Background: Carnosine synthase, the enzyme responsible for carnosine and homocarnosine synthesis, is promiscuous.
Results: We identify PM20D2 as a dipeptidase hydrolyzing β-alanine-lysine and other “wrong” dipeptides made by carnosine synthase.
Conclusion: Specific synthesis of carnosine requires both carnosine synthase and the “metabolite repair enzyme” β-alanyl-lysine dipeptidase (PM20D2).
Significance: Recognizing novel “metabolite-repair” mechanisms is crucial to understanding enzyme specificity and inborn errors of metabolism.
Keywords: Brain Metabolism, Metabolism, Peptidase, Peptides, Skeletal Muscle Metabolism, CARNS1, CNDP1, Carnosine, M20 Metallopeptidase, Homocarnosine
Abstract
Carnosine synthase is the ATP-dependent ligase responsible for carnosine (β-alanyl-histidine) and homocarnosine (γ-aminobutyryl-histidine) synthesis in skeletal muscle and brain, respectively. This enzyme uses, also at substantial rates, lysine, ornithine, and arginine instead of histidine, yet the resulting dipeptides are virtually absent from muscle or brain, suggesting that they are removed by a “metabolite repair” enzyme. Using a radiolabeled substrate, we found that rat skeletal muscle, heart, and brain contained a cytosolic β-alanyl-lysine dipeptidase activity. This enzyme, which has the characteristics of a metalloenzyme, was purified ≈200-fold from rat skeletal muscle. Mass spectrometry analysis of the fractions obtained at different purification stages indicated parallel enrichment of PM20D2, a peptidase of unknown function belonging to the metallopeptidase 20 family. Western blotting showed coelution of PM20D2 with β-alanyl-lysine dipeptidase activity. Recombinant mouse PM20D2 hydrolyzed β-alanyl-lysine, β-alanyl-ornithine, γ-aminobutyryl-lysine, and γ-aminobutyryl-ornithine as its best substrates. It also acted at lower rates on β-alanyl-arginine and γ-aminobutyryl-arginine but virtually not on carnosine or homocarnosine. Although acting preferentially on basic dipeptides derived from β-alanine or γ-aminobutyrate, PM20D2 also acted at lower rates on some “classic dipeptides” like α-alanyl-lysine and α-lysyl-lysine. The same activity profile was observed with human PM20D2, yet this enzyme was ∼100–200-fold less active on all substrates tested than the mouse enzyme. Cotransfection in HEK293T cells of mouse or human PM20D2 together with carnosine synthase prevented the accumulation of abnormal dipeptides (β-alanyl-lysine, β-alanyl-ornithine, γ-aminobutyryl-lysine), thus favoring the synthesis of carnosine and homocarnosine and confirming the metabolite repair role of PM20D2.
Introduction
Enzyme specificity is crucial for life, as it minimizes the formation of useless metabolic byproducts. Recent work indicates, however, that in some cases enzyme specificity is not sufficient and that byproducts would accumulate in the absence of metabolite repair enzymes (also named “metabolite proofreading” enzymes), which convert “abnormal” metabolites to classical metabolites or to breakdown products (1, 2). Deficiency in such enzymes may lead to disease, as exemplified with l-2-hydroxyglutarate dehydrogenase deficiency (3, 4). This enzyme metabolizes l-2-hydroxyglutarate, which is made erroneously from α-ketoglutarate by mitochondrial l-malate dehydrogenase (5, 6). Its absence leads to l-2-hydroxyglutarate accumulation, responsible for a severe neurological disorder characterized by leukoencephalopathy and a possible increase in the incidence of brain tumors (7, 8). Other examples of metabolite repair enzyme are the ATP-dependent dehydratase and epimerase that repair hydrated NAD(P)H (9) and ethylmalonyl-CoA decarboxylase, which decarboxylates a metabolite made erroneously by acetyl-CoA carboxylase (10).
As the recently identified metabolite repair enzymes correspond to proteins whose function was previously unknown, it is likely that numerous putative enzymes encoded by genomes (including mammalian genomes) act as metabolite repair enzymes. In this context we have started to investigate cases of enzymes known to be poorly specific and for which a metabolite repair enzyme could potentially “complement” the principal activity by removing unwanted side products.
A well known example of enzyme-lacking specificity is the ligase that catalyzes the synthesis of carnosine (β-alanyl-histidine; β-Ala-His) in skeletal muscle and homocarnosine (γ-aminobutyryl-histidine; GABA-His) in brain (11–15). Carnosine is an abundant dipeptide present in the skeletal muscle of many vertebrates where it serves as a pH buffer (for review, see Ref. 16) and maybe also as a radical scavenger (17). The GABA derivative homocarnosine is present in brain, where its function is still unknown (18). Carnosine/homocarnosine synthase preferentially ligates β-alanine or GABA to histidine, but it also ligates them to lysine and ornithine with catalytic efficiencies amounting to 2–10% of those observed for the formation of carnosine or homocarnosine (15). Thus, taking into account the relative concentration of histidine and lysine in muscle tissues, one would expect to observe concentrations of β-alanyl-lysine (β-Ala-Lys) amounting to >10% that of the carnosine concentration, yet the concentration of β-Ala-Lys that has been described in rabbit skeletal muscle is only ∼0.1% that of carnosine (13).
This observation led us to hypothesize that there may be a peptidase that hydrolyzes β-Ala-Lys and related basic dipeptides while not acting on carnosine or homocarnosine. In the present work we report the identification of such an enzyme as a protein of unknown function designated PM20D2.4 We characterize this peptidase, and we show that its expression in cells clears them from abnormal dipeptides made by carnosine synthase.
EXPERIMENTAL PROCEDURES
Materials
Most peptides used in this work (radioactive or not) were synthesized in the laboratory as described below. From Sigma we used: 1,10-phenanthroline monohydrate and fluorescamine, which we dissolved in DMSO; lysine oxidase from Trichoderma viride (20–60 units/mg of protein), which was resuspended in 25 mm Hepes, pH 7.2, with 10% glycerol (0.26 ml; ≈100 units·ml−1) and stored at −20 °C; Type I horseradish peroxidase, o-dianisidine dihydrochoride, and l-alanine dehydrogenase from Bacillus subtilis in buffered aqueous glycerol solution (≈56 units·ml−1; 30 units·mg−1). β-[3-3H(N)]Alanine (30–60 Ci/mmol (1.11–2.22 TBq/mmol) and [2,3-3H(N)]GABA 25–60 Ci/mmol (0.925–2.22 TBq/mmol) were from American Radiolabeled Chemical, Inc. (Saint Louis, MO). jetPEI transfection reagent was from Polyplus Transfection. AG50X-4 resin was from Bio-Rad. DMEM cell culture medium, from Lonza Braine SA, was complemented with 10% fetal calf serum, 2 mm glutamine, and 100 units·ml−1 penicillin/streptomycin.
Preparation of [β-3H]Alanyl or [γ-3H]Aminobutyryl Dipeptides
[β-3H]Ala-Lys, -Orn, -Arg, or -His (carnosine) were each prepared in a reaction mixture (1 ml) containing ≈2.108 cpm of purified [β-3H]alanine, 1 μm β-alanine, 50 mm Hepes, pH 7.5, 10 mm KCl, 1 mm DTT, 1 mm EGTA, 1 mm MgCl2, 3 mm ATP-Mg, and 1 mm lysine, ornithine, arginine, or histidine. The reaction was started by the addition of ≈0.02 mg of purified recombinant mouse carnosine synthase (encoded by the carnosine synthase (CARNS1) gene) (15). After a 20-h incubation at 37 °C, the reaction was stopped by heating for 5 min at 80 °C. To separate the [β-3H]alanyl dipeptides from unreacted [β-3H]alanine, the reaction mixture was diluted 3-fold with 5 mm Hepes, pH 7.5, and applied onto a AG50W-X4 (Na+, 1 ml) column equilibrated in the same buffer. After washing unreacted [β-3H]alanine with 10 ml of the equilibration buffer, the [β-3H]alanyl dipeptides were eluted with 5 × 1 ml 1 m NH4OH. Three fractions containing the highest amount of radioactivity were pooled, lyophilized, resuspended in 0.5 ml of H2O, and desalted on Biogel P2 equilibrated in H2O. The yield of the synthesis was ≈65% in the case of [β-3H]Ala-Arg and close to 100% for the other radiolabeled dipeptides.
[3H]GABA-Lys, -Orn, and -His (homocarnosine) were prepared as above in the presence of ≈107 cpm of purified [3H]GABA and 1 μm GABA in the presence of 0.025 mg of purified recombinant human carnosine synthase (for [3H]GABA-Lys and -His), whereas [3H]GABA-Orn was best produced using a similar amount of mouse carnosine synthase. The yield of the synthesis was ≈35% for [3H]GABA-Lys and -His and 40% in the case of [3H]GABA-Orn.
Assays of β-Ala-Lys, GABA-Lys, or Ala-Lys Peptidase Activity
Peptidase activities were determined using four different assays, which measured the production of 1) [β-3H]alanine or [3H]GABA from 3H-labeled radioactive dipeptides, 2) lysine or ornithine from lysine- and ornithine-containing peptides with a lysine oxidase-coupled assay, 3) GABA from GABA-containing peptides with a GABA-transaminase/succinate semialdehyde dehydrogenase-coupled assay, or 4) alanine from alanine-containing peptides with a l-alanine dehydrogenase-coupled assay.
When 3H-labeled radioactive dipeptides were used as substrates, the standard incubation mixture (50 μl) consisted of 50 mm Tris, pH 7.5, [β-3H]Ala or [3H]GABA dipeptides (≈6 × 104 cpm) with (concentrations as stated) or without nonradioactive dipeptides, and the incubation was performed at 37 °C with or without peptidase for the required reaction times (allowing a maximal percentage of conversion of 40% of the total radioactivity). [β-3H]Alanine or [3H]GABA production was linear in all conditions tested both when using recombinant enzyme or tissue proteins. The incubation was stopped (5 min at 85 °C), and denatured proteins were removed by centrifugation (10 min at 13,000 × g). The supernatant was diluted 20-fold with 5 mm Hepes, pH 7.5, and 0.95 ml of this sample was applied to AG50W-X4 columns (1 ml, Na+) equilibrated in 5 mm Hepes, pH 7.5. [β-3H]Alanine or [3H]GABA were eluted from the column with 4 ml of equilibration buffer, and radioactivity was counted (2 ml) after mixing with scintillation mixture (Ultima Gold, PerkinElmer Life Sciences). The unreacted substrate remained bound to the column.
To measure lysine produced by recombinant mouse or human PM20D2, the assay was performed in two steps. First, lysine-containing peptides (0.6 mm unless otherwise stated) were incubated (50 μl) at 37 °C in 50 mm Tris, pH 7.5, containing 0.25 mg·ml−1 BSA. The reaction mixture was prewarmed, and the reaction was started with or without (blank) the addition of 1 or 8 μg of recombinant mouse or human PM20D2, respectively. The reaction was stopped 5 min (mouse PM20D2) or 60 min (human PM20D2) later by adding 40 μl of the incubation mixture to a microplate well containing 2 μl of 0.2 m 1,10-phenanthroline. In the second reaction, lysine (or ornithine, as stated) produced during the first step was measured with a colorimetric assay, which coupled the oxidative deamination of the basic amino acids to the oxidation of o-dianisidine by lysine oxidase and peroxidase. We added 0.166 ml of 0.25 m Tris, pH 8, containing horseradish peroxidase (0.1 mg·ml−1), o-dianisidine (0.1 mg·ml−1), and lysine oxidase (10 milliunits) to each well of the microplate. Absorbance at 420 nm was measured using a microplate reader after 1 h of incubation at 37 °C and compared with a standard curve for the basic amino acid measured.
The two other assays used to measure PM20D2 activity were spectrophotometric tests. One assay measured the production of GABA from GABA-containing dipeptides by coupling it to the change in absorbance at 340 nm that was observed after the reduction of NADP to NADPH in the presence of recombinant GABA-transaminase and succinate semialdehyde dehydrogenase from Escherichia coli. The assay mixture (0.8 ml) contained 50 mm Tris, pH 7.5, 5 μm pyridoxal phosphate, 1 mm α-ketoglutarate, 0.25 mm NADP, 0.5 mg·ml−1 BSA, 0.6 mm GABA-containing dipeptide, purified recombinant E. coli N-terminal His6-tagged GABA transaminase (10 μg), and succinate semialdehyde dehydrogenase (25 μg). The reaction was started by adding 1.2 or 15 μg of recombinant mouse or human PM20D2, and the absorbance at 340 nm was monitored at 37 °C in a Specord 50 from AnalitikJena.
In the other assay alanine that was formed due to PM20D2 activity on alanine-containing peptides was measured in a two-step assay. In the first step the assay mixture (60 μl) contained 25 mm Tris, pH 7.5, 0.5 mg/ml−1 BSA, and variable concentrations of substrate. After preheating at 37 °C, the reaction was started by adding 1 or 8 μg of mouse or human recombinant PM20D2 and stopped after 5 min (mPM20D2) or 60 min (hPM20D2) by heating (5 min at 85 °C). In the second step the alanine that was produced was quantified in an end-point assay performed in a microplate. 50 μl of each sample were placed in each well and mixed with the second assay mixture (150 μl) containing 0.26 m Tris, pH 8.8, 1.6 mm NAD, 0.5 mg·ml−1 BSA, and 2 μl (≈100 milliunits) of l-alanine dehydrogenase from B. subtilis. The absorbance at 340 nm was measured after ≈60 min at 37 °C, once it become stable, and the alanine in the samples could be calculated using the molar extinction coefficient for NADH. Km and Vmax for the enzymatic activities described were calculated using Prism 4.0 GraphPad software using a nonlinear regression.
Purification of β-Ala-Lys Dipeptidase Activity
Skeletal muscle (250 g) from 6 male Wistar rats (200 g) was homogenized in 3 volumes (w/v) of buffer A (50 mm Tris, pH 7, containing 2.5 μg·ml−1 antipain and leupeptin) with an Ultra Turrax homogenizer, and the homogenate was centrifuged at 16,000 × g for 45 min at 4 °C. Poly(ethylene glycol) 6000 (2% w/v) was added to the supernatant, which was stirred for 20 min at 4 °C and centrifuged (15 min at 16,000 × g at 4 °C). The resulting supernatant was filtered through 5 layers of gauze to retain remaining fat particles and was applied on a DEAE-Sepharose column (160-ml bed volume) equilibrated in buffer A. The column was washed with 400 ml of the same buffer, and the retained protein was eluted with a NaCl gradient (0–0.6 m in 800 ml of buffer A). The collected fractions (7 ml) were tested for β-Ala-Lys dipeptidase activity. After pooling the most active fractions (≈70 ml), proteins were concentrated by adding 22% polyethylene glycol (PEG-6000), centrifuging as above, and resuspending the pellet in 22 ml of buffer B (25 mm Tris, pH 8, containing 2.5 μg·ml−1 antipain and leupeptin). The concentrated pool was then applied onto a Q-Sepharose column (15 ml) equilibrated in buffer B. After washing with buffer B (50 ml), the retained proteins were eluted with a NaCl gradient (0–0.6 m NaCl in 340 ml of buffer B) and 3-ml fractions were collected, supplemented with 10% glycerol, and frozen at −80 °C for further assays. The most active fractions (36 ml) were pooled and concentrated by ultrafiltration to ≈2.6 ml using Vivaspin 15 concentrators, and 2 ml were loaded onto a HiLoad 16/600 Superdex 200 pg (GE Healthcare) equilibrated in 25 mm Tris, pH 7, containing 0.15 m NaCl. Proteins were eluted with the same buffer in 0.7-ml fractions and supplemented with 10% glycerol. Three of the most active fractions were pooled and chromatographed again using the same gel filtration column and buffer but supplemented with 0.3 m KSCN. All purification steps were performed at 4 °C, and the active fractions were stored at −80 °C between steps.
During the purification β-Ala-Lys dipeptidase was assayed by incubating the appropriate volume of sample in 50 μl of 50 mm Tris, pH 7.5, in the presence of ≈6 × 104 cpm [β-3H]Ala-Lys for 30 min at 37 °C. 1 unit of activity corresponded to the amount of enzyme required to hydrolyze 1% of the radioactive substrate under these assay conditions.
Identification of β-Ala-Lys Dipeptidase by Mass Spectrometry
An aliquot of each of the analyzed fractions (30 μg of proteins) was precipitated by the addition of trichloroacetic acid to a final concentration of 10% w/v. The resulting pellet was washed with ice-cold acetone, dried in a SpeedVac, resuspended in 0.1 m NH4HCO3, pH 8.0, with 1 μg of sequencing grade trypsin, and digested overnight at 30 °C. The reaction was stopped by adding trifluoroacetic acid (0.1% final concentration) and analyzed by LC-MS/MS (19). Briefly peptides were separated by an acetonitrile gradient on a C18 column, and the MS scan routine was set to analyze by MS/MS the 10 most intense ions of each full MS scan; dynamic exclusion was enabled to ensure detection of co-eluting peptides.
Protein identification was performed with SequestHT, and peak lists were generated using extract-msn (ThermoScientific) within Proteome Discoverer 1.4.1. From raw files, MS/MS spectra were exported with the following settings: peptide mass range, 350–5000 Da; minimal total ion intensity, 500. The resulting peak lists were searched using SequestHT against a target-decoy Rat protein database (57,756 entries comprising forward and reversed sequences) obtained from Uniprot. The following parameters were used: trypsin was selected with proteolytic cleavage only after arginine and lysine, number of internal cleavage sites was set to 1, mass tolerance for precursors and fragment ions was 1.0 Da, and considered dynamic modifications were +15.99 Da for oxidized methionine. Peptide matches were filtered using the q-value, and posterior error probability was calculated by the percolator algorithm ensuring an estimated false positive rate below 5%. The filtered Sequest HT output files for each peptide were grouped according to the protein from which they were derived, and their individual number of peptide spectral matches was taken as an indicator of protein abundance after normalization with the total numbers of peptides spectral matches from all the proteins identified in that particular fraction.
Assay of Carnosine and Homocarnosine
Carnosine (β-Ala-His) or homocarnosine (GABA-His) present in deproteinized cell extracts from transfected HEK293T cells were assayed using a procedure measuring the histidine produced after carnosinase treatment of the samples. This was made possible by cloning, expressing, and purifying N-terminal His6-tagged recombinant histidine ammonia lyase (hutH1) from Pseudomonas aeruginosa PAO1. After carnosinase treatment of the extracts, the deamination of histidine by hutH1 converts it to urocanic acid, which strongly absorbs light at 277 nm (molar extinction coefficient = 18.8 × 103·M−1·cm−1; ≈pH 7). Carnosine and homocarnosine were assayed in a UV-compatible microplate with an assay mixture (150 μl) containing 0.1 m Tris, pH 7.0, 15 μl of deproteinized HEK293T extract, recombinant purified P. aeruginosa hutH1 (6 μg) and 1.5 μg (for carnosine) or 5 μg (for homocarnosine) of human carnosinase. A277 reached a maximum value after ≈15 min (carnosine) or overnight (homocarnosine) incubation at 37 °C.
Cloning, Expression, and Purification of Mouse and Human PM20D2 and Carnosinase in E. coli and HEK293T Cells
Mouse and human PM20D2 and carnosinase (encoded by the CNDP1 gene) were PCR-amplified using cDNA from mouse skeletal muscle or human kidney (PM20D2) and mouse kidney or human brain (carnosinase). In the case of human carnosinase, care was taken to remove the sequence corresponding to the predicted signal peptide. The recombinant protein started at the second conserved methionine (Met-24) and was, therefore, cytosolic. All PCR-amplified sequences were cloned in pET28a, and the clones were confirmed by sequencing, transformed in E. coli BL21 (DE3), and expressed as partially soluble N-terminal His6-tagged fusion proteins in minimal M9 media for 48 h at 18 °C after the addition of 0.4 mm isopropyl-β-d-thiogalactopyranoside (20). Recombinant proteins present in soluble extracts were purified using Histrap HP columns (1 ml), desalted to remove imidazole, and stored at −80 °C in 25 mm Hepes, pH 7.2, containing 50 mm NaCl and 10% glycerol. The yield of recombinant proteins ranged between 2 and 9 mg of homogeneous protein per 0.5 liters of culture.
The same four proteins were also cloned in pEF6/HisB plasmid and expressed in HEK293T cells as N-terminal His6-tagged proteins. When proteins were overexpressed in HEK293T cells (10-cm diameter plate) to be purified and characterized, ≈2 × 106 cells were transfected with 7.5 μg of the appropriate plasmid using 16 μl of jetPEI transfection reagent (21). In the experiments where HEK293T cells were co-transfected with up to three different plasmids, the same number of seeded cells was transfected with 2.5 μg of each plasmid to reach a total of 7.5 μg plasmid DNA. Twenty-four hours after transfection the culture media (7 ml) was supplemented with 0.1 mm β-alanine or GABA in combination or not with 1 mm lysine or ornithine, and the cells were left for an extra 24 h before removing the medium, washing with PBS, and collecting the cells from each plate in 0.4 ml of 25 mm Hepes, pH 7.2, 2 μg·ml−1 antipain and leupeptin, and 0.5 mm PMSF. The cells were lysed by freezing twice in liquid nitrogen, and the lysates were treated with DNase I (125 units·ml−1) before removal of the insoluble material by centrifugation (15,000 × g for 15 min). Part (0.3 ml) of the soluble extract was deproteinized (5 min at 85 °C followed by a 15-min centrifugation at 15,000 × g) and used to assay carnosine, β-Ala-Lys, β-Ala-Orn, homocarnosine, GABA-Lys, or GABA-Orn, and another part (0.1) ml was used for expression analysis of the recombinant proteins by SDS-PAGE and Western blotting (21).
Peptide Synthesis and Quantitation
All peptides that were not commercially available were synthesized in-house on solid phase using standard Fmoc (fluorenylmethoxycarbonyl) chemistry and characterized by mass spectrometry. Before usage, the lyophilized peptides were dissolved in 0.5 ml of PBS and quantitated using fluorescamine. Practically, 10 μl of peptide solutions (≈50 nmol) were mixed with 20 μl of PBS and 90 μl of fluorescamine (3 mg·ml−1 DMSO) in wells of black microplates recommended for fluorescence measurements. After 30 min, the fluorescence was measured using a fluorescence microplate reader set with 355-nm excitation and 460-nm emission wavelengths. The peptide concentration in the unknown samples was calculated by comparing to a standard concentration of similar amino acids. When the peptides contained more than one lysine residue, we used alanine as the standard amino acid and took into account the number of potentially reacting amine groups when estimating their concentration.
RESULTS
Characterization of the β-Ala-Lys Dipeptidase Activity Present in Rat Tissues
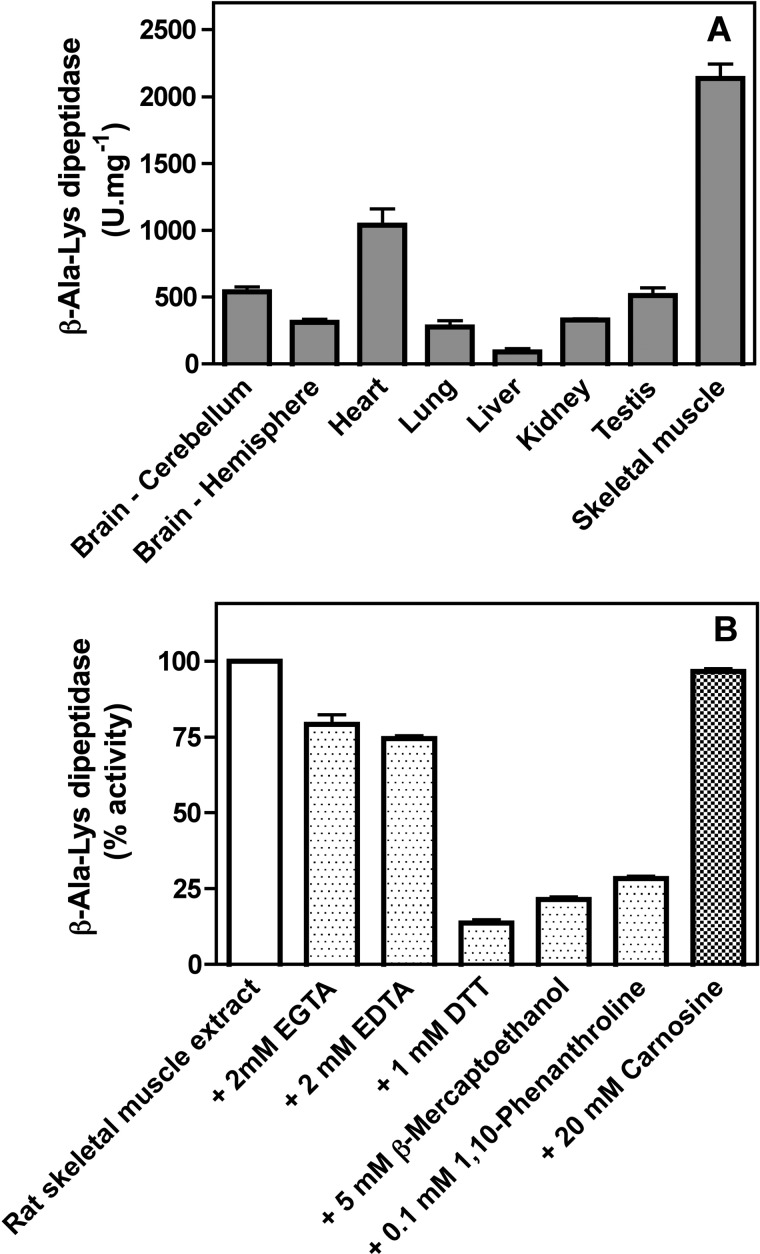
We wanted to search for an enzyme able to hydrolyze β-Ala-Lys in rat tissues. For this purpose we used radioactive [β-3H]Ala-Lys that we prepared by incubating [β-3H]alanine and lysine with recombinant mouse carnosine synthase (which is less specific than the human enzyme; Ref. 15) in the presence of ATP, as described under “Experimental Procedures.” As shown in Fig. 1A, [β-3H]Ala-Lys was hydrolyzed in the soluble extracts of all rat tissues tested, but skeletal muscle, heart, and brain were the ones with the highest activities. It is interesting to note that these tissues are also those that display carnosine synthase activity and accumulate carnosine or homocarnosine (16).
FIGURE 1.
Properties of β-Ala-Lys dipeptidase activity in rat tissue extracts. A, distribution of the activity in various rat tissues. B, effect of metal chelators and carnosine on the β-Ala-Lys dipeptidase activity in skeletal muscle extract. Values are the means ± S.E. (n = 3).
To rule out the possibility that we could have missed part of the enzymatic activity because it would be membrane-bound and eliminated by high speed centrifugation of the muscle extracts, we performed a medium speed (30 min at 16,000 × g) and a high speed (45 min at 100,000 × g) centrifugation of the skeletal muscle whole extract. There was no difference in the total activity measured in the supernatants of the various centrifugations and the non-centrifuged extract. Furthermore, the pellets obtained were also devoid of β-Ala-Lys hydrolyzing activity, suggesting that at least for rat skeletal muscle all enzymatic activity was soluble (not shown).
To characterize the β-Ala-Lys-hydrolyzing activity present in muscle extracts, we studied its metal dependence and saw that adding 100 μm Mg2+, Mn2+, or Cu2+ or 10 μm Zn2+ or Ni2+ did not activate the enzyme, whereas 100 μm Co2+ increased the activity by 20% (not shown). This together with the strong decrease in activity observed when dithiothreitol, β-mercaptoethanol, or 1,10-phenanthroline (Fig. 1B) was added to the assay indicated that the β-Ala-Lys hydrolyzing activity was probably a metalloenzyme and likely a Zn2+-dependent one. Because the carnosine degrading dipeptidases are also Zn2+-dependent enzymes (22), we excluded their contribution to the β-Ala-Lys hydrolyzing activity by showing that the addition of 20 mm carnosine to the assay did not lower the activity (Fig. 1B).
Partial Purification of β-Ala-Lys Dipeptidase Activity from Rat Skeletal Muscle
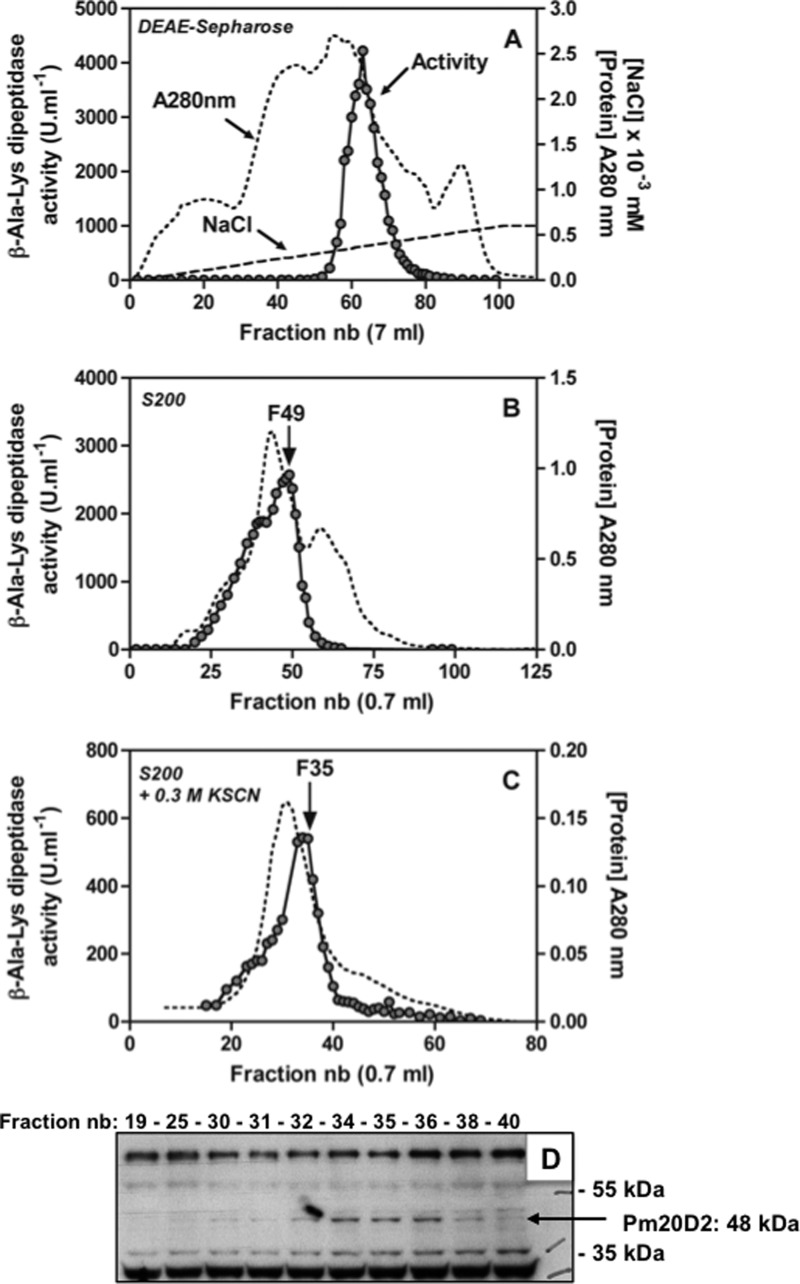
β-Ala-Lys dipeptidase was purified ∼200-fold from rat skeletal muscle (Table 1) with the use of various chromatographic steps (DEAE-Sepharose, Q-Sepharose, and Superdex 200) as described under “Experimental Procedures.” The enzymatic activity was retained on both anion-exchange columns at pH 7 or 8 and was then eluted as one peak of activity with a salt gradient (illustrated in Fig. 2A for the DEAE-Sepharose column), suggesting the presence of a single enzyme hydrolyzing β-Ala-Lys. The gel filtration step on Superdex 200 (Fig. 2B) revealed, however, that the activity peak of the native enzyme was quite wide and asymmetric, particularly when it was compared with the elution of molecular weight standard proteins, which were chromatographed under the same conditions (not shown). This behavior suggested that β-Ala-Lys dipeptidase activity was partially contributed by heterogeneous high molecular weight multimers and/or that it partially interacted with the column matrix. To shed light on this observation, the three most active peak fractions from this purification step (fractions 47–49; see Fig. 2B) were pooled and chromatographed again on the same gel filtration column but in the presence of 0.3 m KSCN, a chaotropic agent known to favor protein dissociation. The activity profile obtained was still heterogeneous, but the enzyme that was present in the peak fractions (33–35) had been further purified (Fig. 2C). The gel filtration disclosed that the size of the native β-Ala-Lys hydrolyzing enzyme in the peak fraction was ≈220 kDa (not shown). The overall yield of the purification was low (<1%, see Table 1) mainly because only the most active fractions from each purification step were used in the next step rather than from loss of activity due to enzyme inactivation. This was most noticeable after the Q-Sepharose/ultrafiltration steps, after which we recovered 22% of the activity with no obvious gain in specific activity. It is likely that this step could have been skipped without great impact on the final purification results.
TABLE 1.
Purification of β-Ala-Lys dipeptidase from rat skeletal muscle
| Volume | Activity | Protein concentration | Total activity | Specific activity | Purification | |
|---|---|---|---|---|---|---|
| ml | Units·ml−1 | mg·ml−1 | Units | Units·mg−1 | -Fold | |
| 16000 × g supernatant | 720 | 412 | 15.4 | 296,640 | 26.8 | 1 |
| DEAE (F58-F68)a | 22 | 10,861 | 11.6 | 238,946 | 938 | 35 |
| Q-Sepharose (F47-F58)b | 2 | 20,418 | 21.1 | 40,837 | 965 | 36 |
| Superdex-200 (F47-F49) | 2.1 | 2,573 | 0.87 | 5,400 | 2,957 | 110 |
| Superdex-200 + KSCN (F35) | 0.7 | 540 | 0.109 | 388 | 4,954 | 185 |
a Proteins were precipitated with 22% PEG and resuspended in the indicated volume.
b The Q-Sepharose fractions were pooled and concentrated to 2 ml by ultrafiltration on Vivaspin concentrators.
FIGURE 2.
Purification of rat muscle β-Ala-Lys dipeptidase on DEAE-Sepharose and Superdex 200 and co-elution of the activity with a protein recognized by anti-PM20D2 antibodies. A, elution of the β-Ala-Lys dipeptidase activity from a DEAE-Sepharose column (see Table 1) with a 0–0.6 m NaCl gradient. B, active fractions from a Q-Sepharose column were further gel-filtered on Superdex-200; the activity peak of the native enzyme was asymmetric, but the peak fraction (F49) co-eluted with marker proteins of ≈220 kDa. C, active fractions F47-F49 from B were gel-filtered in the same Superdex-200 column in the presence of 0.3 KSCN. D, Western blot analysis of the peak fractions from C showing β-Ala-Lys dipeptidase activity co-eluting with a 48-kDa protein that corresponds to PM20D2. nb, number.
Identification of β-Ala-Lys Dipeptidase as PM20D2
The approach used to identify β-Ala-Lys dipeptidase was to determine by (LC-MS/MS) the identity of the most abundant proteins present in the peak fractions after the last three purification steps, select among them those likely to display a peptidase activity and follow the enrichment of proteins with potential peptidase activity. To evaluate this enrichment we calculated the ratio of the number of peptide spectral matches attributable to each candidate peptidase to the total number of peptide spectral matches detected for all identified proteins.
Table 2 shows that among 10 different (putative) peptidases, PM20D2 was the only candidate for which the normalized number of peptide spectral matches increased in parallel with the β-Ala-Lys dipeptidase specific activity (not shown), suggesting that it corresponds to the protein of interest. Thus, the increase in the specific activity of β-Ala-Lys dipeptidase between the first and second gel filtration steps (1.67-fold, Table 2) was in excellent agreement with the enrichment in PM20D2 (1.7-fold) and contrasted with the decrease in relative abundance (0.1–0.73-fold) observed for all other candidate proteins. As the specific activity of β-Ala-Lys dipeptidase is ∼20-fold higher in rat skeletal muscle than in liver, we checked the expression of the mRNA of the 10 candidate proteins in these two tissues in the BioGPS database (last column of Table 2). PM20D2 was, with CAPN1, the only candidate to show a high ratio of expression in skeletal muscle compared with liver, further supporting that PM20D2 was an excellent candidate.
TABLE 2.
Increase in the specific activity of rat skeletal muscle β-Ala-Lys dipeptidase during the last purification step compared with the enrichment of various peptidase/protease-like proteins found by mass spectrometry analysis
β-Ala-Lys dipeptidase was purified as described (see Table I and “Experimental Procedures”), and the peak fractions from the two last purification steps (F49, Superdex-200; F35, Superdex-200 + 0.3 m KSCNl see Fig. 2, B and C) were analyzed by MS/MS. We show, for the detected proteins with potential peptidase/protease-like activities, the ratio of the number of peptide spectral matches (PSM) found in these same fractions after normalization to total PSM. The ratio of β-Ala-Lys dipeptidase activity in rat skeletal muscle and liver (a) is compared with the ratio of mRNAs (b) (in skeletal muscle and liver) coding for the peptidase/protease-like proteins that are shown, according to the mouse data in BioGPS. PM20D2 (IPI00869923.2), peptidase M20 domain containing 2; IDE (IPI00199609.1), insulin-degrading enzyme; NDRG2 (IPI00382069.1), N-myc down-regulated family member 2; NPEPPS (IPI00372700.1), aminopeptidase puromycin-sensitive; CNDP2 (IPI00421899.1), cytosolic nonspecific dipeptidase-metallopeptidase M20 family; BLMH (IPI00231419.3), bleomycin hydrolase; DPYSL2 (IPI00870112.1), dihydropyrimidinase-like 2; XPNPEPL1 (IPI00781123.1), X-prolyl aminopeptidase 1(aminopeptidase P1), soluble; PEPD (IPI00364304.2), peptidase D; CAPN1 (IPI00231610.5), calpain 1, large subunit.
| Enzyme activity | Enrichment F35/F49 | Activity in rat Sk. Muscle/livera | ||
|---|---|---|---|---|
| β-Ala-Lys dipeptidase | 1.67 | 20 |
| Peptidase or hydrolase | Type | Mr monomer/multimer | Normalized PSM F35/F49 | Expression in mouse Sk. muscle/liverb |
|---|---|---|---|---|
| PM20D2 | M20: zinc binding domain | 46.7/≈200 | 1.7 | 7 |
| IDE | Zinc metallopeptidase | 117/≈240 | 0.58 | 0.85 |
| NDRG2 | α/β Hydrolase superfamilly | 40.8 | 0.38 | 1.3 |
| NPEPPS | Zinc metallopeptidase | 103 | 0.4 | 2.6 |
| CNDP2 | M20: zinc metallopeptidase | 52.7/≈100 | 0.5 | |
| BLMH | PepC: cysteine peptidase | 52.6/≈300 | 0.73 | 0.8 |
| DPYSL2 | Hydrolase activity on C-N bonds | 62.2/≈240 | 0.35 | 3.9 |
| XPNPEPL1 | Metalloaminopeptidase | 74.6/≈150 | 0.13 | 1.2 |
| PEPD | Peptidase for di- or tri-peptides with C-terminal (hydroxy)proline residues | 54.7/≈110 | 0.5 | 0.65 |
| CAPN1 | Ca2+-activated intracellular cysteine protease | 82.1/≈110 | 0.1 | 7.7 |
Finally, a Western blot experiment using commercial antibodies raised against PM20D2 showed that a band of the expected size for PM20D2 (48 kDa) co-eluted with β-Ala-Lys dipeptidase activity in the last purification step (Fig. 2D). Taken together these data strongly suggested that PM20D2 corresponded to β-Ala-Lys dipeptidase.
Kinetic Properties of Purified Recombinant Mouse and Human PM20D2 Compared with Those of Carnosinase
To confirm the identity of PM20D2 as β-Ala-Lys dipeptidase, we overexpressed both mouse and human enzymes as fusion proteins with a N-terminal His6 tag in E. coli BL21(DE3). To be able to compare PM20D2 kinetic properties with those of the dipeptidase that is dedicated to carnosine hydrolysis, we used the same approach to produce mouse and human carnosinase, taking care to remove the signal peptide sequence from the human enzyme. All four proteins were produced as soluble proteins and purified from E. coli extracts using His-trap columns.
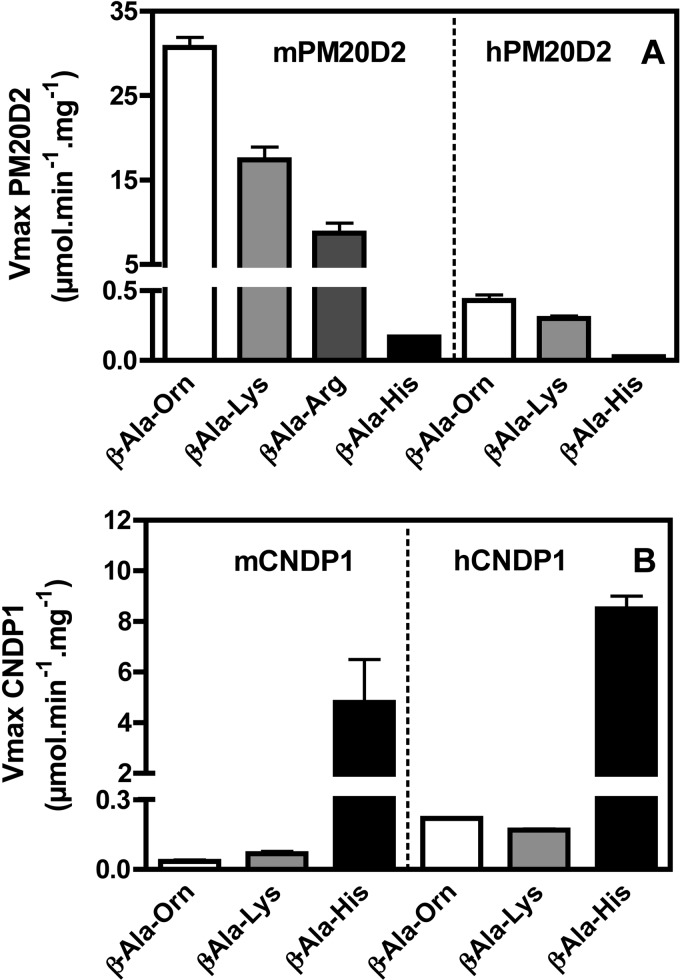
Table 3 shows the activity of all four enzymes on substrates produced by carnosine synthase. Mouse PM20D2 hydrolyzed not only β-Ala-Lys, which confirmed our tentative identification, but also β-Ala-Orn, β-Ala-Arg, GABA-Lys, and GABA-Orn at comparable rates. Remarkably, and in agreement with the lack of inhibition of rat muscle β-Ala-Lys dipeptidase by carnosine, PM20D2 acted almost undetectably on carnosine and homocarnosine.
TABLE 3.
Kinetic properties and specificity of mouse and human dipeptidases PM20D2 and carnosinase
The activities were measured as described using purified recombinant N-terminal His6-tagged enzymes. Values are either the mean ± S.E. (n = 3) or the mean of 2 measurements that did not differ by >10%. When activity was below the detection limit, it is shown as ND (not detected). When the Vmax could not be determined under the experimental conditions used, the activity shown was measured with the substrate concentrations indicated in the footnotes.
| Substrates | Mouse PM20D2 |
Human PM20D2 |
||||
|---|---|---|---|---|---|---|
| Km | Vmax | Kcat/Km | Km | Vmax | Kcat/Km | |
| mm | μmol·min−1·mg−1 | μmol·min−1·mg−1 | mm | μmol·min−1·mg−1 | μmol·min−1·mg−1 | |
| β-Ala-Orn | 1.9 ± 0.16 | 30.7 ± 1.2 | 13.3 × 103 | 6.5 ± 0.9 | 0.43 ± 0.04 | 0.055 × 103 |
| β-Ala-Lys | 1.9 ± 0.31 | 17.4 ± 1.5 | 7.6 × 103 | 5.4 ± 0.7 | 0.30 ± 0.02 | 0.046 × 103 |
| β-Ala-Arg | 6.8 ± 1.4 | 8.7 ± 1.2 | 1.0 × 103 | |||
| β-Ala-His | 0.17a | 0.03a | ||||
| GABA-Orn | 1.35 ± 0.17 | 12.5 ± 0.64 | 7.5 × 103 | 0.24b | ||
| GABA-Lys | 2.6 ± 0.26 | 27 ± 1.4 | 8.4 × 103 | 0.24b | ||
| GABA-His | 0.22a | 0.003a | ||||
| α-Ala-Orn | 1.7 ± 0.34 | 5 ± 0.34 | 2.4 × 103 | 0.12 ± 0.02c | ||
| α-Ala-Lys | 4.7 ± 0.9 | 11.9 ± 1.3 | 2.0 × 103 | 0.09 ± 0.03c | ||
| α-Ala-Arg | 6.3 ± 2.4 | 1.4 ± 0.3 | 0.2 × 103 | |||
| α-Ala-His | NDc | |||||
| Substrates | Mouse carnosinase (CNDP1) |
Human carnosinase (CNDP1) |
||||
|---|---|---|---|---|---|---|
| Km | Vmax | Kcat/Km | Km | Vmax | Kcat/Km | |
| mm | μmol·min−1·mg−1 | μmol·min−1·mg−1 | mm | μmol·min−1·mg−1 | μmol·min−1·mg−1 | |
| β-Ala-Orn | 0.034 ± 0.01b | 0.22 ± 0.002b | ||||
| β-Ala-Lys | 0.068 ± 0.01b | 0.17 ± 0.004b | ||||
| β-Ala-His | 0.28 ± 0.03 | 4.8 ± 1.7 | 16 × 103 | 0.13 ± 0.07 | 8.5 ± 0.5 | 62 × 103 |
| GABA-Orn | 0.008b | 0.003b | ||||
| GABA-Lys | NDb | NDb | ||||
| GABA-His | 1.78 ± 0.94 | 0.086 ± 0.02 | 0.046 × 103 | 8.7 ± 7.1 | 0.36 ± 0.21 | 0.039 × 103 |
a 5 mm.
b 4 mm.
c 10 mm.
Similarly, human PM20D2 hydrolyzed β-Ala-Lys and β-Ala-Orn much better than carnosine and GABA-Lys and GABA-Orn much better than homocarnosine. Human PM20D2, was, however, ∼200-fold less active than the mouse enzyme. This difference in activity was found with all substrates that were tested (Table 3) confirming that it was likely not due to a change in substrate specificity in human PM20D2 but to a decrease in the intrinsic activity of this protein compared with the mouse enzyme. A similar difference in activity between the human and the mouse enzymes was also found when the recombinant proteins were expressed in HEK293T cells (not shown), suggesting that it was not artifactually due to the overexpression of human PM20D2 in E. coli.
As expected, mouse and human carnosinase showed the reverse specificity compared with PM20D2, i.e. they acted on carnosine much better than on β-Ala-Lys and β-Ala-Orn and on homocarnosine much better than on GABA-Lys and GABA-Orn. Taken together these data indicated that PM20D2 was suited to act as a repair enzyme associated with carnosine synthesis (Fig. 3).
FIGURE 3.
Hydrolysis of various β-alanine-containing dipeptides by recombinant mouse and human PM20D2 (β-Ala-Lys dipeptidase) or CNDP1 (carnosinase). Shown are graphic representations of calculated Vmax values (see Table 3) for mouse and human PM20D2 (A) and CNDP1 (B) for the hydrolysis of the indicated dipeptides.
The investigation of the substrate specificity of PM20D2 was extended to peptides that are unlikely to be synthesized by carnosine synthase (Table 3 and 4). The finding that Ala-Lys, Ala-Orn, Ala-Arg, and Lys-Lys (though not Ala-His), are good substrates for the mouse enzyme indicates that the presence of β-alanine or GABA is not mandatory and that these non-standard amino acids can be replaced by standard amino acids. All peptides that are substrates contain a C-terminal basic residue that must be distinct from histidine. This conclusion is also supported by the finding that GABA-Ala and GABA-Leu are not substrates (Table 4) of PM20D2. The absence of activity with GABA-Lys-Ala and amidated GABA-Lys indicate that lysine (and presumably other basic amino acids) must be at the C terminus. The near absence of activity with N(α)-Ac-Lys and with the largest peptides (Ala)4-Lys and (Ala)5-Lys indicates preference for di- or tripeptides, at least if the latter are not too big or too charged (Ala-Ala-Lys is a good substrate, whereas (Lys)3 is not). Remarkably, the human enzyme showed a similar specificity profile but with an activity that was in all cases roughly 2 orders of magnitude lower than that of the mouse enzyme.
TABLE 4.
Substrate specificity of mouse and human recombinant PM20D2
Activity was assayed by measuring the release of the C-terminal lysine using a lysine oxidase-coupled assay or of N-terminal GABA in a NADPH-producing assay coupled to recombinant GABA-transaminase and succinate semialdehyde dehydrogenase from E. coli (see “Experimental Procedures”). mPM20D2, mouse PM20D2; hPM20D2, human PM20D2.
| Substrate (0.5 mm) | mPM20D2 | hPM20D2 |
|---|---|---|
| μmol·min−1·mg−1 | μmol·min−1·mg−1 | |
| GABA-Lys | 4.49 ± 0.22 | 33 ± 1 |
| β-Ala-Lys | 2.73 ± 0.16 | 30 ± 2 |
| Ala-Lys | 1.92 ± 0.13 | 32 ± 1 |
| Ala-Ala-Lys | 2.12 ± 0.10 | 14 ± 1 |
| Ala-Ala-Ala-Lys | 0.01 ± 0.005 | 1.5 ± 0.5 |
| Ala-Ala-Ala-Ala-Lys | 0.02 ± 0.02 | 0.5 ± 0.5 |
| Lys-Lys | 2.39 ± 0.13 | 30 ± 2 |
| Lys-Lys-Lys | 0.05 ± 0.007 | 3 |
| Ala-Lys-Lys | 0.25 ± 0.06 | 1 ± 1 |
| Ala-Ala-Lys-Lys | 0 | 0 |
| Ac-Lys | 0.14 ± 0.03 | 0 |
| γ-Glu-Lys | 0 | 0 |
| GABA-Lys | 3.12 ± 0.14 | 19.0 ± 0.6 |
| GABA-Orn | 4.18 ± 0.12 | 27.3 ± 0.9 |
| GABA-Arg | 0.97 ± 0.09 | 1.8 ± 0.2 |
| GABA-His | 0.05 ± 0.01 | 0.5 ± 0.3 |
| GABA-Ala | 0 | 0 |
| GABA-Leu | 0 | 0 |
| GABA-Lys-Ala | 0 | 0 |
| GABA-Lys-CONH2 | 0 | 0 |
Role of PM20D2 as a Metabolite Repair Enzyme during the Synthesis of Carnosine and Homocarnosine in HEK293T Cells
To investigate the synthesis and degradation of histidine and basic amino acid-containing dipeptides under more physiological conditions, we transfected HEK293T cells with plasmids driving the expression of mouse and human carnosine synthase with or without PM20D2 and/or a cytosolic form of carnosinase. After 24 h of transfection, the media were supplemented with 0.1 mm β-alanine or GABA in combination or not with 1 mm lysine or ornithine, and the cells were further incubated for 24 h. At this stage extracts were prepared, and the concentration of both carnosine or homocarnosine and β-Ala-Lys or -Orn/GABA-Lys or -Orn dipeptides were assayed in deproteinized cell extracts.
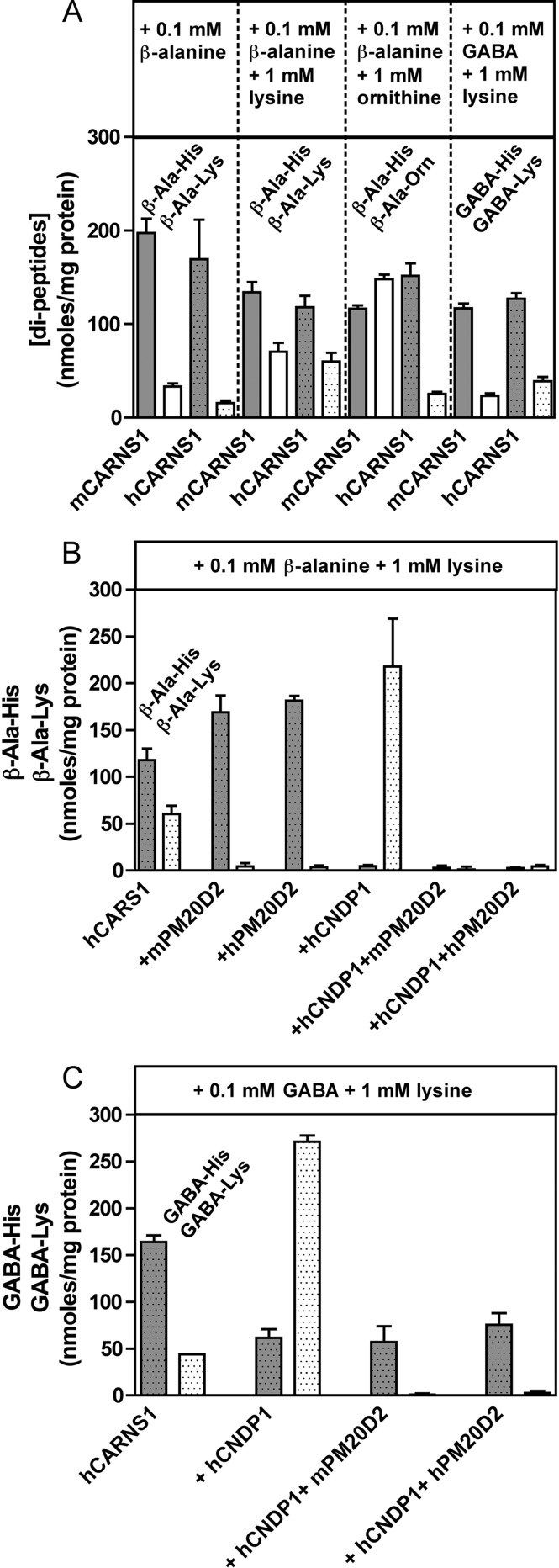
We show in Fig. 4A that HEK293T cells transfected with mouse or human carnosine synthase do indeed synthesize β-Ala-Lys when 0.1 mm β-alanine was added to the culture medium and that this production was further increased if 1 mm lysine was also added. Remarkably, when lysine was replaced by ornithine, mouse carnosine synthase made about as much β-Ala-Orn as carnosine. This was less the case for human carnosine synthase, which is in perfect accordance with the higher specificity of human carnosine synthase compared with the mouse enzyme in studies with purified recombinant proteins (15). Fig. 4A also shows that release Lys was made by carnosine synthase-transfected HEK293T cells when GABA (0.1 mm) and lysine (1 mm) where added to the cell medium.
FIGURE 4.
Synthesis of carnosine, homocarnosine, β-Ala-Lys, and β-Ala-Orn in HEK293T cells transfected with carnosine synthase; effect of the addition of various amino acids in the medium and of cotransfection with mouse or human PM20D2 or human carnosinase. HEK293T cells were transfected for 48h with plasmids expressing various mouse (m) or human (h) proteins. Panel A illustrates the effect of adding β-alanine, lysine, ornithine, and GABA to the cell culture medium on the accumulation of different dipeptides in the cells. Panels B and C illustrate the effect of cotransfection of human carnosine synthase (hCARNS1) with mouse or human PM20D2 and human carnosinase (hCNDP1) in the indicated combinations. The medium was supplemented with β-alanine (panel B) or GABA (panel C) and lysine to favor the formation of carnosine and homocarnosine, respectively. Results are the means ± S.E. for three independent transfections.
Next, we investigated the capacity of both human and mouse PM20D2 to “repair” carnosine synthesis if co-transfected with human carnosine synthase (Fig. 4B) in cells cultured with medium supplemented with β-alanine (0.1 mm) and lysine (1 mm). Strikingly, β-Ala-Lys was undetectable in cells co-transfected with mouse or human PM20D2. Furthermore, neither carnosine nor β-Ala-Lys was detectable in cells in which both carnosinase and human or mouse PM20D2 were co-transfected with human carnosine synthase. Of note is the fact that co-transfection of carnosine synthase with carnosinase led to an increase in β-Ala-Lys content compared with cells that were transfected with carnosine synthase alone and that, reciprocally, co-transfection of carnosine synthase with PM20D2 led to an increase in carnosine. This suggests that β-alanine entry is limiting or that the dipeptides exert feedback inhibition on carnosine synthase.
Fig. 4C shows a similar experiment where β-alanine was replaced by GABA (0.1 mm) and demonstrates that PM20D2 can also repair homocarnosine synthesis in HEK293T cells. Transfection of carnosinase resulted in a partial decrease in the concentration of homocarnosine and to an increase in the concentration of GABA-Lys. Transfection of mouse or human PM20D2 together with carnosine synthase and human carnosinase led to a complete suppression of GABA-Lys accumulation.
PM20D2 Dipeptidase Activity in Mammalian Muscle
Due to the ≈100-fold lower activity of recombinant human PM20D2 compared with the mouse enzyme, we were interested in comparing the activity of this peptidase in tissue extracts of rodents and primates. This activity was determined by measuring the hydrolysis of the best substrate, β-Ala-Orn, at a concentration of 50 μm with a radiochemical assay. Such measurements indicated that mouse and rat muscle showed activities that were 1–2 orders of magnitude higher (respectively, 137 ± 11 and 109 ± 14 pmol·min−1·mg−1) than those observed for two macaque muscles (11 ± 1 and 1 ± 0.1 pmol·min−1·mg−1 protein for splenius capitis and biceps brachii) and for human muscle biopsies (1 ± 0.6 pmol·min−1·mg−1 protein for gastrocnemius). Activities in brain cortex were 24 ± 4, 11 ± 0.6, and 2 ± 0.5 pmol·min−1·mg−1 protein for mouse, rat, and macaque.
DISCUSSION
We report here the molecular identification of an enzyme able to hydrolyze dipeptides containing β-alanine or GABA linked to the basic amino acids lysine, ornithine, or arginine (though not histidine) that we decided to name β-alanyl-lysine dipeptidase. Although this peptidase acts marginally better on other, structurally related dipeptides, this name was chosen because β-Ala-Lys was used as the substrate to measure the enzymatic activity during purification from rat skeletal muscle, which eventually led to the identification of PM20D2. The identification is based on the enrichment of this protein during the purification process, on the co-elution of PM20D2 with β-Ala-Lys dipeptidase, and on the demonstration that recombinant mouse PM20D2 has a similar catalytic action as the enzyme purified from muscle. This enzyme is most likely the same protein as the one described by Kumon et al. (23). Both the purified hog kidney peptidase and recombinant mouse PM20D2 are indeed active on β-Ala-Arg, β-Ala-Lys, β-Ala-Orn, GABA-Lys, and GABA-Orn but display very little or no detectable activity on carnosine or homocarnosine. Furthermore, they are both inhibited with 1,10-phenanthroline and to a lesser extent by EDTA, consistent with their being metalloenzymes with a tight binding divalent cation, most likely Zn2+. They differ from the enzyme described as β-Ala-Arg dipeptidase by Kunze et al. (24) as the latter, contrary to PM20D2 is inhibited by Bestatin (not shown), is dependent on Mn2+, does not act on GABA-Arg, and has only a weak activity on β-Ala-Lys and β-Ala-Orn.
The human enzyme shows similar kinetic properties, yet its activity is lower on β-Ala-Lys and β-Ala-Orn by ≈200-fold while being still higher on β-alanine or GABA-containing dipeptides than on α-alanine containing dipeptides. This low specific activity is found both with PM20D2 produced in bacteria and in mammalian cells, indicating that it is not due to an artifact caused by the expression in bacterial cells. Furthermore, the activity of PM20D2, as assessed with its best substrate, was also found to be much lower in human or macaque muscle or brain than in the same rodent tissues. As the macaque PM20D2 shares ∼97% sequence identity with the human enzyme as compared with 82% with the mouse enzyme, this low specific activity is most likely due to differences in the sequence between the primate and the rodent enzyme.
Protein Family
PM20D2 belongs, as its name implies, to the metalloprotease M20 family. In mammals this family comprises at least two other members that do not act on classical peptides: carnosinase (CNDP1), which acts on carnosine and homocarnosine (22), and aminoacylase 1, which deacetylates free N-acetylamino acids (25) to the exception of N-acetyl aspartate. A fourth member, PM20D1 (very distant from PM20D2), has no known function, and the only protein that acts as a true peptidase is CNDP2, which is a cysteinyl-glycine dipeptidase (26) but also acts on various other true dipeptides (22).
Proteins homologous to PM20D2 comprise HmrA (36% identity), a protein that confers resistance to ampicillin in Staphylococcus and has endopeptidase activity (27), as well as E. coli p-aminobenzoyl-glutamate hydrolase (28) (25% identity) and the auxin-conjugate amidohydrolase (ILL2) from Arabidopsis thaliana (25% identity) involved in auxin metabolism (29). Thus, the PM20D2 homologues appear to exert often functions different from proteolysis.
Physiological Role of PM20D2 in Carnosine/Homocarnosine Synthesis
The mouse enzyme appears to be particularly suited to destroy abnormal peptides made by carnosine synthase. The latter is not very specific, as it may use lysine, ornithine, or arginine instead of histidine. In that respect the mouse enzyme is less specific than the human enzyme, as the relative catalytic efficiencies (compared with histidine) for lysine and ornithine are 1/11 and 1/1.5 in the case of the mouse enzyme compared with 1/20 and 1/40 for the human enzyme. These properties observed in vitro are confirmed by the finding that the ratios of β-Ala-Lys or β-Ala-Orn content to carnosine content are ∼2- and 5-fold higher in cells transfected with mouse instead of human carnosine synthase (Fig. 4A).
Both mouse and human PM20D2 are able to eliminate virtually all the wrong dipeptides from cells transfected with carnosine synthase. This is of course under conditions of strong overexpression of PM20D2, but it should be stressed that there is also a strong expression of carnosine synthase under these conditions. Under physiological conditions, the activity present in mouse muscle is sufficient to decrease the concentration of β-Ala-Lys and β-Ala-Orn by 50% in 1 min as can be calculated from the activity measured in muscle tissue and by considering that the Km of PM20D2 for these peptides is much higher than their physiological concentrations. Such activity is, therefore, largely sufficient to destroy the wrong dipeptides in rodent muscle.
The ∼100-fold lower activity found in human muscle suggests that the half-life of β-Ala-Lys and β-Ala-Orn would be on the order of 100 min, which may seem quite high. However, carnosine synthesis is a very slow process, as indicated by the finding that supplements of β-alanine increase the muscle carnosine content by only 10% per week (30). Thus there is no need for an extremely rapid correction process.
Other Roles of PM20D2
Tissue expression suggests that there is no strict parallelism between the tissue expression of carnosine synthase and PM20D2. Furthermore, homologues of PM20D2 are found in organisms (fungi, several fishes) that do not have carnosine synthase homologues. Thus, PM20D2 may have other functions related or not with its ability to act on non-classical dipeptides. It is important to stress in this respect that what seems to be extremely low activities may have physiological significance. Thus, CNDP1, which is a carnosinase that is much more active on carnosine than on homocarnosine (3000-fold higher catalytic efficiency on the former than on the latter substrate; see Table 3 and Refs. 22, 31, and 32), effectively acts on homocarnosine in vivo, as indicated by the finding that deficiency in carnosinase leads to homocarnosinuria (32, 33).
Conclusion
In conclusion, carnosine synthesis in mouse muscle is a clear cut example of a situation where biosynthetic specificity depends on the presence of two enzymes: a biosynthetic enzyme, which has limited specificity (carnosine synthase), assisted by a second enzyme (PM20D2), which essentially degrades all the wrong synthetic products to leave only the physiologically relevant product. The situation in humans is somewhat different, as carnosine synthase is more specific and PM20D2 less active than its rodent counterparts.
Acknowledgments
We thank Farah Hadi and Jakub Drozak for contributions during the initial steps of this project.
This work was supported by grants from the Fonds National de la Recherche Scientifique (FNRS) and the Interuniversity Attraction Poles Programme, Belgian Science Policy (Networks P7/43).
- PM20D2
- peptidase M20 domain containing 2/β-alanyl-lysine dipeptidase
- CNDP1
- carnosinase.
REFERENCES
- 1. Linster C. L., Van Schaftingen E., Hanson A. D. (2013) Metabolite damage and its repair or pre-emption. Nat. Chem. Biol. 9, 72–80 [DOI] [PubMed] [Google Scholar]
- 2. Van Schaftingen E., Rzem R., Marbaix A., Collard F., Veiga-da-Cunha M., Linster C. L. (2013) Metabolite proofreading, a neglected aspect of intermediary metabolism. J. Inherit. Metab. Dis. 36, 427–434 [DOI] [PubMed] [Google Scholar]
- 3. Rzem R., Veiga-da-Cunha M., Noël G., Goffette S., Nassogne M. C., Tabarki B., Schöller C., Marquardt T., Vikkula M., Van Schaftingen E. (2004) A gene encoding a putative FAD-dependent l-2-hydroxyglutarate dehydrogenase is mutated in l-2-hydroxyglutaric aciduria. Proc. Natl. Acad. Sci. U.S.A. 101, 16849–16854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rzem R., Van Schaftingen E., Veiga-da-Cunha M. (2006) The gene mutated in l-2-hydroxyglutaric aciduria encodes l-2-hydroxyglutarate dehydrogenase. Biochimie 88, 113–116 [DOI] [PubMed] [Google Scholar]
- 5. Rzem R., Vincent M. F., Van Schaftingen E., Veiga-da-Cunha M. (2007) l-2-hydroxyglutaric aciduria, a defect of metabolite repair. J. Inherit. Metab. Dis. 30, 681–689 [DOI] [PubMed] [Google Scholar]
- 6. Van Schaftingen E., Rzem R., Veiga-da-Cunha M. (2009) l-2-Hydroxyglutaric aciduria, a disorder of metabolite repair. J. Inherit. Metab. Dis. 32, 135–142 [DOI] [PubMed] [Google Scholar]
- 7. Haliloglu G., Jobard F., Oguz K. K., Anlar B., Akalan N., Coskun T., Sass J. O., Fischer J., Topcu M. (2008) l-2-Hydroxyglutaric aciduria and brain tumors in children with mutations in the L2HGDH gene: neuroimaging findings. Neuropediatrics 39, 119–122 [DOI] [PubMed] [Google Scholar]
- 8. Kranendijk M., Struys E. A., Salomons G. S., Van der Knaap M. S., Jakobs C. (2012) Progress in understanding 2-hydroxyglutaric acidurias. J. Inherit. Metab. Dis. 35, 571–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marbaix A. Y., Noël G., Detroux A. M., Vertommen D., Van Schaftingen E., Linster C. L. (2011) Extremely conserved ATP- or ADP-dependent enzymatic system for nicotinamide nucleotide repair. J. Biol. Chem. 286, 41246–41252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Linster C. L., Noël G., Stroobant V., Vertommen D., Vincent M. F., Bommer G. T., Veiga-da-Cunha M., Van Schaftingen E. (2011) Ethylmalonyl-CoA decarboxylase, a new enzyme involved in metabolite proofreading. J. Biol. Chem. 286, 42992–43003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kalyankar G. D., Meister A. (1959) Enzymatic synthesis of carnosine and related β-alanyl and γ-aminobutyryl peptides. J. Biol. Chem. 234, 3210–3218 [PubMed] [Google Scholar]
- 12. Horinishi H., Grillo M., Margolis F. L. (1978) Purification and characterization of carnosine synthetase from mouse olfactory bulbs. J. Neurochem. 31, 909–919 [DOI] [PubMed] [Google Scholar]
- 13. Matsuoka M., Nakajima T., Sano I. (1969) Identification of α-(β-alanyl)-lysine in rabbit muscle. Biochim. Biophys. Acta 177, 169–171 [DOI] [PubMed] [Google Scholar]
- 14. Nakajima T., Kakimoto Y., Kumon A., Matsuoka M., Sano I. (1969) α-(γ-Aminobutyryl)-lysine in mammalian brain: its identification and distribution. J. Neurochem. 16, 417–422 [DOI] [PubMed] [Google Scholar]
- 15. Drozak J., Veiga-da-Cunha M., Vertommen D., Stroobant V., Van Schaftingen E. (2010) Molecular identification of carnosine synthase as ATP-grasp domain-containing protein 1 (ATPGD1). J. Biol. Chem. 285, 9346–9356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Boldyrev A. A., Aldini G., Derave W. (2013) Physiology and pathophysiology of carnosine. Physiol. Rev. 93, 1803–1845 [DOI] [PubMed] [Google Scholar]
- 17. Kohen R., Yamamoto Y., Cundy K. C., Ames B. N. (1988) Antioxidant activity of carnosine, homocarnosine, and anserine present in muscle and brain. Proc. Natl. Acad. Sci. U.S.A. 85, 3175–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bauer K. (2005) Carnosine and homocarnosine, the forgotten, enigmatic peptides of the brain. Neurochem. Res. 30, 1339–1345 [DOI] [PubMed] [Google Scholar]
- 19. Arts I. S., Ball G., Leverrier P., Garvis S., Nicolaes V., Vertommen D., Ize B., Tamu Dufe V., Messens J., Voulhoux R., Collet J.-F. (2013) Dissecting the machinery that introduces disulfide bonds in Pseudomonas aeruginosa. MBio 4, e00912–00913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Veiga-da-Cunha M., Hadi F., Balligand T., Stroobant V., Van Schaftingen E. (2012) Molecular identification of hydroxylysine kinase and of ammoniophospholyases acting on 5-phosphohydroxy-l-lysine and phosphoethanolamine. J. Biol. Chem. 287, 7246–7255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Veiga-da-Cunha M., Tyteca D., Stroobant V., Courtoy P. J., Opperdoes F. R., Van Schaftingen E. (2010) Molecular identification of NAT8 as the enzyme that acetylates cysteine S-conjugates to mercapturic acids. J. Biol. Chem. 285, 18888–18898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Teufel M., Saudek V., Ledig J. P., Bernhardt A., Boularand S., Carreau A., Cairns N. J., Carter C., Cowley D. J., Duverger D., Ganzhorn A. J., Guenet C., Heintzelmann B., Laucher V., Sauvage C., Smirnova T. (2003) Sequence identification and characterization of human carnosinase and a closely related non-specific dipeptidase. J. Biol. Chem. 278, 6521–6531 [DOI] [PubMed] [Google Scholar]
- 23. Kumon A., Matsuoka Y., Kakimoto Y., Nakajima T., Sano I. (1970) A peptidase that hydrolyzes Nα-(γ-aminobutyryl)lysine. Biochim. Biophys. Acta 200, 466–474 [DOI] [PubMed] [Google Scholar]
- 24. Kunze N., Kleinkauf H., Bauer K. (1986) Characterization of two carnosine-degrading enzymes from rat brain. Partial purification and characterization of a carnosinase and a β-alanyl-arginine hydrolase. Eur. J. Biochem. 160, 605–613 [DOI] [PubMed] [Google Scholar]
- 25. Sass J. O., Mohr V., Olbrich H., Engelke U., Horvath J., Fliegauf M., Loges N. T., Schweitzer-Krantz S., Moebus R., Weiler P., Kispert A., Superti-Furga A., Wevers R. A., Omran H. (2006) Mutations in ACY1, the gene encoding aminoacylase 1, cause a novel inborn error of metabolism. Am. J. Hum. Genet. 78, 401–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kaur H., Kumar C., Junot C., Toledano M. B., Bachhawat A. K. (2009) Dug1p Is a Cys-Gly peptidase of the γ-glutamyl cycle of Saccharomyces cerevisiae and represents a novel family of Cys-Gly peptidases. J. Biol. Chem. 284, 14493–14502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Botelho T. O., Guevara T., Marrero A., Arêde P., Fluxà V. S., Reymond J. L., Oliveira D. C., Gomis-Rüth F. X. (2011) Structural and functional analyses reveal that Staphylococcus aureus antibiotic resistance factor HmrA is a zinc-dependent endopeptidase. J. Biol. Chem. 286, 25697–25709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Green J. M., Hollandsworth R., Pitstick L., Carter E. L. (2010) Purification and characterization of the folate catabolic enzyme p-aminobenzoyl-glutamate hydrolase from Escherichia coli. J. Bacteriol. 192, 2407–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bitto E., Bingman C. A., Bittova L., Houston N. L., Boston R. S., Fox B. G., Phillips G. N., Jr. (2009) X-ray structure of ILL2, an auxin-conjugate amidohydrolase from Arabidopsis thaliana. Proteins 74, 61–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stellingwerff T., Anwander H., Egger A., Buehler T., Kreis R., Decombaz J., Boesch C. (2012) Effect of two β-alanine dosing protocols on muscle carnosine synthesis and washout. Amino Acids 42, 2461–2472 [DOI] [PubMed] [Google Scholar]
- 31. Jackson M. C., Kucera C. M., Lenney J. F. (1991) Purification and properties of human serum carnosinase. Clin. Chim. Acta 196, 193–205 [DOI] [PubMed] [Google Scholar]
- 32. Lenney J. F., Peppers S. C., Kucera C. M., Sjaastad O. (1983) Homocarnosinosis: lack of serum carnosinase is the defect probably responsible for elevated brain and CSF homocarnosine. Clin. Chim. Acta 132, 157–165 [DOI] [PubMed] [Google Scholar]
- 33. Gjessing L. R., Lunde H. A., Mørkrid L., Lenney J. F., Sjaastad O. (1990) Inborn errors of carnosine and homocarnosine metabolism. J. Neural. Transm. Suppl. 29, 91–106 [DOI] [PubMed] [Google Scholar]