receptors regulates key cellular functions, including cell migration, axon guidance, and angiogenesis, under physiological and pathological conditions, such as cancer (8, 9). Phosphorylation is known to be central to the regulation of Eph receptor function. For example, increased EphA2 tyrosine phosphorylation is a characteristic of basal breast cancer cells (10) and is associated with increased apoptosis of cardiomyocytes (11). Apart from ligand binding, receptor activation involves the phosphorylation of specific residues of the juxtamembrane region and the kinase domain (12–14). In addition, in vivo studies and proteomics surveys have revealed that the tyrosines of the C-terminal SAM4 domain (present in all Eph receptors but none of the other receptor tyrosine kinase subfamilies) also undergo phosphorylation (15–18). The SAM domains are common protein-protein interaction modules that typically form homo- or heterodimers and are present in a diverse set of proteins (19–21). The structures of several SAM domains have been solved, showing a relatively well conserved topology of five α-helices (22, 23). The EphA2 SAM domain has three tyrosines, Tyr921, Tyr930, and Tyr960, of which Tyr921 is absolutely conserved in Eph and many other SAM domains (Fig. 1). By contrast to most receptor tyrosine kinase phosphorylation sites, which occur in relatively unstructured protein domain linker regions or loops (23, 24), the three SAM domain tyrosines are part of the folded protein structure. The tertiary structure may thus provide an additional level of regulation. Biological studies showing the phosphorylation of all of the three tyrosines have already been reported; Tyr921 and Tyr960 were found to be phosphorylated when an EphA2 kinase-SAM domain construct is expressed in Escherichia coli (12, 25), and Tyr960 phosphorylation was identified in a colorectal carcinoma cell line (26). Tyr930 is phosphorylated in mouse lung epithelial cells; furthermore, the Y930F phosphorylation-defective variant inhibited both the kinase activity and vascular assembly (10). Similarly, phosphatase LAR was shown to dephosphorylate Tyr930 (and possibly other tyrosines), an event that appears to abrogate binding to the SH2 domain of the adaptor protein Nck2 and attenuates cell migration (Y930F had the same effect on cell migration) (26). There is no biological information on the role of Tyr921 phosphorylation in EphA2 (although binding to the SH2 domain of Vav3 has been proposed (17)). However, in vivo studies have also shown that the conserved SAM domain tyrosine (Tyr921 in EphA2) is responsible for recruiting SH2 domains of Grb7 and Grb10 to EphB1, and this interaction is deemed essential for the regulation of cell migration (15, 17, 28, 29).
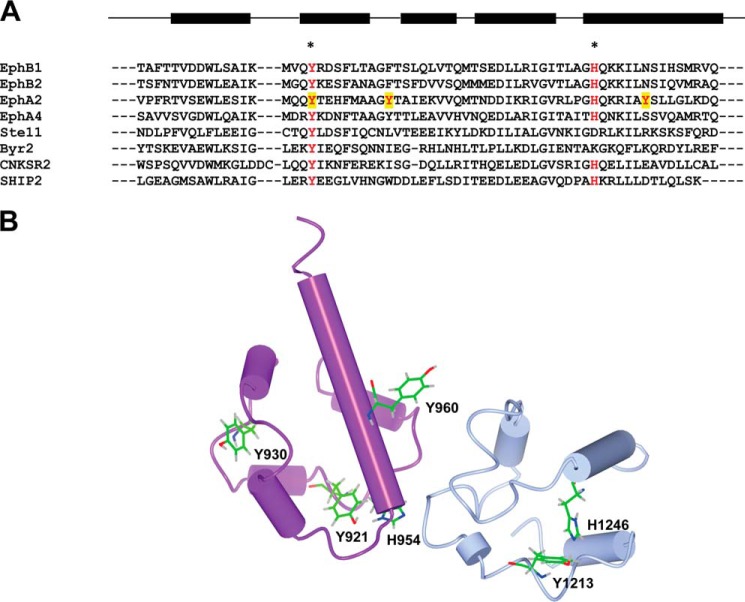
FIGURE 1.
The tyrosines of EphA2 SAM. A, sequence alignment of selected SAM domains; the highly conserved tyrosine and histidine (Tyr921 and His894 in EphA2) residues are highlighted in red type and marked with asterisks. The three tyrosines of the EphA2 SAM domain are highlighted by a yellow background, and the secondary structure of this domain is shown above the alignment. B, location of the tyrosine and histidine residues of EphA2 SAM and SHIP2 SAM in the three-dimensional structure. The structure of the EphA2-SHIP2 SAM-SAM complex (Protein Data Bank entry 2KSO) is drawn using a cylinder representation. EphA2 SAM is shown in purple, SHIP2 SAM is blue, and the tyrosine and histidine residues are shown as sticks. Tyr921 and Tyr930 are partially buried. The conserved tyrosine and histidine side chains of both EphA2 SAM and SHIP2 SAM (Tyr921/His954 and Tyr1213/His1246, respectively) are involved in hydrogen bonds.
The extent of phosphorylation of the tyrosine residues of proteins in cells is typically not easy to ascertain or to manipulate in a site-specific manner. Thus, the experiments reported above rely on the expression of proteins in which a single tyrosine has been mutated to phenylalanine (a side chain that mimics the unmodified residue and cannot be phosphorylated). Although these biological findings suggest the importance of particular sites for the interactions, this strategy can deliver false negatives, because an interaction may still persist upon mutating a single site if interactions with several phosphorylated tyrosines are possible. Similarly, it may be noted that the previous reports were not accompanied by a molecular level framework, which involves consideration of protein conformational changes and competing binding processes. Biophysical studies in vitro, as reported here, can provide deeper insight and propose models for investigation at the cellular level. Specifically, the EphA2 SAM domain forms a heterodimer with the SAM domain of SH2 domain-containing inositol-5′-phosphatase (SHIP2) (23, 30, 31). Binding of EphA2 SAM to SHIP2 SAM inhibits receptor endocytosis and enhances activation of Eph kinase (31). In vivo studies have also shown (using Tyr to Phe mutations in the EphA2 SAM domain) that tyrosine phosphorylation is not required for SHIP2 recruitment (31); however, it is not clear whether phosphorylation could, in fact, be detrimental to SHIP2 binding. Here we studied directly whether the phosphorylation adds another level of complexity to the regulation of Eph receptors by controlling SAM domain-mediated interactions.
Using synthetic domains, we studied the effect of phosphorylation of the EphA2 SAM domain on its structure and interactions with SHIP2 SAM. Further, stimulated by reports on EphB1 recruiting the SH2 domain of Grb7 (15, 17), we examined interactions of the phosphorylated domains with Grb7 SH2. Unexpectedly, we show that phosphorylation of the tyrosines of the EphA2 SAM domain has little effect on the overall structure of the domain. EphA2 SAM phosphorylated at Tyr930 could simultaneously engage the Grb7 SH2 and SHIP2 SAM domains. In contrast, Tyr921 is located near the SHIP2 binding region, and Grb7 SH2 and SHIP2 SAM compete for binding. Surprisingly, EphA2 SAM phosphorylated at Tyr960 does not interact with Grb7 SH2 but also has no effect on SHIP2 SAM binding. We discuss how this phosphorylation-dependent specificity could give rise to different signaling platforms, regulating the function of EphA2 receptors.
EXPERIMENTAL PROCEDURES
Protein Cloning, Expression, and Purification
cDNA for human Grb7 was a gift from Prof. Jun-Lin Guan (University of Michigan). Residues 425–532, corresponding to the SH2 domain, were amplified by PCR and subsequently cloned into a pET30 Xa/LIC vector using ligation-independent cloning (EMD Biosciences). The plasmid containing Grb7 SH2 was transformed into E. coli BL21 (DE3) cells and grown at 310 K in either Luria-Bertini (LB) medium or M9 minimal medium supplemented with 15NH4Cl. Cultures were grown to an A600 of 0.8 for both media and then induced with 0.2 mm isopropyl 1-thio-β-d-galactopyranoside. Bacterial cells were harvested after 24 h of induction at 289 K.
Purification of the Grb7 SH2 domain was carried out using Ni2+ affinity chromatography. In brief, the harvested cells were resuspended in lysis/binding buffer (50 mm sodium phosphate, pH 7.4, 500 mm NaCl, 30 mm imidazole, 1 mm TCEP-HCl) supplemented with EDTA-free CompleteTM protease inhibitors (Roche Applied Science). Cells were disrupted by sonication. After centrifugation, the cleared lysate was applied to nickel-nitrilotriacetic acid beads (Qiagen). Following washing with binding buffer containing 45 mm imidazole, the bound protein was eluted with 50 mm sodium phosphate buffer (pH 7.4) with 250 mm imidazole and 1 mm TCEP-HCl. The eluted protein was concentrated and buffer-exchanged into the NMR buffer (20 mm Tris, pH 6.8, 100 mm NaCl, 1 mm TCEP-HCl). The EphA2 and SHIP2 SAM domain constructs and their expression and purification have been described previously (20). The EphA2 and SHIP2 SAM proteins were also exchanged into the same NMR buffer.
Preparation of Tyrosine-phosphorylated Peptides
Phosphorylated peptides corresponding to residues 910SEWLESIKMQQpYTEHFMAAGFT931 (denoted pep.pY921),916WKMQQFTEHFMAAGpYTAIEVVQ937 (pep.pY931), and 951LPGHQKRIApYSLLGLKDQVNTV972 (pep.pY960) as well as the equivalent unphosphorylated peptides were purchased from GenScript. The peptides were dissolved into the NMR buffer without further purification. Three domain-length EphA2 SAM peptides (residues 901–976) were synthesized (United Peptide, Inc.), also referred to as simply EphA2 below (experiments were carried out only with the SAM domains in this paper). Each domain peptide has phosphorylated side chains at Tyr921 (EphA2.pY921), Tyr930 (EphA2.pY930), or Tyr960 (EphA2.pY960). These full-length phosphorylated peptides initially had poor solubility in water and were refolded by incubation in 8 m urea (20 mm Tris, pH 6.8, 100 mm NaCl, 1 mm TCEP-HCl) overnight and then were dialyzed extensively against the NMR buffer. Peptide and protein concentrations were determined by UV absorbance with reference to predicted extinction coefficients.
Circular Dichroism (CD) Spectroscopy
The secondary structure and the thermal stability of the phosphorylated domains were examined by CD spectroscopy using established protocols (32). Spectra were recorded on a 20 μm sample using a cuvette with a path length of 4 mm on an Aviv (model 215) instrument. The temperature scans were carried out in the range of 293–363 K, at 222 nm, with a step size of 2 K and a 30-s equilibration period and a 30-s recording time. All of the experiments were carried out in triplicate, and signal from the buffer was subtracted.
NMR Spectroscopy
All experiments were run at 298 K on an 800-MHz spectrometer equipped with a TCI probe (Bruker Avance). One-dimensional 1H NMR (using WATERGATE) and homonuclear two-dimensional 1H NOESY experiments (mixing time of 300 ms) were recorded with 300 μm samples of the SAM domains. 15N-1H HSQC experiments on Grb7 SH2 were recorded on the 15N-labeled protein itself or on a 1:1 mixture with unlabeled EphA2 domains or after the further addition of 2 molar eq of unlabeled SHIP2 SAM. The data were processed using nmrPipe (33), and the two-dimensional spectra were visualized using Sparky (Goddard TD, Kneller DG, SPARKY3, University of California, San Francisco). The one-dimensional 1H NMR spectra were plotted using the software Origin (OriginLab, Northampton, MA).
Isothermal Titration Calorimetry (ITC)
Interaction of the short (unphosphorylated and phosphorylated) peptides with Grb7 SH2 and of the domain-length phosphorylated and refolded EphA2 peptides with Grb7 SH2 and/or the SHIP2 SAM domain was measured by ITC (MicroCal iTC200, GE Healthcare) with established protocols. Typically a 40 μm concentration of the EphA2 protein was used in the chamber, and a 400 μm concentration of the SHIP2 SAM domain was used in the syringe for titration. ITC experiments with Grb7 SH2 were performed using Grb7 SH2 in the chamber and titrating in the corresponding binding partner. The data were analyzed using Origin (OriginLab).
Solvent-accessible Surface Area
The solvent-accessible surface area for the tyrosine residues of EphA2 was calculated using the algorithm SURFACE of the CCP4 suite (34). The complex structure of EphA2 SAM-SHIP2 SAM (Protein Data Bank entry 2KSO) was used, and calculations were set up with a probe of 1.4-Å radius.
RESULTS
Chemically Synthesized Polypeptides Share Native-like Folds and Are Stable
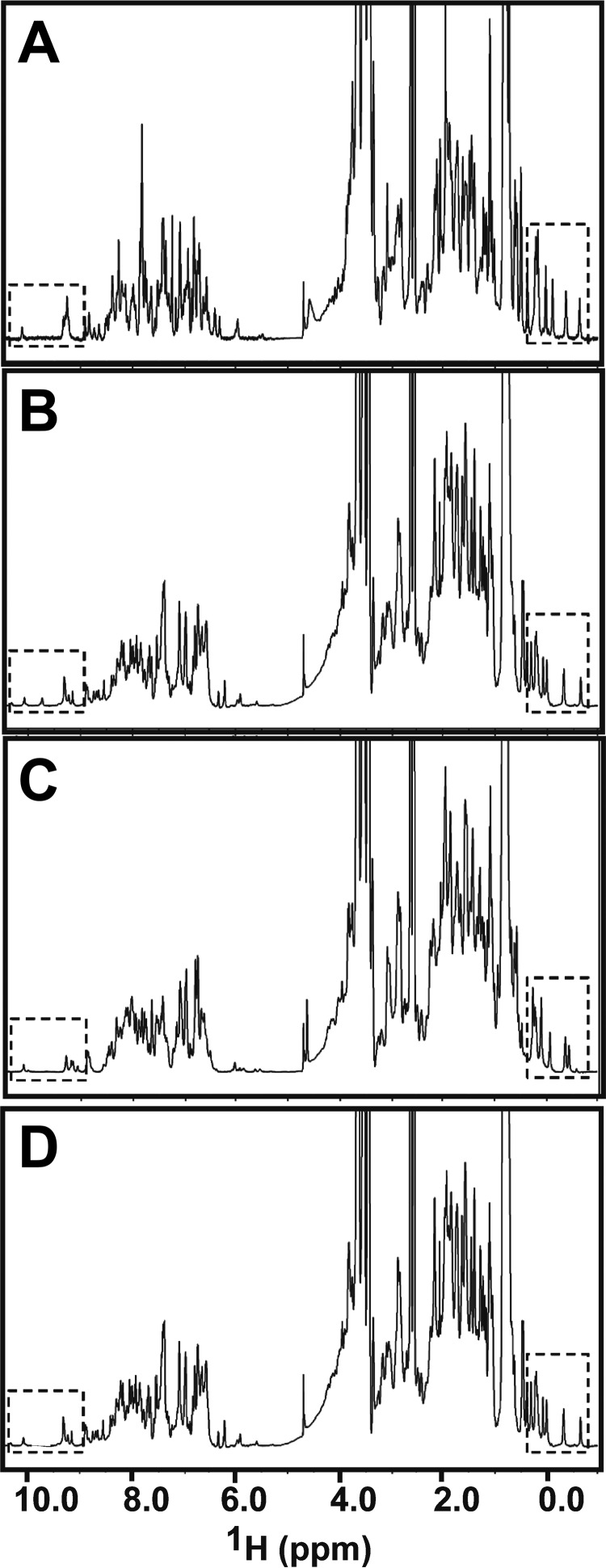
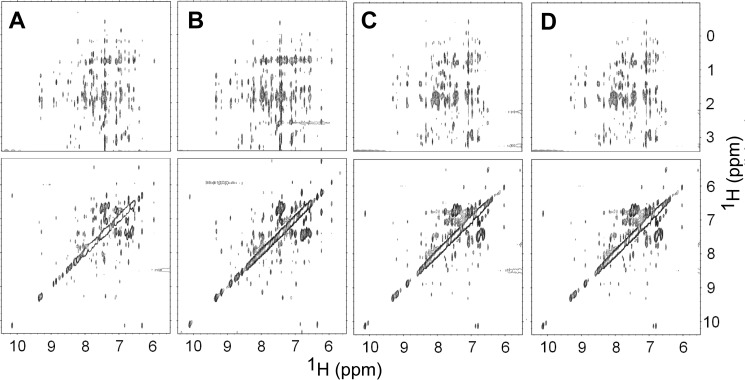
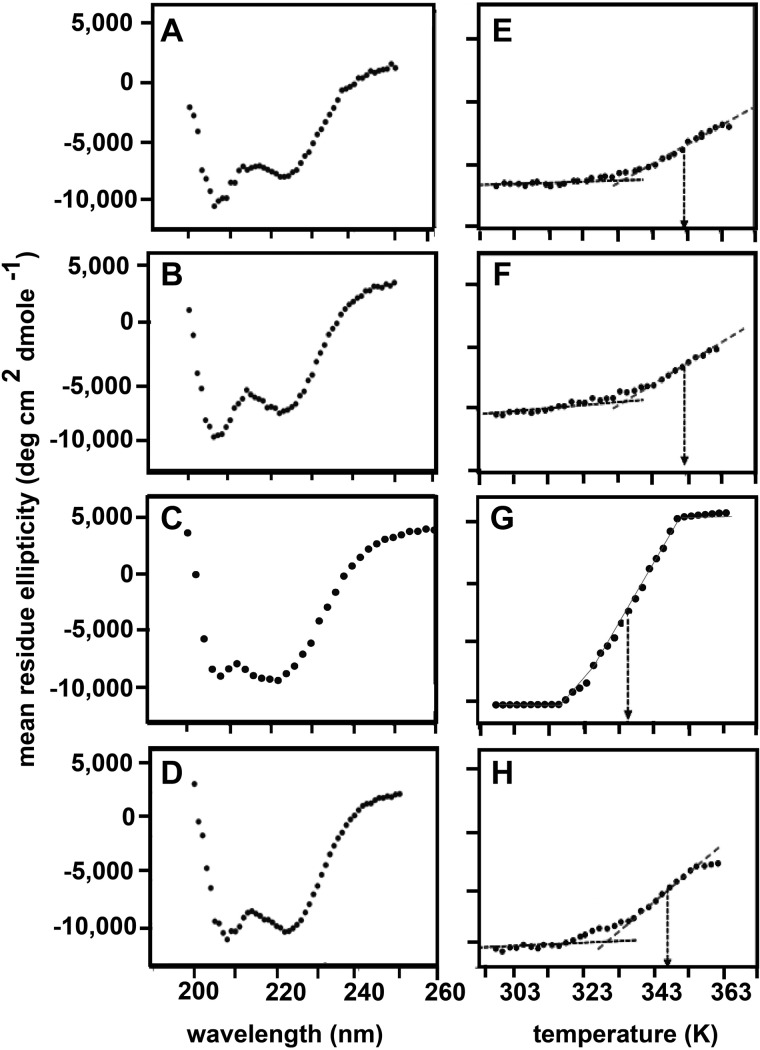
Biophysical studies demand large quantities of pure proteins. It is difficult to obtain proteins that are fully phosphorylated at one specific site by exposure to kinases in vitro. Here, we obtained SAM domain polypeptides that have specific sites fully and stably phosphorylated during their chemical synthesis (United Peptides Inc.). Initial NMR experiments with the synthesized domains dissolved in buffer showed that these proteins are not properly folded (data not shown). We carried out a chemical denaturation, followed by a refolding step to correctly fold the domains. In order to confirm that a nearly native SAM domain structure is obtained, we analyzed the conformations of the refolded proteins by both one-dimensional 1H NMR (Fig. 2) and homonuclear two-dimensional 1H NOESY experiments (Fig. 3). The NMR spectra show that all three specifically phosphorylated SAM domains (referred to as EphA2.pY921, EphA2.pY930, and EphA2.pY960) are well folded, as is evident from the dispersed amide signals, resonances for the tryptophan side chains, and up-field shifted methyl signals (highlighted with boxes in Fig. 2). The spectra show that the peptides adopt a structure very similar to that of the recombinant protein. Subtle differences are apparent in EphA2.pY921 and EphA2.pY930, the two tyrosines that are partially buried in the wild type protein (with 17.6 and 32.9% solvent exposure calculated for Tyr921 and Tyr930, respectively) and probably have become more exposed upon phosphorylation. In addition, we characterized the secondary structure of the phosphorylated proteins by far-UV CD spectroscopy and found that they share within ±15% an α-helical content similar to that of the recombinant EphA2 SAM domain. We also assessed the thermal stabilities of the phosphorylated proteins and of the recombinant EphA2 SAM by measuring the signal at 222 nm as a function of temperature; phosphorylation at any one of its three tyrosines does not dramatically destabilize the SAM domain fold (Fig. 4 and Table 1).
FIGURE 2.
The phosphorylated EphA2 SAM domains are well folded. One-dimensional 1H NMR spectra of EphA2 SAM domains: EphA2.pY921 (A), EphA2.pY930 (B), EphA2.pY960 (C), and unphosphorylated/recombinant EphA2 (D). The chemically synthesized, phosphorylated EphA2 polypeptides and the recombinant EphA2 domain share a similar global fold.
FIGURE 3.
The phosphorylation of EphA2 SAM domains is not accompanied by large conformational changes. Shown are two-dimensional homonuclear 1H NOESY spectra of unphosphorylated EphA2 SAM (A), EphA2.pY921 (B), EphA2.pY930 (C), and EphA2.pY960 (D); the phosphorylated domains adopt nearly native-like global folds.
FIGURE 4.
Phosphorylated SAM domains share similar secondary structure with the recombinant EphA2 SAM domain and are thermally stable. A–D, far-UV circular dichroism (CD) spectra of the phosphorylated and unphosphorylated SAM domains; all of the proteins are α-helical. E–H, thermal unfolding of the domains monitored at 222 nm; the approximate midpoint of unfolding (Tm) is shown by arrows. Phosphorylation did not significantly destabilize the domains.
TABLE 1.
Thermal stabilities of the recombinant and phosphorylated EphA2 SAM domains
| Protein | Thermal stability (Tm) |
|---|---|
| K | |
| EphA2.pY921 | 351 ± 2.0 |
| EphA2.pY930 | 352 ± 1.6 |
| EphA2.pY960 | 337 ± 3.2 |
| Recombinant EphA2 | 345 ± 2.6 |
The Phosphorylated Proteins Still Interact with SHIP2 SAM
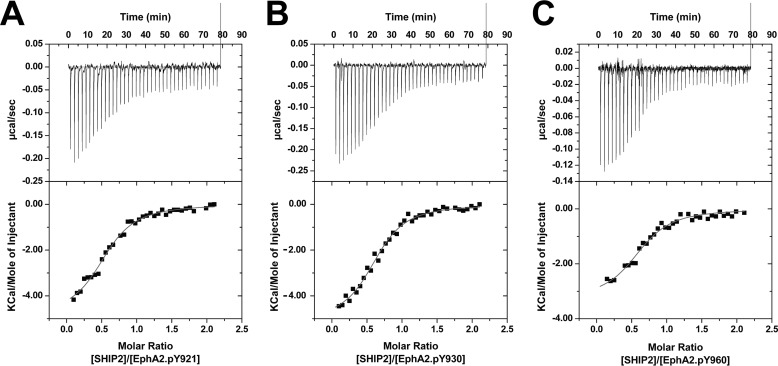
ITC measurements have shown that the EphA2 SAM domain binds the SHIP2 SAM domain with a micromolar affinity (23). The phosphorylated EphA2 SAM domains (phosphorylated at Tyr921, Tyr930, or Tyr960) bind SHIP2 SAM with an affinity that is comparable with that of the recombinant unphosphorylated protein (representative ITC interaction data are shown in Fig. 5). We report the dissociation constants (KD) and the derived thermodynamic contributions of the individual interactions in Table 2. The change in binding enthalpy is slightly more favorable for the phosphorylated proteins compared with the unphosphorylated protein, but the binding entropy is lower. It is likely that the change in enthalpy for this process is a systematic effect because the synthetic proteins have shorter termini, compared with the recombinant protein. Thus, remarkably, the binding of EphA2 to SHIP2 SAM is essentially insensitive to tyrosine phosphorylation. We, therefore, decided to study the interaction of the phosphorylated domains with Tyr(P)-binding proteins.
FIGURE 5.
Phosphorylation of EphA2 SAM does not affect its binding to SHIP2 SAM domain. Interactions of EphA2.pY921 (A), EphA2.pY930 (B), and EphA2.pY960 (C) with SHIP2 SAM were measured by ITC. The synthetic domain bind SHIP2 SAM with micromolar affinities (KD ∼4 μm) similar to the recombinant EphA2 SAM (KD ∼5 μm). The derived thermodynamic parameters are listed in Table 1.
TABLE 2.
Thermodynamics of binding of phosphorylated and unphosphorylated EphA2 SAM domains and peptides to SHIP2 SAM and Grb7 SH2
| Protein in ITC cell | Titrant | KD | ΔH | TΔS | ΔG | Comment |
|---|---|---|---|---|---|---|
| μm | kcal/mol | kcal/mol/deg | kcal/mol | |||
| EphA2.pY921 | SHIP2 | 4.1 ± 0.5 | −4.9 | 2.5 | −7.4 | |
| EphA2.pY930 | SHIP2 | 3.4 ± 0.4 | −5.1 | 2.4 | −7.5 | |
| EphA2.pY960 | SHIP2 | 3.9 ± 0.2 | −4.7 | 2.7 | −7.4 | |
| Recombinant EphA2 | SHIP2 | 5.2 ± 0.3 | −2.5 | 4.7 | −7.2 | |
| Grb7 SH2 | SHIP2 | 3.5 ± 0.1 | −1.95 | 18.4 | −7.3 | |
| Grb7 SH2 | EphA2.pY921 | 2.6 ± 0.7 | −8.0 | −0.3 | −7.7 | |
| Grb7 SH2 | EphA2.pY930 | 8.6 ± 4.3 | −2.5 | 4.4 | −6.9 | |
| Grb7 SH2 | EphA2.pY960 | No interaction | ||||
| Grb7 SH2 | Recombinant EphA2 | No interaction | ||||
| Grb7 SH2 | pep.pY921 | 3.2 ± 0.6 | −14.7 | −7.2 | −7.5 | |
| Grb7 SH2 | pep.pY930 | 2.6 ± 0.4 | −4.8 | 2.8 | −7.6 | |
| Grb7 SH2 | pep.pY960 | 3.0 ± 0.6 | −15.4 | −7.9 | −7.5 | |
| Grb7 SH2 | All 3 of the unphosphorylated short peptides | No interaction |
Binding of EphA2 SAM to Grb7 SH2 Is Phosphorylation Site-specific
SH2 domains of the Grb family are known to bind the conserved phosphorylated tyrosine of Eph SAM domains (Tyr921 for EphA2 SAM) (29). We tested the binding of several Grb SH2 domains with EphA2 SAM and carried out a complete study by ITC and NMR for the Grb7 SH2-EphA2 SAM interaction. Neither the unphosphorylated EphA2 SAM nor EphA2 SAM phosphorylated at Tyr960 interacts appreciably with Grb7 SH2. In contrast, both EphA2.pY921 and EphA2.Y930 bound Grb7 SH2 with similar affinities (Table 2). The binding of EphA2.pY921 is almost entirely enthalpic, whereas EphA2.pY930 binding is largely driven by a favorable entropic contribution. This is the first study to report interaction of phosphorylated EphA2 SAM domains with Grb7 SH2.
In order to examine the possibility whether the conformational restraints imparted by the protein fold play a role in the interactions, we measured the interaction of short phosphorylated peptides with Grb7 SH2 (Table 2). All of the short phosphorylated peptides, including pep.pY960, interact with Grb7 SH2 with a similar affinity. Because the Tyr(P)960 within the folded SAM domain did not bind Grb7 SH2, this observation suggests that binding at this site is conformation-dependent. We also carried out binding experiments with unphosphorylated short peptides, none of which bound.
Differential Effects of EphA2.pY Complex Formation with Grb7 SH2 on the Interaction with SHIP2 SAM
Our ITC data show that the phosphorylated proteins, EphA2.pY921 and EphA2.pY930, can bind both Grb7 SH2 and SHIP2 SAM with similar affinities. The question arises whether SHIP2 SAM and Grb7 SH2 can bind EphA2.pY921 or EphA2.pY930 simultaneously or whether the binding is mutually exclusive (and competitive). To answer these questions, we carried out ITC and NMR experiments to examine the possibility of a trimolecular interaction. ITC experiments (Table 3) show a slight decrease in binding affinity of EphA2.pY921 and EphA2.pY930 for SHIP2 SAM in the presence of Grb7 SH2, suggesting that Grb7 SH2 influences the EphA2-SHIP2 interaction. Because the binding affinities between Grb7 SH2 and SHIP2 SAM are similar, the equilibrium cannot be shifted substantially unless one protein is in large excess concentration. In the case of EphA2.pY960, it is possible that this domain only interacts with Grb7 SH2 in the presence of SHIP2 SAM. However, the binding affinity and thermodynamic contributions are identical (within the error limits) for SHIP2 SAM binding to EphA2.pY960 whether Grb7 SH2 is present or not, underscoring the fact that EphA2.pY960 does not bind Grb7 SH2 (Table 3).
TABLE 3.
Thermodynamics of SHIP2 SAM competing for phosphorylated EphA2 SAM bound to Grb7 SH2 in comparison with the phosphorylated domains binding to SHIP2 SAM
| In ITC cell | Titrant | KD | ΔH | TΔS | ΔG |
|---|---|---|---|---|---|
| μm | kcal/mol | kcal/mol/deg | kcal/mol | ||
| EphA2.pY921-Grb7 SH2 | SHIP2 | 6.5 ± 4.0 | −4.1 | 3.0 | −7.1 |
| EphA2.pY930-Grb7 SH2 | SHIP2 | 6.8 ± 3.2 | −4.4 | 2.7 | −7.1 |
| EphA2.pY960-Grb7 SH2 | SHIP2 | 4.5 ± 0.4 | −5.2 | 2.2 | −7.4 |
To gather additional support for these observations, we acquired 15N-1H HSQC spectra of labeled Grb7 SH2 in the presence of unlabeled EphA2 with or without SHIP2 SAM proteins (Fig. 6). Binding of both EphA2.pY921 and EphA2.pY930 to Grb7 SH2 is characterized by a decrease of resonance intensity of Grb7 SH2. This change arises due to the formation of a larger molecular weight complex because Grb7 SH2 is a dimer and the Tyr(P) binding interface and the dimerization interface are different (35, 36) (data not shown). However, it is not clear to what extent, if any, Tyr(P) binding alters the dimerization of Grb7 SH2 (35, 36, 37). Upon the addition of SHIP2 SAM to the premixed complex of Grb7 SH2 (labeled)-EphA2.pY921, we saw a change in intensity of several but not all of the dispersed resonances compared with the spectrum of Grb7 SH2 bound to Eph.pY921 (Fig. 6A). The changes occur at the Tyr(P) binding interface (38, 39), suggesting that some of the EphA2.pY921-Grb7 SH2 complex is dissociating, so that EphA2 can form a complex with SHIP2. When we added SHIP2 SAM to the EphA2.pY930/Grb7 SH2 (labeled) premixed complex, we observed significant line broadening of most of the Grb7 SH2 resonances (Fig. 6B); this is consistent with the formation of a large complex (the Grb7 domains would still dimerize).
FIGURE 6.
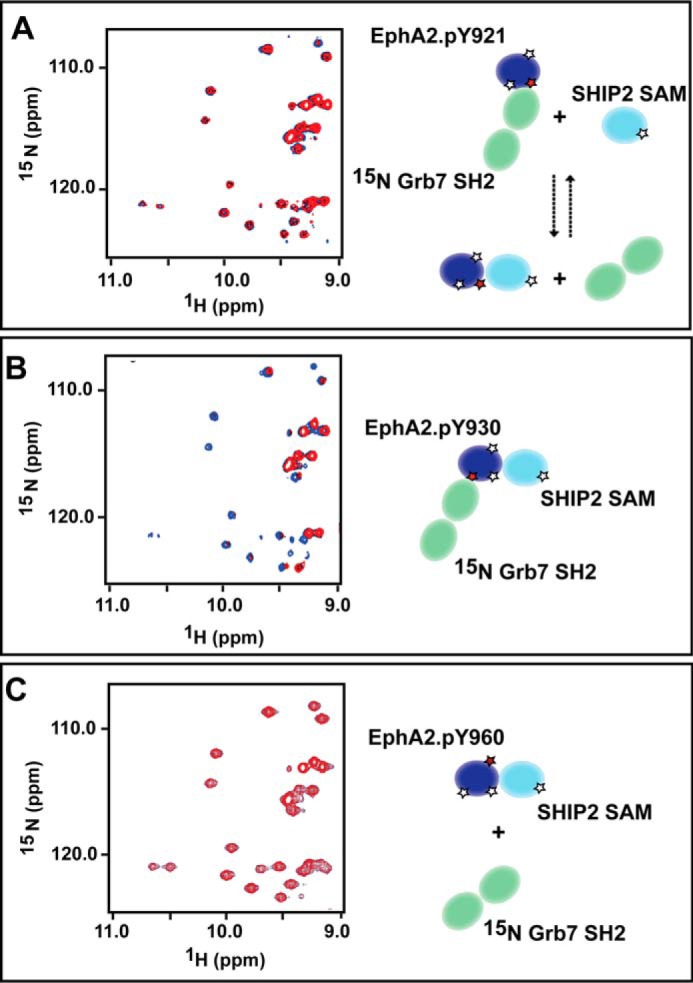
Grb7 SH2 competes with SHIP2 SAM for binding to the EphA2 SAM domain phosphorylated at Tyr930. Left, an overlay of part of the 15N, 1HN HSQC spectrum of a Grb7 SH2 (15N-labeled)/EphA2 phosphorylated protein mixture (blue) and in the presence of SHIP2 (red) is shown in the left-hand panels. The right-hand panels show schematic representations of the complexes formed. A, SHIP2 SAM competes with Grb7 SH2 for binding to EphA2.pY921; the overlaid spectra are similar, suggesting that EphA2.pY921 bound to Grb7 SH2 cannot bind SHIP2 SAM simultaneously. However, broadening of only some resonances corresponding to the Tyr(P)-binding residues of Grb7 SH2 are observed due to intermediate NMR time scale exchange that occurs in the competition. B, EphA2.pY930 can bind both Grb7 SH2 and SHIP2 SAM simultaneously, as evidenced by extensive line broadening of essentially all but the most flexible residues. This broadening occurs due to the formation of a large trimolecular complex; because Grb7 SH2 is a dimer, the complex would be even larger. C, the spectrum of EphA2.pY960 premixed with Grb7 SH2 (15N-labeled) shows no significant changes upon the addition of SHIP2 SAM, demonstrating that this SAM domain does not bind Grb7 SH2.
The addition of unphosphorylated EphA2 SAM domain or EphA2.pY960 did not alter the spectrum of Grb7 SH2 (not shown), consistent with the ITC data showing that these SAM domains do not interact with the SH2 domain. Furthermore, when we added SHIP2 SAM to the premixed complexes of Grb7 SH2 and EphA2.pY960, we did not see any significant changes to the Grb7 SH2 resonances (Fig. 6C), highlighting that Grb7 SH2 does not bind EphA2.pY960 even when the latter is bound to SHIP2. The differential signaling output that results from these selective interactions is discussed below (and in the legend to Fig. 7).
FIGURE 7.
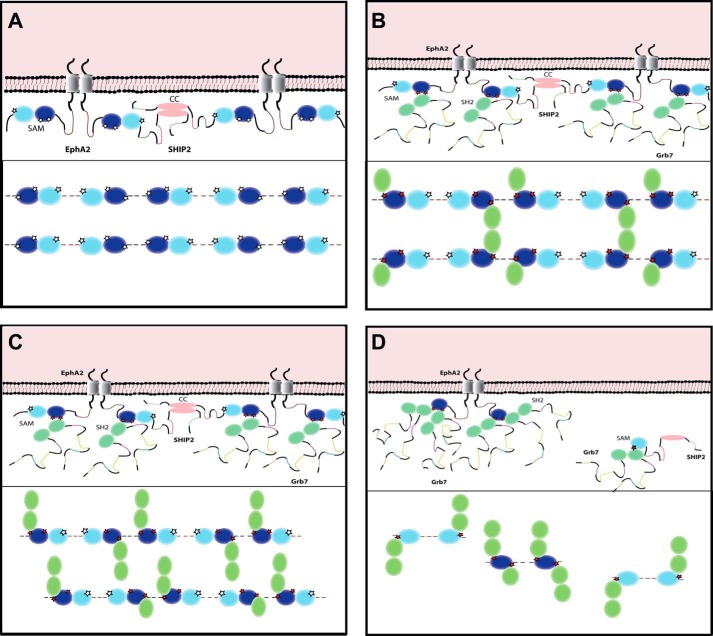
The proposed model for the differential regulation of the EphA2 receptor and SHIP2 SAM localization by Grb7 SH2 bound to phosphorylated EphA2 SAM. A, in the absence of Grb7 and irrespective of phosphorylation of the SAM domain, EphA2 SAM (dark blue) is bound to the SAM domain of SHIP2 (blue). Interaction of EphA2 SAM-SHIP2 SAM domains localizes SHIP2 to the plasma membrane. The extracellular and the transmembrane regions are also likely to be dimerized throughout, as shown. Given EphA2 and Ship2 are dimers, linear assemblies are predicted to be formed, as shown in the panel below. EphA2 and SHIP2 are drawn as dimers, and only the SAM and CC domains (pink) are depicted. B, phosphorylation of Tyr921 and Tyr930 and EphA2-SHIP2-Grb7 complex at substoichiometric Grb7 with respect to an EphA2/SHIP2 1:1 concentration; Grb7 SH2 dimer (green) binds to EphA2 at Tyr(P)930 and provides maximum cross-linking forming arrays of EphA2-SHIP2 (bottom). C, when Grb7 SH2 is present at stoichiometric concentration, less cross-linking of EphA2-SHIP2 occurs via Grb7 dimers, giving rise to linear chains. D, for excess Grb7 SH2, there would be a competitive binding of the adaptor protein to Tyr(P)921 of EphA2 (and also Tyr(P)1213 SHIP2 SAM), which displaces the SHIP2 protein from the membrane. This results in endocytosis and down-regulation of the receptor.
DISCUSSION
The detailed characterization of posttranslational modifications, such as tyrosine phosphorylation, and their role in specific protein-protein interactions is a prerequisite to understanding the mechanistic basis of signaling processes that in turn regulate the great majority of cellular functions. We took advantage of the recent progress in peptide synthesis technology to obtain domain-length polypeptides with specific tyrosine phosphorylation. Following a refolding procedure, the NMR and CD spectroscopic studies of the phosphorylated SAM domains (EphA2.pY921, EphA2.pY930, and EphA2.pY960) demonstrate that the chemically synthesized domains adopt native-like structures that are stable. Our finding that phosphorylation is not accompanied by a large conformational change in the domain structure was initially surprising, given that both Tyr921 and Tyr930 are partially buried. However, both of the tyrosine residues are probably capable of maintaining interactions with the neighboring residues even after phosphorylation. For example, the tyrosine hydroxyl of Tyr921 is exposed to the solvent and makes hydrogen bond contacts with the side chains of the conserved His954 (Fig. 1); the phosphate group of Tyr921 may interact with His954 similarly and help to maintain the overall conformation of the domain. Taken together, our observations establish that the domain-length phosphorylated peptides are a good model system to study the impact of EphA2 SAM phosphorylation on the domain's interaction with other proteins.
Unphosphorylated EphA2 SAM binds SHIP2 SAM (23, 31, 32); phosphorylation might alter the affinity of this interaction. Unexpectedly, ITC measurements show that both the phosphorylated and unphosphorylated EphA2 SAM domains share a similar affinity for SHIP2 SAM. We anticipated an effect with phosphorylation in the case of phosphorylated Tyr921 and Tyr960 because these are located close to the binding interface with SHIP2 SAM. Adding negative charge to the EphA2 interface (which by itself is dominated by positively charged residues) would be expected to weaken binding of the negatively charged SHIP2 SAM interface. However, our recent refinement of the structure of the complex suggests that the complex can sample alternate configurations (23, 40). The equilibrium between these different configurations may be shifted in the EphA2.pY921- and EphA2.pY960-SHIP2 complexes, but assessing this possibility is beyond the scope and interest of the current study. Overall, we can conclude that phosphorylation of the EphA2 SAM domain by itself is not involved in the regulation of EphA2 SAM-SHIP2 SAM domain interactions. However, phosphorylation could affect the interactions of the domain with other proteins, which would influence EphA2-SHIP2 interaction indirectly.
Tyrosine phosphorylation of receptor tyrosine kinases and the subsequent recruitment of Src homology 2 (SH2) domain-containing adaptor proteins is a central event in the signaling (26, 41, 42). Here, we report that the phosphorylated Tyr921 and Tyr930 of EphA2 SAM recruit Grb7 SH2. A 23-residue peptide containing phosphorylated Tyr960 binds Grb7 SH2 just as well as the other two peptides, but surprisingly, the Tyr960-phosphorylated folded domain has no affinity for Grb7 SH2. This observation suggests that binding at this site is conformation-dependent. Grb7 family SH2 domains bind to peptides in extended or hairpin conformations (43); pep.Y960 (and the other short peptides) is unstructured/only very weakly structured by themselves in solution, as indicated by AGADIR prediction (44), and is therefore able to bind the Grb7 SH2. In the folded protein, Tyr960 is located in the helix α5 of the EphA2 SAM domain, which is unlikely to undergo the unfolding that would be required to allow SH2 binding. Thus, protein conformational features can override the binding affinity that unstructured Tyr(P)-containing polypeptides may have for SH2 proteins (43). This is in accordance with observations on other systems (45, 46) and emphasizes the need for caution in the interpretation of data obtained using peptide libraries/protein fragments in the elucidation of cell signaling mechanisms.
Our study of EphA2 SAM and Grb7 SH2 domains should translate to other Eph-like SAM domains because Tyr921 is highly conserved in Eph-like SAM domains. Furthermore, the SAM domain structures and the topology of its interaction/location of the interacting surfaces are similar across Eph-like SAM domains (21). Indeed, our ITC data show that a SHIP2 SAM-derived peptide in which Tyr1213 is phosphorylated (the equivalent of the highly conserved EphA2 Tyr921) also binds to Grb7 SH2 (Table 1). Binding partners specific for SHIP2.pY1213 are yet to be identified in vivo, but proteomics studies have found this tyrosine to be phosphorylated in myelogenous leukemia. Thus, it is likely that phosphorylation of the highly conserved tyrosine (Tyr921 in EphA2) has a similar function throughout the Eph receptor family, by itself maintaining interactions with a SAM domain binding partner and, at the same time, allowing a competition for it with Grb SH2 adaptor proteins.
Although future studies are needed to examine the specificity of SH2 domain binding (e.g. Grb7 versus other family members or other adaptor proteins like Vav and the Nck family), our in vitro study presents an important finding with respect to SH2 binding site selection on EphA SAM domains. In the absence of other binding partners, both Tyr(P)930 and the highly conserved Tyr(P)921 in EphA2 bind the SH2 domain equally well in vitro, but in cells, only binding to Tyr(P)930 was inferred (17). However, we can rationalize this finding in the context of a competition of the SH2 domain with SHIP2 binding near the Tyr(P)921 site. Because SHIP2 binds several other, if not all, EphA SAM domains in the same region (close to Tyr921), the site preference of SH2 binding for the distal Tyr(P)930 site may be common to 8 of the 10 EphA isoforms that have this second tyrosine. Conversely, Grb7 has been reported to bind at the highly conserved tyrosine in EphB1 because EphB family SAM domains may not bind SHIP2 (23).
Because opposite surfaces are involved, the ability of EphA2.pY930 to bind SHIP2 SAM and Grb7 SH2 simultaneously could lead to the formation of extended networks after binding of Grb7 SH2 dimers to the dimerized EphA2 receptor that is still bound to SHIP2 SAM. In addition, SHIP2 SAM is expected to form a homodimer/trimer through a coiled-coil region that is located in the middle of the protein from predictions (47, 48), thus allowing further cross-linking of SHIP2 SAM-bound EphA2 receptors. Continued association with SHIP2 is likely to be important because this interaction has been shown to inhibit EphA2 receptor endocytosis (31). By contrast, Tyr921 is close to the predominant SHIP2 binding site, and our results show that Grb7 and SHIP2 compete with one another for the same binding region on EphA2 SAM.
If indeed the affinities are similar, the level of network formation involving Grb7 SH2 would depend both on SAM domain phosphorylation and on the concentration of Grb7, leading to the proposal of a stepwise mechanism, as shown in Fig. 7. The local concentrations of adaptor proteins of receptor tyrosine kinases are often increased upon receptor activation due to their recruitment by clustered receptors at the plasma membrane (49). Grb7 (and other members of the Grb family) may also be localized to the plasma membrane via their pleckstrin homology domain (50) (not shown in the model). Excess Grb7 would dissociate SHIP2 from EphA2 SAM and may help to release SHIP2 from the membrane, leading to receptor endocytosis and down-regulation. However, the overall system is likely to be complex because, in addition to the EphA2-mediated localization, there are also several other mechanisms for localization of SHIP2 to the membrane (51). A similar model of receptor clustering and signaling has been proposed for the LAT-Grb2-SOS1 system (52), and for another receptor tyrosine kinase, FRGFR2, the concentration of Grb2 also plays a regulatory role (27, 53).
In vivo experiments are required to test our model regarding these differential roles of EphA2 SAM phosphorylation in the context of different cellular concentrations of Grb7 and SHIP2 and the formation of ternary complexes. The present study has established that chemically synthesized, full-length protein domains are valid, if not better, alternatives to proteins expressed using recombinant systems because posttranslational modifications can be introduced fully and in a site-specific manner. This opens up avenues to probe other signaling systems and provide detailed molecular insight into their mechanisms of signaling.
In summary, our study shows that the binding of adaptor protein, Grb7 SH2, to EphA2 SAM is dependent upon the phosphorylation state of specific tyrosine residues of the SAM domain. Further, binding of Grb7 to phosphorylated Tyr930 EphA2 SAM does not affect SHIP2 SAM binding (Fig. 8). By contrast, phosphorylated Tyr921 cannot bind Grb7 and SHIP2 simultaneously.
FIGURE 8.
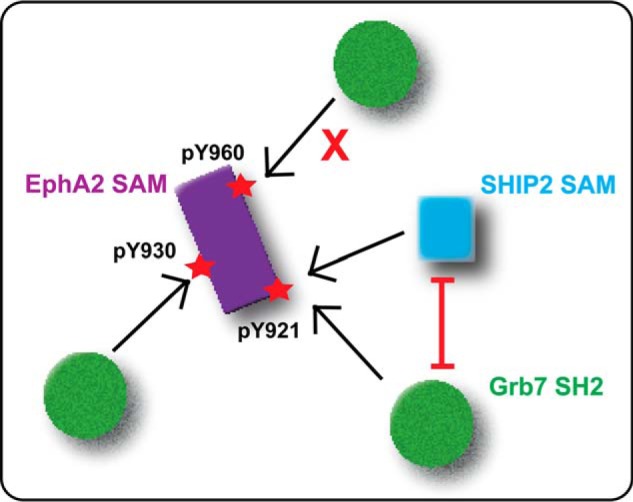
Recruitment of Grb7 SH2 by EphA2 is specific to the phosphorylation of tyrosine residues of the SAM domain. The phosphorylated Tyr930 of the SAM domain of EphA2 can interact with Grb7 SH2 and the SAM domain of SHIP2 simultaneously, whereas Grb7 SH2 and SHIP2 SAM domains compete for the phosphorylated Tyr921. EphA2 SAM phosphorylated at Tyr960 does not bind Grb7 SH2.
Acknowledgment
We thank Prof. Jun-Lin Guan (University of Michigan) for the gift of Grb7 cDNA.
This work was supported, in whole or in part, by National Institutes of Health Grants R01GM092851 and R01CA152371 (to M. B.). This work was also supported by a postdoctoral fellowship from the American Heart Association (to S. B.).
The atomic coordinates and structure factors (code 2KSO) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- SAM
- sterile α motif
- SH2
- Src homology 2
- SHIP2
- SH2 domain-containing inositol-5′-phosphatase
- ITC
- isothermal titration calorimetry.
REFERENCES
- 1. Pawson T. (1995) Protein modules and signaling networks. Nature 373, 573–580 [DOI] [PubMed] [Google Scholar]
- 2. Mochly-Rosen D. (1995) Localization of protein-kinases by anchoring proteins: a theme in signal-transduction. Science 268, 247–251 [DOI] [PubMed] [Google Scholar]
- 3. Schlessinger J. (2000) Cell signaling by receptor tyrosine kinases. Cell 103, 211–225 [DOI] [PubMed] [Google Scholar]
- 4. Pawson T. (2004) Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116, 191–203 [DOI] [PubMed] [Google Scholar]
- 5. Lemmon M. A., Schlessinger J. (2010) Cell signaling by receptor tyrosine kinases. Cell 141, 1117–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kalo M. S., Pasquale E. B. (1999) Signal transfer by Eph receptors. Cell Tissue Res. 298, 1–9 [PubMed] [Google Scholar]
- 7. Singla N., Erdjument-Bromage H., Himanen J. P., Muir T. W., Nikolov D. B. (2011) A semisynthetic Eph receptor tyrosine kinase provides insight into ligand-induced kinase activation. Chem. Biol. 18, 361–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Murai K. K., Pasquale E. B. (2003) “Eph”ective signaling: forward, reverse and crosstalk. J. Cell Sci. 116, 2823–2832 [DOI] [PubMed] [Google Scholar]
- 9. Wang H. U., Chen Z. F., Anderson D. J. (1998) Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell 93, 741–753 [DOI] [PubMed] [Google Scholar]
- 10. Hochgräfe F., Zhang L., O'Toole S. A., Browne B. C., Pinese M., Porta Cubas A., Lehrbach G. M., Croucher D. R., Rickwood D., Boulghourjian A., Shearer R., Nair R., Swarbrick A., Faratian D., Mullen P., Harrison D. J., Biankin A. V., Sutherland R. L., Raftery M. J., Daly R. J. (2010) Tyrosine phosphorylation profiling reveals the signaling network characteristics of basal breast cancer cells. Cancer Res. 70, 9391–9401 [DOI] [PubMed] [Google Scholar]
- 11. Jehle J., Staudacher I., Wiedmann F., Schweizer P., Becker R., Katus H., Thomas D. (2012) Regulation of apoptosis in HL-1 cardiomyocytes by phosphorylation of the receptor tyrosine kinase EphA2 and protection by lithocholic acid. Br. J. Pharmacol. 167, 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Davis S., Gale N. W., Aldrich T. H., Maisonpierre P. C., Lhotak V., Pawson T., Goldfarb M., Yancopoulos G. D. (1994) Ligands for Eph-related receptor tyrosine kinases that require membrane attachment or clustering for activity. Science 266, 816–819 [DOI] [PubMed] [Google Scholar]
- 13. Himanen J. P., Nikolov D. B. (2003) Eph receptors and ephrins. Int. J. Biochem. Cell B 35, 130–134 [DOI] [PubMed] [Google Scholar]
- 14. Wiesner S., Wybenga-Groot L. E., Warner N., Lin H., Pawson T., Forman-Kay J. D., Sicheri F. (2006) A change in conformational dynamics underlies the activation of Eph receptor tyrosine kinases. EMBO J. 25, 4686–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kalo M. S., Yu H. H., Pasquale E. B. (2001) In vivo tyrosine phosphorylation sites of activated ephrin-B1 and EphB2 from neural tissue. J. Biol. Chem. 276, 38940–38948 [DOI] [PubMed] [Google Scholar]
- 16. Kalo M. S., Pasquale E. B. (1999) Multiple in vivo tyrosine phosphorylation sites in EphB receptors. Biochemistry 38, 14396–14408 [DOI] [PubMed] [Google Scholar]
- 17. Fang W. B., Brantley-Sieders D. M., Hwang Y., Ham A. J., Chen J. (2008) Identification and functional analysis of phosphorylated tyrosine residues within EphA2 receptor tyrosine kinase. J. Biol. Chem. 283, 16017–16026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Balasubramaniam D., Paul L. N., Homan K. T., Hall M. C., Stauffacher C. V. (2011) Specificity of HCPTP variants toward EphA2 tyrosines by quantitative selected reaction monitoring. Protein Sci. 20, 1172–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schultz J., Ponting C. P., Hofmann K., Bork P. (1997) SAM as a protein interaction domain involved in developmental regulation. Protein Sci. 6, 249–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lackmann M., Oates A. C., Dottori M., Smith F. M., Do C., Power M., Kravets L., Boyd A. W. (1998) Distinct subdomains of the EphA3 receptor mediate ligand binding and receptor dimerization. J. Biol. Chem. 273, 20228–20237 [DOI] [PubMed] [Google Scholar]
- 21. Ramachander R., Bowie J. U. (2004) SAM domains can utilize similar surfaces for the formation of polymers and closed oligomers. J. Mol. Biol. 342, 1353–1358 [DOI] [PubMed] [Google Scholar]
- 22. Stapleton D., Balan I., Pawson T., Sicheri F. (1999) The crystal structure of an Eph receptor SAM domain reveals a mechanism for modular dimerization. Nat. Struct. Biol. 6, 44–49 [DOI] [PubMed] [Google Scholar]
- 23. Lee H. J., Hota P. K., Chugha P., Guo H., Miao H., Zhang L., Kim S. J., Stetzik L., Wang B. C., Buck M. (2012) NMR structure of a heterodimeric SAM:SAM complex: characterization and manipulation of EphA2 binding reveal new cellular functions of SHIP2. Structure 20, 41–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schlessinger J., Lemmon M. A. (2003) SH2 and PTB domains in tyrosine kinase signaling. Sci. STKE 2003, RE12. [DOI] [PubMed] [Google Scholar]
- 25. Paavilainen S., Grandy D., Karelehto E., Chang E., Susi P., Erdjument-Bromage H., Nikolov D., Himanen J. (2013) High-level expression of a full-length Eph receptor. Protein Expr. Purif. 92, 112–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lee H., Bennett A. M. (2013) Receptor tyrosine phosphatase-receptor tyrosine kinase substrate screen identifies EphA2 as a target for LAR in cell migration. Mol. Cell Biol. 33, 1430–1441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lin C. C., Melo F. A., Ghosh R., Suen K. M., Stagg L. J., Kirkpatrick J., Arold S. T., Ahmed Z., Ladbury J. E. (2012) Inhibition of basal FGF receptor signaling by dimeric Grb2. Cell 149, 1514–1524 [DOI] [PubMed] [Google Scholar]
- 28. Stein E., Cerretti D. P., Daniel T. O. (1996) Ligand activation of ELK receptor tyrosine kinase promotes its association with Grb10 and Grb2 in vascular endothelial cells. J. Biol. Chem. 271, 23588–23593 [DOI] [PubMed] [Google Scholar]
- 29. Han D. C., Shen T. L., Miao H., Wang B., Guan J. L. (2002) EphB1 associates with Grb7 and regulates cell migration. J. Biol. Chem. 277, 45655–45661 [DOI] [PubMed] [Google Scholar]
- 30. Leone M., Cellitti J., Pellecchia M. (2008) NMR Studies of a Heterotypic Sam-Sam domain association: the interaction between the lipid phosphatase Ship2 and the EphA2 receptor. Biochemistry 47, 12721–12728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhuang G., Hunter S., Hwang Y., Chen J. (2007) Regulation of EphA2 receptor endocytosis by SHIP2 lipid phosphatase via phosphatidylinositol 3-kinase-dependent Rac1 activation. J. Biol. Chem. 282, 2683–2694 [DOI] [PubMed] [Google Scholar]
- 32. Kelly S. M., Price N. C. (2000) The use of circular dichroism in the investigation of protein structure and function. Curr. Protein Pept. Sci. 1, 349–384 [DOI] [PubMed] [Google Scholar]
- 33. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) Nmrpipe: a multidimensional spectral processing system based on Unix pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 34. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 35. Porter C. J., Matthews J. M., Mackay J. P., Pursglove S. E., Schmidberger J. W., Leedman P. J., Pero S. C., Krag D. N., Wilce M. C., Wilce J. A. (2007) Grb7 SH2 domain structure and interactions with a cyclic peptide inhibitor of cancer cell migration and proliferation. BMC Struct. Biol. 7, 58–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Peterson T. A., Benallie R. L., Bradford A. M., Pias S. C., Yazzie J., Lor S. N., Haulsee Z. M., Park C. K., Johnson D. L., Rohrschneider L. R., Spuches A., Lyons B. A. (2012) Dimerization in the Grb7 protein. J. Mol. Recognit. 25, 427–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pias S., Peterson T. A., Johnson D. L., Lyons B. A. (2010) The intertwining of structure and function: proposed helix-swapping of the SH2 domain of Grb7, a regulatory protein implicated in cancer progression and inflammation. Crit. Rev. Immonol. 30, 299–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ivancic M., Daly R. J., Lyons B. A. (2003) Solution structure of the human Grb7-SH2 domain/erbB2 peptide complex and structural basis for Grb7 binding to ErbB2. J. Biomol. NMR 27, 205–219 [DOI] [PubMed] [Google Scholar]
- 39. Ivancic M., Spuches A. M., Guth E. C., Daugherty M. A., Wilcox D. E., Lyons B. A. (2005) Backbone nuclear relaxation characteristics and calorimetric investigation of the human Grb7-SH2/erbB2 peptide complex. Protein Sci. 14, 1556–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhang L., Buck M. (2013) Molecular simulations of a dynamic protein complex: role of salt-bridges and polar interactions in configurational transitions. Biophys. J. 105, 2412–2417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stein D., Wu J., Fuqua S. A., Roonprapunt C., Yajnik V., D'Eustachio P., Moskow J. J., Buchberg A. M., Osborne C. K., Margolis B. (1994) The Sh2 domain protein Grb-7 is co-amplified, overexpressed and in a tight complex with Her2 in breast-cancer. EMBO J. 13, 1331–1340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Janes P. W., Lackmann M., Church W. B., Sanderson G. M., Sutherland R. L., Daly R. J. (1997) Structural determinants of the interaction between the erbB2 receptor and the Src homology 2 domain of Grb7. J. Biol. Chem. 272, 8490–8497 [DOI] [PubMed] [Google Scholar]
- 43. Waksman G., Kominos D., Robertson S. C., Pant N., Baltimore D., Birge R. B., Cowburn D., Hanafusa H., Mayer B. J., Overduin M., Resh M. D., Rios C. B., Silverman L., Kuriyan J. (1992) Crystal structure of the phosphotyrosine recognition domain SH2 of v-src complexed with tyrosine-phosphorylated peptides. Nature 358, 646–653 [DOI] [PubMed] [Google Scholar]
- 44. Muñoz V., Serrano L. (1994) Elucidating the folding problem of helical peptides using empirical parameters. Nat. Struct. Biol. 1, 399–409 [DOI] [PubMed] [Google Scholar]
- 45. Torres E., Rosen M. K. (2003) Contingent phosphorylation/dephosphorylation provides a mechanism of molecular memory in WASP. Mol. Cell 11, 1215–1227 [DOI] [PubMed] [Google Scholar]
- 46. Torres E., Rosen M. K. (2006) Protein-tyrosine kinase and GTPase signals cooperate to phosphorylate and activate Wiskott-Aldrich syndrome protein (WASP)/neuronal WASP. J. Biol. Chem. 281, 3513–3520 [DOI] [PubMed] [Google Scholar]
- 47. Armstrong C. T., Vincent T. L., Green P. J., Woolfson D. N. (2011) SCORER 2.0: an algorithm for distinguishing parallel dimeric and trimeric coiled-coil sequences. Bioinformatics 27, 1908–1914 [DOI] [PubMed] [Google Scholar]
- 48. McDonnell A. V., Jiang T., Keating A. E., Berger B. (2006) Paircoil2: improved prediction of coiled coils from sequence. Bioinformatics 22, 356–358 [DOI] [PubMed] [Google Scholar]
- 49. Kholodenko B. N., Hoek J. B., Westerhoff H. V. (2000) Why cytoplasmic signalling proteins should be recruited to cell membranes. Trends Cell Biol. 10, 173–178 [DOI] [PubMed] [Google Scholar]
- 50. Depetris R. S., Wu J., Hubbard S. R. (2009) Structural and functional studies of the Ras-associating and pleckstrin-homology domains of Grb10 and Grb14. Nat. Struct. Mol. Biol. 16, 833–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Edimo W. E., Janssens V., Waelkens E., Erneux C. (2012) Reversible Ser/Thr SHIP phosphorylation: a new paradigm in phosphoinositide signalling?: targeting of SHIP1/2 phosphatases may be controlled by phosphorylation on Ser and Thr residues. Bioessays 34, 634–642 [DOI] [PubMed] [Google Scholar]
- 52. Nag A., Monine M. I., Faeder J. R., Goldstein B. (2009) Aggregation of membrane proteins by cytosolic cross-linkers: theory and simulation of the LAT-Grb2-SOS1 system. Biophys. J. 96, 2604–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Timsah Z., Ahmed Z., Lin C. C., Melo F. A., Stagg L. J., Leonard P. G., Jeyabal P., Berrout J., O'Neil R. G., Bogdanov M., Ladbury J. E. (2014) Competition between Grb2 and Plcγ1 for FGFR2 regulates basal phospholipase activity and invasion. Nat. Struct. Mol. Biol. 21, 180–188 [DOI] [PubMed] [Google Scholar]