Abstract
We provide here a protocol for the preparation of cap-analysis gene expression (CAGE) libraries, which allow measuring the expression of eukaryotic capped RNAs and simultaneously map the promoter regions. The presented protocol simplified the previously published ones and moreover produces tags that are 27 nucleotides long, which facilitates mapping to the genome. The protocol takes less than 5 days to complete and presents a notable improvement compared to previously published versions.
Keywords: Cap-analysis gene expression, RNAseq, transcriptome, sequencing, RNA
1. Introduction
CAGE (Cap Analysis of Gene Expression) is based on a series of full-length cDNA technologies previously developed at RIKEN. The purpose of the technology is to comprehensively map the vast majority of human transcription starting sites and hence their promoters, and simultaneously decipher the expression of the RNAs produced at each promoter. Thus, CAGE allows high-throughput gene expression profiling with simultaneous identification of the tissue/cell/condition specific transcriptional start sites (TSS), including promoter usage analysis. CAGE has various advantages over microarray-based expression analysis. The identification of the promoters used in the analyzed biological phenomena (tissue, cells, treatments, time courses, etc), together with the determination of the expression level at each promoter, allows identifying regulatory elements, such as core promoters and the transcription factor binding sites (TFBS) that are responsible for transcription. Bioinformatic analysis allows analysis of promoters having similar expression profiles are analyzed for the presence of common TFBS. Coupled to the determination of the expression of transcription factors, whichdrive the gene transcription, this analysis allows to reconstruct the networks that drive gene expression (1). By counting the number of CAGE tags for each promoter within a gene, we can determine not only the RNA expression level (this is a digital detection of frequency), but also from which of the various alternative promoters the RNA is transcribed, allowing comprehensive mapping of promoters in mammalian genomes (2). Additionally, sequencing based methods like CAGE allow to identify the transcriptome of expressed retrotransposon elements, a task not previously possible by microarrays hybridization (3) Comprehensive examples of applications have been published elsewhere (4). Here we present an updated protocol for the Illumina GA2X sequencer, in which the CAGE tags are 27 nucleotides long thanks to the use of EcoP15I, as compared with the previously used MmeI, which allows preparing only 20~21 bp long tags. Longer CAGE tags contribute to high efficiency mapping, if compared to previous versions of CAGE. To prepare a CAGE library, cDNA complementary strands are synthesized from total RNA extracted from cells or tissues, generally by using random primers, or a mixture of random and oligo-dT primers. The 5′ end of cDNA is then selected by using the cap-trapper method (5). Next, a biotinylated linker is attached to the 5′ end of single-strand cDNA. The linker contains the recognition site of the endonuclease EcoP15I. After the second cDNA strand is synthesized, the cDNA is cleaved 27 nucleotides away from the EcoP15I recognition site, which isolates the DNA derived from the 5′ end of the original RNA, to produce CAGE tags for the molecules present in the reactions. Next, a linker is attached to the 3′ end, in order to amplify and apply the sample into the Illumina GA2X sequencer to produce up to 20 million tags or more per sequencing lane. By changing the primers, the user may adapt the technology to other sequencing platforms.
2. Materials
2.1 Preparation of Sorbitol-Trehalose (3.3 M / 0.66 M) mix
Saturated trehalose solution: dissolve 7.27 g D-trehalose (Sigma-Aldrich, Cat#90208) in 10 ml water. Autoclave at 121°C, 30 min.
4.9 M sorbitol: dissolve 17.8 g D-Sorbitol (Wako, Cat#198-03755) in 20 ml water. Autoclave at 121°C, 30 min.
Mix saturated trehalose and 4.9 M sorbitol in 50 ml tube. To remove traces of heavy metals, which cleave nucleic acids, add about one gram of Chelex 100 Resin (Sigma, Cat#C7901) (do not use metal spoons), Mix well and incubate for 3 hrs at room temperature. At the end, collect the supernatant by centrifugation and store at room temperature.
2.2. 250 mM NaIO4 for Oxidation of the diol groups
Dissolve 530.47 μg NaIO4 (ICN Bio. Inc, Cat#152577) in 10 μl water. Store room temperature with avoid light. Prepare freshly before use.
2.3. 15 mM Biotin (Long Arm) Hydrazide for Biotinylation
Dissolve 75.06 μg Biotin (Long Arm) Hydrazide (VECTOR Lab, Cat# SP-1100, 50mg) in 13.5 μl water. Prepare freshly before use.
2.4. Preparation of 20 μg/μl E.coli tRNA
Dissolve 30 mg E.coli tRNA (Ribonucleic acid, transfer from Escherichia coli Type XX, Strain W, lyophilized powder (Sigma, Cat# R1753) in 400 μl water and add 45 μl of 10×RQ1 DNase buffer and 30 μl of RQ1 RNase-Free DNase (Promega, Cat# 9PIM610). Incubate at 37 °C, 2 hrs.
Add 10 μl of 0.5 M EDTA (pH 8.0), 10 μl of 10% SDS and 10 μl of 10 ng/ml Protenase K (Invitrogen, Cat#25530-049) to tRNA solution. Incubate at 45 °C, 30 min.
Add 500 μl of phenol/chloroform and 500 μl chloroform to incubate solution. Centrifuge at 15,000 rpm for 3 min. at room temperature.
Collect supernatant and add 25 μl of 5M NaCl, 525 μl of Isopropanol. Centrifuge at 15,000 rpm for 5 min. at room temperature.
Remove supernatant and add 900 μl of 80 % Ethanol to tRNA pellet. Centrifuge at 15,000 rpm for 5 min. at room temperature. Repeat this step.
Dissolve the tRNA pellet in 1.5 ml water. Store in aliquots at −20 °C,
2.5. Preparation of wash buffer for MPG beads
Wash buffer 1: Mix 45 ml of 5 M NaCl and 5 ml of 0.5 M EDTA (pH 8.0). Store at room temperature.
Wash buffer 2: Mix 3 ml of 5M NaCl, 100 μl of 0.5 M EDTA (pH 8.0) and 46.9 ml of water. Store at room temperature.
Wash buffer 3: Mix 1 ml of 1M Tris-HCl (pH 8.5), 100 μl of 0.5 M EDTA (pH 8.0), 25 ml of 1 M NaOAc (pH 6.1), 2 ml of 10% SDS and 21.9 ml of water. Store at room temperature.
Wash buffer 4: Mix 500 μl of 1M Tris-HCl (pH 8.5), 100 μl of 0.5 M EDTA (pH 8.0), 25 ml of 1M NaOAc (pH 6.1) and 24.4 ml of water. Store at room temperature.
2.6. Preparation of 3′ linker ligation buffer (5×)
Mix 50 μl of 250 mM Tris-HCl (pH 7.0), 10 μl of 100 mM ATP and 0.5 μl of 10 mg/ml BSA (Invitrogen, Cat#15561-020). Adjust to 200 μl in water. Store at room temperature.
Other reagents
PrimeScript Reverse Transcriptase (TAKARA, Cat# 2680A, 10000 U)
Agencourt RNAClean XP Kit (BECKMAN COULTER, Cat# A63987, 40 ml)
Agencourt AMPure XP Kit (BECKMAN COULTER, Cat# A63881, 60 ml)
RNase ONE Ribonuclease (Promega, Cat# M4261, 1000 U)
MPG Streptavidin (TAKARA, Cat# 6124A, 2ml)
OliGreen ssDNA Quantitation Kit (Molecular Probes, Cat#O11492)
Agilent RNA Pico-kit (Agilent, Cat#5067-1513)
Agilent DNA1000 kit (Agilent, Cat# 5067-1504)
DNA Ligation Kit <Mighty Mix> (TAKARA, Cat# 6023, 1 kit)
T4 DNA ligase (NEB, Cat# M0202S, 20000 U)
TaKaRa LA Taq (TAKARA, Cat# RR002A, 125 U)
Antarctic Phosphatase (NEB, Cat# M0289L, 5000 U)
EcoP15I (NEB, Cat# R0646S, 500 U)
Sinefungin (Calbiochem-Novabiochem international, Cat# 567051, 2 mg)
Phusion™ High-Fidelity DNA Polymerase (FINNZYMES, Cat# F-530S, 100 U)
Exonuclease I (E. coli) (NEB, Cat# M0293S, 3000 U)
MinElute PCR Purification Kit (QIAGEN, Cat# 28004, 50 columns)
Ethanol (70%)
10% SDS
10 mM dNTPs (Invitrogen, Cat#18427-088)
1M NaOAc (pH 4.5)
1M NaCitrate (pH 6.0)
0.5M EDTA (pH 8.0)
40% glycerol
1M Tris-HCl (pH 8.5)
1M Tris-HCl (pH 7.0)
50 mM NaOH
0.4M MgCl2
Nuclease-free water (Invitrogen Corp, Cat #10977-015)
Equipment
Micro pipettes
Multi pipetters
Pipette Tips (Low binding tips)
1.5 ml SnapLock Microtube, Non-Sterile, MaxyClear, Maxymum Recovery (AXYGEN, Cat# MCT-150-L-C, 250 tube)
96 well PCR Plate, 0.2 ml, Non-Sterile, Clear (AXYGEN, Cat# PCR-96-C, 50 plate)
Agilent 2100 Bioanalyzer (Agilent Technologies, Cat # G2928B)
NanoDrop 1000 spectrophotometer (Thermo Fisher Inc., Cat #S09NND360)
Dynal Magnetic stand (Invitriogen, MPC-96S)
Centrifugal Concentrator (TOMY Digital Biology Co., Ltd., Cat #35041048)
Thermal cycler
Genome Analyzer 2-X (Illumina)
Oligonucleotides
Reverse transcription: RT-N15-EcoP primer (EcoP15I site=Italic) 5′- AAGGTCTATCAGCAGNNNNNNNNNNNNNNNN -3′
2nd SOL primer 5′- Bio CCACCGACAGGTTCAGAGTTCTACAG -3′
PCR Forward primer 5′- AATGATACGGCGACCACCGACAGGTTCAGAGTTC -3′
PCR Reverse primer 5′- CAAGCAGAAGACGGCATACGA -3′
Sequencing Primer 5′- CGGCGACCACCGACAGGTTCAGAGTTCTACAG -3′
5′ linkers
- 5′-N6 upper linker, (barcode=Bold, EcoP15I site=Italic)
- AGA: 5′- CCACCGACAGGTTCAGAGTTCTACAGAGACAGCAGNNNNNN Phos- 3′
- CTT: 5′- CCACCGACAGGTTCAGAGTTCTACAGCTTCAGCAGNNNNNN Phos- 3′
- GAT: 5′- CCACCGACAGGTTCAGAGTTCTACAGGATCAGCAGNNNNNN Phos- 3′
- ACA: 5′- CCACCGACAGGTTCAGAGTTCTACAGACACAGCAGNNNNNN Phos- 3′
- ACT: 5′- CCACCGACAGGTTCAGAGTTCTACAGACTCAGCAGNNNNNN Phos- 3′
- ACG: 5′- CCACCGACAGGTTCAGAGTTCTACAGACGCAGCAGNNNNNN Phos- 3′
- ATC: 5′- CCACCGACAGGTTCAGAGTTCTACAGATCCAGCAGNNNNNN Phos- 3′
- ATG: 5′- CCACCGACAGGTTCAGAGTTCTACAGATGCAGCAGNNNNNN Phos- 3′
- AGC: 5′- CCACCGACAGGTTCAGAGTTCTACAGAGCCAGCAGNNNNNN Phos- 3′
- AGT: 5′- CCACCGACAGGTTCAGAGTTCTACAGAGTCAGCAGNNNNNN Phos- 3′
- TAG: 5′- CCACCGACAGGTTCAGAGTTCTACAGTAGCAGCAGNNNNNN Phos- 3′
- TGG: 5′- CCACCGACAGGTTCAGAGTTCTACAGTGGCAGCAGNNNNNN Phos- 3′
- GTA: 5′- CCACCGACAGGTTCAGAGTTCTACAGGTACAGCAGNNNNNN Phos- 3′
- GAC: 5′- CCACCGACAGGTTCAGAGTTCTACAGGACCAGCAGNNNNNN Phos- 3′
- GCC: 5′- CCACCGACAGGTTCAGAGTTCTACAGGCCCAGCAGNNNNNN Phos- 3′
- 5′-GN5 upper linker, (barcode=Bold, EcoP15I site=Italic)
- AGA: 5′- CCACCGACAGGTTCAGAGTTCTACAGAGACAGCAGGNNNNN Phos- 3′
- CTT: 5′- CCACCGACAGGTTCAGAGTTCTACAGCTTCAGCAGGNNNNN Phos- 3′
- GAT: 5′- CCACCGACAGGTTCAGAGTTCTACAGGATCAGCAGGNNNNN Phos- 3′
- ACA: 5′- CCACCGACAGGTTCAGAGTTCTACAGACACAGCAGGNNNNN Phos- 3′
- ACT: 5′- CCACCGACAGGTTCAGAGTTCTACAGACTCAGCAGGNNNNN Phos- 3′
- ACG: 5′- CCACCGACAGGTTCAGAGTTCTACAGACGCAGCAGGNNNNN Phos- 3′
- ATC: 5′- CCACCGACAGGTTCAGAGTTCTACAGATCCAGCAGGNNNNN Phos- 3′
- ATG: 5′- CCACCGACAGGTTCAGAGTTCTACAGATGCAGCAGGNNNNN Phos- 3′
- AGC: 5′- CCACCGACAGGTTCAGAGTTCTACAGAGCCAGCAGGNNNNN Phos- 3′
- AGT: 5′- CCACCGACAGGTTCAGAGTTCTACAGAGTCAGCAGGNNNNN Phos- 3′
- TAG: 5′- CCACCGACAGGTTCAGAGTTCTACAGTAGCAGCAGGNNNNN Phos- 3′
- TGG: 5′- CCACCGACAGGTTCAGAGTTCTACAGTGGCAGCAGGNNNNN Phos- 3′
- GTA: 5′- CCACCGACAGGTTCAGAGTTCTACAGGTACAGCAGGNNNNN Phos- 3′
- GAC: 5′- CCACCGACAGGTTCAGAGTTCTACAGGACCAGCAGGNNNNN Phos- 3′
- GCC: 5′- CCACCGACAGGTTCAGAGTTCTACAGGCCCAGCAGGNNNNN Phos- 3′
- 5′-lower linker, (barcode=Bold, EcoP15I site=Italic)
- AGA: 5′-Phos CTGCTGTCTCTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
- CTT: 5′-Phos CTGCTGAAGCTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
- GAT: 5′-Phos CTGCTGATCCTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
- ACA: 5′-Phos CTGCTGTGTCTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
- ACT: 5′-Phos CTGCTGAGTCTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
- ACG: 5′-Phos CTGCTGCGTCTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
- ATC: 5′-Phos CTGCTGGATCTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
- ATG: 5′-Phos CTGCTGCATCTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
- AGC: 5′-Phos CTGCTGGCTCTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
- AGT: 5′-Phos CTGCTGACTCTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
- TAG: 5′-Phos CTGCTGCTACTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
- TGG: 5′-Phos CTGCTGCCACTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
- GTA: 5′-Phos CTGCTGTACCTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
- GAC: 5′-Phos CTGCTGGTCCTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
- GCC: 5′-Phos CTGCTGGGCCTGTAGAACTCTGAACCTGTCGGTGG NH2 -3′
Preparation of 5′ linkers by annealing oligonucleotides (see Note 1)
Dissolve the purified oligonucleotides to 2 μg/μl in 1mM Tris-HCl (pH 7.5) and 0.1 mM EDTA (pH 8.0).
N6 linker reaction solution: Mix 1.5 μl of each specific 5′-N6 upper linker (3.0 μg), 1.5 μl of each specific 5′-lower linker (3.0 μg), 0.75 μl of 1 M NaCl and 3.25 μl of water.
GN5 linker reaction solution: Mix 6 μl of each specific 5′-N6 upper linker (12 μg), 6 μl of each specific 5′-lower linker (12 μg), 3 μl of 1 M NaCl and 15 μl of water.
The annealing reaction is carried at the following conditions: 95°C, 5 min gradient 0.1°C/sec, 83°C, 5 min, gradient 0.1°C/sec, 71°C 5 min, gradient 0.1°C/sec, 59°C 5 min, gradient 0.1°C/sec, 59°C 5 min, gradient 0.1°C/sec, 47°C 5 min, gradient 0.1°C/sec, 35°C 5 min, gradient 0.1°C/sec, 23°C 5 min, gradient 0.1°C/sec and 11°C Hold.
The final annealed linker solution can be kept on hold at 4°C, but for long term storage should be frozen at −20°C.
The “N6” and “GN5” linkers carrying the same barcode should be mixed at this stage. The total volume is 37.5 μl (0.8 μg/μl), using a final ratio of N6:GN5 = 1:4. This was found effective to maximize the ligation efficiency of the cDNAs with cap-trapped cDNA ends. These linkers are used at the concentration of 200 ng/μl when it’s starting from 5 μg of total RNA.
3′ linkers
3′ upper linker 5′-NNTCGTATGCCGTCTTCTGCTTG -3′
3′ lower linker 5′-CAAGCAGAAGACGGCATACGA -3′
Preparation of 3′ linkers by annealing oligonucleotides
Dissolve the purified oligonucleotides to 2 μg/μl in 1mM Tris-HCl pH 7.5 and 0.1 mM EDTA (pH 8.0).
Linker reaction solution: Mix 2.5 μl of 3′ upper linker (5.0 μg), 2.5 μl of 3′ lower linker (5.0 μg), 1.25 μl of 1 M NaCl and 6.25 μl of water.
The annealing reaction is carried at the following conditions: 95°C, 5 min gradient 0.1°C/sec, 83°C, 5 min, gradient 0.1°C/sec, 71°C 5 min, gradient 0.1°C/sec, 59°C 5 min, gradient 0.1°C/sec, 59°C 5 min, gradient 0.1°C/sec, 47°C 5 min, gradient 0.1°C/sec, 35°C 5 min, gradient 0.1°C/sec, 23°C 5 min, gradient 0.1°C/sec and 11°C Hold.
The final annealed linker concentration is 0.8 μg/μl. These linkers are used at the concentration of 100 ng/μl when it’s starting from 5 μg of total RNA. It can be kept on hold at 4°C, but for long term storage should be frozen at −20°C.
Samples (5 μg total RNA, polyA plus RNA or polyA minus RNA)
Advisable to use RNAs that isolated Trizol LS (Invitrogen, Cat#10296-010) or RNeasy kit (Quiagen, Cat#74104) have RIN value over 7 by measure Agilent RNA nano kit. This protocol RNA amounts are set for 5 μg total RNA. However we could prepare CAGE library with 5 μg polyA minus RNA and 1 μg polyA plus RNA which separated by Poly(A)Purist mRNA Purification Kits (Ambion, Cat# AM1916).
3. Methods
3.1. First-strand: Reverse transcription (RT) (see Note 2 and Note 3)
Preparation of RNA / primer mix: Mix 5 μg total RNA and 2.2 μl of 210 μM RT-N15-EcoP primer. Adjust volume to 7.5 μl in water. Incubate at 65°C, 5min and then cool on ice immediately.
Preparation of Enzyme mix: Mix 7.5 μl of 5× PrimeScript buffer, 1.87 μl of 10 mM dNTPs, 7.5 μl of Sorbitol / Trehalose mix solution, 3.75 μl of PrimeScript Reverse Transcriptase and 9.38 μl of water (see Note 4).
Add Enzyme mix to RNA/primer mix tube and carefully mix by pipetting on ice (total volume 37.5 μl).
Incubate in a thermal cycler as follows: 25°C, 30 sec; 42°C, 30 min; 50°C, 10 min; 56°C, 10 min; 60°C, 10 min; keep on hold on ice (see Note 5).
3.2. cDNA clean-up with the RNAClean XP kit (see Note 6)
Fragments shorter than 100 nt (like the first strand cDNA primers) are removed by this purification step.
Add 65 μl of RNA clean XP to 37.5 μl of RT reaction solution. And thoroughly mix by pipetting. Incubate at room temperature for 30 min, mixing every 10 min by pipetting.
Set the tube on the magnetic stand for 5 min and remove the supernatant.
Keep the sample on the magnetic stand and wash the beads with ethanol by pouring 150 μl of 70% EtOH, washing the beads and the tube walls. After checking that the beads are settled on the tube wall, remove the supernatant. Repeat this washing step once more.
To the rinsed beads, add 40 μl of water (pre-heated at 37°C) and extensively pipette (the manufactures suggest pipetting at least 20 times) to elute the cDNA / DNA hybrid.
Incubate at 10 min, 37°C and then set on the magnetic stand for 5 min to separate the beads. Collect the supernatant (40 μl).
Keep the remainder of cDNA on ice.
3.3. Oxidation of the diol groups, including the cap-site
The cap is modified in a two step reactions, the first is constituted by oxidation with NaIO4.
Mix 40 μl of cDNA, 2 μl of 1 M NaOAc (pH 4.5) and 2 μl of 250 mM NaIO4 by 10 times pipetting on ice. Proceed the reaction in the dark by putting immediately aluminum foil to cover the samples and leave on ice for 45 min.
Stop the reaction by adding 2 μl of 40% glycerol and mix thoroughly. Add 14 μl of 1M Tris-HCl (pH8.5) to bring the pH above 5.6. Subsequently, purify the sample with the RNAClean XP as in Step 3.2. Notice that the volume of the cDNA is different: to keep the ratio of RNA clean-up XP solution/cDNA at 1.8 fold, add 105 μl of RNA clean XP reagents to the 60 μl of cDNA obtained from the oxidation reaction above. At the end, collect the supernatant in 40 μl of water.
3.4. Biotinylation of the RNA (see Note 7)
Mix 40 μl of Oxidate cDNA, 4 μl of 1M Na-Citrate (pH 6.0) and 13.5 μl of 15 mM biotin hydrazide (Long Arm) (total volume 57.5 μl) by pipetting for 10 times and incubate at 23°C for 14-15 hrs (overnight) (see Note 8).
3.5. RNase I treatment (see Note 9)
Add 6 μl of 1M Tris-HCl (pH 8.5), 1 μl of 0.5M EDTA (pH 8.0) and 5 μl of RNase ONE Ribonuclease to 57.5 μl of Biotinylated solution (total volume 69.5 μl). Mix by pipetting and incubate at 37°C, 30 min and at 65°C, 5 min (see Note 10).
Cool the cDNA on ice for 2 min and proceed with the RNAClean XP purification using 125 μl of the Agencourt reagent with 69.5 μl of cDNA (1.8 fold ratio) and perform all the steps as in section 3.2. Redissolve the cDNA in 40 μl of water.
3.6. Cap-trapping using the MPG Streptavidin beads
Cap-trapping is achieved by capturing the cDNA that have reached the cap-site with MPG streptavidin beads. The vast majority of the truncated cDNAs are left in the solution and eliminated.
Prepare the beads by blocking them with tRNAs, to diminish non-specific interactions. Add 1.5 μl of 20 μg/μl E. coli tRNA mix to 100 μl of MPG beads and incubate at room temperature for 30-60 min, mixing every 10 min by pipetting. Separate the beads on a magnetic stand and remove supernatant. Wash the beads with 50 μl of Wash buffer 1 (2 times) and resuspend in 80 μl of Wash buffer 1.
Add 40 μl of RNase I treated cDNA to the 80 μl of washed MPG beads.
Incubate at room temperature for 30 min (mix by 10 times pipetting or moderate vortexing every 5 min.). Separate on the magnetic stand for 3 min. Next, remove the supernatant.
Extensively wash the beads by multiple pipetting with 150 μl of the washes below, followed by capture with a magnetic stand. Wash buffer 1 (1 time), Wash buffer 2 (1 time) Wash buffer 3 (2 times) and Wash buffer 4 (2 times) (see Note 11).
3.7. Release cDNA from beads
Before proceeding with the next reaction, cDNAs has to be removed from the magnetic beads with alkali, which denatures RNA / cDNA hybrids and simultaneously fragments the RNAs. To do this, add 65 μl of 50 mM NaOH to the RNA / cDNA-washed beads tube and incubate at room temperature for 10 min, with occasional mixing.
Separate the beads on the magnetic stand for 3 min and collect the supernatant to a new tube.
To this new tube, add 12 μl of 1M Tris-HCl (pH7.0) to neutralize the alkali solution. The total collected volume will consist of 72 μl. Keep the cDNA on ice before the next step.
Subsequently purify the sample using the AMPure XP using 130 μl of the reagents to 72 μl of eluted cDNA as in step 3.2 RNA clean XP kit. Resuspend the cDNA in 34 μl of water, and keep 3 μl for quality control (QC). QC consists in measuring the concentration of the cDNAs with Oligreen and the size of captured cDNAs with the RNA Pico Kit (see Note 12). The cDNA is subsequently concentrated by centrifugal concentrator at room temperature in a siliconized tube, and finally redissolved in 4 μl water. It is preferable to avoid complete drying of the pellet by measuring the remaining volume of the water during the concentrate operation, although this may be a tedious operation.
3.8. Ligation of a linker to the single stranded (ss) cDNA (see Note 13)
In a separate tube, prepare 1μl of the 5′ linker (200 ng/μl) for each sample, and incubate at 37°C, 5 min (see Note 14).
At the end, cool the linker on ice for 2 min, and add 4 μl of cDNA and 10 μl of DNA ligation Mighty Mix to 5′ linker tubes.
After extensive mixing, incubate overnight at 16°C
Add 55 μl of water to the 5′ linker ligated cDNA In case of mixing cDNA, add 10 μl of water to mixed cDNA (total volume 70 μl) (see Note 15). Purification cDNA with AMPure XP kit as in step 3.2. Resuspend the cDNA in 30.5 μl of water.
To avoid remaining of 5′ linkers, repeat twice of this purification step.
3.9. Second strand cDNA synthesis
At this stage, the second strand cDNA is prepared by priming the sequences added in the previous stage. The enzyme for the synthesis is LA-Taq, a thermostable DNA polymerase that is able to amplify long cDNA fragments. The reaction is set up as follows for each sample:
Add 5 μl of 10× LA Taq buffer, 5 μl of 25mM MgCl2, 8 μl of 2.5mM dNTPs, 1 μl of 2nd SOL primer (200 ng/μl, 24 μM) and 0.5 μl of LA Taq (5U/μl) to 30.5 μl of 5′ linker ligated sscDNA (total volume 50 μl) and gently mix by pipetting on ice.
Incubate at 94°C for 3 min, 42°C for 5 min to anneal the primer, 68°C for 20 min, 72°C for 2 min and then hold at 4°C.
3.10. Antarctic Phosphatase
Remove phosphate of 5′ lower linker (see Note 16).
Add 6 μl of 10 × Antarctic Phospatase reaction buffer, 4 μl of Antarctic Phospatase (5 U/μl) to 3.9. reaction solution and gently mix by 10 times pipetting.
Incubate at 37°C for 1 hr. Then inactivate enzyme reaction at 65°C for 5 min and cool on ice for 2 min.
The sample is then purified again using the AMPure XP kit as in step 3.2, adding 90 μl of beads to 50 μl of the second strand cDNA reaction, and redissolving the cDNA in 30 μl of water.
3.11. EcoP15I digestion
At this stage, cDNAs is cleaved with EcoP15I, from the end of second strand primer 27 nt into the cDNA. (see Note 17)
Preparation of premix solution: Mix 4 μl of 10 × NEBuffer 3, 0.4 μl of 10 mg/ml (100 ×) BSA, 4 μl of 10 mM (10 ×) ATP, 0.4 μl of 10 mM Sinefungin, 0.1 μl of EcoP15I (10 U/μl) and 1.1 μl of water (total volume 10 μl) by pipetting on ice.
Add 30 μl of the double strand cDNA from step 3.10. to 10 μl of premix solution. Incubate at 37°C for 3 hrs.
Add 1 μl of 0.4 M MgCl2 (to 10 mM final concentration) to stabilize the short tags and prevent their denaturation.
Incubate at 65°C, 20 min to inactivate the restriction enzyme. The digested cDNA can be kept on ice until the next step.
3.12. Addition of a 3′ linker to the cleaved tags
This step provides to the 5′ cDNA tags a 3′ end linker, suitable for the subsequent PCR for the final preparation of the CAGE tags suitable for sequencing.
Add 16 μl of 5 × 3′ linker ligation buffer (2.6), 1 μl of 3′linker (100 ng/μl), 3 μl of T4 DNA ligase (400 U/μl) and 19 μl of water to 41 μl of EcoP15I digested cDNA (total volume 80 μl) by pipetting on ice and incubate overnight at 16°C.
3.13. Removal of excess of 3′ linker (see Note 18)
Preparation the beads by mixing 10 μl of MPG beads and 1 μl of 20 μg/μl) E. coli tRNA, followed by moderate vortexing and incubation at room temperature for 30-60 min to coat the surface to avoid non-specific binding.
Separate the beads on a magnetic stand as in step 3.6 and was with Wash buffer 1 twice as above. Finally redissolve the beads in 25 μl of Wash buffer 1.
Add the beads to 80 μl of the cDNA ligated from Step 3.12, and incubate at room temperature for 30 min with occasional mixing by pipetting or mild vortexing.
Wash the beads as in Step 3.6, followed by a final quick wash with 100 μl of water. At this step, avoid heating the sample and perform as quickly as possible, to avoid losing the tags due to denaturation. After separating the beads, redissolve them with water, in a final volume of 20 μl. This will be the template for subsequent PCR reactions. Beads/CAGE tags may be kept at −20°C.
3.14. Pilot PCR experiments
An aliquot of the beads is tested by PCR to verify the number of cycles necessary to amplify the bulk PCR reaction in the subsequent stage. We test multiple cycles.
At first, set up 3 reactions to check the number of PCR cycles (e.g. 8, 10, 12 cycles). Depending on the experiment the amount of cDNAs may be lower, so a different number of PCR cycles is recommended when starting.
Preparation of PCR premix reaction solution: Mix 10 μl of 5× High-Fidelity buffer, 4 μl of 2.5 mM dNTPs, 0.5 μl of 100 μM PCR Forward primer, 0.5 μl of 100 μM PCR Reverse primer, 0.5 μl of Phusion polymerase (2 U/μl), 32.5 μl of water and 2 μl of cDNA from Step 3.13 (total volume 50 μl) by pipetting on ice.
Perform the PCR at the following conditions: 98°C for 30 sec, followed by (98°C for 10 sec, 60°C for 10 sec times the number of PCR cycles as required, hold at 4°C.
Check the product on the Bioanalyzer DNA1000 to measure the concentration and verify the size of the amplified product. The desired product is 96 bp long. Appearance of other contaminants should be minimal (see Note 19).
3.15. Bulk PCR amplification of the CAGE library
After selecting the best PCR cycle number at step 3.14, the bulk PCR (6 PCR tubes) is performed for the large part of the 12 μl remaining samples (see Note 20).
3.16. Purification of primers by Exonuclease I
Rather than performing tedious purification steps, excess of PCR primers are removed by Exonuclease I treatment, which cleaves only single stranded RNA. The CAGE tags are thus protected, being double strand.
Pool the PCR reaction solutions into one 1.5 ml siliconized tube. For the equivalent of each 3 PCR reactions.
Add 1 μl of ExonucleaseI (20 U/μl) to the 150 μl of PCR reaction solution and mix by pipetting on ice, then incubate at 37°C for 0.5-1 hr.
At the end, the CAGE tag sample is purified with the MinElute PCR Purification Kit, using the 151 μl of above product for each column, following the manufacturer’s instructions.
At the end, the CAGE tag is eluted in 10 μl EB for 1 column.
An aliquot is used to check the DNA concentration with the Agilent Bioanalyzer DNA1000. An example result is shown in Fig. 2D. While the remaining sample is ready for the Illumina GA2-X sequencing, using the 36 nt reads cycle. The standard protocol required a DNA concentration of 10 nM (0.67 ng/μl) (10 μl for a 96 bp CAGE tags), while the final DNA concentration in the sequencing reaction should be in the order of 5.0-7.0 pM.
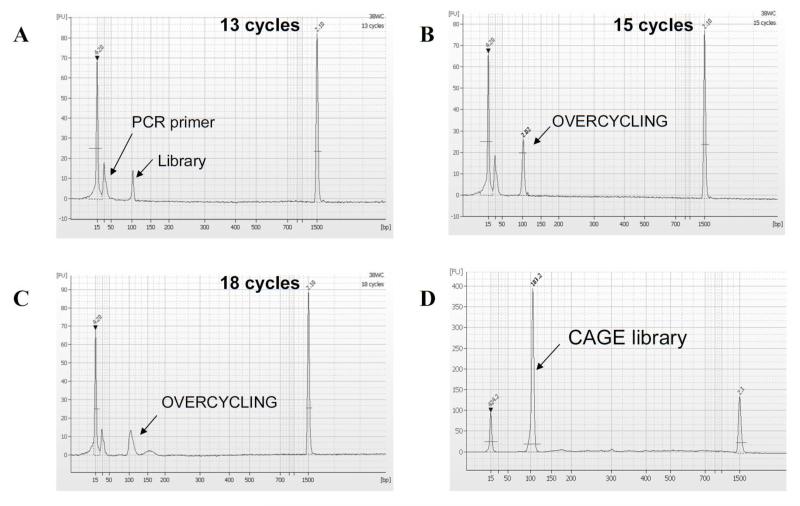
Fig. 2. Measurment of PCR products.
PCR cycle check results with Agilent DNA 1000 kit. The amount of loaded PCR pro duct is 1 μl. (A) Result is shown for 13 cycles, (B) 15 cycles and (C) 18 cycles. In case of 13 cycles, the single peak (103 bp) has a FU value between 5 to 10 (molarity: ~10 nmol/l), which is usable for bulk PCR. In case of 15 cycles, the FU exceeds 20 (molarity: ~30 nmol/l) and with 18 cycles the reactions shows an additional broad peaks, due to overcycling. (D) Final product is measured with the Agilent DNA 1000 kit. The molarity of the single peak at 103 bp was estimated to be ~183.2 nmol/l. PCR primers are removed before the assay during the step 3.15, consisting of an ExoI treatment and Minelute column purification. The single peak products are ready for sequencing.
4. Notes
The 5′ linkers are barcoded for pooling the CAGE libraries. This helps to (1) pool multiple libraries in the same sequencing lane; and (2) run the PCR reaction treating all the samples equally, to avoid differences due to different PCR conditions in different tubes. The latter is particularly important when comparing different samples, like in a time course. There are linkers labeled N6 (where the random fraction that will anneal on the cDNA will contain simply a random hexamer) or a GN5, where one of the bases is a G, which preferentially anneals with the first base of the cDNA that is often a C, which is added frequently by the RT in correspondence of the cap site. “Phos” stands for a phosphate group, NH2 for an amino-link that prevent ligation of 3′ ends. Upper and lower linkers are combined together according to their barcode sequence to form a double strand with partial single strand random protruding ends, which ligates on the terminal end of the cDNA. Linkers are mixed at GN5 to N6 at ratio of 4:1, to an equimolar amount of lower linker. Annealing should take place by slowly cooling the linkers as described temperature.
The purpose of this step is to convert the RNA to cDNA. The use of random primers allows reverse transcription of all RNAs including poly-A minus RNAs. In fact, a large amount of non polyadenylated RNAs are constituted by capped molecules, including long non-coding RNAs (6). Additionally, random priming minimizes the risk of under representation of long polyadenylated RNAs in the libraries, which may be due to differences in reverse transcription efficiency of small differences in quality of RNA. Altogether, random priming minimizes the chances to introduce biases in the library that are due to the mRNA size or potential mRNA truncation.
It is very important to work under RNase free conditions from steps 3.1 to 3.4. Damaging the RNA even after the synthesis of the cDNA may interfere with cap-trapping method.
The amount is given for a single tube, but the reaction can be scaled up depending on the number of samples. Add the enzyme to the mixture at the last moment before mixing.
The incubation at 25°C is essential to anneal the random primer and the RNA, which is extended at 42°C at first, followed by further extension at higher temperature. In presence of trehalose and sorbitol (7), the RT preserves its activity at higher temperatures and further extends the cDNA by reverse transcribing structured RNA regions, which are often present at the 5′ UTRs (untranslated regions) of the mRNAs.
It is important to clean-up the cDNA reaction and change the buffer before proceeding with the cap-oxidation and biotinylation reactions. Traces of Tris buffer, as well as saccharides or glycerol interfere with the oxidation and biotinylation, as diol groups are reactive at step 3.3. Additionally, fragments shorter than 100 nt (like the first strand cDNA primers) are removed by this purification steps.
This step adds a biotin group to the cap (and the 3′ end of the RNAs).
The reaction might alternatively be kept at 37°C for about 3 hrs, but some biotin hydrazide batches have shown to degrade nucleic acids at 37°C due to some impurities; at room temperature we have not observed issues in many years of experience with this reaction.
At this stage, the cap (and the 3′ ends of the cDNAs) are biotinylated. To perform the cap-trap and eliminate the cDNAs that did not reach the cap-site, it is mandatory to cleave with RNAse digesting the single strand RNAs at the 3′ ends of the cDNAs and at the 5′ ends, when the cDNAs do not reach the cap site. The RNAseI is an RNAse that cleaves at every base and it is relatively easy to inactivate by high temperature or SDS, thus is an ideal reagent for a RNA-dedicated laboratory.
This is important to remove, by denaturation, partially digested/nicked RNA from cDNA molecules. This happens at the random priming sites. Multiple random primers may produce multiple cDNA on the same RNA, only one of which would reach the cap site. Since random primers are long, they prime with several mismatches, cleaved by RNAse I. Heat treatment denatures these double strand nucleic acids where the RNA is nicked, preventing the capture of multiple cDNA hybridized to the same cDNA. Using total RNAs, this step has been important to reduce the ribosomal contamination below 1%.
Although multiple washes may seem tedious, we found that this helps to prevent contamination of non-capped molecules in the final library.
The cDNA concentration with Oligreen result is around 3.5 ng which measures is actual concentration of double strand cDNA. We expect to see a relatively broad cDNA size range by Agilent RNA pico kit. An example result is shown in Fig. 1.
This step is essential to ligate a linker at the 5′ ends, which will later be used to prime the second strand cDNA. This is obtained with the SSLLM abbreviation (8), which exploits the ability of a double strand linker with a protruding single strand random fraction to single strand cDNAs with DNA ligase. 200 ng of linkers are added to the 4 μl of cDNA solution, incubated at 65°C for 5 min to denature the cDNA secondary structures, followed by cooling on ice for 2 min. Sequences of various linkers with barcode sequencer are listed in the methods section and they should be annealed to each other separately before the step of ligation with the cDNA.
Since the linker contains regions of random sequences, they may have annealed during storage of the linker. This step thus helps to melt any dimer structure formed by linkers and make them fully available for the reaction.
It is possible at this stage to pool different cDNA mixtures if the 5′ ends linkers, used for each different cDNA, are barcoded. Sequencing through the barcode will allow distinguishing, after sequencing, the origin of the sample (9). Since the concentration of the linker dimer is high, repeat twice the purification of the cDNAs to avoid linker dimers in the final library. During the twice of purification step, we can pool another pooled cDNA after binding (just before 70% Ethanol wash) step.
Second strand synthesis primer can anneal and synthesis remaining free 5′ lower linkers. These anneled double stranded free linkers have phosphate at 3′ end. They are easy to ligate 3′ linker at step 3.13. To avoid occurring 5′ to 3′ linker dimer, we remove phosphate of anneled double stranded free linkers.
Also the first strand primer contains a EcoP15I sequence: having two sequences in opposite orientation has been found important to increase cleavage by EcoP15I, as well as the introduction of sinefungin to the reaction. We calculate carefully the amount of the enzyme, avoiding over digestion of the cDNA. Some class IIS restriction enzymes have been reported to inhibit the reaction when too large amount of enzymes were used.
The excess of 3′ linkers must be removed before performing the final PCR, otherwise the dimers produced by their ligation would heavily contaminate the CAGE library. To do this, we take advantage of the biotinylated primer that was used to prime the second strand (see reagents). The cDNA is retained on the beads, while the 3′ linkers are washed away.
When the protocol works properly, the CAGE tags can be applied on the Illumina sequencer without any size fractionation. To minimize contamination, we usually select the lowest number of PCR cycles that produce an acceptable amount of PCR product for the Illumina sequencer. Depending on the number of sequencing runs, or the desire to repeat the run multiple times or at a later time, the number of PCR cycles can be moderately increased. An example results is shown in Fig. 2A, B, C.
Do not amplify in a single tube a large amount of beads, as they may be inhibitory for the PCR reaction (in general, do not amplify more than 2 μl of beads for a 50 μl PCR reaction).
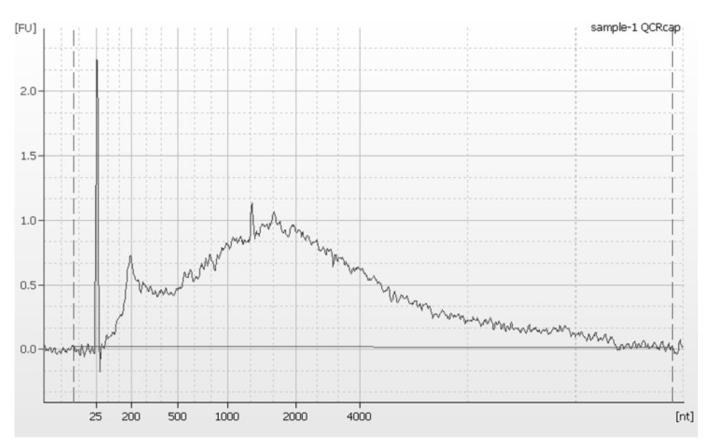
Fig. 1. Length and shape of cDNA.
cDNA quality check result of after Cap-trapping cDNA with Agilent RNA pico kit. One μl of the reaction was applied. Expected size ranges from few hundred nt to above 1-2 Kb. cDNA concentration is also measured by Oligreen (see Note 12 for the details).
Conclusions
Using this protocol, we routinely sequence 10-20 million tags per library we sequence with the current Illumina GA2-X sequencer. Depending on the user needs and future developments, it will be possible to sequence even more deeply to detect very rare transcriptional events, which take place only in few cells or cell compartments. Sequencing technology progress is relentless. We foresee that by changing primer sequences, this method will be suitable for other platforms or other versions of the Illumina sequencers. High throughput will allow pooling multiple, barcoded CAGE libraries for each lane of the Illumina sequencer, allowing to profile RNA by sequencing their 5′ end at a fraction of the cost of a microarray experiment, notably enhancing our capacity to interpret the genome and the significance of expression analysis. Bioinformatics analysis still poses big challenges, starting from the storage of progressively larger amount of data, to the development of all the interpretation tools. This part goes beyond the scope of this chapter. There are other publications on the CAGE (1, 4). At RIKEN we have prepared a web site (http://www.osc.riken.jp/english/activity/cage/) which contains the outline of the various versions of the CAGE technology, an updated list of publications and software that can be used to analyze the CAGE data.
We are convinced that this type of analysis, taking into account only the 5′ end of the cDNA, will allow maximizing the cost/performance of sequencing RNAs to study biology, with the further strength to analyze TSSs and thus the promoters that are responsible for gene expression.
Acknowledgments
This work was founded by a Research Grant for RIKEN Omics Science Center from MEXT and by the National Human Genome Research Institute grants U54 HG004557. We thank all the colleagues at the OSC for the precious feedbacks during the development of the methodology.
References
- 1.Suzuki H, Forrest ARR, van Nimwegen E, Daub CO, Balwierz PJ, Irvine KM, Lassmann T, Ravasi T, Hasegawa Y, de Hoon MJL, Katayama S, Schroder K, Carninci P, Tomaru Y, Kanamori-Katayama M, Kubosaki A, Akalin A, Ando Y, Arner E, Asada M, Asahara H, Bailey T, Bajic VB, Bauer D, Beckhouse AG, Bertin N, Bjorkegren J, Brombacher F, Bulger E, Chalk AM, Chiba J, Cloonan N, Dawe A, Dostie J, Engstrom PG, Essack M, Faulkner GJ, Fink JL, Fredman D, Fujimori K, Furuno M, Gojobori T, Gough J, Grimmond SM, Gustafsson M, Hashimoto M, Hashimoto T, Hatakeyama M, Heinzel S, Hide W, Hofmann O, Hornquist M, Huminiecki L, Ikeo K, Imamoto N, Inoue S, Inoue Y, Ishihara R, Iwayanagi T, Jacobsen A, Kaur M, Kawaji H, Kerr MC, Kimura R, Kimura S, Kimura Y, Kitano H, Koga H, Kojima T, Kondo S, Konno T, Krogh A, Kruger A, Kumar A, Lenhard B, Lennartsson A, Lindow M, Lizio M, MacPherson C, Maeda N, Maher CA, Maqungo M, Mar J, Matigian NA, Matsuda H, Mattick JS, Meier S, Miyamoto S, Miyamoto-Sato E, Nakabayashi K, Nakachi Y, Nakano M, Nygaard S, Okayama T, Okazaki Y, Okuda-Yabukami H, Orlando V, Otomo J, Pachkov M, Petrovsky N, Plessy C, Quackenbush J, Radovanovic A, Rehli M, Saito R, Sandelin A, Schmeier S, Schonbach C, Schwartz AS, Semple CA, Sera M, Severin J, Shirahige K, Simons C, Laurent GS, Suzuki M, Suzuki T, Sweet MJ, Taft RJ, Takeda S, Takenaka Y, Tan K, Taylor MS, Teasdale RD, Tegner J, Teichmann S, Valen E, Wahlestedt C, Waki K, Waterhouse A, AWells C, Winther O, Wu L, Yamaguchi K, Yanagawa H, Yasuda J, Zavolan M, Hume DA, Arakawa T, Fukuda S, Imamura K, Kai C, Kaiho A, Kawashima T, Kawazu C, Kitazume Y, Kojima M, Miura H, Murakami K, Murata M, Ninomiya N, Nishiyori H, Noma S, Ogawa C, Sano T, Simon C, Tagami M, Takahashi Y, Kawai J, Hayashizaki Y, Consortium F, Ctr ROS. Nature Genetics. 2009;41:553–62. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, Forrest AR, Alkema WB, Tan SL, Plessy C, Kodzius R, Ravasi T, Kasukawa T, Fukuda S, Kanamori-Katayama M, Kitazume Y, Kawaji H, Kai C, Nakamura M, Konno H, Nakano K, Mottagui-Tabar S, Arner P, Chesi A, Gustincich S, Persichetti F, Suzuki H, Grimmond SM, Wells CA, Orlando V, Wahlestedt C, Liu ET, Harbers M, Kawai J, Bajic VB, Hume DA, Hayashizaki Y. Nature Genetics. 2006;38:626–35. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 3.Faulkner GJ, Kimura Y, Daub CO, Wani S, Plessy C, Irvine KM, Schroder K, Cloonan N, Steptoe AL, Lassmann T, Waki K, Hornig N, Arakawa T, Takahashi H, Kawai J, Forrest ARR, Suzuki H, Hayashizaki Y, Hume DA, Orlando V, Grimmond SM, Carninci P. Nature Genetics. 2009;41:563–71. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 4.Carninci P. Cap-analysis gene expression (CAGE): the science of decoding gene transcription. Pan Stanford; Singapore: 2010. [Google Scholar]
- 5.Carninci P, Kvam C, Kitamura A, Ohsumi T, Okazaki Y, Itoh M, Kamiya M, Shibata K, Sasaki N, Izawa M, Muramatsu M, Hayashizaki Y, Schneider C. Genomics. 1996;37:327–36. doi: 10.1006/geno.1996.0567. [DOI] [PubMed] [Google Scholar]
- 6.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, Kodzius R, Shimokawa K, Bajic VB, Brenner SE, Batalov S, Forrest ARR, Zavolan M, Davis MJ, Wilming LG, Aidinis V, Allen JE, Ambesi-Impiombato X, Apweiler R, Aturaliya RN, Bailey TL, Bansal M, Baxter L, Beisel KW, Bersano T, Bono H, Chalk AM, Chiu KP, Choudhary V, Christoffels A, Clutterbuck DR, Crowe ML, Dalla E, Dalrymple BP, de Bono B, Della Gatta G, di Bernardo D, Down T, Engstrom P, Fagiolini M, Faulkner G, Fletcher CF, Fukushima T, Furuno M, Futaki S, Gariboldi M, Georgii-Hemming P, Gingeras TR, Gojobori T, Green RE, Gustincich S, Harbers M, Hayashi Y, Hensch TK, Hirokawa N, Hill D, Huminiecki L, Iacono M, Ikeo K, Iwama A, Ishikawa T, Jakt M, Kanapin A, Katoh M, Kawasawa Y, Kelso J, Kitamura H, Kitano H, Kollias G, Krishnan SPT, Kruger A, Kummerfeld SK, Kurochkin IV, Lareau LF, Lazarevic D, Lipovich L, Liu J, Liuni S, McWilliam S, Babu MM, Madera M, Marchionni L, Matsuda H, Matsuzawa S, Miki H, Mignone F, Miyake S, Morris K, Mottagui-Tabar S, Mulder N, Nakano N, Nakauchi H, Ng P, Nilsson R, Nishiguchi S, Nishikawa S, Nori F, Ohara O, Okazaki Y, Orlando V, Pang KC, Pavan WJ, Pavesi G, Pesole G, Petrovsky N, Piazza S, Reed J, Reid JF, Ring BZ, Ringwald M, Rost B, Ruan Y, Salzberg SL, Sandelin A, Schneider C, Schonbach C, Sekiguchi K, Semple CAM, Seno S, Sessa L, Sheng Y, Shibata Y, Shimada H, Shimada K, Silva D, Sinclair B, Sperling S, Stupka E, Sugiura K, Sultana R, Takenaka Y, Taki K, Tammoja K, Tan SL, Tang S, Taylor MS, Tegner J, Teichmann SA, Ueda HR, van Nimwegen E, Verardo R, Wei CL, Yagi K, Yamanishi H, Zabarovsky E, Zhu S, Zimmer A, Hide W, Bult C, Grimmond SM, Teasdale RD, Liu ET, Brusic V, Quackenbush J, Wahlestedt C, Mattick JS, Hume DA, Kai C, Sasaki D, Tomaru Y, Fukuda S, Kanamori-Katayama M, Suzuki M, Aoki J, Arakawa T, Iida J, Imamura K, Itoh M, Kato T, Kawaji H, Kawagashira N, Kawashima T, Kojima M, Kondo S, Konno H, Nakano K, Ninomiya N, Nishio T, Okada M, Plessy C, Shibata K, Shiraki T, Suzuki S, Tagami M, Waki K, Watahiki A, Okamura-Oho Y, Suzuki H, Kawai J, Hayashizaki Y, Consortium F, S RGERG. Science. 2005;309:1559–63. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 7.Carninci P, Shiraki T, Mizuno Y, Muramatsu M, Hayashizaki Y. Biotechniques. 2002;32:984–5. doi: 10.2144/02325bm01. [DOI] [PubMed] [Google Scholar]
- 8.Shibata Y, Carninci P, Watahiki A, Shiraki T, Konno H, Muramatsu M, Hayashizaki Y. Biotechniques. 2001;30:1250–54. doi: 10.2144/01306st01. [DOI] [PubMed] [Google Scholar]
- 9.Maeda N, Nishiyori H, Nakamura M, Kawazu C, Murata M, Sano H, Hayashida K, Fukuda S, Tagami M, Hasegawa A, Murakami K, Schroder K, Irvine K, Hume D, Hayashizaki Y, Carninci P, Suzuki H. Biotechniques. 2008;45:95–7. doi: 10.2144/000112814. [DOI] [PubMed] [Google Scholar]