Abstract
Recent cancer therapies have focused on targeting biology networks through a single regulatory protein. Heat shock protein 90 (hsp90) is an ideal oncogenic target as it regulates over 400 client proteins and cochaperones. However, clinical inhibitors of hsp90 have had limited success; the primary reason being that they induce a heat shock response. We describe the synthesis and biological evaluation of a new hsp90 inhibitor, SM253. The previous generation on which SM253 is based (SM145) has poor overall synthetic yields, low solubility, and micromolar cytotoxicity. By comparison SM253 has relatively high overall yields, good aqueous solubility, and is more cytotoxic than its parent compound. Verification that hsp90 is SM253’s target was accomplished using pull-down and protein folding assays. SM253 is superior to both SM145 and the clinical candidate 17-AAG as it decreases proteins related to the heat shock response by 2-fold, versus a 2–4-fold increase observed when cells are treated with 17-AAG.
Keywords: SM253, heat shock protein 90, heat shock response, SM145, macrocycle, conformation, peptide, natural product
Cancer cells utilize numerous pathways in order to maintain their propagation. Such rapid oncogenic proliferation produces an abundance of mutated and mis-folded proteins, which require molecular chaperones such as heat shock proteins (hsps) to fold and stabilize these proteins. Specifically, hsps 90, 70, 40, and 27 are required to maintain the cell’s function under stressed conditions by deaggregating and folding over 400 proteins (Figure 1).1,2 Heat shock factor 1 (HSF1) transcribes and up-regulates the protein levels of hsp90, 70, 40, and 27 under stressed conditions, and the increase in HSF-1 and hsp levels is a primary feature of many cancers (Figure 1).3−8 High levels of hsps and HSF-1 are usually referred to as a heat shock response (HSR), whereby these up-regulated proteins block apoptosis, promote oncogenic growth, and reverse effects of a chemotherapeutic drug all by maintaining and stabilizing proteins in the cell.3,6,7,9−12 A high level of HSF-1 is also associated with highly malignant tumors and indicates poor patient prognosis in breast, colon, and lung cancer.13,14 Since cancer cells have an abundance of mis-folded and mutated proteins, they are also significantly more dependent on these hsps than normal cells, making them prime anticancer targets.15
Figure 1.
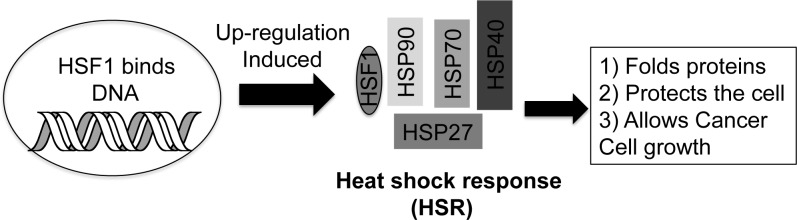
Heat shock response involves HSF1 up-regulating HSF1 protein, hsp90, hsp70, hsp40, and hsp27.
Current clinical hsp90 inhibitors bind to the ATP binding pocket in the amino terminus of hsp90. Over 40 clinical trials were initiated using amino terminal inhibitors; however, the entire class of compounds induces a significant HSR,16 which led to antiapoptotic and chemo-resistance mechanisms and their subsequent removal from the clinic as a monotherapy.17,18 In an attempt to circumvent the HSR, clinical hsp90 inhibitors have been combined with alternative cancer therapies, which has proven successful in preclinical and clinical settings.19−23 Finding an hsp90 inhibitor that does not induce a HSR would be a significant achievement. The development of specific C-terminal hsp90 inhibitors has been reported as a promising alternative to the N-terminal clinical hsp90 inhibitors, as they do not induce the heat shock response.24,25 However, these inhibitors have micromolar and millimolar IC50 values, and only one reported inhibitor had data discussing the HSR.25
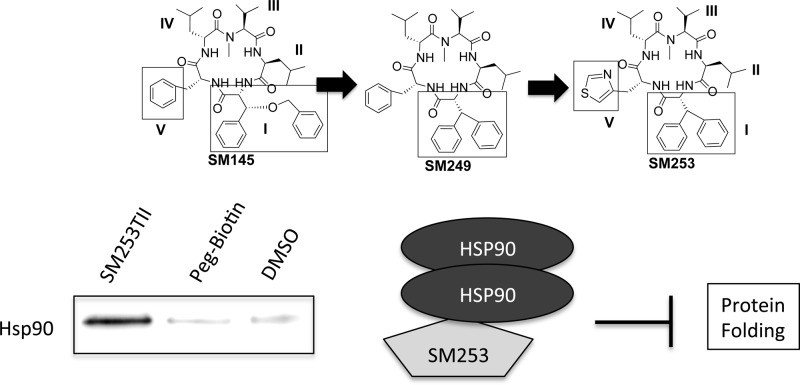
Our previous work describes the hsp90 inhibition activity of SM145 (Figure 2).26−30 This molecule binds to a unique site on hsp90 and blocks hsp90 from interacting with multiple proteins associated with its C-terminus. Furthermore, SM145 does not induce the HSR survival mechanism.26−30 However, as noted in our previous work, there are three significant issues associated with SM145: its low yielding synthesis (overall final yield 1–7%), its hydrophobicity (ClogP = ∼9), and its micromolar potency.
Figure 2.
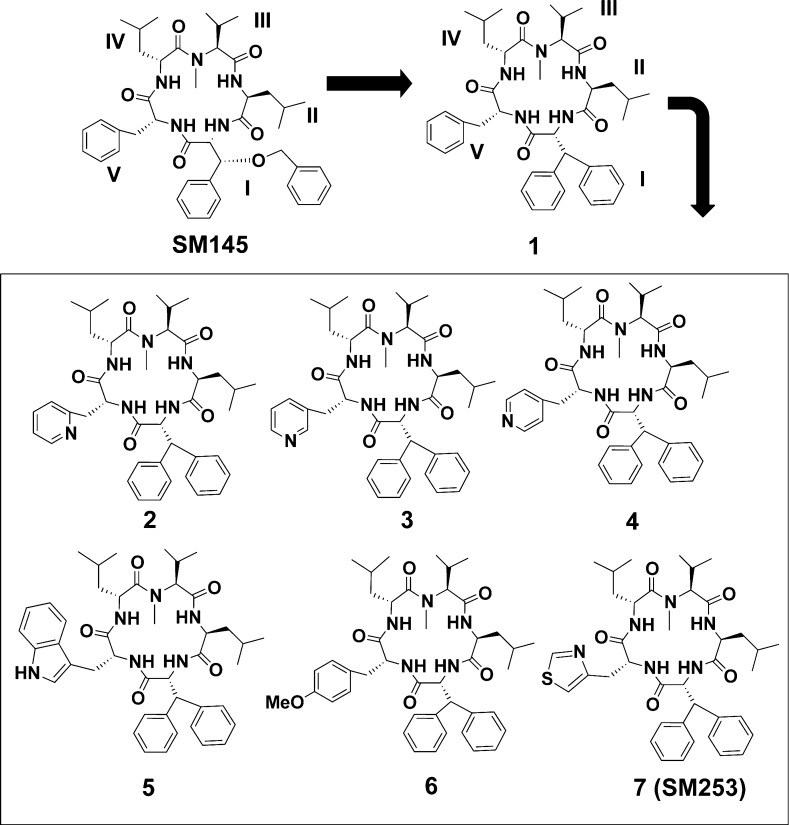
Structures of SM145 and new compounds 1–7.
Herein we describe the design, synthesis, and characterization of 7 new molecules, compounds 1–7, derived from the hsp90 inhibitor SM145 (Figure 2). The first analogue involved a substitution of a biphenyl moiety in place of the benzylated phenyl serine (compound 1), which reduced synthetic complexity and increased the synthetic yield dramatically, while not disrupting the cytotoxicity or binding affinity for hsp90 (Table 1, Figure 2, and Supplemental Figures S5 and S7). This scaffold was used to create a series of analogues. Our early structure–activity relationship (SAR) work indicated that position V did not have significant impact on binding affinity for hsp90; thus, we modified this position in order to improve the pharmacological properties.31−35 The modifications involved incorporating aromatic heteroatom containing side chains with the goal of increasing hydrogen-bonding capabilities, improving solubility, and improving cytotoxicity while not impacting the macrocycles overall conformation.
Table 1. Comparison of SM145 and Compounds 1–7.
| compound | IC50 HCT-116 (μM) | IC50 MiaPaCa-2 (μM) | % yield | ClogP | saturated solution of 80% EtOH/20% H2O |
|---|---|---|---|---|---|
| SM145 | 7.4 ± 0.5 | 7.8 ± 0.5 | 1–7% | 9.02 | 5.2 mg/mL |
| 1 | 7.6 ± 0.4 | 12.8 ± 2.3 | 40% | 9.09 | 2.1 mg/mL |
| 2 | 18.1 ± 0.4 | 23.6 ± 2.5 | 25% | 7.60 | N.D.a |
| 3 | 20.8 ± 2.5 | 23.0 ± 2.8 | 28% | 7.60 | N.D. |
| 4 | 15.7 ± 2.9 | 18.5 ± 5.3 | 30% | 7.60 | N.D. |
| 5 | 3.9 ± 0.2 | 4.5 ± 0.9 | 32% | 9.02 | 1.9 mg/mL |
| 6 | 5.1 ± 0.6 | 4.4 ± 0.7 | 30% | 9.09 | 1.5 mg/mL |
| 7 (SM253) | 5.0 ± 0.5 | 5.5 ± 0.2 | 40% | 7.44 | 9.5 mg/mL |
N.D.: not determined.
The inclusion of ortho-, meta-, and para-pyridal groups (compounds 2–4) decreased the ClogP value (an estimate for solubility); however, they have dramatically reduced potency compared to compound 1 (>15 μM, Table 1). In addition, when a tryptophan or methylated tyrosine was placed at position V (compounds 5 and 6), the cytotoxicity was improved over compound 1, but the aqueous solubility of these two molecules is poor (<2 mg/mL, Table 1). One derivative, compound 7, which incorporated a thiazole moiety, had both improved cytotoxicity and solubility over compound 1 (Table 1).
Overall, compound 7 (from here on referred to as SM253) shows improved properties over the parent compound SM145. SM253, which contains a d-3-(4-thiazoyl)alanine at position V and the biphenyl moiety at position I, is (1) significantly easier to synthesize than SM145 (overall yields = 40%); (2) more soluble than both SM145 and compound 1 (9.5 mg/mL); (3) has improved cytotoxicity against 2 cancer cell lines relative to both SM145 and compound 1 (Table 1); and (4) reduces the HSR relative to the clinical inhibitor 17-AAG and SM145 (Figure 4).
Figure 4.
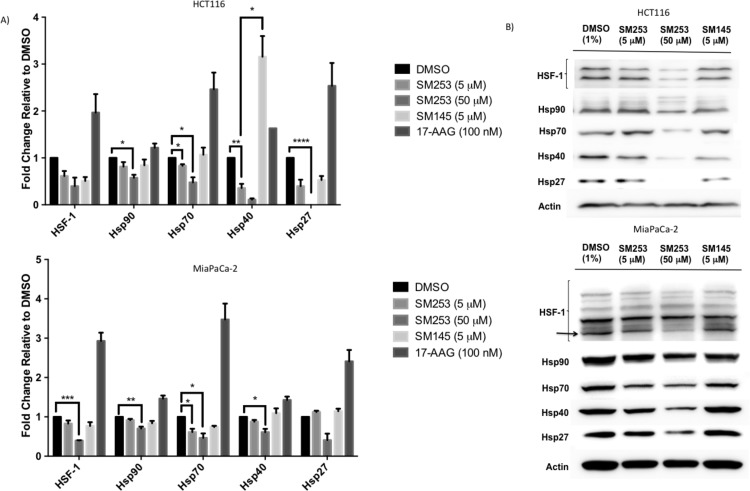
(A) Protein levels of HSR proteins in HCT-116 and MiaPaCa-2 cells after 24 h with the indicated treatment; density values were normalized to actin and then compared to DMSO. Graph of mean ± SEM, n = 3. (B) Representative Western blots of protein levels in HCT-116 and MiaPaCa-2 cells (black arrow indicates HSF1 band that was quantitated, *p < 0.02, **p < 0.005, ***p < 0.0005, ****p < 0.0001). 17-AAG Western blots for MiaPaCa-2 appear in Supplemental Figure S6.
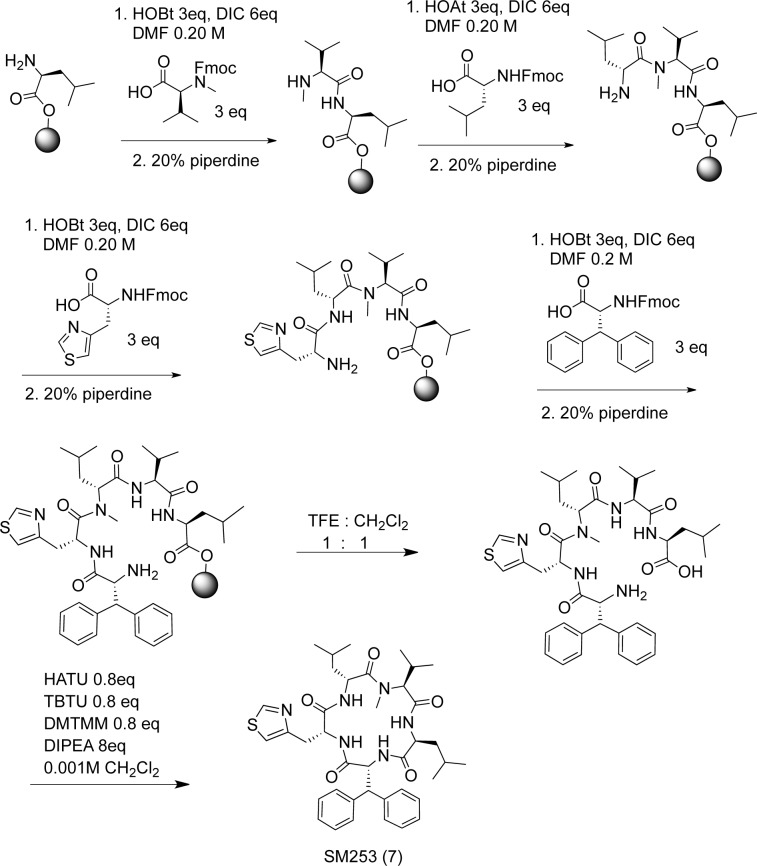
All the analogues were made via a combination of solid-phase and solution phase chemistry (Scheme 1).27−30,36 Using a preloaded 2-chlorotrityl-leucine resin (CTC) (Supporting Information) and then sequentially coupling fluorenylmethyloxy carbonyl (Fmoc) protected amino acids followed by deprotection of the amine produced the desired linear pentapeptide. The peptide was cleaved from the resin with 50% trifluoroethanol in dichloromethane, which generated the double deprotected linear pentapeptide. The linear peptide was then cyclized using three coupling reagents under dilute conditions (Scheme 1 and Supplemental Schemes 1 and 4).37 To explore the role of SM253 in inducing cytotoxicity, we examined its effects in two human cancer cell lines, HCT116 colon carcinoma and MiaPaCa-2 pancreatic carcinoma cells. The cytotoxicity of compound 1 and SM253 toward HCT-116 and MiaPaCa-2 cells were analyzed using a cell-counting kit 8 (CCK8). Compound 1 demonstrated similar cytotoxicity to that of SM145, while SM253 had a lower IC50 value compared to SM145 in both HCT-116 and MiaPaCa-2 cells lines (Table 1 and Supplemental Figure S7). In general, hsp90 inhibitors have been reported to display remarkable selectivity for tumor cells as compared to healthy cells, and the inhibitors have been shown to accumulate in tumor tissue while being rapidly cleared from circulation and normal tissue.15,38,39 Analogues of this series have shown an average of 3-fold greater selectivity of cancer cells over normal cells (WS1) (Supplemental Figure S7).31,33,35,40,41 In addition to its enhanced potency, SM253 is 2-fold more soluble than SM145 and 5-fold more soluble than compound 1, which is attributed to the inclusion of the d-3-(4-Thiazoyl)alanine. This improved solubility and cytotoxicity warranted further mechanistic studies of SM253.
Scheme 1. Synthesis of SM253 (Compound 7).
Abbreviations: 1-hydroxybenzotriazole (HOBt), 1-hydroxy-7-azabenzotriazole (HOAt), dimethylformamide (DMF), N,N′-diisopropylcarbodiimide (DIC), 2,2,2-trifluoroethanol (TFE), 2-(1H-7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HATU), O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium tetrafluoroborate (TBTU), 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholin-4-ium chloride (DMTMM), N,N-diisopropylethylamine (DIPEA); sphere indicates CTC resin.
Since the parent molecule SM145 was already identified as an hsp90 inhibitor,26,28 we used the same approach to determine whether SM253 bound to hsp90. Our previous work with SM145 had shown that tagging the molecule at position II did not impact its ability to bind to hsp90.26,42,43 Using the same conditions as described in Scheme 1, SM253 was synthesized with a tag at position II (Scheme 2) via the Boc-lysine derivative of SM253.
Scheme 2. Synthesis of SM253T-II.
Abbreviations: tert-butoxycarbonyl (Boc), trifluoroacetic acid (TFA), N-hydroxysuccinimide ester (NHS), poly(ethylene glycol) (PEG).
Briefly, using a pre-loaded 2-chlorotrityl-d-leucine resin and then sequentially coupling the Fmoc protected amino acids followed by removal of the Fmoc group and cleavage from the resin with trifluoroethanol produced the linear precursor (Scheme 2).44 After cyclization of the linear precursor, the lysine was deprotected using 25% trifluoroacetic acid, and then coupled to N-hydroxysuccinimydyl-d-biotin-15-amino-4,7,10,13-tetraoxapentadecylate (NHS-Peg-4-biotin), thus completing the synthesis of SM253TII.28,29,36,45,7
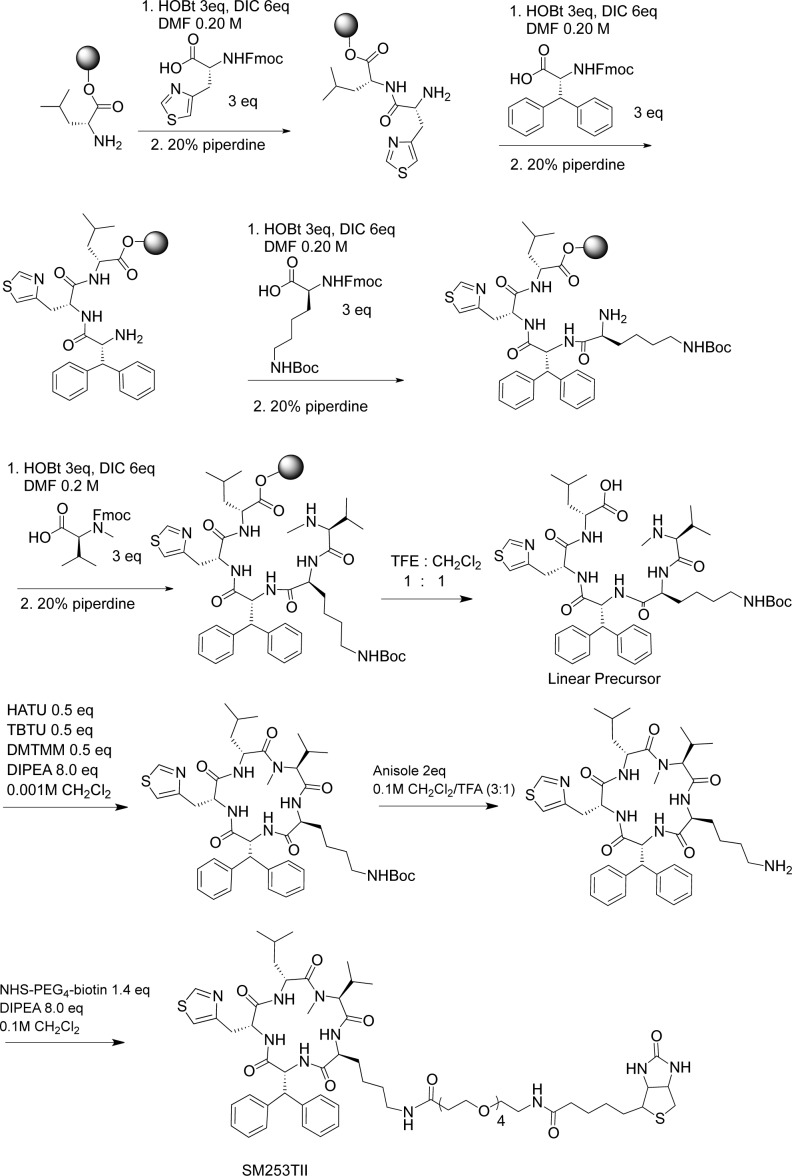
The biotinylated SM253TII was then used in a pull-down assay.28 Briefly, 0.1 mg of the biotinylated analogue was incubated in HCT-116 or MiaPaCa-2 lysates, and then the compound was retrieved from the mixture using the biotin tag by adding NeutrAvadin beads. Proteins bound to the compound were then eluted off the beads with sample buffer and visualized using coomassie blue or Western blotting and were sequenced using nano-LC/MS/MS (Figure 3A and Supplemental Figures S4 and S8). Like its parent compound SM145, SM253 bound to hsp90 in both HCT-116 and MiaPaCa-2 cells lysates as demonstrated by the dark band identified as hsp90 in the SM253TII lane (Figure 3A). In addition, sequencing of the proteins bound to SM253 revealed that hsp90 was bound to SM253 (Supplementary Figure S8). These data together demonstrate that SM253 interacts with hsp90 in both HCT-116 and MiaPaCa-2 cells.
Figure 3.
(A) Pull-down with SM253TII in HCT-116 and MiaPaCa-2 cell lysates, representative image shown of 3 independent assays. (B) Luciferase refolding assay. Graph of mean ± SEM, n = 3.
Since SM253 binds to hsp90 like its parent compound, it should inhibit the function of hsp90 in a similar manner to SM145.
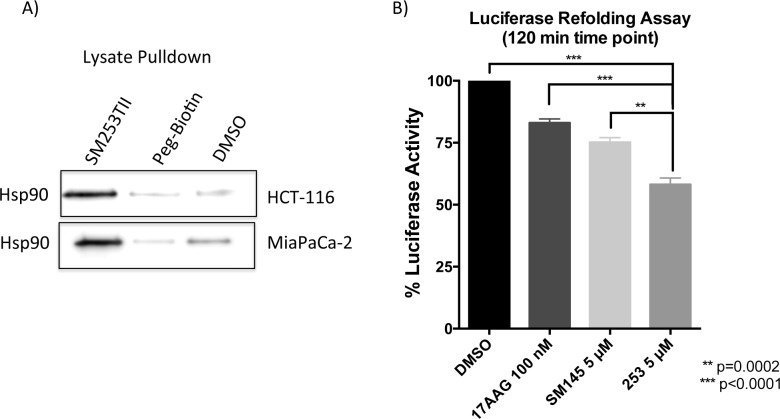
Confirmation that SM253 inhibits the function of hsp90 was accomplished using a protein-folding assay, involving rabbit reticulocyte lysate (RRL, Promega) and denatured firefly luciferase. Denatured luciferase was incubated in RRL, which was pretreated with the approximate IC50 values of 17-AAG (a clinical hsp90 inhibitor), SM145, and SM253.46,47 After 2 h of incubation, SM253 had the greatest impact on inhibiting the refolding activity of the hsp90 machinery, reducing the luciferase activity by 42% relative to control (Figure 3B). In contrast, 17-AAG and SM145 only reduced luciferase activity by 17% and 25%, respectively. Significantly, SM253 demonstrates a greater ability to inhibit hsp90 machinery protein folding over SM145 and prevents hsp90 from completing its role as a molecular chaperone.
A key factor to consider when designing new hsp90 inhibitors is the ability to block hsp90 function without inducing the HSR since it initiates a cascade that can protect the cell from chemotherapy drugs and induces cell survival mechanisms.16,48 Like the other clinical candidates, the HSR induced by 17-AAG was a major reason behind its failure in the clinic.26 Given the success of SM253 at blocking the protein folding function of hsp90, we explored whether SM253 did so without inducing the HSR. Treating the cells with the IC50 value of SM253 and evaluating the protein levels of HSF1, hsp90, hsp70, hsp40, and hsp27 in two cell lines (HCT-116 and MiaPaCa-2) showed that SM253 decreased these HSR proteins (Figure 4). In contrast to SM253, treatment with the IC50 value of 17-AAG induced a significant heat shock response increasing the antiapoptotic proteins HSF-1, hsp70, and hsp27 by greater than 2-fold in both cells lines (Figure 4 and Supplemental Figure S6).26 Thus, SM253 is clearly acting via a different mechanism to 17-AAG, as treatment of cells with 10-fold over the IC50 value for SM253 not only failed to induce a HSR, but significantly decreased all levels of these heat shock proteins by approximately 2–4-fold (Figure 4).
Our previous work shows that 10-fold over 17-AAG’s IC50 value (1 μM) induces hsp90 by 3-fold, hsp70 by 2.5-fold, and HSF1 by 4-fold.26 Perhaps the most remarkable aspect of SM253 is the complete erasure of hsp27 protein in HCT-116 cells (Figure 4A) versus the 2.5-fold increase seen in cells treated with 17-AAG. Since hsp27 is linked to the development of chemo-resistance, the complete removal of this protein by SM253 is novel and highlights that hsp90 can be inhibited without inducing a resistance protein. Interestingly, SM145 has a similar although less desirable profile to that of SM253. The two molecules specifically diverge in their effect on hsp40. Although hsp40’s role in cancer is poorly understood, it is known to play a key role in delivering the unfolded proteins to hsp70, which are then transferred to hsp90 for final folding and activation. Therefore, SM253 may prove a highly relevant tool for delineating the activity of hsp40 in colon and pancreatic cancer.49 In summary, SM253’s ability to block hsp90 activity and inhibit the HSR makes it a novel hsp90 inhibitor that overcomes the resistance mechanisms associated with current and past clinical hsp90 inhibitors.
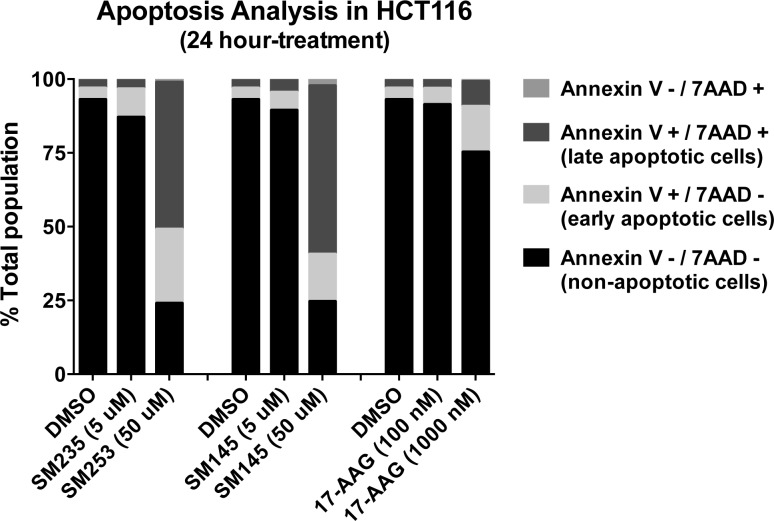
It is already established that inhibition of hsp90 leads to cell death via apoptosis and that both 17-AAG and SM145 induce apoptosis.28 Using annexin V and 7-AAD we demonstrated that SM253 also induces apoptosis (Figure 5). After 24 h of treatment with 10-fold over the IC50 value of SM253 and 17-AAG, 75% versus 25% of cells were in apoptosis, respectively (Figure 5 and Supplemental Figure S2). Analysis of the apoptotic mechanism was made by examining the impact of drug treatment on the caspase3/7 pathway. Treatment with SM253 significantly increases caspase 3/7 activity (4-fold) compared to 17-AAG (2-fold) in both HCT-116 and MiaPaCa-2 cells after 24 h (p < 0.01, Supplemental Figure S3). This is similar to the parent compound SM14526,28,36 (Supplemental Figure S3). It is interesting to note that treatment with 10-fold over the IC50 values for both SM253 and SM145 induced significant apoptosis (Figure 5), likely a result of suppressing the HSR. Consistent with this hypothesis is that, even at 10-fold over its IC50 value, 17-AAG does not induce significant apoptosis. SM253 also arrested cells in G0/G1 after 24 h, consistent with previous reports on this class of compound (Supplemental Figure S1).50
Figure 5.
Apoptosis analysis of HCT-116 cells treated with SM253, SM145, and 17-AAG. Graph shows the average of 3 separate assays (p = 0.004 for all apoptotic cells of SM253 (50 μM) compared to DMSO; raw data and table of mean ± SD appears in Supplemental Figure S2).
In conclusion, we have developed a new hsp90 inhibitor, SM253, which has significantly improved properties over our current unique lead structure SM145. Specifically, SM253 is synthesized in high yields, is relatively soluble, and is more potent than our previous lead. Furthermore, we show that it pulls-down hsp90 and inhibits the protein folding machinery in an hsp90-regulated luciferase assay. We have also shown that SM253 is superior to clinical inhibitors as it does not induce a heat shock response, and contrary to clinical inhibitors, SM253 reduces the proteins associated with the heat shock response by ∼2-fold. Finally, we show that SM253 rapidly induces apoptosis. Our success in designing and synthesizing an improved lead structure over 17-AAG and SM145 indicates that hsp90 can be an effective oncogenic target when the appropriate inhibitor is utilized.
Acknowledgments
We thank C. Brownlee and Biological Resources Imaging Laboratory (BRIL) and UNSW for support of S.R.M. Y.C.K. thanks the Australian government for providing the Endeavour scholarship fund. Finally we thank James Gardiner from CSIRO for helpful discussions.
Supporting Information Available
Synthetic procedures, characterization data, and biological assay procedures. This material is available free of charge via the Internet at http://pubs.acs.org.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Fierro-Monti I.; Echeverria P.; Racle J.; Hernandez C.; Picard D.; Quadroni M. Dynamic impacts of the inhibition of the molecular chaperone hsp90 on the T-cell proteome have implications for anti-cancer theapy. PLoS One 2013, 8, e80425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taipale M.; Krykbaeva I.; Koeva M.; Kayatekin C.; Westover K. D.; Karras G. I.; Lindquist S. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 2012, 150, 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohonowych J. E.; Gopal U.; Isaacs J. S. Hsp90 as a gatekeeper of tumor angiogenesis: clinical promise and potential pitfalls. J. Oncol. 2010, 2010, 412–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama S.; Reed J. C.; Homma S. Heat-shock proteins as regulators of apoptosis. Oncogene 2003, 22, 9041–7. [DOI] [PubMed] [Google Scholar]
- Neckers L. Heat shock protein 90: the cancer chaperone. J. Biosci. 2007, 32, 517–30. [DOI] [PubMed] [Google Scholar]
- Neckers L. Hsp90 inhbitors as novel cancer chemotherapeutic agents. Trends Mol. Med. 2002, 8, S55–S61. [DOI] [PubMed] [Google Scholar]
- Jolly C.; Morimoto R. I. Role of the heat shock protein response and molecular chaperones in oncogenesis and cell death. J. Natl. Cancer Inst. 2000, 92, 1564–1572. [DOI] [PubMed] [Google Scholar]
- Mayer T. U.; Kapoor T. M.; Haggarty S. J.; King R. W.; Schreiber S. L.; Mitchison T. J. Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen. Science 1999, 286, 971–974. [DOI] [PubMed] [Google Scholar]
- Neckers L.; Ivy S. P. Heat shock protein 90. Curr. Opin. Oncol. 2003, 15, 419–424. [DOI] [PubMed] [Google Scholar]
- Sarto C.; Binz P. A.; Mocarelli P. Heat shock proteins in human cancer. Electrophoresis 2000, 21, 1218–1226. [DOI] [PubMed] [Google Scholar]
- Morimoto R. I.; Kline M. P.; Bimston D. N.; Cotto J. J. The heat-shock response: regulation and function of heat-shock proteins and molecular chaperones. Essays Biochem. 1997, 32, 17–29. [PubMed] [Google Scholar]
- Hartl F. U.; Hayer-Hartl M. Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 2002, 295, 1852–1858. [DOI] [PubMed] [Google Scholar]
- Santagata S.; Hu R.; Lin N. U.; Mendillo M. L.; Collins L. C.; Hankinson S. E.; Schnitt S. J.; Whitesell L.; Tamimi R. M.; Lindquist S.; Ince T. A. High levels of nuclear heat-shock factor 1 (HSF1) are associated with poor prognosis in breast cancer. Proc. Natl. Acad. Sci. U.S.A. 2011, 108, 18378–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendillo M. L.; Santagata S.; Koeva M.; Bell G. W.; Hu R.; Tamimi R. M.; Fraenkel E.; Ince T. A.; Whitesell L.; Lindquist S. HSF1 drives a transcriptional program distinct from heat shock to support highly malignant human cancers. Cell 2012, 150, 549–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A.; Thao L.; Sensintaffar J.; Zhang L.; Boehm M. F.; Fritz L. C.; Burrows F. J. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature 2003, 425, 407. [DOI] [PubMed] [Google Scholar]
- Hsu H. S.; Lin J. H.; Huang W. C.; Hsu T. W.; Su K.; Chiou S. H.; Tsai Y. T.; Hung S. C. Chemoresistance of lung cancer stemlike cells depends on activation of Hsp27. Cancer 2011, 117, 1516–28. [DOI] [PubMed] [Google Scholar]
- Powers M. V.; Workman P. Inhibitors of the heat shock response: biology and pharmacology. FEBS Lett. 2007, 581, 3758–69. [DOI] [PubMed] [Google Scholar]
- Whitesell L.; Bagatell R.; Falsey R. The stress response: implications for clinical development of hsp90 inhibitors. Curr. Cancer Drug Targets 2003, 3, 349–358. [DOI] [PubMed] [Google Scholar]
- Lamoureux F.; Thomas C.; Yin M. J.; Kuruma H.; Beraldi E.; Fazli L.; Zoubeidi A.; Gleave M. E. Clusterin inhibition using OGX-011 synergistically enhances Hsp90 inhibitor activity by suppressing the heat shock response in castrate-resistant prostate cancer. Cancer Res. 2011, 71, 5838–49. [DOI] [PubMed] [Google Scholar]
- Zaarur N.; Gabai V. L.; Porco J. A.; Calderwood S.; Sherman M. Y. Targeting heat shock response to sensitize cancer cells to proteasome and Hsp90 inhibitors. Cancer Res. 2006, 66, 1783–91. [DOI] [PubMed] [Google Scholar]
- Jhaveri K.; Modi S. HSP90 inhibitors for cancer therapy and overcoming drug resistance. Adv. Pharmacol. 2012, 65, 471–517. [DOI] [PubMed] [Google Scholar]
- Jhaveri K.; Taldone T.; Modi S.; Chiosis G. Advances in the clinical development of heat shock protein 90 (Hsp90) inhibitors in cancers. Biochim. Biophys. Acta 2012, 1823, 742–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www.clinicaltrials.gov.
- Shelton S. N.; Shawgo M. E.; Matthews S. B.; Lu Y.; Donnelly A. C.; Szabla K.; Tanol M.; Vielhauer G. A.; Rajewski R. A.; Matts R. L.; Blagg B. S. J.; Robertson J. D. KU135, a Novel Novobiocin-Derived C-Terminal Inhibitor of the 90-kDa Heat Shock Protein, Exerts Potent Antiproliferative Effects in Human Leukemic Cells. Mol. Pharmacol. 2009, 76, 1314–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eskew J. D.; Sadikot T.; Morales P.; Duren A.; Dunwiddie I.; Swink M.; Zhang X.; Hembruff S.; Donnelly A.; Rajewski R. A.; Blad B.; Manjarrez J. R.; Matts R. L.; Holzbeierlein J. M.; Vielhauer G. A. Development and characeterizatrion of a novel C-terminal inhibitor of Hsp90 in androgen dependent and independent prostate cancer cells. BMC Cancer 2011, 11, 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell J. R.; Alexander L. D.; McAlpine S. R. A heat shock protein inhibitor that modulates immunophilins and regulates hormone receptors. Bioorg. Med. Chem. Lett. 2014, 24, 661–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis M. R.; Singh E. K.; Wahyudi H.; Alexander L. D.; Kunicki J.; Nazarova L. A.; Fairweather K. A.; Giltrap A. M.; Jolliffe K. A.; McAlpine S. R. Synthesis of Sansalvamide A peptiodmimetics: trizaole, oxazole, thiazole, and psuedoproline containing compounds. Tetrahedron 2012, 68, 1029–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardi V. C.; Alexander L. D.; Johnson V. A.; McAlpine S. R. Macrocycles that inhibit the binding between heat shock protein 90 and TPR-containing proteins. ACS Chem. Biol. 2011, 6, 1357–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasko R. C.; Rodriguez R. A.; Cunningham C. N.; Ardi V. C.; Agard D. A.; McAlpine S. R. Mechanistic studies of Sansalvamide A-Amide: an allosteric modulator of Hsp90. ACS Med. Chem. Lett. 2010, 1, 4–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers R. P.; Alexander L. D.; Johnson V. A.; Lin C.-C.; Savage J.; Corral R.; Moss J.; Slugocki T. S.; Singh E. K.; Davis M. R.; Ravula S.; Spicer J. E.; Oelrich J. L.; Thornquist A.; Pan C.-M.; McAlpine S. R. A third generation of Sansalvamide A derivatives: Design and synthesis of Hsp90 Inhibitors. Bioorg. Med. Chem. 2010, 18, 6822–6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otrubova K.; Lushington G. H.; Vander Velde D.; McGuire K. L.; McAlpine S. R. A comprehensive study of Sansalvamide A derivatives and their structure–activity relationships against drug-resistant colon cancer cell lines. J. Med. Chem. 2008, 51, 530–544. [DOI] [PubMed] [Google Scholar]
- Rodriguez R. A.; Pan P.-S.; Pan C.-M.; Ravula S.; Lapera S. A.; Singh E. K.; Styers T. J.; Brown J. D.; Cajica J.; Parry E.; Otrubova K.; McAlpine S. R. Synthesis of second generation Sansalvamide A derivatives: Novel templates as potent anti-tumor agents. J. Org. Chem. 2007, 72, 1980–2002. [DOI] [PubMed] [Google Scholar]
- Pan P. S.; McGuire K.; McAlpine S. R. Indentification of compounds potent against pancreatic cancers. Bioorg. Med. Chem. Lett. 2007, 17, 5072–5077. [DOI] [PubMed] [Google Scholar]
- Styers T. J.; Kekec A.; Rodriguez R. A.; Brown J. D.; Cajica J.; Pan P.-S.; Parry E.; Carroll C. L.; Medina I.; Corral R.; Lapera S.; Otrubova K.; Pan C.-M.; McGuire K. L.; McAlpine S. R. Synthesis of Sansalvamide A derivatives and their cytotoxicity in the colon cancer cell line HT-29. Bioorg. Med. Chem. 2006, 14, 5625–5631. [DOI] [PubMed] [Google Scholar]
- Otrubova K.; Styers T. J.; Pan P.-S.; Rodriguez R.; McGuire K. L.; McAlpine S. R. Synthesis and novel structure–activity relationships of potent Sansalvamide A derivatives. Chem. Commun. 2006, 1033–1034. [DOI] [PubMed] [Google Scholar]
- Kunicki J. B.; Petersen M. N.; Alexander L. D.; Ardi V. C.; McConnell J. R.; McAlpine S. R. Synthesis and evaluation of biotinylated Sansalvamide A analogs and their modulation of Hsp90. Bioorg. Med. Chem. Lett. 2011, 21, 4716–4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styers T. J.; Rodriguez R. A.; Pan P.-S.; McAlpine S. R. High-yielding macrocyclization conditions used in the synthesis of novel Sansalvamide A derivatives. Tetrahedron Lett. 2006, 47, 515–517. [Google Scholar]
- Sydor J. R.; Normant E.; Pien C. S.; Porter J. R.; Ge J.; Grenier L.; Pak R. H.; Ali J. A.; Dembski M. S.; Hudak J.; Patterson J.; Penders C.; Pink M.; Read M. A.; Sang J.; Woodward C.; Zhang Y.; Grayzel D. S.; Wright J.; Barrett J. A.; Palombella V. J.; Adams J.; Tong J. K. Development of 17-allylamino-17-demethoxygeldanamycin hydroquinone hydrochloride (IPI-504), an anti-cancer agent directed against Hsp90. Proc. Natl. Acad. Sci. U.S.A. 2006, 103, 17408–17413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter J. R.; Fritz C. C.; Depew K. M. Discovery and development of Hsp90 inhibitors: a promising pathway for cancer therapy . Curr. Opin. Chem. Biol. 2010, 14, 412–420. [DOI] [PubMed] [Google Scholar]
- Pan P. S.; Vasko R. C.; Lapera S. A.; Johnson V. A.; Sellers R. P.; Lin C.-C.; Pan C.-M.; Davis M. R.; Ardi V. C.; McAlpine S. R. A comprehensive study of Sansalvamide A derivatives: their structure–activity relationships and their binding mode to Hsp90. Bioorg. Med. Chem. 2009, 17, 5806–5825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otrubova K.; McGuire K. L.; McAlpine S. R. A scaffold targeting drug-resistant colon cancers. J. Med. Chem. 2007, 50, 1999–2002. [DOI] [PubMed] [Google Scholar]
- Wahyudi H.; Wang Y.; McAlpine S. R. Utilizing a dimerization strategy to inhibit the dimer protein Hsp90: Synthesis and biological activity of a sansalvamide A dimer. Org. Biomol. Chem. 2014, 12, 765–773. [DOI] [PubMed] [Google Scholar]
- Kim S. J.; Ramsey D. M.; Boyer C.; Davis T.; McAlpine S. R. Effectively delivering a drug using star polymers: Improving solubility of a unique hsp90 inhibitor. ACS Med. Chem. Lett. 2013, 4, 915–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahyudi H.; Tantisantisom W.; Liu X.; Ramsey D. M.; Singh E. K.; McAlpine S. R. Synthesis, structure–activity analysis, and biological evaluation of structurally related conformational isomers. J. Org. Chem. 2012, 77, 10596–10616. [DOI] [PubMed] [Google Scholar]
- Ramsey D. M.; McConnell J. R.; Alexander L. D.; Tanaka K. W.; Vera C. M.; Mcalpine S. R. A new Hsp90 inhibitorthat exhibits a novel biological profile. Bioorg. Med. Chem. Lett. 2012, 22, 3287–3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmacher R. J.; Hansen W. J.; Freeman B. C.; Alnemri E.; Litwack G.; Toft D. O. Cooperative action of Hsp70, Hsp90, and DnaJ proteins in protein renaturation. Biochemistry 1996, 35, 148–149. [DOI] [PubMed] [Google Scholar]
- Wilsen S.; Gestwicki J. E. Identification of small molecules that modify the protein folding activity of heat shock protein 70 (Hsp70). Anal. Biochem. 2008, 374, 371–376. [DOI] [PubMed] [Google Scholar]
- Wei L.; Liu T.; Wang H.; Hong H.; Yu A.; Feng H.; Chang W. Hsp27 participates in the maintenance of breast cancer stem cells through regulation of epithelial mesenchymal transition and nuclear factor-kB. Breast Cancer Res. 2011, 13, R101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra A.; Shevde L. A.; Samant R. S. Multi-faceted role of HSP40 in cancer. Clin. Exp. Metastasis 2009, 26, 559–67. [DOI] [PubMed] [Google Scholar]
- Heiferman M. J.; Salabat M. R.; Ujiki M. B.; Strouch M. J.; Cheon E. C.; Silverman R. B.; Bentrem D. J. Sansalvamide induces pancreatic cancer growth arrest through changes in the cell cycle. Anticancer Res. 2010, 30, 73–78. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.