Abstract
Sleeping Beauty (SB) transposon-mediated integration has been shown to achieve long-term transgene expression in a wide range of host cells. In this study, we improved the SB transposon-mediated gene transfer system for transduction of human CD34+ stem/progenitor cells by two approaches: (1) to increase the transposition efficacy, a hyperactive mutant of SB, HSB, was used; (2) to improve the expression of the SB transposase and the transgene cassette carried by the transposon, different viral and cellular promoters were evaluated. SB components were delivered in trans into the target cells by Nucleoporation. The SB transposon-mediated integration efficacy was assessed by integrated transgene (enhanced green fluorescent protein [eGFP]) expression both in vitro and in vivo. In purified human cord blood CD34+ cells, HSB achieved long-term transgene expression in nearly 7-fold more cells than the original SB transposase. Significantly brighter levels of eGFP expression (5-fold) were achieved with the human elongation factor 1α (EF1-α) promoter in Jurkat human T cells, compared with that achieved with the modified myeloproliferative sarcoma virus long terminal repeat enhancer–promoter (MNDU3); in contrast, the MNDU3 promoter expressed eGFP at the highest level in K-562 myeloid cells. In human CD34+ cord blood cells studied under conditions directing myeloid differentiation, the highest transgene integration and expression were achieved using the EF1-α promoter to express the SB transposase combined with the MNDU3 promoter to express the eGFP reporter. Stable transgene expression was achieved at levels up to 27% for more than 4 weeks of culture after improved gene transfer to CD34+ cells (average, 17%; n = 4). In vivo studies evaluating engraftment and differentiation of the SB-modified human CD34+ cells demonstrated that SB-modified human CD34+ cells engrafted in NOD/SCID/γ chainnull (NSG) mice and differentiated into multilineage cell types with eGFP expression. More importantly, secondary transplantation studies demonstrated that the integrated transgene was stably expressed in more primitive CD34+ hematopoietic stem cells (HSCs) with long-term repopulating capability. This study demonstrates that an improved HSB gene transfer system can stably integrate genes into primitive human HSCs while maintaining the pluripotency of the stem cells, which shows promise for further advancement of non-virus-based gene therapy using hematopoietic stem cells.
Introduction
Hematopoietic stem cells (HSCs) represent a major focus for efforts at gene therapy for genetic diseases, cancer and leukemia, and AIDS. HSCs produce all the blood cells, essentially for a lifetime, and can be isolated from bone marrow, genetically modified ex vivo, and retransplanted into a patient to provide a long-lasting source of blood cells engineered to replace a congenital deficiency, or to afford new properties, such as specific anticancer immunoreactivity or resistance to infection with HIV-1. There has been significant progress in the development of methods for gene modification of human HSCs, and results from a select number of clinical trials are promising (Kohn and Candotti, 2009).
The most effective method for gene modification of HSCs has been with the use of integrating retroviral and lentiviral vectors. These can be used to transduce relatively high percentages of HSCs with minimal acute cytotoxicity. Clinical trials using retroviral vectors to add to HSCs the cDNA for genes involved in the pathogenesis of congenital immune deficiencies (adenosine deaminase [ADA]-deficient and X-linked forms of severe combined immunodeficiency [SCID] and chronic granulomatous disease [CGD]) have led to clear-cut clinical benefits, with the sustained production of genetically corrected lymphocytes and neutrophils, respectively, and restoration of protective immunity in a majority of treated patients (Cavazzana-Calvo et al., 2000; Horwitz et al., 2001; Aiuti et al., 2002; Gaspar et al., 2006; Ott et al., 2006). These immune deficiencies were chosen as initial disease candidates for clinical trials of gene therapy because they can be ameliorated by only a relatively low fraction of gene correction of stem cells; the genetically normal lymphocytes have selective survival and accumulate to clinically relevant levels from a low number of corrected HSCs in SCID patients.
However, 5 of 20 X-SCID patients and 2 of 3 CGD subjects developed leukemia-like hyperproliferative disorders, due to insertional oncogenesis by integrated copies of the retroviral vectors trans-activating cellular proto-oncogenes (Hacein-Bey-Abina et al., 2003; Kaiser, 2003; Ott et al., 2006; Metais and Dunbar, 2008). Novel methods to genetically modify HSCs that retain the beneficial effects of the retroviral vectors but with better safety profiles and greater gene transfer efficiency are needed to allow these methods to be applied to a broader range of diseases lacking the strong selective advantage seen in the immune deficiencies, such as hemoglobinopathies, lysosomal storage diseases, leukodystrophies, and cancer and AIDS applications.
As an alternative to integrating viral vectors, delivery of plasmids to cells may have several advantages. Plasmids may carry larger gene expression units than viral vectors, allowing more sophisticated patterns of gene expression rather than the constitutive, ubiquitous expression that is often achieved with viral vectors and strong enhancer/promoters. Genomic elements, such as extended upstream, downstream, and intronic sequences affecting transcription, locus control regions, insulators, and so on, can be included more readily in plasmids than in viral vectors. Plasmids by themselves would not persist in HSCs once the HSCs begin their massive proliferation for blood cell production. Several approaches may be used to make the results of plasmid-mediated gene delivery have long-lasting effects (Porteus and Baltimore, 2003; Dorigo et al., 2004; Porteus and Carroll, 2005; Hollis et al., 2006).
The Sleeping Beauty (SB) transposon system, as one of the plasmid-mediated gene delivery approaches, is a recombinase-based gene integration system with the ability to stably integrate genes in various target host cells (Izsvak et al., 2000; Yant et al., 2000; Izsvak and Ivics, 2004; Essner et al., 2005; Hackett et al., 2005). The SB transposon belongs to the Tc1/mariner family of transposable elements and was reconstructed from the fish (salmonid) genome (Ivics et al., 1997). In its natural configuration, the SB transposase consists of a DNA-binding domain at the N terminus, a nuclear localization signal (NLS), and a catalytic core domain at the C terminus of the transposase (Ivics et al., 1997; Yant et al., 2004). There are two components in the SB-mediated gene integration system: a transposon that carries the therapeutic gene and a transposase that catalyzes the mobilization of the transposon. The components are encoded by two separate plasmids that are codelivered to the target cells. In this two-plasmid system (Walisko et al., 2007), one plasmid carrying an enhanced green fluorescent protein (eGFP) reporter cassette (or a therapeutic gene) flanked by the SB inverted/direct repeat (IR/DR) sequences for permanent integration is coelectroporated with a second plasmid carrying the SB transposase expression cassette for transient production of the SB transposase enzyme. SB-mediated gene integration is based on a cut-and-paste mechanism that integrates the transgene into the chromosomal target site (Walisko and Ivics, 2006). SB first binds to the IR/DR sequences on both ends of the transposon and excises the transposon from the donor site. The excised transposon is then integrated into TA dinucleotide base pairs of the host chromosomal DNA. Several studies have characterized the integrations site preferences of the SB transposon system and found that it has a significantly lower frequency of integration into expressed genes, compared with retroviral and lentiviral vectors, which may afford a decreased risk for causing insertional oncogenesis, although this has not been formally proven (Liu et al., 2005; Yant et al., 2005; Berry et al., 2006).
The major hindrance to using nonviral gene modifications in HSCs has been the great difficulty in getting them into HSCs, compared with the more efficient gene delivery by viral vectors. Methods such as transfection, lipofection, or electroporation suffer from low efficiency and/or high cytotoxicity. More recently, stable gene transfer and expression in primary human T cells, neurons, embryonic cells, and HSCs (CD34+) by the SB transposon system in vitro have been reported by using the Amaxa Nucleofector (an electroporation-based device; Amaxa/Lonza Walkersville, Walkersville, MD) as the SB carrier in SB-mediated gene therapy (Hollis et al., 2006; Huang et al., 2006; Wilber et al., 2007; Zeitelhofer et al., 2007, 2009; Hohenstein et al., 2008). Amaxa Nucleoporation has been developed as a more efficient and gentle approach, delivering SB components via electroporation.
Despite the success of SB gene transfer, the efficacy of gene transfer by the SB-mediated system to primary cell populations for HPC gene therapy still remains to be optimized. On the basis of the data reported by Hollis and colleagues, suboptimal stable gene expression was achieved. The level of eGFP expression in CD34+ cells (enriched for HPCs) by the SB transposon system drastically decreased from 30–70 to 1–6% by 2 weeks postelectroporation (Hollis et al., 2006). The efficiency of gene transfer into CD34+ cells electroporated with the SB plasmids was also evaluated in vivo. Nonobese diabetic/severe combined immunodeficiency/β2-microglobulin deficient (NOD/SCID/B2mnull) mice were transplanted with electroporated human CD34+ cells to demonstrate human cell engraftment and reporter gene transduction. Unfortunately, the results were inconclusive because of significant impairment of cell survival and engraftment due to cytotoxicity in electroporated cells. Therefore, further improvement remains necessary to verify and improve the efficacy of the SB transposon gene transfer system for HSC gene therapy.
In this study, we improved the SB transposon-mediated gene transfer system to achieve higher stable transgene expression in K-562 human erythroleukemia cells, Jurkat human T-lymphoid cells, and primary human CD34+ hematopoietic stem/progenitor cells. A hyperactive mutant of SB, HSB (Baus et al., 2005), with higher transposition efficiency than the original SB transposase, was used to increase the transposition efficacy in target cells. In addition, expression of the SB transposase and transgene cassette carried by the transposon were also enhanced on analysis of three different viral and cellular promoters: the modified myeloproliferative sarcoma virus (MPSV) long terminal repeat enhancer–promoter (MNDU3), the human cytomegalovirus (CMV) immediate-early region enhancer–promoter, and the human elongation factor 1α (EF1-α) promoter. The stable gene transfer efficiency of the improved SB-mediated gene delivery system was also evaluated in vivo in a NOD/SCID/γ chainnull (NSG) neonatal transplant model. The improved SB transposon system in primary human CD34+ hematopoietic progenitors reported here has improved the stable gene transfer efficiency by ∼30-fold, compared with our prior published data (Hollis et al., 2006). Furthermore, this study is the first report demonstrating that SB-modified human CD34+ progenitor cells can be engrafted and differentiated into multilineage cell types in vivo, which shows promise for further advancement of non-virus-based gene therapy using hematopoietic stem cells.
Materials and Methods
Plasmid construction
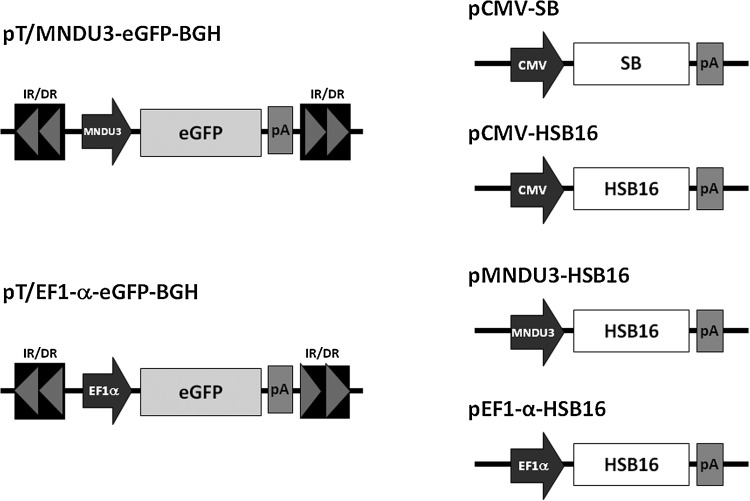
The transposon plasmid pT/MNDU3-eGFP-BGH (Hollis et al., 2006), used in this study, contains the Sleeping Beauty (SB) inverted repeat (IR) sequences flanking the eGFP expression cassette. The pT/MNDU3-eGFP-BGH plasmid was modified from plasmid pT/MC containing the IRs from SB flanking a multicloning site (kind gift from M. Kay, Stanford University, Stanford, CA) as described previously (Hollis et al., 2006). The transient expression plasmids pCMV-SB and pCMV-mSB, containing regular and defective versions of the SB transposase, respectively, were also obtained from M. Kay (Yant et al., 2000). The plasmid pCMV-HSB16, containing the “hyperactive” version of the SB transposase, HSB16, was obtained from B. Fletcher (University of Florida, Gainesville, FL) (Baus et al., 2005).
The plasmid pT/EF1-α-eGFP-BGH was created from pT/MNDU3-eGFP-BGH (Hollis et al., 2006) and pCCL-EF1-α-GFP (Uetsuki et al., 1989; Mizushima and Nagata, 1990; Nakai et al., 1998; Ramezani et al., 2000; Haas et al., 2003; Nakai et al., 2003). The MNDU3 fragment was removed from pT/MNDU3-eGFP-BGH with BgIII. The human EF1-α promoter was removed from pCCL-EF1-α-GFP with PvuI and BamHI, blunt ended, and inserted into the BgIII site of pT/X-eGFP-BGH to create pT/EF1-α-eGFP-BGH. The human EF1-α promoter fragment generated with PvuI and BamHI was also used to create pEF1-α-HSB16. The CMV promoter was removed from plasmid pCMV-HSB16 (Baus et al., 2005) with SpeI and BamHI to create pX-HSB16. The EF1-α promoter was then inserted into the blunted SpeI site to obtain plasmid pEF1-α-HSB16. Plasmid pMNDU3-HSB16 was created by inserting the MNDU3 promoter into pX-HSB16. The MNDU3 promoter was obtained from pT/MNDU3-eGFP-BGH by means of BgIII and SpeI, blunt ended, and ligated into the blunted BamHI site of pX-HSB16. The plasmids used in this study are illustrated in Fig. 1.
FIG. 1.
Vectors for Sleeping Beauty-mediated gene transfer. The pTransposon transposon plasmids include the Sleeping Beauty inverted/direct repeat (IR/DR) sequences flanking the expression cassette, which consists of the promoter, the enhanced green fluorescent protein (eGFP) coding sequence, and a bovine growth hormone (BGH) polyadenylation site (pA). The pSB transposase transient expression plasmids contain the Sleeping Beauty (SB) or hyperactive Sleeping Beauty (HSB16) transposase (or the mutant inactive SB, not shown) and the BGH poly(A) sequence driven by various viral and cellular promoters including human cytomegalovirus (CMV), the myeloproliferative sarcoma virus long terminal repeat enhancer–promoter (MNDU3), and human elongation factor 1α (EF1-α).
Cells
Human K-562 myeloid leukemia and Jurkat T cells were both obtained from the American Type Culture Collection (ATCC, Manassas VA) and cultured in RPMI 1640, 10% fetal calf serum, with 2 mM glutamine and penicillin–streptomycin (100 U/ml). CD34+ cells were isolated from human cord blood (CB) obtained from normal deliveries, using Ficoll-Paque PLUS (GE Healthcare Life Sciences, Piscataway, NJ) density gradient centrifugation followed by passage through MidiMACs separation columns (Miltenyi Biotech, Sunnyvale, CA). Use of CB was approved by the Committee on Clinical Investigations at Childrens Hospital Los Angeles (Los Angeles, CA) (Hao et al., 1995).
Electroporation
Cells were electroporated with the Amaxa Nucleofector according to the manufacturer's instructions. An Amaxa Nucleoporation cell line kit with program T-16 was used to electroporate 1 × 106 cells (K-562 cells), and program C-16 was used to electroporate 2 × 106 cells (Jurkat cells). For human CD34+ cells, 2 × 106 cells were used with Amaxa program U-08. Before electroporation, freshly isolated CD34+ cells were stimulated overnight in X-VIVO 15 medium (Cambrex, East Rutherford, NJ) containing 2 mM l-glutamine with stem cell factor (SCF, 50 ng/ml; R&D Systems, Minneapolis, MN), Flt-3 ligand (50 ng/ml; R&D Systems), and thrombopoietin (50 ng/ml; R&D Systems). After electroporation, CD34+ cells for in vitro analyses were cultured for 4 weeks under conditions inducing myeloid differentiation in RetroNectin-coated plates (Takara Mirus Bio, Madison, WI) in Iscove's modified Dulbecco's medium with 20% fetal calf serum, 0.5% bovine serum albumin (BSA), 2 mM glutamine, and penicillin–streptomycin (100 U/ml) plus human interleukin-3 (IL-3, 5 ng/ml; Biosource International, Camarillo, CA), IL-6 (10 ng/ml; Biosource International), and SCF (25 ng/ml; R&D Systems).
Human CD34+ cell transplantation into neonatal immune-deficient mice
NOD/SCID/γ chainnull (NSG) mice (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, stock no. 005557; Jackson Laboratory, Bar Harbor, ME) were housed in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Saban Research Institute of Childrens Hospital Los Angeles and the National Institutes of Health (Bethesda, MD). All animals were handled in laminar flow hoods and housed in microinsulator cages in a pathogen-free colony.
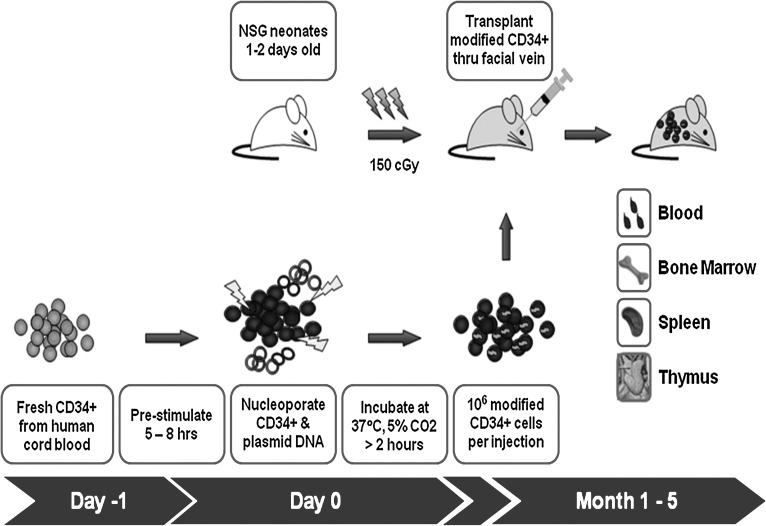
On the day of transplantation, CD34+ cells were electroporated after 5–8 hr of prestimulation, counted by trypan blue exclusion to determine numbers of live cells, and resuspended in phosphate-buffered saline (PBS) with heparin (10 U/ml) for transplantation at a concentration of 1 × 106 live cells per 50 μl. Before transplantation, NSG mice received 150 cGy of sublethal total body irradiation from a 137Ce source with attenuator. Neonatal NSG mice (1 to 2 days old) received approximately 1 × 106 modified human CD34+ cells by facial vein injection, using a 28-gauge needle. Transplanted pups were housed with nursing mothers and weaned to separate cages at 3 weeks. Blood samples were collected via the retro-orbital venous plexus under general anesthesia with isoflurane at 6 and 10 weeks posttransplantation to evaluate engraftment efficiency. Bone marrow, thymus, spleen, and blood were harvested from each animal 5 months posttransplantation for flow cytometric and quantitative polymerase chain reaction (PCR) analyses.
Secondary bone marrow transplantation
Adult (6- to 10-week-old) NSG mice were used as recipients for the secondary transplants of marrow from the mice that had received primary transplants of human CD34+ cells as neonates 5 months earlier. Five months after transplantation, the primary recipient mice were killed by CO2 narcosis. Femurs and tibias were harvested and washed in PBS with 5% fetal bovine serum (FBS; Omega Scientific, Tarzana, CA). Marrow was flushed with a 23-gauge needle and a 1-ml syringe filled with PBS with 5% FBS. Cells were counted by trypan blue exclusion to determine numbers of live cells and were resuspended in 100 μl of PBS with 5% FBS for transplantation. Recipient mice received 270 cGy of sublethal irradiation from a 137Ce source with attenuator 1 hr pretransplantation. Approximately 2–7 × 107 bone marrow cells were transplanted into each recipient mouse via the retro-orbital venous plexus under general anesthesia with isoflurane. Bone marrow cells of the secondary recipients were analyzed by flow cytometry for human CD45 and eGFP expression 2 months posttransplantation.
Flow cytometry
In vitro transgene expression analysis
For in vitro studies, electroporated cells were analyzed by flow cytometry performed with a FACSCalibur (BD Biosystems, San Jose, CA) using CellQuest software for eGFP transgene expression weekly for up to 4 weeks. Initial eGFP expression and regimen-related toxicity in the electroporated cultures were analyzed by fluorescence-activated cell sorting (FACS) on day 3 postelectroporation. Toxicity was determined by propidium iodide (PI; Sigma-Aldrich, St. Louis, MO) staining followed by FACS analysis. Levels of eGFP expression obtained after week 4 of culture were defined as an indication of stable long-term transgene integration in this study.
In vivo hematopoietic chimerism analysis
To detect human cells in NSG mice, multicolor cytometric analysis was performed with the FACSCalibur. Peripheral blood (PB) was taken from the retro-orbital venous plexus 6 and 10 weeks after transplantation under general anesthesia to assess the level of engraftment. Blood was collected through heparinized calibrated pipettes (Drummond Scientific, Broomall, PA) and transferred to 50 μl of FBS for FACS analysis. Five months posttransplantation, the mice were killed by CO2 narcosis. The PB, femurs, thymus, and spleens were harvested from each animal. Cell suspensions from each organ were made by pressing tissue through a 70-μm pore size sterile nylon cell strainer (BD Biosciences) and were subjected to FACS analysis and quantitative PCR. Samples were depleted of erythrocytes with BD FACS lysing solution (BD Biosciences) and approximately 3 × 106 cells from each sample were incubated in 10% FBS for 30 min at 4°C to block nonspecific staining. The remaining cells were used for PCR analysis (see the section Quantitative PCR Analysis). Appropriate volumes of the indicated antibodies (BD Biosciences) were added to the blocked samples for 30 min in the dark at room temperature. Human T cells were examined by triple staining with peridinin–chlorophyll protein (PerCP)-conjugated anti-human CD45 antibody, allophycocyanin (APC)-conjugated anti-human CD4 antibody, and phycoerythrin (PE)-conjugated anti-human CD8 antibody. To detect human B and natural killer (NK) cells in each sample, three-color cytometric analysis with PerCP-conjugated anti-human CD45 antibody, APC-conjugated anti-human CD19 antibody, and PE-conjugated anti-human CD56 antibody was also performed. Last, human granulocytes were detected by double staining with PerCP-conjugated anti-human CD45 antibody and PE-conjugated anti-human CD14 antibody.
Hematopoietic repopulation analysis
Bone marrow was harvested from adult NSG mice 2 months after secondary transplantation. To detect repopulating human eGFP+ cells in the recipients, bone marrow cells were stained with PerCP-conjugated anti-human CD45 antibody (BD Biosciences) and analyzed by FACS for human CD45+ and eGFP+ cell expression.
Quantitative PCR analysis
eGFP DNA copy numbers were determined by real-time quantitative PCR (qPCR). DNA from in vitro experiments was extracted with a DNeasy tissue kit (Qiagen, Valencia, CA). DNA from tissues and cell populations purified from organs was extracted with phenol–chloroform and resuspended in Tris–ethylenediaminetetraacetate (TE). All DNA was quantified by fluorimetry with a DNA-specific dye (Hoechst dye; Sigma-Aldrich). qPCR was performed with primers and probe designed to amplify integrated eGFP sequence: sense primer (ctg ctg ccc gac aac ca), antisense primer (gaa ctc cag cag gac cat gtg), and the TAMRA probe sequence (ccc tga gca aag acc cca acg aga). The primer concentrations (sense and antisense) were 400 nM and the probe concentration was 50 nM in all reactions. All reactions used universal master mix (Applied Biosystems, Foster City, CA) and were run under default conditions in a 7900HT fast real-time PCR system (Applied Biosystems). Each of the wells contained 350 ng of template DNA and was compared with a standard curve made by diluting DNA from a cell line containing one copy of an integrated eGFP gene diluted into the DNA of nontransduced cells, yielding a detection sensitivity of 1:100,000 vector-containing cells.
Statistical analysis
The Student t test was used to determine statistical significance, and p < 0.05 was considered significant.
Results
HSB is a more efficient enzyme in comparison with the original SB
Baus and colleagues derived a hyperactive mutant version of the SB transposase (HSB), which they reported to increase SB-mediated transposition up to 17-fold over the original SB transposase in HeLa cells (Baus et al., 2005). To evaluate the transposition efficacy of HSB in our target cell types, namely human hematopoietic cells, HSB was compared with the original SB using a two-plasmid system (Hollis et al., 2006) in K-562 human erythroleukemia cells, Jurkat human T-lymphoid cells, and primary human CD34+ hematopoietic progenitor/stem cells. SB components were delivered in trans into the target cells by Nucleoporation. One plasmid carrying a transposon consisting of the eGFP reporter gene under the transcriptional control of the MNDU3 retroviral LTR promoter and flanked by the SB IR sequences (pT/MNDU3-eGFP-BGH) was codelivered with a transposase-expressing plasmid containing the original SB (CMV-SB), the hyperactive SB (CMV-HSB), or an inactive mutant SB transposase (CMV-mutSB) (Fig. 1). In these studies, 10 μg of the transposon plasmid (pT/MNDU3-eGFP-GBH) and 1 μg of the transposase-expressing plasmid were used.
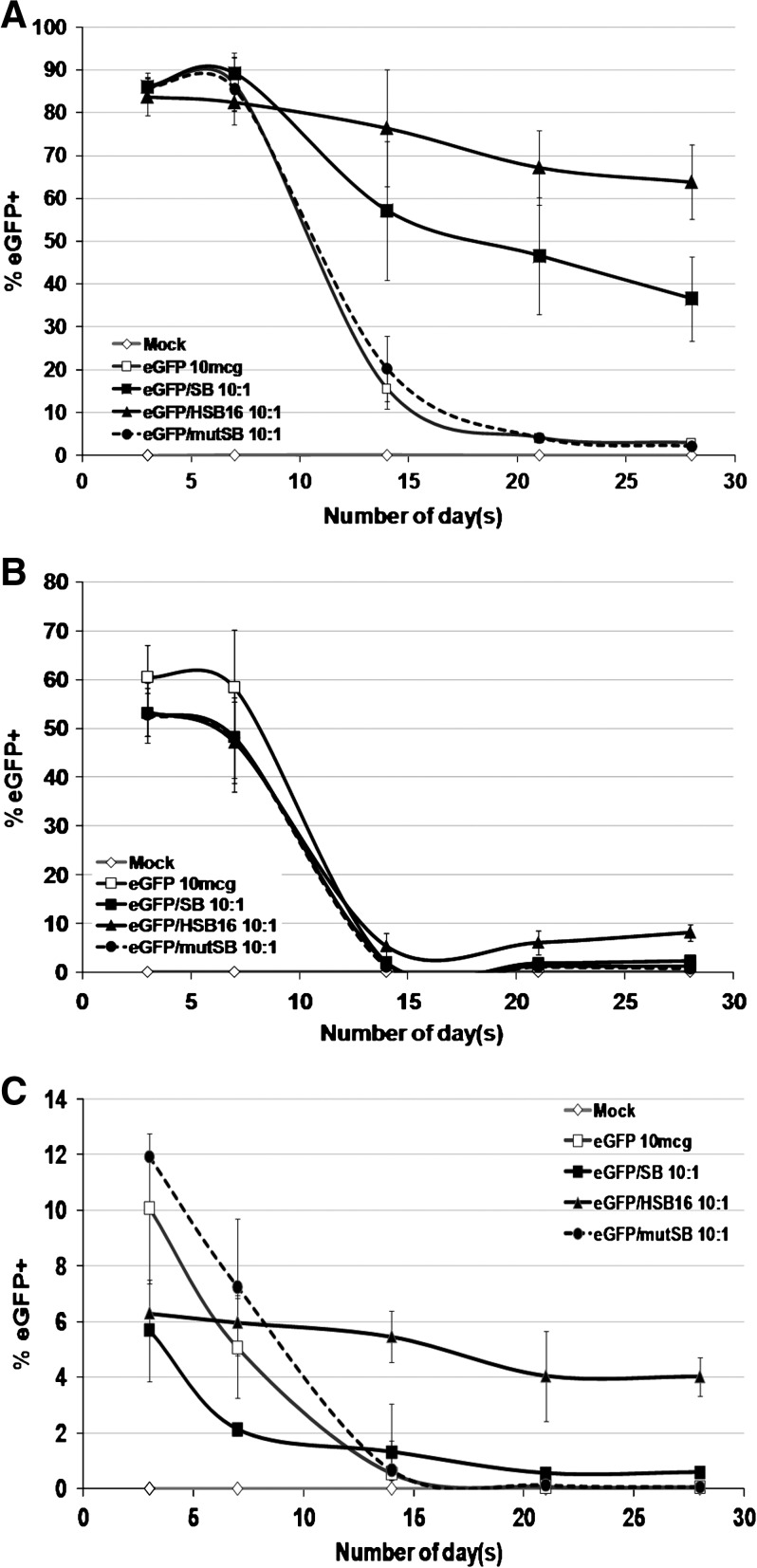
Initial levels of GFP-expressing cells 3 days after Nucleoporation were consistent for each cell type, independent of which SB plasmid was used (Fig. 2). K-562 cells showed the highest percentages of GFP-expressing cells (:80%) and Jurkat cells had >50% expressing eGFP. Primary human CD34+ cells had a lower level of expression (6–12%). Four weeks postelectroporation, the percentages of eGFP+ cells detected in Jurkat cells and human CD34+ cells were decreased from the initial levels. In all three cell types, cells that were electroporated with the mutant inactive SB transposase plasmid (CMV-mutSB) or with no transposase plasmid did not show any stable eGFP reporter gene expression, with GFP-expressing cells declining to background levels by 4 weeks postelectroporation (Fig. 2). In contrast, cells receiving the SB or HSB plasmid persistently expressed GFP out to 4 weeks. This observation indicated that a functional SB transposase was required to achieve stable transgene persistence.
FIG. 2.
Stable long-term HSB- and SB-mediated gene transfer. Transgene expression over time in (A) K-562, (B) Jurkat, and (C) primary human CD34+ cells isolated from cord blood. Ten micrograms of the transposon plasmid (pT/MNDU3-eGFP-BGH) and 1 μg of either pCMV-SB (solid squares), pCMV-HSB (solid triangles), or pCMV-mutSB (solid circles) transposase-expressing plasmid were coelectroporated into each cell population. Cells were also electroporated with pT/MNDU3-eGFP-BGH alone (open squares) or with Amaxa Nucleoporation solution only, without added DNA (open diamonds), as controls. Aliquots of cells were analyzed by flow cytometry at each time point to determine the percentage of GFP-expressing cells and the percentage of propidium iodide (PI)+ cells. Each experiment was repeated three times (n = 3) with two or three replicates per experiment. Error bars represent the standard error of the mean (SEM).
The functional assay based on stable eGFP expression showed that HSB was a more efficient enzyme compared with SB under the same delivery conditions in K-562, Jurkat, and primary human CD34+ cells (Fig. 2). K-562 cells electroporated with the HSB-expressing plasmid showed a 2-fold higher level of stable eGFP expression in comparison with cells electroporated with the original SB at 4 weeks postelectroporation (64 vs. 37%). Jurkat human T cells that received the HSB plasmid showed a 4.5-fold increase in stable transgene expression compared with cells receiving the original SB plasmid (9 vs. 2%). The stable transgene expression detected in primary human CD34+ cells electroporated with HSB plasmid was 6.6-fold higher (4 vs. <1%) when compared with CD34+ cells electroporated with the original SB plasmid (Table 1). These findings demonstrated that HSB was a superior SB transposase leading to higher transposition efficacy than the original SB in these human hematopoietic cell types.
Table 1.
Summary of HSB- versus SB-Mediated Gene Transfer in K562, Jurkat, and Primary Human CD34+ Cells
| Cell type | SB-mediated gene transfer (% eGFP ± SEM) | HSB-mediated gene transfer (% eGFP ± SEM) | Fold increase |
|---|---|---|---|
| K-562 (n = 3) | 36.6 ± 9.9 | 63.8 ± 8.7 | 2 (p = 0.0014) |
| Jurkat (n = 3) | 1.2 ± 0.8 | 8.0 ± 1.7 | 4.5 (p = 0.013) |
| LTC-CD34+ (n = 4) | 0.6 ± 0.1 | 4.1 ± 0.6 | 6.9 (p = 0.0012) |
Abbreviations: eGFP, enhanced green fluorescent protein; HSB, hyperactive mutant of Sleeping Beauty transposon; LTC-CD34+, long-term-cultured CD34+ cells; SB, Sleeping Beauty transposon.
In addition, the results from the stable transgene expression time-course study suggested that there might be cell type-specific differences in SB integration efficiency as reported by other studies (Izsvak et al., 2000; Berry et al., 2006). K-562 cells showed the highest levels of long-term GFP expression; SB transposition efficiency was significantly lower in Jurkat human T cells, and lowest in primary human CD34+ cells. Over a series of experiments, the overall level of stable eGFP expression in K-562 cells, using the HSB plasmid, was approximately 10- to 16-fold higher than in Jurkat T cells and primary human CD34+ cells (Table 1).
Improvement of transposon-mediated gene transfer
Fresh versus frozen CD34+ cells
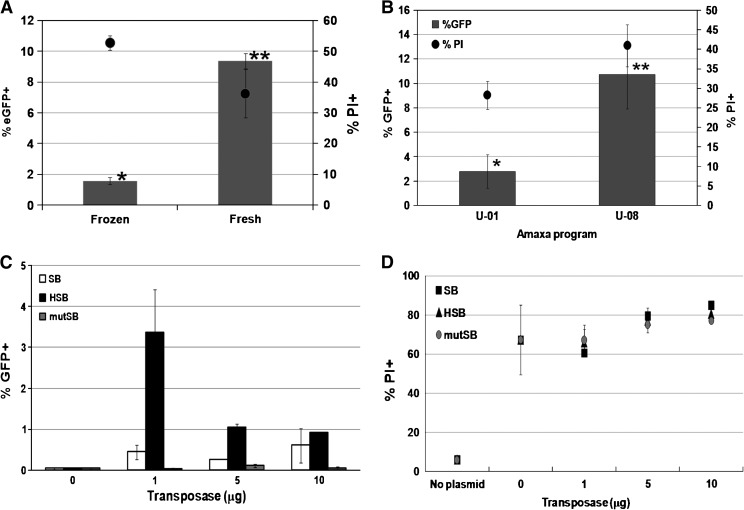
In our previously published studies of gene transfer with SB in human hematopoietic cells, SB-mediated gene transfer to primary human CD34+ hematopoietic stem/progenitor cells by electroporation was relatively inefficient, caused significant cytotoxicity, and was not able to achieve any stable transgene expression in vivo (Hollis et al., 2006). Those studies were done with human umbilical cord blood (CB) CD34+ cells that were cryopreserved and thawed before electroporation. One of the possible ways to improve stable transgene expression could be by using fresh human CD34+ cells to avoid any possible alterations in cell integrity that might have occurred during the freeze–thaw process. To investigate the transposition efficiency of the HSB transposon system using previously frozen human CD34+ cells, equal numbers of viable frozen–thawed or fresh primary human CD34+ cells were coelectroporated with the transposon-containing plasmid (pT/MND-eGFP-BGH) and the HSB-expressing plasmid. Fresh CD34+ cells showed 6-fold higher transgene expression than frozen CD34+ cells under the same conditions (Fig. 3A). This finding suggested that the HSB transposon system delivered by electroporation occurs more effectively in fresh human CD34+ cells. Therefore, subsequent experiments using primary human CD34+ cells in this study were all done with freshly isolated human CD34+ cells from CB and were not frozen before the experiment.
FIG. 3.
Improvement of transposon-mediated gene transfer. (A) Viable frozen–thawed or freshly isolated human CD34+ cells (2 × 106) isolated from cord blood (CB) were electroporated with 10 μg of pT/MNDU3-eGFP-BGH with 1 μg of HSB transposase-expressing plasmid. Flow cytometric analysis was performed 4 weeks postelectroporation for stable transgene expression detection. Asterisks (* and **) indicate significant differences (p < 0.01; n = 3) between the data points. (B) Cells were electroporated with 10 μg of pT/MNDU3-eGFP-BGH according to either program U-08 or U-01 of the Amaxa Nucleofector. The percentages of eGFP+ cells (columns) and the percentages of PI+ cells (circles) were determined by flow cytometric analysis 3 days postelectroporation for transgene expression and cell viability, respectively. Asterisks (* and **) indicate significant differences (p < 0.05; n = 4) between the data points marked. (C) Primary human CD34+ cells were coelectroporated with 10 μg of pT/MNDU3-eGFP-BGH and increasing quantities of pCMV-SB (open columns), pCMV-HSB (solid columns), or pCMV-mutSB (gray columns). Flow cytometric analysis was performed 4 weeks postelectroporation for stable transgene expression (n = 2). (D) Ten micrograms of pT/MNDU3-eGFP-BGH was coelectroporated with increasing quantities of pCMV-SB (squares), pCMV-HSB (triangles), or pCMV-mutSB (circles) (n = 2). The percentages of PI+ cells were determined by flow cytometric analysis 3 days postelectroporation. Error bars represent the standard error of the mean (SEM).
Nucleoporation program comparison
Stable gene transfer and expression in primary human T cells and hematopoietic progenitor/stem cells (CD34+ cells), using the SB transposon system, have been reported with the Amaxa Nucleofector (Amaxa/Lonza Walkersville) as the nonviral deliver system for the SB-mediated gene transfer approach (Hollis et al., 2006; Huang et al., 2006; Von Levetzow et al., 2006; Nakata et al., 2007; Wiehe et al., 2007). The Amaxa Nucleofector is an electroporation-based technique that augments transfer of DNA into the nucleus of the target cell. An ideal electrical parameter for each specific cell type has been preprogrammed into the Nucleofection device and cell-specific solutions are provided by the manufacturer. In an attempt to reduce cell mortality occurring during electroporation to primary human CD34+ cells, two different recommended Nucleofection programs (U-01 and U-08) were compared. Primary human CD34+ cells were electroporated with 10 μg of pT/MNDU3-eGFP-BGH, using either program U-01 or U-08 of the Amaxa Nucleofector, and analyzed by flow cytometry for initial eGFP reporter gene expression and cell viability (by PI staining) 3 days after electroporation. Cells electroporated according to program U-08 showed 4-fold higher transfection efficiency compared with cells treated according to program U-01 (10.7 vs. 2.8%), with only minimally higher cytotoxicity (32 vs. 28%) (Fig. 3B). This suggested that whereas program U-01 was a gentler electroporation program, it was also less efficient in delivering DNA into human CD34+ cells. Subsequent studies (and those previously discussed) were done with the U-08 program.
Improvement of the transposon-to-transposase plasmid ratio
The quantities of transposon and transposase plasmids have been known to be one of the major factors that determines the transposition efficiency of the SB transposon system. An optimal ratio of transposase to transposon is needed to achieve high transposition levels (Yant et al., 2000; Liu and Visner, 2007). On the basis of the previously published quantities of SB plasmids used to achieve stable transgene expression in human CD34+ cells in vitro (10 μg of the transposon and 10 μg of the transposase SB plasmid) (Hollis et al., 2006), the amount of pCMV-HSB was varied from 1 to 10 μg, with the amount of transposon plasmid (pT/MND-eGFP-BGH) kept constant at 10 μg. Populations of human CD34+ cells were electroporated and then analyzed by flow cytometry to measure eGFP expression after 4 weeks, without any selection. A transposon-to-transposase plasmid ratio of 10 to 1 μg was determined to be the optimal ratio for the HSB transposon system to achieve the highest level of gene transfer into human CD34+ cells (Fig. 3C). Lower levels of gene transfer were achieved with the SB plasmid at each of the amounts used. The greater activity of HSB allows the total amount of DNA used in a Nucleoporation reaction to be decreased from 20 μg (10 μg of transposon and 10 μg of SB transposase) to 11 μg (10 μg of transposon and 1 μg of HSB transposase).
Interestingly, the levels of gene marking in human CD34+ cells were inversely proportional to the quantity of HSB transposase plasmid used, declining from 3.4% when using 1 μg of pCMV-HSB to less than 1% with 10 μg of pCMV-HSB. The decrease in the level of gene expression observed for the largest quantity of transposase could be due either to toxicity caused by the DNA plasmid (Hollis et al., 2006) or by the activity of the SB transposase per se, or may represent the phenomenon of “overproduction inhibition,” wherein excessive amounts of transposase activity lead to inhibition of transposition, presumably by increasing the rate of the reverse, excision reaction (Yant et al., 2000; Geurts et al., 2003; Hollis et al., 2006; Hackett, 2007).
When cells were mocked electroporated without any plasmid added to the reaction, low levels of PI+ cells were seen (<10%) (Fig. 3D). Addition of 10 μg of only the eGFP-expressing transposon plasmid, but no transposase plasmid, led to a large increase in cytotoxicity, with more than 60% PI+ cells. Addition of increasing amounts of any of the transposase-expressing plasmids led to further dose-related increases in the percentage of PI+ cells, but these were essentially the same with any of the HSB, SB, and mutSB plasmids, indicating that the presence of functional HSB transposase did not affect cell viability. Large quantities of plasmid added to the Nucleoporation, in contrast, were responsible for the cytotoxicity, as reported previously (Hollis et al., 2006). Thus, the ability of the HSB to achieve greater gene transfer with lower amounts of plasmid also moderately reduces acute cytotoxicity.
HSB transposase expressed by human EF1-α promoter achieved the highest stable transgene expression
Promoters used in expression plasmids display different expression strengths, depending on the specific cell type (Fitzsimons et al., 2002; Weber and Cannon, 2007). The human CMV early region enhancer–promoter, for example, expresses strongly in nonhematopoietic cells (Boshart et al., 1985) but performs poorly in vivo and in hematopoietic cells (Scharfmann et al., 1991; Kay et al., 1992; Baskar et al., 1996; Miyoshi et al., 1999; An et al., 2000). In contrast to the CMV promoter, the human EF1-α promoter has been identified as a strong promoter in primary human hematopoietic cells (Ye et al., 1998; Chang et al., 1999) and exhibits high transcription activity in vivo (Taboit-Dameron et al., 1999). The MNDU3 LTR promoter, derived from the MPSV retrovirus, has been shown to express well in murine and human hematopoietic and lymphoid cells (Halene et al., 1999; Haas et al., 2003).
To further improve both the activity of the HSB transposase to mediate stable gene persistence and the expression level of the reporter eGFP transgene carried by the transposon in human hematopoietic cells, we evaluated these three different promoters. Each promoter was cloned into an individual construct to express either the HSB transposase or the eGFP reporter gene carried by the transposon. The improvement of HSB-mediated gene transfer efficacy involved two parts. First, expression of the HSB transposase by the various promoters was evaluated, using the pT/MNDU3-eGFP-BGH reporter transposon plasmid. Second, with the improved HSB transposase construct identified, expression of the eGFP reporter gene contained in the transposon construct using the various promoters was then examined.
In the comparison of different promoters driving expression of the HSB, K-562 cells were much more permissive to HSB-mediated gene integration than Jurkat T cells and primary human CD34+ cells with any of the promoters used to express HSB (Table 2). Long-term stable transgene expression in K-562 cells under all conditions remained approximately 80% by 4 weeks postelectroporation. Both Jurkat and human CD34+ cells showed lower levels (5- to 14-fold, respectively) of HSB-mediated stable transgene expression than that observed in K-562 cells.
Table 2.
Comparison of Stable Transgene Expression Achieved with HSB Transposase Expressed by NMDU3, CMV, or EF1-α Promoter in K-562, Jurkat, and Primary Human CD34+ Cells
| Cell type | CMV-HSB-mediated gene expression (% eGFP ± SEM) | MNDU3-HSB-mediated gene expression (% eGFP ± SEM) | EF1-α-HSB-mediated gene expression (% eGFP ± SEM) | Fold increase |
|---|---|---|---|---|
| K-562 (n = 2) | 76.0 ± 1.1 | 68.1 ± 13.6 | 85.6 ± 0.7 | 1.1 (p = 0.18) |
| Jurkat (n = 2) | 8.0 ± 2.1 | 7.6 ± 1.6 | 16.6 ± 2.3 | 2.1 (p = 0.006) |
| LTC-CD34+ (n = 3) | 3.8 ± 0.8 | 2.1 ± 1.2 | 6.9 ± 1.8 | 1.8 (p = 0.05) |
Abbreviations: CMV, cytomegalovirus; EF1-α, elongation factor 1α; MNDU3, modified myeloproliferative sarcoma virus long terminal repeat enhancer–promoter.
The highest level of stable transgene integration was achieved when using the EF1-α promoter to express HSB in all three cell types, compared with either the human CMV or MNDU3 promoter (Table 2). Although this effect was not significantly above the already high level of gene transfer with the CMV-HSB plasmid in K-562 cells, there was a 2-fold increase in the percentages of GFP-expressing Jurkat and primary CD34+ cells when the EF1-α promoter was used to direct expression of the HSB transposase.
eGFP reporter gene expression from the integrated transposon was influenced by the promoter and the target cell types
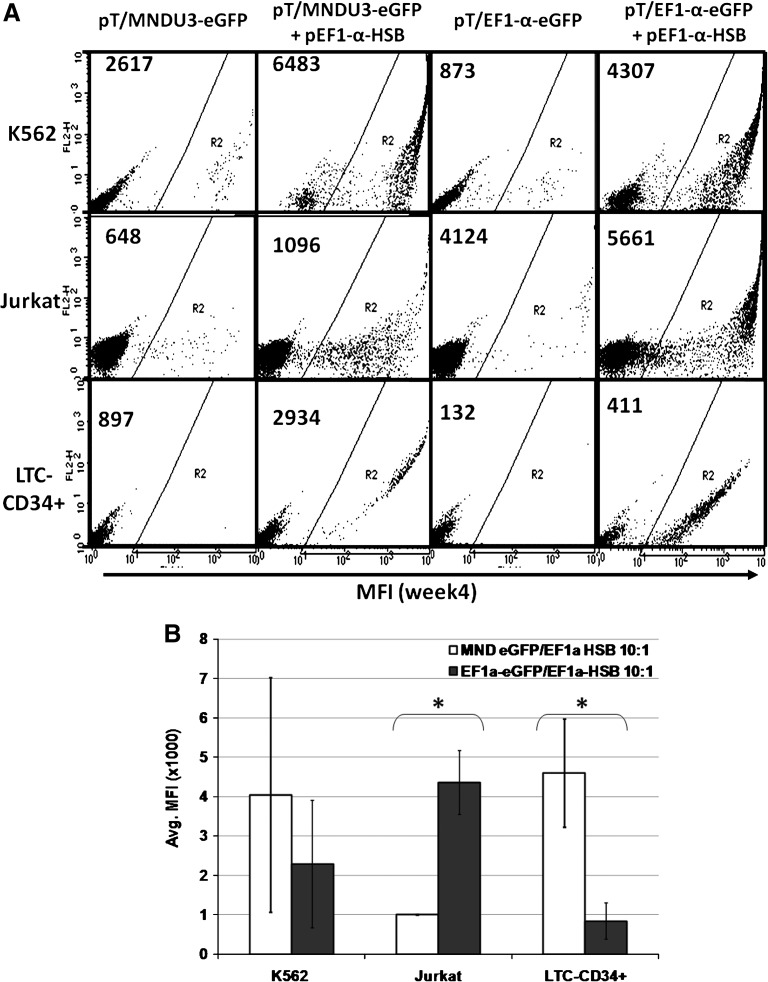
Next, we evaluated the MNDU3 and EF1-α promoters for expression of the eGFP reporter gene from the integrated transposon in K-562, Jurkat, and primary human CD34+ cells (Fig. 4A). The mean fluorescence intensity (MFI) of eGFP expression by the MNDU3 promoter was 2-fold higher than by the EF1-α promoter in K-562 cells (Fig. 4A and B). In primary human CD34+ cells (cultured under conditions directing myeloid differentiation; LTC-CD34+), the MFI of eGFP expression was significantly higher with the MNDU3 promoter than with the EF1-α promoter (p < 0.01). In contrast, the highest MFI of GFP expression in Jurkat human T cells occurred when the EF1-α promoter was used to express the GFP reporter (Fig. 4A). Thus, the MND promoter was more active in myeloid-type cells (K-562 erythroleukemia cells and cultured CD34+ cells), whereas the EF1-α promoter was more active in the T-lymphoid cells.
FIG. 4.
Promoter analysis for expression of transposase and transposon reporter. (A) Representative FACS plots showing stable HSB-mediated eGFP expression after 4 weeks in K-562, Jurkat, and primary human CD34+ cells (LTC-CD34+) cultured under conditions directing myeloid differentiation. Average mean fluorescence intensity (MFI) values are indicated in the top left corner of each plot. (B) Comparison of the average MFI of eGFP-expressing cells. LTC-CD34+ refers to long-term cultured human CD34+ cells under myeloid differentiation conditions. Ten micrograms of pT/MNDU3-eGFP-BGH or pT/EF1-α-eGFP-BGH transposon plasmid was coelectroporated with 1 μg of pEF1-α-HSB plasmid in this study. Cells were analyzed at week 4 postelectroporation for detection of HSB-mediated stable eGFP reporter gene expression. Each experiment was done with two or three replicates per condition. Error bars represent the SEM. The Student t test was performed for statistical analyses and p values are as indicated. *p < 0.01.
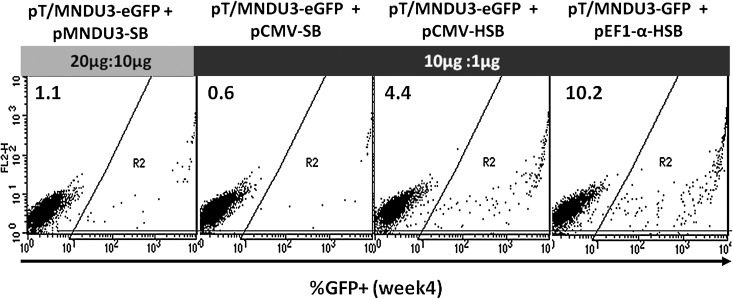
By incrementally enhancing each element of the SB transposon system we have been able to significantly increase the efficiency of stable gene delivery to human hematopoietic cells, including primary human CD34+ hematopoietic progenitor/stem cells. The level of stable gene expression increased 20-fold by combining the use of the HSB transposase and the EF1-α promoter to direct HSB expression and the MND promoter to express the GFP transgene (Fig. 5).
FIG. 5.
Overall SB transposon system improvement in primary human CD34+ hematopoietic progenitor/stem cells in vitro. Shown are representative FACS plots elucidating the progression of improvement of the SB-mediated gene integration system in this study. The percentages of eGFP-positive cells 4 weeks postelectroporation are indicated in the top left corner of each plot. The transposon–transposase plasmid DNA combinations used for each condition are also denoted at the top of the FACS plots.
HSB-modified human CD34+ progenitor cells were engrafted and reconstituted in NSG mice after neonatal transplantation
Primitive human hematopoietic cells can be assayed on the basis of their ability to repopulate immune-deficient NOD/SCID mice. The efficiency of HSB-mediated gene transfer to human CD34+ hematopoietic cells was therefore evaluated in vivo. Fresh primary CD34+ hematopoietic progenitor cells were isolated from human cord blood, electroporated with the best HSB plasmids, and transplanted into nonobese diabetic NOD/SCID/γ chainnull (NSG) neonatal mice (Fig. 6). To assess the transduction of primitive normal human stem cells (SCID repopulating cells, or SRCs), human cell engraftment (percent CD45+) and gene transduction (percent eGFP+) were determined in peripheral blood from NSG mice 6, 10, and 20 weeks posttransplantation by flow cytometric analysis.
FIG. 6.
Schematic diagram of the experimental timeline for in vivo analysis of SB-mediated gene transfer to human CD34+ cells by NOD/SCID/γ chainnull neonatal transplantation. NSG, NOD/SCID/γ chainnull mouse.
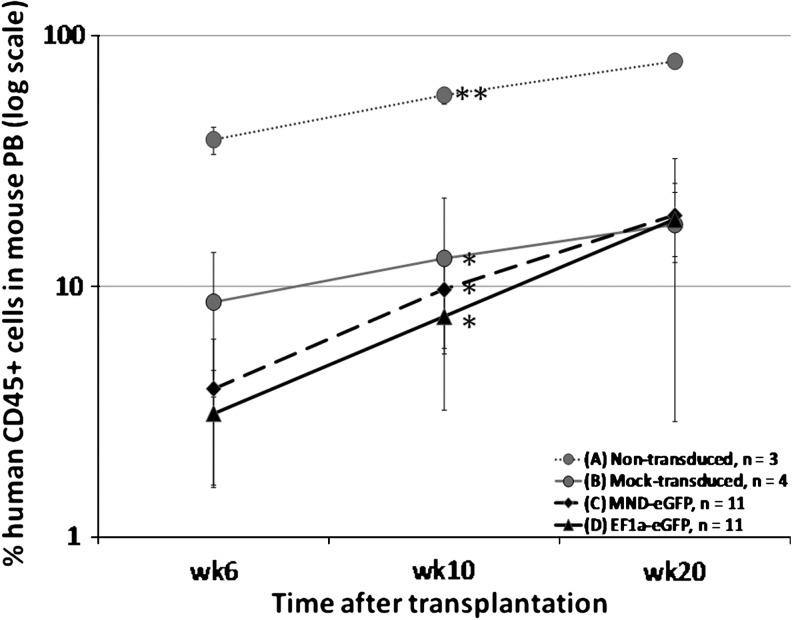
Seven NSG litters (n = 49) received neonatal bone marrow transplantation in four different treatment groups. Twenty of 49 transplanted neonatal NSG mice did not survive to 1 week posttransplantation, because of poor nurturing by their birth parents. The successfully transplanted animals were divided among treatment groups receiving the following CD34+ cells: group A, nontransduced (n = 3); group B, mock transduced (n = 4); group C, electroporated with pT/MNDU3-eGFP-BGH transposon plasmid and the pEF1-α-HSB plasmid (n = 11); and group D, electroporated with pT/EF1-α-eGFP-BGH transposon plasmid and the pEF1-α-HSB plasmid (n = 11). Table 3 summarizes the level of engraftment percent human CD45+ in mouse peripheral blood and bone marrow at 10 and 20 weeks posttransplantation. Differences were observed in the engraftment efficiencies of the nontransduced CD34+ cells (group A), mock-transduced human CD34+ cells (group B), and human CD34+ cells electroporated with either MNDU3- or EF1-α-expressed eGFP in the presence of HSB (groups C and D, respectively). When the cells were electroporated with or without plasmid DNA, the engraftment efficiency of the transplanted CD34+ cells was significantly reduced compared with engraftment of CD34+ cells that were not electroporated (Fig. 7; p < 0.05). At 10 weeks posttransplantation, an average engraftment level of 60% was observed in animals transplanted with nontransduced human CD34+ cells (group A). In comparison, mice that received electroporated human CD34+ cells (groups B, C, and D) showed significantly (5- to 8-fold) lower engraftment levels, at a range of 8–12%. This lower level of human cells in mice receiving CD34+ cells that had been electroporated suggested that the use of electroporation as a DNA delivery method to primary human CD34+ cells is detrimental to their engraftment and normal hematopoietic ability. The level of engrafted human cells from the groups receiving the transposon and transposase plasmids (groups C and D) increased to a level comparable (∼20%) to that detected in the mock-transduced group (group B) by 5 months posttransplantation, despite the lower initial engraftment levels of the HSB-modified human CD34+ cells (average, 3 vs. 7%, respectively). This finding suggested that the significant decrease in the engraftment level of HSB-modified CD34+ cells was due mainly to toxic effects of the electroporation and not to the HSB transposase or the integration process.
Table 3.
Summary of Engraftment Level and Transgene Expression in Primary Transplanted NSG Mice
| % Human CD45+ | % Human CD45+ eGFP+ | Gene marking (copies/human cell) | |||||
|---|---|---|---|---|---|---|---|
| Treatment | Animal no. | PBMCs (week 10) | BM (week 20) | Spleen (week 20) | BM (week 20) | BM (week 20) | Spleen (week 20) |
| (A) Nontransduced; n = 3 | 1 | 48.8 | 4.7 | 48.5 | 0.0 | 0 | 0 |
| 2 | 61.2 | 59.0 | 55.1 | 0.0 | 0 | 0 | |
| 3 | 63.0 | ND | ND | ND | ND | ND | |
| (B) Mock transduced; n = 4 | 1 | 8.6 | 1.7 | 38.4 | 0.0 | 0 | 0 |
| 2 | 1.6 | 8.4 | 11.6 | 0.0 | 0 | 0 | |
| 3 | 0.0 | 0.3 | 12.0 | 0.0 | 0 | 0 | |
| 4 | 41.3 | 48.0 | 89.0 | 0.0 | 0.001 | 0 | |
| (C) pT/MNDU3-eGFP + pEF1-α-HSB; n = 11 | 1 | 3.5 | 58.2 | 23.0 | 0.0 | 0.001 | ND |
| 2* | 24.9 | 51.2 | 92.1 | 0.6 | 0.006 | 0.160 | |
| 3 | 0.2 | 0.0 | 0.1 | 0.0 | 0 | 0 | |
| 4 | 0.0 | 0.3 | 3.0 | 0.0 | 0.109 | 0.010 | |
| 5 | 0.3 | ND | ND | ND | ND | ND | |
| 6 | 30.2 | 14.2 | 80.6 | 0.2 | 0.011 | 0.001 | |
| 7 | 2.9 | 71.1 | 79.5 | 0.0 | 0 | 0 | |
| 8 | 3.8 | 55.2 | 80.7 | 0.0 | 0.001 | 0 | |
| 9 | 5.7 | 8.5 | 4.1 | 0.1 | 0.006 | 0.012 | |
| 10 | 0.5 | 1.4 | 3.0 | 0.2 | 0.037 | 0.010 | |
| 11 | 34.5 | 46.1 | 57.9 | 0.0 | 0.004 | ND | |
| (D) pT/EF1-α-eGFP + pEF1-α-HSB; n = 11 | 1 | 16.3 | 46.1 | 48.1 | 0.0 | 0.001 | 0.001 |
| 2 | 5.5 | ND | ND | ND | ND | ND | |
| 3 | 0.2 | 1.8 | 1.2 | 0.0 | 0.017 | 0.025 | |
| 4 | 2.3 | 66.9 | 63.0 | 0.0 | 0.001 | 0 | |
| 5* | 16.4 | 48.9 | 71.4 | 10.2 | 0.164 | 0.102 | |
| 6 | 1.2 | 42.9 | 9.6 | 0.0 | 0.001 | 0.032 | |
| 7 | 21.1 | ND | ND | ND | 0.002 | 0.002 | |
| 8 | 4.0 | 53.6 | 63.8 | 0.0 | 0.001 | 0 | |
| 9 | 0.7 | 0.5 | 1.8 | 0.1 | 0.065 | 0.018 | |
| 10 | 7.7 | 21.3 | 62.5 | 0.1 | 0.005 | 0 | |
| 11 | 7.8 | 19.1 | 52.3 | 0.0 | 0.002 | 0.001 | |
Abbreviations: BM, bone marrow; ND, not done; PBMCs, peripheral blood mono nuclear cells.
eGFP expression was detected in human repopulating cells of transplanted NSG mice by flow cytometric analysis.
FIG. 7.
Comparison of engraftment levels of human CD34+ progenitor cells in NSG mice. At the indicated times after neonatal transplantation of human CD34+ cells, human CD45+ cells in mouse peripheral blood (PB) were assayed by flow cytometric analysis. Freshly isolated human CD34+ cells isolated from CB were coelectroporated with 10 μg of pT/MNDU3-eGFP-BGH and 1 μg of pEF1-α-HSB plasmid (group C; n = 11) or with 10 μg of pT/EF1-α-eGFP-BGH and 1 μg of pEF1-α-HSB plasmid (group D; n = 11). As study controls, NSG mice received human CD34+ cells that were not transduced (group A; n = 3) or that were “mock transduced” (electroporated without plasmid DNA) (group B; n = 4). Asterisks (* and **) indicate significant differences (p < 0.01) between the data points marked.
Transgene expression detected in multiple organs of NSG mice transplanted with HSB-modified human CD34+ cells
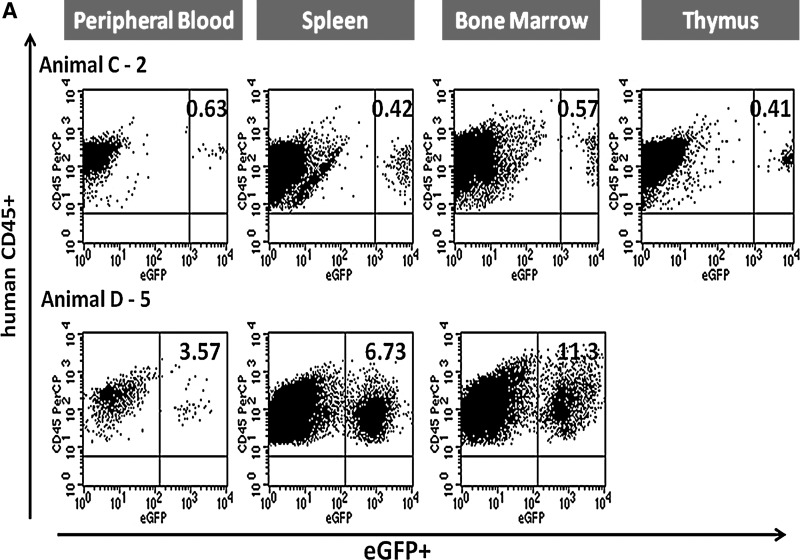
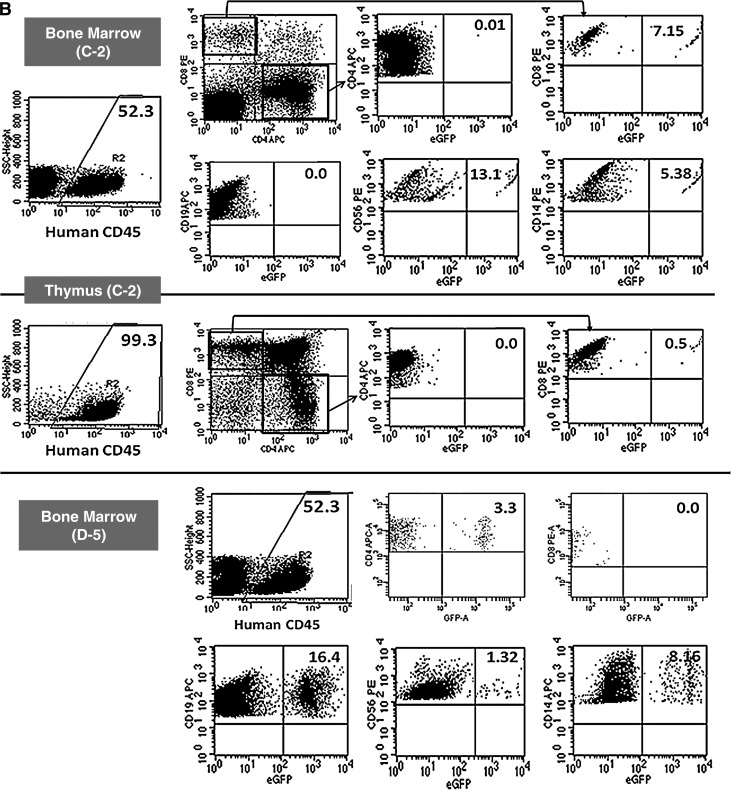
To evaluate the efficiency of HSB-mediated stable gene transfer to human CD34+ cells in vivo, eGFP expression in human cells (percent CD45+eGFP+) was determined (Table 3). Stable transgene expression in up to 2.5% of the cells was first observed by flow cytometric analysis at week 10 posttransplantation in the peripheral blood of mice that received HSB-modified human CD34+ cells. Five months after transplantation, high eGFP expression levels (MFI range, from 1000 to 6500; data not shown) were detected in peripheral blood, spleen, bone marrow, and thymus of two animals, C-2 and D-5 (Fig. 8A). Up to 11.3% human CD45+eGFP+ cells were detected in the bone marrow of the transplanted NSG mice 5 months posttransplantation.
FIG. 8.
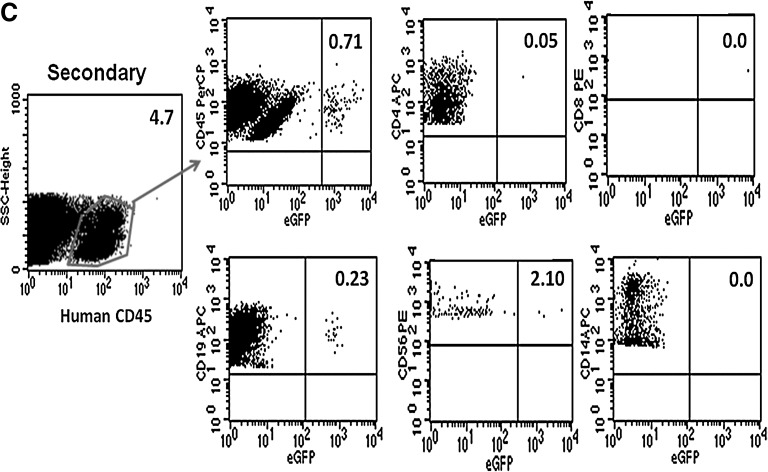
Representative flow cytometric analysis of eGFP-expressing human cells from NSG mice engrafted with HSB-modified human CD34+ cells at 5 months posttransplantation. (A) Stable eGFP expression was detected in peripheral blood, bone marrow, thymus, and spleen from NSG mice transplanted with human CD34+ cells modified by HSB-mediated gene transfer. (B) Stable eGFP expression was detected in CD4+ or CD8+ T cells, CD19+ B cells, CD56+ NK cells, and CD14+ myeloid cells from all organs. Data shown here are FACS plots of the BM and thymus harvested from C-2 and D-5 NSG mice. The thymus of mouse D-5 was not sufficiently populated for immunostaining analysis. (C) Total bone marrow cells harvested from NSG mouse D-5 were successfully engrafted into secondary adult NSG recipients. eGFP transgene expression was also detected in NSG human repopulation cells. Each number represents the percentages of human CD45+eGFP+ cells in the specific cell lineage.
eGFP expression detected in human cells in NSG mice 5 months after neonatal bone marrow transplantation
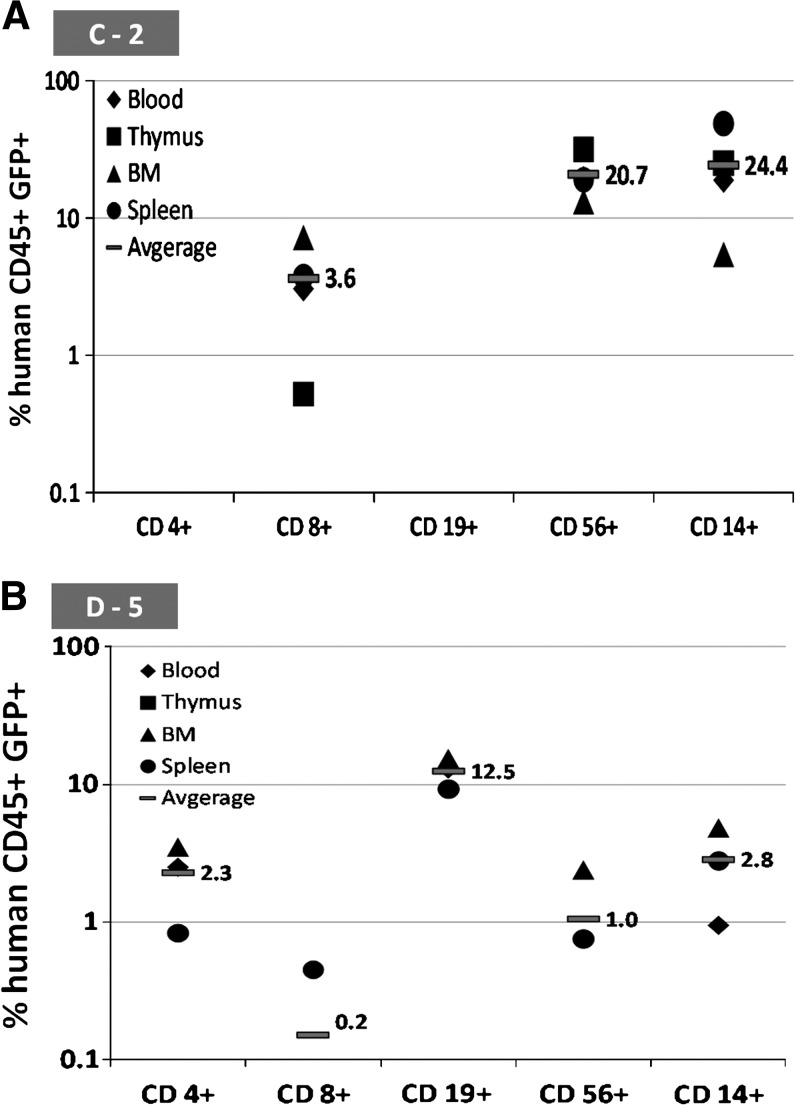
eGFP expression in total human leukocytes was determined in peripheral blood (PB), bone marrow (BM), thymus, and spleen harvested from NSG mice at 5 months posttransplantation. Distinct GFP+ populations were detected by flow cytometry 5 months posttransplantation in two NSG mice that had been transplanted with HSB-modified human CD34+ cells. With an average engraftment level of 50% human CD45+ cells in the BM, stable eGFP expression was detected in T cells (CD4 and CD8, single positive), B cells (CD19+), NK cells (CD56+), and myeloid cells (CD14+) in both C-2 and D-5 animals (Fig. 8B). Normal T cell development was observed in the bone marrow of both eGFP+ animals by 10 weeks posttransplantation. In animal C-2 bone marrow, eGFP expression was detected in CD8+ T cells, CD19+ B cells, CD56+ NK cells, and also CD14+ myeloid cells with the highest percentage of eGFP-expressing cells (13.1%) found in the CD56+ NK cell population (Fig. 9A). Normal thymopoiesis was also observed in mouse C-2 with 0.5% CD8+eGFP+ T cells. No eGFP expression was detected in the CD4+ or CD19+ populations of the BM and thymus of mouse C-2. In comparison with animal C-2, animal D-5 showed a high percentage (16.4%) of eGFP+ cells in the CD19+ B cell population of the bone morrow and 3.3% eGFP expression in its CD4+ T cells (Fig. 9B).
FIG. 9.
Summary of multilineage eGFP expression levels determined in peripheral blood, bone marrow, thymus, and spleen harvested from NSG mice 5 months after neonatal transplantation of HSB-modified human CD34+ cells. Shown is the percentage of human CD45+eGFP+ cells detected in CD4+ or CD8+ T cells, CD19+ B cells, CD56+ NK cells, and CD14+ myeloid cells from all organs harvested from (A) NSG mouse C-2 and (B) NSG mouse D-5.
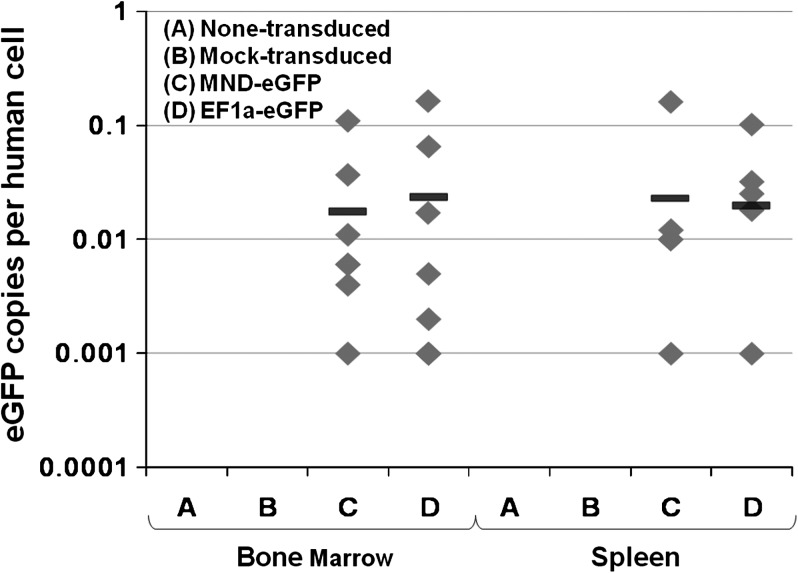
eGFP gene marking (copies per cell) in human CD45+ cells in mouse bone marrow and spleen at 20 weeks posttransplantation is also summarized in Table 3 and Fig. 10. DNA from total cells harvested from the BM of each transplanted mouse was isolated and analyzed for eGFP copy number by quantitative PCR analysis. There were no significant differences in vector copy numbers between groups C and D when the reporter gene was expressed by either the MNDU3 or EF1-α promoter (geometric mean, 6.1 × 10−4 vs. 7.3 × 10−4 copies per cell, respectively). Mouse C-2 with 0.6% human CD45+eGFP+ cells was found to have 0.006 eGFP copies per human cell in the BM. Mouse D-5 with 11.3% human CD45+eGFP+ cells was found to have 0.164 eGFP copies per human cell in the BM. Up to 0.16 vector copies per cell was detected in cells from the spleen.
FIG. 10.
Summary of eGFP gene marking in human CD45+ cells isolated from the BM and spleen of NSG mice 5 months posttransplantation. The geometric mean of the eGFP marking, calculated on the basis of the values listed in Table 3, is represented by the horizontal bars. Zero values were not included in the determination of geometric means.
eGFP expression levels in each cell lineage were also determined in PB, BM, thymus, and spleen harvested from transplanted NSG mice (Fig. 9). The highest eGFP expression was detected in the NK cell (21%) and B cell (25%) populations in mouse C-2 (group C, MNDU3 treatment group). In contrast, the majority of eGFP+ cells (12.5%) were found in the B cell lineage of mouse D-5 (group D, EF1-α treatment group).
To characterize human hematopoietic stem cells in the human cells engrafted in the NSG mice, secondary transplantations were conducted. Donor animals were selected on the basis of the engraftment level of human CD45+ cells detected in the peripheral blood 10 weeks posttransplantation. After euthanasia, total bone marrow cells from two femurs and tibias from each primary mouse were injected via the retro-orbital venous plexus into individual secondary mice, after 350 cGy of total body irradiation. Eight adult NSG mice were transplanted with bone marrow cells harvested from primary mice (Table 4). At 8 weeks posttransplantation, substantial secondary grafts with eGFP transgene expression (0.7% human CD45+eGFP+) were detected in one secondary murine recipient that had received bone marrow cells from mouse D-5. This observation indicates that the HSB transposon system could achieve stable gene transfer to a primitive human HSC population with long-term repopulating capacity (Fig. 8C).
Table 4.
Engraftment Level and Transgene Expression in Secondary Transplanted NSG Mice
| Recipient/donor | No. of transplanted cells | Estimated transplanted human CD45+ | Engraftment (% human CD45+) | % human CD45+ eGFP+ |
|---|---|---|---|---|
| A. Nontransduced | ||||
| 1/A-2 | 5.3 × 107 | 3.1 × 107 | 7.0 | 0 |
| B. Mock transduced | ||||
| 2/B-4 | 2.8 × 107 | 1.3 × 107 | 22.0 | 0 |
| C. pT/MNDU3-eGFP-BGH + pEF1-α-HSB | ||||
| 3/C-8 | 3.1 × 107 | 1.7 × 107 | 12.3 | 0 |
| 4/C-6 | 4.3 × 107 | 0.6 × 107 | 0.9 | 0 |
| 5/C-9 | 7.1 × 107 | 0.6 × 107 | ND | ND |
| D. pT/EF1-α-eGFP-BGH + pEF1-α-HSB | ||||
| 6/D-10 | 3.6 × 107 | 0.8 × 107 | 0.3 | 0 |
| 7/D-5 | 1.9 × 107 | 1.0 × 107 | 4.7 | 0.7 |
| 8/D-11 | 3.9 × 107 | 0.7 × 107 | 0.1 | 0 |
Abbreviation: ND, not determined.
Discussion
This study reports improved efficacy using the Sleeping Beauty transposon system for stable gene transduction of human CD34+ hematopoietic stem/progenitor cells achieved by incrementally improving elements of the system. By combining multiple modifications to the Nucleoporation delivery methods we used previously, we improved gene transduction to CD34+ cells so that we could readily detect expression of the GFP reporter gene in cells transplanted into immune-deficient mice and beyond into secondarily transplanted recipients.
We first assessed benefits from using a “hyperactive” version of the SB transposase (HSB). This modified transposase had been created by a phylogenetically based comparison approach to generate a combination of mutations in the N-terminal DNA-binding domain, with triple alanine substitutions (K33A, T83A, and L91A) (Geurts et al., 2003; Yant et al., 2004; Baus et al., 2005), and also in the C-terminal domain with mutations in the integrase and also the C1 and C2 proteins. We observed that the level of stable gene transfer to human hematopoietic and lymphoid cells was consistently higher when the HSB was used, compared with the original SB transposase. The greater activity of the HSB allowed us to reduce the total amount of plasmid DNA used for Nucleoporation, with 1 μg of HSB plasmid leading to greater gene transfer than 10 μg of the parental SB plasmid. Using the HSB transposase, we defined an effective ratio of transposase to transposon plasmids in the Nucleoporation system. Under the conditions we used, the most effective ratio of transposon plasmid to HSB transposase plasmid was 10 to 1 μg. These numbers may vary for different cell types, different electroporation conditions, and with different plasmids.
With the greater activity of the HSB transposase, we observed the phenomenon of overexpression inhibition, which has been one of the major limitations of the SB transposon system for efficient gene delivery. Only a narrowly defined ratio of transposon to transposase will allow maximal transposition activity of the SB transposase (Geurts et al., 2003). We observed higher transduction levels with 1 μg of the HSB plasmid than with 5 or 10 μg. SB transposition efficiency depends greatly on the relative availability of transposase molecules per transposon. Studies demonstrated that a total of four transposase molecules (two per IR binding site on each end of a transposon) were required to complete a stable integration of one transposon molecule. The transposase molecules can bind to each other in a criss-cross manner to juxtapose the two ends of the transposon (Hartl et al., 1997; Cui et al., 2002; Geurts et al., 2003). Insufficient levels of transposase will not effectively form a functional synaptic complex for transgene transposition. However, surplus levels of transposase will also inhibit SB transposition activity (Lohe et al., 1996; Hartl et al., 1997; Lampe et al., 1998; Geurts et al., 2003; Wilson et al., 2005) by preventing the juxtaposition of transposon ends and inhibiting the transposition process. In this study, an overexpression inhibition effect was observed when the transposon-to-transposase ratio increased from 10:1 to 2:1.
As another strategy to increase the efficiency of HSB transposition activity, we also investigated the effect of various promoters used for the expression of the HSB transposase and the GFP reporter cassette in the transposon. The promoter from the ubiquitously expressed cellular elongation factor 1α (EF1-α) gene, used to direct expression of the HSB transposase, led to the highest level of stable transgene integration in each of the cell types examined. Among the promoters tested, the best promoter for expression of the transgene from the integrated transposon varied in the different target cell types studied. We observed that the MND promoter, from the myeloproliferative sarcoma virus, was more active for expressing the transgene in human myeloid cells (K-562 and primary myeloid cells derived from transduced CD34+ cells). In contrast, the EF1-α promoter was more active in T-lymphoid cells. It is important to consider the different expression needs for the transposase, in the immediate target cells, and the transposon expression cassette, in the ultimate cell type where transgene expression is intended.
Other important factors that we examined included the quality of the CD34+ cells used for electroporation, and the specific electroporation parameters encoded by the Amaxa Nucleofector. Freshly isolated human CD34+ cells were shown to allow higher levels of initial and stable transgene expression than cryopreserved cells. No higher immediate cytotoxicity to the frozen cells was observed, and therefore the basis for their poorer transduction is not known. The Amaxa Nucleoporation program setting U-08 led to 6-fold higher transgene expression than did program U-01; there was somewhat more acute cytotoxicity with the U-08 program than with the more gentle U-01 program, but U-08 nonetheless led to higher DNA delivery efficiency in human CD34+ HSCs.
With the significantly improved gene delivery efficiency achieved in primary human CD34+ cells in vitro, these improved HSB transposon methods were evaluated in a more stringent model for evaluating transduction of human HSCs, by xenografting the transduced CD34+ cells in the NSG neonatal transplantation model. In our prior study with the Sleeping Beauty system and human CD34+ cells, we were not able to obtain engraftment of transduced cells in an immune-deficient mouse model, primarily because of severe cytotoxicity from the Nucleoporation (Hollis et al., 2006). For the studies reported here, we made several changes to our approach. First, we used the NSG mouse strain for these studies, whereas we had used NOD/SCID/β2-microglobulin−/− mice in the previous studies. We have found that the NSG model affords significantly greater human HSC engraftment (and life expectancy) and supports vigorous T cell production (Ito et al., 2002; Ishikawa et al., 2005). In addition, the NSG mice were transplanted with human cells by neonatal transplantation, which also leads to greater engraftment and T cell production than in postnatal transplants. The ability of newborn NSG pups to engraft human hematopoietic cells and support the presence of circulating human cells in the peripheral blood makes the neonatal transplant model more applicable than the adult transplant model to investigate the development of rare human cell populations in the transplanted mice (Sands et al., 1993; Ishikawa et al., 2005; Park et al., 2008).
In addition, the length of culture with cytokines (stem cell factor, Flt-3 ligand, and thrombopoietin [SCF/Flt-3L/TPO]) for the ex vivo gene modification was also significantly reduced to less than 12 hr compared with our previous study (Hollis et al., 2006). These culture conditions have commonly been used for human HSC priming before retroviral vector transduction to facilitate HSC proliferation and transduction (Murphy et al., 1992; Murray et al., 1999; Ueda et al., 2000; Wu et al., 2001a). In the context of transfection via electroporation, prestimulation with cytokines has also been shown to be necessary and beneficial for transfection efficiency (Wu et al., 2001a; Nightingale et al., 2006). Shorter cytokine exposure time may better maintain the stem cell characteristics of HSCs (Ailles et al., 2002) Last, because of the possible electroporation- and/or plasmid DNA-related toxicity involved in our system (Hollis et al., 2006), the cell dose used for transplantation was increased to 1 × 106, which was 5-fold higher than the cell dose used in our previous study.
These modifications of the transplantation protocol augmented the engraftment level of HSB-modified human CD34+ cells in the transplanted NSG mice. Multiorgan and multilineage stable transgene expression was detected in the repopulating human cells in the NSG mice 5 months posttransplantation. In addition, persistent transgene expression was also observed in secondarily transplanted NSG mice. These results demonstrated that the improved HSB transposon protocol delivered by electroporation could transfer transgenes into primitive human long-term hematopoietic stem cells that could engraft and sustain expression of the transgene in vivo. However, the low absolute levels of transduction and engraftment preclude definitive demonstration that pluripotent HSCs were transduced.
During the preparation of this manuscript, similar findings were published using another version of hyperactive SB transposase mutant (SB100X) (Xue et al., 2009). In their report, the CMV promoter was used to express the SB transposase and a hybrid CMV enhancer/chicken β-actin promoter (CAGGS) was used to express the transgene encoded in the transposase cassette. Efficient gene transfer and stable transgene expression were achieved in human CD34+ progenitor cells (HPCs) and also in multiple cell lineages when differentiated both in vitro and in vivo. However, the “stem cell” characteristics of the transgene-expressing SRCs were not addressed by serial transplantation and the SCID repopulating cells (SRCs) detected in their in vivo model could be more mature, committed hematopoietic progenitor cells.
Furthermore, on the basis of the promoter identification data shown in our study, use of the CMV promoter to express SB100X and of the CAGGS promoter to express transgene may be suboptimal. We found that the EF1-α promoter was more effective to increase SB transposition activity when compared with the CMV promoter (Table 2). The CAGGS promoter has been shown to have lower promoter strength than the EF1-α promoter in lymphoid cells and lower strength than the MND promoter in myeloid cells (Weber and Cannon, 2007). Therefore, the promoter analysis approaches and findings described in our study may further improve the stable gene transfer efficiency of SB100X. Nevertheless, the study reported here, together with the work published by Xue and colleagues (2009), further strengthen the possibility of using the SB transposon system as a nonviral gene transfer system for HSC gene therapy.
Despite the significant progress made in these studies, limitations still remain to be overcome in order for the efficacy of the SB transposon system to be more clinically relevant for HSC gene therapy. One of the major limitations in the approach is cytotoxicity to human CD34+ cells from the electroporation per se, which is augmented by the presence of plasmid DNA. In the in vitro studies, toxicity observed during electroporation of human CD34+ cells was due mainly to the quantity of plasmid DNA and occurred in a concentration-dependent manner, as reported previously (Wu et al., 2001b; Hollis et al., 2006). However, electroporation alone seems to be detrimental to the engraftment and differentiation ability of human hematopoietic cells in vivo; we observed a significant decline in the engraftment efficiency of electroporated cells even in the absence of plasmid DNA. Therefore, other methods of DNA delivery with lower toxicity should be explored. One possible alternative is the use of nonintegrating lentiviral (NIL) vectors as a carrier for SB component delivery. NIL vectors are designed to disable vector integration but to retain the normal viral entry functions. (Nightingale et al., 2006) NIL vectors have been shown to achieve gene delivery and to mediate transient gene expression in various cell types, including primary human HPCs and embryonic stem cells (Yanez-Munoz et al., 2006; Lombardo et al., 2007). In combination with the SB transposon, an NIL/HSB transposon hybrid gene transfer system could potentially provide not only the benefit of viral transduction (high-level, minimally cytotoxic transient gene expression) but also SB-mediated permanent transgene integration. Other delivery methods for SB plasmids such as hydrodynamic injection, lipofection, and cationic polymer polyethyleneimine (PEI) have also been actively investigated (Yant et al., 2000; Bell et al., 2007; Belur et al., 2007; Liu et al., 2009).
The efficiency of SB-mediated transposition also depends on the size of the transposon being delivered. Use of alternative nonviral transposon systems with higher transgene carrying capacity, such as the PiggyBac transposon derived from the cabbage looper moth Trichoplusia ni or the bacteriophage ϕC31 integrase, may be more effective to deliver larger DNA fragments. In addition to their larger carrying capacity, the lack of overexpression inhibition observed in the PiggyBac transposon system and the site-specific integration capacity of the ϕC31 integrase may be advantageous (Wilson et al., 2007; Keravala and Calos, 2008).
Although we were able to achieve multilineage transgene expression in NSG repopulating cells, the engraftment of electroporated HSB-modified human CD34+ cells showed large variability (0.01 to 60% in BM at 5 months), with only about 10% of the transplanted mice achieving high-level engraftment of human cells with persistent transgene expression. Because of the low numbers of well-engrafted mice, we were not able to draw any conclusions about the relative efficacy of the MNDU3 or EF1-α promoter on transgene expression in specific cell lineages in vivo; further improvement in transduction and engraftment is needed to make these studies feasible. To improve the inconsistency of this NSG neonatal transplant model, several modifications could be made to potentially better evaluate the efficacy of stem cell survival and transduction in vivo. For example, because cells are relatively fragile immediately after electroporation, the 1–2 hr of incubation we allowed before transplantation may not be optimal to allow electroporated cells to recover fully and may subsequently lead to further cells being lost during transplantation. A longer incubation time after electroporation (i.e., 4–5 hr) may be able to reduce cell death and consequently increase engraftment of stably transduced human CD34+ cells. In addition, the radiation dose given to the animals may be increased to facilitate engraftment ability of the NSG neonates, although neonatal mice may not tolerate much higher levels of total body irradiation. Alternatively, the electroporated CD34+ cells that express eGFP could be sorted by flow cytometry and transplanted into the mice to increase the level of cells expressing the transgene in vivo.
In conclusion, the improved SB transposon system for transduction of primary human CD34+ hematopoietic progenitors reported here has improved the stable gene transfer efficiency by at least 20-fold, compared with our prior published data. Furthermore, this study demonstrated that SB-modified human CD34+ progenitor/stem cells can be engrafted and differentiated into multilineage cell types in vivo, which shows promise for further advancement of non-virus-based HSC gene therapy.
References
- Ailles L. Schmidt M. Santoni De Sio F.R. Glimm H. Cavalieri S. Bruno S. Piacibello W. Von Kalle C. Naldini L. Molecular evidence of lentiviral vector-mediated gene transfer into human self-renewing, multi-potent, long-term NOD/SCID repopulating hematopoietic cells. Mol. Ther. 2002;6:615–626. [PubMed] [Google Scholar]
- Aiuti A. Slavin S. Aker M. Ficara F. Deola S. Mortellaro A. Morecki S. Andolfi G. Tabucchi A. Carlucci F. Marinello E. Cattaneo F. Vai S. Servida P. Miniero R. Roncarolo M.G. Bordignon C. Correction of ADA-SCID by stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- An D.S. Wersto R.P. Agricola B.A. Metzger M.E. Lu S. Amado R.G. Chen I.S. Donahue R.E. Marking and gene expression by a lentivirus vector in transplanted human and nonhuman primate CD34+ cells. J. Virol. 2000;74:1286–1295. doi: 10.1128/jvi.74.3.1286-1295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskar J.F. Smith P.P. Nilaver G. Jupp R.A. Hoffmann S. Peffer N.J. Tenney D.J. Colberg-Poley A.M. Ghazal P. Nelson J.A. The enhancer domain of the human cytomegalovirus major immediate-early promoter determines cell type-specific expression in transgenic mice. J. Virol. 1996;70:3207–3214. doi: 10.1128/jvi.70.5.3207-3214.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baus J. Liu L. Heggestad A.D. Sanz S. Fletcher B.S. Hyperactive transposase mutants of the Sleeping Beauty transposon. Mol. Ther. 2005;12:1148–1156. doi: 10.1016/j.ymthe.2005.06.484. [DOI] [PubMed] [Google Scholar]
- Bell J.B. Podetz-Pedersen K.M. Aronovich E.L. Belur L.R. McIvor R.S. Hackett P.B. Preferential delivery of the Sleeping Beauty transposon system to livers of mice by hydrodynamic injection. Nat. Protocols. 2007;2:3153–3165. doi: 10.1038/nprot.2007.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belur L.R. Podetz-Pedersen K. Frandsen J. McIvor R.S. Lung-directed gene therapy in mice using the nonviral Sleeping Beauty transposon system. Nat. Protocols. 2007;2:3146–3152. doi: 10.1038/nprot.2007.460. [DOI] [PubMed] [Google Scholar]
- Berry C. Hannenhalli S. Leipzig J. Bushman F.D. Selection of target sites for mobile DNA integration in the human genome. PLoS Comput. Biol. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boshart M. Weber F. Jahn G. Dorsch-Hasler K. Fleckenstein B. Schaffner W. A very strong enhancer is located upstream of an immediate early gene of human cytomegalovirus. Cell. 1985;41:521–530. doi: 10.1016/s0092-8674(85)80025-8. [DOI] [PubMed] [Google Scholar]
- Cavazzana-Calvo M. Hacein-Bey S. De Saint Basile G. Gross F. Yvon E. Nusbaum P. Selz F. Hue C. Certain S. Casanova J.L. Bousso P. Deist F.L. Fischer A. Gene therapy of human severe combined immunodeficiency (SCID)-X1 disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- Chang L.J. Urlacher V. Iwakuma T. Cui Y. Zucali J. Efficacy and safety analyses of a recombinant human immunodeficiency virus type 1 derived vector system. Gene Ther. 1999;6:715–728. doi: 10.1038/sj.gt.3300895. [DOI] [PubMed] [Google Scholar]
- Cui Z. Geurts A.M. Liu G. Kaufman C.D. Hackett P.B. Structure–function analysis of the inverted terminal repeats of the sleeping beauty transposon. J. Mol. Biol. 2002;318:1221–1235. doi: 10.1016/s0022-2836(02)00237-1. [DOI] [PubMed] [Google Scholar]
- Dorigo O. Gil J.S. Gallaher S.D. Tan B.T. Castro M.G. Lowenstein P.R. Calos M.P. Berk A.J. Development of a novel helper-dependent adenovirus–Epstein-Barr virus hybrid system for the stable transformation of mammalian cells. J. Virol. 2004;78:6556–6566. doi: 10.1128/JVI.78.12.6556-6566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essner J.J. McIvor R.S. Hackett P.B. Awakening gene therapy with Sleeping Beauty transposons. Curr. Opin. Pharmacol. 2005;5:513–519. doi: 10.1016/j.coph.2005.04.015. [DOI] [PubMed] [Google Scholar]
- Fitzsimons H.L. Bland R.J. During M.J. Promoters and regulatory elements that improve adeno-associated virus transgene expression in the brain. Methods. 2002;28:227–236. doi: 10.1016/s1046-2023(02)00227-x. [DOI] [PubMed] [Google Scholar]
- Gaspar H.B. Bjorkegren E. Parsley K. Gilmour K.C. King D. Sinclair J. Zhang F. Giannakopoulos A. Adams S. Fairbanks L.D. Gaspar J. Henderson L. Xu-Bayford J.H. Davies E.G. Veys P.A. Kinnon C. Thrasher A.J. Successful reconstitution of immunity in ADA-SCID by stem cell gene therapy following cessation of PEG-ADA and use of mild preconditioning. Mol. Ther. 2006;14:505–513. doi: 10.1016/j.ymthe.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Geurts A.M. Yang Y. Clark K.J. Liu G. Cui Z. Dupuy A.J. Bell J.B. Largaespada D.A. Hackett P.B. Gene transfer into genomes of human cells by the Sleeping Beauty transposon system. Mol. Ther. 2003;8:108–117. doi: 10.1016/s1525-0016(03)00099-6. [DOI] [PubMed] [Google Scholar]
- Haas D.L. Lutzko C. Logan A.C. Cho G.J. Skelton D. Jin Yu X. Pepper K.A. Kohn D.B. The Moloney murine leukemia virus repressor binding site represses expression in murine and human hematopoietic stem cells. J. Virol. 2003;77:9439–9450. doi: 10.1128/JVI.77.17.9439-9450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacein-Bey-Abina S. Von Kalle C. Schmidt M. McCormack M.P. Wulffraat N. Leboulch P. Lim A. Osborne C.S. Pawliuk R. Morillon E. Sorensen R. Forster A. Fraser P. Cohen J.I. De Saint Basile G. Alexander I. Wintergerst U. Frebourg T. Aurias A. Stoppa-Lyonnet D. Romana S. Radford-Weiss I. Gross F. Valensi F. Delabesse E. Macintyre E. Sigaux F. Soulier J. Leiva L.E. Wissler M. Prinz C. Rabbitts T.H. Le Deist F. Fischer A. Cavazzana-Calvo M. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- Hackett P.B. Integrating DNA vectors for gene therapy. Mol. Ther. 2007;15:10–12. doi: 10.1038/sj.mt.6300065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett P.B. Ekker S.C. Largaespada D.A. McIvor R.S. Sleeping Beauty transposon-mediated gene therapy for prolonged expression. Adv. Genet. 2005;54:189–232. doi: 10.1016/S0065-2660(05)54009-4. [DOI] [PubMed] [Google Scholar]
- Halene S. Wang L. Cooper R.M. Bockstoce D.C. Robbins P.B. Kohn D.B. Improved expression in hematopoietic and lymphoid cells in mice after transplantation of bone marrow transduced with a modified retroviral vector. Blood. 1999;94:3349–3357. [PMC free article] [PubMed] [Google Scholar]
- Hao Q.L. Shah A.J. Thiemann F.T. Smogorzewska E.M. Crooks G.M. A functional comparison of CD34+ CD38− cells in cord blood and bone marrow. Blood. 1995;86:3745–3753. [PubMed] [Google Scholar]
- Hartl D.L. Lohe A.R. Lozovskaya E.R. Regulation of the transposable element mariner. Genetica. 1997;100:177–184. [PubMed] [Google Scholar]
- Hohenstein K.A. Pyle A.D. Chern J.Y. Lock L.F. Donovan P.J. Nucleofection mediates high-efficiency stable gene knockdown and transgene expression in human embryonic stem cells. Stem Cells. 2008;26:1436–1443. doi: 10.1634/stemcells.2007-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis R.P. Nightingale S.J. Wang X. Pepper K.A. Yu X.J. Barsky L. Crooks G.M. Kohn D.B. Stable gene transfer to human CD34+ hematopoietic cells using the Sleeping Beauty transposon. Exp. Hematol. 2006;34:1333–1343. doi: 10.1016/j.exphem.2006.05.023. [DOI] [PubMed] [Google Scholar]
- Horwitz M.E. Barrett A.J. Brown M.R. Carter C.S. Childs R. Gallin J.I. Holland S.M. Linton G.F. Miller J.A. Leitman S.F. Read E.J. Malech H.L. Treatment of chronic granulomatous disease with nonmyeloablative conditioning and a T-cell-depleted hematopoietic allograft. N. Engl. J. Med. 2001;344:881–888. doi: 10.1056/NEJM200103223441203. [DOI] [PubMed] [Google Scholar]
- Huang X. Wilber A.C. Bao L. Tuong D. Tolar J. Orchard P.J. Levine B.L. June C.H. McIvor R.S. Blazar B.R. Zhou X. Stable gene transfer and expression in human primary T cells by the Sleeping Beauty transposon system. Blood. 2006;107:483–491. doi: 10.1182/blood-2005-05-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F. Yasukawa M. Lyons B. Yoshida S. Miyamoto T. Yoshimoto G. Watanabe T. Akashi K. Shultz L.D. Harada M. Development of functional human blood and immune systems in NOD/SCID/IL2 receptor γ chainnull mice. Blood. 2005;106:1565–1573. doi: 10.1182/blood-2005-02-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M. Hiramatsu H. Kobayashi K. Suzue K. Kawahata M. Hioki K. Ueyama Y. Koyanagi Y. Sugamura K. Tsuji K. Heike T. Nakahata T. NOD/SCID/γcnull mouse: An excellent recipient mouse model for engraftment of human cells. Blood. 2002;100:3175–3182. doi: 10.1182/blood-2001-12-0207. [DOI] [PubMed] [Google Scholar]
- Ivics Z. Hackett P.B. Plasterk R.H. Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Izsvak Z. Ivics Z. Sleeping beauty transposition: Biology and applications for molecular therapy. Mol. Ther. 2004;9:147–156. doi: 10.1016/j.ymthe.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Izsvak Z. Ivics Z. Plasterk R.H. Sleeping Beauty, a wide host-range transposon vector for genetic transformation in vertebrates. J. Mol. Biol. 2000;302:93–102. doi: 10.1006/jmbi.2000.4047. [DOI] [PubMed] [Google Scholar]
- Kaiser J. Gene therapy: Seeking the cause of induced leukemias in X-SCID trial. Science. 2003;299:495. doi: 10.1126/science.299.5606.495. [DOI] [PubMed] [Google Scholar]
- Kay M.A. Baley P. Rothenberg S. Leland F. Fleming L. Ponder K.P. Liu T. Finegold M. Darlington G. Pokorny W. Woo S.L.C. Expression of human α1-antitrypsin in dogs after autologous transplantation of retroviral transduced hepatocytes. Proc. Natl. Acad. Sci. U.S.A. 1992;89:89–93. doi: 10.1073/pnas.89.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keravala A. Calos M.P. Site-specific chromosomal integration mediated by ϕC31 integrase. Methods Mol. Biol. 2008;435:165–173. doi: 10.1007/978-1-59745-232-8_12. [DOI] [PubMed] [Google Scholar]
- Kohn D.B. Candotti F. Gene therapy fulfilling its promise. N. Eng. J. Med. 2009;360:518–521. doi: 10.1056/NEJMe0809614. [DOI] [PubMed] [Google Scholar]
- Lampe D.J. Grant T.E. Robertson H.M. Factors affecting transposition of the Himar1 mariner transposon in vitro. Genetics. 1998;149:179–187. doi: 10.1093/genetics/149.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G. Geurts A.M. Yae K. Srinivasan A.R. Fahrenkrug S.C. Largaespada D.A. Takeda J. Horie K. Olson W.K. Hackett P.B. Target-site preferences of Sleeping Beauty transposons. J. Mol. Biol. 2005;346:161–173. doi: 10.1016/j.jmb.2004.09.086. [DOI] [PubMed] [Google Scholar]
- Liu H. Visner G.A. Applications of Sleeping Beauty transposons for nonviral gene therapy. IUBMB Life. 2007;59:374–379. doi: 10.1080/15216540701435722. [DOI] [PubMed] [Google Scholar]
- Liu L. Liu H. Mah C. Fletcher B.S. Indoleamine 2,3-dioxygenase attenuates inhibitor development in gene-therapy-treated hemophilia A mice. Gene Ther. 2009;16:724–733. doi: 10.1038/gt.2009.13. [DOI] [PubMed] [Google Scholar]
- Lohe A.R. Sullivan D.T. Hartl D.L. Subunit interactions in the mariner transposase. Genetics. 1996;144:1087–1095. doi: 10.1093/genetics/144.3.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardo A. Genovese P. Beausejour C.M. Colleoni S. Lee Y.L. Kim K.A. Ando D. Urnov F.D. Galli C. Gregory P.D. Holmes M.C. Naldini L. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat. Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- Metais J.Y. Dunbar C.E. The MDS1–EVI1 gene complex as a retrovirus integration site: Impact on behavior of hematopoietic cells and implications for gene therapy. Mol. Ther. 2008;16:439–449. doi: 10.1038/sj.mt.6300372. [DOI] [PubMed] [Google Scholar]
- Miyoshi H. Smith K.A. Mosier D.E. Verma I.M. Torbett B.E. Transduction of human CD34+ cells that mediate long-term engraftment of NOD/SCID mice by HIV vectors. Science. 1999;283:682–686. doi: 10.1126/science.283.5402.682. [DOI] [PubMed] [Google Scholar]
- Mizushima S. Nagata S. pEF-BOS, a powerful mammalian expression vector. Nucleic Acids Res. 1990;18:5322. doi: 10.1093/nar/18.17.5322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M. Reid K. Williams D.E. Lyman S.D. Bartlett P.F. Steel factor is required for maintenance, but not differentiation, of melanocyte precursors in the neural crest. Dev. Biol. 1992;153:396–401. doi: 10.1016/0012-1606(92)90124-y. [DOI] [PubMed] [Google Scholar]
- Murray L.J. Young J.C. Osborne L.J. Luens K.M. Scollay R. Hill B.L. Thrombopoietin, Flt3, and Kit ligands together suppress apoptosis of human mobilized CD34+ cells and recruit primitive CD34+ Thy-1+ cells into rapid division. Exp. Hematol. 1999;27:1019–1028. doi: 10.1016/s0301-472x(99)00031-4. [DOI] [PubMed] [Google Scholar]
- Nakai H. Herzog R.W. Hagstrom J.N. Walter J. Kung S.H. Yang E.Y. Tai S.J. Iwaki Y. Kurtzman G.J. Fisher K.J. Colosi P. Couto L.B. High K.A. Adeno-associated viral vector-mediated gene transfer of human blood coagulation factor IX into mouse liver. Blood. 1998;91:4600–4607. [PubMed] [Google Scholar]
- Nakai H. Fuess S. Storm T.A. Meuse L.A. Kay M.A. Free DNA ends are essential for concatemerization of synthetic double-stranded adeno-associated virus vector genomes transfected into mouse hepatocytes in vivo. Mol. Ther. 2003;7:112–121. doi: 10.1016/s1525-0016(02)00034-5. [DOI] [PubMed] [Google Scholar]
- Nakata Y. Shetzline S. Sakashita C. Kalota A. Rallapalli R. Rudnick S.I. Zhang Y. Emerson S.G. Gewirtz A.M. c-Myb contributes to G2/M cell cycle transition in human hematopoietic cells by direct regulation of cyclin B1 expression. Mol. Cell. Biol. 2007;27:2048–2058. doi: 10.1128/MCB.01100-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nightingale S.J. Hollis R.P. Pepper K.A. Petersen D. Yu X.J. Yang C. Bahner I. Kohn D.B. Transient gene expression by nonintegrating lentiviral vectors. Mol. Ther. 2006;13:1121–1132. doi: 10.1016/j.ymthe.2006.01.008. [DOI] [PubMed] [Google Scholar]
- Ott M.G. Schmidt M. Schwarzwaelder K. Stein S. Siler U. Koehl U. Glimm H. Kuhlcke K. Schilz A. Kunkel H. Naundorf S. Brinkmann A. Deichmann A. Fischer M. Ball C. Pilz I. Dunbar C. Du Y. Jenkins N.A. Copeland N.G. Luthi U. Hassan M. Thrasher A.J. Hoelzer D. Von Kalle C. Seger R. Grez M. Correction of X-linked chronic granulomatous disease by gene therapy, augmented by insertional activation of MDS1-EVI1, PRDM16 or SETBP1. Nat. Med. 2006;12:401–409. doi: 10.1038/nm1393. [DOI] [PubMed] [Google Scholar]
- Park C.Y. Majeti R. Weissman I.L. In vivo evaluation of human hematopoiesis through xenotransplantation of purified hematopoietic stem cells from umbilical cord blood. Nat. Protocols. 2008;3:1932–1940. doi: 10.1038/nprot.2008.194. [DOI] [PubMed] [Google Scholar]
- Porteus M.H. Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Porteus M.H. Carroll D. Gene targeting using zinc finger nucleases. Nat. Biotechnol. 2005;23:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- Ramezani A. Hawley T.S. Hawley R.G. Lentiviral vectors for enhanced gene expression in human hematopoietic cells. Mol. Ther. 2000;2:458–469. doi: 10.1006/mthe.2000.0190. [DOI] [PubMed] [Google Scholar]
- Sands M.S. Barker J.E. Vogler C. Levy B. Gwynn B. Galvin N. Sly W.S. Birkenmeier E. Treatment of murine mucopolysaccharidosis type VII by syngeneic bone marrow transplantation in neonates. Lab. Invest. 1993;68:676–686. [PubMed] [Google Scholar]
- Scharfmann R. Axelrod J.H. Verma I.M. Long-term in vivo expression of retrovirus-mediated gene transfer in mouse fibroblast implants. Proc. Natl. Acad. Sci. U.S.A. 1991;88:4626–4630. doi: 10.1073/pnas.88.11.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taboit-Dameron F. Malassagne B. Viglietta C. Puissant C. Leroux-Coyau M. Chereau C. Attal J. Weill B. Houdebine L.M. Association of the 5′ HS4 sequence of the chicken β-globin locus control region with human EF1α gene promoter induces ubiquitous and high expression of human CD55 and CD59 cDNAs in transgenic rabbits. Transgenic Res. 1999;8:223–235. doi: 10.1023/a:1008919925303. [DOI] [PubMed] [Google Scholar]
- Ueda T. Tsuji K. Yoshino H. Ebihara Y. Yagasaki H. Hisakawa H. Mitsui T. Manabe A. Tanaka R. Kobayashi K. Ito M. Yasukawa K. Nakahata T. Expansion of human NOD/SCID-repopulating cells by stem cell factor, Flk2/Flt3 ligand, thrombopoietin, IL-6, and soluble IL-6 receptor. J. Clin. Invest. 2000;105:1013–1021. doi: 10.1172/JCI8583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetsuki T. Naito A. Nagata S. Kaziro Y. Isolation and characterization of the human chromosomal gene for polypeptide chain elongation factor-1α. J. Biol. Chem. 1989;264:5791–5798. [PubMed] [Google Scholar]
- Von Levetzow G. Spanholtz J. Beckmann J. Fischer J. Kogler G. Wernet P. Punzel M. Giebel B. Nucleofection, an efficient nonviral method to transfer genes into human hematopoietic stem and progenitor cells. Stem Cells Dev. 2006;15:278–285. doi: 10.1089/scd.2006.15.278. [DOI] [PubMed] [Google Scholar]
- Walisko O. Ivics Z. Interference with cell cycle progression by parasitic genetic elements: Sleeping Beauty joins the club. Cell Cycle. 2006;5:1275–1280. doi: 10.4161/cc.5.12.2888. [DOI] [PubMed] [Google Scholar]
- Walisko O. Schorn A. Rolfs F. Devaraj A. Miskey C. Izsvak Z. Ivics Z. Transcriptional activities of the Sleeping Beauty transposon and shielding its genetic cargo with insulators. Mol. Ther. 2007;16:359–369. doi: 10.1038/sj.mt.6300366. [DOI] [PubMed] [Google Scholar]
- Weber E.L. Cannon P.M. Promoter choice for retroviral vectors: Transcriptional strength versus trans-activation potential. Human Gene Ther. 2007;18:849–860. doi: 10.1089/hum.2007.067. [DOI] [PubMed] [Google Scholar]
- Wiehe J.M. Ponsaerts P. Rojewski M.T. Homann J.M. Greiner J. Kronawitter D. Schrezenmeier H. Hombach V. Wiesneth M. Zimmermann O. Torzewski J. mRNA-mediated gene delivery into human progenitor cells promotes highly efficient protein expression. J. Cell. Mol. Med. 2007;11:521–530. doi: 10.1111/j.1582-4934.2007.00038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilber A. Linehan J.L. Tian X. Woll P.S. Morris J.K. Belur L.R. McIvor R.S. Kaufman D.S. Efficient and stable transgene expression in human embryonic stem cells using transposon-mediated gene transfer. Stem Cells. 2007;25:2919–2927. doi: 10.1634/stemcells.2007-0026. [DOI] [PubMed] [Google Scholar]
- Wilson M.H. Kaminski J.M. George A.L., Jr. Functional zinc finger/sleeping beauty transposase chimeras exhibit attenuated overproduction inhibition. FEBS Lett. 2005;579:6205–6209. doi: 10.1016/j.febslet.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Wilson M.H. Coates C.J. George A.L., Jr. PiggyBac transposon-mediated gene transfer in human cells. Mol. Ther. 2007;15:139–145. doi: 10.1038/sj.mt.6300028. [DOI] [PubMed] [Google Scholar]
- Wu M.H. Smith S.L. Danet G.H. Lin A.M. Williams S.F. Liebowitz D.N. Dolan M.E. Optimization of culture conditions to enhance transfection of human CD34+ cells by electroporation. Bone Marrow Transplant. 2001a;27:1201–1209. doi: 10.1038/sj.bmt.1703054. [DOI] [PubMed] [Google Scholar]
- Wu M.H. Smith S.L. Dolan M.E. High efficiency electroporation of human umbilical cord blood CD34+ hematopoietic precursor cells. Stem Cells. 2001b;19:492–499. doi: 10.1634/stemcells.19-6-492. [DOI] [PubMed] [Google Scholar]
- Xue X. Huang X. Nodland S.E. Mates L. Ma L. Izsvak Z. Ivics Z. Lebien T.W. McIvor R.S. Wagner J.E. Zhou X. Stable gene transfer and expression in cord blood-derived CD34+ hematopoietic stem and progenitor cells by a hyperactive Sleeping Beauty transposon system. Blood. 2009;114:1319–1330. doi: 10.1182/blood-2009-03-210005. [DOI] [PubMed] [Google Scholar]
- Yanez-Munoz R.J. Balaggan K.S. MacNeil A. Howe S.J. Schmidt M. Smith A.J. Buch P. MacLaren R.E. Anderson P.N. Barker S.E. Duran Y. Bartholomae C. Von Kalle C. Heckenlively J.R. Kinnon C. Ali R.R. Thrasher A.J. Effective gene therapy with nonintegrating lentiviral vectors. Nat. Med. 2006;12:348–353. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- Yant S.R. Meuse L. Chiu W. Ivics Z. Izsvak Z. Kay M.A. Somatic integration and long-term transgene expression in normal and haemophilic mice using a DNA transposon system. Nat. Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- Yant S.R. Park J. Huang Y. Mikkelsen J.G. Kay M.A. Mutational analysis of the N-terminal DNA-binding domain of Sleeping Beauty transposase: Critical residues for DNA binding and hyperactivity in mammalian cells. Mol. Cell. Biol. 2004;24:9239–9247. doi: 10.1128/MCB.24.20.9239-9247.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant S.R. Wu X. Huang Y. Garrison B. Burgess S.M. Kay M.A. High-resolution genome-wide mapping of transposon integration in mammals. Mol. Cell. Biol. 2005;25:2085–2094. doi: 10.1128/MCB.25.6.2085-2094.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Z.Q. Qiu P. Burkholder J.K. Turner J. Culp J. Roberts T. Shahidi N.T. Yang N.S. Cytokine transgene expression and promoter usage in primary CD34+ cells using particle-mediated gene delivery. Hum. Gene Ther. 1998;9:2197–2205. doi: 10.1089/hum.1998.9.15-2197. [DOI] [PubMed] [Google Scholar]
- Zeitelhofer M. Vessey J.P. Xie Y. Tubing F. Thomas S. Kiebler M. Dahm R. High-efficiency transfection of mammalian neurons via nucleofection. Nat. Protocols. 2007;2:1692–1704. doi: 10.1038/nprot.2007.226. [DOI] [PubMed] [Google Scholar]
- Zeitelhofer M. Karra D. Vessey J.P. Jaskic E. Macchi P. Thomas S. Riefler J. Kiebler M. Dahm R. High-efficiency transfection of short hairpin RNAs-encoding plasmids into primary hippocampal neurons. J. Neurosci. Res. 2009;87:289–300. doi: 10.1002/jnr.21840. [DOI] [PubMed] [Google Scholar]