Background: Abl family kinases bind different targets despite highly similar sequences.
Results: Two residues in the Src homology (SH) 2 domain regulate binding to phosphorylated cortactin and modulate cell protrusion.
Conclusion: The Arg SH2 domain binds with higher affinity than the Abl SH2 domain to phosphorylated cortactin.
Significance: Slight sequence changes can cause affinity differences, leading to important functional changes in cellular interactions.
Keywords: Actin, Crystal Structure, Phosphotyrosine, Src Homology 2 Domain (SH2 Domain), Tyrosine-Protein Kinase (Tyrosine Kinase)
Abstract
The closely related Abl family kinases, Arg and Abl, play important non-redundant roles in the regulation of cell morphogenesis and motility. Despite similar N-terminal sequences, Arg and Abl interact with different substrates and binding partners with varying affinities. This selectivity may be due to slight differences in amino acid sequence leading to differential interactions with target proteins. We report that the Arg Src homology (SH) 2 domain binds two specific phosphotyrosines on cortactin, a known Abl/Arg substrate, with over 10-fold higher affinity than the Abl SH2 domain. We show that this significant affinity difference is due to the substitution of arginine 161 and serine 187 in Abl to leucine 207 and threonine 233 in Arg, respectively. We constructed Abl SH2 domains with R161L and S187T mutations alone and in combination and find that these substitutions are sufficient to convert the low affinity Abl SH2 domain to a higher affinity “Arg-like” SH2 domain in binding to a phospho-cortactin peptide. We crystallized the Arg SH2 domain for structural comparison to existing crystal structures of the Abl SH2 domain. We show that these two residues are important determinants of Arg and Abl SH2 domain binding specificity. Finally, we expressed Arg containing an “Abl-like” low affinity mutant Arg SH2 domain (L207R/T233S) and find that this mutant, although properly localized to the cell periphery, does not support wild type levels of cell edge protrusion. Together, these observations indicate that these two amino acid positions confer different binding affinities and cellular functions on the distinct Abl family kinases.
Introduction
Abelson (Abl)7 family non-receptor tyrosine kinases are important regulators of actin dynamics and coordinate changes in cell morphology and migration (1). The N-terminal halves of Abl and the Abl-related gene (Arg), containing their Src homology 3 (SH3), Src homology 2 (SH2), and kinase domains, are nearly 90% identical, whereas their C-terminal extensions, which contain cytoskeletal-binding domains and scaffolding functions, are much less conserved (1, 2). Despite sequence similarities and known roles in actin cytoskeletal regulation, Abl and Arg regulate differential zones of actomyosin contractility in fibroblasts (3). Moreover, Abl and Arg have differential roles in neuronal dendrite maintenance and in the support of invadopodial function in invasive breast cancer cells (4, 5).
In addition to their differences in expression, subcellular localization, and function, Arg and Abl interact with different substrates in vivo (3, 6–9). For example, we have previously shown that whereas both Abl and Arg phosphorylate the actin polymerization regulator cortactin in vitro, Arg alone interacts with cortactin via a set of scaffolding interactions that are essential for both proteins to promote cell edge protrusion during cell adhesion (10, 11). Furthermore, Arg, but not Abl, is critical for cortactin phosphorylation during actin-based protrusion in breast cancer cells (5). This selectivity can be explained in part by the high-affinity binding of the cortactin SH3 domain to an extended Pro-X-X-Pro-X-X-Pro motif that occurs in Arg but not Abl (10). A recent crystal structure suggests that the cortactin SH3 domain engages the first three of these prolines along an extended type II polyproline helix (12).
During cell adhesion, cortactin SH3 domain binding to Arg is essential for subsequent phosphorylation of cortactin by Arg at one or more of three previously identified tyrosine phosphory10, 13, 14). This phosphorylation creates a binding site for the Arg SH2 domain, a discrete second interaction interface between the two proteins, although the exact phosphotyrosines required for binding were previously unknown (15). This finding raised the question of whether the Arg SH2 domain might also confer additional specificity to the Arg-cortactin interactions. SH2 domains bind phosphotyrosines with a high degree of selectivity, which is dictated by surrounding residues. In particular, three residues C-terminal to the phosphotyrosine determine SH2 domain binding specificity (16, 17). Within the SH2 domain itself, there are three binding pockets that confer specificity for each of these binding target residues as well as several loops, which govern binding pocket accessibility (18, 19).
We report here that of the three previously described cortactin tyrosine phosphorylation sites (Tyr421, Tyr466, Tyr482), only peptides corresponding to the phosphorylated Tyr421 and Tyr466 sites bind with significant affinities to the Arg SH2 domain. We also find that despite nearly 90% domain sequence identity, the Arg SH2 domain binds the phosphorylated cortactin Tyr421 peptide with over 10-fold greater affinity than the Abl SH2 domain. We show that this binding specificity is due to two residues that differ between Abl and Arg, arginine 161 and serine 187 in Abl, which correspond to leucine 207 and threonine 233 in Arg, respectively (Fig. 2A). We provide the first crystal structure of the Arg SH2 domain and show that Leu207 is located near the P + 1 binding pocket and Thr233 is on the EF loop previously shown to mediate access to the P + 3 binding pocket (18). Finally, we show that although these residues do not affect Arg localization to the cell periphery, they are critical for mediating actin-based cell edge protrusion in fibroblasts.
FIGURE 2.
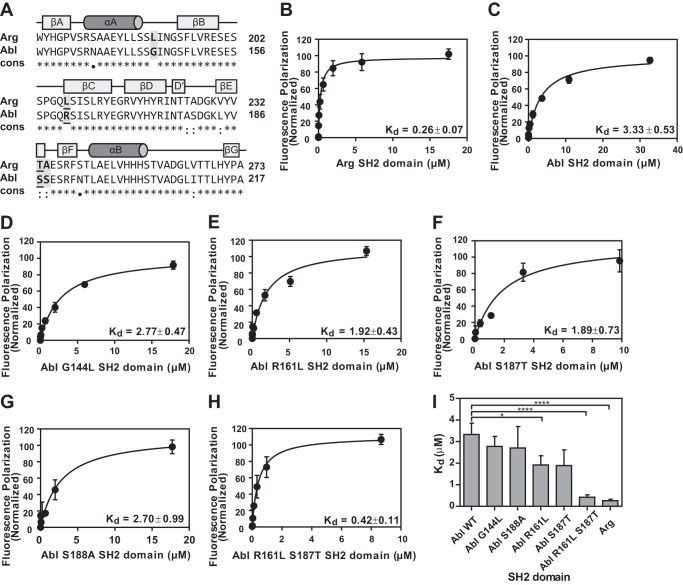
Sequence alignment and binding analysis of SH2 domains for cortactin Tyr(P)421 peptide. A, sequence alignment of Arg SH2 and Abl SH2 domains using ClustalW. ClustalW conservation: *, identical; :, conserved; ●, semi-conserved. The secondary structure elements are assigned based on the Arg SH2 crystal structure with β-strands indicated as boxes, and α-helices as cylinders. Residues mutated in this study are highlighted with a gray box and the two residues important for binding affinity (Leu207/Arg161 and Thr233/Ser187) are underlined. B–H, quantification of binding between Arg SH2 domain (B), Abl SH2 domain (C), Abl G144L SH2 domain (D), Abl R161L SH2 domain (E), Abl S187T SH2 domain (F), Abl S188A SH2 domain (G), or Abl R161L/S187T SH2 domain (H) and cortactin Tyr(P)421 phosphopeptide. Experiments were performed in triplicate and points represent mean ± S.E. (error bars may be obscured by data points). Data were fit using a one-site specific binding curve (GraphPad Prism) normalized to saturation (Bmax). All binding affinities (Kd) are given in micromolar. I, statistical analysis of binding affinities determined from binding curves in GraphPad Prism. Values are mean ± S.E., *, p < 0.1; **, p < 0.01.
EXPERIMENTAL PROCEDURES
Molecular Cloning of Recombinant SH2 Domains
Human Arg (Val131–Tyr238) and Abl (Asn121–Val226) SH2 domains were subcloned into pGEX6p-1 (GE Healthcare) using BamHI and EcoRI restriction sites. Abl SH2 G144L, R161L/S187T, and S188A single and double mutants were generated by PCR and also cloned into pGEX6p-1. Arg L207R/T233S-EYFP was generated using site-directed mutagenesis of wild type Arg N1-EYFP (Clontech Laboratories, Inc.). EYFP, wild type Arg EYFP, and Arg L207R/T233S-EYFP were subcloned into the retroviral expression vector PK1 (10).
Protein Purification for Crystallization
Initial protein expression and purification were carried out in Escherichia coli BL21(DE3) cells and induced at 20 °C overnight with 0.1 mm isopropyl β-d-1-thiogalactopyranoside. After induction, the cells were pelleted, resuspended, and lysed using a freeze-thaw protocol and sonication with the presence of 1 mg/ml of lysozyme. Following sonication and centrifugation (20,000 × g) at 4 °C for 30 min, the resulting supernatant was incubated with glutathione-Sepharose 4B beads (GE Healthcare) for 4 h at 4 °C. 20 bead volumes of PBS buffer were then used to wash the resin before being incubated with PreScission protease (GE Healthcare) at 4 °C overnight to remove the GST tag. The Arg SH2 protein was further purified on a Superdex 75 column in 20 mm Hepes (pH 7.5) buffer containing 100 mm NaCl, 1 mm EDTA, 1 mm DTT. The protein was concentrated to 20 mg/ml for crystallization.
Peptide Synthesis and 5-Carboxyfluorescein Labeling
Peptides corresponding to phosphorylated tyrosine sites 421 (SSPIpYEDAA), 466 (SEPVpYETTE), or 482 (EDDTpYDGYE) of mouse cortactin were synthesized on a 30-μmol scale with a Liberty 12-channel microwave synthesizer (CEM Corp., Matthews, NC) using standard Fmoc chemistry. Following removal of the final Fmoc protecting group, the resin was washed with dimethylformamide and methylene chloride alternatively for a total of 16 washes and dried for 20 min under nitrogen gas. The peptides were N terminally labeled overnight with a mixture comprised of 5-carboxyfluorescein, succinimidyl ester (8 mg, 0.017 mmol), and diisopropylethylamine (24 μl, 17.8 mg, 0.138 mmol) in 1.5 ml of dimethylformamide.
Labeled peptide was treated with a cleavage mixture of 2.5% (v/v) 3,6-dioxa-1,8-octanedithiol, 2.5% (v/v) H2O, 2.5% (v/v) triisopropylsilyl in TFA (50% power at 400 W maximum, 38 °C ramp for 2 min, hold for 30 min; followed by treatment with fresh mixture at 50% power, 400 W maximum, 38 °C, ramp 2 min, hold 5 min). Crude peptide was lyophilized and reconstituted in acetonitrile/water (1:1). Synthesis efficiency was assessed by MALDI-TOF analysis of the crude reaction mixture followed by purification to homogeneity by reverse phase HPLC. A second round of MALDI-TOF reconfirmed peak sample homogeneity. Following purification, peptide was lyophilized and stored in the dark at −20 °C.
Fluorescence Polarization Measurements
SH2 domains were dialyzed into binding buffer (50 mm Hepes, pH 7.25, 150 mm NaCl, 0.01% Nonidet P-40, 5% glycerol) and a 30-μl volume serially diluted into pre-chilled 384-well low flange black flat bottom non-binding microplates (Corning) while on ice. 5 μl of 35 nm fluorescent peptide dissolved in binding buffer was added to each well (final concentration 5 nm), mixed 5 times by pipetting, and plates were incubated at 4 °C for 40 min. Fluorescence polarization experiments were performed with an Analyst AD (Molecular Devices, Sunnyvale, CA) spectrofluorimeter. Each well was excited using 485 nm light and emission was read at 530 nm. Fluorescence polarization was measured 1 mm from the bottom of each well with an integration time of 560 ms. All experiments were conducted in triplicate. The change in fluorescence polarization was normalized such that maximum change for each condition (Bmax) was set at an arbitrary value of 100 for ease of comparison (Figs. 1 and 2). Normalized values were graphed and fit to a single binding site hyperbola using GraphPad Prism.
FIGURE 1.
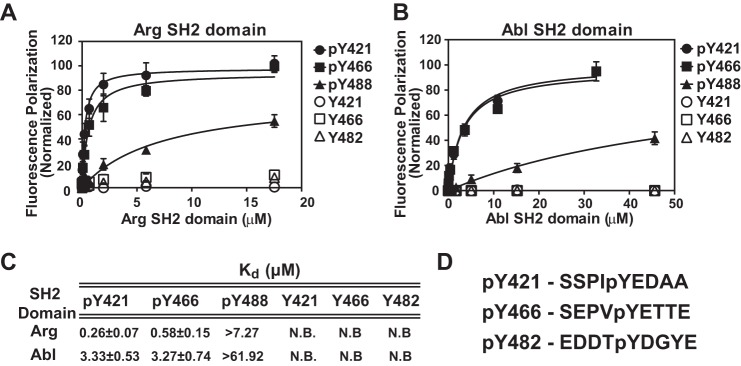
Arg and Abl SH2 domains bind phosphorylated cortactin peptides Tyr(P)421 and Tyr(P)466, but not Tyr(P)482. Quantification of binding between Arg (A) and Abl (B) SH2 domains and peptides corresponding to phosphorylated (pY) and non-phosphorylated (Y) cortactin tyrosines 421, 466, and 482. Experiments were performed in triplicate and points represent mean ± S.E. (error bars may be obscured by data points). Data were fit using a one-site specific binding curve (GraphPad Prism) normalized to saturation (Bmax). C, table of binding affinities and mean ± S.E. from binding curves in A and B, peptides that showed no binding are labeled as N.B. D, sequences of cortactin peptides used in A and B.
Crystallization and Structure Determination
Arg SH2 domain crystals were grown in conditions containing 15–17% PEG 4000, 0.2 m sodium acetate, and 100 mm Tris-HCl, pH 8.5. Crystals were grown within 3 days at room temperature by hanging drop vapor diffusion. Before data collection, the crystals were flash frozen in liquid nitrogen using the crystallization conditions supplemented with 20% PEG 400. Crystallographic data were collected at beamline X6A of the National Synchrotron Light Source (Table 1), and data were processed using HKL2000 (20).
TABLE 1.
Crystallographic data collection and refinement statistics
| Data collection | |
| Space group | C2221 |
| Cell dimensions, a, b, c (Å) | 56.3, 81.9, 37.7 |
| Resolution (Å)a | 20.0-1.2 (1.24-1.20) |
| Unique reflectionsa | 25,814 |
| Completeness (%) | 93.9 (63.9) |
| Rsym (%)a | 4.6 (23.7) |
| Mn(I/σ)a | 29.0 (5.3) |
| Rfactor (%) | 16.1 |
| Free Rfactor (%) | 18.1 |
| Residues built | 167–267 |
| Free R reflections (%) | 5 |
| Free R reflections No. | 1,302 |
| No. non-hydrogen protein atoms | 819 |
| No. water molecules | 100 |
| Model quality | |
| Root mean square deviation bond length (Å) | 0.014 |
| Root mean square deviation bond angles (°) | 1.54 |
| Mean B-factors | |
| Overall (Å2) | 29.6 |
| Protein atoms (Å2) | 23.8 |
| Water (Å2) | 47.8 |
| Solvent (Å2) | 22.7 |
| Ramachandran plot (%) favored/allowed/disallowed | 94.4/5.6/0 |
a Parentheses indicate the highest resolution shell.
The structure of the Arg SH2 domain was determined by molecular replacement using the program PHASER (21). The SH2 domain from Abl (Protein Data Bank code 3K2M (22)) was used as a search model and yielded a translation Z-score of 17.2. Automated model building using ARP/wARP (23) built 95 residues, thus avoiding potential problems with model bias. Manual model building and refinement were then conducted using COOT (24) and Refmac5 (25) from the CCP4 suite, respectively. Restrained refinement with anisotropic B-factors and automatic water placement with ARP_waters (26) were used in later rounds of refinement. A total of 100 water molecules were added during refinement. The final structure was validated using PROCHECK (27) and MolProbity (28) and the secondary structure was assigned using the computer program, DSSP (29). All residues fall within the favored or allowed regions of the Ramachandran plot. Arg SH2 maintains a typical SH2-fold with 2 major α-helices (αA and αB) and 7 β-strands (βA to βG). The structure is deposited in the Protein Data Bank under accession code 4EIH.
Cell Culture and Retroviral Expression
Wild type and arg−/− fibroblasts were maintained as previously described (30). EYFP, Arg-EYFP, and Arg L207R/T233S-EYFP were expressed via retroviral expression and 1.0 μg/ml of puromycin selection as previously described (10). Expression levels of wild type and retrovirally expressed Arg proteins were determined by immunoblotting 75 μg of cell lysate as determined with a BCA kit (Pierce) with a mouse anti-Arg antibody and control mouse monoclonal antibody against HSP70 (3A3) (Santa Cruz). Expression levels were quantified using Quantity One software (Bio-Rad).
Immunoprecipitation and Western Blotting
Fibroblasts were serum starved for 1 h then plated on 10 μg/ml of bovine fibronectin-coated plates (Sigma), blocked with 1% BSA, and allowed to adhere for 45 min. Cells were washed with PBS and lysed in 50 mm Hepes, pH 7.5, 1% Nonidet P-40, 150 mm KCl, 5% glycerol, 0.5 mm metavanadate, 0.5 mm orthovanadate, 2 mm NaF, and complete protease inhibitors. 0.75 mg/ml of soluble lysate was incubated with protein A/G beads and anti-cortactin 4F11 antibody for 24 h at 4 °C. Supernatant and precipitant samples were immunoblotted with anti-phosphotyrosine 4G10 antibody and anti-cortactin 4F11 antibody. Phosphorylation levels were quantified and normalized to total cortactin levels using Quantity One software (Bio-Rad).
Immunofluorescence Microscopy
Fibroblasts were plated on 10 μg/ml of bovine fibronectin-coated coverslips (Sigma), blocked with 1% BSA, and allowed to adhere for 30 min. Cells were fixed with 4% PFA for 20 min at room temperature and permeabilized with 0.5% Triton X-100 for 10 min. Cells were then stained with a mouse anti-GFP monoclonal antibody (Rockland), which cross-reacts with EYFP, followed with Alexa Fluor 647 goat anti-mouse antibody (Invitrogen), as well as Alexa Fluor 488 phalloidin (Invitrogen). Cells were imaged on a model TE2000-S Nikon microscope with a ×20 or 100 objective with NIS Elements (Nikon).
To control for possible bleed-through of the Arg/Arg mutant-YFP signal into the actin channel, arg−/− fibroblasts expressing EYFP were stained only with antibodies to EYFP and Alexa 647 goat anti-mouse secondary antibody or only with phalloidin-Alexa 488 alone and were imaged at both 501 and 654 nm. No bleed-through of the Arg/Arg mutant-YFP signal was observed in the 501 channel, and no actin staining was observed in the 654 nm channel. These experiments indicate that the EYFP signal from the expressed protein is disrupted during fixation and processing.
Quantification of Cell Circularity
Cells stained for phalloidin and imaged at ×100 were traced peripherally in ImageJ. Circularity was quantified using the analyze shape descriptors function in the ImageJ Software package, which can be described as a function of cell area and cell perimeter.
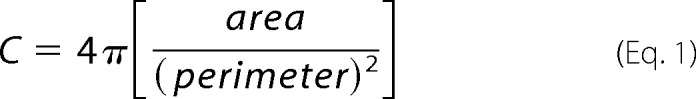 |
Adhesion-dependent Cell Edge Protrusion Assays and Kymography
Adhesion-dependent fibroblast cell edge protrusion assays were performed 30 min after cells were plated on 10 μg/ml of bovine fibronectin (Sigma)-coated MatTek dishes (MatTek Corp.) and imaged as described previously (10). Kymographic analysis of cell edge protrusion was performed as described previously using ImageJ software and GraphPad Prism (10, 30).
Measurements of Cell Spreading
Fibroblasts were plated on 10 μg/ml of bovine fibronectin (Sigma)-coated MatTek dishes (MatTek Corp.). Time-lapse images of spreading cells were recorded for 30 min. The cell area was measured at 0, 10, 20, and 30 min by peripherally tracing and quantifying each cell in ImageJ.
Statistics
Unless otherwise noted, grouped samples were subjected to two-way analysis of variance followed by post-hoc Student's t tests where described (* = p < 0.05; ** = p < 0.01; *** = p < 0.001; **** = p < 0.0001). All graphs display mean ± S.E.
RESULTS
Arg and Abl Bind Cortactin Phosphotyrosines 421 and 466, but Not 482
We used fluorescence anisotropy to measure the binding affinity of wild type Abl and Arg SH2 domains for fluoresceinated cortactin peptides corresponding to the sequences surrounding phosphorylated Tyr421, Tyr466, and Tyr482 (Fig. 1D). We found that the Arg SH2 domain binds with highest affinity to the Tyr(P)421 peptide, binds with a 2-fold lower affinity to Tyr(P)466 peptide, and does not bind detectably to Tyr(P)482 peptide (Fig. 1, A and C). The Abl SH2 domain showed a similar trend in binding affinity for the three phosphorylated cortactin peptides (Fig. 1, B and C). Neither SH2 domain binds unphosphorylated cortactin peptides with any detectable affinity.
Arg and Abl SH2 Domains Differ in Only a Few Non-conserved Amino Acids
Despite nearly identical sequences, the Arg SH2 domain shows a greater than 10-fold higher affinity in comparison to the Abl SH2 domain for the cortactin Tyr(P)421 peptide (Fig. 1). ClustalW was used to perform sequence alignment of Arg and Abl SH2 domains. These two SH2 domain sequences are 90% identical, with two non-conserved residues, two semi-conserved residues, and six conserved residues (Fig. 2A). Of these 10 residues, three are located in regions within or near the binding pocket region of the SH2 domain (Fig. 3A). These residues are Leu207, Thr233, and Ala234 in Arg, which correspond to Arg161, Ser187, and Ser188 in Abl. Leu190 in Arg and the corresponding residue in Abl, Gly144, are also non-conserved but are located outside of the binding pocket and we therefore predicted this substitution would have little effect on the binding affinity.
FIGURE 3.
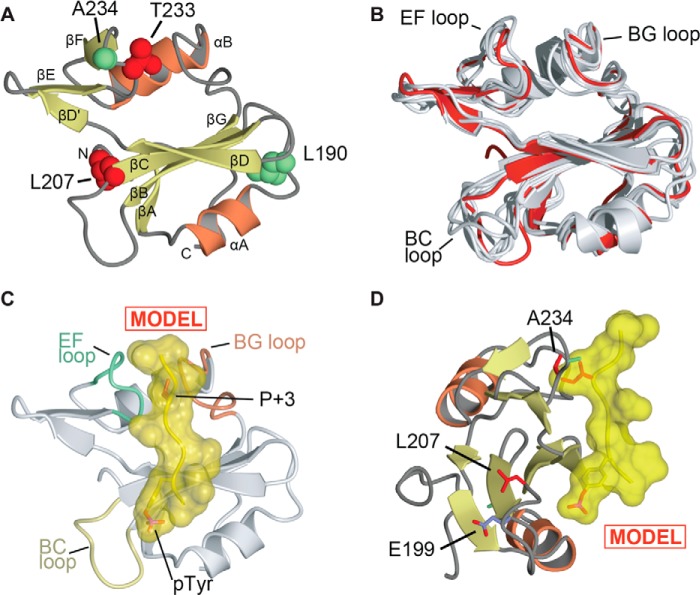
Structural analysis of Arg SH2 domain. A, overall view of the crystal structure of the Arg SH2 domain. α-Helices and β-strands are labeled. Arg residues Leu190 and Ala234 are shown as spheres and colored green. Arg residues Leu207 and Thr233 are shown as spheres and colored red. The structure is deposited in the Protein Data Bank under accession code 4EIH. B, superposition of determined structures of Arg and Abl SH2 domains. Arg SH2 domain (this study) is colored red, previously determined structures are shown in gray (PDB codes 1OPK, 1OPL, 1AB2, 2ABL, 2ECD, 2FO0, and 3K2M) (22, 31–34). The superposition was conducted using Topp (25). C and D, model of phosphotyrosine peptide binding to Arg SH2 domain. The model was generated by superposition of Arg SH2 with the Lck SH2-peptide (EPQpYEEIPIYL) complex (35). C, shows only the Arg SH2 domain and the location of the superposed peptide (in yellow). Structural diagrams generated using CCP4MG (48). D, shows an orientation rotated 90° with Arg residues Leu207, Thr233, and Glu199 shown in stick format.
Two Amino Acid Residues in the Arg and Abl SH2 Domains Confer Different Binding Affinities for a Phosphorylated Cortactin Peptide
We measured the binding affinity of Abl SH2 mutants with substitutions at the residues that differ from the Arg SH2 domain for the cortactin Tyr(P)421 peptide. The Abl SH2 single mutants, G144L and S188A, showed no change in affinity from that of wild type Abl SH2 domain (Fig. 2, C, D, G, and I). However, both Abl SH2 R161L and S187T showed intermediate affinities for the Tyr(P)421 peptide (Fig. 2, E, F, and I). In light of this partial increase in affinity, we hypothesized that the double mutation (R161L/S187T) of the Abl SH2 domain may confer the strong affinity observed in the wild type Arg SH2 domain. As hypothesized, the Abl double mutant (R161L/S187T) SH2 domain is able to bind the cortactin Tyr(P)421 peptide with similar affinity to that observed for the wild type Arg SH2 domain (Fig. 2, B, H, and I). These data suggest that these two residues determine the relative binding affinities of Arg and Abl SH2 domains for the Tyr(P)421 cortactin peptide.
Structural Analysis of Arg SH2 Domain
The structure of the Arg SH2 domain has never been solved crystallographically. In light of our affinity measurements (Fig. 2) we wondered whether there were structural explanations for both Arg SH2 domain specificity for Tyr(P)421 and Tyr(P)466 over Tyr(P)482, and the higher affinity of Arg SH2 domain for cortactin when compared with Abl SH2 domain. We therefore conducted a structural analysis of the Arg SH2 domain. We first purified the Arg SH2 domain and crystallized this domain in space group C2221. We then determined the structure of the Arg SH2 domain to 1.2-Å resolution (Fig. 3A, Table 1). This allowed us to directly compare the Arg SH2 domain to the Abl SH2 domain (22, 31–34) and other previously determined type IA SH2 domain structures (35–39).
First, we compared the structure of the Arg SH2 domain with previously determined Abl crystal structures as well as an NMR structure of Arg SH2 domain (PDB codes 1OPK, 1OPL, 1AB2, 2ABL, 2ECD, 2FO0, and 3K2M) (22, 31–34). The crystal structure of the Arg SH2 domain shows root mean square deviations for Cα atoms with these structures ranging from 0.75 to 1.55 Å (Fig. 3B). Overall these structures, including the NMR structure of the Arg SH2 domain, are extremely similar, however, there is distinct conformational flexibility for the BC loop, which is observed in conformations that vary by as much as 9 Å.
We next compared the Arg SH2 domain to other type 1A SH2 domain structures that have been determined in complex with phosphotyrosine peptides. These structures (e.g. for Lck (35, 40) and Src (39)) show that the peptide specificity is determined at the P + 3 location by a Tyr(P)[−][−]Ψ motif, where Ψ is a hydrophobic residue. This binding site is maintained in the Arg SH2 domain by formation of the phosphotyrosine binding pocket by residues Arg180, Arg198, Gln206, Ser208, Tyr218, His219, Tyr220, and Arg221, and formation of the P + 3 hydrophobic binding pocket by residues Val232, Thr233, Gly254, and Leu255. We conducted structural modeling by superposition of the Arg SH2 domain with the structure of Lck bound to an 11-residue phosphopeptide (EPQpYEEIPIYL) derived from the hamster polyoma middle-T antigen (PDB 1LCJ) (35) (Fig. 3C). The structural superposition clearly shows that a large residue at the P + 3 location would be incompatible with binding. This correlates well with the undetectable binding for the cortactin Tyr(P)482 peptide, which has a tyrosine residue in the P + 3 location as compared with the Tyr(P)421 and Tyr(P)466 peptides (Fig. 1), which have alanine and threonine residues, respectively.
We then analyzed the locations of the residues that diverge between the Arg and Abl SH2 domains. We found that Arg Leu190/Abl Gly144 residues are distal from the peptide binding cleft correlating well with our data showing that mutagenesis of these residues does not alter peptide binding specificity. Thr233 is located in the Arg kinase EF loop and is proximal to the P + 3 peptide binding location, consistent with our data showing that the amino acid identity at this position affects peptide binding affinity. Notably, Leu207 is part of the BC loop, but points away from the phosphotyrosine binding site. In structures of the Abl SH2 domain, the corresponding residue, Arg161, can make a salt bridge to Glu153, potentially impacting the conformation of the BC loop. However, in the Arg SH2 domain structure the presence of Leu207 in this location results in a loss of the salt bridge and an orientation change in the side chain of the corresponding Glu199 (Fig. 3D). We hypothesize that for the Abl double mutant (R161L/S187T), the loss of this salt bridge formation and potential alteration in BC loop conformation combined with an altered P + 3 pocket work in concert to allow increased binding affinity toward phosphorylated cortactin.
Wild Type Arg-EYFP and Arg L207R/T233S-EYFP Both Localize to the Cell Periphery
Wild type fibroblasts display dynamic cell edge protrusions as they adhere to fibronectin-coated surfaces (10, 30, 41). We have shown that the Arg SH2 domain binding to phosphorylated cortactin is critical for this process (10). To test if this high affinity interaction supported by residues Leu207 and Thr233 is relevant in cells we retrovirally expressed wild type Arg-EYFP or Arg L207R/T233S-EYFP in mouse arg−/− fibroblasts (Fig. 4, A–C). Cells fixed and stained for actin, and EYFP reveal that localization of Arg L207R/T233S-EYFP is similar to that of wild type Arg-EYFP in arg−/− fibroblasts and is enriched at the cell periphery (Fig. 4E), suggesting that these residues do not impact localization within the cell. Control experiments (described under “Experimental Procedures”) indicated that channel bleed-through of the Arg/Arg mutant and actin staining was not an issue (Fig. 4F).
FIGURE 4.
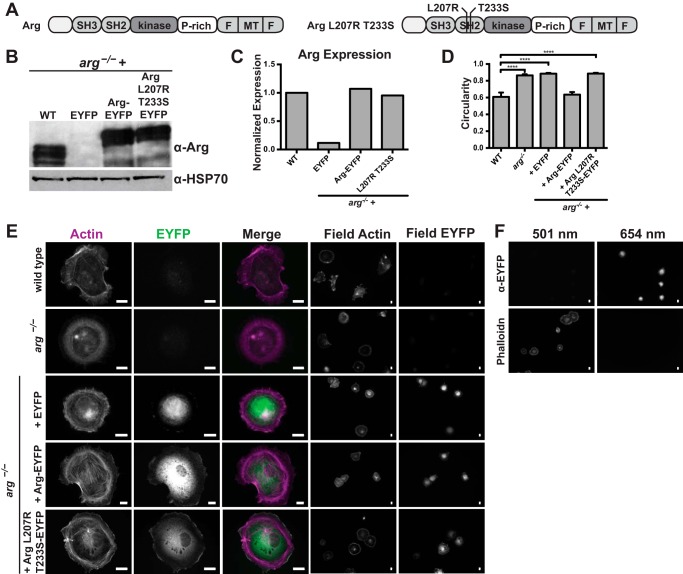
Arg-EYFP and Arg L207R/T233S-EYFP localize to the cell periphery. A, domain structure of full-length wild type Arg and mutant Arg L207R/T233S. B, immunoblot of cell lysates from wild type fibroblasts and arg−/− fibroblasts expressing EYFP, wild type Arg-EYFP, or Arg L207R/T233S-EYFP. Western blot with α-Arg antibody shows Arg expression levels compared with the control α-HSP70 blot. C, quantification of Arg expression from B. D, circularity of cells quantified from phalloidin-stained fibroblasts plated on fibronectin as shown in E. Values are mean ± S.E. for a minimum 15 cells per cell type, ****, p < 0.0001. E, immunostaining of wild type fibroblasts, arg−/− fibroblasts, and arg−/− fibroblasts expressing EYFP, wild type Arg-EYFP, or Arg L207R/T233S-EYFP for actin (first column) and EYFP (second column). Merged images (third column) show colocalization. The fourth and fifth columns show field images of multiple cells immunostained for actin and EYFP, respectively. F, control for channel bleed-through arg−/− fibroblasts expressing EYFP stained only with antibodies to EYFP and Alexa 647 secondary antibodies (top row) or with phalloidin-Alexa 488 alone (bottom row) and imaged at both 501 (left column) and 654 (right column) nm. Scale bar indicates 10 μm.
Overall morphology, which we quantified as cell circularity, shows similarity in cell shape between wild type fibroblasts and arg−/− fibroblasts expressing wild type Arg-EYFP. Cells expressing mutant Arg L207R/T233S-EYFP appear similar in circularity to arg−/− fibroblasts and arg−/− fibroblasts expressing EYFP, suggesting that despite its proper localization, the mutant Arg does not support normal cell morphology (Fig. 4D).
Arg Residues Leu207 and Thr233 Support Wild Type Level Cell Edge Protrusion Fibroblasts
Wild type fibroblasts undergo dynamic cell edge protrusions as they adhere and spread on fibronectin-coated glass and Arg is critical for this behavior (10, 30, 41). To test if residues Leu207 and Thr233 are required to support adhesion-dependent cell edge protrusion, we recorded time-lapse images of wild type fibroblasts and arg−/− fibroblasts expressing EYFP, wild type Arg-EYFP, and Arg L207R/T233S-EYFP adhering to fibronectin-coated coverslips (Fig. 5). We quantified cell edge protrusive behavior using kymography as previously described (30) and find that wild type fibroblasts and arg−/− fibroblasts expressing wild type Arg-EYFP exhibit a high number of protrusions and retractions (Fig. 5, A, C, E, and F). In contrast, arg−/− fibroblasts expressing EYFP alone or expressing mutant Arg L207R/T233S-EYFP show a significant reduction in the number of protrusions and retractions (Fig. 5, B, D, E, and F), confirming that these residues are important for cell edge protrusion in fibroblasts. We do not see a significant difference in the overall spreading area of cells, indicating that whereas dynamic cell edge protrusions are disrupted, overall area of the spread is not affected (Fig. 5H).
FIGURE 5.
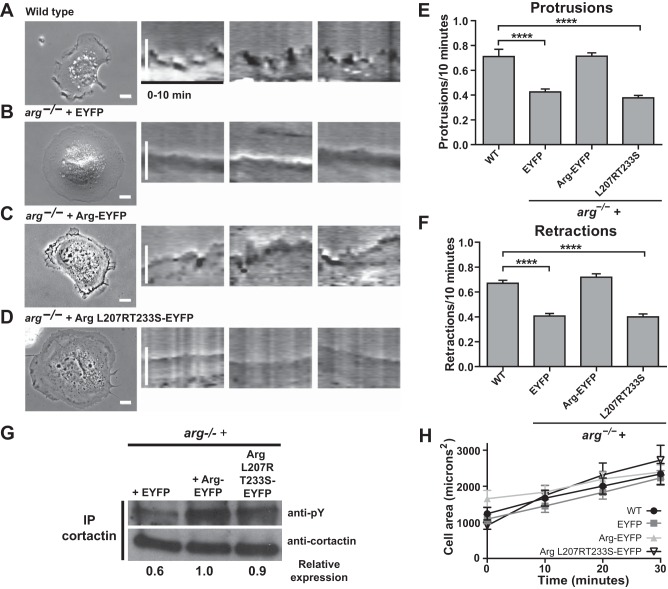
Arg L207R/T233S-EYFP is unable to rescue cell edge protrusion in fibroblasts adhering to fibronectin. A–D, phase images and 10-min kymographs of wild type fibroblasts (A) and arg−/− fibroblasts expressing EYFP (B), wild type Arg-EYFP (C), or Arg L207R/T233S-EYFP (D). Scale bars indicate 10 μm. Quantification of protrusions (E) and retractions (F) were measured over 10 min. Values are mean ± S.E., ****, p < 0.0001. G, phosphorylation levels of cortactin in arg−/− fibroblasts expressing EYFP, Arg-EYFP, or Arg L207R/T233S-EYFP. Cortactin was immunoprecipitated (IP) and Western blotted for phosphotyrosine. Cortactin phosphorylation is reduced in arg−/− + EYFP fibroblasts, but similar in arg−/− + Arg-EYFP and arg−/− + Arg L207R/T233S-EYFP cells. H, quantification of cell spreading. Cell area was measured at 0, 10, 20, and 30 min of spreading on fibronectin.
Efficient phosphorylation of cortactin by Arg relies on the Arg PXXP-cortactin SH3 domain interaction as well as an intact Arg kinase domain. These interactions should not be compromised in the Arg L207R/T233S-EYFP mutant. We immunoprecipitated cortactin from arg−/− fibroblasts, or arg−/− fibroblasts expressing EYFP, Arg-EYFP, or mutant Arg L207R/T233S-EYFP and immunoblotted for phosphotyrosine. Cortactin phosphorylation levels are reduced in arg−/− + EYFP fibroblasts, but arg−/− fibroblasts were reconstituted with Arg-YFP and Arg L207R/T233S-EYFP exhibited similar levels of cortactin phosphorylation (Fig. 5G).
We also hypothesized that a chimera of Arg containing the Abl R161L/S187T SH2 domain would rescue cell edge protrusion in a manner similar to wild type Arg. Unfortunately, these constructs did not express well in fibroblasts despite repeated attempts (data not shown).
DISCUSSION
We show that Arg and Abl SH2 domains bind with high affinity to phosphorylated cortactin residues Tyr421 and Tyr466, but not Tyr482. Despite nearly identical sequences, we find that the Arg and Abl SH2 domains bind phosphorylated cortactin with a nearly 10-fold difference in affinity. Interestingly, we find that two specific residues, Arg161 and Ser187 in Abl and Leu207 and Thr233 in Arg, mediate the difference in affinity of these SH2 domains for phosphorylated cortactin. We provide a crystal structure of the Arg SH2 domain, which shows that these two residues are located near the binding pocket. Finally, we demonstrate that these two SH2 domain residues are functionally important to support cell edge protrusion.
Phosphorylation of cortactin residues Tyr421 and Tyr466 is required for actin polymerization in breast cancer cell invadopodia, whereas phosphorylation of the previously identified Tyr482 is not (42). Furthermore, we previously showed that Arg phosphorylates cortactin on two of these tyrosines, Tyr421 and Tyr466 (10, 11). We support these observations by demonstrating that Tyr(P)421 and Tyr(P)466, but not Tyr(P)482, interact with Arg and Abl SH2 domains. This is likely due to the inability of the Arg and Abl SH2 domain binding pocket to accommodate the bulky tyrosine residue found in the P + 3 site of the Tyr(P)482 peptide. Similarly, only Tyr421 and Tyr466 are conserved in and important for regulation of the cortactin homolog HS1 (43).
We show that Arg and Abl SH2 domains bind phosphorylated cortactin with dramatically different affinities. Through biochemical and structural approaches, we identified Arg residues Leu207 and Thr233 as key mediators of this high affinity interaction. Although the difference in affinity does not impact Arg localization, it is important for proper cell edge protrusion. Arg localization to the cell periphery is conferred by its C-terminal cytoskeletal binding domains and its SH2 domain does not impact subcellular localization (44). Our data agree with these previous studies. These findings further reinforce the fact that despite sequence similarity, Arg and Abl have unique roles within the cell. Evolving interaction specificity through affinity is one mechanism by which kinases target specific substrates, and swapping SH2 domains between kinases has been shown to alter the substrate profile from the wild type kinase (45). Our research suggests that this difference in affinity between Arg and Abl SH2 domains contributes to the different roles these two kinases play in the cell.
SH2 domains also play an important role in regulating processive phosphorylation of substrates with multiple phosphorylation sites (46) and SH2 domain binding preference has been shown to correlate well with phosphorylation site preference of the associated kinase (45, 47). It is possible that the Arg SH2 domain specificity for Tyr(P)421 in cortactin potentiates efficient phosphorylation of a subsequent tyrosine residue, such as Tyr466, leading to increased actin polymerization in invadopodia. This increased cortactin phosphorylation would lead to the up-regulation of actin polymerization in invadopodia. This mechanism may contribute to the fact that Arg, but not Abl, localizes to invadopodia in breast cancer cells and is essential for cortactin-mediated actin polymerization at these sites (5).
Previous studies show that selective interactions between Arg and cortactin underlie the ability of Arg to serve as a scaffold to support cortactin function during cell edge protrusion. Loss of the Arg SH2 domain-cortactin phosphotyrosine interaction leads to a reduction in cell edge protrusion (10). In relationship to these studies, we show that affinity of these interactions is an important determinant of whether or not they are able to perform their intended function in the cell. By expressing the mutant Arg L207R/T233S-EYFP in cells, we decrease the affinity between the Arg SH2 domain and phosphorylated cortactin and this decrease in affinity results in a failure of Arg to support wild type levels of cell edge protrusion. We also find that the circularity of cells expressing Arg L207R/T233S-EYFP is altered compared with wild type cells, reflecting a disruption of overall cytoskeletal structure and cell shape. Together, our results demonstrate the importance of two residues in the binding pocket of Arg and Abl SH2 domains for high affinity binding to phosphorylated cortactin and further highlight the non-redundant function of these two related kinases in cellular processes.
Acknowledgments
Vivian Stojanoff and Jean Jakoncic of NSLS beamline X6A are thanked. We also thank Dr. Martin Schiller at University of Connecticut Health Center for insight into the structure of SH2 domains.
This work was supported, in whole or in part, by National Institutes of Health Grants GM100411 and AI075133 (to T. J. B.), NS39475, GM100411, and CA133346 (to A. J. K.), and pilot grants from the Connecticut Breast Health Initiative and Women's Health Research at Yale (to T. J. B. and A. J. K.).
- Abl
- Abelson
- Arg
- Abl-related gene
- SH2
- Src homology 2
- SH3
- Src homology 3
- Fmoc
- N-(9-fluorenyl)methoxycarbonyl
- pY
- phosphotyrosine.
REFERENCES
- 1. Bradley W. D., Koleske A. J. (2009) Regulation of cell migration and morphogenesis by Abl-family kinases: emerging mechanisms and physiological contexts. J. Cell Sci. 122, 3441–3454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kruh G. D., Perego R., Miki T., Aaronson S. A. (1990) The complete coding sequence of Arg defines the Abelson subfamily of cytoplasmic tyrosine kinases. Proc. Natl. Acad. Sci. U.S.A. 87, 5802–5806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peacock J. G., Couch B. A., Koleske A. J. (2010) The Abl and Arg non-receptor tyrosine kinases regulate different zones of stress fiber, focal adhesion, and contractile network localization in spreading fibroblasts. Cytoskeleton 67, 666–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Moresco E. M., Donaldson S., Williamson A., Koleske A. J. (2005) Integrin-mediated dendrite branch maintenance requires Abelson (Abl) family kinases. J Neurosci 25, 6105–6118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mader C. C., Oser M., Magalhaes M. A., Bravo-Cordero J. J., Condeelis J., Koleske A. J., Gil-Henn H. (2011) An EGFR-Src-Arg-cortactin pathway mediates functional maturation of invadopodia and breast cancer cell invasion. Cancer Res. 71, 1730–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koleske A. J., Gifford A. M., Scott M. L., Nee M., Bronson R. T., Miczek K. A., Baltimore D. (1998) Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron 21, 1259–1272 [DOI] [PubMed] [Google Scholar]
- 7. Müller R., Slamon D. J., Tremblay J. M., Cline M. J., Verma I. M. (1982) Differential expression of cellular oncogenes during pre- and postnatal development of the mouse. Nature 299, 640–644 [DOI] [PubMed] [Google Scholar]
- 8. Renshaw M. W., Capozza M. A., Wang J. Y. (1988) Differential expression of type-specific c-abl mRNAs in mouse tissues and cell lines. Mol. Cell Biol. 8, 4547–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Perego R., Ron D., Kruh G. D. (1991) Arg encodes a widely expressed 145 kDa protein-tyrosine kinase. Oncogene 6, 1899–1902 [PubMed] [Google Scholar]
- 10. Lapetina S., Mader C. C., Machida K., Mayer B. J., Koleske A. J. (2009) Arg interacts with cortactin to promote adhesion-dependent cell edge protrusion. J. Cell Biol. 185, 503–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boyle S. N., Michaud G. A., Schweitzer B., Predki P. F., Koleske A. J. (2007) A critical role for cortactin phosphorylation by Abl-family kinases in PDGF-induced dorsal-wave formation. Curr. Biol. 17, 445–451 [DOI] [PubMed] [Google Scholar]
- 12. Liu W., MacGrath S. M., Koleske A. J., Boggon T. J. (2012) Lysozyme contamination facilitates crystallization of a heterotrimeric cortactin-Arg-lysozyme complex. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 68, 154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Martin K. H., Jeffery E. D., Grigera P. R., Shabanowitz J., Hunt D. F., Parsons J. T. (2006) Cortactin phosphorylation sites mapped by mass spectrometry. J. Cell Sci. 119, 2851–2853 [DOI] [PubMed] [Google Scholar]
- 14. Huang C., Liu J., Haudenschild C. C., Zhan X. (1998) The role of tyrosine phosphorylation of cortactin in the locomotion of endothelial cells. J. Biol. Chem. 273, 25770–25776 [DOI] [PubMed] [Google Scholar]
- 15. MacGrath S. M., Koleske A. J. (2012) Cortactin in cell migration and cancer at a glance. J. Cell Sci. 125, 1621–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang H., Li L., Wu C., Schibli D., Colwill K., Ma S., Li C., Roy P., Ho K., Songyang Z., Pawson T., Gao Y., Li S. S. (2008) Defining the specificity space of the human SRC homology 2 domain. Mol. Cell Proteomics 7, 768–784 [DOI] [PubMed] [Google Scholar]
- 17. Songyang Z., Shoelson S. E., Chaudhuri M., Gish G., Pawson T., Haser W. G., King F., Roberts T., Ratnofsky S., Lechleider R. J. (1993) SH2 domains recognize specific phosphopeptide sequences. Cell 72, 767–778 [DOI] [PubMed] [Google Scholar]
- 18. Kaneko T., Huang H., Zhao B., Li L., Liu H., Voss C. K., Wu C., Schiller M. R., Li S. S. (2010) Loops govern SH2 domain specificity by controlling access to binding pockets. Sci. Signal. 3, ra34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liu B. A., Jablonowski K., Shah E. E., Engelmann B. W., Jones R. B., Nash P. D. (2010) SH2 domains recognize contextual peptide sequence information to determine selectivity. Mol. Cell Proteomics 9, 2391–2404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Macromol. Crystallogr. A 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 21. McCoy A. J., Grosse-Kunstleve R. W., Storoni L. C., Read R. J. (2005) Likelihood-enhanced fast translation functions. Acta Crystallogr. D Biol. Crystallogr. 61, 458–464 [DOI] [PubMed] [Google Scholar]
- 22. Wojcik J., Hantschel O., Grebien F., Kaupe I., Bennett K. L., Barkinge J., Jones R. B., Koide A., Superti-Furga G., Koide S. (2010) A potent and highly specific FN3 monobody inhibitor of the Abl SH2 domain. Nat. Struct. Mol. Biol. 17, 519–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perrakis A., Harkiolaki M., Wilson K. S., Lamzin V. S. (2001) ARP/wARP and molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 57, 1445–1450 [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 26. Perrakis A., Morris R., Lamzin V. S. (1999) Automated protein model building combined with iterative structure refinement. Nat. Struct. Biol. 6, 458–463 [DOI] [PubMed] [Google Scholar]
- 27. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 28. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kabsch W., Sander C. (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 [DOI] [PubMed] [Google Scholar]
- 30. Miller A. L., Wang Y., Mooseker M. S., Koleske A. J. (2004) The Abl-related gene (Arg) requires its F-actin-microtubule cross-linking activity to regulate lamellipodial dynamics during fibroblast adhesion. J. Cell Biol. 165, 407–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nagar B., Hantschel O., Seeliger M., Davies J. M., Weis W. I., Superti-Furga G., Kuriyan J. (2006) Organization of the SH3-SH2 unit in active and inactive forms of the c-Abl tyrosine kinase. Mol. Cell 21, 787–798 [DOI] [PubMed] [Google Scholar]
- 32. Nagar B., Hantschel O., Young M. A., Scheffzek K., Veach D., Bornmann W., Clarkson B., Superti-Furga G., Kuriyan J. (2003) Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell 112, 859–871 [DOI] [PubMed] [Google Scholar]
- 33. Nam H. J., Haser W. G., Roberts T. M., Frederick C. A. (1996) Intramolecular interactions of the regulatory domains of the Bcr-Abl kinase reveal a novel control mechanism. Structure 4, 1105–1114 [DOI] [PubMed] [Google Scholar]
- 34. Overduin M., Rios C. B., Mayer B. J., Baltimore D., Cowburn D. (1992) Three-dimensional solution structure of the Src homology 2 domain of c-abl. Cell 70, 697–704 [DOI] [PubMed] [Google Scholar]
- 35. Eck M. J., Shoelson S. E., Harrison S. C. (1993) Recognition of a high-affinity phosphotyrosyl peptide by the Src homology-2 domain of p56lck. Nature 362, 87–91 [DOI] [PubMed] [Google Scholar]
- 36. Metzler W. J., Leiting B., Pryor K., Mueller L., Farmer B. T., 2nd (1996) The three-dimensional solution structure of the SH2 domain from p55blk kinase. Biochemistry 35, 6201–6211 [DOI] [PubMed] [Google Scholar]
- 37. Schindler T., Sicheri F., Pico A., Gazit A., Levitzki A., Kuriyan J. (1999) Crystal structure of Hck in complex with a Src family-selective tyrosine kinase inhibitor. Mol. Cell 3, 639–648 [DOI] [PubMed] [Google Scholar]
- 38. Arold S. T., Ulmer T. S., Mulhern T. D., Werner J. M., Ladbury J. E., Campbell I. D., Noble M. E. (2001) The role of the Src homology 3-Src homology 2 interface in the regulation of Src kinases. J. Biol. Chem. 276, 17199–17205 [DOI] [PubMed] [Google Scholar]
- 39. Gilmer T., Rodriguez M., Jordan S., Crosby R., Alligood K., Green M., Kimery M., Wagner C., Kinder D., Charifson P. (1994) Peptide inhibitors of Src SH3-SH2-phosphoprotein interactions. J. Biol. Chem. 269, 31711–31719 [PubMed] [Google Scholar]
- 40. Tong L., Warren T. C., King J., Betageri R., Rose J., Jakes S. (1996) Crystal structures of the human p56lck SH2 domain in complex with two short phosphotyrosyl peptides at 1.0 Å and 1.8 Å resolution. J. Mol. Biol. 256, 601–610 [DOI] [PubMed] [Google Scholar]
- 41. Miller M. M., Lapetina S., MacGrath S. M., Sfakianos M. K., Pollard T. D., Koleske A. J. (2010) Regulation of actin polymerization and adhesion-dependent cell edge protrusion by the Abl-related gene (Arg) tyrosine kinase and N-WASp. Biochemistry 49, 2227–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Oser M., Mader C. C., Gil-Henn H., Magalhaes M., Bravo-Cordero J. J., Koleske A. J., Condeelis J. (2010) Specific tyrosine phosphorylation sites on cortactin regulate Nck1-dependent actin polymerization in invadopodia. J. Cell Sci. 123, 3662–3673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Butler B., Kastendieck D. H., Cooper J. A. (2008) Differently phosphorylated forms of the cortactin homolog HS1 mediate distinct functions in natural killer cells. Nat. Immunol. 9, 887–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang Y., Miller A. L., Mooseker M. S., Koleske A. J. (2001) The Abl-related gene (Arg) nonreceptor tyrosine kinase uses two F-actin-binding domains to bundle F-actin. Proc. Natl. Acad. Sci. U.S.A. 98, 14865–14870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mayer B. J., Baltimore D. (1994) Mutagenic analysis of the roles of SH2 and SH3 domains in regulation of the Abl tyrosine kinase. Mol. Cell Biol. 14, 2883–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mayer B. J., Hirai H., Sakai R. (1995) Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Curr. Biol. 5, 296–305 [DOI] [PubMed] [Google Scholar]
- 47. Pellicena P., Stowell K. R., Miller W. T. (1998) Enhanced phosphorylation of Src family kinase substrates containing SH2 domain binding sites. J. Biol. Chem. 273, 15325–15328 [DOI] [PubMed] [Google Scholar]
- 48. Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]