Abstract
Inherited mutations in human PALB2 are associated with a predisposition to breast and pancreatic cancers. The tumor-suppressing capability of PALB2 is thought to be based on its ability to enable BRCA2 function in homologous recombination. However, the biochemical properties of PALB2 are unknown. Here we show that human PALB2 binds DNA, preferentially D-loop structures, and directly interacts with the RAD51 recombinase to strongly stimulates strand invasion, a vital step of homologous recombination. Such stimulation occur by reinforcing biochemical mechanisms as PALB2 alleviates the inhibitory role of RPA and stabilizes the RAD51 filament. Moreover, PALB2 can function synergistically with a BRCA2 chimera (termed piccolo) to further promote strand invasion. Finally, we show that PALB2-deficient cells are sensitive to PARP inhibitors. Collectively, our studies provide the first biochemical insights into the homologous recombination mediator functions of PALB2 with piBRCA2 in DNA double-strand break repair.
Keywords: RAD51, PALB2, BRCA2, homologous recombination
Introduction
Breast and ovarian cancer is estimated to be responsible for 20% of cancer mortality among woman 1. Familial breast cancer can result from impaired genome stability control following mutations in homologous recombinational repair proteins such as BRCA2 2,3. In mammalian cells, homologous recombination (HR) has emerged as the major mechanism for the error-free homology-directed repair of DSBs during S and G2 phase of the cell cycle. The central activity of HR is conferred by the RAD51 protein, which catalyzes the invasion of resected ends of the DSB into the intact sister chromatids 4. BRCA2 plays a pivotal role in controlling the function and localization of RAD51. The BRC repeats located at the center of BRCA2 confer a very complicated mode of regulation. The BRC repeats bind RAD51 and control RAD51 nucleoprotein filament formation in conjunction with the BRCA2 DNA-binding domain, whereas another, BRC-unrelated RAD51-binding motif at the C-terminus of BRCA2 appears to stabilize the filament 5–8. Importantly, point mutations in BRC repeats are found in individuals predisposed to breast cancer (http://research.nhgri.nih.gov/bic/).
Homologous recombination can be divided in three major steps: the presynaptic stage involves RAD51 nucleoprotein filament assembly, the synaptic stage is characterized by strand invasion and branch migration, and the postsynaptic stage includes Holliday junction formation and resolution. Once a DSB is created, the nuclease activity of MRE11/CtIP/EXO1 9–11 are required for the processing of DNA double-strand breaks (DSBs) to generate the replication protein A (RPA)-coated ssDNA that is needed for ATR recruitment and subsequent phosphorylation and activation of CHK1 12. Replication protein A, possess high avidity for these 3′ single-stranded DNA tails, leading to an inhibitory effect for RAD51 loading and HR. Recombination mediators, such as the BRCA2 homolog Brh2, overcomes the inhibitory effect of RPA and allows RAD51 to nucleate on the ssDNA to promote the presynaptic filament assembly 13.
PALB2 was identified as a major partner of BRCA2 and shown to be an essential upstream regulator of BRCA2 intranuclear localization and its HR function 14. Furthermore, multiple independent studies have established PALB2 as the central core of a BRCA1-PALB2-BRCA2 complex essential for HR 15–17. To date, about a dozen PALB2 truncating mutations have been found in multiple countries to be associated with 2–6 fold increased risk of breast cancer 18,19. Like BRCA2 (FANCD1), PALB2 (also known as FANCN) is mutated in a subgroup of Fanconi anemia (FA) 20,21. Moreover, PALB2 was also recently identified as a pancreatic susceptibility gene, being the second most frequently mutated after BRCA2 22. Thus, similar to BRCA2, PALB2 is at the crossroads of breast cancer, pancreatic cancer and FA. Like other FA cells, PALB2/FANCN cells display increased chromosome breakage after treatment with drugs causing interstrand crosslinks consistent with a role in DNA repair. Although PALB2 affects homologous recombination and genome stability, biochemical insights into PALB2 are lacking.
Herein, we have purified the PALB2 tumor suppressor and demonstrate that PALB2 is a DNA binding protein that physically interacts with RAD51. Analysis of the DNA binding properties of PALB2 revealed a preference for the D-loop structure. PALB2 strongly stimulates DNA strand invasion activity of RAD51 and stabilizes the nucleoprotein filament against a disruptive BRC3/4 polypeptide. Moreover, PALB2 can work in concert with a BRCA2 chimera to further promote D-loop formation. Consistent with a role in homologous recombination, PALB2-deficient cells, like BRCA2-deficient cells, are sensitive to a PARP inhibitor. Our findings establish PALB2 as an important regulator of homologous recombination.
Results
Purified PALB2 binds DNA
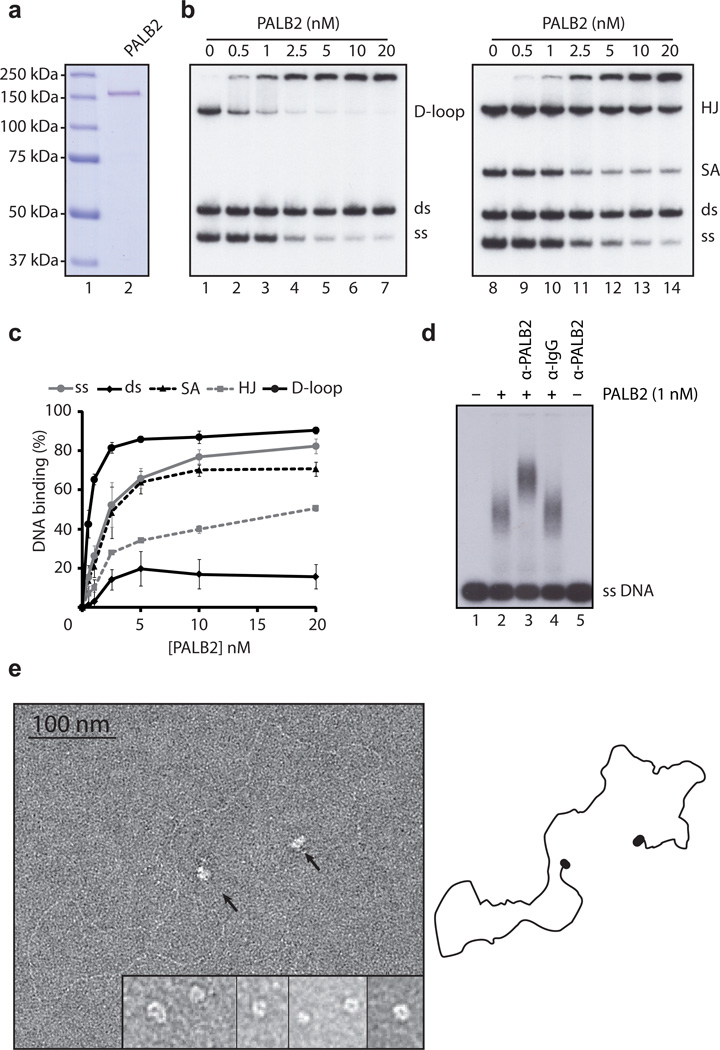
siRNA knockdown of PALB2 leads to a decrease in homologous recombination 14 and also MMC-induced RAD51 foci formation (Supplementary Fig. 1A-C). To better understand the role of PALB2 in HR, we purified the PALB2 protein from insect cells (Fig. 1A). Since PALB2 is a chromatin-bound protein 14, we tested whether PALB2 could bind DNA directly. DNA mobility shift assays revealed that PALB2 bound ssDNA, dsDNA, splayed arms, Holliday junctions, and D-loop structures with different affinities (Supplementary Fig. 2A-G). Competitions assays were carried out to clearly establish the preferred substrate. PALB2 bound D-loop preferentially over ssDNA, splayed arms, Holliday junctions, and dsDNA (Fig. 1B). At 0.5 nM PALB2, 40% of D-loop was bound, followed by ssDNA and splayed arms, compared with 1–15 % for all other DNA substrates (Fig. 1C). PALB2-DNA complexes resolved on an agarose gel were supershifted with a PALB2 antibody suggesting that the DNA binding activity is PALB2-dependent (Fig. 1D). The preferential PALB2 binding to D-loop structures could not be competed with an excess of ssDNA of different lengths (60 nt, 91 nt or 4300 bases) suggesting that this was the most preferred substrate. The binding to dsDNA was vastly reduced in the presence of ssDNA (60 and 91 nucleotides) while ssDNA binding is poorly competed by dsDNA (4300 bp) (Supplementary Fig 2H). We further explored PALB2-DNA association using linear tailed DNA that comprises both single-stranded and double-stranded DNA regions by electron microscopy. First, isolated PALB2 particles were observed as ring or open-ring like structures (Fig. 1E, insert bottom right). Second, PALB2 was found to be completely concentrated at the ends of the tailed DNA, while the remaining DNA was protein-free. These results demonstrate that PALB2 associates preferentially with single-stranded DNA portion of the substrate over dsDNA.
Figure 1. Purified PALB2 binds DNA.
(a) SDS-PAGE of purified PALB2 protein. Left: lane 1, Precision plus molecular weight markers (BioRad); lane 2, purified PALB2 (500 ng). (b) PALB2 binds D-loops preferentially. Competitions electrophoretic mobility shift assays were performed with PALB2 (0.5–20 nM) and D-loop, dsDNA (ds), and ssDNA (ss) (lanes 2–7) or Holliday junctions (HJ), splayed arms (SA), dsDNA and ssDNA substrates (lanes 9–14) on 8% acrylamide gel. Lanes 1 and 8, controls without protein. (c) Quantification of electrophoretic mobility shift assays. (d) PALB2-ssDNA protein complexes are supershifted with a PALB2 antibody (lane 3) or a control IgG (lane 4) and are analyzed on 0.8% agarose gel. Lane 1 and 5, controls with DNA alone or incubated with the PALB2 antibody respectively. (e) Electron microscopy of complexes made by PALB2 (40 nM) on a linear DNA molecule (~3900 bp) containing 3’single-strand tails at each end (~200 nt). Insert : closeup view of PALB2 in the absence of DNA. 86% of the DNA molecules observed had both ends bound while 14% had one end bound.
PALB2 enhances homologous recombination and interacts with RAD51
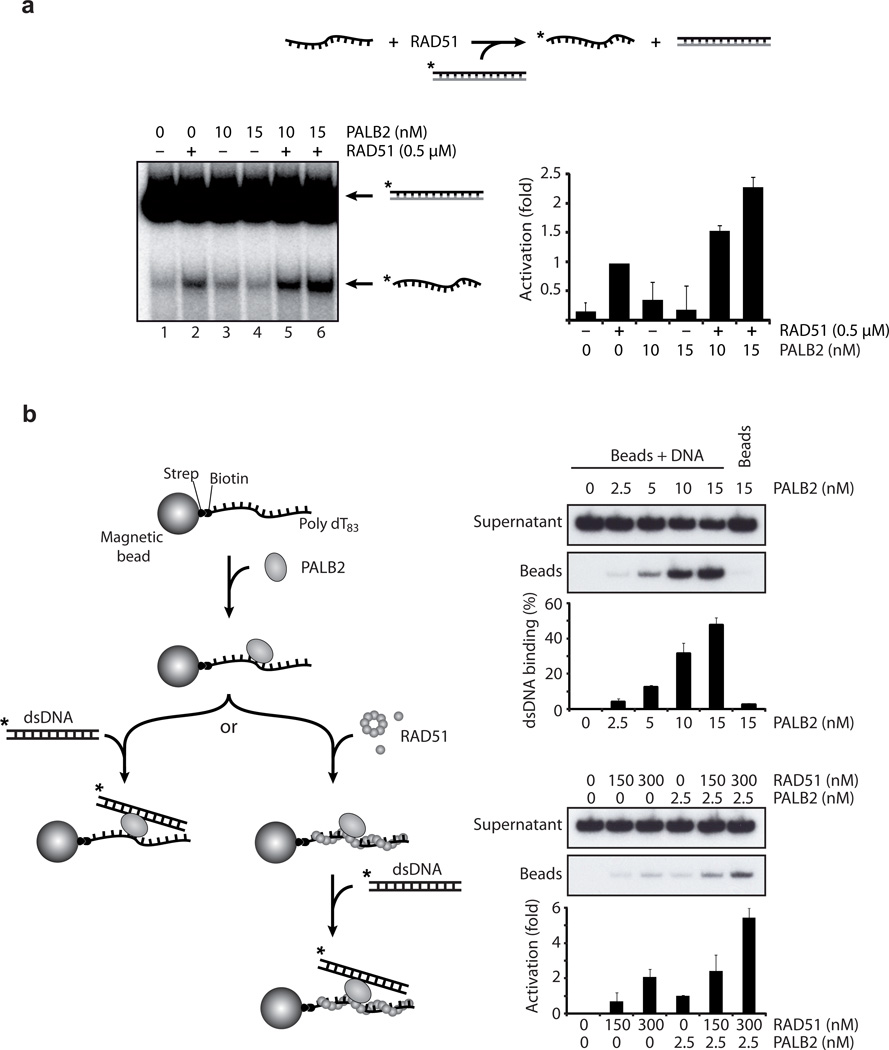
The specific binding of PALB2 towards D-loop structures led us to propose that PALB2 is involved in the invasion step of HR. Therefore we tested whether PALB2 could possess biochemical activities related to HR. First, we tested whether PALB2 could enhance strand exchange in vitro by using a linear ssDNA oligonucleotide and a homologous linear duplex DNA 23 (Fig. 2A). Indeed an addition of PALB2 to RAD51, at a saturating concentration of RAD51, led to further enhancement of strand exchange (compare lines 5 and 6 with 2) as measured by the release of ssDNA (lane 2). In contrast, PALB2 alone did not promote strand exchange efficiently (lanes 3–4). Second, PALB2 also acted on another critical step of HR by promoting the conjoining of DNA molecules through the capture of double-stranded DNA by PALB2–ssDNA complexes (Fig. 2B). Moreover, PALB2-dependent DNA capture was increased synergistically with RAD51.
Figure 2. PALB2 stimulates RAD51 strand exchange and DNA capture.
(a) PALB2 stimulates RAD51-mediated strand exchange. RAD51 and /or PALB2 were mixed with 1.5 µM 63-mer ssDNA. DNA strand exchange was initiated by addition of homologous 3 µM of 63-bp dsDNA in which the strand that would be displaced during DNA strand exchange was 32P-labeled. (b) Left: scheme of the second capture strategy; right: second capture assays with PALB2 and /or RAD51. Indicated concentration of PALB2 or/and RAD51 were incubated with beads containing 1 µM of 5’-biotinylated ssDNA poly dT. Then 300 nM of non homologous dsDNA was added with the DNA-protein complex. Beads and supernatant were separated, washed and treated with SDS to elute the bound proteins and analyzed by polyacrylamide gel.
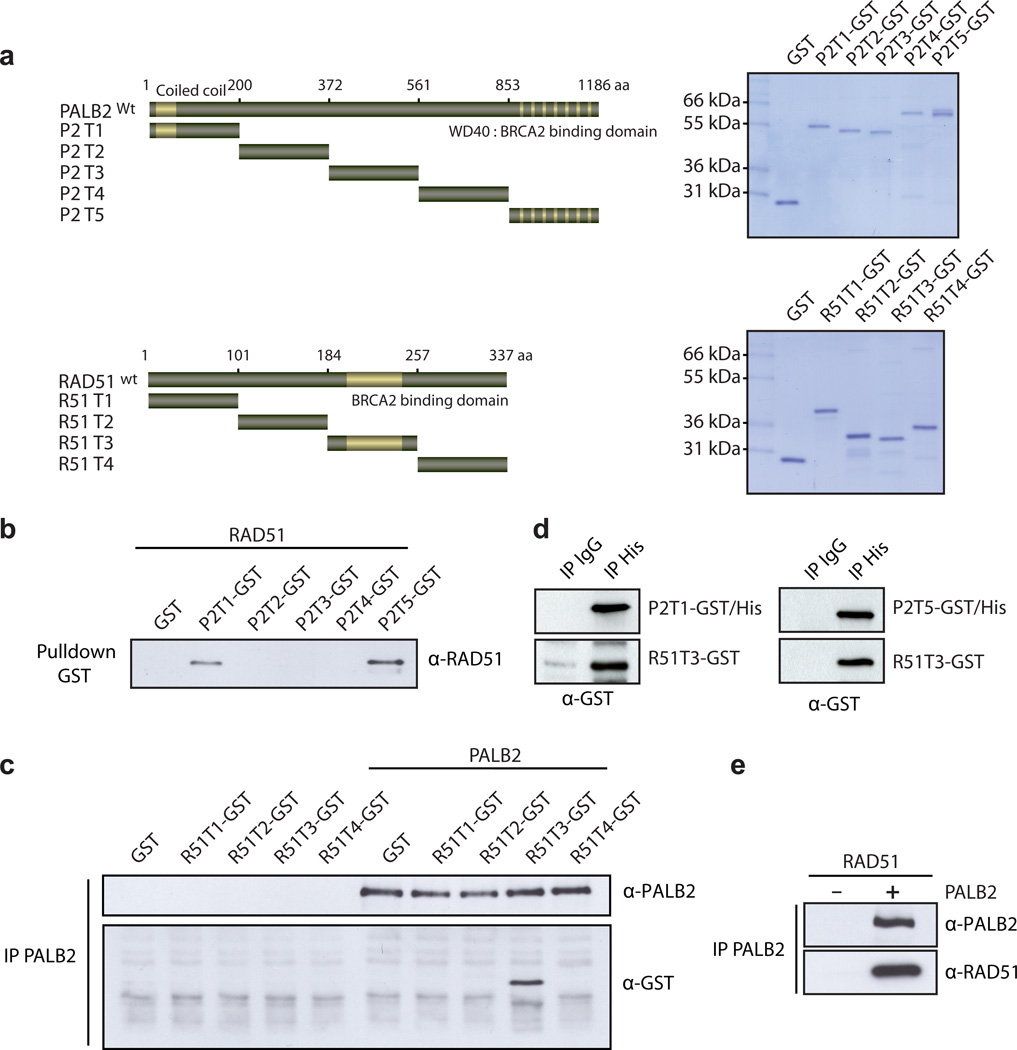
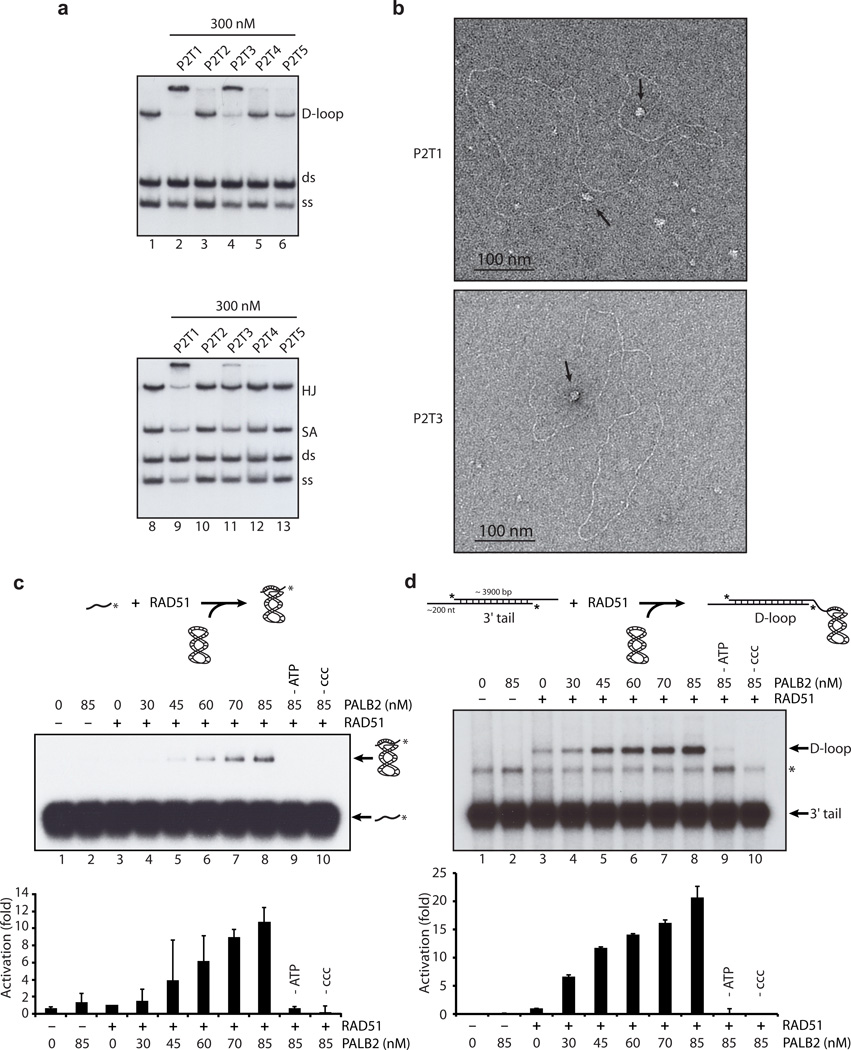
Since PALB2 exhibited the above activities in the absence of BRCA2, we tested whether PALB2 enhances HR by interacting with RAD51 directly. A series of non-overlapping glutathione S-transferase (GST) fusion proteins (designated P2T1 to P2T5, for PALB2 truncations 1-5, and R51T1 to R51T4, for RAD51 truncations 1-4, Fig. 3A) were used to define the regions of PALB2 that interact with RAD51 and vice-versa. GST-pull down assays revealed that RAD51 interacted with the N-terminus (1-200) of PALB2 and also its C-terminus (amino acids 853-1186) (Fig. 3B). Using immunoprecipitation (IP) analyses, the opposite experiment revealed that purified PALB2 bound RAD51 at amino acids 184-257 (Fig. 3C). Collectively, these experiments demonstrated that amino acids 184-257 of RAD51 interact with PALB2 at its C- and N-terminus (Fig. 3D). In addition, co-IP of purified RAD51 and PALB2 proteins was observed (Fig. 3E). Having established the domains responsible for the interaction, we asked which region(s) of PALB2 could bind DNA. The N-terminal coiled-coil domain of PALB2 (1-200) and a region encompassing amino acids 372-561 bound D-loop very efficiently (Fig. 4A-B, Supplementary Fig. 3). The N-terminal part of PALB2 contains a coiled-coil domain while the function of amino acids 372-561 is unknown. We noticed that unlike the full-length protein or P2T1, P2T3 did not bind Holliday Junctions, suggesting that P2T1 has more affinity for these structures. Interestingly, electron microscopy visualization revealed that P2T1 and P2T3 truncations bound DNA suggesting that PALB2 has at least 2 DNA binding domains (Fig. 4B).
Figure 3. PALB2 interacts directly with RAD51.
(a) Right: Scheme of the PALB2 deletion variants fused to GST and the RAD51 GST fusions. Left: and SDS-PAGE of the corresponding purified proteins (b) GST alone or GST-tagged PALB2 fragments purified from bacteria were incubated with RAD51 and glutathione sepharose beads were used to capture protein complexes. The beads were washed and treated with SDS to elute the bound proteins, and revealed by western blotting against RAD51. (c) GST alone or GST-tagged RAD51 fragments were incubated with/without PALB2 followed by immunoprecipitation analysis using PALB2 antibodies and revealed by western blotting against PALB2 and GST. (d) Co-immunoprecipitations of purified PALB2 truncations (P2T1 or P2T5) and RAD51 truncation (R51T3) were performed with anti-IgG or anti-Histidine tag antibodies followed by western blotting using an anti-GST antibody. (e) Co-immunoprecipitation of purified PALB2 and RAD51. Immunoprecipitations were conducted with a polyclonal antibody against PALB2 and blotted against PALB2 and RAD51 as indicated.
Figure 4. PALB2 binds DNA by two distinct domains and stimulates RAD51-mediated D-loop formation.
(a) Competitions electrophoretic mobility shift assays were performed with GST-cleaved PALB2 truncations (300 nM) and D-loop, dsDNA (ds), and ssDNA (ss) (lanes 2–6) or Holliday junctions (HJ), splayed arms (SA), dsDNA and ssDNA substrates (lanes 9–13) on 8% acrylamide gel. (b) Electron microscopy of complexes made by PALB2 truncation P2T1 and P2T3 (40 nM) on linear DNA molecule containing 3’single-strand tails of ~200 nucleotides at each end. (c,d) D-loop reactions mediated by RAD51 (300 nM (c) or 400 nM (d)) or PALB2 alone (lanes 2–3), or combinations of RAD51 and PALB2 (lanes 4 to 8) with (a) 1 µM of 100-mer single-stand DNA oligonucleotide. Bottom: quantification of the results. (d) D-loop reactions with 1 µM of linear DNA molecule containing 3’-single-strand tails of ~200 nucleotides at each end. Bottom: quantification of the results. The band indicated with an asterisk corresponds to annealed tailed molecules.
A key step in homologous recombination is the formation of a D-loop structure, characterized by the invasion of an ssDNA substrate into a homologous duplex DNA. We tested whether PALB2 enhances RAD51-mediated strand invasion using a ssDNA oligonucleotide. In this experiment, RAD51 was used at saturating concentration (300 nM) and practically no D-loop products were observed (Fig. 4C, lane 3). While PALB2 could not promote strand invasion by itself (lane 2), adding increasing amounts of PALB2 to the RAD51 reaction led to a 10-fold increase in D-loop product formation (lane 8). The stimulation of RAD51 activity by PALB2 was dependent on the presence of ATP and supercoiled DNA (lane 9 and 10, respectively). To further demonstrate that PALB2 stimulates strand invasion by RAD51 we used a D-loop assay that uses substrates more relevant to those found in eukaryotes in vivo 24. In this assay, linear duplex substrate (4.3 kb) with an exposed 3'-tail (approximately 200 nucleotides) are created by treatment of linear duplex DNA with exonuclease T7. The protein of interest are incubated with this substrate, followed by the addition of homologous supercoiled DNA. The use of such a large substrate (4.3 kb) completely eliminates possible artifacts caused by oligonucleotides annealling to transitory single strand regions in the supercoiled DNA during strand invasion assays. Similar to reactions with oligonucleotides, PALB2 stimulated RAD51 D-loop formation (~ 20-fold activation compared to RAD51 alone) (Fig. 4D, compare lane 3 and 8). PALB2 also displayed annealing activity in the absence of supercoiled DNA (Fig. 4D, asterisk). These results show that PALB2 can strongly enhance RAD51 function in D-loop assays.
PALB2 and piBRCA2 stimulate D-loop formation
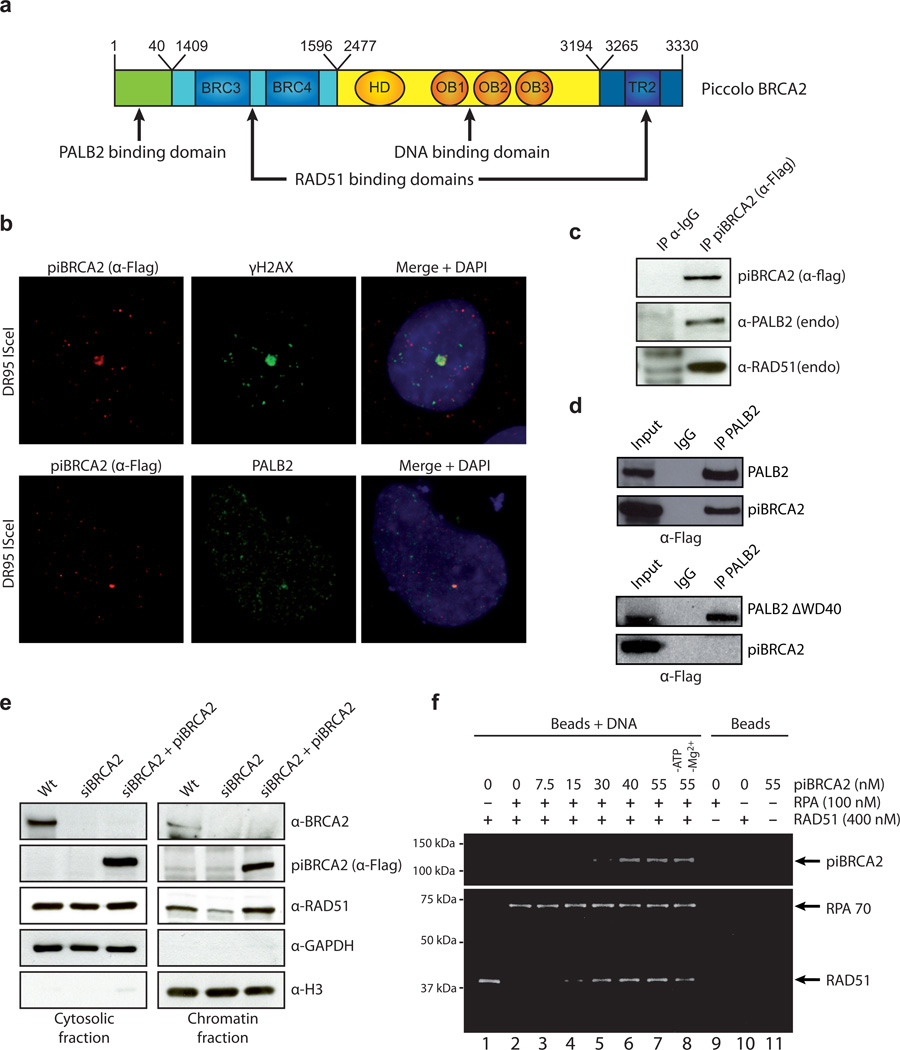
As BRCA2 is a close binding partner of PALB2 14, we therefore investigated if BRCA2 could work synergistically with PALB2. Since purification of BRCA2 poses significant technical challenges due to its large size (3418 amino acids), we purified a BRCA2 chimera (termed piccolo, italian for small) containing the PALB2 binding domain, BRC3 and BRC4 repeats, the three OB-folds, and the TR2 RAD51-binding region (Fig. 5A). Several approaches were used to test if piBRCA2 is functional in vivo and in vitro. First, we investigated whether piBRCA2 localized to a unique DNA double-strand break in vivo (Fig. 5B). Following I-SceI cleavage and DSB formation in DR95 cells 25, piBRCA2 co-localized with ©–H2AX, a DSB marker 26–28. Moreover, piBRCA2 co-localized with endogenous PALB2 following DSB cleavage by I-SceI. Notably, cells which were not transformed with I-SceI did not show co-localization of piBRCA2 with γ-H2AX or PALB2 at the unique DSB (data not shown). Second, piccolo BRCA2 interacted with endogenous RAD51 and PALB2 (Fig. 5C). The deletion of PALB2 WD40, which is important for BRCA2 binding to PALB2 14, inactivated the interaction between piBRCA2 and PALB2 (Fig. 5D). Third, consistent with BRCA2 controlling the nuclear localization of RAD51 5, expression of piBRCA2 in BRCA2 knockdown cells restored the nuclear accumulation of RAD51 as judged with cell fractionation assays (Fig. 5E), and RAD51 foci formation (Supplementary Fig. 4). The RAD51 foci appeared smaller and more diffuse than in normal cells, suggesting that additional portions of BRCA2 might help to sustain RAD51 foci formation. In contrast, piBRCA2 could not restore nuclear abundance of RAD51 in siPALB2 cells (Supplementary Fig. 4C). Fourth, similar to the Brh2 protein of Ustilago maydis 13, piBRCA2 could overcome the inhibitory effect of RPA and act as a recombination mediator (Fig. 5F). Fifth, piBRCA2 promoted dsDNA capture (Supplementary Fig. 5A). Collectively, these results show that piBRCA2 displays key functions of full length BRCA2.
Figure 5. A BRCA2 chimera protein (piccolo BRCA2) recapitulates human BRCA2 properties.
(a) The BRCA2 chimera contains the PALB2 binding domain (amino acids 1-40), BRC3 and BRC4 repeats (amino acids 1409-1596), the three OB-folds (amino acids 2477 to 3194), and TR2 repeat (amino acids 3265 to 3330). (b) piBRCA2 colocalizes with γ-H2AX and PALB2 to a unique I-SceI DNA double-strand break. DR95 cells containing DR-GFP were transfected with pCBASce (encoding I-SceI) and immunofluorescence was conducted with the indicated antibodies. All piBRCA2 transfected cells showed co-localization with γ-H2AX. (c-d) piBRCA2 interacts with PALB2 and RAD51. (c) HEK293T cells were transfected with a plasmid expressing piBRCA2-Flag or (d) plasmids expressing piBRCA2-Flag and PALB2-Flag or PALB2 lacking the interaction domain with BRCA2 (WD40 repeat). Whole cell extracts were prepared and immunoprecipitations were conducted using IgG alone, anti-flag or anti-PALB2 antibody, followed by western blotting with the indicated antibodies. (e) The nuclear localization of RAD51 is restored in presence of piBRCA2. Hela cells were transfected with a siRNA againt BRCA2 and with or without a plasmid expressing piBRCA2-Flag. Cells were fractionated into cytosolic and chromatin fractions and analyzed by Western blotting for the presence of piBRCA2 and endogenous BRCA2 and RAD51. Immunoblots against GAPDH and Histone H3 were positive controls for cytosolic and chromatin fractions respectively. (f) piBRCA2 overcomes RPA inhibition to promote RAD51 assembly. RPA bound to a ssDNA oligonucleotide prevents RAD51 assembly (lane 2) whereas addition of piBRCA2 stimulates RAD51 filament formation in presence of RPA (lanes 3 to 7). The stimulation was weaker in absence of ATP and Mg2+ (lane 8). Lanes 9–11, interaction of RPA, RAD51 and PALB2 on beads alone.
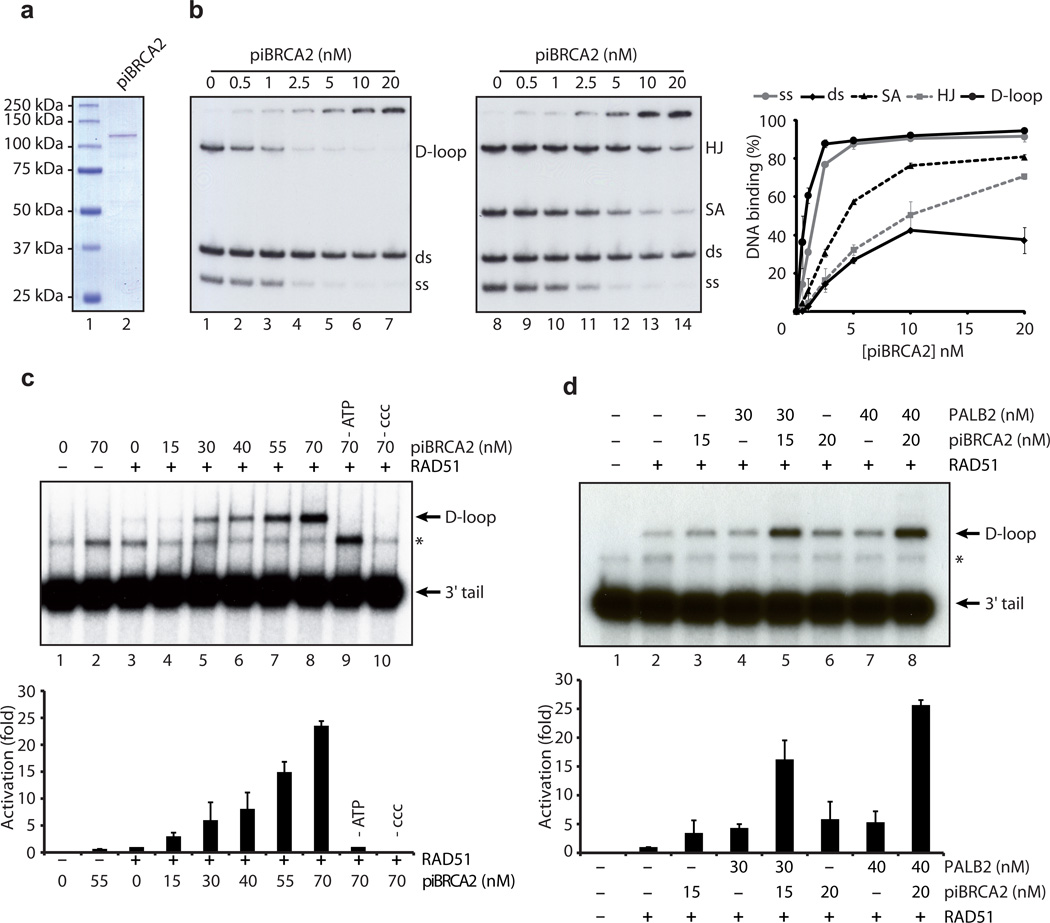
Having established that piBRCA2 is functional in vitro and in vivo, we tested whether piBRCA2 could bind DNA and promote D-loop formation. purified piBRCA2 bound D-loops and ssDNA efficiently (Fig. 6A-B). piBRCA2 failed to promote D-loop formation on its own (Fig. 6C, lane 2, 70 nM). Rather, piBRCA2 promoted the annealing of 3’-tailed DNA molecules (lane 2, asterisk). However, it stimulated RAD51-mediated homologous pairing by more than 20-fold (lane 8) in an ATP-dependent manner (lane 9). Similar results were obtained with ssDNA oligonucleotides (Supplementary Fig. 5B). Next, we investigated whether PALB2 and piBRCA2 could function together in D-loop assays. Adding piBRCA2 (15–20 nM) and PALB2 (30–40 nM) to a RAD51 reaction led to a highly synergistic stimulation of RAD51 (25-fold activation, Fig. 6D).
Figure 6. A BRCA2 chimera stimulates RAD51-mediated D-loop formation and function with PALB2 in a synergistic manner to promote D-loop formation.
(a) SDS-PAGE of the purified piBRCA2 protein (300 ng). Left: lane 1, Precision plus molecular weight markers (BioRad); lane 2, purified piBRCA2. (b) Electrophoretic mobility shift assays were performed with piBRCA2 (0.5–20 nM) and D-loop, dsDNA (ds), and ssDNA (ss) (lanes 2–7) or Holliday junctions (HJ), splayed arms (SA), dsDNA (ds) and ssDNA (ss) substrates (lanes 9–14) and migrated on a 8% acrylamide gel. Lanes 1 and 8, controls without protein. Far right : quantification of percentage of DNA binding. (c) Effect of piBRCA2 in RAD51-catalyzed D-loop formation. Reactions contained DNA alone (lane 1); piBRCA2 (70 nM, lane 2); RAD51 (400 nM, lane 3); or a combination of both proteins (lanes 4–10) with 1 µM on linear DNA molecule containing 3’single-strand tails. Right: Quantification of the results (d) PALB2 (30–40 nM) works in concert with piBRCA2 (15–20 nM) to stimulate RAD51 (400 nM) in D-loop reactions. Right: Quantification of the results. The band indicated with an asterisk in panels c and d corresponds to annealed tailed molecules.
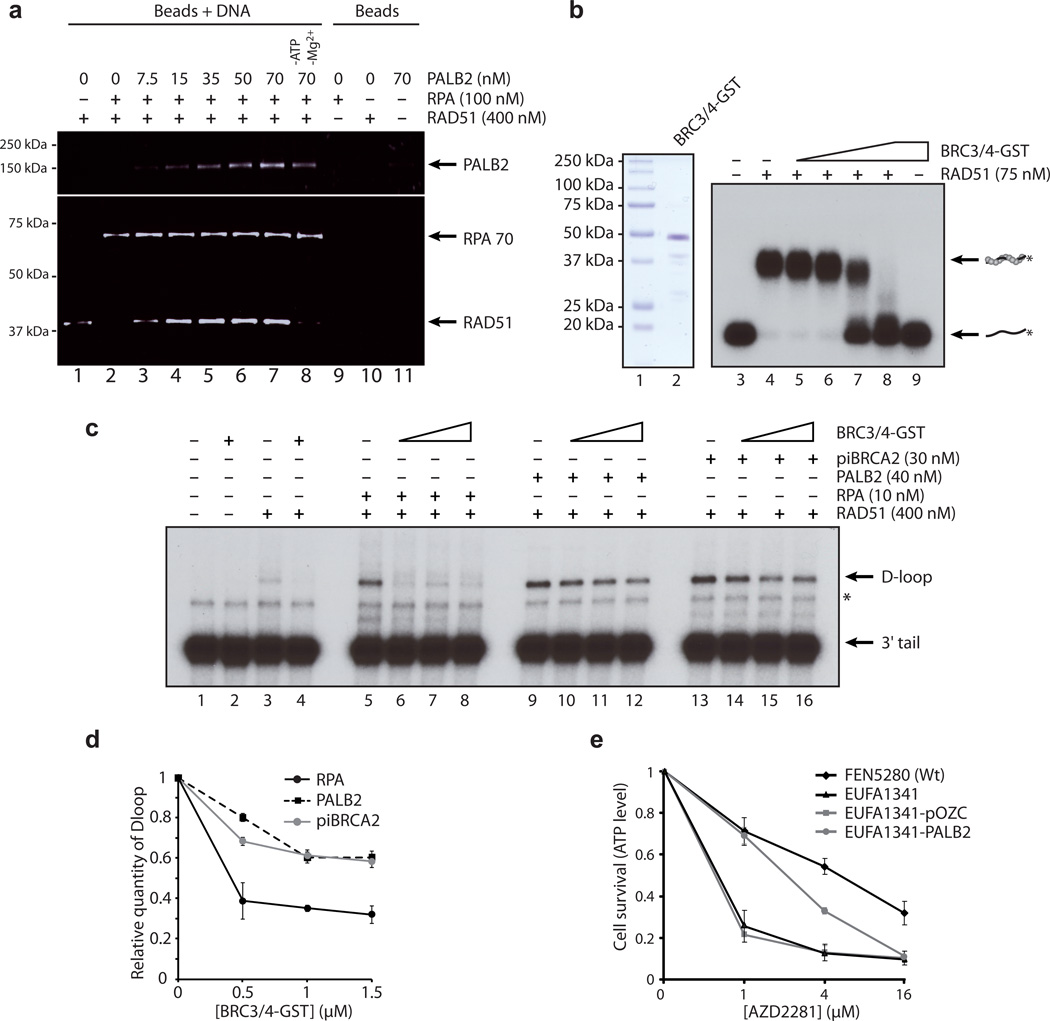
Next, we investigated the biochemical mechanisms by which PALB2 stimulates homologous recombination. Although RPA is essential for recombination, it also has an inhibitory effect on RAD51 filament formation. Therefore, HR requires accessory factors, termed mediators, to overcome the inhibitory effect of RPA to allow RAD51 binding on ssDNA 29. Thus, we verified whether PALB2 could perform such a function. The addition of 100 nM RPA completely prevented the association of RAD51 to ssDNA-beads (Fig. 7A, lane 2). When PALB2 was added in increasing concentration after RAD51 and RPA, RAD51 gained access to the ssDNA as judged by the increasing amount of RAD51 bound to ssDNA beads (lanes 3–7). Under these conditions, RPA70 was still bound to DNA, suggestive of a ternary complex containing RAD51, PALB2 and RPA on ssDNAs. These results show that PALB2 helps RAD51 to overcome the suppressive effect of RPA, and functions as a recombination mediator.
Figure 7. PALB2 is a HR mediator protecting the RAD51 filament from disassembly.
(a) RPA bound to a ssDNA oligonucleotide prevents RAD51 assembly (lane 2) whereas addition of PALB2 stimulates RAD51 filament formation in presence of RPA (lanes 3 to 7) but not in absence of ATP and Mg2+ (lane 8). Lanes 9–11, interaction of RPA, RAD51 and PALB2 on beads alone (b) Left, SDS-PAGE of purified BRC3/4-GST polypeptide (1 µg) corresponding to the BRC3 and BRC4 region of BRCA2 (amino acids 1409-1596). Lane 1, Precision plus molecular weight markers (BioRad); lane 2, purified BRC3/4-GST (1 µg). Right, inhibition of RAD51–DNA complex formation by BRC3/4-GST polypeptide. Purified RAD51 protein was incubated with a 100-mer single-strand DNA oligonucleotide (lane 4) prior to the addition of BRC3/4-GST (lanes 5–8). Lane 3, DNA alone; lane 9, BRC3/4-GST incubated with DNA. (c) PALB2 or piBRCA2 protect D-loop formation from a BRC3/4-GST polypeptide. RPA (lanes 5 to 8), PALB2 (lanes 9 to 12) and piBRCA2 (lanes 13 to 16) were incubated with RAD51-3’tail DNA complex prior addition of BRC3/4-GST polypeptide. The band indicated with an asterisk corresponds to annealed tailed molecules. (d) Relative D-loop formation in presence of RPA, PALB2 or piBRCA2 with BRC3/4-GST polypeptide. (e) Survival curves of wild-type lymphoblasts (FEN5280), EUFA1341 (expressing PALB2 Y551X), and EUFA1341 complemented with vector alone (pOZC) or PALB2 to PARP inhibitor (AZD2281). Cell survival was analyzed by quantification of ATP levels.
It was previously shown that BRC3 and BRC4 at high concentration can disrupt RAD51 filament formation by blocking nucleation on DNA 5. In vivo, the expression of BRC3 reduces HR and RAD51 foci formation consistent with an inhibitory role 30. Hence, we purified a fusion protein bearing the BRCA2 BRC3 and BRC4 repeats (Fig. 7B left panel). In the absence of the BRC3/4-GST protein, RAD51 bound to the ssDNA, forming distinct protein–ssDNA complexes (Fig. 7B, right panel, lane 2). However, the addition of BRC3/4-GST led to a reduction of DNA binding (lanes 3–6). Next, we performed D-loop reactions in presence of the disruptive BRC3/4-GST to monitor whether the addition of PALB2 would overcome this effect. Control reactions showed that RAD51 mediated D-loop formation in the presence of RPA (Fig. 7C, lane 5) was inhibited when BRC3/4-GST was added to the reaction. In contrast, addition of purified PALB2, before the addition of BRC3/4-GST, elicited a protective effect (lanes 9–12). This result demonstrates that PALB2, can stabilize the RAD51 filament in the presence of disrupting BRC3/4-GST (Fig. 7C and D).
BRCA2-deficient cells have been found to be exquisitely sensitive to potent poly (ADP-ribose) polymerase (PARP) inhibitors 31,32. The inhibition of PARP leads to the persistence of ssDNA lesions which collapse DNA replication forks yielding DSBs that are normally repaired by HR. In the absence of BRCA1/2, a severe HR deficit would profoundly sensitize cells deficient for BRCA2 and BRCA1 to such agents. Given the common functions between PALB2 and BRCA2 as mediators of homologous recombination, we tested whether PARP inhibitors could efficiently kill PALB2-deficient cells. Indeed, PARP inhibitor (AZD2281) was cytotoxic for PALB2-deficient cells (EUFA1341) as compared with the control cell line FEN5280 (Fig. 7E). This result indicates that tumors harboring PALB2 mutations are likely to be sensitive to PARP-inhibition.
Discussion
Cellular DNA is constantly exposed to endogenous and exogenous DNA damage, which, if unrepaired or misrepaired, will compromise genome integrity and promote tumorigenesis. These deleterious effects of DNA damage are limited by the action of certain tumor suppressor proteins, such as BRCA1, BRCA2 and PALB2. It is generally believed that these proteins exert their tumor suppression function, at least in part, by promoting HR-based, faithful DSBR. In this study, we have investigated the biochemical functions of the PALB2 protein in HR. During this process, the RAD51 recombinase polymerizes as a right-handed helical filament on ssDNA and forms the presynaptic filament. This filament binds a recipient duplex DNA and searches for homology leading to the formation of a joint molecule between the invading ssDNA and the recombining DNA, termed the D-loop structure. Using electrophoretic mobility shift assays, we have established that PALB2 is a DNA binding protein which has very high affinity for D-loop, which comprises single-strand, double-strand and branched DNA structures. The affinity for D-loop structures was very strong compared to ssDNA and dsDNA. The higher affinity for ssDNA compared to dsDNA was confirmed by electron microscopy visualization of PALB2 complexes on 3’-resected DNA ends. However, the higher affinity for ssDNA compared to dsDNA might result from DNA secondary structures or the sequences of the substrates used. The results reported in the companion manuscript by Dray et al using other experimental conditions 33, suggest that PALB2 has high affinity for dsDNA.
The DNA binding properties of PALB2 are very complex. Consistent with the identification of at least two DNA binding sites (P2T1 and P2T3), the ratios of protein/oligonucleotide used in binding assays, and PALB2 homo-oligomerizes 34, PALB2 appears able to bind multiple DNA molecules simultaneously. DNA capture assays support this claim as single and double-strand DNA were both efficiently captured from ssDNA-PALB2 beads (Supplementary Fig. 6). Both P2T1 and P2T3 domains seem to be important for ssDNA binding as fragments P2T1 and P2T3 exhibit relatively strong D-loop substrate binding, but, as on contrary to the full-length protein, they showed reduced ssDNA binding. P2T3 did not bind Holliday Junctions, suggesting that P2T1 has the ability to bind these structures. The multiple DNA binding sites of PALB2 might allow some flexibility to discriminate damaged DNA in the context of collapsed replication forks, which might produce unusual DNA structures needed to be recognized for DNA repair and genome stability.
PALB2 stimulates RAD51 for strand invasion and homologous recombination, similar to studies reported by Dray and colleagues in the accompanying paper 33. The stimulatory effect was dependent on the presence of ATP and supercoiled DNA isolated in native conditions. It was surprising to find that PALB2 could mediate this effect in the absence of BRCA2. This result prompted us to ask if PALB2 can directly bind RAD51. Indeed, pull-down experiments were used to show that PALB2 interacts directly with RAD51 via two distinct segments present in its N- (amino acids 1-200) and C- (amino acids 853-1186) termini. Conversely, RAD51 binds to PALB2 through a region encompassing amino acids 184-257. Therefore, the enhancement of RAD51-mediated strand invasion by PALB2 might occur through interactions and stabilization of the RAD51 filament. A role for PALB2 in the regulation of RAD51 presynaptic filament through protein-protein interactions is consistent with the observation that PALB2 is required for RAD51 foci formation 21. Although in vitro PALB2 can interact with RAD51 directly and stimulate RAD51-mediated strand invasion without BRCA2, in cells this stimulation likely occur on DNA, after BRCA2 has enabled RAD51 loading on resected DNA.
The fact that PALB2 stabilized RAD51 nucleoprotein filament in the presence of BRC3/4 (Fig. 7C) is also notable. Given that PALB2 interacts directly with RAD51, this effect may be brought about by a direct protein-protein interaction, as PALB2 antibodies pulled-down PALB2, BRC3/4 and RAD51 (Supplementary Fig 7A). Hence, the interactions between PALB2, BRC3/4 and RAD51 are not mutually exclusive. Although there is no isolated BRC3/4 in cells to antagonize RAD51 filament formation in the cell, other proteins such as Srs2 35, BLM 36, or RecQL5 37 have been suggested to function as anti-recombination factors by disrupting the RAD51 filament. Therefore, this PALB2 activity may be important for HR in vivo. Further enhancement of HR may also be provided by the high affinity of PALB2 for D-loop structures.
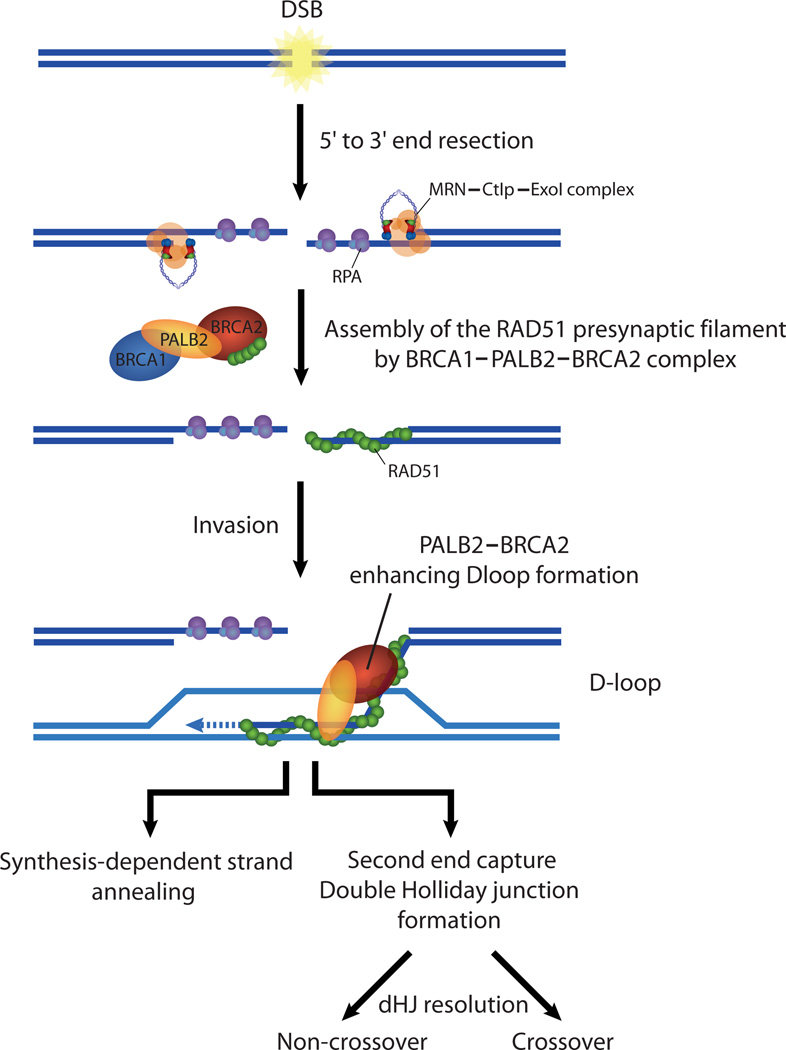
Previous studies have identified PALB2 as a BRCA2 interacting factor that binds to the extreme N-terminus of BRCA2 14,38. The PALB2-BRCA2 interaction is disrupted in breast and ovarian cancers by mutations in BRCA2 14,38. Given the close association of PALB2 and BRCA2 in the cell, we sought to test if they could function together in strand invasion. Purification of BRCA2 is technically difficult, so we purified a BRCA2 chimera containing the PALB2 binding domain, RAD51 binding sites in BRCA2, three OB-folds for ssDNA binding, and the TR2-RAD51 binding region. Importantly, piccolo BRCA2 displayed known properties of BRCA2, as it localized to DNA damage sites and enhanced the accumulation of RAD51 in the nucleus and interacted with RAD51 and PALB2 in co-immunoprecipitation assays. Moreover, piBRCA2 also stimulated the RAD51-mediated D-loop formation similar to the BRCA2 homolog Brh2 from Ustilago maydis 39 or the C. elegans protein CeBRC-2 40. These results suggest that piBRCA2 represent a functional alternative to full length BRCA2. When added together in the same reaction, piBRCA2 acted in concert with PALB2 to promote strand invasion, suggesting that piBRCA2 and PALB2 do not impede each other but rather can work synergistically in stimulating RAD51 during the repair of DNA double-strand breaks (Fig. 8).
Figure 8.
Model. PALB2, and BRCA2 work synergistically to stimulate homologous recombination. Following DNA damage and resection by MRN/CtIP, PALB2-BRCA2 activates RAD51 to promote the invasion of an undamaged template, leading to synthesis-dependant strand annealing or second end capture and double Holliday junction formation to allow DSB repair.
Striking parallels can be drawn between BRCA2 and PALB2. Two regions of BRCA2, the BRC repeats and the TR2 6, bind RAD51 and regulates RAD51-mediated HR 5. Similarly, two regions of PALB2 bind RAD51 and PALB2 stimulates RAD51 for HR. Interestingly, both proteins bind RAD51 at the same region 41 suggestive of a complex regulation of HR. Both proteins can stimulate RAD51-mediated strand invasion. As such, our data indicate that PALB2, at the functional level, might be more closely related to BRCA2 than previously anticipated. Hence, PALB2 function is not only important to target BRCA2 to chromatin but also is directly involved in the regulation of homologous recombination. However, since BRCA2-mutant cells (containing wild-type PALB2) are severely impaired in HR, it is unlikely that PALB2 can function adequately by itself in vivo as a proportion of the RAD51 pool will not be nuclear. Therefore, in BRCA2-proficient cells, PALB2 likely acts together with BRCA2 to first overcome the inhibitory effect of RPA and recruit RAD51 to resected DSB ends and then stimulate strand invasion into the undamaged sister chromatid. In BRCA2-deficient cells, PALB2 by itself may help maintain a basal, albeit limited, HR function through its direct RAD51 binding and stimulating activity. This is consistent with the fact that PALB2 A1025R, which cannot bind BRCA2, interacted with RAD51 at lower levels than wild-type PALB2 (Supplementary Fig. 7B). Such a basal level of HR, although not sufficient to suppress tumorigenesis, may be critical to sustain cell viability.
One of the most successful outcome of DNA repair studies at the clinical level is the use of PARP inhibitors on BRCA1 and BRCA2-deficient cells 42–44. PALB2-deficient cells, as a result of their deficiency in HR, are sensitive to PARP inhibitors most likely because resultant collapsed replication forks are not efficiently repaired. Taken together, our findings establish PALB2 as a critical HR mediator in human cells, thus providing a plausible mechanism for the development of cancer. Moreover, the fundamental understanding of the function of PALB2 presented here may be beneficial for the design of potential synthetic lethal therapeutic strategy for cancer treatment using PARP inhibitors.
Methods
Protein purification
Human RAD51 45 and human RPA were purified as described 46. GST-PALB2-His was purified from baculovirus-infected Sf9 cells. Sf9 insect cells (1 L) were infected with the PALB2 baculovirus for 2 days at 27°C. The cell pellet was resuspended in 40 mL of GST buffer (10 mM Na2HPO4, 2 mM KH2PO4, 300 mM NaCl, 2,7 mM KCl, 1mM EDTA and 1 mM DTT) containing protease inhibitors (Roche). The suspension was lysed using a Dounce homogenizer (10 strokes), sonicated three times for 20 seconds, and then homogenized a second time. Insoluble material was removed by centrifugation (47,893 g for 20 minutes in a Sorvall SS34). 2 mL of glutathione sepharose (GE Healthcare) was added to the supernatant and incubated during 2 hours at 4°C. The beads were washed three times with GST washing buffer (10 mM Na2HPO4, 2 mM KH2PO4, 500 mM NaCl, 2,7 mM KCl, 1 mM EDTA and 1 mM DTT) and three times with PreScission washing buffer (50 mM Tris HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, 0.04 % Tween 20). The PALB2 protein was eluted by cleavage with PreScission protease (60 U mL−1, GE Healthcare) overnight at 4°C. The supernatant was dialyzed against Talon buffer (20 mM NaHPO4 pH 7.4, 500 mM NaCl, 10 % glycerol, 0,02 % Triton, 5 mM Imidazole). Then, 300 µL of Talon resin (Clontech) was added and incubated for 2 hours at 4°C. The resin was washed three times with 10 mL of Talon washing buffer (20 mM NaHPO4 pH 7.4, 1 M NaCl, 10 % glycerol, 0,02 % Triton, 30 mM imidazole). PALB2 protein eluted with in buffer containing 500 mM imidazole and dialysed in storage buffer (20 mM Tris-Acetate, pH 8.0, 200 mM KOAc, 10% glycerol, 1 mM EDTA, 0.5 mM DTT).
GST-BRC3/4-His was purified from 2 L of BL21(DE3) RP (Stratagene), grown at 37°C in Luria broth medium supplemented with 100 µg mL−1 ampicillin, and 25 µg mL−1 chloramphenicol. At OD600 = 0.4, 0.05 mM IPTG was added to the culture and incubated at 15°C overnight (16 hours). The cell pellet was resuspended in 40 mL of GST buffer (10 mM Na2HPO4, 2 mM KH2PO4, 297 mM NaCl, 2.7 mM KCl, 1 mM EDTA and 1 mM DTT) containing protease inhibitors (Roche). The protein was then purified by affinity on glutathione sepharose and Talon beads as described for the PALB2 purification scheme.
Piccollo BRCA2 was purified from 2.4 L of Sf9 cells infected with the piBRCA2 baculovirus. Briefly, cells were processed as above using 120 mL of P buffer (50 mM NaHPO4 pH 7.0, 500 mM NaCl, 10% glycerol, 0.05% Triton X-100) containing 5 mM imidazole and EDTA-free protease inhibitors (Roche). The cell extract was loaded on a 5 mL Talon column (BD Biosciences). The column was washed successively with 100 mL, 30 mL and 15 mL of P buffer containing 30 mM, 40 mM, and 50 mM imidazole, respectively, before piBRCA2 was eluted a step elution with P buffer containing 500 mM imidazole. Fractions containing piBRCA2, were identified by SDS-PAGE, pooled and dialyzed against K buffer (20 mM KH2PO4, pH 7.4, 0.5 mM EDTA, 1 mM DTT, and 10% glycerol) and loaded on a 1 mL Hydroxylapatite column (Biorad). piBRCA2 was found in the flow-through and concentrated with a centricon (Millipore), dialysed in storage buffer and stored at −80°C.
GST truncation purifications
Recombinant PALB2 GST fusions (T1-T5) or RAD51 GST fusions (T1-T4) were purified from BL21(DE3) RP (Stratagene), grown at 37°C in Luria broth medium supplemented with 100 µg mL−1 ampicillin, and 25 µg/mL chloramphenicol. At OD600 = 0.4, 80 µg/mL ampicillin was added to the media, and At OD600 = 0.8, 0.1 mM IPTG was added to the culture and incubated at 15°C overnight (16 hours). The cells were harvested by centrifugation, frozen on dry ice and stored at −80°C. GST fusions were purified according to the manufacturer’s protocol (GE Healthcare) and cleaved with PreScission protease when required (GE Healthcare).
DNA binding assays
Reactions (10 µL) contained 32P-labelled DNA oligonucleotides (25 nM in nucleotides of each substrate) and PALB2 or piBRCA2, at the indicated concentrations, in binding buffer (20 mM Triethanolamine-HCl pH 7.5, 40 mM KOAc, 0.5 mM Mg(OAc)2, 0.5 mM DTT, and 100 µg mL−1 BSA). In competition reactions, proteins were added to reactions containing all oligonucleotides. Reaction mixtures were incubated at 37°C for 10 minutes, followed by 15 minutes of fixation in 0.2% glutaraldehyde. Reactions were loaded on a 0.8% TBE1X agarose gel or 8% TBE1X/acrylamide gel.
D-loop assays
The D-loop assay with 3’tail DNA was conducted essentially as described 24. Linear duplex DNA was prepared by restriction digestion of pPB4.3 DNA (4.3 kb) with Sma I. Linear DNA with 3′-tails (approximately 200 nucleotides in length) were generated by treatment of linearised pPB4.3 DNA (100 µg) with T7 gene 6 exonuclease (830 units, one minute at 25°C) in an 800 µL reaction volume containing 20 mM Tris-Acetate pH 7.9, 50 mM KOAc pH 7.9, 10 mM MgOAc and 1 mM DTT. Exonuclease digestion was stopped by extraction with phenol/chloroform. Finally, the resected DNA was purified by gel electrophoresis through a 0.8 % (w/v) agarose gel and recovered by electroelution. 32P-labeled 3’tail DNA substrate (1 µM nucleotides) was incubated for 5 minutes with the indicated concentration of RAD51 and/or PALB2 and/or chiBRCA2 in 50 mM TEA pH 7.5, 60 mM KOAc, 500 µM CaCl2, 300 µM EDTA, 1 mM DTT, 2 mM ATP and 100 µg mL−1 BSA in 9 µL. CsCl-purified pPB4.3 replicative form I DNA (300 µM) was added and the reaction was incubated for 1 h 30 min followed by of one-fifth volume of stop buffer (10% SDS and 10 mg ml−1 proteinase K) followed by 30 min incubation at 37°C. Labeled DNA products were analyzed by electrophoresis through a 0.8 % TAE1X/agarose gel run at 65V, dried onto DE81 filter paper and visualized by autoradiography.
Supplementary Material
Acknowledgments
We are grateful to Eloise Dray, Patrick Sung, Stéphane Richard, Isabelle Brodeur, Eric Paquet, Marc Tishkowitz, and Julien Vignard for helpful discussions, and Gilles Hamel for technical help. We thank also Isabelle Brodeur, Julie Birraux and Amélie Rodrigue for critical reading of the manuscript. R.B. is supported by a FQRNT doctoral scholarship. This work was supported by funds from the Swiss National Science Foundation (grant 3100A0-103962 to A.S.), the Cancer Institute of New Jersey (B.X.) and the National Cancer Institute (B.X.), and The Cancer Research Society (J.-Y.M.).
Footnotes
Author contributions
R.B., A.-M.D.C, Y.C., H.L., H.C., and A.Z.S. conceived and carried out experiments. A.S. and B.X. contributed expertise. R.B. conceived the figures and J.-Y.M. provided guidance throughout and wrote the paper.
References
- 1.Jemal A, et al. Cancer Statistics, 2009. CA Cancer J Clin. 2009 [Google Scholar]
- 2.Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- 3.Venkitaraman AR. Linking the cellular functions of BRCA genes to cancer pathogenesis and treatment. Annu Rev Pathol. 2009;4:461–487. doi: 10.1146/annurev.pathol.3.121806.151422. [DOI] [PubMed] [Google Scholar]
- 4.West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4:435–445. doi: 10.1038/nrm1127. [DOI] [PubMed] [Google Scholar]
- 5.Davies AA, et al. Role of BRCA2 in control of the RAD51 recombination and DNA repair protein. Mol Cell. 2001;7:273–282. doi: 10.1016/s1097-2765(01)00175-7. [DOI] [PubMed] [Google Scholar]
- 6.Esashi F, et al. CDK-dependent phosphorylation of BRCA2 as a regulatory mechanism for recombinational repair. Nature. 2005;434:598–604. doi: 10.1038/nature03404. [DOI] [PubMed] [Google Scholar]
- 7.Esashi F, Galkin VE, Yu X, Egelman EH, West SC. Stabilization of RAD51 nucleoprotein filaments by the C-terminal region of BRCA2. Nat Struct Mol Biol. 2007;14:468–474. doi: 10.1038/nsmb1245. [DOI] [PubMed] [Google Scholar]
- 8.Carreira A, et al. The BRC repeats of BRCA2 modulate the DNA-binding selectivity of RAD51. Cell. 2009;136:1032–1043. doi: 10.1016/j.cell.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sartori AA, et al. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nimonkar AV, Ozsoy AZ, Genschel J, Modrich P, Kowalczykowski SC. Human exonuclease 1 and BLM helicase interact to resect DNA and initiate DNA repair. Proc Natl Acad Sci U S A. 2008;105:16906–16911. doi: 10.1073/pnas.0809380105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair (Amst) 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jazayeri A, et al. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 13.Yang H, Li Q, Fan J, Holloman WK, Pavletich NP. The BRCA2 homologue Brh2 nucleates RAD51 filament formation at a dsDNA-ssDNA junction. Nature. 2005;433:653–657. doi: 10.1038/nature03234. [DOI] [PubMed] [Google Scholar]
- 14.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 15.Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A. 2009;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang F, et al. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res. 2009;7:1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahman N, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tischkowitz M, et al. Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci U S A. 2007;104:6788–6793. doi: 10.1073/pnas.0701724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reid S, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 21.Xia B, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 22.Jones S, et al. Exomic sequencing identifies PALB2 as a pancreatic cancer susceptibility gene. Science. 2009;324:217. doi: 10.1126/science.1171202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ploquin M, et al. Stimulation of fission yeast and mouse Hop2-Mnd1 of the Dmc1 and Rad51 recombinases. Nucleic Acids Res. 2007;35:2719–2733. doi: 10.1093/nar/gkm174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McIlwraith MJ, et al. Reconstitution of the strand invasion step of double-strand break repair using human Rad51 Rad52 and RPA proteins. J Mol Biol. 2000;304:151–164. doi: 10.1006/jmbi.2000.4180. [DOI] [PubMed] [Google Scholar]
- 25.Rodrigue A, et al. Interplay between human DNA repair proteins at a unique double-strand break in vivo. EMBO J. 2006;25:222–231. doi: 10.1038/sj.emboj.7600914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine-139. J. Biol. Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 27.Rogakou EP, Boon C, Redon C, Bonner WM. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J Cell Biol. 1999;146:905–916. doi: 10.1083/jcb.146.5.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Celeste A, et al. Genomic instability in mice lacking histone H2AX. Science. 2002;296:922–927. doi: 10.1126/science.1069398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sung P. Mediating repair. Nat Struct Mol Biol. 2005;12:213–214. doi: 10.1038/nsmb0305-213. [DOI] [PubMed] [Google Scholar]
- 30.Saeki H, et al. Suppression of the DNA repair defects of BRCA2-deficient cells with heterologous protein fusions. Proc Natl Acad Sci U S A. 2006;103:8768–8773. doi: 10.1073/pnas.0600298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bryant HE, et al. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- 32.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 33.Dray E, et al. Enhancement of the RAD51 Recombinase by the Tumor Suppressor PALB2. Submitted. 2010 doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sy SM, Huen MS, Zhu Y, Chen J. PALB2 Regulates Recombinational Repair through Chromatin Association and Oligomerization. J Biol Chem. 2009;284:18302–18310. doi: 10.1074/jbc.M109.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colavito S, et al. Functional significance of the Rad51-Srs2 complex in Rad51 presynaptic filament disruption. Nucleic Acids Res. 2009;37:6754–6764. doi: 10.1093/nar/gkp748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bugreev DV, Yu X, Egelman EH, Mazin AV. Novel pro- and anti-recombination activities of the Bloom's syndrome helicase. Genes Dev. 2007;21:3085–3094. doi: 10.1101/gad.1609007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y, et al. RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments. Genes Dev. 2007;21:3073–3084. doi: 10.1101/gad.1609107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliver AW, Swift S, Lord CJ, Ashworth A, Pearl LH. Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep. 2009;10:990–996. doi: 10.1038/embor.2009.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mazloum N, Zhou Q, Holloman WK. D-loop formation by Brh2 protein of Ustilago maydis. Proc Natl Acad Sci U S A. 2008;105:524–529. doi: 10.1073/pnas.0707031105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petalcorin MI, Sandall J, Wigley DB, Boulton SJ. CeBRC-2 stimulates D-loop formation by RAD-51 and promotes DNA single-strand annealing. J Mol Biol. 2006;361:231–242. doi: 10.1016/j.jmb.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 41.Pellegrini L, et al. Insights into DNA recombination from the structure of a RAD51-BRCA2 complex. Nature. 2002;420:287–293. doi: 10.1038/nature01230. [DOI] [PubMed] [Google Scholar]
- 42.Haince JF, Rouleau M, Hendzel MJ, Masson JY, Poirier GG. Targeting poly(ADP-ribosyl)ation: a promising approach in cancer therapy. Trends Mol Med. 2005;11:456–463. doi: 10.1016/j.molmed.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 43.Fong PC, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 44.Audeh MW, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. doi: 10.1016/S0140-6736(10)60893-8. [DOI] [PubMed] [Google Scholar]
- 45.Baumann P, Benson FE, Hajibagheri N, West SC. Purification of human Rad51 protein by selective spermidine precipitation. Mut. Res. DNA Repair. 1997;384:65–72. doi: 10.1016/s0921-8777(97)00028-1. [DOI] [PubMed] [Google Scholar]
- 46.Henricksen LA, Umbricht CB, Wold MS. Recombinant replication protein A: Expression, complex formation, and functional characterization. J. Biol. Chem. 1994;269:11121–11132. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.