Abstract
The prevalence of porcine sapoviruses (SaVs) and noroviruses (NoVs) in nursing piglets on three pig farms in Ohio was studied. Fecal samples (n = 139) were collected from individual pigs and screened for caliciviruses by RT-PCR. Phylogenetic analysis was conducted using partial sequences of the RNA polymerase region. Three different SaV genogroups, including a newly emerging one (DO19 Korea-like) were detected. No NoVs were detected. Kobuviruses, emerging members of the family Picornaviridae, were detected by primers designed for SaV. To our knowledge, this is the first report of porcine DO19 Korea-like SaV and kobuvirus in the United States.
Noroviruses (NoVs) and sapoviruses (SaVs) belong to the genera Norovirus and Sapovirus, respectively, within the family Caliciviridae. Human NoVs and SaVs cause foodand water-borne gastroenteritis outbreaks worldwide. NoVs have an approximately 7.3- to 7.8-kb positive-sense, single-stranded RNA genome with three open reading frames (ORFs). NoVs are genetically diverse and currently classified into five genogroups (GI to GV) based on the complete capsid sequence [1]. The SaV genome is 7.3 to 7.5 kb in length and contains two to three ORFs. Sapoviruses are classified into at least five distinct genogroups (GI to GV) based on the complete capsid sequence [2, 3]. Human SaVs belong to genogroups GI, GII, GIV, and GV, whereas the first porcine SaV identified (Cowden strain) belongs to GIII [4]. The Cowden strain was isolated from feces of a nursing piglet in the US [5]. New porcine sapovirus genogroups (GVI, GVII, GVIII) have also been proposed based on phylogenetic analysis of the complete capsid VP1 sequences [6-8]. Potentially new genogroups of porcine SaVs based on partial RNA-dependent RNA polymerase (RdRp) sequences have been reported [9, 10]. Recombinant SaVs have been described in both human and swine hosts [6, 11]. Sapovirus infection of pigs has been described in American, Asian and European countries [5-7, 10, 12-14]. Porcine GIII SaV was the predominant genogroup, with the highest prevalence in postweaning pigs and the lowest in nursing pigs [15, 16].
In the past two decades, a number of enteric viruses have emerged in the swine population, including porcine kobuvirus [17]. Kobuviruses are recently identified, nonenveloped, single-stranded, positive-sense RNA viruses in the family Picornaviridae [18-20]. Currently Aichi, bovine and porcine kobuvirus genomes have been characterized. The prototype porcine kobuvirus strain (S-1-HUN, EU787450) was first identified in 2008, in Hungary [21]. Porcine kobuvirus has been detected at high frequency in healthy pigs. However, a recent study found an association between porcine kobuvirus infection and diarrhea in pigs [22].
Fecal samples (n = 139) were collected from individual nursing pigs (up to 30 days old) from three different commercial swine farms in Ohio, US, using sterile swabs and containers. Thirty-four, 37 and 68 samples were collected during April 2011-January 2012 from farms A, B and C, respectively (Table 1). Fecal consistency (diarrhea or not) information was not provided for all the samples. Ten-percent (w/v) fecal suspensions were prepared in phosphate-buffered saline (PBS, 0.01 M, pH 7.2) and centrifuged (2000 × g, 20 min, 4 °C). Viral RNA was extracted from the supernatant using an RNeasy Mini Kit (QIAGEN, Valencia, CA, USA). Extracted RNA was treated with 5 U DNase I (Invitrogen, Carlsbad, CA, USA) to remove contaminating DNA. cDNA was synthesized using SuperScript III reverse transcriptase (Invitrogen, USA) with a random hexamer according to the manufacturer’s instructions. The cDNA obtained was used for the subsequent PCR.
Table 1.
Prevalence of porcine SaVs in pigs detected using RT-PCR and sequencing
| Swine farm | Month of collection | No. tested | No. of pigs positive by RT-PCR (%) | No. SaV-positive strains by RT-PCR (primers used) | No. sequenced (genogroup) |
|---|---|---|---|---|---|
| Farm A | April 2011 | 34 | 2 (5.8) | 1 (PEC66/65) | 1 (GIII) |
| 1 (p290/110) | 1 (GIII) | ||||
| Farm B | May 2011 | 7 | 1 (14.7) | 1 (p290/110) | 1 (GIII) |
| June 2011 | 30 | 11 (36.7) | 7 (PEC66/65) | 3 (GIII) | |
| 4 (p290/110) | 4 (GIII) | ||||
| 6 PSV11M/14M | 2 (GVII) | ||||
| 6 (SaVXF/R) | 5 (Unknown G?) | ||||
| Farm C | July 2011 | 40 | 0 | 0 | 0 |
| December 2011 | 20 | 0 | 0 | ||
| January 2012 | 8 | 0 | 0 | 0 | |
| Total | 139 | 14 (10.1) | 26 (18.7) | 17 |
Porcine SaVs and NoVs are detected by RT-PCR using specific primer sets [23] or using calicivirus universal primers followed by sequencing [6, 24]. In this study, we employed genogroup-specific primer pairs targeting the RdRp region and universal primer pairs to detect SaVs and NoVs in the swine population. The primer pairs used to detect porcine SaVs and NoVs are summarized in Table 2. The PCR conditions were optimized for all primers with the plasmid DNA of calicivirus strains available in our lab including Po/SaV/GIII/Cowden, Po/SaV/GVI/JJ681, Po/SaV/GVII/LL26, Po/SaV/GVIII/QW19, Po/NoV/GII.11/QW48, Po/NoV/GII.18/QW125, Po/NoV/GII.11 (RdRp)GII.19(capsid)/QW218, Hu/NoV/GII.4/HS194, and Bo/NoV/GIII.2/CV186 [6, 24]. The PCR products were separated by electrophoresis in an agarose gel, and representative samples from each farm were purified using a QIAquick Gel Extraction Kit (QIAGEN) before direct sequencing or cloning into the pCR2.1-TOPO (T/A) vector (Invitrogen) prior to sequencing.
Table 2.
Primers used to detect SaVs and NoVs by RT-PCR
| Primer | Sequence (5′–3′) | Polarity | Virus specificity | Target gene | Size (bp) | Location (nt) | Reference |
|---|---|---|---|---|---|---|---|
| p290 | GATTACTCCAAGTGGGACTCCAC | F | NoV, SaV | RdRp | 317 or 329 | 4568-4590a | [25] |
| P110 | DATYTCATCATCACCATA | R | NoV, SaV | RdRp | 4865–4884a | [26] | |
| PEC66 | GACTACAGCAAGTGGGATTCC | F | Po SaV | RdRp | 330 | 4327-4347b | [23] |
| PEC65 | ATACACACAATCATCCCCGTA | R | Po SaV | RdRp | 4636-4656b | ||
| PEC68M | AYY TRY TGG GTG AGT TTG TG | F | Po SaV | RdRp | 233 | 33-52c | This study |
| PEC67M | RAAY ACA TTG CCC TGG TAC | R | Po SaV | RdRp | 247-265c | ||
| PSV6M | CGG TCA TTY TGT GTR GAY TG | F | Po SaV | RdRp | 219 | 40-59d | This study |
| PSV7M | A TTVCCCGTRTAAGGMRCA | R | Po SaV | RdRp | 240-258d | ||
| PSV11M | CAC CCR GAG GGG ATC WCA | F | Po SaV | RdRp | 224 | 5-22e | This study |
| PSV14M | TAA CAV TSV AGC ACA CAA CAT G | R | Po SaV | RdRp | 207-228e | ||
| SV-F13 | GAYYWGGCYCTCGCYACCTAC | F | Hu SaV | Capsid | 803 | 5074-5094f | [32] |
| SV-R13 | GGTGANAYNCCATTKTCCAT | R | Hu SaV | Capsid | 5857-5876f | ||
| SV-F14 | GAACAAGCTGTGGCATGCTAC | F | Hu SaV | Capsid | 803 | 5074-5094f | |
| SV-R14 | GGTGAGMMYCCATTCTCCAT | R | Hu SaV | Capsid | 5857-5876f | ||
| SV-F22 | SMWAWTAGTGTTTGARATG | F | Hu SaV | Capsid | 438 | 5154-5172f | |
| SV-R2 | GWGGGRTCAACMCCWGGTGG | R | Hu SaV | Capsid | 5572-5591f | ||
| SaV PoVF | CATATGGTGATGATTGCCTCTATG | F | Po SaV | RdRp | 514 | 1133-1156g | This study |
| SaV PoVR | TCCATCTCAAACACTAATAGCCCA | R | Po SaV | RdRp | 1623-1646g | ||
| SaV XF | ATATGATGAGGGCTTTTGGCAT | F | Po SaV | RdRp | 425 | 326-347h | This study |
| SaV XR | CCCCTCCATGACATACACTACTG | R | Po SaV | RdRp | 728-750h | ||
| SaV XF2 | TGGAATTCGTGGTTGAAGACGACC | F | Po SaV | RdRp | 200 | 17-40h | This study |
| SaV XR2 | GTTGAACCTCTGGTACACTCCCAA | R | Po SaV | RdRp | 193-216h | ||
| SaV XIIF | AAGTTGGCCATTGACACCTTGTCG | F | Po SaV | RdRp | 265 | 20-43i | This study |
| SaV XIIR | CAACAACACGCTCATGCTGGAACA | R | Po SaV | RdRp | 261-284i | ||
| SaV XIIIF | CCAAATGTGCTGGCACAAGCTACT | F | Po SaV | Capsid | 144 | 115-138j | This study |
| SaV XIIIR | GCCAATCAAAGTGTTGGGTGCTGA | R | Po SaV | Capsid | 235-258j | ||
| G2SKFM-Po | CGTGGGARGGCGATCGCAA | F | Po NoV | Capsid | 344 | 5025-5043k | This study |
| G2SKRM-Po | CCVCCHGCRTANSCRTTRTACAT | R | Po NoV | Capsid | 5346-5368k | ||
| JV12Y | YATACCACTATGATGCAGAYTA | F | NoV | RdRp | 347 | 4551-4572a | [27] |
| JV13I | CATCATCACCATAGAAIGAG | R | NoV | RdRp | 4858-4897a |
Reference NoV and SaV strains with accession numbers (in parentheses) were used for primer design
Hu/NoV/GI.1/Norwalk (M87661)
Po/SaV/GIII/Cowden (AF182760)
Po/SaV/GVI/OH-JJ681 (AY974192)
F19-10 (FJ498786)
Po/SaV/GVII/ OH-LL26 (AY974195)
Hu/SaV/Manchester (X86560)
Po/SaV/TYMPo31 (AB521772)
Po/SaV/F2-4 (GU230161)
Po/SaV/K8/JP (AB222999)
Po/SaV/Brazil/2053P4 (DQ359100)
Po/NoV/GII.18/OH-QW125 (AY823305)
Cloning was done only for a few PCR-positive samples that had multiple and weak bands on the agarose gel. For most of the samples, direct sequencing was done. DNA sequencing was performed with using BigDye Terminator Cycle Sequencing Kit with a 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA). Sequence editing was performed using the Lasergene software package (v8, DNASTAR Inc., Madison, WI, USA). The Basic Local Alignment Search Tool (BLAST http://www.ncbi.nlm.nih.gov/BLAST) was used to find homologous hits. Multiple sequence alignments of SaVs were generated by Clustal W, and bootstrapped phylogenetic trees were constructed by the neighbor-joining method with 1,000 bootstrap replicates using the Lasergene software.
Primer pair PEC66/65 detected eight GIII SaVs from the different farms; four of these were selected for sequence analysis and were confirmed as GIII SaV (Table 1). Primer pair PSV11M/14M detected six SaV strains, and two were sequenced and confirmed as GVII SaV. No SaVs were detected using the primer pairs PSV6M/7M or PEC68M/67M. Primer pairs designed based on human SaVs were used to detect human SaVlike RNA in swine; however, no human-like SaVs were detected. In this study, different newly designed primer pairs were used to detect the potential newly emerging porcine SaV genogroups GV?, GX?, GXII? and GXIII?. Primer pair SaVXF/R detected six SaVs, of which five were sequenced and confirmed as potential DO19 Korealike SaVs (unclassified genogroup, G?). For samples that were negative using the SaV genogroup-specific primer pairs, we next employed the calicivirus universal primer pair p290/110, which was designed based on the conserved motifs found in the RdRp genes of caliciviruses [25, 26]. Six samples were positive and were identified as GIII SaVs by sequence analysis. No porcine or humanlike NoVs were detected in the 139 samples tested using primer pairs G2SKFM-Po/G2SKRM-Po and JV12Y/JV13I [27].
Overall, the different primers used in this study detected 26 SaVs by RT-PCR, from 14 different pigs (Table 1). Of these, six pigs in farm B showed multiple SaV infections detected using primer pairs PEC66/65, PSV11M/14M and SaVXF/R. Most of the porcine SaVs detected were from farm B, where 32 % of pigs (12/37) were positive for at least one of the SaV genogroups.
A total of 17 samples from 26 PCR products that were positive for the partial RdRp genes of porcine SaVs were sequenced (Table 1). For phylogenetic analysis, 286 nucleotides from the RdRp gene were examined for all samples (Fig. 1A), except those amplified with primer pair SaVXF/R (425 nt), which were from a different region of the RdRp. The phylogenetic analysis divided the SaV sequences identified in this study into three different genogroups. Most SaVs belonged to GIII, which originally prevailed in swine populations worldwide. Genogroup GVII was also confirmed for two samples (Fig. 1A, Table 1). Furthermore, strains belonging to a potentially new sapovirus DO19 Korea-like genogroup [13], tentatively unclassified (G?), were also detected (Table 1). The porcine GIII SaVs identified in this study demonstrated nucleotide sequence identities of 77.5-100 % to one another in the RdRp region, and 77.1-100 % identity to the Cowden strain, the prototype porcine SaV. The potential GVII sequences from this study had 82.6-89.3 % sequence identity to our previously reported OH-LL26/2002/US strain from Ohio swine [6]. Moreover, the RdRp sequences of the potentially new genogroup (G?) (KC242615–242619) showed 98.4-100 % nucleotide sequence identity among themselves and 85.9-86.2 % nucleotide sequence identity to the strain DO19 Korea (HM346630) [13]. Additionally, in this study, primer pair SaVXF/R also detected porcine kobuviruses (KC242620–242624) with a similar amplicon size to that of SaV, with 78.3-98.4 % sequence identity among the detected strains and 81.6-87.4 % nucleotide identity to the prototype strain, S-1-HUN/2007 (EU787450) (Fig. 1B).
Fig. 1.
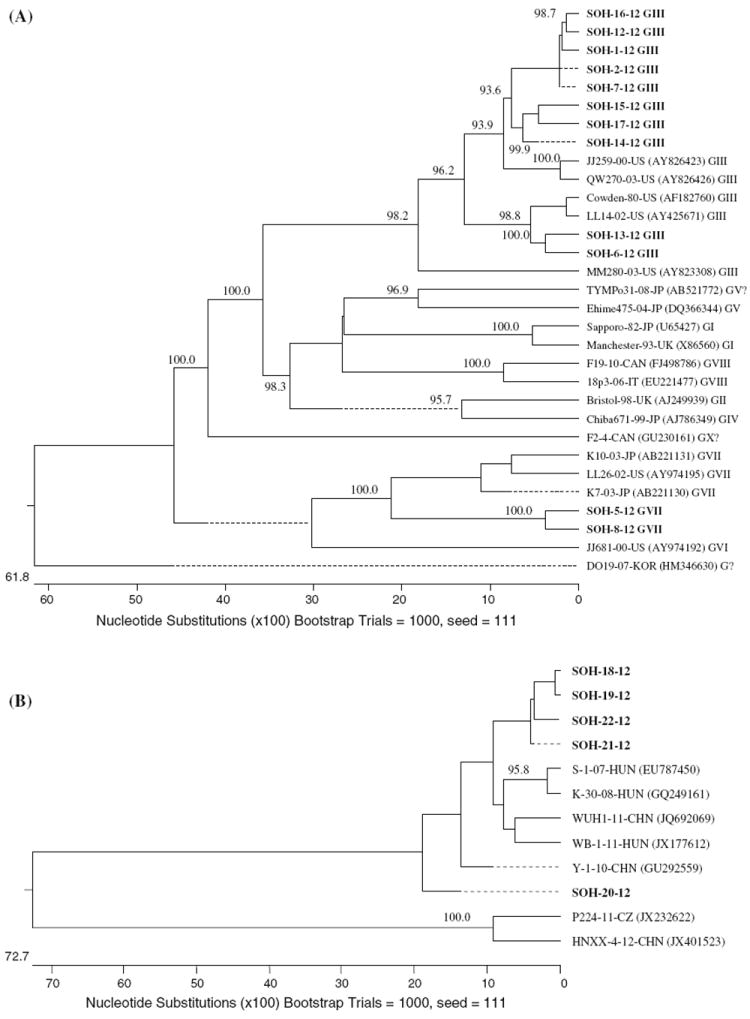
Neighbor-joining phylogenetic trees of the porcine enteric viruses detected. A. Tree based on 286 nucleotides of the RdRP gene of SaVs. B. Tree based on 390 nucleotides of the L and VP0 regions of porcine kobuvirus. Only bootstrap values greater than 85 % are shown. The bold prefix SOH indicates strains from the current study
Sapoviruses are genetically diverse. However, there are few studies of the molecular epidemiology of SaVs in nursing pigs in the US. Based on our data, different genogroups of SaVs were detected in 14 (10.1 %) of the nursing pigs tested from farms in Ohio, US. The porcine strains clustered into two known genogroups, GIII being the predominant genogroup sequenced (10/17, 58.8 %), and one potentially new unclassified DO19 Korea-like genogroup. In the present study, the presence of humanlike SaVs in swine was also tested to detect potential interspecies transmission; however, no human-like SaV was found in the pigs tested. Moreover, no human-like or porcine NoVs were detected in this study, which is in agreement with previous reports that NoVs were detected exclusively in adult pigs [15].
The potential SaV GIII (14/139, 10.1 %) and GVII (6/139, 4.3 %) positive rates in our study were lower than in our previous studies of nursing pigs in the US: 21 % and 20 %, respectively [15]. For GIII SaVs, this is probably due to the low sensitivity of the current primer set PEC66/65, which was designed based on the Po/SaV/GIII/Cowden strain detected in 1980 [5]. The calicivirus universal primer set p290/p110 detected six more GIII SaVs that were missed by primer set PEC66/65 (Table 1). Another similar study of nursing pigs (less than 4 weeks of age) from the Czech Republic reported a GIII positive rate of 3 % [16]. Multiple infections with different genogroups of SaVs were also observed in six diarrheic pigs in Farm B, which were put on milk replacer rather than milk from the sow. These particular piglets were also positive for rotaviruses [28]. The multiple infections suggest a lack of maternal antibodies due to artificial milk feeding or lower hygienic standards in the pens of these pigs, both of which could play a role in infection by multiple pathogens among the piglets. Wide variation in the rate of SaV detection among farms was also observed (0-32.4 %), with no SaVs detected in farm C, although it was sampled in both summer and winter. These data suggest that SaV infection rates may be influenced by the management or sanitary conditions of the farms where pigs were kept or that the prevalence of SaV maternal antibodies may vary greatly. A seasonality pattern for SaVs could not be defined from this study, as the samples were collected from each farm in various months.
In this study, kobuviruses were also detected using the newly designed primer pair SaVXF/R, which indicates a low specificity of the primer pair. Interestingly, the first porcine kobuvirus was also detected using the calicivirus universal primer set p290/289 [21]. Kobuviruses are a relatively newly recognized group of viruses in humans and animals [18, 19, 21]. Since its discovery in Hungary [21], porcine kobuvirus has been detected in 30 %, 99 %, 45 % and 52 % of domestic pigs tested in China [29], Thailand [30], Japan [31] and Korea [22], respectively. Recently, porcine kobuvirus has also been detected in the Czech Republic [16], which indicates the increased geographic distribution of porcine kobuviruses in pig farms. To our knowledge, this is the first report of porcine kobuvirus detection in five nursing pig samples from the US.
In summary, we detected SaVs in nursing pigs in two of three swine farms in Ohio, including a newly emerging genogroup. Continued surveillance of SaVs and other enteric viruses in swine is important to define their epidemiology, disease association and zoonotic potential.
Acknowledgments
This work was supported in part by a National Institutes of Health-Fogarty Grant (project GRT00117993/60023878). Salaries and research support were provided by state and federal funds provided to the Ohio Agricultural Research and Development Center (OARDC), The Ohio State University. Special thanks to Kelly Scheuer, for technical assistance and assistance in manuscript preparation, and Anastasia Vlasova and Kwonil Jung for their critical review of this manuscript. We also thank Joshua Amimo for help in sample preparation.
Contributor Information
Zufan Sisay, Department of Veterinary Preventive Medicine, Food Animal Health Research Program, Ohio Agricultural Research and Development Center, The Ohio State University, Wooster, OH 44691, USA; Department of Microbiology and Immunology, Aklilu Lemma Institute of Pathobiology, Addis Ababa University, P. O. Box 1176, Addis Ababa, Ethiopia.
Qiuhong Wang, Email: wang.655@osu.edu, Department of Veterinary Preventive Medicine, Food Animal Health Research Program, Ohio Agricultural Research and Development Center, The Ohio State University, Wooster, OH 44691, USA.
Tomoichiro Oka, Department of Veterinary Preventive Medicine, Food Animal Health Research Program, Ohio Agricultural Research and Development Center, The Ohio State University, Wooster, OH 44691, USA.
Linda Saif, Email: saif.2@osu.edu, Department of Veterinary Preventive Medicine, Food Animal Health Research Program, Ohio Agricultural Research and Development Center, The Ohio State University, Wooster, OH 44691, USA.
References
- 1.Zheng DP, Ando T, Fankhauser RL, Beard RS, Glass RI, Monroe SS. Norovirus classification and proposed strain nomenclature. Virology. 2006;346(2):312–323. doi: 10.1016/j.virol.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 2.Farkas T, Zhong WM, Jing Y, Huang PW, Espinosa SM, Martinez N, Morrow AL, Ruiz-Palacios GM, Pickering LK, Jiang X. Genetic diversity among sapoviruses. Arch Virol. 2004;149(7):1309–1323. doi: 10.1007/s00705-004-0296-9. [DOI] [PubMed] [Google Scholar]
- 3.Oka T, Mori K, Iritani N, Harada S, Ueki Y, Iizuka S, Mise K, Murakami K, Wakita T, Katayama K. Human sapovirus classification based on complete capsid nucleotide sequences. Arch Virol. 2012;157(2):349–352. doi: 10.1007/s00705-011-1161-2. [DOI] [PubMed] [Google Scholar]
- 4.Guo M, Chang KO, Hardy ME, Zhang Q, Parwani AV, Saif LJ. Molecular characterization of a porcine enteric calicivirus genetically related to sapporo-like human caliciviruses. J Virol. 1999;73(11):9625–9631. doi: 10.1128/jvi.73.11.9625-9631.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saif LJ, Bohl EH, Theil KW, Cross RF, House JA. Rotavirus-like, calicivirus-like, and 23-nm virus-like particles associated with diarrhea in young pigs. J Clin Microbiol. 1980;12(1):105–111. doi: 10.1128/jcm.12.1.105-111.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang QH, Han MG, Funk JA, Bowman G, Janies DA, Saif LJ. Genetic diversity and recombination of porcine sapoviruses. J Clin Microbiol. 2005;43(12):5963–5972. doi: 10.1128/JCM.43.12.5963-5972.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin Y, Tohya Y, Ogawa Y, Numazawa D, Kato K, Akashi H. Genetic analysis of calicivirus genomes detected in intestinal contents of piglets in Japan. Arch Virol. 2006;151(9):1749–1759. doi: 10.1007/s00705-006-0750-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martella V, Lorusso E, Banyai K, Decaro N, Corrente M, Elia G, Cavalli A, Radogna A, Costantini V, Saif LJ, Lavazza A, Di Trani L, Buonavoglia C. Identification of a porcine calicivirus related genetically to human sapoviruses. J Clin Microbiol. 2008;46(6):1907–1913. doi: 10.1128/JCM.00341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reuter G, Zimsek-Mijovski J, Poljsak-Prijatelj M, Di Bartolo I, Ruggeri FM, Kantala T, Maunula L, Kiss I, Kecskemeti S, Halaihel N, Buesa J, Johnsen C, Hjulsager CK, Larsen LE, Koopmans M, Bottiger B. Incidence, diversity, and molecular epidemiology of sapoviruses in swine across Europe. J Clin Microbiol. 2010;48(2):363–368. doi: 10.1128/JCM.01279-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakamura K, Saga Y, Iwai M, Obara M, Horimoto E, Hasegawa S, Kurata T, Okumura H, Nagoshi M, Takizawa T. Frequent detection of noroviruses and sapoviruses in swine and high genetic diversity of porcine sapovirus in Japan during fiscal year 2008. J Clin Microbiol. 2010;48(4):1215–1222. doi: 10.1128/JCM.02130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dos Anjos K, Lima LM, Silva PA, Inoue-Nagata AK, Nagata T. The possible molecular evolution of sapoviruses by interand intra-genogroup recombination. Arch Virol. 2011;156(11):1953–1959. doi: 10.1007/s00705-011-1079-8. [DOI] [PubMed] [Google Scholar]
- 12.Kim HJ, Cho HS, Cho KO, Park NY. Detection and molecular characterization of porcine enteric calicivirus in Korea, genetically related to sapoviruses. J Vet Med B Infect Dis Vet Public Health. 2006;53(4):155–159. doi: 10.1111/j.1439-0450.2006.00939.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Song YJ, Yu JN, Nam HM, Bak HR, Lee JB, Park SY, Song CS, Seo KH, Choi IS. Identification of genetic diversity of porcine norovirus and sapovirus in Korea. Virus Genes. 2011;42(3):394–401. doi: 10.1007/s11262-011-0588-6. [DOI] [PubMed] [Google Scholar]
- 14.L’Homme Y, Brassard J, Ouardani M, Gagne MJ. Characterization of novel porcine sapoviruses. Arch Virol. 2010;155(6):839–846. doi: 10.1007/s00705-010-0651-y. [DOI] [PubMed] [Google Scholar]
- 15.Wang QH, Souza M, Funk JA, Zhang W, Saif LJ. Prevalence of noroviruses and sapoviruses in swine of various ages determined by reverse transcription-PCR and microwell hybridization assays. J Clin Microbiol. 2006;44(6):2057–2062. doi: 10.1128/JCM.02634-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dufkova L, Scigalkova I, Moutelikova R, Malenovska H, Prodelalova J. Genetic diversity of porcine sapoviruses, kobuviruses, and astroviruses in asymptomatic pigs: an emerging new sapovirus GIII genotype. Arch Virol. 2012 doi: 10.1007/s00705-012-1528-z. [DOI] [PubMed] [Google Scholar]
- 17.Meng XJ. Emerging and re-emerging swine viruses. Transboundary Emerg Dis. 2012;59(suppl 1):85–102. doi: 10.1111/j.1865-1682.2011.01291.x. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita T, Kobayashi S, Sakae K, Nakata S, Chiba S, Ishihara Y, Isomura S. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J Infect Dis. 1991;164(5):954–957. doi: 10.1093/infdis/164.5.954. [DOI] [PubMed] [Google Scholar]
- 19.Yamashita T, Ito M, Kabashima Y, Tsuzuki H, Fujiura A, Sakae K. Isolation and characterization of a new species of kobuvirus associated with cattle. J Gen Virol. 2003;84(Pt 11):3069–3077. doi: 10.1099/vir.0.19266-0. [DOI] [PubMed] [Google Scholar]
- 20.Reuter G, Boldizsar A, Pankovics P. Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch Virol. 2009;154(1):101–108. doi: 10.1007/s00705-008-0288-2. [DOI] [PubMed] [Google Scholar]
- 21.Reuter G, Boldizsar A, Kiss I, Pankovics P. Candidate new species of kobuvirus in porcine hosts. Emerg Infect Dis. 2008;14(12):1968–1970. doi: 10.3201/eid1412.080797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park SJ, Kim HK, Moon HJ, Song DS, Rho SM, Han JY, Nguyen VG, Park BK. Molecular detection of porcine kobuviruses in pigs in Korea and their association with diarrhea. Arch Virol. 2010;155(11):1803–1811. doi: 10.1007/s00705-010-0774-1. [DOI] [PubMed] [Google Scholar]
- 23.Wang QH, Chang KO, Han MG, Sreevatsan S, Saif LJ. Development of a new microwell hybridization assay and an internal control RNA for the detection of porcine noroviruses and sapoviruses by reverse transcription-PCR. J Virol Methods. 2006;132(1–2):135–145. doi: 10.1016/j.jviromet.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Wang QH, Han MG, Cheetham S, Souza M, Funk JA, Saif LJ. Porcine noroviruses related to human noroviruses. Emerg Infect Dis. 2005;11(12):1874–1881. doi: 10.3201/eid1112.050485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiang X, Huang PW, Zhong WM, Farkas T, Cubitt DW, Matson DO. Design and evaluation of a primer pair that detects both norwalk- and sapporo-like caliciviruses by RT-PCR. J Virol Methods. 1999;83(1–2):145–154. doi: 10.1016/s0166-0934(99)00114-7. [DOI] [PubMed] [Google Scholar]
- 26.Le Guyader F, Estes MK, Hardy ME, Neill FH, Green J, Brown DW, Atmar RL. Evaluation of a degenerate primer for the PCR detection of human caliciviruses. Arch Virol. 1996;141(11):2225–2235. doi: 10.1007/BF01718228. [DOI] [PubMed] [Google Scholar]
- 27.Vennema H, de Bruin E, Koopmans M. Rational optimization of generic primers used for norwalk-like virus detection by reverse transcriptase polymerase chain reaction. J Clin Virol. 2002;25(2):233–235. doi: 10.1016/s1386-6532(02)00126-9. [DOI] [PubMed] [Google Scholar]
- 28.Amimo JO, Vlasova AN, Saif LJ. Detection and genetic diversity of porcine group A rotaviruses in historic (2004) and recent (2011/12) swine fecal samples in Ohio, USA: Predominance of G9P[13] genotype in nursing piglets (accepted) 2013 doi: 10.1128/JCM.03193-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu JM, Jin M, Zhang Q, Li HY, Li DD, Xu ZQ, Li JS, Cui SX, Yang SH, Liu N, Duan ZJ. Candidate porcine kobuvirus, China. Emerg Infect Dis. 2009;15(5):823–825. doi: 10.3201/eid1505.081518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khamrin P, Maneekarn N, Kongkaew A, Kongkaew S, Okitsu S, Ushijima H. Porcine kobuvirus in piglets, Thailand. Emerg Infect Dis. 2009;15(12):2075–2076. doi: 10.3201/eid1512.090724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khamrin P, Maneekarn N, Hidaka S, Kishikawa S, Ushijima K, Okitsu S, Ushijima H. Molecular detection of kobuvirus sequences in stool samples collected from healthy pigs in Japan. Infection, genetics and evolution. J Mol Epidemiol Evol Genet Infect Dis. 2010;10(7):950–954. doi: 10.1016/j.meegid.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Okada M, Yamashita Y, Oseto M, Shinozaki K. The detection of human sapoviruses with universal and genogroupspecific primers. Arch Virol. 2006;151(12):2503–2509. doi: 10.1007/s00705-006-0820-1. [DOI] [PubMed] [Google Scholar]


