Background: DsbA enzymes assemble bacterial virulence factors and are targets for an entirely new drug class.
Results: Proteus mirabilis DsbA was characterized and its structure determined with a peptide bound non-covalently at the active site.
Conclusion: The structure provides an important basis for future inhibitor design.
Significance: New drugs to treat superbugs are urgently needed. DsbA inhibitors could have antivirulence activity against bacterial pathogens.
Keywords: Crystal Structure, Enzyme Catalysis, Enzyme Structure, Peptide Interaction, Structural Biology, Dithiol Oxidase, Oxidative Folding, Protein·Peptide Complex, Thioredoxin fold, Virulence
Abstract
The disulfide bond forming DsbA enzymes and their DsbB interaction partners are attractive targets for development of antivirulence drugs because both are essential for virulence factor assembly in Gram-negative pathogens. Here we characterize PmDsbA from Proteus mirabilis, a bacterial pathogen increasingly associated with multidrug resistance. PmDsbA exhibits the characteristic properties of a DsbA, including an oxidizing potential, destabilizing disulfide, acidic active site cysteine, and dithiol oxidase catalytic activity. We evaluated a peptide, PWATCDS, derived from the partner protein DsbB and showed by thermal shift and isothermal titration calorimetry that it binds to PmDsbA. The crystal structures of PmDsbA, and the active site variant PmDsbAC30S were determined to high resolution. Analysis of these structures allows categorization of PmDsbA into the DsbA class exemplified by the archetypal Escherichia coli DsbA enzyme. We also present a crystal structure of PmDsbAC30S in complex with the peptide PWATCDS. The structure shows that the peptide binds non-covalently to the active site CXXC motif, the cis-Pro loop, and the hydrophobic groove adjacent to the active site of the enzyme. This high-resolution structural data provides a critical advance for future structure-based design of non-covalent peptidomimetic inhibitors. Such inhibitors would represent an entirely new antibacterial class that work by switching off the DSB virulence assembly machinery.
Introduction
Proteus mirabilis is a significant Gram-negative extra-intestinal human pathogen that belongs to the Enterobacteriaceae family, which also includes other important pathogens such as Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae. Together, the Enterobacteriaceae account for the vast majority of community-acquired and nosocomial urinary tract infections (1, 2). P. mirabilis is a frequent cause of both complicated and catheter-associated urinary tract infections (3, 4). Multidrug-resistant strains of clinical pathogenic P. mirabilis have been reported for several decades (1). For example, clinical isolates of P. mirabilis resistant to streptomycin, tetracycline, kanamycin, chloramphenicol, and polymyxin B were described in the 1960s (5, 6) and since then reports of antibiotic resistance have increased (7–16).
DsbA enzymes from Gram-negative bacteria are targets for the development of anti-virulence drugs that could represent an entirely new class of antimicrobial agents (17, 18). Drugs that target bacterial virulence could minimize the selective pressure that generates antibiotic resistance and simultaneously preserve the endogenous host microbiome (19). DsbA is a key target for such drugs because it is essential for the correct folding or assembly of multiple virulence factors, including toxins, fimbrial adhesins, flagella, and type II and type III secretion systems (17). The mutation of dsbA results in the attenuation of virulence factor production in multiple pathogens (17), some examples, include P. mirabilis (20), uropathogenic E. coli (21), and Burkholderia pseudomallei (21, 22), Vibrio cholerae (23), Shigella flexneri (24), and Salmonella enterica serovar Typhimurium (25).
DsbA enzymes are thioredoxin-fold proteins (26) localized to the periplasm of Gram-negative bacteria where they catalyze oxidative folding (27). This reaction involves the transfer of a disulfide bond from the active site 30CXXC33 motif of DsbA to cysteine thiols in newly translocated proteins (28). Through this disulfide-exchange reaction, the active site cysteines of DsbA become reduced. The enzymatic cycle is completed through oxidation of DsbA by the inner membrane partner protein, DsbB (29). DsbA from E. coli (EcDsbA) is a highly promiscuous enzyme that catalyzes disulfide bond formation in many cysteine-containing proteins (28). In contrast, EcDsbB has a very strict binding specificity for EcDsbA (29). The interaction between EcDsbA and EcDsbB involves the formation of a mixed disulfide between Cys30 of the 30CXXC33 motif of EcDsbA and Cys104 of EcDsbB in the P2 periplasmic loop, which has the sequence 98PSPFATCDF106 (30). The crystal structure of the complex between EcDsbA and EcDsbB also revealed non-covalent binding of EcDsbB P2 loop residues to the EcDsbA hydrophobic groove (30). The P2 periplasmic loop sequence of P. mirabilis DsbB (PmDsbB) is identical to that of EcDsbB, suggesting a similar interaction occurs between PmDsbA and PmDsbB.
Here we characterize PmDsbA from P. mirabilis, which shares 59% sequence identity with EcDsbA, and confirm its activity as a DsbA enzyme. We demonstrate that the heptapeptide PWATCDS, derived from the DsbB P2 loop sequence, binds to wild type PmDsbA and to a C30S active site variant (PmDsbAC30S). We report three high-resolution crystal structures including the PmDsbAC30S·PWATCDS complex, which represents to our knowledge the first reported example of a peptide bound non-covalently to a DsbA structure. This non-covalent complex provides the critical starting point for the design of non-covalent drugs that act as anti-virulence agents targeting Enterobacteriaceae DsbAs.
EXPERIMENTAL PROCEDURES
Protein Production and Molecular Biology
Codon-optimized wild type P. mirabilis dsbA (GenBank® accession number CAR45574), lacking the sequence coding for the predicted signal peptide (amino acids 1–19), was cloned into a modified pMCSG7 (31) vector using ligation-independent cloning. The cytoplasmic expressed PmDsbA contained an N-terminal His6 affinity tag followed by a linker region including a tobacco etch virus (TEV)6 protease cleavage site. The PmDsbAC30S variant was generated from the wild type construct using QuikChange® (Agilent Technologies). The following primers were used to introduce the point mutation, forward, GAATTTTTCTCATTTTATTCTCCGCATTGTTACC and reverse, GGTAACAATGCGGAGAATAAAATGAGAAAAATTC. Transformed BL21(DE3)pLys cells containing plasmids for either PmDsbA or PmDsbAC30S were grown (1 liter) at 30 °C in 2.5-liter baffled shaker flasks for 16–20 h using autoinduction media (32). Cells were resuspended in 25 mm Tris, 150 mm NaCl (10 g of cells/100 ml of buffer), and protease inhibitor mixture (diluted 1 in 1000 into the lysate) (BioPioneer Inc., San Diego, CA) and DNase (1300 units/100 ml of lysate) (Roche Applied Science) were added. Lysis was performed in a Cell Disruptor (TS-Series, Constant Systems LTD., UK) applying a single run with a constant pressure of 25 Kpsi. Cell debris was removed by centrifugation (18,500 rpm, 30 min, 4 °C, rotor JM-25.5, Beckman Coulter, Brea, CA). The proteins were further purified by immobilized metal ion affinity chromatography using 12 ml/1-liter cells of equilibrated Talon (Clontech) and eluted with 25 mm Tris, 150 mm NaCl, 200 mm imidazole after incubation of 30 min at room temperature. Total protein was determined at 280 nm with a NanoDropTM 2000c (Thermo Fisher Scientific). The His6 affinity tag was removed by TEV cleavage using a 50:1 (w/w) ratio (protein/TEV-protease) incubated in a 50-ml Falcon tube including 1 mm β-mercaptoethanol for 2 h on a rotary mixer at room temperature. After cleavage, the PmDsbA proteins had an additional two non-native residues (Ser−2–Asn−1) at the N terminus. To remove imidazole, the mixture was rapidly buffer exchanged into 25 mm Tris, 150 mm NaCl using a Sephadex G-25 fine 16/60 column connected to an ÄKTA system (GE Healthcare). The His6-tagged TEV protease was removed by reverse metal ion affinity chromatography using Talon resin (0.5 ml of resin/2 mg of TEV protease) (Clontech, Australia) and the PmDsbA proteins were recovered in the flow through. The final step of purification was performed using a Superdex S75 gel-filtration column (GE Healthcare). Peak fractions were then combined and the protein was oxidized using copper(II)/1,10-phenanthroline at 1.7 mm final concentration or reduced using DTT at ×25 molar excess. To remove oxidizing or reducing agent, the mixture was then buffer-exchanged into 10 mm HEPES pH 7.4 using a Sephadex G-25 fine 16/60 column. Finally, the protein was concentrated to 100 mg/ml using Amicon Ultra filter devices with a 10-kDa cutoff (Millipore). Yield was generally 120–150 mg/liter of culture. Protein quality was assessed by SDS-PAGE (NuPAGE® system, 4–12% BisTris gel, Invitrogen, Australia). Molar protein concentrations were determined from calculated extinction coefficients derived from ProtParam (33).
Purified EcDsbA (GenBank accession number CAA56736) and EcDsbC (GenBank accession number AAA83074), lacking the periplasmic leader signal were expressed and purified as described above for PmDsbA. Yields were generally 120 and 80 mg/liter of culture, respectively. E. coli membrane extracts containing over-expressed EcDsbB (GenBank accession number AAC74269) were prepared as described previously (34), and resuspended in PBS buffer containing 10% glycerol.
EcDsbA Complementation
E. coli ΔdsbA (JCB817) and ΔdsbA/ΔdsbB (JCB818) non-motile strains were used for motility assays as described previously (35). The gene sequence coding for mature PmDsbA (lacking the periplasmic signal sequence) was cloned into pBAD33 under an arabinose-inducible promoter with an N-terminal EcDsbA signal sequence (36). As a positive control, EcDsbA was expressed within the same pBAD33 vector background. 2 × 106 non-motile E. coli ΔdsbA (JCB817) and ΔdsbA/ΔdsbB (JCB818) double-mutant (JCB818) (27) cells harboring pBAD33(EcDsbA) or pBAD33(PmDsbA) were spotted onto the center of a soft M63 minimal agar plate containing 40 mg/ml of each amino acid (except l-cysteine) in the absence or presence of 0.1% arabinose (negative control). Plates were incubated at 37 °C and cell motility was monitored after 4–5 h using a Molecular Imager® Gel DocTM system from Bio-Rad. Complementation experiments were performed as biological triplicates.
Cysteine Thiol Oxidation Assay
A synthetic peptide substrate of EcDsbA (CQQGFDGTQNSCK) with a europium 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid group amide coupled to the N terminus and a methylcoumarin amide coupled to the ϵ-amino group of the C-terminal lysine was purchased from AnaSpec (Fremont, CA) and prepared as previously reported (37).
Assays were performed using a Synergy H1 multimode plate reader (BioTek) as described previously (38). In brief, fluorescence (excitation λ = 340 nm and emission λ = 615 nm) corresponding to disulfide formation in the substrate peptide was measured in a white 384-well plate (PerkinElmer Life Sciences OptiPlate-384, number 6007290) in 50 mm MES, 50 mm NaCl, and 2 mm EDTA at pH 5.5 buffer. The total reaction volume in each well was 50 μl, containing 40, 80, or 160 nm PmDsbA or EcDsbA, 1.6 μm EcDsbB membrane, and 8 μm peptide substrate (added last to initiate the reaction). Controls used were: PmDsbA or EcDsbA + buffer + substrate (no DsbB); EcDsbB + buffer + substrate (no DsbA); buffer + substrate (no DsbA or DsbB); and no components (fluorescence background of the plate, which was subtracted from all other measurements). Three independent experiments (biological replicates) were performed, using three technical replicates for each measurement. The reaction rate was estimated by calculating the increase in fluorescence over the first 8 min of reaction. The fluorescence rates were plotted including the standard deviation calculated from the three biological replicates.
Thermal Stability of PmDsbA
Temperature-induced unfolding of native PmDsbA was recorded by UV circular dichroism as previously reported for other DsbA enzymes (39) using a Jasco J-810 spectropolarimeter (Jasco). The initial redox state of PmDsbA was confirmed by Ellman's assay (40). The maximum difference in CD signal was determined by subtraction of the CD spectra of the folded protein (25 °C) from the unfolded (95 °C) protein, for the oxidized and the reduced forms of the protein. Unfolding of oxidized PmDsbA (210 nm) and reduced PmDsbA (213 nm) was monitored in a 1-mm quartz cuvette using a heating rate of 1 °C/min from 25 to 95 °C. Measurements were carried out using 10 μm protein in a buffer containing 100 mm NaH2PO4/Na2HPO4, 1 mm EDTA, pH 7.0. To ensure PmDsbA remained reduced throughout the entire measurement, reduced samples contained 0.75 mm DTT. Data were fitted to a two-state unfolding model and errors were calculated using Prism 6 (GraphPad) as described previously (41).
Redox Properties of PmDsbA
The standard redox potential of PmDsbA was measured utilizing the intrinsic fluorescence of tryptophan residues in PmDsbA, similar to the method used for EcDsbA (42). In brief, oxidized PmDsbA was equilibrated for 3 h at 25 °C in degassed 100 mm NaH2PO4/Na2HPO4, pH 7.0, 1 mm EDTA, containing 1 mm oxidized glutathione (GSSG) and a range of reduced glutathione (GSH) concentrations (0–2000 μm). 200 μl of PmDsbA from each redox condition was dispensed into a 96-well plate (TPP AG, Switzerland, number 92096) and tryptophan fluorescence was measured (excitation wavelength 280 nm, emission 332 nm) using a Synergy H1 microplate reader and Gen5 2.0 software (Biotek). Data were analyzed in Prism 6 (GraphPad) and the redox potential was calculated as described previously for EcDsbA (42).
Determination of Cys30 pKa
Absorbance at 240 nm of the catalytic thiolate anion is pH-dependent allowing the equilibrium between protonated and deprotonated Cys30 to be measured (43) using a UV-visible spectrophotometer (CARY 50, Agilent Technologies). Absorbance at 240 and 280 nm were measured over pH values starting at 6.5 and decreasing to 2.0, in 0.25 pH unit increments. Samples contained either oxidized or reduced PmDsbA (40 μm) in composite buffer (10 mm Tris, 10 mm sodium citrate, 10 mm K2HPO4, 10 mm KH2PO4, 200 mm KCl, and 1 mm EDTA) at 22 °C. The pKa value was calculated from the fitted curves using the Henderson Hasselbalch equation (pH = pKa − log ([A240/A280]red/[A240/A280]ox)). Average and mean ± S.D. from triplicate measurements are plotted.
Disulfide Reductase Activity
DsbA enzymes can reduce the intermolecular disulfide bonds between insulin chains A and B under mild reducing conditions (27). Disulfide bond reduction of insulin can be followed spectrophotometrically at λ = 650 nm (A650 nm). The A650 nm value gives a measure of turbidity, which occurs as a result of the increase in production of insoluble B chain of insulin (44). Samples were prepared in 1-cm cuvettes containing 10 μm protein (PmDsbA, EcDsbA, or EcDsbC), 0.33 mm DTT, and 2 mm EDTA in 100 mm NaH2PO4/Na2HPO4, pH 7.0. Catalysis was initiated by the addition of 0.131 mm insulin (I0516, Sigma) to the mixture. The assay was repeated three times and the mean ± S.D. of the measurement at each time point was calculated.
Peptide Synthesis
Peptides were synthesized using solid-phase peptide synthesis on rink-amide (4-methyl)benzhydrylamine resin (ChemImpex International, Wood Dale, IL) with a loading of 0.65 mmol/g. De-protection of the resin and amino acids was performed using an 80:20 (v/v) mixture of dimethylformamide/piperidine (Rci Labscan, Bangkok Thailand/Auspep, Australia) for 2 × 5 min. l-Amino acids (ChemImpex International) were activated using 4 resin equivalents (eq) and 4 eq of HbtU (O-benzotriazole-N,N,N′,N′-tetramethyluronium hexafluorophosphate, ChemImpex International) (500 mm) and 5 eq of N,N-diisopropylethylamine (Auspep, Australia) for 5 min before coupling to the de-protected resin for 60 min. After the final coupling, all peptides were acetylated at the N terminus using 4 eq of acetic acid (Chem Supply, Australia), 4 eq of HbtU, and 5 eq of N,N-diisopropylethylamine for 30 min. Cleaving was executed using a 95:2.5:1.25:1.25 (v/v) mixture of trifluoroacetic acid (TFA)/ethanedithiol/triisopropylsilane/water (H2O) (chemicals from Sigma). Cleaved peptides resulted in an amidated C terminus from the resin rink-amide and dried using N2 gas, washed with diethyl ether (DEE, Ajax Finechem, Sydney Australia), and dissolved in a 80:20 (v/v) mixture of acetonitrile/H2O (RCI Labscan, Bangkok, Thailand) before purification on HPLC. Purification was executed on a C18 column (Phenomenex, Torrance CA) using a gradient from 20 to 80% of acetonitrile and TFA 0.1%. Fractions were analyzed by mass spectrometry (Waters Micromass LCT, Milford, CT) and the purified peptide was lyophilized using a freeze drier (Christ, Osterode am Harz, Germany).
Peptide-induced Thermal Shift Measurements
25 μm PmDsbA in phosphate-buffered saline (PBS), pH 7.4, was incubated with peptide PWATCDS at concentrations ranging from 125 μm to 4 mm for 1 h. Then Sypro Orange (S-6650, Invitrogen) was added to a 5× final concentration (stock concentration ×5000). Controls contained either no peptide, no DsbA, or the dye alone. Measurements were conducted in a white 384-well plate (Perkin Elmer OptiPlate-384, number 6007290) with 5 replicates. Fluorescence emission from Sypro Orange binding to unfolded protein was measured following a temperature time course increasing from 25 to 95 °C (heat rate = 0.05 °C/s) using a VIAA7 Real-time PCR system (Invitrogen) with a λ = 585 ± 15 nm wavelength filter. Raw data were analyzed using Prism 6 (GraphPad). Fluorescence emission was fitted to a classic Boltzmann sigmoidal curve, and the inflection point was used as the melting temperature, Tm. To determine ΔTm (the shift in melting temperature in the presence of peptide), the Tm value for the wild type protein was subtracted from the Tm value for PmDsbA-PWATCDS and the Tm value for the variant PmDsbAC30S was subtracted from the Tm value for PmDsbAC30S-PWATCDS. ΔTm values are presented as the mean ± S.D. from 5 replicates. A significant ΔTm is considered to be greater than two times the S.D. of the Tm value for the protein in the absence of ligand (45).
Isothermal Titration Calorimetry
Evaluation of affinity and thermodynamics of binding between PWATCDS and PmDsbA or PmDsbAC30S were assessed by isothermal titration calorimetry (ITC) using an Auto-iTC200 instrument (MicroCalTM, GE Healthcare). The sample cell was loaded with 200 μl of purified protein (oxidized PmDsbA or untreated PmDsbAC30S) at 100 μm concentration in 25 mm HEPES, pH 7.4, 50 mm NaCl, 0.8% dimethyl sulfoxide (ITC buffer). The syringe was filled with purified peptide PWATCDS in ITC buffer at a concentration of 4 mm. Titrations were conducted at 25 °C using 19 consecutive injections of 2 μl each delayed by 180 s with a stirring speed of 1000 rpm. In every experiment an initial 0.5 μl of peptide was injected to avoid slow leakage of titrant and this data point was discarded for binding analysis. As a control for background noise, titration of PWATCDS into a solution containing ITC buffer only was performed. The association constant (Ka = 1/Kd), free energy (ΔG), and enthalpy change (ΔH) were calculated by fitting the data to a single-site binding model using the MicroCal Origin software (Origin 7.0 SR4 version 7.0552β) and correcting peptide concentrations to adjust the stoichiometry parameter close to 1.0. Entropy change (ΔS) was deduced from the standard free energy equation ΔG = ΔH − TΔS. Parameters reported include the mean ± S.D. across three replicates. The calculated c-value for these measurements is 12.
Crystallization, Structure Determination, and Structural Analysis
Crystallization screening was performed at the University of Queensland ROCX diffraction facility. Protein crystallization trials were performed in 96-well plates using the hanging drop vapor diffusion method at 8 or 20 °C. In general, purified protein (200 nl) was mixed with crystallization solution (200 nl) using a Mosquito crystallization robot (TTP Labtech, UK), and trays were incubated and imaged in a RockImager 1000 (Formulatrix). Wild type PmDsbA was crystallized by mixing a 1:3.3 molar ratio of the protein·peptide complex PmDsbA·PSPWATCDF (3 mm protein, 10 mm peptide, 2% dimethyl sulfoxide incubated on ice for 1 h) with 0.5 μl of 15% PEG 3350 and 0.4 m sodium malonate, pH 5.0, and incubating the drop over the same solution at 20 °C. The presence of the peptide permitted crystallization but no density was evident for the peptide in the phased data; the final refined structure from this condition is referred to as native PmDsbA. Crystals were harvested in cryo solution (25% PEG 3350, 0.4 m sodium malonate, pH 5.0, 20% PEG 400) and immediately flash frozen in liquid nitrogen.
PmDsbAC30S crystals appeared overnight at 8 °C in 0.2 m KSCN, 23% PEG 3350 using a 1:10 molar ratio of the protein·peptide complex (1.5 mm protein and 15 mm PWATCDS, final concentrations of both, incubated on ice for 1 h); again the presence of the peptide permitted crystallization but there was no evidence of peptide binding in the electron density maps. The cryo solution used was 0.2 m KSCN, 25% PEG 3350, and 20% PEG 400. The structure of the protein derived from this condition is referred to as PmDsbAC30S.
PmDsbAC30S·PWATCDS co-crystals grew at 20 °C using a 1:10 molar ratio of the protein·peptide complex (3 mm protein, 30 mm peptide, final concentrations of both), with the two components incubated on ice for 1 h prior to crystallization. The protein·peptide solution was then mixed with an equal volume of crystallization solution, which contained 0.2 m KSCN, 22% PEG 3350, 0.2 m non-detergent sulfobetaine (NDSB-221). Differences in crystallization conditions in comparison to the PmDsbAC30S crystals that did not yield bound peptide included higher concentrations of protein and peptide (but the same molar ratio), the use of NDSB-221 as an additive and an incubation temperature of 20 °C (rather than 8 °C). Crystals were flash frozen in cryo solution consisting of 0.3 m KSCN, 30% PEG 3350, and 20% PEG 400.
Diffraction data for all three crystal structures were measured at the Australian Synchrotron MX2 beamline at a wavelength of 0.9537 Å, and recorded with an ADSC Quantum 315r detector controlled by BLU-ICE (46). Reflections were indexed and integrated in Mosflm (47) or XDS (48), analyzed in Pointless (49), and scaled in SCALA (49) from the CCP4 suite (50). Phases for PmDsbA were obtained by molecular replacement using PHASER (51) with EcDsbA as a template (PDB code 1FVK, sequence identity 59%). PmDsbAC30S·PWATCDS were solved using the wild type PmDsbA structure as the template. Initial electron density maps from PHASER were improved by cycles of iterative refitting of the model using the program COOT (52) and PHENIX.refine (53). The refinement of the complex structure was stalled at R-factor/R-free: 30/33%. Phenix.xtriage analysis indicated that the diffraction data were twinned with a twinning fraction of 0.49. The twin target function implemented in PHENIX was applied in further refinement cycles with the twinning operator −h+k, k, −l. The final R-factor/R-free was 17.2/19.9%. For PmDsbAC30S·PWATCDS, the density corresponding to the bound peptide was modeled for all seven residues including N-terminal acetylation and C-terminal amidation. In addition, a malonate anion was modeled into the crystal structure of PmDsbA and four SCN molecules were modeled into the PmDsbAC30S·PWATCDS crystal structure with one pair in each protomers A and C. One thiocyanate molecule is buried between α1 and β2 of protomers A/C, whereas the second molecule binds between Pro and PmDsbAC30S helix α1, possibly forming a weak hydrogen bond with the Pro backbone carbonyl (distance N:O = 3.4 Å). Thiocyanate molecules do not appear to influence peptide binding, because the peptide binding site and conformation is identical in protomer B, which lacks a bound thiocyanate.
Data processing and refinement statistics for all three crystal structures are provided in Table 1. Molecular figures were generated using PyMOL (The PyMOL Molecular Graphics System, version 1.6.0.0 Schrödinger, LLC) and figures of the electrostatic potential were generated using APBS (54). r.m.s. deviation calculations and structural alignments were conducted in PyMOL and FATCAT (55). Interaction analyses were conducted with the program COOT (52) and the PDBePISA (56) and Cocomaps servers (57).
TABLE 1.
X-ray data collection and refinement statistics
| Data collection | PmDsbA (4OCE) | PmDsbAC30S (4OCF) | PmDsbAC30S · PWATCDS (4OD7) |
|---|---|---|---|
| Wavelength (Å) | 0.95369 | 0.95369 | 0.95370 |
| Resolution range (Å) | 42.64–1.77 | 57.22–1.98 | 64.13–1.60 |
| Highest resolution shell (Å) | 1.86–1.77 | 2.09–1.98 | 1.68–1.60 |
| Space group | P 43212 | P21 | P32 |
| Unit cell dimensions | |||
| a (Å) | 37.8 | 37.9 | 74.06 |
| b (Å) | 37.8 | 80.2 | 74.06 |
| c (Å) | 298.4 | 114.4 | 93.27 |
| α, β, γ (°) | 90, 90, 90 | 90, 91, 90 | 90, 90, 120 |
| Total reflections | 303398 | 176891 | 839517 |
| Unique reflections | 22817 | 47865 | 74969 |
| Multiplicity | 13.3 (13.6)a | 3.7 (3.6)a | 11.2 (10.2)a |
| Completeness (%) | 100 (100) | 99.7 (98.9) | 98.6 (93.9) |
| Mean 〈I〉/〈σI〉 | 16.1 (4.8) | 14.3 (8.4) | 16.3 (2.4) |
| Rmergea | 0.127 (0.587) | 0.073 (0.155) | 0.080 (0.973) |
| Rp.i.m. | 0.035 (0.157) | 0.044 (0.095) | 0.026 (0.327) |
| Refinement statistics | |||
| Rfree (%) | 16.9 (21.9) | 21.1 (24.0) | 19.9 (29.2) |
| Rwork (%) | 14.8 (17.3) | 15.2 (15.2) | 17.2 (30.3) |
| Unique reflections | 22686 | 47846 | 74969 |
| Number of non-H atoms | |||
| Protein | 1492 | 5841 | 4506 |
| Water | 268 | 810 | 542 |
| Protein residues | 189 | 749 | 561 |
| r.m.s. deviation bond | |||
| Lengths (Å) | 0.009 | 0.012 | 0.017 |
| Angles (°) | 1.152 | 1.240 | 1.555 |
| Ramachandran | |||
| Favored (%) | 98.4 | 98.5 | 98.9 |
| Outliers (%) | 0 | 0 | 0 |
| Average B-factor (Å2) | 14.6 | 12.8 | 25.7 |
| Wilson B-factor (Å2) | 16.0 | 10.3 | 22.1 |
| Molprobityb | |||
| Clashscore (percentile) | 1.03 [100th, (839)] | 2.32 [100th, (714)] | 4.25 [97th, (699)] |
| Score (percentile) | 0.80 [100th, (11193)] | 1.01 [100th, (12309)] | 1.21 [98th, (6780)] |
a The values in parentheses refer to the highest resolution shell.
b 100th Molprobity (74) percentile is the best among the structures of comparable resolution. The percentile and the number of structures included in the comparison (N) are given in parentheses within the square bracket.
RESULTS
PmDsbA Catalyzes Disulfide Formation in Vitro and in Vivo
DsbA enzymes catalyze oxidative folding, or the introduction of disulfide bonds into proteins. To assess whether PmDsbA has dithiol oxidase activity we assessed its activity in an in vitro peptide oxidation assay. A europium-labeled peptide (CQQGFDGTQNSCK) fluoresces when the two cysteines are oxidized, but not when the cysteines are reduced (37). We found that PmDsbA, like EcDsbA, catalyzed peptide thiol oxidation as evident by an increase in the fluorescent signal over time (Fig. 1A). The fluorescence rate increased with increasing concentrations of PmDsbA and EcDsbA (Fig. 1A). PmDsbA catalysis was a little slower than that of EcDsbA at the same enzyme concentrations. This slightly reduced activity may reflect the use of a peptide derived from an EcDsbA substrate. Nonetheless, the results establish that PmDsbA catalyzes dithiol oxidation in a model substrate.
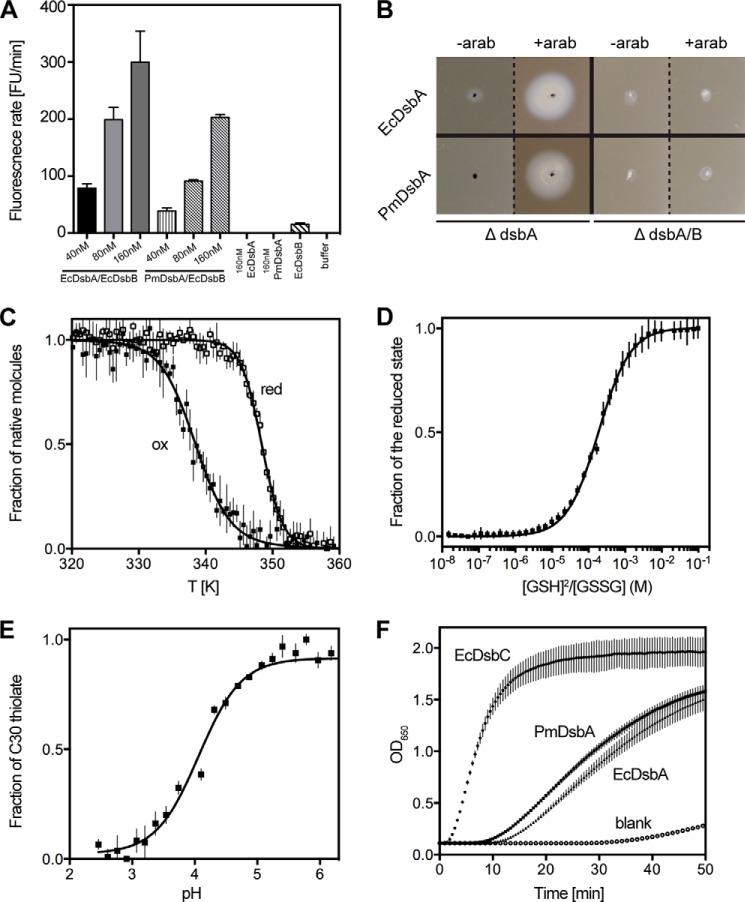
FIGURE 1.
Redox properties of PmDsbA. A, in vitro disulfide catalysis. Plot shows increase in fluorescence as a consequence of peptide disulfide formation catalyzed by EcDsbA or PmDsbA in the presence of EcDsbB. B, in vivo disulfide catalysis. ΔdsbA knock-out or ΔdsbA/B double knock-out cells are non-motile due to their inability to fold FlgI. Expression of PmDsbA or EcDsbA restores motility in ΔdsbA, but not in ΔdsbA/B. C, thermal melting curves of oxidized and reduced PmDsbA shows that reduced PmDsbA (Tmred 348.4 ± 0.1 K) is more stable than its oxidized counterpart (Tmox 338.4 ± 0.2 K). D, measurement of PmDsbA redox potential. PmDsbA was equilibrated in glutathione (GSSG/GSH) redox buffers to measure the equilibrium constant Keq (187. 5 ± 6 μm), which corresponds to a redox potential of −129 mV. E, absorbance of the catalytic thiolate anion is pH-dependent and this property was used to determine that the pKa of PmDsbA Cys30 is 4.0. F, disulfide reductase activity measured by following A650 nm. PmDsbA has activity similar to that of EcDsbA, and much lower than that of the isomerase EcDsbC. For panels A and C–F, data are presented as mean ± S.D. from three biological replicates. The error bars for the DsbA-only and buffer-only controls in panel A are essentially 0 as there was no increase in fluorescence over the period of the experiment.
We also assessed the ability of PmDsbA to complement EcDsbA in vivo. E. coli strains deficient in the P-ring protein FlgI fail to assemble functional flagella (58). Moreover, E. coli strains lacking EcDsbA, EcDsbB, or both, have the same phenotype (59) because FlgI requires a disulfide bond to function. The non-motile strains E. coli ΔdsbA (JCB817) and ΔdsbA/ΔdsbB (JCB818) were therefore used for in vivo DsbA complementation experiments. When EcDsbA or PmDsbA were expressed in JCB817 following arabinose induction, full rescue of motility was observed in JCB817 (Fig. 1B). However cells from the double knock-out (JCB818) remained non-motile for both PmDsbA and EcDsbA. These results demonstrate that PmDsbA can replace EcDsbA functionally in vivo in the context of flagella assembly. Because EcDsbB was also essential for complementation in this experiment, the results confirm that PmDsbA and EcDsbB can form a functionally competent system. This was expected because the P2 loop sequence from PmDsbB is identical to that of EcDsbB. Furthermore, the P. mirabilis FlgI homologue shares 74% sequence identity with EcFlgI including two conserved cysteine residues.
PmDsbA Shares the Same Characteristic Redox Properties as EcDsbA
DsbA enzymes exhibit unique properties that contribute to their ability to catalyze disulfide bond formation (28, 60). We next established whether these characteristics are shared by PmDsbA. First, we investigated the relative thermostability of the oxidized and reduced forms of the enzyme. In most DsbAs, the reduced form of the CXXC active site is more stable than the oxidized form. For instance, the melting temperature of reduced EcDsbA is almost 10 K higher than that of the oxidized form (39). Similarly, we found that reduced PmDsbA (Tmred 348.4 ± 0.1 K) is 10 K more stable than oxidized PmDsbA (Tmox 338.4 ± 0.2 K) (Fig. 1C).
DsbAs are also highly oxidizing. The redox potential of EcDsbA is −122 mV (42) and the range of values reported for other DsbAs varies from −80 mV for NmDsbA1 (37) to −163 mV for WpDsbA (41). Nevertheless, closely related (>80% sequence identity) homologues of EcDsbA such as SeDsbA and KpDsbA have redox potential values very similar to that of EcDsbA (−126 and −116 mV, respectively). We determined the redox potential of PmDsbA to be −129 mV (Fig. 1D), which is consistent with those of other Enterobacteriaceae DsbAs.
The oxidizing nature of DsbA proteins is thought to be a consequence of the highly acidic cysteine in the CXXC active site motif. The pKa of 3.3 for this nucleophilic cysteine Cys30 for EcDsbA is unusually low for a cysteine (pKa 9.0) (61). Values reported for other DsbAs vary from 3.0 (NmDsbA1) (62) to 5.1 (VcDsbA) (63). We measured the pKa of the equivalent cysteine in PmDsbA, by pH-dependent specific absorbance at λ = 240 nm, and determined the value to be 4.0 (Fig. 1E). Thus, the nucleophilic cysteine of PmDsbA, like that of other DsbAs, is likely to be in the thiolate form at physiological pH when the enzyme is reduced.
Although DsbA proteins are dithiol oxidases they can catalyze disulfide reduction in the presence of mild reducing agents such as DTT. Typically, insulin is used as a substrate to study in vitro disulfide bond reduction. The two chains of insulin are linked by three disulfide bonds, which can be rapidly reduced by disulfide reductases such as the disulfide isomerase EcDsbC. This reduction leads to separation of the insulin A and B chains, and precipitation of the insoluble B chain. Reduction can be followed by measuring the increase in turbidity of the solution over time. We found that PmDsbA is able to reduce the disulfide bonds of insulin as rapidly as EcDsbA (Fig. 1F), but more slowly than the specialist reductant EcDsbC. In summary, the redox properties of PmDsbA reported here place it in the same class as EcDsbA and other Enterobacteriaceae DsbAs such as S. enterica DsbA (SeDsbA) and K. pneumoniae DsbA (KpDsbA) (18).
PWATCDS Binding to PmDsbA and PmDsbAC30S
We were interested to understand how peptides interact with PmDsbA, as the basis for future peptidomimetic inhibitor development. The low resolution crystal structures and NMR characterization of the EcDsbB·EcDsbA complex revealed that the EcDsbB periplasmic loop P2 forms a mixed disulfide with the nucleophilic cysteine of EcDsbA and binds to a hydrophobic groove near the active site (29, 30, 64, 65). The peptide sequence of the EcDsbB P2 loop 98PSPFATCDF106 is conserved in PmDsbB (99PSPFATCDF107) suggesting that a similar interaction occurs between PmDsbA and PmDsbB. From the sequence of this P2 loop peptide, we developed a shorter, modified peptide PWATCDS optimized for solubility, binding affinity, and inhibition of EcDsbA.7 Briefly, alanine scanning, peptide length scouting, and substitution of specific residues showed that the peptide PWATCDS had a good affinity to mass ratio for binding to EcDsbA. We hypothesized that this peptide would also interact with PmDsbA, and therefore evaluated its binding using thermal shift assay and ITC.
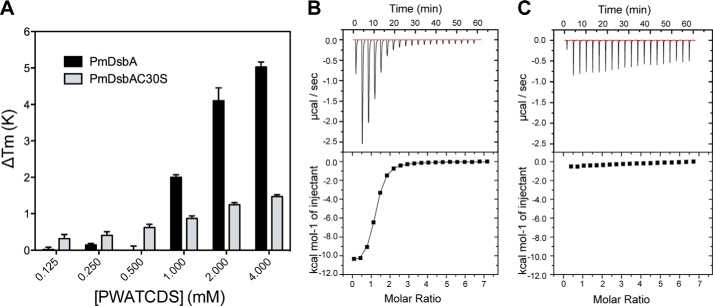
Tm values for PmDsbA and PmDsbAC30S (a variant in which the nucleophilic cysteine was replaced by serine) were measured in the presence and absence of the peptide PWATCDS, with varying peptide concentrations. For PmDsbA, Tm values of +2.0, +4.1, and +5.0 K were observed at peptide concentrations of 1, 2, and 4 mm, respectively, suggesting that PWATCDS binds to PmDsbA. These data show that the Tm shift continued with increasing peptide concentration beyond saturation, as is commonly observed in this assay (66). When the variant PmDsbAC30S was used, thermal shifts (ΔTm > +0.3 K) were detectable at concentrations of PWATCDS ranging from 250 μm to 4 mm. However, the maximum ΔTm at 4 mm was significantly lower than that at the same concentration for PmDsbA (ΔTm +1.5 K, compared with 5.0 K, respectively) (Fig. 2A). This difference in ΔTm suggests the possibility of different binding modes.
FIGURE 2.
Peptide PWATCDS interacts with PmDsbA. A, values of ΔTm upon addition of increasing PWATCDS peptide for PmDsbA and PmDsbAC30S. Data are shown as mean ± S.D. from 5 replicates. B, ITC data titrating PWATCDS into PmDsbA. The reaction was exothermic suggesting a dominant enthalpic contribution to binding. C, ITC data titrating PWATDCS into PmDsbAC30S. Panels B and C show a representative example from three replicates.
We also investigated the interaction using ITC. An exothermic reaction was observed when titrating PWATCDS into PmDsbA (Fig. 2B). Analysis of the raw data using a 1:1 binding model revealed a binding affinity KD of 8.3 ± 0.4 μm with a high enthalpic contribution (ΔH = −13.7 ± 0.3 kcal/mol) and an unfavorable entropy of binding (ΔS = −22.3 ± 1.1 cal/mol/deg). When PmDsbAC30S was used, there was no evidence of PWATCDS binding by ITC under these same conditions (Fig. 2C). Clearly, the C30S mutation reduced the affinity of peptide binding, as evidenced by both thermal shift and ITC. Nevertheless, we were able to generate a crystal structure of this complex as described below. These differing outcomes for the apparently weak complex of PmDsbAC30S·PWATCDS may be a consequence of different experimental design (thermal shift and crystallization used a 1-h preincubation of peptide with PmDsbAC30S, ITC methodology does not allow this; ITC was performed at 25 °C, whereas preincubation was done at 4 °C and crystallization at 20 °C) or different experimental conditions (buffer, pH, and concentration of peptide as outlined under “Experimental Procedures”).
Crystal Structure of PmDsbA
To investigate the different binding modes further, we attempted to determine the crystal structures of PmDsbA and PmDsbAC30S, in the presence of the peptide PWATCDS. We were unable to generate a crystal of a stable complex of PmDsbA with PWATCDS. However, crystals of PmDsbA and PmDsbAC30S without peptide, and PmDsbAC30S with peptide yielded high-resolution diffraction data (PmDsbA, 1.77-Å resolution; PmDsbAC30S, 1.98-Å resolution; and PmDsbAC30S-PWATCDS, 1.6-Å resolution).
PmDsbA crystallized in a tetragonal crystal system and was solved (PDB code 4OCE) by molecular replacement using the EcDsbA structure (PDB code 1FVK) as a template. One protein chain is present in the asymmetric unit, and this has a typical DsbA-like architecture, comprising a TRX core domain (β1, β2, α1, β3, C-terminal region of α6, β4, β5, and α7), interrupted by an α-helical domain (α2-α5 and the N-terminal region of α6). This PmDsbA structure is structurally similar to other Enterobacteriaciae DsbAs including EcDsbA (PDB code 1FVK (67)), KpDsbA (PDB code 4MCU (18)), and SeDsbA (PDB code 3L9S (68)) (Fig. 3A). This is reflected in the r.m.s. deviation of 1.0–1.2 Å comparing 177 Cα atoms from equivalent positions of these enzymes. Fig. 3B shows a structure-based sequence alignment of PmDsbA with homologous DsbA proteins from clinically relevant pathogens that share >59% sequence identity. Sequences are highly conserved, especially at the active site CPHC motif. Sequence identity translates into overall structural similarity between DsbA proteins from these pathogens.
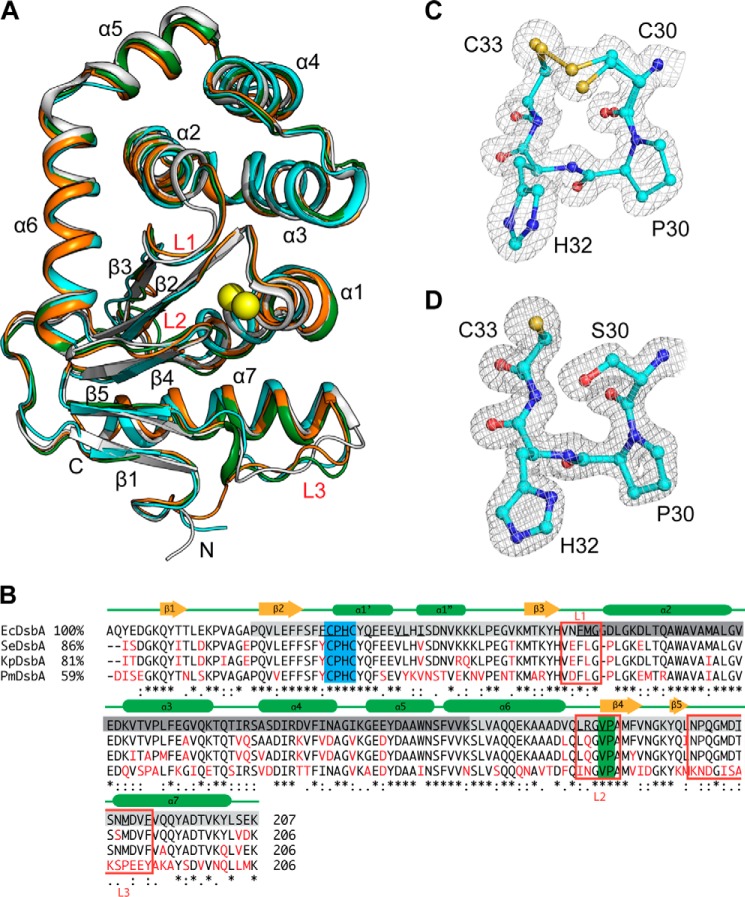
FIGURE 3.
Crystal structure of PmDsbA and its comparison with close homologues. A, structural comparison of four closely related DsbA homologues, PmDsbA (4OCE) in cyan, EcDsbA (1FVK protomer B) in white, KpDsbA (4MCU protomer E) in orange, and SeDsbA in green. Conserved structural regions are annotated; the catalytic cysteines are shown as yellow spheres. B, structure based sequence alignment of EcDsbA (1FVK, chain A), SeDsbA (3L9S, chain A), KpDsbA (4MCU, chain F), and PmDsbA (4OCE, chain A). Sequence identity with EcDsbA is shown on the left, conserved residues are black, differing residues are red. The TRX domain is highlighted in light gray, and the helical domain in dark gray. Red squares mark the three loop regions L1/2/3. Amino acid conservation code is shown beneath the sequences. The active site CXXC motif is highlighted in blue, and the cis-Pro region in green. Secondary structure elements are indicated above the sequences. Residues underlined in EcDsbA bind to the EcDsbB P2 loop peptide (PISA server analysis). C, CXXC active site of PmDsbA. D, CXXC active site of PmDsbAC30S:PWATCDS. For panels C and D, the electron density is from 2F0 − Fc FFT maps generated in Phenix (53) and contoured at 1.0 σ.
The sequence and structure of the active site motif (CPHC) and the cis-Pro loop are identical in these DsbA homologues. However, helix α1 and the loops connecting β1 and β2, and α3 and α4 differ in their relative positions. Most importantly loop L3 that connects β3 and helix α7 varies across the structures. The EcDsbA L3 residues Asp167, Thr168, and Ser169 are positioned closer to the active site helix α1 than the equivalent residues (Ala167, Lys168, and Ser169) in PmDsbA. This region is relatively hydrophobic in both PmDsbA and EcDsbA, although more basic in PmDsbA than in EcDsbA due to the presence of Lys159 in the former and Gln160 in the latter. Overall these differences in structure are relatively minor compared with the structures of other DsbA proteins, such as Pseudomonas aeruginosa DsbA (25% identity with PmDsbA, r.m.s. deviation 2.2 Å, 175 Cα, PDB code 3H93 (35)), and Wolbachia pipientis DsbA1 (16% identity with PmDsbA, r.m.s. deviation 3.6 Å, 151 Cα, PDB code 3F4R (41)).
Crystal Structure Determination of PmDsbAC30S and PmDsbAC30S·PWATCDS
We solved the structure of the active site mutant PmDsbAC30S (PDB code 4OCF). The crystals grew in the presence of peptide, but no bound peptide was evident in the electron density map. We therefore used this structure of PmDsbAC30S to assess whether replacement of Cys30 with Ser30 induced any structural changes. The mutant crystallized in a monoclinic crystal system containing 4 protomers in the asymmetric unit. The structure was solved by molecular replacement using the coordinates of PmDsbA described above. The average r.m.s. deviation for comparison of the 6 combinations of the 4 protomers range from 0.3 to 0.7 Å for 177 equivalent Cα atoms (residues 6–181).
PmDsbAC30S in complex with peptide PWATCDS crystallized in a trigonal crystal system, containing 3 protomers in the asymmetric unit. The complex was solved by molecular replacement using PmDsbA (PDB code 4OD7) as the template. All three protomers (A, B, and C) reveal strong electron density corresponding to bound PWATCDS. The average r.m.s. deviation for the 3 comparisons of the protomers in this crystal structure range from 0.4 to 0.6 Å for 177 equivalent Cα atoms (residues 6–181). The r.m.s. deviation for comparison of PmDsbAC30S with PmDsbAC30S·PWATCDS range from 0.5 to 0.9 Å (177 Cα) for the 12 combinations. Similarly, comparison of the wild type PmDsbA structure with PmDsbAC30S and PmDsbAC30S·PWATCDS gave r.m.s. deviation values of 0.3–0.9 Å (177 Cα). This result indicates there is no major reorganization of the PmDsbAC30S structure upon interaction with PWATCDS. However, there is evidence of rigid body shifts for helix 1 and side chain adjustments for residues on helix 1 (His32, Tyr34, Gln35, Phe36, Ser37) and flexible loop 3 (Ile165 and Ser166) that together form part of the peptide binding site.
Active Site Structure Is Conserved after Peptide Binding
A comparison of the active site motifs (CPHC and CPHS) and the Val150/Pro151 residues of the cis-Pro loop across all the protomers in the PmDsbA, PmDsbAC30S, and PmDsbAC30S·PWATCDS crystal structures reveals a high degree of structural conservation. The active site cysteines Cys30 and Cys33 in the native PmDsbA structure were modeled as a mixture of oxidized and reduced (ratio 0.7:0.3). In the oxidized state the two sulfurs (Fig. 3C) are 2.2 Å apart, reflecting the typical length of a covalent disulfide bond found in other oxidized DsbA structures (e.g. EcDsbA and SeDsbA, S-S distance 2.0 Å) (26, 68). The dithiol form of the cysteines is likely a consequence of radiation-induced disulfide reduction. The distance between the dithiol sulfurs is 3.4 Å. In the PmDsbAC30S and PmDsbAC30S·PWATCDS structures, the distance between the hydroxyl oxygen of Ser30 and the sulfur atom of Cys33 varies between 3.3 and 3.5 Å (Fig. 3D). These distances are in agreement with those of other reduced DsbA structures (e.g. KpDsbA 3.3–3.8 Å) (18). Overall, there is no apparent change in the active site upon binding of the peptide.
Peptide Binding Mode and Interaction with PmDsbAC30S
The binding mode of the peptide (Fig. 4, A and B) is highly conserved across the three protomers (r.m.s. deviation 0.1–0.2 Å over all 82 atoms). The electron density from this high-resolution structure provides strong evidence for both the position and conformation of the bound peptide (Fig. 4C). The binding site for PWATCDS (residues 1 to 7) includes the hydrophobic groove and the active site regions (CXXC and cis-Pro motif) of the enzyme. As expected, the hydrophobic residues Pro1 and Trp2 (italics indicate peptide residues) interact with the hydrophobic groove, and the C-terminal residues interact with the cis-Pro region of the active site.
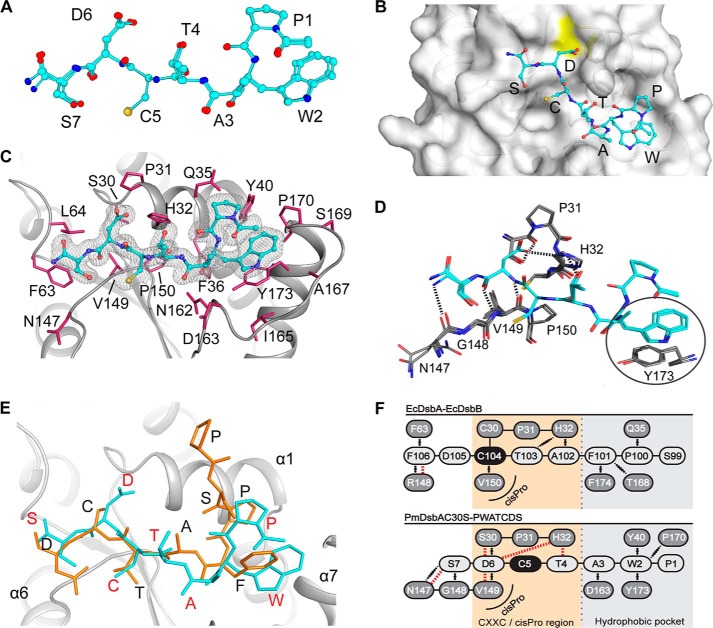
FIGURE 4.
Analysis of the interaction between peptide PWATCDS and PmDsbAC30S. A, superposition of the PWATCDS peptides from all three protomer·peptide complexes in the asymmetric unit. Note the peptide includes an N-terminal acetyl and a C-terminal amide group. Carbon atoms are shown in cyan, oxygens in red, nitrogens in blue, and sulfurs in yellow. B, location of the bound PWATCDS peptide on the surface of PmDsbAC30S (protomer A shown in gray). Peptide residues are labeled and colored as for panel A. The yellow patch indicates the location of Ser30. C, electron density map of PWATCDS (chain F) at the interface with PmDsbAC30S (chain B). The 2F0 − Fc map was generated in Phenix (53) and is contoured at 1.0 σ. PmDsbAC30S residues forming the binding site are labeled. D, interactions between the peptide (D–F, in cyan) and protein (chains A-C, in gray) are shown: hydrogen bonds are indicated as black dashed lines; a circle highlights the stacking interaction between Trp2 and PmDsbAC30S Tyr173. E, superposition of the EcDsbB P2 periplasmic loop (PSPFATCD, orange and black letters, PDB code 2ZUP) with PmDsbAC30S-PWATCDS (red letters, backbone in cyan). F, schematic representation of the interactions formed between EcDsbB P2 loop binding to EcDsbA (top) in comparison to PWATCDS binding to PmDsbAC30S (bottom), showing the comparative shift in register. Covalent bonds are shown as black lines, hydrogen bonds are indicated with red dashed lines, and hydrophobic interactions with black dotted lines.
However, the observed binding mode was not entirely as predicted. On the basis of the EcDsbA·EcDsbB covalent complex, Cys5 of the peptide should interact with PmDsbAC30S residue 30 (mutated from Cys to Ser). This was not the case. In all three complexes in the asymmetric unit, Asp6 and not Cys5 interacts with PmDsbAC30S Ser30 (Fig. 4D). The acidic side chain of Asp6 is within hydrogen bond contact distance of Ser30 and His32. Moreover, the backbone amide of Asp6 forms hydrogen bonds with the backbone amide of Val149 of the PmDsbAC30S cis-Pro loop (Fig. 4D).
Comparison of Peptide Binding Mode with DsbA-DsbB Complexes
The EcDsbB P2 loop (sequence PSPFATCDF) and the synthetic peptide we used (PWATCDS) share the same binding location on EcDsbA and PmDsbAC30S, respectively. The EcDsbA-EcDsbB complex (3.7-Å resolution (30)), revealed one possible hydrogen bond between P2 and the enzyme (backbone oxygen of Arg148 of the EcDsbA cis-Pro loop with the backbone nitrogen of EcDsbB Phe106). All other interactions with the P2 loop, aside from the covalent disulfide, are hydrophobic (28).
As indicated above, no interaction was observed between Cys5 and Ser30 of PmDsbAC30S. Instead, hydrogen bond interactions with Asp6 and hydrophobic interactions with Trp2 appear to dominate the PWATCDS binding mode. Residues between these two anchor points, Cys5, Thr4, and Ala3, protrude out of the binding site relative to the equivalent EcDsbB loop residues (Fig. 4, E and F).
The r.m.s. deviation for comparison of PWATCDS with the EcDsbB P2 loop conformation is relatively high (2.2 Å for 28 backbone atoms), because of the bulge in the peptide conformation. Binding of PWATCDS results in an average buried surface area (i.e. surface that becomes inaccessible to solvent) of 955 ± 12 Å or 9.0 ± 0.1% of the total surface of PmDsbAC30S (values are mean ± S.D. generated from the three molecules in the asymmetric unit). This is similar in area to the EcDsbA buried surface upon binding of the EcDsbB P2 loop (924 Å2) (30). In both structures a major feature is the binding of an aromatic residue in the hydrophobic groove. EcDsbB P2 loop residue Phe101 forms a T-shaped π-π stacking interaction with EcDsbA Phe174, whereas Trp2 of the PWATCDS peptide forms a parallel π-π stacking interaction with PmDsbAC30S Tyr173 and possible edge interactions with enzyme residues Pro170 and Tyr40.
DISCUSSION
The increasing incidence of infections caused by multidrug-resistant pathogens represents a serious global human health issue. Indeed, the emergence of carbapenem-resistant Enterobacteriaceae threatens to make common infections such as urinary tract infections untreatable (69). Antibiotic resistance is spreading rapidly and treatment options are becoming increasingly limited. One possible approach to address the paucity of new antimicrobials in the developmental pipeline lies in the generation of novel anti-virulence drugs (19, 70).
The Gram-negative DsbA/B system has been proposed as a target for the development of novel anti-virulence drugs (17). Targeting DsbA/B for development of inhibitors could be beneficial in many ways. First, DsbA is not essential for bacterial survival (27) although it plays an essential role in virulence (17). Thus, inhibiting DsbA/B would not kill bacteria and this property may reduce the selective pressure to develop resistance. Second, structures of several DsbA homologues have been solved (18, 28, 71), providing a framework for structure-based drug design. Third, DsbA structures and properties are more highly conserved than the structures and sequences of virulence factors across pathogens (18). Therefore, inhibitors that target one DsbA enzyme within a subclass are likely to block DsbAs within the subclass offering the possibility of medium-spectrum inhibitors (18).
Designing an inhibitor to block a protein-protein interface, such as that of a DsbA, requires a comprehensive knowledge of its binding interactions. Here we have used an innovative approach to define the interaction surface of PmDsbA by using knowledge from the low resolution crystal structure of EcDsbA-EcDsbB. We characterized a peptide derived from the sequence of DsbB, showed that it bound to PmDsbA and co-crystallized it in a non-covalent complex with PmDsbAC30S. This provides the first example of a high-resolution crystal structure of a DsbA in complex with a non-covalently bound peptide. Two previously published structures of DsbA in complex with bound peptides include (i) EcDsbA bound covalently with a substrate SigA-derived peptide; and (ii) Xyfella fastidiosa DsbA also bound covalently with a peptide that co-crystallized fortuitously.
P. mirabilis associated infections are often difficult to treat due to its propensity to form biofilms (4). Two cell surface organelles associated with P. mirabilis biofilm formation, mannose-resistant Proteus-like (MR/P) fimbriae (72) and flagella (which mediate swarming) (4), require DsbA for their correct assembly. Taken together, our data show that PmDsbA exhibits redox, functional, and structural properties typical of the DsbA class Ia enzymes (73), which extends to all DsbAs characterized to date from Enterobacteriaceae (18). We expect that the structural details of the peptide binding mode and the interactions observed will likely hold true for all of these enzymes. Specifically, the hydrophobic groove and the cis-Pro loop provide key points of interaction that could be exploited further. The high-resolution crystal structure of the non-covalent complex between PmDsbAC30S and peptide may provide an important platform for the development of peptidomimetic antivirulence compounds targeting PmDsbA and by extension the DsbAs from all Enterobacteriaceae.
Acknowledgments
We are grateful to Brett Collins and Makrina Totsika for helpful advice and Stephanie Tay for preparing EcDsbB membranes for the cysteine thiol oxidation assay. We thank the beam-line staff at the Australian Synchrotron for their assistance. We acknowledge use of the UQ ROCX Diffraction Facility.
The atomic coordinates and structure factors (codes 4OCE, 4OCF, and 4OD7) have been deposited in the Protein Data Bank (http://wwpdb.org/).
W. Duprez, L. Premkumar, M. Halili, F. Lindahl, R. Reid, D. P. Fairlie, and J. L. Martin, unpublished data.
- TEV
- tobacco etch virus
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- HbtU
- O-benzotriazole-N,N,N′,N′-tetramethyluronium hexafluorophosphate
- ITC
- isothermal titration calorimetry
- PDB
- Protein Data Bank
- r.m.s.
- root mean square.
REFERENCES
- 1. Guay D. R. (2008) Contemporary management of uncomplicated urinary tract infections. Drugs 68, 1169–1205 [DOI] [PubMed] [Google Scholar]
- 2. Ronald A. (2003) The etiology of urinary tract infection: traditional and emerging pathogens. Disease-a-month: DM 49, 71–82 [DOI] [PubMed] [Google Scholar]
- 3. Jacobsen S. M., Stickler D. J., Mobley H. L., Shirtliff M. E. (2008) Complicated catheter-associated urinary tract infections due to Escherichia coli and Proteus mirabilis. Clin. Microbiol. Rev. 21, 26–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jacobsen S. M., Shirtliff M. E. (2011) Proteus mirabilis biofilms and catheter-associated urinary tract infections. Virulence 2, 460–465 [DOI] [PubMed] [Google Scholar]
- 5. Coetzee J. N., Sacks T. G. (1960) Transduction of streptomycin resistance in Proteus mirabilis. J. Gen. Microbiol. 23, 445–455 [DOI] [PubMed] [Google Scholar]
- 6. Sabath L. D. (1969) Drug resistance of bacteria. N. Engl. J. Med. 280, 91–94 [DOI] [PubMed] [Google Scholar]
- 7. Pearson M. M., Sebaihia M., Churcher C., Quail M. A., Seshasayee A. S., Luscombe N. M., Abdellah Z., Arrosmith C., Atkin B., Chillingworth T., Hauser H., Jagels K., Moule S., Mungall K., Norbertczak H., Rabbinowitsch E., Walker D., Whithead S., Thomson N. R., Rather P. N., Parkhill J., Mobley H. L. (2008) Complete genome sequence of uropathogenic Proteus mirabilis, a master of both adherence and motility. J. Bacteriol. 190, 4027–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hitchings G. H. (1973) Mechanism of action of trimethoprim-sulfamethoxazole. I. J. Infect. Dis. 128, 433–436 [DOI] [PubMed] [Google Scholar]
- 9. Wellington E. M., Boxall A. B., Cross P., Feil E. J., Gaze W. H., Hawkey P. M., Johnson-Rollings A. S., Jones D. L., Lee N. M., Otten W., Thomas C. M., Williams A. P. (2013) The role of the natural environment in the emergence of antibiotic resistance in Gram-negative bacteria. Lancet Infect. Dis. 13, 155–165 [DOI] [PubMed] [Google Scholar]
- 10. Wang M., Guo Q., Xu X., Wang X., Ye X., Wu S., Hooper D. C., Wang M. (2009) New plasmid-mediated quinolone resistance gene, qnrC, found in a clinical isolate of Proteus mirabilis. Antimicrob. Agents Chemother. 53, 1892–1897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Elsea S. H., Osheroff N., Nitiss J. L. (1992) Cytotoxicity of quinolones toward eukaryotic cells: identification of topoisomerase II as the primary cellular target for the quinolone CP-115,953 in yeast. J. Biol. Chem. 267, 13150–13153 [PubMed] [Google Scholar]
- 12. Tibbetts R., Frye J. G., Marschall J., Warren D., Dunne W. (2008) Detection of KPC-2 in a clinical isolate of Proteus mirabilis and first reported description of carbapenemase resistance caused by a KPC β-lactamase in P. mirabilis. J. Clin. Microbiol. 46, 3080–3083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mehtar S., Tsakris A., Pitt T. L. (1991) Imipenem resistance in Proteus mirabilis. J. Antimicrob. Chemother. 28, 612–615 [DOI] [PubMed] [Google Scholar]
- 14. Franklin T. J., Rownd R. (1973) R-factor-mediated resistance to tetracycline in Proteus mirabilis. J. Bacteriol. 115, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coetzee J. N. (1975) High frequency transduction of resistance to ampicillin and kanamycin in Proteus mirabilis. J. Gen. Microbiol. 87, 173–176 [DOI] [PubMed] [Google Scholar]
- 16. Zhao W. H., Hu Z. Q. (2013) Epidemiology and genetics of CTX-M extended-spectrum β-lactamases in Gram-negative bacteria. Crit. Rev. Microbiol. 39, 79–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Heras B., Shouldice S. R., Totsika M., Scanlon M. J., Schembri M. A., Martin J. L. (2009) DSB proteins and bacterial pathogenicity. Nat. Rev. Microbiol. 7, 215–225 [DOI] [PubMed] [Google Scholar]
- 18. Kurth F., Rimmer K., Premkumar L., Mohanty B., Duprez W., Halili M. A., Shouldice S. R., Heras B., Fairlie D. P., Scanlon M. J., Martin J. L. (2013) Comparative sequence, structure and redox analyses of Klebsiella pneumoniae DsbA show that anti-virulence target DsbA enzymes fall into distinct classes. PLoS One 8, e80210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rasko D. A., Sperandio V. (2010) Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 9, 117–128 [DOI] [PubMed] [Google Scholar]
- 20. Burall L. S., Harro J. M., Li X., Lockatell C. V., Himpsl S. D., Hebel J. R., Johnson D. E., Mobley H. L. (2004) Proteus mirabilis genes that contribute to pathogenesis of urinary tract infection: identification of 25 signature-tagged mutants attenuated at least 100-fold. Infect. Immun. 72, 2922–2938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Totsika M., Heras B., Wurpel D. J., Schembri M. A. (2009) Characterization of two homologous disulfide bond systems involved in virulence factor biogenesis in uropathogenic Escherichia coli CFT073. J. Bacteriol. 191, 3901–3908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ireland P. M., McMahon R. M., Marshall L. E., Halili M., Furlong E., Tay S., Martin J. L., Sarkar-Tyson M. (2014) Disarming Burkholderia pseudomallei: structural and functional characterization of a disulfide oxidoreductase (DsbA) required for virulence in vivo. Antioxid. Redox Signal. 20, 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Peek J. A., Taylor R. K. (1992) Characterization of a periplasmic thiol:disulfide interchange protein required for the functional maturation of secreted virulence factors of Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 89, 6210–6214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yu J. (1998) Inactivation of DsbA, but not DsbC and DsbD, affects the intracellular survival and virulence of Shigella flexneri. Infect Immun. 66, 3909–3917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin D., Rao C. V., Slauch J. M. (2008) The Salmonella SPI1 type three secretion system responds to periplasmic disulfide bond status via the flagellar apparatus and the RcsCDB system. J. Bacteriol. 190, 87–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Martin J. L., Bardwell J. C., Kuriyan J. (1993) Crystal structure of the DsbA protein required for disulphide bond formation in vivo. Nature 365, 464–468 [DOI] [PubMed] [Google Scholar]
- 27. Bardwell J. C., McGovern K., Beckwith J. (1991) Identification of a protein required for disulfide bond formation in vivo. Cell 67, 581–589 [DOI] [PubMed] [Google Scholar]
- 28. Shouldice S. R., Heras B., Walden P. M., Totsika M., Schembri M. A., Martin J. L. (2011) Structure and function of DsbA, a key bacterial oxidative folding catalyst. Antioxid. Redox Signal. 14, 1729–1760 [DOI] [PubMed] [Google Scholar]
- 29. Inaba K., Murakami S., Suzuki M., Nakagawa A., Yamashita E., Okada K., Ito K. (2006) Crystal structure of the DsbB-DsbA complex reveals a mechanism of disulfide bond generation. Cell 127, 789–801 [DOI] [PubMed] [Google Scholar]
- 30. Inaba K., Murakami S., Nakagawa A., Iida H., Kinjo M., Ito K., Suzuki M. (2009) Dynamic nature of disulphide bond formation catalysts revealed by crystal structures of DsbB. EMBO J. 28, 779–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Eschenfeldt W. H., Lucy S., Millard C. S., Joachimiak A., Mark I. D. (2009) A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol. Biol. 498, 105–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Studier F. W. (2005) Protein production by autoinduction in high density shaking cultures. Protein Expr. Purif. 41, 207–234 [DOI] [PubMed] [Google Scholar]
- 33. Gasteiger E. H. C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., Bairoch A. (2005) Protein identification and analysis tools on the ExPASy server. in The Proteomics Protocols Handbook (Walker J. M., ed) pp. 571–607, Humana Press, New York [Google Scholar]
- 34. Bader M., Muse W., Zander T., Bardwell J. (1998) Reconstitution of a protein disulfide catalytic system. J. Biol. Chem. 273, 10302–10307 [DOI] [PubMed] [Google Scholar]
- 35. Shouldice S. R., Heras B., Jarrott R., Sharma P., Scanlon M. J., Martin J. L. (2010) Characterization of the DsbA oxidative folding catalyst from Pseudomonas aeruginosa reveals a highly oxidizing protein that binds small molecules. Antioxid. Redox Signal. 12, 921–931 [DOI] [PubMed] [Google Scholar]
- 36. Guzman L. M., Belin D., Carson M. J., Beckwith J. (1995) Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177, 4121–4130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vivian J. P., Scoullar J., Rimmer K., Bushell S. R., Beddoe T., Wilce M. C., Byres E., Boyle T. P., Doak B., Simpson J. S., Graham B., Heras B., Kahler C. M., Rossjohn J., Scanlon M. J. (2009) Structure and function of the oxidoreductase DsbA1 from Neisseria meningitidis. J. Mol. Biol. 394, 931–943 [DOI] [PubMed] [Google Scholar]
- 38. Walden P. M., Heras B., Chen K. E., Halili M. A., Rimmer K., Sharma P., Scanlon M. J., Martin J. L. (2012) The 1.2-Å resolution crystal structure of TcpG, the Vibrio cholerae DsbA disulfide-forming protein required for pilus and cholera toxin production. Acta Crystallogr. D Biol. Crystallogr. 68, 1290–1302 [DOI] [PubMed] [Google Scholar]
- 39. Heras B., Kurz M., Jarrott R., Shouldice S. R., Frei P., Robin G., Cemazar M., Thöny-Meyer L., Glockshuber R., Martin J. L. (2008) Staphylococcus aureus DsbA does not have a destabilizing disulfide: a new paradigm for bacterial oxidative folding. J. Biol. Chem. 283, 4261–4271 [DOI] [PubMed] [Google Scholar]
- 40. Ellman G. L. (1959) Tissue sulfhydryl groups. Arch. Biochem. Biophys. 82, 70–77 [DOI] [PubMed] [Google Scholar]
- 41. Kurz M., Iturbe-Ormaetxe I., Jarrott R., Shouldice S. R., Wouters M. A., Frei P., Glockshuber R., O'Neill S. L., Heras B., Martin J. L. (2009) Structural and functional characterization of the oxidoreductase α-DsbA1 from Wolbachia pipientis. Antioxid. Redox Signal. 11, 1485–1500 [DOI] [PubMed] [Google Scholar]
- 42. Wunderlich M., Glockshuber R. (1993) Redox properties of protein disulfide isomerase (DsbA) from Escherichia coli. Protein Sci. 2, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nelson J. W., Creighton T. E. (1994) Reactivity and ionization of the active site cysteine residues of DsbA, a protein required for disulfide bond formation in vivo. Biochemistry 33, 5974–5983 [DOI] [PubMed] [Google Scholar]
- 44. Holmgren A. (1979) Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. J. Biol. Chem. 254, 9627–9632 [PubMed] [Google Scholar]
- 45. Kranz J. K., Schalk-Hihi C. (2011) Protein thermal shifts to identify low molecular weight fragments. Methods Enzymol. 493, 277–298 [DOI] [PubMed] [Google Scholar]
- 46. McPhillips T. M., McPhillips S. E., Chiu H. J., Cohen A. E., Deacon A. M., Ellis P. J., Garman E., Gonzalez A., Sauter N. K., Phizackerley R. P., Soltis S. M., Kuhn P. (2002) Blu-Ice and the distributed control system: software for data acquisition and instrument control at macromolecular crystallography beamlines. J. Synchrotron Radiat. 9, 401–406 [DOI] [PubMed] [Google Scholar]
- 47. Battye T. G., Kontogiannis L., Johnson O., Powell H. R., Leslie A. G. (2011) iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kabsch W. (2010) Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 50. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems: application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ye Y., Godzik A. (2003) Flexible structure alignment by chaining aligned fragment pairs allowing twists. Bioinformatics 19, ii246–255 [DOI] [PubMed] [Google Scholar]
- 56. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 57. Vangone A., Spinelli R., Scarano V., Cavallo L., Oliva R. (2011) COCOMAPS: a web application to analyze and visualize contacts at the interface of biomolecular complexes. Bioinformatics 27, 2915–2916 [DOI] [PubMed] [Google Scholar]
- 58. Ohnishi K., Homma M., Kutsukake K., Iino T. (1987) Formation of flagella lacking outer rings by flaM, flaU, and flaY mutants of Escherichia coli. J. Bacteriol. 169, 1485–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dailey F. E., Berg H. C. (1993) Mutants in disulfide bond formation that disrupt flagellar assembly in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 90, 1043–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pogliano J., Lynch A. S., Belin D., Lin E. C., Beckwith J. (1997) Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 11, 1169–1182 [DOI] [PubMed] [Google Scholar]
- 61. Huber-Wunderlich M., Glockshuber R. (1998) A single dipeptide sequence modulates the redox properties of a whole enzyme family. Folding Design 3, 161–171 [DOI] [PubMed] [Google Scholar]
- 62. Lafaye C., Iwema T., Carpentier P., Jullian-Binard C., Kroll J. S., Collet J. F., Serre L. (2009) Biochemical and structural study of the homologues of the thiol-disulfide oxidoreductase DsbA in Neisseria meningitidis. J. Mol. Biol. 392, 952–966 [DOI] [PubMed] [Google Scholar]
- 63. Ruddock L. W., Hirst T. R., Freedman R. B. (1996) pH-dependence of the dithiol-oxidizing activity of DsbA (a periplasmic protein thiol:disulphide oxidoreductase) and protein disulphide-isomerase: studies with a novel simple peptide substrate. Biochem. J. 315, 1001–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Malojći G., Owen R. L., Grimshaw J. P., Glockshuber R. (2008) Preparation and structure of the charge-transfer intermediate of the transmembrane redox catalyst DsbB. FEBS Lett. 582, 3301–3307 [DOI] [PubMed] [Google Scholar]
- 65. Sperling L. J., Tang M., Berthold D. A., Nesbitt A. E., Gennis R. B., Rienstra C. M. (2013) Solid-state NMR study of a 41 kDa membrane protein complex DsbA/DsbB. J. Phys. Chem. B 117, 6052–6060 [DOI] [PubMed] [Google Scholar]
- 66. Cimmperman P., Baranauskiene L., Jachimoviciūte S., Jachno J., Torresan J., Michailoviene V., Matuliene J., Sereikaite J., Bumelis V., Matulis D. (2008) A quantitative model of thermal stabilization and destabilization of proteins by ligands. Biophys. J. 95, 3222–3231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Guddat L. W., Bardwell J. C., Glockshuber R., Huber-Wunderlich M., Zander T., Martin J. L. (1997) Structural analysis of three His32 mutants of DsbA: support for an electrostatic role of His32 in DsbA stability. Protein Sci. 6, 1893–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Heras B., Totsika M., Jarrott R., Shouldice S. R., Guncar G., Achard M. E., Wells T. J., Argente M. P., McEwan A. G., Schembri M. A. (2010) Structural and functional characterization of three DsbA paralogues from Salmonella enterica serovar typhimurium. J. Biol. Chem. 285, 18423–18432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Totsika M., Moriel D. G., Idris A., Rogers B. A., Wurpel D. J., Phan M. D., Paterson D. L., Schembri M. A. (2012) Uropathogenic Escherichia coli mediated urinary tract infection. Curr. Drug Targets 13, 1386–1399 [DOI] [PubMed] [Google Scholar]
- 70. Clatworthy A. E., Pierson E., Hung D. T. (2007) Targeting virulence: a new paradigm for antimicrobial therapy. Nat. Chem. Biol. 3, 541–548 [DOI] [PubMed] [Google Scholar]
- 71. Premkumar L., Heras B., Duprez W., Walden P., Halili M., Kurth F., Fairlie D. P., Martin J. L. (2013) Rv2969c, essential for optimal growth in Mycobacterium tuberculosis, is a DsbA-like enzyme that interacts with VKOR-derived peptides and has atypical features of DsbA-like disulfide oxidases. Acta Crystallogr. D Biol. Crystallogr. 69, 1981–1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jansen A. M., Lockatell V., Johnson D. E., Mobley H. L. (2004) Mannose-resistant Proteus-like fimbriae are produced by most Proteus mirabilis strains infecting the urinary tract, dictate the in vivo localization of bacteria, and contribute to biofilm formation. Infect. Immun. 72, 7294–7305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. McMahon R. M., Premkumar L., Martin J. L. (2014) Four structural subclasses of the antivirulence drug target disulfide oxidoreductase DsbA provide a platform for design of subclass-specific inhibitors. Biochim. Biophys. Acta 1844, 1391–1401 [DOI] [PubMed] [Google Scholar]
- 74. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]