Background: TpsB transporters secrete large exoproteins across the outer membrane of Gram-negative bacteria.
Results: Exchanging the polypeptide transport-associated (POTRA) domains between TpsBs switches their substrate specificity.
Conclusion: POTRA domains select and bind TpsB substrates to initiate secretion.
Significance: These findings increase our understanding of how bacteria secrete proteins that increase their virulence, using a transporter of a protein family found in pro- and eukaryotes.
Keywords: Bacterial Pathogenesis, Membrane Protein, Membrane Transport, Protein Secretion, Virulence Factor, Omp85 Protein Family, POTRA Domains, Two-partner Secretion System
Abstract
The two-partner secretion (TPS) systems of Gram-negative bacteria secrete large TpsA exoproteins by a dedicated TpsB transporter in the outer membrane. TpsBs contain an N-terminal module located in the periplasm that includes two polypeptide transport-associated (POTRA) domains. These are thought to initiate secretion of a TpsA by binding its N-terminal secretion signal, called the TPS domain. Neisseria meningitidis encodes up to five TpsA proteins that are secreted via only two TpsB transporters: TpsB1 and TpsB2. Of these two, the TpsB2 recognizes the TPS domains of all TpsAs, despite their sequence diversity. By contrast, the TpsB1 shows a limited recognition of a TPS domain that is shared by two TpsAs. The difference in substrate specificity of the TpsBs enabled us to investigate the role of the POTRA domains in the selection of TPS domains. We tested secretion of TPS domains or full-length TpsAs by TpsB mutants with deleted, duplicated, and exchanged POTRA domains. Exchanging the two POTRA domains of a TpsB resulted in a switch in specificity. Furthermore, exchanging a single POTRA domain showed that each of the two domains contributed to the cargo selection. Remarkably, the order of the POTRA domains could be reversed without affecting substrate selection, but this aberrant order did result in an alternatively processed secretion product. Our results suggest that secretion of a TpsA is initiated by engaging both POTRA domains of a TpsB transporter and that these select the cognate TpsAs for secretion.
Introduction
Proteins of the type V secretion pathway of Gram-negative bacteria cross the cell envelope consisting of the inner membrane, the peptidoglycan-containing periplasmic space, and the outer membrane in a series of consecutive steps (1). Within the type V secretion pathway, the two-partner secretion (TPS)4 systems form a distinct subclass (1, 2). TPS systems comprise a secreted TpsA protein and an outer membrane-embedded TpsB transporter. TpsAs are large exoproteins of more than 100 kDa and function as bacterial adhesins, as toxins for bacterial or eukaryotic targets, and in obtaining nutrients from the environment. Both TpsA and TpsB proteins are synthesized with an N-terminal signal peptide and are transported across the inner membrane via the Sec complex. The TpsB then inserts into the outer membrane and binds and secretes the TpsA across this membrane. TpsAs target their TpsB translocator via a TPS domain located at the N terminus of the processed TpsA.
The TpsB proteins belong to the Omp85 family of proteins that also includes the BamA protein involved in the biogenesis of outer membrane-based β-barrel proteins and its eukaryotic homologs (3, 4). The family is characterized by a C-terminal 16-stranded β-barrel and a soluble module of 1–5 polypeptide transport-associated (POTRA) domains. POTRA domains are also found outside the Omp85 family (e.g. for example in the cell division protein FtsQ) (5–7). TpsB proteins contain two POTRA domains, as shown by the crystal structure of FhaC, the TpsB of filamentous hemagglutinin of Bordetella pertussis (FHA) (8). POTRA domains adopt a conserved βααββ configuration that folds into a three-stranded β-sheet overlaid with two anti-parallel α-helices, although the α2 is missing in the POTRA1 of FhaC (4, 8–11) (Fig. 1). Several lines of evidence indicate that POTRA domains interact directly with the TpsB substrates. Two POTRA domains of BamA of Escherichia coli were shown by NMR to change conformation when incubated with peptides derived from a β-barrel OMP (12). Surface plasmon resonance measurements as well as pull-down and overlay experiments indicated binding of TPS domains to isolated POTRA domains of their cognate TpsBs (8, 13–15) or full-length TpsB in the case of FhaC (14, 15), whereas deletion of POTRA domains abolished the secretion of truncated FHA constructs (8). By contrast, deletion of POTRA domains of BamA proteins of E. coli and Neisseria meningitidis rendered the proteins partially functional (9, 16). In N. meningitidis, it was possible to delete four of the five POTRA domains of BamA without affecting the viability, although it affected the efficiency of β-barrel protein assembly progressively (16). In E. coli, single POTRA domains 1 and 2 could be deleted from the array of five, but not 3–5 (9). A particular feature of FhaC and other TpsBs that is not shared with other members of the Omp85 family is that the channel inside the β-barrel domain is occupied by both an α-helix that precedes the two POTRA domains in the sequence and an extended external loop (loop 6) that folds inward. In the recent crystal structures of BamA (17), the β-barrel channel is not blocked but closed off by a dome formed by the extracellular loops.
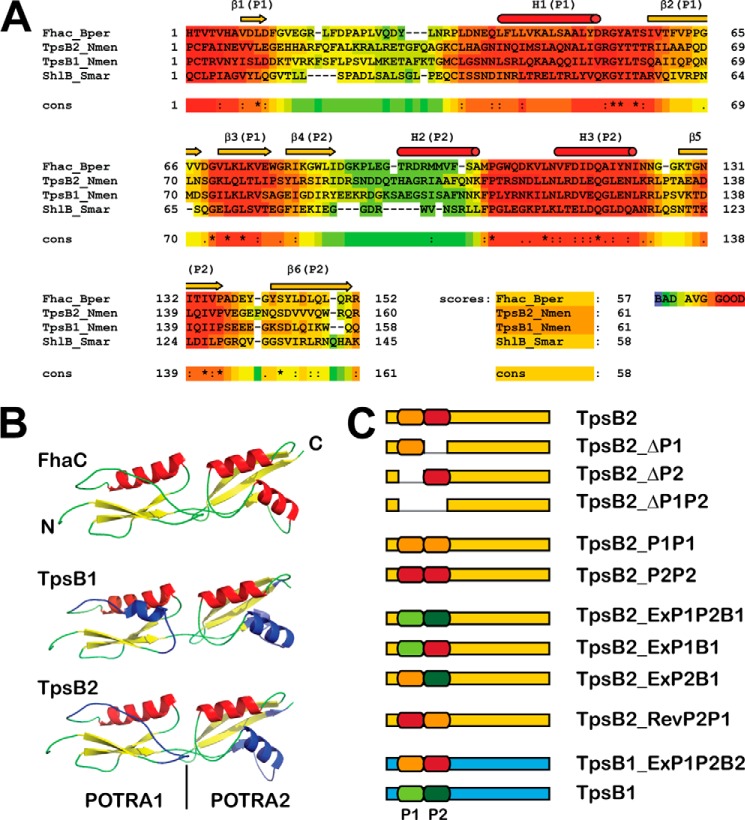
FIGURE 1.
A, alignment of the TpsB1 and TpsB2 of N. meningitidis H44/76 with FhaC of B. pertussis and ShlB of S. marcescens using M-Coffee (30). The color coding represents the quality of the alignment from bad (blue/green) to good (red), and the percentage scores are given. P1, POTRA1; P2, POTRA2. B, schematic models of the crystallized POTRA domains of FhaC and the modeled POTRA domains of TpsB1 and TpsB2 obtained using the Phyre2 program (31). The areas that showed a lower score in the alignment are colored blue in the models. C, graphic representation of the various TpsB2 and TpsB1 constructs used in the article. The absent parts in the deletion mutants of TpsB2 are depicted as gaps in the sequences. For expression, they are combined with either TPS1 or TPS2 domains, as indicated in Table 2 and under “Results.”
The Gram-negative diplococcus N. meningitidis (meningococcus) is a major cause of meningitis and sepsis worldwide (18). N. meningitidis genomes encode up to five TpsA proteins organized into three TPS systems that have been implicated in pathogenesis (19, 20). Of these three, TPS system 1 is ubiquitous, whereas systems 2 and 3 are more prevalent among hyperinvasive clonal complexes. Furthermore, antibodies binding to the TPS domains of the latter two systems have been detected in sera of patients recovering from meningococcal disease. Functions have only been attributed to system 1. TpsA1 (HrpA) promotes adherence to and the intracellular survival and escape from cultured human epithelial cell lines (21, 22), is involved in biofilm formation (23), and acts as contact-dependent toxin involved in bacterial fratricide against other meningococcal strains (24).
Remarkably, the genomes encode only two tpsB genes (i.e. tpsB1 and tpsB2), which are located in an operon with either the system 1 tpsA1a ORF or system 2 tpsA2a ORF, respectively (19, 25). Both systems also encode and express a second tpsA (i.e. tpsA1b and tpsA2b, respectively) that is not part of the operon. The system 3 TpsA is encoded on a genetic island that lacks a gene encoding a dedicated TpsB. Our previous study showed that the TPS domain of TpsA3 is efficiently secreted by TpsB2 (26). In fact, the TpsB2 showed a relaxed system specificity and was able to recognize and secrete TPS domains of N. meningitidis and Neisseria lactamica. In contrast, the TpsB1 of system 1 transported only cognate TPS domains, with the exception of one TPS domain of N. lactamica. Here, we have used this difference in substrate specificity between TpsB2 and TpsB1 to investigate the function of the POTRA domains in the recognition and selection of TPS domains of secreted TpsA proteins. We found that both POTRA domains are crucial for and define this specificity. Interestingly, the order of the POTRA domains seems irrelevant for secretion.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Growth Conditions
The N. meningitidis strains HB-1 tpsB1::kan/tpsB2::gen (26), BB-1, BB-1 ΔtpsA-C, and BB-1 ΔtpsB (24) were grown on GC agar (Oxoid) supplemented with Vitox (Oxoid) at 37 °C, 5% CO2, supplemented with 8 μg/ml chloramphenicol, 60 μg/ml gentamycin, or 15 μg/ml rifampicin when needed. Liquid cultures of N. meningitidis strains were grown at 37 °C in tryptic soy broth (Gibco-BRL). E. coli strains Top10F′ (Invitrogen) and DH5a were grown on lysogeny broth (LB) or LB agar plates supplemented with 100 μg/ml ampicillin or 30 μg/ml chloramphenicol for plasmid maintenance and with 0.5% glucose for full repression of the lac operator when appropriate.
Plasmid Construction
The wild-type tpsB ORFs analyzed here were obtained by PCR using chromosomal DNA obtained from lysed N. meningitidis HB-1 cells as template and Phusion DNA polymerase (Finnzymes) as the synthesizing enzyme according to the recommendations of the manufacturer. The ORFs of mutant tpsBs were obtained by using either chromosomal DNA or pGEM-T plasmids (Promega) containing wild-type or mutant tpsB ORFS as the template. The primers, templates, and PCRs used are listed in Table 1, and the constructed ORFs are depicted in Fig. 1C. The final PCR amplicons were cloned into the pGEM-T cloning vector and confirmed by sequencing (Macrogen). Cloning involved 1–3 PCR steps, and the primers contained overlapping sequences to enable extensions and overlapping PCRs (Table 1). Single-step PCRs were used to obtain the wild-type ORFs of TpsB1 and TpsB2. Two-step PCRs in which the first PCR amplicon was used as a megaprimer in the second PCR were used to obtain mutants TpsB2_ΔP1, TpsB2_ΔP2, and TpsB2_ΔP1P2. Three-step PCRs yielded the ORFs of TpsB2_ExP1P2B1. The first PCR amplicon was extended with a POTRA-encoding sequence in the second PCR, and the resulting amplicon then served as a megaprimer in the third PCR. Another three-step PCR strategy yielded ORFs for TpsB2_P1P1, TpsB2_P2P2, TpsB2_ExP1B1, TpsB2_ExP2B1, and TpsB2_RevP2P1 and involved two PCRs to obtain the 5′- and 3′- regions, which were then joined in a third PCR combining the overlapping amplicons.
TABLE 1.
Primers and cloning strategies used for tpsB ORFs
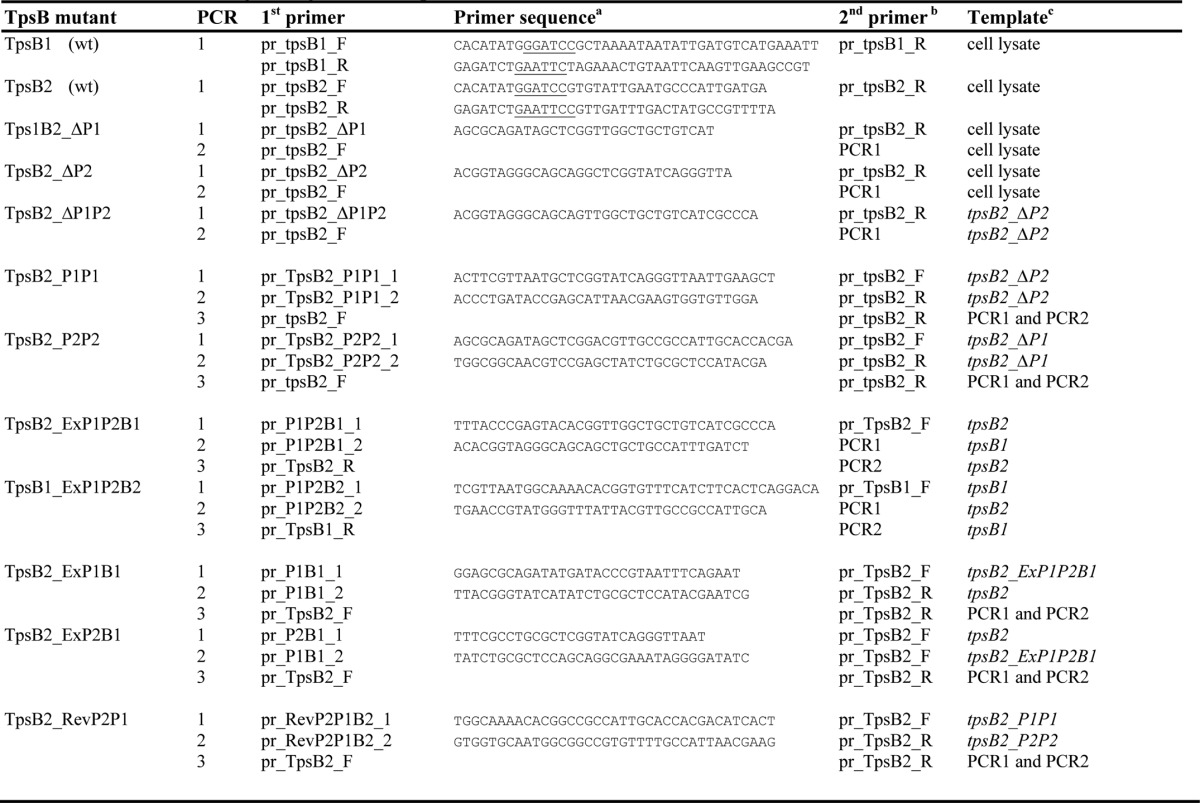
a Primer sequences are given when the primer is mentioned for the first time; restriction sites used for cloning are underlined.
b PCR1 and PCR2 indicate the use of an amplicon of a PCR from a previous step in the cloning procedure.
c Chromosomal DNA came from cell lysates of N. meningitidis strain H44/76; the ORFs mentioned are inserted in pGEM-T vectors; the amplicons mentioned derive from previous steps in the cloning procedure.
The tpsB ORFs were cloned as a single gene or in combination with truncated tpsA ORFs into the pEN vector, which is a neisserial expression vector (27) (Table 2). The constructs of wild-type tpsB2 and tpsB1 ORFs in combination with truncated tpsA1 or tpsA2 ORFs have been described earlier (26). Mutated ORFs were first subcloned downstream of the truncated tpsA ORF in pPU1000 (tpsA1a), pPU1200 (tpsA2a), or pPU1300 (tpsA2b) (19) using the EcoRI and BamHI restriction sites inserted in the primers. Then the combination of the mutant tpsB ORF and the truncated tpsA was cloned into the pEN vector using NdeI and AatII. The tpsB2_ExP1P2B1 and tpsB1_ExP1P2B2 ORFs were cloned into pEN as single genes using NdeI and AatII.
TABLE 2.
Expression and cloning vectors used in this study
| Construct | Plasmid name | TPS ORFa | Source/Reference |
|---|---|---|---|
| TPS2a with TpsB2 and its derivatives | |||
| TPS2a + TpsB2 | pEN1220 | tpsA2a-tr | Ref. 26 |
| TPS2a + TpsB2_ΔP1 | pEN1222 | tpsA2a-tr | This study |
| TPS2a + TpsB2_ΔP2 | pEN1223 | tpsA2a-tr | |
| TPS2a + TpsB2_ΔP1P2 | pEN1224 | tpsA2a-tr | |
| TPS2a + TpsB2_P1P1 | pEN1226 | tpsA2a-tr | |
| TPS2a + TpsB2_P2P2 | pEN1227 | tpsA2a-tr | |
| TPS2a + TpsB2_ExP1P2B1 | pEN1225 | tpsA2a-tr | |
| TPS2a + TpsB2_ExP1B1 | pEN1232 | tpsA2a-tr | |
| TPS2a + TpsB2_ExP2B1 | pEN1233 | tpsA2a-tr | |
| TPS2a + TpsB2_RevP2P1 | pEN1234 | tpsA2a-tr | |
| TPS2a with TpsB1 and its derivatives | |||
| TPS2a + TpsB1 | pEN1250 | tpsA2a-tr | Ref. 26 |
| TPS2a + TpsB1_ExP1P2B2 | pEN1229 | tpsA2a-tr | This study |
| TPS2b with TpsB2 and its derivatives | |||
| TPS2b + TpsB2 | pEN1320 | tpsA2b-tr | Ref. 26 |
| TPS2b + TpsB2P2P1 | pEN1334 | tpsA2b-tr | This study |
| TPS1 with TpsB1, TpsB2, and their derivatives | |||
| TPS1 + TpsB1 | pEN1030 | tpsA1a-tr | Ref. 26 |
| TPS1 + TpsB2 | pEN1050 | tpsA1a-tr | Ref. 26 |
| TPS1 + TpsB1_ExP1P2B2 | pEN1029 | tpsA1a-tr | This study |
| TPS1 + TpsB2_ExP1P2B1 | pEN1028 | tpsA1a-tr | |
| TPS1 + TpsB2_ExP1B1 | pEN1032 | tpsA1a-tr | |
| TPS1 + TpsB2_ExP2B1 | pEN1033 | tpsA1a-tr | |
| TPS1 + TpsB2_RevP2P1 | pEN1034 | tpsA2b-tr | |
| Not combined with TPS construct | |||
| TpsB2_ExP1P2B1 | pEN_tpsB2_ExP1P2B1 | This study | |
| TpsB1_ExP1P2B2 | pEN_tpsB2_ExP1P2B2 | ||
| Cloning vectors | |||
| TPS1 | pPU1000 | tpsA1a-tr | Ref. 19 |
| TPS2a | pPU1200 | tpsA2a-tr | Ref. 19 |
| TPS2b | pPU1300 | tpsA2b-tr | Ref. 19 |
| pGEM-T | Finnzymes | ||
| pEN300 | Ref. 27 | ||
a The suffix “-tr” indicates that the construct comprises a truncated tpsA ORF that encodes the signal peptide and the TPS domain (26).
SDS-PAGE and Western Blotting
All procedures were carried out as described earlier (19, 26). Briefly, N. meningitidis HB-1 cultures were grown for 4–6 h to an optical density at 600 nm (A600) of ∼2.5–3.5 in the presence or absence of 0.1 or 0.25 mm IPTG. Cells were harvested by centrifugation (4,500 × g, 5 min), and the pellet was resuspended in PBS, pH 7.4, to a final A600 of 10. The whole cell lysates were obtained by adding an equal volume of 2× sample buffer (125 mm Tris-HCl, pH 6.8, 20% (v/v) glycerol, 0.02% (w/v) bromphenol blue, 40 mm DTT, and 4% (w/v) SDS and boiling the samples for 10 min. Culture supernatants were centrifuged (16,000 × g, 10 min) to remove residual cells, and the culture supernatant was then subjected to ultracentrifugation (200,000 × g, 1 h) in a bench top ultracentrifuge (Beckman and Coulter). Proteins were precipitated from the supernatants with 5% TCA and dissolved in a volume of PBS corresponding to a cell density of A600 100 (10× concentrated compared with cell samples), further diluted in 2× sample buffer, and boiled for 10 min. Protein samples were separated on 7.5–10% or 12% SDS-polyacrylamide gels and stained with Coomassie Brilliant Blue G250, or proteins were blotted onto nitrocellulose for Western blot analyses.
Blots were preincubated in blocking buffer (PBS with 0.5% skim milk powder (Fluka) and 0.1% (v/v) Tween 20 (Merck)) for at least 4 h. Sera were diluted 1:5,000 (anti-TPS1 and anti-TPS2) or 1:10,000 (anti-TpsB1 and anti-TpsB2) in blocking buffer and incubated for 1–2 h. Blots were washed and incubated for 1 h with goat anti-rabbit immunoglobulin G serum conjugated to horseradish peroxidase (BIOSOURCE) diluted 1:10,000 in blocking buffer. The binding of antibodies to the blots was visualized using Lumilight normal or Plus (Roche Applied Science). The indicated relative molecular weight of the proteins was deduced from the Precision Plus protein standard (Bio-Rad), which was included in each SDS-polyacrylamide gel.
Outer Membrane Isolation and Heat Modifiability
Outer membrane fractions were isolated according to Ref. 28. Cells expressing the tpsB gene to be analyzed were harvested by centrifugation (4,500 × g, 5 min). The pellet was stored in the freezer (>18 h) to kill the bacteria and washed. Cells were resuspended in a 50 mm Tris-HCl, pH 8.0, and 2 mm EDTA buffer and were passed two times through a One Shot cell disrupter (Constant Systems Ltd.) at 30,000 p.s.i. Unbroken cells were pelleted by centrifuging the lysate (4,500 × g, 5 min). The supernatant was then subjected to ultracentrifugation (200,000 × g, 30 min). The resulting pellet containing a crude outer membrane fraction was resuspended in PBS to represent a culture of A600 ∼20. The denatured samples that were further diluted in 2× sample buffer and boiled for 10 min. The native samples were diluted in 2× seminative sample buffer (i.e. 2× sample buffer with 0.4% SDS and lacking DTT) and kept at room temperature for 10 min. Samples were loaded on seminative SDS-polyacrylamide gels prepared without SDS in the gel and run at 12 mA for at least 3 h while cooled in ice. Blotting was performed as described above.
Growth Inhibition Assay
Gentamycin-resistant BB-1 and BB-1 ΔtpsB1 carrying pEN_TpsB1_ExP1P2B2 or pEN_TpsB2_ExP1P2B1 (the killer strains) were mixed with BB-1 ΔtpsA-C, which carries pFP10 as empty vector, and was selected as a spontaneous rifampicin-resistant mutant (the bait) (24). The strains were cultured in TSB without antibiotics to an A600 of ∼3.0, after which cultures were mixed in a 1:1 ratio. Drops were spotted onto GC agar plates with chloramphenicol when needed for plasmid maintenance and containing 0.25 mm IPTG when induction of the tpsB mutants from plasmid was desired. After two nights of incubation, the cells were taken from the plate, resuspended in fresh TSB, and then plated in serial dilutions on GC agar plates containing either rifampicin or gentamycin to determine the colony-forming units. All strains showed equal growth and remained resistant or sensitive against the antibiotics used when cultured independently. Results are expressed as the ratio of the ΔtpsA-tpsC mutant over BB-1 or BB-1 ΔtpsB strains carrying the respective plasmids and are given as the mean and S.D. of three independent experiments.
In Silico Sequence Analyses
Sequence analyses were performed with amino acid sequences of mature TpsBs, which lack the sequence of the signal peptide. The signal peptides were identified by analyzing the full-length sequence with SignalP 4.0 (29). Initial pairwise sequence alignments of TpsB proteins were performed using the Bl2Seq server, using standard parameters. Additional multiple sequence alignments were performed with the mature TpsB sequence and the sequences encompassing the POTRA domains by using the M-Coffee program (30). The encoded protein sequence models of the mature TpsBs of N. meningitidis and their POTRA domains were generated by the Phyre2 Web site (31). The quality of the obtained model was assessed by comparing it with the crystallized FhaC, using the TM-align program (32).
RESULTS
POTRA Domain Assignment in TpsB1 and TpsB2
To construct various POTRA mutants of TpsB1 and TpsB2 derived from N. meningitidis strain H44/76, we used the crystal structure of FhaC (8) to build structural models. TpsB1 and TpsB2 show a reasonable similarity to FhaC with an identity/similarity of 26%/42% for TpsB1 and 27%/44% for TpsB2, respectively. Sequence alignments and structural modeling identified amino acid residues 69–148 and 149–226 of TpsB1 and 85–164 and 165–244 of TpsB2 as POTRA1 and POTRA2, respectively (Fig. 1, A and B). Differences between the POTRA domains cluster in two specific regions of the POTRA domains and include α-helix 3 of the structural elements, which is not part of the proposed POTRA binding surface for TPS domains (15). When we mention POTRA1 or -2 in the remainder of the study, we refer to these assigned sequences (Fig. 1C and Table 2). We focused primarily on the TpsB2 protein because this protein showed a relaxed specificity in the secretion of minimal TPS constructs that consist of the signal peptide and TPS domain of one of the TpsAs of the neisserial TPS systems (26).
Deletion of POTRA Domains from TpsB2 Prevents Secretion
To investigate the role of the POTRA domains in secretion, we first deleted POTRA1, POTRA2, or both POTRA domains of TpsB2 (Fig. 1C). The ORFs encoding these mutated TpsB proteins were cloned in a neisserial expression vector downstream of a truncated tpsA2a gene under the control of a lac promoter (26). This truncated tpsA2a includes the TPS domain and represents the minimal TpsA protein that is still secreted. Plasmids carrying the wild-type tpsB2 or the truncated derivatives were used to transform H44/76 derivative tpsB1kan tpsB2::gen (19, 26). In these strains, the tpsB2 and/or tpsB1 genes are replaced by kanamycin and gentamycin resistance cassettes, respectively. Cultures were grown in the presence or absence of 0.1 mm IPTG (Fig. 2). Samples of whole cell lysates and concentrated culture supernatants were analyzed by Western blotting using anti-TpsB2 and TPS2a antisera. As expected, the combination of the TPS2a construct with the wild-type tpsB2 resulted in an efficient secretion of TPS2a (26), which is detected in the culture supernatant (Fig. 2A). Truncated TpsB2, however, was not able to secrete the TPS2a construct because there was no TPS2a detected in the concentrated culture supernatant. The non-secreted TPS2a did not accumulate inside the cells but appeared to be degraded, similar to what we observed previously (26). The growth curves of the cultures did not change upon the induction of gene expression with IPTG (results not shown).
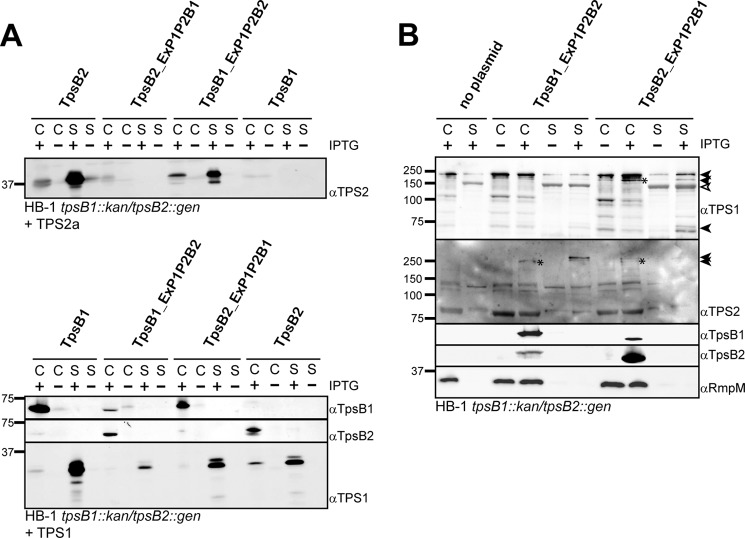
FIGURE 2.
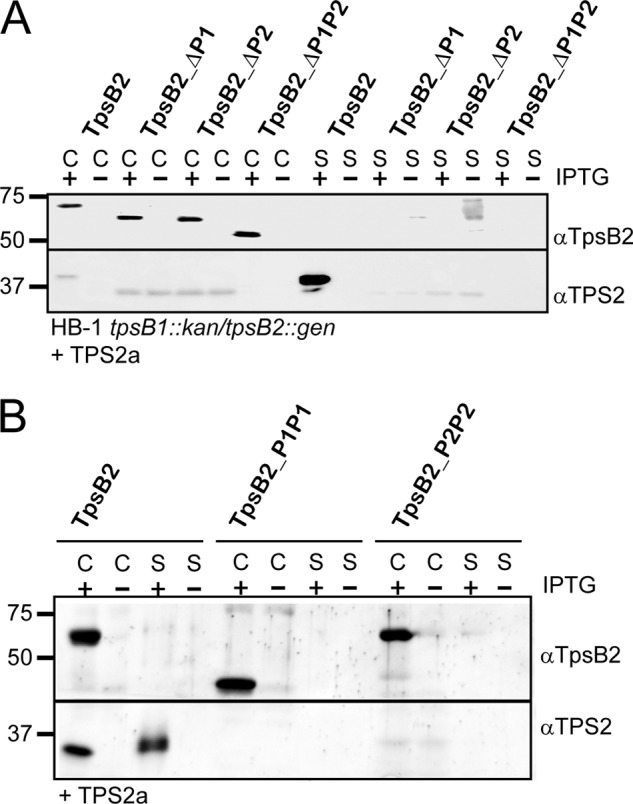
The influence of deletion or duplication of the POTRA domains of TpsB2 on TPS2a secretion. A, immunoblots of whole cell lysates (C) and culture supernatants (S) of N. meningitidis HB-1 tpsB1::kan/tpsB2::gen cells carrying plasmids encoding TpsB2 or its mutant derivatives with either one or two POTRA domains deleted, as indicated above the lanes, in combination with the TPS2a construct. B, immunoblots of whole cell lysates (C) and culture supernatants (S) of N. meningitidis HB-1 tpsB1::kan/tpsB2::gen cells carrying plasmids encoding TpsB2 or its mutant derivatives with either POTRA1 or POTRA2 duplicated as indicated above the lanes in combination with the TPS2a construct. The cells were grown in the presence (+) or absence (−) of 0.01 mm IPTG for expression. The blots were incubated with antisera against the TpsB2 and TPS2 domains, as indicated on the right. Indicated on the left are the molecular weight markers.
The secretion defect was not the result of the absence or improper localization of the TpsB2 mutants because these mutants were detected in whole cell lysates at comparable levels (Fig. 2A). Furthermore, wild-type and truncated TpsB2 variants showed a similar running behavior on seminative polyacrylamide gels when heated (10 min at 100 °C) and non-heated (10 min at ambient temperature) outer membrane preparations were compared (Fig. 3). The non-heated samples showed increased mobility of the TpsB protein on gel, at a position of ∼47 kDa for wild-type TpsB2, which shifted toward the position corresponding to the expected molecular mass of ∼64 kDa for wild-type TpsB2 when samples were heated. This heat modifiability is considered to be indicative of folding of a β-barrel protein (33–35). Both the wild-type TpsB and the truncated variants lacking one POTRA domain showed this behavior. The samples incubated at room temperature already contained some protein running at the position of denatured protein in the gel, probably indicating some instability of the protein under these mild conditions. All TpsB2 variants migrated as a folded protein, although the relative amounts of denatured and folded material suggested that truncated TpsBs were a bit less stable. In particular, the mutant that lacks two POTRA domains showed a decreased amount of the faster running band, when compared with wild-type TpsB2. Overall, the results indicated that deleting a POTRA domain resulted in an inactive TpsB2 transporter, suggesting that two POTRA domains are needed for secretion.
FIGURE 3.
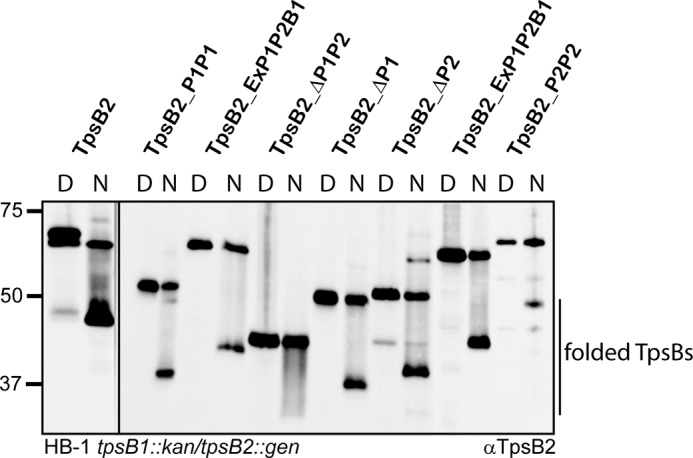
Heat modifiability of TpsB2 and mutant derivatives of TpsB2. Immunoblot of outer membrane preparations of N. meningitidis HB-1 tpsB1::kan/tpsB2::gen cells expressing the TpsB variants indicated above the lanes. Samples were either heated for 10 min at 100 °C (D) or kept at room temperature (N). The blot was incubated with anti-TpsB2. The positions of the bands representing folded TpsBs are indicated on the left side, and the molecular weight markers are shown on the right. The panels were taken from the same blot. The TpsB2_ExP1P2B1 was co-expressed with either the TPS2a or the TPS1 construct, as indicated.
TpsB2 with a Duplicated POTRA2 Domain Are Not Functional
We then tested whether secretion required the presence of a specific set of POTRA domains or two random POTRA domains. To test this, we constructed tpsB2 mutants that encoded TpsB2 with either a duplicated POTRA1 (TpsB2-P1P1) or a duplicated POTRA2 (TpsB2-P2P2) and cloned them downstream of the TPS2a construct in the neisserial expression vector (Fig. 2B). Both mutants were expressed (Fig. 2B) and appeared properly localized judged from the heat modifiability of the proteins (Fig. 3). However, the denatured sample of the TpsB2-P1P1 variant ran at a position in the gel of ∼52 kDa, which is much lower than the calculated ∼63 kDa. Apparently, a periplasmic part of the TpsB was cleaved off, resulting in an inactive protein (Fig. 2B). Indeed, we could not detect TPS2a in the culture supernatants of strains carrying TpsB2-P1P1 (Fig. 2B). The TpsB2_P2P2 variant, despite having similar expression and folding characteristics as wild type, was also inactive. The results led us to conclude that a tandem of POTRA domains is not sufficient for secretion but that secretion requires a specific couple of POTRA domains.
Swapping POTRA Domains between TpsB2 and TpsB1 Changes Substrate Specificity
We previously observed that the meningococcal TpsB1 operates system-specifically (i.e. only on cognate TPS domains). By contrast, TpsB2 is able to secrete TPS domains of all neisserial TpsAs tested (26). Isolated POTRA domains have been shown to bind TPS domains in vitro but with a rather low affinity, and this has only been tested for cognate TpsBs (14, 15). We therefore investigated the role of the POTRA domains in substrate selection by exchanging sequence encoding the POTRA domains between the two meningococcal tpsBs, yielding hybrids TpsB2_ExP1P2B1 and TpsB1_ExP1P2B2, respectively (Fig. 1C). The two hybrids were cloned downstream of the TPS2a or the TPS1 construct, to allow us to analyze the effect on secretion of system 1- and system 2-derived proteins, and introduced into N. meningitidis HB-1 tpsB1::kan tpsB2::gen. The hybrid proteins were difficult to detect when the corresponding sera were applied (Fig. 4). Apparently, the POTRA domains are immunodominant regions in the proteins, because the sera did clearly detect the hybrids carrying the POTRA domains of the protein it was raised against. Overall levels of hybrid and wild-type TpsBs appeared comparable, and the hybrid TpsBs showed the characteristic heat modifiability on seminative gels indicative of proper localization and folding (see Fig. 3 for TpsB2_ExP1P2B1).
FIGURE 4.
The influence of swapping the POTRA domains of TpsB1 and TpsB2 on the secretion of TPS2a and TPS1 constructs and full-length TpsA1 and TpsA2 proteins. A, immunoblots of whole cell lysates (C) and culture supernatants (S) of N. meningitidis HB-1 tpsB1::kan/tpsB2::gen cells carrying plasmids encoding a wild-type or mutated TpsB as indicated above the lanes in combination with the TPS1 or TPS2a construct. B, immunoblots of whole cell lysates and culture supernatants of N. meningitidis HB-1 tpsB1::kan/tpsB2::gen cells carrying plasmids encoding a wild-type or mutated TpsB without TPS construct to analyze secretion of full-length TpsAs. The cells were grown in the presence (+) or absence (−) of 0.01 mm IPTG for expression of the TpsB. The blots were incubated with antisera against a TpsB or a TPS domain, as indicated on the right. On the left, the molecular weight markers are indicated. The full-length TpsA-derived bands in B are indicated by closed arrowheads (∼240, 200, and 75 kDa for TpsA1 and ∼250 and 260 kDa for TpsA2, respectively). A distinct background band detected by the TPS1 antiserum is indicated by an open arrowhead (19).
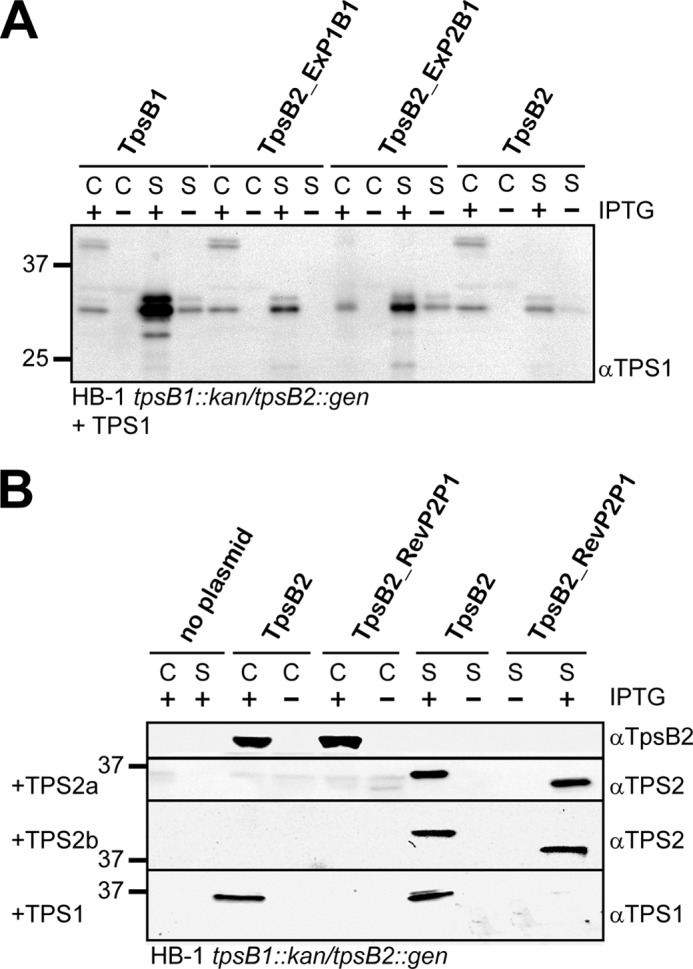
As shown previously, co-expression of TPS2a and wild-type TpsB2 resulted in secretion of TPS2a, whereas TPS2a secretion was absent in the presence of TpsB1 (Fig. 4A). Simultaneous production of TPS2a and the TpsB2-ExP1P2B1 hybrid did not result in secreted TPS2a in the medium. Interestingly, co-expression of the reciprocal TpsB1-ExP1P2B2 hybrid with TPS2a resulted in secretion of significant amounts of TPS2a. We concluded that the recognition of the TPS2a construct by the TpsB2 POTRA domains is sufficient for secretion, whereas the lack of recognition by the TpsB1 POTRA domains prevents secretion.
We then analyzed the secretion of TPS1 (Fig. 4A). As expected, co-expression of TPS1 together with TpsB2 resulted in secretion of TPS1, albeit with a reduced efficiency (26). In line with the results obtained for TPS2, the TpsB1_ExP1P2B2 hybrid secreted only very limited amounts of TPS1, again suggesting a major role for the POTRA domains in substrate selection and the initiation of secretion. The expression of TPS1 with TpsB2-ExP1P2B1 resulted in secretion of TPS1 to a level comparable with secretion of TPS1 by wild-type TpsB2 but lower than that of wild-type TpsB1. This could be caused by the absence of other TpsB1 domains that may contribute to TPS1 secretion or suboptimal functioning of the chimeric protein.
The decisive role of the POTRA domains in the secretion of truncated TPS constructs prompted us to investigate whether the hybrids could select and support secretion of full-length TpsAs. We cloned the ORFs encoding TpsB1_ExP1P2B2 and TpsB2_ExP1P2B1 into the neisserial expression vector as singular genes and introduced them in HB-1 tpsB1::kan tpsB2::gen. This double knock-out strain mutant lacks the neisserial TpsBs, and as a result, the full-length TpsAs remain intracellular, where they accumulate as a ∼240 kDa band (TpsA1s) or are degraded (TpsA2s) (26). Upon induction with IPTG, the complemented strains express the hybrid tpsBs in comparable amounts (Fig. 4B). The presence of the hybrid TpsB2_ExP1P2B1 results in the secretion of TpsA1, as judged from the appearance of bands of ∼240, ∼200, and ∼75 kDa on blots that were also found in wild-type HB-1, albeit in different relative amounts (19). The presence of these proteins in the supernatants was not the result of leakage, as judged from the absence of the marker protein RmpM in the concentrated culture supernatants (Fig. 4B). Furthermore, in the whole cell lysates, a cell-associated band at ∼200 kDa is detected that was also observed in wild-type cells (19). Similar to what we observed for the truncated TPS2a construct, the full-length TpsA2a and TpsA2b proteins do not appear to be secreted by the TpsB2_ExP1P2B1 hybrid. Note that the antiserum detects both full-length system 2 TpsAs (19). Analogous to what we observed for the truncated TPS constructs, the expression of the TpsB1_ExP1P2B2 hybrid resulted in the detection of full-length TpsA2a- or TpsA2b-derived bands in the whole cell lysates and the culture supernatant samples, but the cell-associated or secreted TpsA1 bands that result from an active TpsB1 were not detected. Overall, the results clearly indicate that the POTRA domains have an important and decisive role in substrate recognition, but they also indicate that other domains within the complete TpsB are needed for efficient completion of the subsequent steps in secretion.
The TpsA1 functions as a contact-dependent toxin against other N. meningitidis cells (24). To test whether secretion of TpsA1 by the TpsB1_ExP1P2B2 and TpsB2_ExP1P2 mutants influenced TpsA1 toxicity, we performed growth inhibition assays in unencapsulated N. meningitidis strain BB-1 (24). This strain contains only a single operon of tpsB1 and tpsA1 and no other meningococcal TPS systems (24). Comparison of TpsA1 of BB-1 and the two TpsAs of system 1 of HB-1 showed an overall amino acid identity of ∼70%. TpsAs are known to show mosaic homology patterns (19, 20, 24), and the highest divergence between the HB-1 and BB-1 TpsAs is found in the central region (24), whereas the TPS domains are almost identical. Interestingly, the C-terminal region of the BB-1 TpsA1, involved in the killing activity is identical to one of the two TpsA1s (TpsA1b) of HB-1 (24). The TpsB1s of the two strains are 98% similar. The plasmids carrying the mutated tpsB ORFs were introduced into BB-1 ΔtpsB, and Western blot analysis indicated that the TpsB1, TpsB1_ExP1P2B2, and TpsB2_ExP1P2B1 proteins are expressed (results not shown), with higher expression levels of the plasmid-encoded tpsB variants (26). Similar to HB-1, blots of BB-1 cell and concentrated culture supernatant samples showed TpsA1 protein species of ∼240 and ∼200 kDa, but a ∼75-kDa protein species appeared absent (Fig. 5A). Secretion and processing was blocked in BB-1 ΔtpsB, and a ∼240-kDa TpsA1 species accumulated in the cell samples. However, the secretion of TpsA1 was very efficient in the presence of the TpsB2_ExP1P2B1 mutant. Furthermore, the presence of TpsB1_ExP1P2B2 resulted in a reduced but clearly detectable secretion of the TpsA1, as judged by the detection of the ∼240- and ∼200-kDa TpsA1 species when IPTG was added (Fig. 5A). Apparently, secretion of full-length TpsA1 by the TpsB1_ExP1P2B2 mutant in BB-1 is more efficient than in HB-1 (see Fig. 4B), which could be caused by the differences in sequence. However, the reduced secretion mirrored the reduced but detectable secretion of the TPS1 construct in HB-1 (Fig. 4A).
FIGURE 5.

Secretion of TpsA1 of N. meningitidis BB-1 by TpsB1_ExP1P2B2 or TpsB2_ExP1P2B1 and its effect on TpsA1-mediated growth inhibition. A, immunoblot containing whole cell lysates and culture supernatants of N. meningitidis BB-1 and its tpsB1::kan knock-out derivative, the latter without plasmid or carrying plasmids encoding the mutated TpsB variant indicated. The cells were grown in the presence (+) or absence (−) of 0.01 mm IPTG for expression of the TpsB from plasmid. The blot was incubated with an antiserum against the TPS1 domain. On the left, the molecular weight markers are indicated. The full-length TpsA1-derived bands are indicated by closed arrowheads at ∼240 and ∼200 kDa. A distinct background band detected by the TPS1 antiserum is indicated by an open arrowhead (19). B, growth inhibition assay performed with the killer cell line N. meningitidis BB-1 and its ΔtpsB1 derivative grown in the presence (+IPTG) or absence of 0.25 mm IPTG (+IPTG) to induce the production of TpsB1_ExP1P2B2 or TpsB2_ExP1P2B1. BB-1 ΔtpsA-C served as bait, and the amount of killing is expressed as the ratio of bait over killer cells in colony-forming units (log10 CFU). The values are the mean of three independent experiments, with the S.D. indicated (error bars).
Next the growth inhibition by the TpsA1 of N. meningitidis BB-1 was measured. Secretion of TpsA1 by both TpsB1_ExP1P2B2 and TpsB2_ExP1P2 in BB-1 ΔtpsB1 resulted in similar levels of killing of the bait N. meningitidis cells (Fig. 5B), indicating that, although secretion of the TpsA1 by mutant TpsB1_ExP1P2B2 appeared less efficient (Fig. 5A), the levels of TpsA1 at the cell surface were sufficient for killing activity. Apparently, once TpsA1 secretion has occurred, the TpsA1 protein is active, irrespective of the TpsB used for secretion.
Single POTRA Domains Contribute to Substrate Selection
Next, we investigated whether a single POTRA domain within the POTRA pairs was decisive in target selection. For that, we constructed two-hybrid TpsB2s in which a single POTRA domain was swapped for the corresponding POTRA of TpsB1, yielding TpsB2_ExP1B1 and TpsB2_ExP2B1, respectively (Fig. 1C). Subsequently, these hybrids were expressed in combination with either the TPS1 or the TPS2a constructs. The hybrid TpsB2 proteins were expressed and showed heat-modifiable running behavior on seminative PAGE (results not shown). In contrast to TpsB2_ExP1P2B1 hybrid that was unable to secrete the TPS2a construct, the single-POTRA exchange hybrids TpsB2_ExP1B1 and TpsB2_ExP2B1 were able to secrete TPS2a, although the levels were lower than with wild-type TpsB2. The two single-POTRA hybrids were also able to secrete TPS1 to levels that were higher than that obtained by the double-POTRA hybrid TpsB2_ExP1P2B1 or wild-type TpsB2 (Fig. 6A). Overall, these results indicate that both POTRA domains contribute to the binding site of TPS domains and that there is some flexibility in binding for secretion to occur. Nevertheless, the differences in efficiency also suggest that the binding is influenced by the context of the POTRA domains and the interaction between these two domains.
FIGURE 6.
The influence of replacing POTRA1 or POTRA2 of TpsB2 with the corresponding POTRA domain of TpsB1 (A) or reversing the order of the POTRA domains in TpsB2 (B) on the secretion of TPS1 and TPS2 constructs. Shown are immunoblots of whole cell lysates (C) and culture supernatants (S) of N. meningitidis HB-1 tpsB1::kan/tpsB2::gen cells carrying plasmids encoding the TpsB variants in combination with either the TPS2a, TPS2b, or TPS1, as indicated on the right. The cells were grown in the presence (+) or absence (−) of 0.01 mm IPTG for expression. The blots were incubated with antisera against the TpsBs and TPS domains, as indicated on the right. Indicated on the left are the molecular weight markers.
TpsB2 Is Functional When the Order of POTRA Domains Is Switched
Because our results indicated that the combined POTRA domains determine substrate specificity of the TpsB transporter and contribute to the binding site for the TPS domain, we next wondered whether the order of the two POTRA domains is important for the initiation of secretion. To test this, we cloned a tpsB2 variant that encodes a TpsB2 with its POTRA domains in reversed order (TpsB2_RevP2P1; Fig. 1C) and placed it downstream of the TPS2a and TPS1 constructs. In contrast to wild-type TpsB2, this TpsB2_RevP2P1 mutant was not able to secrete TPS1 (Fig. 6B). Co-expression of the TPS2a or TPS2b construct with TpsB2_RevP2P1 resulted in efficient secretion of TPS2a and TPS2b to levels comparable with wild type, clearly indicating that the TpsB2 mutant is functional. Remarkably, however, the secreted TPS2a and TPS2b proteins run at a lower position in the gel when secreted by the TpsB2_RevP2P1. Our previous work already suggested that the TPS2a and TPS2b constructs undergo a processing step during translocation (26). Apparently, the interaction of the system 2 TPS domains with the POTRA domains in reversed order corrupts the concomitant processing step. This could suggest a different alignment of the TPS domain in the TpsB2 protein during secretion.
DISCUSSION
A distinctive feature of the TPS systems of N. meningitidis and N. lactamica is that these include multiple tpsA copies that appear secreted by a single TpsB. Furthermore, both species encode a homologous TPS system that lacks a specific tpsB (19, 20). Recently, we have reported on the redundancy of the meningococcal TpsB transporters and showed that the TpsB1 binds primarily its system-specific substrates, whereas the TpsB2 shows a more promiscuous binding of TPS domains (26). Here we have used this difference in secretion specificity to investigate the role of the periplasmic POTRA domains of the TpsB transporters in the recognition and secretion of TpsAs. By exchanging POTRA domains between the neisserial TpsB1 and TpsB2, we could swap the specificity of the respective transporters for their substrates. Our results show clearly not only that the POTRA domains of TpsB transporters contribute to binding of the substrates but that they select the substrates that are secreted by the TpsB. These results were reciprocal between TpsB1 and TpsB2 and, therefore, appear to be a general feature of the POTRA domains. The relaxed specificity of the TpsB2 transporter seems also to be determined by its POTRA domains, because the TpsB2_ExP1P2B1 mutant carrying the POTRA domains of B1 did not secrete the TPS2 construct, whereas the TpsB1_ExP1P2B2 mutant carrying those of TpsB2 still secreted some TPS1 (Fig. 4). Importantly, the secretion of full-length TpsAs strictly depended on the selection of its TPS domain by the POTRA domains of the TpsB, because secretion of full-length TpsAs in the presence of the hybrid TpsB switched to the mutant carrying the corresponding POTRA domains. The sequences of the TpsAs of N. meningitidis differ considerably (19), and apparently, the TPS domains fully determine the specificity for the TpsB transporter they use for secretion. However, the growth inhibition assay indicated that once the TpsA1 is secreted, regardless of the TpsB variant used, it remained active as a toxin (Fig. 5).
All members of the Omp85 protein family carry one or more POTRA domains (3, 7), and our results add to the notion that POTRA domains are the initial binding sites of the substrates of these transporters (4, 12, 15). Deletion of either POTRA domain of the meningococcal TpsB2 abolishes secretion of the truncated TPS2a construct (Fig. 2), similar to what was found for FhaC of B. pertussis (8). Furthermore, the results with the TpsB mutants that carried duplicated and swapped POTRA domains show that efficient secretion requires the presence of two POTRA domains. Each POTRA appears to have a specific role in the selection process, because a mere duplication of the POTRA2 domain of TpsB2 did not result in a functional protein. However, their order seemed less relevant, because placing them in reversed order did not affect secretion, although it did change the concomitant modification of the TPS domain. Furthermore, both POTRA domains appeared to contribute to system specificity, because exchanging only one POTRA domain between TpsB2 and TpsB1 resulted in intermediate substrate specificities (Fig. 5). Overall, these results indicate that the binding site for TPS domains stretches over the two POTRA domains.
The FhaC transporter of FHA is the most studied TpsB, and its crystal structure has been solved (8). Mutational studies of FhaC targeted a continuous hydrophobic groove that extends over the two POTRA domains and involves α-helix 1 and β-strand 2 for POTRA1 and α-helix 3 and β-strand 5 for POTRA2 (see Fig. 1). The results indicated that this region is involved in substrate binding and secretion (8, 15). However, the mutated residues are in the conserved regions of the POTRA domains, suggesting that they have a general role in TPS domain binding (Fig. 1A). Based upon our results with the hybrid TpsB mutants, we hypothesize that the specificity of the binding is determined by the non-conserved regions of the domains (e.g. the region comprising β-strand 2 and α-helix 2 (Fig. 1). This surface is close to the hydrophobic groove in the POTRA domains but may constitute a second binding site. Of note, TPS domains interact with TpsBs in an unfolded fashion (14, 15), making a stretched interface likely. Ongoing work in our laboratory focuses on the role of the non-conserved regions in system-specific secretion of TpsAs.
Surprisingly, when we reversed the order of the POTRA domains, we observed fairly normal secretion of TPS2 domains. An explanation could be that the two POTRA domains have a symmetric role in the binding of the TPS domain, but this seems less likely, because duplicating POTRA2 did not result in a functional TpsB2. Alternatively, the POTRA domains may bind the TPS domains in two steps, first at a nonspecific general level and then second in a more specific and intricate way. Such a bimodal interaction complies with the observation that the exchange of a single POTRA domain also resulted in a switch in binding specificity.
Reversing the order of the POTRA domains changed the modification of the secreted TPS domains. Previously, we had observed that the position on the blot of the secreted TPS2 constructs was lower than what was expected from the sequence, already suggesting a modification event (26). The results presented here clearly indicate that, indeed, a modification step occurs, which depends on how the TPS domain interacts with the POTRA domains. Two totally different types of modifications of TPS domains have been described. First, proteolytic cleavage of the N-terminal TPS domain of HMW1A of Haemophilus influenzae results in the release of the N-terminal end of the protein from the cell surface (36). Second, the TPS domain of ShlA of Serratia marcescens undergoes a conformational switch when secreted by ShlB (37). This switch is needed to activate the hemolysin activity of ShlA and, for the TPS domain, results in an altered mobility on gels and blots. However, the different sizes that we observe point to a proteolytic cleavage event and seem incompatible with a conformational change. This cleavage must occur during secretion, while the protein interacts with the TpsB, and the switch of the POTRA domains, apparently, changes the position of the TPS domain in the TpsB so that a new site is cleaved.
Additional binding of TpsAs to other regions of the TpsBs (e.g. the β-barrel or the preceding linker region) could contribute to the formation of the secreting complex. Mutations in the FhaC POTRA domains that abolish binding in vitro still supported secretion of the FhaB TPS domain in vivo (15). Furthermore, full-length TpsA2 in our experiments appeared protected from degradation by binding to the hybrid TpsB2_ExP1P2B1. In Pseudomonas aeruginosa, a hybrid secretion system exists in which a TpsA is secreted by an usher-like OMP that carries POTRA domains (38). This P-usher can also transport and assemble pilin subunits into a pilus. Deletion of the P-usher POTRA domains blocked TpsA secretion but not its binding to the usher. As a result, the pilus assembly was blocked, whereas pili normally assembled when TpsA was absent.
In conclusion, our results clearly indicate that interaction between the POTRA domains and the TPS domain is the decisive step for secretion to occur. The POTRA domains of a TpsB thereby act as a specificity filter, and their interaction with the TPS domain initiates the secretion process.
Acknowledgments
We thank Joen Luirink and Wilbert Bitter for critically reading the manuscript.
Footnotes
- TPS
- two-partner secretion
- POTRA
- polypeptide transport-associated
- FHA
- filamentous hemagglutinin of B. pertussis
- IPTG
- isopropyl 1-thio-β-d-galactopyranoside.
REFERENCES
- 1. van Ulsen P., ur Rahman S., Daleke M., Jong W. S. P., Luirink J. (2013) Type V secretion: from Biogenesis to biotechnology. Biochim. Biophys. Acta. Mol. Cell Res. 10.1016/j.bbamcr.2013.11.006 [DOI] [PubMed] [Google Scholar]
- 2. Jacob-Dubuisson F., Guérin J., Baelen S., Clantin B. (2013) Two-partner secretion: as simple as it sounds? Res. Microbiol. 164, 583–595 [DOI] [PubMed] [Google Scholar]
- 3. Jacob-Dubuisson F., Villeret V., Clantin B., Delattre A. S., Saint N. (2009) First structural insights into the TpsB/Omp85 superfamily. Biol. Chem. 390, 675–684 [DOI] [PubMed] [Google Scholar]
- 4. Arnold T., Zeth K., Linke D. (2010) Omp85 from the thermophilic cyanobacterium Thermosynechococcus elongatus differs from proteobacterial Omp85 in structure and domain composition. J. Biol. Chem. 285, 18003–18015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. D'Ulisse V., Fagioli M., Ghelardini P., Paolozzi L. (2007) Three functional subdomains of the Escherichia coli FtsQ protein are involved in its interaction with the other division proteins. Microbiology 153, 124–138 [DOI] [PubMed] [Google Scholar]
- 6. Robson S. A., King G. F. (2006) Domain architecture and structure of the bacterial cell division protein DivIB. Proc. Natl. Acad. Sci. U.S.A. 103, 6700–6705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sánchez-Pulido L., Devos D., Genevrois S., Vicente M., Valencia A. (2003) POTRA: a conserved domain in the FtsQ family and a class of β-barrel outer membrane proteins. Trends Biochem. Sci. 28, 523–526 [DOI] [PubMed] [Google Scholar]
- 8. Clantin B., Delattre A. S., Rucktooa P., Saint N., Méli A. C., Locht C., Jacob-Dubuisson F., Villeret V. (2007) Structure of the membrane protein FhaC: a member of the Omp85-TpsB transporter superfamily. Science 317, 957–961 [DOI] [PubMed] [Google Scholar]
- 9. Kim S., Malinverni J. C., Sliz P., Silhavy T. J., Harrison S. C., Kahne D. (2007) Structure and function of an essential component of the outer membrane protein assembly machine. Science 317, 961–964 [DOI] [PubMed] [Google Scholar]
- 10. Gatzeva-Topalova P. Z., Walton T. A., Sousa M. C. (2008) Crystal structure of YaeT: conformational flexibility and substrate recognition. Structure 16, 1873–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Koenig P., Mirus O., Haarmann R., Sommer M. S., Sinning I., Schleiff E., Tews I. (2010) Conserved properties of polypeptide transport-associated (POTRA) domains derived from cyanobacterial Omp85. J. Biol. Chem. 285, 18016–18024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Knowles T. J., Jeeves M., Bobat S., Dancea F., McClelland D., Palmer T., Overduin M., Henderson I. R. (2008) Fold and function of polypeptide transport-associated domains responsible for delivering unfolded proteins to membranes. Mol. Microbiol. 68, 1216–1227 [DOI] [PubMed] [Google Scholar]
- 13. Surana N. K., Grass S., Hardy G. G., Li H., Thanassi D. G., Geme J. W., 3rd (2004) Evidence for conservation of architecture and physical properties of Omp85-like proteins throughout evolution. Proc. Natl. Acad. Sci. U.S.A. 101, 14497–14502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hodak H., Clantin B., Willery E., Villeret V., Locht C., Jacob-Dubuisson F. (2006) Secretion signal of the filamentous haemagglutinin, a model two-partner secretion substrate. Mol. Microbiol. 61, 368–382 [DOI] [PubMed] [Google Scholar]
- 15. Delattre A.-S., Saint N., Clantin B., Willery E., Lippens G., Locht C., Villeret V., Jacob-Dubuisson F. (2011) Substrate recognition by the POTRA domains of TpsB transporter FhaC. Mol. Microbiol. 81, 99–112 [DOI] [PubMed] [Google Scholar]
- 16. Bos M. P., Robert V., Tommassen J. (2007) Functioning of outer membrane protein assembly factor Omp85 requires a single POTRA domain. EMBO Rep. 8, 1149–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Noinaj N., Kuszak A. J., Gumbart J. C., Lukacik P., Chang H., Easley N. C., Lithgow T., Buchanan S. K. (2013) Structural insight into the biogenesis of β-barrel membrane proteins. Nature 501, 385–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stephens D. S., Greenwood B., Brandtzaeg P. (2007) Epidemic meningitis, meningococcaemia, and Neisseria meningitidis. Lancet 369, 2196–2210 [DOI] [PubMed] [Google Scholar]
- 19. van Ulsen P., Rutten L., Feller M., Tommassen J., van der Ende A. (2008) Two-partner secretion systems of Neisseria meningitidis associated with invasive clonal complexes. Infect. Immun. 76, 4649–4658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. van Ulsen P., Tommassen J. (2006) Protein secretion and secreted proteins in pathogenic Neisseriaceae. FEMS Microbiol. Rev. 30, 292–319 [DOI] [PubMed] [Google Scholar]
- 21. Schmitt C., Turner D., Boesl M., Abele M., Frosch M., Kurzai O. (2007) A functional two-partner secretion system contributes to adhesion of Neisseria meningitidis to epithelial cells. J. Bacteriol. 189, 7968–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tala A., Progida C., De Stefano M., Cogli L., Spinosa M. R., Bucci C., Alifano P. (2008) The HrpB-HrpA two-partner secretion system is essential for intracellular survival of Neisseria meningitidis. Cell Microbiol. 10, 2461–2482 [DOI] [PubMed] [Google Scholar]
- 23. Neil R. B., Apicella M. A. (2009) Role of HrpA in biofilm formation of Neisseria meningitidis and regulation of the hrpBAS transcripts. Infect. Immun. 77, 2285–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arenas J., Schipper K., van Ulsen P., van der Ende A., Tommassen J. (2013) Domain exchange at the 3′ end of the gene encoding the fratricide meningococcal two-partner secretion protein A. BMC Genomics 14, 622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. van Ulsen P., Adler B., Fassler P., Gilbert M., van Schilfgaarde M., van der Ley P., van Alphen L., Tommassen J. (2006) A novel phase-variable autotransporter serine protease, AusI, of Neisseria meningitidis. Microbes Infect. 8, 2088–2097 [DOI] [PubMed] [Google Scholar]
- 26. ur Rahman S., van Ulsen P. (2013) System specificity of the TpsB transporters of coexpressed two-partner secretion systems of Neisseria meningitidis. J. Bacteriol. 195, 788–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Ulsen P., van Alphen L., ten Hove J., Fransen F., van der Ley P., Tommassen J. (2003) A Neisserial autotransporter NaIP modulating the processing of other autotransporters. Mol. Microbiol. 50, 1017–1030 [DOI] [PubMed] [Google Scholar]
- 28. van Ulsen P., van Alphen L., Hopman C. T., van der Ende A., Tommassen J. (2001) In vivo expression of Neisseria meningitidis proteins homologous to the Haemophilus influenzae Hap and Hia autotransporters. FEMS Immunol. Med. Microbiol. 32, 53–64 [DOI] [PubMed] [Google Scholar]
- 29. Petersen T. N., Brunak S., von Heijne G., Nielsen H. (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786 [DOI] [PubMed] [Google Scholar]
- 30. Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J. M., Taly J. F., Notredame C. (2011) T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kelley L. A., Sternberg M. J. (2009) Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 4, 363–371 [DOI] [PubMed] [Google Scholar]
- 32. Zhang Y., Skolnick J. (2005) TM-align: a protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 33, 2302–2309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Nakamura K., Mizushima S. (1976) Effects of heating in dodecyl sulfate solution on the conformation and electrophoretic mobility of isolated major outer membrane proteins from Escherichia coli K-12. J. Biochem. 80, 1411–1422 [DOI] [PubMed] [Google Scholar]
- 34. Oomen C. J., van Ulsen P., van Gelder P., Feijen M., Tommassen J., Gros P. (2004) Structure of the translocator domain of a bacterial autotransporter. EMBO J. 23, 1257–1266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Saurí A., Oreshkova N., Soprova Z., Jong W. S. P., Sani M., Peters P. J., Luirink J., van Ulsen P. (2011) Autotransporter β-domains have a specific function in protein secretion beyond outer-membrane targeting. J. Mol. Biol. 412, 553–567 [DOI] [PubMed] [Google Scholar]
- 36. Grass S., St Geme J. W. (2000) Maturation and secretion of the non-typable Haemophilus influenzae HMW1 adhesin: roles of the N-terminal and C-terminal domains. Mol. Microbiol. 36, 55–67 [DOI] [PubMed] [Google Scholar]
- 37. Walker G., Hertle R., Braun V. (2004) Activation of Serratia marcescens hemolysin through a conformational change. Infect. Immun. 72, 611–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ruer S., Ball G., Filloux A., de Bentzmann S. (2008) The “P-usher”, a novel protein transporter involved in fimbrial assembly and TpsA secretion. EMBO J. 27, 2669–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]