ABSTRACT
The basic tenets of germ cell development are conserved among metazoans. Following lineage commitment in the embryo, germ cells proliferate, transition into meiosis, and then differentiate into gametes capable of fertilization. In lower organisms such as Drosophila and C. elegans, germline stem cells make the decision to proliferate or enter meiosis based in large part on the regulated expression of genes by translational control. This study undertakes a direct characterization of mRNAs that experience translational control and their involvement in similar decisions in the mammalian testis. We previously showed that translation of mRNA encoding the germ cell-specific gene Rhox13 was suppressed in the fetal and neonatal testis. By investigating changes in message utilization during neonatal testis development, we found that a large number of mRNAs encoding both housekeeping and germ cell-specific proteins experience enhanced translational efficiency, rather than increase in abundance, in the testis as quiescent gonocytes transition to mitotic spermatogonia. Our results indicate that translational control is a significant regulator of the germ cell proteome during neonatal testis development.
Keywords: gonocyte, spermatogenesis, spermatogonia, testis, translation
A developmental increase in cap-dependent translation in neonatal male germ cells accompanies enhanced translation of specific mRNAs.
INTRODUCTION
The sequence of gene regulatory events that drive fetal and neonatal germ cell development in the mammalian testis remains somewhat of a mystery. Primordial germ cells become gonocytes (also called prospermatogonia) after sex determination, proliferate briefly, and then become arrested in G0/G1 from 12.5 to 14.5 days postcoitum until after birth [1, 2]. By 2–3 days postpartum (dpp), gonocytes resume mitotic divisions and migrate to the periphery of the testis cords as they transition to spermatogonia [3]. These spermatogonia then divide asynchronously, either remaining as stem cell spermatogonia (type As) or dividing (Apr, Aal) to differentiate (A1–4) into type B spermatogonia that will become preleptotene spermatocytes by 10 dpp to enter meiosis [4]. Currently, little is known about the signaling pathways and gene expression changes responsible for the developmental transition from quiescent gonocytes to proliferating spermatogonia. Microarray studies revealed that ∼50 genes were upregulated ≥2-fold in the testis from 0 to 3 dpp [5]. The relatively constant mRNA pool in these rapidly changing cells suggests that changes in transcription do not play a major role in this transition.
Once in the cytoplasm, mRNAs become translated, stored, or degraded. The decision among these fates provides the cell with an important level of posttranscriptional control over gene expression that allows for rapid and large-scale responses to stimuli during periods of change such as proliferation or differentiation. In response to differentiation cues, mRNAs readily switch fates and become mobilized from stored messenger ribonucleoprotein particles (mRNPs) into a translated state depending on the needs of the cell [6, 7]. Changes in global translation are typically mediated through phosphorylation-mediated availability of initiation factor EIF2A, which mediates all protein synthesis initiation events equally [8]. Greater availability of EIF4E, the mRNA cap-binding protein, preferentially stimulates “cap-dependent” initiation events that recruit mRNAs with long, complex 5′ untranslated regions (UTRs). In the case of EIF4E regulation, the repressor binding protein EIF4EBP1 binds to EIF4E and prevents its association with the scaffolding protein EIF4G. Phosphorylation of EIF4EBP1 releases EIF4E, which binds to EIF4G and recruits the 43S ribosomal complex to mRNA during cap-dependent translation, generally in response to mitogenic stimuli [9]. EIF2A phosphorylation results in an overall decrease in protein synthetic activity due to a paucity of tRNA-Met delivery. Under these restrictive conditions, however, certain mRNAs experience increased relative translation by enhancing ribosome-scanning activity through 5′ UTRs that contain upstream open reading frames (uORFs) in order to reach the authentic initiation codon. Hence, the inverse regulation of EIF2A and EIF4E by separate phosphorylation events provides broad regulation of translational control at both a global and mRNA-specific level.
One further mode of translational control resides in the mRNPs themselves. A program of translational repression of mRNAs by UTR binding proteins is prevalent in many meiotic cell types [10]. The role of mRNA repression in meiotic and postmeiotic spermatogenic cells in adult mice has been studied for many years [11–14]. However, several independent lines of evidence implicate translational control in fetal and neonatal male germ cell development. First, a number of essential RNA binding proteins (RBPs) have been identified (NANOS2, NANOS3, DAZL, TIAR/TIAL1, PIWIL2/MILI, PIWIL4/MIWI2, and DDX4/VASA), with roles in either repression or activation of mRNA translation during germ cell development and differentiation [15–19]. Loss of these translational control events has dire consequences for germ cell progression. Similar RBPs have been shown to control C. elegans germline stem cell proliferation, including NOS-3, FOG-1, FOG-3, ATX-2, EGO-1, PRG-1, PRG-2, and multiple members of the FBF, MOG, and GLD families [20]. Specific derepression of mRNA targets for such RBPs at later stages of germ cell development occurs in response to unique translation–initiation-factor isoforms and mRNA polyadenylation [20–23]. Second, fetal and neonatal mouse gonocytes contain numerous electron-dense perinuclear “nuage,” functionally similar to germ cell granules in Drosophila, Xenopus, and C. elegans, which contain both RNA and protein components. These are present in germ cells of at least 80 different species [24, 25]. These large mRNPs are involved in mRNA metabolism, including genomic defense against transposable elements, which are demethylated and transcribed in gonocytes [26–28]. Third, we previously found that mRNAs from the X-linked reproductive homeobox gene 13 (Rhox13) remain translationally repressed in the fetal and neonatal testis through an interaction with the essential RNA binding protein NANOS2, but that this repression was relieved by 3–4 dpp [29]. Each of these evidences implicates translational control as a means to regulate germ cell gene expression. Together they suggest that translational control regulates molecular events that promote gonocyte differentiation in fetal testes.
In this study, we examine translational control as a regulator of gene expression during the gonocyte-to-spermatogonia transition in mouse neonatal testis. We identify a number of mRNAs encoding both germ cell-specific and housekeeping mRNAs that become more efficiently translated when recruited into polyribosomes (polysomes) from 1 to 4 dpp. These changes accompany increased phosphorylation of EIF4EBP1 and a decreased phosphorylation of EIF2A, which are hallmarks of stimulated cap-dependent translation. The increase in EIF4EBP1 phosphorylation is most evident in germ cells. Our results suggest that the neonatal germ cell proteome is determined in large part by changes in mRNA translation efficiency. We identify a number of mRNAs encoding both germ cell-specific and housekeeping genes that become more efficiently translated by recruitment to polysomes during neonatal testis development.
MATERIALS AND METHODS
Animal Care
All animal procedures were performed in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals and approved by the Animal Care and Use Committee of East Carolina University (AUPs A178, A181). CD-1 mice (Charles River Laboratories) were used for all analyses. Euthanasia was performed by decapitation, and testes were snap frozen in liquid nitrogen and stored at −80°C.
Preparation of Cytoplasmic Lysates
Testes were harvested and washed with 1× ice-cold PBS containing cycloheximide (100 μg/ml) to freeze polysomes, immediately snap frozen in liquid nitrogen, and stored at −80°C. Testes were pulverized under liquid nitrogen and lysed in 1 ml polysome lysis buffer (100 mM KCl, 5 mM MgCl2, 10 mM HEPES [pH 7.4], 0.5% NP-40 [Sigma], and 100 μg/ml cycloheximide). Lysates were homogenized by passing the sample five times through a 25-gauge needle, and nuclei were then pelleted by centrifugation at 16 000 × g for 5 min at 4°C. Supernatants were transferred to clean 1.5-ml tubes and stored on ice. A minimum of 28, 22, 20, and 10 testes were used from mice euthanized at 1, 3, 4, and 8 dpp, respectively.
Polysome Gradient Analysis
Separation of polysomes, ribosomal subunits, and initiation complexes was accomplished by sucrose gradient sedimentation. Cytoplasmic extracts (2.5–3.0 mg of protein, measured with BCA Protein Assay; Pierce) were layered onto a 15%–45% linear sucrose density gradient in polysome gradient buffer (100 mM KCl, 5 mM MgCl2, and 10 mM HEPES [pH 7.4]) and then centrifuged at 210 000 × g at 4°C for 2 h without braking. Gradients were then fractionated using an ISCO gradient-fractionation system equipped with a UA-6 detector (ISCO). RNP fractions, 40S, 60S, 80S, light polysomes, and heavy polysomes were monitored by continuous ultraviolet absorption profiles at 254 nm. Fourteen successive 750-μl fractions were collected and stored at −80°C. Total RNA was isolated from each polysome fraction by TRIzol extraction (Invitrogen) using the manufacturer's instructions. Polysome gradients using testis lysates from 1, 3, 4, and 8 dpp mice were done in triplicate.
Quantitative RT-PCR
Quantitative RT-PCR (qRT-PCR) analyses were performed with RNA isolated from the polysome fractions. RNA (100 ng) was subjected to reverse transcription and qPCR in the same reaction tube using iScript One-Step RT-PCR kit with SYBR green (BioRad). Amplification and detection of specific gene products were performed using the iCycler IQ real-time PCR detection system (BioRad). Threshold temperatures were selected automatically, and all amplifications were followed by melt-curve analysis. The sequences of the specific primers are provided in Table 1.
TABLE 1.
Quantitative RT-PCR primers.

The following comparative quantitation equation was used to calculate relative mRNA levels per fraction: mRNA level = (2dCt), where dCt = (Ctgene of interest) – (Ctmin), where Ctmin = the minimum Ct value within the gene set and controls. For qRT-PCR on total RNA, Ct values were normalized to Rpl19 and relative fold changes presented (with value from 1 dpp set to 1).
Immunoblotting
Immunoblotting of polysome fractions was carried out by conventional methods. Ten microliters of each fraction was added directly to a modified sample buffer (final concentration of 50 mM dithiothreitol, 2% SDS, and 10% glycerol). Primary antibodies used were against DDX6 (1:1000; 14632-1-AP; ProteinTech), RPS6 (1:2000; 2217S; Cell Signaling Technology), RHOX13 (1:1,000) [30], phospho-EIF4EBP1 (1:3000; 2855; Cell Signaling Technology), EIF4EBP1 (1:1000; 9452; Cell Signaling Technology), and phospho-EIF2A (1:1000; 9721; Cell Signaling Technology) and were incubated overnight at 4°C. Secondary anti-rabbit-horseradish peroxidase (1:3000; 7074S; Cell Signaling Technology) was incubated for 1 h at room temperature. Blots were developed using LumiGlo detection reagent (Cell Signaling Technology).
Immunohistochemistry and Indirect Immunofluorescence
Immunohistochemistry (IHC) was performed as described previously [29]. Experiments were repeated at least thrice, and at least two gonads were analyzed from different mice. Cell types were identified based on characteristic morphological differences, and germ cell identities were verified in testis sections stained for either POU5F1 (0 and 3 dpp) or RHOX13 (4 and 8 dpp). Anti-POU5F1 (1:500; SC-8629; Santa Cruz Biotechnology, Inc.) and anti-RHOX13 (1:500) [30] were applied for 1 h at room temperature without antigen retrieval. Primary antibody was omitted as a negative control. Detection was performed using a Vectastain ABC kit (Vector Laboratories).
Immunofluorescence (IIF) was performed using standard methods. Briefly, 8 μm of 4% paraformaldehyde-fixed and compound-embedded frozen sections were blocked for 30 min at room temperature, incubated in primary antibody (1:500) overnight at 4°C, and incubated in secondary antibody (1:1000; Alexa Fluor-488 donkey anti-rabbit IgG; Invitrogen) plus phalloidin-594 (1:1000; Invitrogen) for 1 h at room temperature. Blocking and antibody incubations were done in 1× PBS containing 3% bovine serum albumin + 0.1% Triton X-100, and stringency washes were done with 1× PBS + 0.1% Triton X-100. Primary antibody was omitted as a negative control. Coverslips were mounted with Vectastain containing DAPI (4′,6-diamidino-2-phenylindole; Vector Laboratories), and images were obtained using a Fluoview FV1000 confocal laser scanning microscope (Olympus America).
Translation State Array Analysis
Extracts of testes from 1 dpp and 4 dpp CD-1 mice were fractionated over a 15%–45% sucrose gradient in triplicate as described above. The position of the 80S peak defined the boundary between translating and nontranslating mRNAs. Pooled nonpolysomal (nontranslating, fractions 1–3) and heavy polysomal (efficiently translating, fractions 7–13) RNAs were isolated from sucrose gradient fractions by TRIzol extraction (Invitrogen) as per the manufacturer's suggested protocol, with a few modifications. Following the initial phase separation, the aqueous layer was transferred to a clean microfuge tube, and 200 μl of chloroform was added and mixed. Samples were centrifuged at 12 000 × g for 10 min at 4°C, and the aqueous layers were transferred to a clean microfuge tube. This chloroform wash was repeated twice, and the TRIzol extraction was then resumed. RNA was recovered by ethanol precipitation. Biotin-labeled amplified antisense RNA was synthesized from 5 μg of RNA by the standard Affymetrix 3′ IVT procedure and hybridized to the Affymetrix 430 2.0 mouse expression array (Affymetrix) at the core facility at the University of North Carolina, Neuroscience Research Center, Chapel Hill, NC. Microarray raw data were normalized using the robust multichip average method. The raw intensity values were background adjusted, log2 transformed, and then quantile normalized [31]. The data are summarized as signal intensities for each probe set on each array. Increased RNA polysome distribution between 1 dpp and 4 dpp mouse testes was determined by first averaging the triplicate array values for each probe set. Then, the resultant mean signal intensities were analyzed to determine fold change of 4 dpp heavy polysomal-1 dpp heavy polysomal RNA. Genes with mRNA signal intensity differences of ≥2 were analyzed by Ingenuity Pathway Analysis (Ingenuity Systems).
Statistics
Statistical analyses of the qRT-PCR results were performed using the Student t-test, and the level of significance was set at P < 0.01.
RESULTS
Polysome Gradient Analysis Reveals Translational Activation During Neonatal Testis Development
We previously discovered that mRNAs for the germ cell-specific Rhox13 gene were present in the fetal and neonatal testis without readily detectable protein [29]. There are several possibilities that might account for the suppressed translation of Rhox13 mRNAs in the neonatal testis until 3–4 dpp: 1) sequestration in RNP particles, which would preclude association with ribosomes; 2) association with very few ribosomes, indicating inefficient translation; and 3) efficient production of RHOX13 protein that is then rapidly degraded. To distinguish between these possibilities, we assessed the translational efficiency of specific mRNAs during neonatal testis development by direct assay on polysome gradients. Testis lysates from 1, 3, 4, and 8 dpp mice were resolved on sucrose gradients, fractionated, and protein and RNA extracted (n = 3 different gradients). These lysates were prepared from the testis at developmental times when Rhox13 mRNAs are present but RHOX13 protein 1) is not readily detectable (1 dpp), 2) is detectable in a subset of germ cells (3–4 dpp), and 3) is present in a majority of germ cells (8 dpp) [29]. To confirm resolution of the nontranslating from ribosome-containing fractions, immunoblotting for DDX6, an RNA helicase present in RNPs, and RPS6, a 40S ribosomal protein, was conducted across all fractions. As expected, DDX6 was present in ribosome-free fractions 1 and 2 [32], while RPS6 was concentrated in fractions 3–7 that contain ribosomal subunits, and was less abundant in the polysome fractions 8–13 (Fig. 1).
FIG. 1.
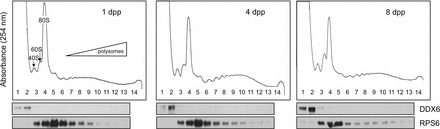
Absorbance (A254) profiles for linear sucrose gradient fractionation of 1, 4, and 8 dpp mouse testis lysates. The monosome (80S) is the highest resolved peak, and each peak to the right represents the addition of one ribosome to mRNAs (polysomes). Immunoblots below each profile were performed using antibodies against DDX6 and RPS6, with total protein isolated from the corresponding fraction noted above each lane (1–13).
The distribution of individual mRNAs across the polysome gradient provides a direct measure of their absolute translation efficiency. We quantified specific mRNAs in each gradient fraction encoding housekeeping genes (Pgk1 and Actb), well-characterized representative germ cell-specific genes (Pou5f1/Oct4 and Stra8), and Rhox13, and compared the distribution for each gene's mRNA separately. The majority of Pgk1 and Actb mRNAs were present in the medium-density or light-polysome fractions in testes from 1 dpp mice, respectively, but were noticeably redistributed to heavy polysomes in 3, 4, and 8 dpp testes (Fig. 2), indicating enhanced efficiency of translation of these housekeeping mRNAs. Pou5f1 encodes a transcription factor expressed in gonocytes and undifferentiated spermatogonia [33]. Approximately 80% of its mRNA was associated with light polysomes at all stages examined, indicating consistently low translational efficiency. Stra8 is first transcribed in response to retinoic acid in the neonatal testis [34–36]. Its mRNA was nearly undetectable at 1 and 3 dpp, but in 4 and 8 dpp testes, the majority of its newly expressed mRNA was associated with heavy polysomes, indicating efficient translation. Nearly 60% of Rhox13 mRNA was found to translate inefficiently on very light polysomes (monosomes) at 1 dpp. By 3 dpp, however, the majority of Rhox13 mRNA was concentrated in light and medium polysomes (52% and 30% relative mRNA levels, respectively, with peaks over fractions 7 and 9). By 4 and then 8 dpp, the majority of Rhox13 messages shifted into heavy polysomes, indicting very efficient translation. This approximately 3-fold redistribution of Rhox13 mRNAs into the heavy polysomal fraction from 1 to 4 dpp corresponds precisely with detection of de novo RHOX13 protein in the neonatal testis at 3–4 dpp [29]. Similar mRNA recruitments from light to heavy polysomes after 1 dpp were observed for Gata2 and Taf7l mRNAs (data not shown), which are also repressed in association with NANOS2 in the fetal testis [37].
FIG. 2.
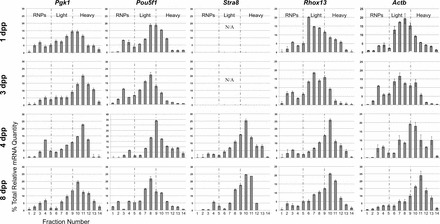
Quantitative RT-PCR analysis of translational status of mRNAs during neonatal testis development. RNA was isolated from sucrose gradient fractions from 1, 3, 4, and 8 dpp mouse testis lysates (ages indicated to the left), and Pgk1, Pou5f1, Stra8, Rhox13, and Actb mRNA levels were measured by qRT-PCR. Each graph is partitioned into three translationally distinct parts (from left to right): RNPs (not translated) and light (inefficiently translated) and heavy (efficiently translated) polysome fractions, based on the corresponding absorbance profiles (see Fig. 1). N/A indicates that Stra8 mRNA was not detectable from these gradient fractions. Error bars represent standard deviation.
Assessment of Total mRNA Levels for Representative Genes in the Neonatal Testis
Several of the mRNAs (Pgk1, Actb, and Rhox13) became progressively more efficiently translated over the interval from 1 to 4 dpp. To determine whether this increased polysome occupancy might arise from substantial de novo increase in gene transcription and immediate recruitment, we quantified each mRNA in total RNA from 1, 3, 4, and 8 dpp testes. Pgk1, Actb, and Pou5f1 mRNA levels did not change appreciably over the time course, although there was slightly more Pou5f1 mRNA present at 4 dpp (Fig. 3, A–C). By contrast, Stra8 mRNA was nearly undetectable until 4 dpp, but was strongly expressed by 8 dpp (Fig. 3D), as has been shown by others [29, 36]. Increases in Rhox13 mRNA levels were not statistically significant (P = 0.02) until 8 dpp, when Stra8 mRNA levels rose nearly 15-fold (P = 0.0001; Fig. 3E).
FIG. 3.
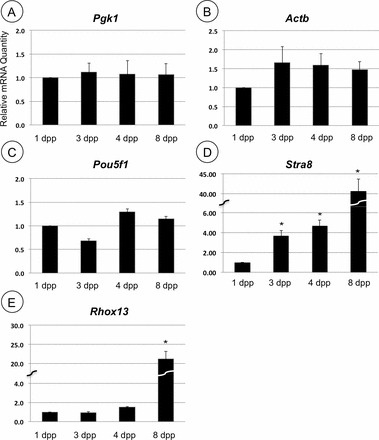
Assessment of mRNA abundance during neonatal testis development. Total relative mRNA levels were analyzed by qRT-PCR from 1, 3, 4, and 8 dpp mouse testis RNA for the genes indicated. Asterisks indicate statistically significant difference when compared to 1 dpp (P < 0.01). Error bars represent standard error.
Assessment of Germ Cell Population from 1 to 4 dpp
Having established that Rhox13 mRNA becomes translationally activated while Stra8 mRNA is newly transcribed early on (1 to 4 dpp), we followed the fate of these developmental mRNAs into later spermatogonial development. Both Rhox13 and Stra8 mRNA levels increased substantially from 4 to 8 dpp. The increase might result from either increased transcription or expansion of the spermatogonial population, which would increase the germ cell-somatic cell ratio in the juvenile testis. The ratios of germ cells to Sertoli cells in stages from 6 dpp through adulthood were estimated many years ago [38]. We manually counted the number of germ cells and Sertoli cells within 40 randomly selected testis cords from mice at age 0 and 3 dpp (Fig. 4, A and B, germ cells stained for POU5F1) as well as 4 and 8 dpp (Fig. 4, C and D, germ cells stained for RHOX13). Sertoli cells were identified based on their distinct morphology (small ovoid nuclei, punctate heterochromatin) along with their consistent darker staining by hematoxylin in Bouin-fixed sections. The percentage of germ cells in the testis cords increased from 9% to 42% between birth and 8 dpp, and the percentage of Sertoli cells decreased accordingly (Fig. 4E). Expansion of the germ cell population between 4 and 8 dpp reflects a burst in their mitotic activity and accumulation of spermatogonia [4]. Thus, expansion of the germ cell population, rather than substantially enhanced transcriptional activity per cell, would explain the increase in mRNA levels for Rhox13 and Stra8 that we observed during this interval (Fig. 3, D and E).
FIG. 4.
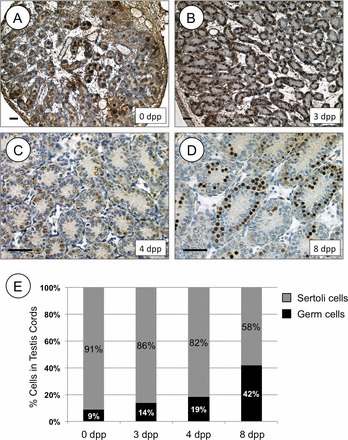
Germ cell-Sertoli cell ratio increases during neonatal testis development. IHC was performed on testis sections from 0 and 3 dpp mice using anti-POU5F1 (A, B) or on testis sections from 4 and 8 dpp mice using anti-RHOX13 (C, D). Ages are indicated on each section. Brown staining indicates a positive signal, and each section was counterstained with hematoxylin. E) The relative numbers of germ cells and Sertoli cells counted from stained testis cords shown in A–D are represented for each age from 0 to 8 dpp. Bars = 100 μm.
Translational Recruitment of Large mRNA Populations
It was evident from the proportion of ribosomes in actively translating complexes that general protein synthetic activity was also increasing over the 8-day period of neonatal testis development. Overlaying the polysome profiles from 1, 4, and 8 dpp testes (from Fig. 1) revealed a decrease in absorbance in fractions 2–5, which represent 40S, 60S, and 80S ribosome subunits, and a significant increase in absorbance in fractions 6–12, which represent 3–9 ribosomes on mRNAs (polysomes) in 4 and 8 dpp testes (Fig. 5A). This indicates a progressive overall increase in efficient recruitment of the mRNA population to polysomes from 1 dpp to 4 and 8 dpp.
FIG. 5.
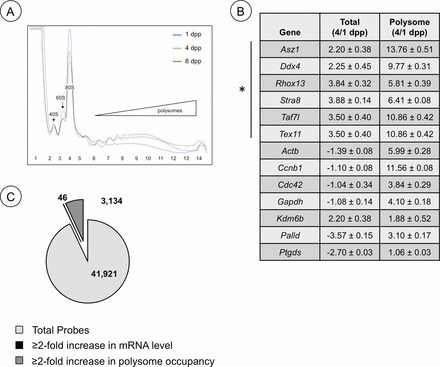
Changes in mRNA and ribosome utilization during neonatal testis development. A) Overlay of polysome gradient profiles from 1, 4, and 8 dpp testis lysates. Blue line, 1 dpp; red line, 4 dpp; green line, 8 dpp polysome profile. B) Quantitative RT-PCR was used to assess mRNA levels for the genes indicated. The data is represented as a ratio of message abundance from total RNA (Total) or RNA isolated from heavy polysome fractions (Polysome) from testes of 4 dpp/1 dpp mice and from RA/dimethyl sulfoxide-injected mice. C) A representation of the results from TSAA and previous microarray results [5] depicting relative numbers of testicular mRNAs that increased ≥2-fold in abundance, became enriched ≥2-fold in heavy polysomes, or were unchanged from 1 to 4 dpp.
To assess whether progressive upregulation of translation is more widespread during this period, we took a genome-wide approach to assess changes in translation efficiency (translation state array analysis [TSAA]) of the mouse testis transcriptome. Sucrose gradients were used to separate mRNAs from 1 and 4 dpp testes into nontranslated (RNPs) and efficiently translated (heavy polysomes) pools, which were then subjected to microarray analysis. By comparative polysomal microarray analysis, we found that mRNAs encoding 3134 genes became enhanced ≥2-fold within heavy polysomes from 1 to 4 dpp (Fig. 5C and Supplemental Table S1; all supplemental data are available online at www.biolreprod.org). This data has been deposited in the NCBI GEO database (GSE49283; http://www.ncbi.nlm.nih.gov/geo/). This indicates ∼7.5% of mRNAs present in the testis at these stages are undergoing dynamic translational control through recruitment or activation that exceeds general increases in protein synthesis activity. We employed Ingenuity Pathway Analysis software to identify cellular signaling pathways and networks where the products of these 3134 genes are predicted to function. Top categories include: cell cycle (e.g., Gspt1, Ccnb1, Cdc42), cellular movement and morphology (e.g., Tuba3b, Actb), and mitochondria function (e.g., Cisd1, Ndufc2, Cycs, Atp5g3). In addition, a number of germ cell mRNAs undergo translational control (Tex11, Dazl, Ddx4, Rbmy, and Asz1).
To validate the results of the TSAA, we used qRT-PCR to measure the abundance and polysome occupancy (indicating efficient translation) of a panel of mRNAs from genes with predicted housekeeping, germ cell-specific, or tissue-specific functions. As shown in the table in Fig. 5C, there were substantial increases (3- to 13-fold) in polysome occupancy from 1 to 4 dpp for mRNAs from genes with housekeeping (Gapdh, Actb, and Palld), cell cycle regulatory (Ccnb1 and Cdc42), and germ cell-specific (Ddx4, Tex11, Asz1, and Rhox13) functions. In each case, the fold recruitment to ribosomes was substantially greater than the increase in abundance for each message over the interval. By contrast, there was little change in translation of Ptgds and Kdm6b mRNA, which show predominant tissue-restricted expression outside of the postnatal testis [39, 40].
Translation Efficiency Changes Are Accompanied by Phosphorylation of EIF4EBP1 and RPS6KB1
More efficient use of ribosomes from 1 to 8 dpp is indicative of enhanced translation initiation activity through steps catalyzed by initiation factors EIF2A and EIF4. Coincidentally, specific translational control is also generally exerted at the initiation step [8]. Three modes can be envisioned whereby specific mRNAs would become more efficiently translated in the neonatal testis: release from compartmentalization, increased cap-dependent translation (role of EIF4), and/or enhanced scanning to identify the bona fide AUG (role of EIF2A). Our results revealed that specific mRNAs that became recruited to heavy polysomes from 1 to 4 dpp were not particularly abundant in the ribosome-free mRNP fractions of our polysome gradients (Fig. 2 and TSAA results), but rather were simply translating poorly. Therefore, release of mRNAs from compartmentalization does not appear to be a predominant mechanism regulating their translation. We next assessed the phosphorylation state of EIF4EBP1 and EIF2A. Increased phosphorylation of 4EBP1 would suggest that an increase in cap-dependent initiation was responsible for increased translation in the 4 dpp testis, whereas decreased EIF2A phosphorylation might suggest that ribosome scanning was responsible. By densitometry, we detected an ∼20% increase in phosphorylated EIF4EBP1 without a change in total EIF4EBP1 protein in testis lysates over this interval, while phosphorylation of EIF2A was reduced by ∼30% (Fig. 6A). We next used IIF to assess changes in EIF4EBP1 phosphorylation in situ in the germ cell and somatic cell population. The signal representing phospho-EIF4EBP1 was enhanced in cytoplasm of spermatogonia at 4 dpp, but did not appear to change in somatic Sertoli or Leydig cells (Fig. 6, B and C). Taken together, our data suggest increased cap-dependent translation initiation, along with some enhanced mRNA scanning, is mediating the mRNA translational control we observe in male germ cells in the neonatal testis as they transition from gonocytes to spermatogonia.
FIG. 6.
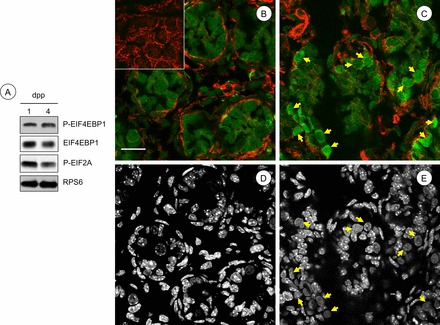
Increases in phosphorylation of EIF4EBP1 accompany the gonocyte-to-spermatogonia transition in the neonatal testis. A) Immunoblots were performed with total protein isolated from 1 and 4 dpp testes using antibodies against the indicated phosphorylated (P-) and total proteins indicated to the right. B, C) IIF was performed to detect phospho-EIF4EBP1 (green signal) in testis sections from 1 dpp (B) and 4 dpp (C) mice. No primary antibody was omitted from the negative control (inset in B). Yellow arrows indicate spermatogonia with increased cytoplasmic signal for phosphorylated EIF4EBP1. F-actin is labeled by phalloidin, in red. D, E) DNA staining (DAPI) from the corresponding color images (B and C, above) was converted to black and white for ease of identification of germ cell nuclei (yellow arrows). Bar = 50 μm.
DISCUSSION
Mouse germ cells undergo a dramatic transition from quiescent gonocytes to proliferating spermatogonia in the first few days after birth. A first and important step toward understanding the pathways and cellular changes involved in this transition was to define changes in the transcriptome of the neonatal testis. Surprisingly, however, very few genes were identified for which mRNA levels changed significantly over this interval [5]. Furthermore, the interpretation of transcriptome microarray results relies on the assumption that all mRNAs are translated with equivalent efficiency. This is not the case, as highlighted by a recent study revealing that mRNA translational efficiency is a more accurate predictor of the proteome than mRNA levels [41]. In this study, we found that more than 3000 mRNAs encoding both germ cell-specific (e.g., Asz1, Ddx4, Gata2, Rhox13, Taf7l, Tex11) and housekeeping (e.g., Actb, Ccnb1, Cdc42, Gapdh, Palld) proteins underwent dramatic changes in translational efficiency from 1 to 4 dpp. In addition, our results provide an important advance in understanding how the newly expressed proteome is determined during neonatal testis development.
Translation is the most energy-consumptive process in nearly all cell types, particularly those undergoing proliferation or differentiation [42]. Predictably, translational control occurs most commonly during the initiation step, which saves energy, avoids the accumulation of incompletely synthesized proteins, and preserves the limited supply of active ribosomes. Three modes can be envisioned whereby mRNAs become more efficiently translated in the neonatal testis, and these are not mutually exclusive. In the first, compartmentalization of inactive mRNAs (repression) would prevent an interaction with translation initiation factors or ribosomes. This scenario appears to affect a small proportion of mRNAs, which were detected in fractions containing the nonpolysomal mRNPs, for each gene we analyzed (Fig. 2 and TSAA results). In the second mode, which is mediated by EIF2A, enhanced scanning of 5′ UTRs that results in a high translation initiation rate at upstream initiation codons would prevent active initiation complexes from reaching the authentic AUG codon for the subset of mRNAs containing uORFs. Phosphorylation of EIF2A reduces the availability of active ternary complex (eIF2-tRNAi-GTP), which allows ribosomes to scan farther down such mRNAs to reach authentic AUGs, thus increasing mRNA partitioning into heavy polysomes. We detected a decrease in EIF2A phosphorylation from 1 to 4 dpp, which suggests overall enhanced initiation activity, but likewise indicates a diminishment of this mode of regulation as a significant contributor to translational control in the neonatal testis. In the third mode, mRNAs are recruited to ribosomes through EIF4E, which can be repressed through an interaction with EIF4EBP1 [43, 44]. Greater EIF4E activity is most commonly due to phosphorylation EIF4EBP1, which releases EIF4E, allowing it to associate with eIF4G and the ribosome, and results in enhanced cap-dependent translation initiation activity that often accompanies developmental progression [10]. Sequences in regulated mRNAs can reduce translation efficiency, including complex 5′ UTR secondary structures and the presence of binding sites for repressor proteins, most commonly found in the 3′ UTR [45]. Future analyses will identify common features among this set of ∼3000 mRNAs that become more able to effectively compete for ribosome binding during neonatal testis development.
Examples of global increases in translation efficiency have been documented during cellular transitions in other systems, including the epithelial-to-mesenchymal transition [46], T-cell activation [47], and oocyte maturation [48, 49]. A particularly relevant example is found in mouse embryonic stem cells (ESCs). Mouse ESCs share the expression of many genes with primordial germ cells and gonocytes and can be induced to form male gametes in vitro [50–52]. During the differentiation of mouse ESCs into embryoid bodies, there is a global increase in translation initiation accompanied by increased phosphorylation of EIF4EBP1 and decreased phosphorylation of EIF2A [53]. In addition, the cytoplasmic-nuclear ratio of the cells increases, along with the appearance of prominent Golgi bodies and rough endoplasmic reticulum [53]. These features are suggestive of increased biosynthetic activity of differentiating cells. It is worth noting that these same changes have been documented in gonocytes as they transition to spermatogonia in the neonatal testis [54, 55].
Our results suggest that enhanced cap-dependent translation is occurring primarily, if not exclusively, in germ cells. Coincidentally, a significant number of the mRNAs that are recruited to heavy polysomes from 1 to 4 dpp are germ cell-specific (Fig. 5B and Supplemental Table S1). Also, germ cells undergo significant changes as they differentiate during this interval, in contrast to the somatic cell population, which merely expands modestly in number. It is noteworthy that a large number of mRNAs that became more efficiently translated are involved in processes linked to gonocyte changes such as cell division, cell movement, and cell morphology.
We detected an apparent decrease in free ribosomes during neonatal testis development, which implies that their numbers are not rate limiting for the protein synthetic activity necessary to drive the gonocyte-differentiation program. This suggests that ribosomes (and the corresponding initiation factors) may be awaiting a differentiation signal to direct mRNA recruitment. These studies highlight the importance of integrating changes in mRNA utilization and translational control mechanisms with existing transcriptome data in order to predict the utilitarian proteome. An understanding of translational control will also further our pursuit of novel gene products involved in developmental processes such as spermatogenesis.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Dr. Phil Pekala and Joani Zary-Oswald for technical assistance, and Drs. E. Mitch Eddy and Karen Adelman (National Institute of Environmental Health Sciences) for critically reading the manuscript.
Footnotes
Supported by a grant from the National Institutes of Health National Institute of Child Health and Human Development (HD072552 to C.B.G.).
REFERENCES
- Western P, Miles D, van den Bergen J, Burton M, Sinclair A. Dynamic regulation of mitotic arrest in fetal male germ cells. Stem Cells 2008; 26: 339 347. [DOI] [PubMed] [Google Scholar]
- Vergouwen RP, Jacobs SG, Huiskamp R, Davids JA, de Rooij DG. Proliferative activity of gonocytes, Sertoli cells and interstitial cells during testicular development in mice. J Reprod Fertil 1991; 93: 233 243. [DOI] [PubMed] [Google Scholar]
- Nagano R, Tabata S, Nakanishi Y, Ohsako S, Kurohmaru M, Hayashi Y. Reproliferation and relocation of mouse male germ cells (gonocytes) during prespermatogenesis. Anat Rec 2000; 258: 210 220. [DOI] [PubMed] [Google Scholar]
- de Rooij DG. Proliferation and differentiation of spermatogonial stem cells. Reproduction 2001; 121: 347 354. [DOI] [PubMed] [Google Scholar]
- Shima JE, McLean DJ, McCarrey JR, Griswold MD. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 2004; 71: 319 330. [DOI] [PubMed] [Google Scholar]
- Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 2005; 310: 486 489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keiper BD, Rhoads RE. Translational recruitment of Xenopus maternal mRNAs in response to poly(A) elongation requires initiation factor eIF4G-1. Dev Biol 1999; 206: 1 14. [DOI] [PubMed] [Google Scholar]
- Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol 2004; 5: 827 835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meric F, Hunt KK. Translation initiation in cancer: a novel target for therapy. Mol Cancer Ther 2002; 1: 971 979. [PubMed] [Google Scholar]
- Mendez R, Richter JD. Translational control by CPEB: a means to the end. Nat Rev Mol Cell Biol 2001; 2: 521 529. [DOI] [PubMed] [Google Scholar]
- Chennathukuzhi V, Morales CR, El-Alfy M, Hecht NB. The kinesin KIF17b and RNA-binding protein TB-RBP transport specific cAMP-responsive element modulator-regulated mRNAs in male germ cells. Proc Natl Acad Sci U S A 2003; 100: 15566 15571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold B, Fujimoto H, Kramer JM, Erickson RP, Hecht NB. Haploid accumulation and translational control of phosphoglycerate kinase-2 messenger RNA during mouse spermatogenesis. Dev Biol 1983; 98: 392 399. [DOI] [PubMed] [Google Scholar]
- Iguchi N, Tobias JW, Hecht NB. Expression profiling reveals meiotic male germ cell mRNAs that are translationally up- and down-regulated. Proc Natl Acad Sci U S A 2006; 103: 7712 7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Medvedev S, Yu J, Tang LC, Agno JE, Matzuk MM, Schultz RM, Hecht NB. Absence of the DNA-/RNA-binding protein MSY2 results in male and female infertility. Proc Natl Acad Sci U S A 2005; 102: 5755 5760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck AR, Miller IJ, Anderson P. RNA binding protein TIAR is essential for primordial germ cell development. Proc Natl Acad Sci U S A 1998; 95: 2331 2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, Lin H, Matsuda Y, et al. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 2004; 131: 839 849. [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, Totoki Y, Shibata T, et al. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev 2010; 24: 887 892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, Hata K, Li E, et al. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 2008; 22: 908 917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggiu M, Speed R, Taggart M, McKay SJ, Kilanowski F, Saunders P, Dorin J, Cooke HJ. The mouse Dazla gene encodes a cytoplasmic protein essential for gametogenesis. Nature 1997; 389: 73 77. [DOI] [PubMed] [Google Scholar]
- Kimble J, Crittenden SL. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu Rev Cell Dev Biol 2007; 23: 405 433. [DOI] [PubMed] [Google Scholar]
- Amiri A, Keiper BD, Kawasaki I, Fan Y, Kohara Y, Rhoads RE, Strome S. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development 2001; 128: 3899 3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson MA, Cronland E, Dunkelbarger S, Contreras V, Strome S, Keiper BD. A germ line-specific isoform of eIF4E (IFE-1) is required for efficient translation of stored mRNAs and maturation of both oocytes and sperm. J Cell Sci 2009; 122: 1529 1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song A, Labella S, Korneeva NL, Keiper BD, Aamodt EJ, Zetka M, Rhoads RE. A. C. elegans eIF4E-family member upregulates translation at elevated temperatures of mRNAs encoding MSH-5 and other meiotic crossover proteins. J Cell Sci 2010; 123: 2228 2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy EM. Fine structural observations on the form and distribution of nuage in germ cells of the rat. Anat Rec 1974; 178: 731 757. [DOI] [PubMed] [Google Scholar]
- Eddy EM. Germ plasm and the differentiation of the germ cell line. Int Rev Cytol 1975; 43: 229 280. [DOI] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 2009; 5: e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P. The chromatoid body of male germ cells: similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci U S A 2006; 103: 2647 2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CP, Chaillet JR, Bestor TH. Transcription of IAP endogenous retroviruses is constrained by cytosine methylation. Nat Genet 1998; 20: 116 117. [DOI] [PubMed] [Google Scholar]
- Geyer CB, Saba R, Kato Y, Anderson AJ, Chappell VK, Saga Y, Eddy EM. Rhox13 is translated in premeiotic germ cells in male and female mice and is regulated by NANOS2 in the male. Biol Reprod 2012; 86: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer CB, Eddy EM. Identification and characterization of Rhox13, a novel X-linked mouse homeobox gene. Gene 2008; 423: 194 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003; 19: 185 193. [DOI] [PubMed] [Google Scholar]
- Lachke SA, Alkuraya FS, Kneeland SC, Ohn T, Aboukhalil A, Howell GR, Saadi I, Cavallesco R, Yue Y, Tsai AC, Nair KS, Cosma MI, et al. Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science 2011; 331: 1571 1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesce M, Wang X, Wolgemuth DJ, Scholer H. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech Dev 1998; 71: 89 98. [DOI] [PubMed] [Google Scholar]
- Snyder EM, Davis JC, Zhou Q, Evanoff R, Griswold MD. Exposure to retinoic acid in the neonatal but not adult mouse results in synchronous spermatogenesis. Biol Reprod 2011; 84: 886 893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder EM, Small C, Griswold MD. Retinoic acid availability drives the asynchronous initiation of spermatogonial differentiation in the mouse. Biol Reprod 2010; 83: 783 790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Nie R, Li Y, Friel P, Mitchell D, Hess RA, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin A-sufficient postnatal murine testes. Biol Reprod 2008; 79: 35 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrios F, Filipponi D, Pellegrini M, Paronetto MP, Di Siena S, Geremia R, Rossi P, De Felici M, Jannini EA, Dolci S. Opposing effects of retinoic acid and FGF9 on Nanos2 expression and meiotic entry of mouse germ cells. J Cell Sci 2010; 123: 871 880. [DOI] [PubMed] [Google Scholar]
- Bellve AR, Cavicchia JC, Millette CF, O'Brien DA, Bhatnagar YM, Dym M. Spermatogenic cells of the prepuberal mouse. Isolation and morphological characterization. J Cell Biol 1977; 74: 68 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 2007; 130: 1083 1094. [DOI] [PubMed] [Google Scholar]
- Urade Y, Hayaishi O. Biochemical, structural, genetic, physiological, and pathophysiological features of lipocalin-type prostaglandin D synthase. Biochim Biophys Acta 2000; 1482: 259 271. [DOI] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature 2011; 473: 337 342. [DOI] [PubMed] [Google Scholar]
- Wagner A. Energy constraints on the evolution of gene expression. Mol Biol Evol 2005; 22: 1365 1374. [DOI] [PubMed] [Google Scholar]
- Keiper BD, Gan W, Rhoads RE. Protein synthesis initiation factor 4G. Int J Biochem Cell Biol 1999; 31: 37 41. [DOI] [PubMed] [Google Scholar]
- Morley SJ, Jeffrey I, Bushell M, Pain VM, Clemens MJ. Differential requirements for caspase-8 activity in the mechanism of phosphorylation of eIF2alpha, cleavage of eIF4GI and signaling events associated with the inhibition of protein synthesis in apoptotic Jurkat T cells. FEBS Lett 2000; 477: 229 236. [DOI] [PubMed] [Google Scholar]
- Andreassi C, Riccio A. To localize or not to localize: mRNA fate is in 3′UTR ends. Trends Cell Biol 2009; 19: 465 474. [DOI] [PubMed] [Google Scholar]
- Jechlinger M, Grunert S, Tamir IH, Janda E, Ludemann S, Waerner T, Seither P, Weith A, Beug H, Kraut N. Expression profiling of epithelial plasticity in tumor progression. Oncogene 2003; 22: 7155 7169. [DOI] [PubMed] [Google Scholar]
- Mikulits W, Pradet-Balade B, Habermann B, Beug H, Garcia-Sanz JA, Mullner EW. Isolation of translationally controlled mRNAs by differential screening. FASEB J 2000; 14: 1641 1652. [DOI] [PubMed] [Google Scholar]
- Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, Conti M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev 2011; 25: 755 766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Braun RE. Pathway to totipotency: lessons from germ cells. Cell 2006; 127: 891 904. [DOI] [PubMed] [Google Scholar]
- Geijsen N, Horoschak M, Kim K, Gribnau J, Eggan K, Daley GQ. Derivation of embryonic germ cells and male gametes from embryonic stem cells. Nature 2004; 427: 148 154. [DOI] [PubMed] [Google Scholar]
- Nayernia K, Nolte J, Michelmann HW, Lee JH, Rathsack K, Drusenheimer N, Dev A, Wulf G, Ehrmann IE, Elliott DJ, Okpanyi V, Zechner U, et al. In vitro-differentiated embryonic stem cells give rise to male gametes that can generate offspring mice. Dev Cell 2006; 11: 125 132. [DOI] [PubMed] [Google Scholar]
- Toyooka Y, Tsunekawa N, Akasu R, Noce T. Embryonic stem cells can form germ cells in vitro. Proc Natl Acad Sci U S A 2003; 100: 11457 11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, Murry CE. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell 2008; 2: 448 460. [DOI] [PubMed] [Google Scholar]
- Baillie AH. The histochemistry and ultrastructure of the genocyte. J Anat 1964; 98: 641 645. [PMC free article] [PubMed] [Google Scholar]
- Franchi LL, Mandl AM. The ultrastructure of germ cells in foetal and neonatal male rats. J Embryol Exp Morphol 1964; 12: 289 308. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


