Abstract
In this study, swine fecal specimens (n = 251) collected from nursing and weaned piglets raised under smallholder production systems were screened for the presence of kobuviruses by RT-PCR. Porcine kobuviruses were detected in 13.1 % (33/251) of the samples. We demonstrated that porcine kobuvirus infections exist in indigenous pigs in Kenya and Uganda and that the prevalence was higher in young piglets than older pigs: nursing piglets (15 %), post-weaning (3-month-old) pigs (17 %), 4-month-old pigs (10 %). Genetic analysis of the partial RNA-dependent RNA polymerase (RdRp) region (690 nt) revealed that kobuviruses circulating in East Africa are diverse, sharing nucleotide sequence identities ranging from 89.7 to 99.1 % and 88 to 92.3 % among them and with known porcine kobuviruses, respectively. The nucleotide sequence identities between our kobuvirus strains and those of human, bovine and canine kobuviruses were 69.4-70.7 %, 73.1-74.4 % and 67-70.7 %, respectively. Additionally, upon sequencing selected samples that showed consistent 720-bp RT-PCR bands while using the same primer set, we detected porcine astroviruses in our samples belonging to type 2 and type 3 mamastroviruses. To our knowledge, this study reports the first detection and molecular analysis of both porcine kobuviruses and astroviruses in an African region. Further studies are required to determine the role of these viruses in gastrointestinal infections of pigs in this region and to determine the genetic diversity of the circulating strains to develop accurate diagnostic tools and implement appropriate control strategies.
Keywords: Fecal Sample, Reverse Transcription Polymerase Chain Reaction, RdRp Gene, Reverse Transcription Polymerase Chain Reaction Product, Aichi Virus
Introduction
Small-scale pig production (free range or small backyard herds) constitutes >70 % of the total pig farms in East Africa. Pigs in this region often live in close proximity to humans, causing public health concerns and a need for increasing health standards. East Africa is also known for the large population of wild (bush) pigs and warthogs that are in close contact with domestic pigs, creating favorable conditions for intermixing and spread of viral strains. Emerging viruses represent a threat to human and food animal health, as evidenced by sporadic outbreaks of influenza and coronavirus infection. Hence, knowledge about the diversity of viruses present in reservoir animals can lead to a better understanding of the origin of emerging pathogens. Kobuviruses, which are members of the genus Kobuvirus, family Picornaviridae, are small, non-enveloped viruses with single-stranded, positive-sense genomic RNA. The genus Kobuvirus includes three officially recognized species: Aichivirus A, Aichivirus B (bovine kobuvirus), and Aichivirus C (porcine kobuvirus) [1]. The kobuvirus genome is approximately 8.3 kb long and is organized into three structural (VP0, VP3 and VP1) and seven non-structural (2A-2C and 3A-3D) regions with a leader protein (L). The 3D gene region encodes a viral RNA-dependent RNA polymerase (RdRp) and represents a region that is conserved among kobuviruses [2]. Nucleotide and amino acid sequence identities of the 3D RdRp coding region among porcine kobuvirus, bovine kobuvirus and Aichi virus vary from 74.0 % to 81.0 % [3]. Aichi virus was first detected in Japan in 1989 from a human patient with acute gastroenteritis [4]. Since then, Aichi virus has been detected in Asia, Europe, South America and Tunisia [5–8]. Bovine kobuvirus was first recognized in 2003 as a cytopathic contaminant in cell culture medium derived from bovine serum in Japan. Later, it was found in fecal samples of clinically healthy cattle [9]. Bovine kobuvirus has also been detected in domestic sheep in Hungary [10, 11]. Porcine kobuvirus (S-1-HUN/2007: EU787450) was first identified from fecal samples of domestic pigs in 2008 in Hungary [1]. Thereafter, porcine kobuvirus was reported in additional countries, including Asian countries [12, 13], the Netherlands and Brazil [14], and the USA [15]. The prevalence of kobuvirus infection in pigs ranges from 30 % to 99 %. This large variation could be a result of different ages within the populations evaluated, diarrheal status, regional differences, and other factors. Studies showing association of porcine kobuvirus infection with clinical disease are limited; however, a recent study in Korea found an association between porcine kobuvirus detection and diarrhea in pigs [13]. An association between kobuvirus infection and age has been reported in cattle and in pigs [16].
Astroviruses belong to the family Astroviridae, which consists of two genera: Avastrovirus and Mamastrovirus, whose members are associated with gastroenteritis in avian and mammalian hosts, respectively. Astroviruses are generally associated with either mild or severe enteric disease symptoms such as diarrhea and vomiting in a number of mammalian species [17]. Bridger in the UK and Saif et al. [18, 19] in the USA reported the first porcine astroviruses (PAstV) detected in pigs by electron microscopy. Later in 1990, PAstV was isolated in Japan [20]. Recently, Mor and co-workers also reported PAstV in pigs in the USA with a prevalence of 62 % [21]. To date, five PAstV types (PAstV1–PAstV5) have been identified from different countries, including the Czech Republic, Colombia, Canada, the USA, China and Hungary [22–27]. Among the five types, PAstV1, PAstV2 and PAstV3 were identified in fecal samples from healthy pigs.
Epidemiological information on the geographical distribution, incidence and genetic diversity of kobuviruses in African swine populations is not available. In this study, we report the first detection of porcine kobuviruses in samples collected from nursing and weaned pigs in East Africa and analyze the phylogenetic relationships between the East African porcine kobuvirus strains and known kobuvirus strains as well as other representative picornaviruses. This study also reports the first detection of porcine astrovirus in this region.
Accession numbers of nucleotide sequences strains described in this study were deposited in GenBank under the following accession numbers: kobuviruses, KF494340-KF494343 and KF597279; astroviruses, KF597280-KF597282.
Materials and methods
Sample collection
A total of 251 fecal samples were obtained from nursing and post-weaning piglets raised in small-scale farms (n = 1 to 25 pigs per farm) with different management systems in western Kenya (n = 140) and eastern Uganda (n = 111). The management system consisted of pigs being housed, housed and tethered, housed and free-range, tethered, tethered and free-range and free-range. The pigs were mostly fed swill from hotels and leftover household food. The sample collection consisted of 41 samples from nursing pigs (less than 4 weeks of age), 90 samples from post-weaning pigs (12 weeks of age) and 120 samples from growers (16 weeks of age). All pigs appeared clinically healthy at the time of sampling. Fresh samples were collected from individual pigs, placed into a sterile specimen container and stored on dry ice before being transported to the International Livestock Research Institute (ILRI) laboratory in Busia, where the samples were stored at −70 °C until use. The samples were then shipped to the BecA-ILRI laboratories in Nairobi for analysis.
RNA extraction and reverse transcription polymerase chain reaction (RT-PCR)
Fecal samples were prepared as 10 % (v/v) fecal suspensions in minimum essential medium (MEM). The prepared sample suspensions were vortexed and centrifuged for 30 min at 1,800×g, 4 °C. RNA was extracted from a 250-μl starting volume of centrifuged 10 % fecal suspensions using an RNeasy Mini Kit (QIAGEN, Valencia, CA, USA) according to the manufacturer’s instructions. The total RNA recovered was suspended in 40 μl of nuclease-free water and stored at −70 °C until used. cDNA was synthesised using a Maxima First Strand cDNA Synthesis Kit (Thermo scientific®) according to manufacturer’s instructions, and the cDNA was immediately used for amplification or stored at −20 °C.
Previous studies using the calicivirus primer pair p110/p290 [28, 29] targeting the calicivirus RdRp gene (317 nt for norovirus and 329 nt for sapovirus) also amplified the 3D RdRp regions of porcine kobuvirus (~1100 nt) [15], which was confirmed by sequence analysis of five representative suspect samples in the present report. This same primer pair was used for the detection of porcine kobuviruses from East African pigs. RT-PCR was carried out using Accupower® PCR premix (Bioneer Co, S. Korea) according to manufacturer’s instructions. Accupower® PCR premix is a ready-to-use lyophilized premix of dNTPs, Taq DNA Polymerase, reaction buffer, a tracking dye, and a stabilizer. Briefly, in each 20-μl reaction premix tube, 2 μl (5-50 ng) of cDNA template was mixed with 1 μl (10 pMol) each of forward and reverse primers, and 16 μl of nuclease-free water. PCR was performed at 94 °C for 5 min, followed by 40 cycles of 94 °C for 30 s, 48 °C of 30 s, and 72 °C for 30 s, followed by a final extension at 72 °C for 10 min. The temperature was then held at 4 °C. The RT-PCR products were analyzed by 1.5 % agarose gel electrophoresis and visualized by ultraviolet illumination after staining with Gel Red™ nucleic acid gel stain (Biotium, Hayward, CA). When bands were found in the region of 1,100 bp on the agarose gel, we tentatively considered the results kobuvirus positive. The samples with the correct band size were purified using a QIAquick PCR Purification Kit (QIAGEN, Valencia, CA USA) according to the manufacturer’s instructions for sequencing. In other fecal samples, an approximately 720-nt-long weak, nonspecific PCR product was also observed. The nucleotide (nt) sequence of the PCR products of three selected samples from three different farms were determined by direct sequencing.
Sequencing and molecular analysis
To confirm the RT-PCR results and to obtain genetic information on virus diversity, the partial 3D RdRp amplicons of five kobuvirus-positive samples were sequenced directly using forward (p110) and reverse (p290) primers using BigDye Terminator cycle chemistry (Applied Biosystems, Foster City, CA, USA). The resulting RdRP gene sequences were edited using Bioedit version 7.1.9 (http://www.mbio.ncsu.edu/BioEdit/bioedit.html) and compared with sequences of kobuvirus reference strains in the GenBank database by BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). Sequence similarity analysis was performed for the aligned nucleotide and amino acid sequences using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/). Phylogenetic analysis of the East African porcine kobuviruses with kobuvirus reference strains based on the nucleotide and amino acid sequence alignments was conducted using the neighbor-joining method supported with a bootstrap test of 1000 replicates in MEGA 5 software [30]. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Tamura 3-parameter method and are in units of base substitutions per site.
To analyze porcine kobuvirus infection in pigs of different ages with different management systems, the piglet fecal samples were divided according to the age of the animals into three groups: nursing, ≤1-month-old (n = 47), weaned, (3-month-old, n = 90) and growers (>4-month-old, n = 120). Statistical analysis was performed with SAS® to compare the proportions of kobuvirus-positive samples among the four districts, the two countries, the three age groups evaluated and the six different management systems. The analysis was performed using the chi-square (χ2) test or Fisher’s exact test. The confidence limit for the statistical tests was set at 95 % (P < 0.05).
Results and discussion
Incidence of kobuvirus infection
Kobuvirus RNA was detected in 13.1 % (33/251) of the pig fecal samples analyzed using primer pair p110/p290 [28, 29]. Kobuvirus was detected in both Kenya (14.9 %) and Uganda (15.5 %) in equal proportions, and in all four districts sampled (Table 1), an indication of a general circulation and endemicity of the virus on the tested farms. Among the age groups, younger piglets shed more virus than older pigs, and a higher incidence (15 of 90, 16.7 %) of kobuvirus was reported in post-weaning (3-month-old) piglets compared to nursing piglets (6 of 41, 14.6 %) and grower pigs (12 of 120, 10 %). This could possibly be due to an inefficient immune response, diminishing maternal immunity post-weaning, or other intrinsic age-related factors. Higher rates of infection in young piglets has also been reported in other studies [3, 13, 14]. Based on management systems in the study area, confinement of pigs presented a higher risk of kobuvirus infection, with housed pigs shedding more virus than free-ranging pigs (Table 1). This could be the result of viral accumulation in the pig houses or the places where pigs are tethered. Additionally, kobuvirus was more prevalent in the farms with larger herd size (>10 pigs, 20 %) than farms with small herds (<10 pigs, 12.6 %).
Table 1.
Porcine kobuvirus infection status in pigs of different ages, raised under different management systems in East Africa
| Details | Groups | N | Positive (%) |
|---|---|---|---|
| Overall | 251 | 33(13.1) | |
| Country | Kenya | 139 | 18 (14.9) |
| Uganda | 112 | 15 (15.5) | |
| Districts | Busia_Ke | 69 | 9 (13.0) |
| Teso | 70 | 9 (12.9) | |
| Busia_Ug | 64 | 10 (15.6) | |
| Tororo | 48 | 5 (10.4) | |
| Age group | Nursing-<1 mo | 41 | 6 (14.6) |
| Weaner-3 mo | 90 | 15 (16.7) | |
| Grower-4 mo | 120 | 12 (10.0) | |
| Management systems | Free range | 8 | 1 (12.5) |
| Housed | 6 | 2 (33.3) | |
| Housed & free range | 17 | 3 (17.6) | |
| Housed & tethered | 3 | 0 (0) | |
| Tethered | 115 | 14 (12.2) | |
| Tethered & free range | 102 | 13 (12.7 | |
| No. of pigs per household | 1 to 5 | 215 | 27 (12.6) |
| 6 to 10 | 16 | 2 (12.5) | |
| >10 | 20 | 4 (20.0) |
The incidence of porcine kobuvirus infection reported in this study (13.1 %) was lower than that reported in other countries, which ranged between 30.1 % in China and 99 % in Thailand [3, 31–33]. A recent study in Korea detected porcine kobuvirus in 84.5 % of diarrheic pigs and 19.3 % of healthy pigs [13]. Similarly, a study in Japan using samples from healthy pigs reported 45.4 % prevalence of porcine kobuvirus [12]. This difference may be attributable to variations in the samples due to factors like sampling time, sampling place (distribution of sampling farms), sample size, age of pigs, and clinical background of the tested animal population (diarrhea or clinically healthy). The association of this agent with enteric diseases in pigs in the study area remains unclear, since we only analyzed samples from asymptomatic pigs, and no data were available from tests for kobuvirus in pigs with gastroenteritis from the same farms; however, some of the positive samples came from farms with a history of diarrhea. This study confirmed that infection with kobuvirus in asymptomatic pigs is common, as has been reported in other studies [14, 32, 34]. Further studies on risk factors for this infection and the association of this virus with gastroenteritis are necessary, especially in developing countries where a majority of pigs are raised in smallholder production systems with varied management.
Genetic and phylogenetic analysis of East African porcine kobuviruses
To gain more information on the genetic heterogeneity of porcine kobuvirus strains circulating in swine in the East African farms, five kobuvirus-positive samples were selected, and their partial 3D RdRP region was sequenced. Genetic analysis of the partial sequence revealed that the kobuviruses circulating in the East African region are more variable, sharing nucleotide sequence identity ranging from 89.7 from 99.1 % among them. At the nucleotide and amino acid level, they were 69.4–70.7 % and 70.7–71.9 % identical, respectively, to Aichi virus, 73.1–74.4 % and 78.8–80.5 % to bovine kobuvirus, 70.8-71.9 % and 78.5-79.5 % to kobuvirus in sheep, 67-70.7 % and 67.8-69.1 % to canine kobuviruses and 88-92.3 % and 90.4-97.5 % to other porcine kobuviruses (Table 2).
Table 2.
Nucleotide and amino acid sequence identities (%) of partial 3D RdRP genes of selected field strains compared with similar gene sequences for known kobuviruses and other picornaviruses in the GenBank database
| Strain | Nucleotide sequence identity (%) | Amino acid sequence identity (%) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Damaris-3 | K-118 | Kuoba-3 | K-460 | K-1033 | Damaris-3 | K-118 | Kuoba-3 | K-460 | K-1033 | |
| Swine/S-1-HUN/2007-EU787450 | 92.0 | 91.9 | 92.0 | 89.9 | 92.3 | 97.5 | 96.5 | 97.2 | 92.7 | 95.3 |
| Swine/K-30-HUN/2008-GQ249161 | 91.9 | 91.7 | 91.9 | 89.7 | 92.2 | 97.5 | 96.5 | 97.2 | 92.7 | 95.3 |
| Swine/CH/HNXX-4/2012-JX401523 | 91.1 | 90.2 | 91.7 | 88.5 | 91.1 | 97.2 | 95.1 | 96.8 | 92.7 | 94.5 |
| Swine/CH/HZ/2011-JX827598 | 90.9 | 90.2 | 91.4 | 88.0 | 90.9 | 97.2 | 96.5 | 96.8 | 93.6 | 95.3 |
| swine/MF8047/2009/KOR-GU723956 | 91.2 | 91.2 | 91.2 | 88.9 | 90.9 | 96.8 | 95.8 | 96.5 | 92.7 | 95.3 |
| Swine-WUH1/2011-CHN-JQ692069 | 91.1 | 92.0 | 91.4 | 89.4 | 91.1 | 97.2 | 96.8 | 96.8 | 94.1 | 95.3 |
| Swine/XX/CHN/2011-KC204684 | 90.5 | 92.2 | 90.8 | 88.6 | 90.5 | 97.2 | 96.8 | 96.8 | 93.2 | 94.9 |
| Swine/KOR/2010-94DA1-HQ400962 | 90.6 | 91.6 | 91.6 | 91.8 | 91.0 | 92.6 | 91.4 | 92.6 | 92.0 | 96.9 |
| Swine/KOR/2010-95DA2-HQ400963 | 91.0 | 91.6 | 92.0 | 92.2 | 91.4 | 92.6 | 91.4 | 92.6 | 92.0 | 96.9 |
| Swine/P224/11/CZ-JX232622 | 89.8 | 89.4 | 90.4 | 89.2 | 90.2 | 92.2 | 90.4 | 92.2 | 91.6 | 97.0 |
| Swine/P46/11/CZ-JX232619 | 90.6 | 90.0 | 91.2 | 89.6 | 91.0 | 92.2 | 90.4 | 92.2 | 91.6 | 97.0 |
| Swine/P5/11/CZ-JX232616 | 90.6 | 90.6 | 91.2 | 89.6 | 91.0 | 92.2 | 90.4 | 92.2 | 91.6 | 97.0 |
| Aichivirus-AB040749 | 70.7 | 70.4 | 70.5 | 69.7 | 70.5 | 71.4 | 70.7 | 71.0 | 70.8 | 71.9 |
| Aichivirus-1-NC_001918 | 70.4 | 70.1 | 70.2 | 69.4 | 70.2 | 71.4 | 70.7 | 71.0 | 70.8 | 71.9 |
| Bo-U-1-AB084788 | 74.0 | 73.9 | 74.0 | 73.1 | 74.4 | 80.2 | 78.8 | 79.9 | 79.9 | 80.5 |
| Sheep/TB3/HUN/2009-GU245693 | 71.3 | 71.9 | 71.4 | 70.8 | 71.3 | 79.5 | 78.5 | 79.2 | 78.5 | 79.3 |
| Dog/AN211D/USA/2009-JN387133 | 69.7 | 69.0 | 69.6 | 67.7 | 70.1 | 68.6 | 67.8 | 68.2 | 69.0 | 69.1 |
| Canine-US-PC0082-JN088541 | 70.4 | 69.4 | 70.2 | 67.0 | 70.7 | 68.6 | 67.8 | 68.2 | 69.0 | 69.1 |
| Ljungan-87-012-AF327920-Parechovirus | 39.3 | 39.5 | 39.2 | 40.0 | 39.2 | 25.6 | 26.3 | 25.3 | 24.0 | 25.2 |
| Eq-Rhinovirus-X96871-Erbovirus | 49.4 | 49.2 | 48.3 | 48.6 | 48.9 | 37.3 | 37.3 | 37.3 | 37.5 | 36.5 |
| FMDV-O1K-X00871-Aphthovirus | 49.8 | 49.1 | 50.2 | 49.4 | 50.0 | 34.5 | 34.2 | 34.2 | 35.5 | 35.0 |
| HRV14-K02121-Enterovirus | 41.4 | 41.1 | 41.6 | 41.6 | 41.1 | 34.6 | 34.6 | 34.6 | 32.7 | 33.1 |
| Po-UKG/410/73-AF363453-enterovirus | 43.4 | 43.6 | 43.9 | 44.2 | 43.2 | 34.1 | 34.1 | 34.1 | 33.2 | 32.5 |
| Damaris-3 - KF494340 | 100.0 | 92.0 | 98.2 | 89.9 | 99.1 | 100.0 | 96.8 | 99.7 | 95.4 | 96.1 |
| K-118 - KF494342 | 100.0 | 91.7 | 89.7 | 92.0 | 100.0 | 96.5 | 92.2 | 94.9 | ||
| Kuoba-3 - KF494341 | 100.0 | 90.9 | 98.5 | 100.0 | 95.4 | 95.7 | ||||
| K-460 - KF494343 | 100.0 | 89.9 | 100.0 | 91.7 | ||||||
| K-1033 - KF597279 | 100.0 | 100.0 | ||||||||
The strains that were detected and discussed in this study are in bold
Phylogenetic analysis of partial 3D RdRp nucleotide and amino acid sequences of porcine kobuvirus strains detected in this study, together with published sequences of kobuvirus reference strains (porcine kobuviruses, aichi viruses and bovine kobuvirus) and representatives of other picornaviruses, revealed that three of the detected strains (Damaris-3-KF494340, Kuoba-3-KF494341 and K-1033-KF597279) grouped together and formed their own cluster, while one of the strains (K-118-KF494342) clustered with Chinese strains, and another strain (K-460-KF494343) clustered with Korean strains (Fig. 1). This indicates that porcine kobuviruses circulating in the East African region are genetically diverse. Damaris-3-KF494340, Kuoba-3-KF494341 and K-1033-KF597279 samples were collected in the same geographical location, indicating that porcine kobuviruses in the East African region may be geographically restricted; however, more strains from different parts of East Africa need to be sequenced to ascertain the geographical distribution of this virus. Genetic diversity among geographically separated porcine kobuviruses have been reported in other studies [13].
Fig. 1.
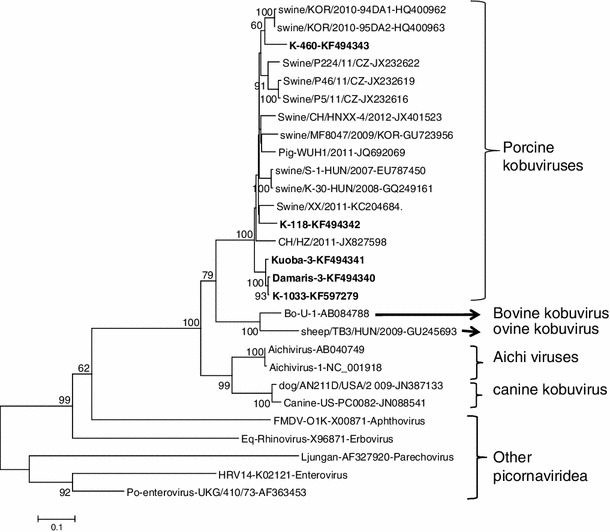
Phylogenetic dendrogram of the partial (690-bp) nucleotide sequence of the 3D RdRp gene of field kobuvirus strains (bold) compared with similar gene sequences for known kobuviruses and other picornaviruses in the GenBank database. The horizontal branch lengths are proportional to the genetic distance calculated by the neighbor-joining method. The evolutionary distances were computed using the Tamura-Nei method [36] and are in the units of base substitutions per site
Phylogenetic analysis also confirmed that the five strains detected were more closely related to the kobuviruses, notably porcine kobuvirus, than to any other picornaviruses, and this was supported by the 100 % bootstrap value (Fig. 1). Evidence of a close genetic relationship or interspecies transmission among different viruses in the genus Kobuvirus has been documented. A sheep kobuvirus strain has high nucleotide sequence identity to bovine kobuvirus [10], and one porcine kobuvirus is more closely related to bovine kobuvirus than to porcine strains [12]. However, in this study, we did not find kobuvirus strains that were similar to members of kobuviruses from species other than pigs.
Genetic and phylogenetic analysis of porcine astroviruses
In other fecal samples, an approximately 720-nt-long weak, nonspecific RT-PCR product was also observed. The nucleotide (nt) sequences of the RT-PCR products of three selected samples were determined by direct sequencing, and the result showed that one of the strains (K-268-KF597282) had 86.1 % nt sequence identity to porcine astrovirus type 2 in the RdRp gene. The other two strains (K-006-KF597280 and K-332-KF597281) had 96.7 % identity to porcine astrovirus type 3. This was confirmed by phylogenetic analysis, in which K-006-KF597280 and K-332-KF597281 strains clustered with astrovirus type 3 (PAstV-3-US-MO123-JX556691) detected in the USA, while the K-268-KF597282 strain clustered with astrovirus type 2 (PAstV-2/2007/HUN-GU562296) detected in Hungary (Fig. 2). Recent studies in the USA revealed high genetic heterogeneity of PAstV strains [35]. The importance of co-circulation of PAstVs and the role pigs may play in the transmission and recombination of this virus need further investigation.
Fig. 2.
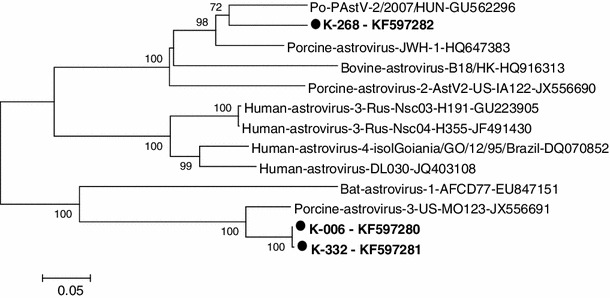
Phylogenetic dendrogram of the partial nucleotide sequence of 3D RdRp gene (541 nt) of field astrovirus strains (bold) compared with similar gene sequences for known astroviruses in the GenBank database. The horizontal branch lengths are proportional to the genetic distance calculated by the neighbor-joining method. The evolutionary distances were computed using the Tamura 3-parameter method [36] and are in the units of base substitutions per site
In conclusion, the findings of this study demonstrate that porcine kobuviruses and astroviruses are present in the swine population in East Africa, and to our knowledge, this study reports the first detection of porcine kobuvirus and astrovirus in this African region and the first molecular analysis of the detected strains. The presence of these gastroenteritis-producing viruses in clinically healthy pigs represents a source of infection of pigs, and possibly to humans, and hence, further studies are required to determine their role in gastrointestinal infections of pigs in this region and to determine their genetic diversity in order to develop accurate diagnostic tools and implement appropriate control strategies.
Acknowledgements
We gratefully acknowledge support of this project to A.J.O. through the African Bioscience Challenge Fund (ABCF) Fellowship of Biosciences of East and Central Africa (BecA). We also thank VPH-Biotech East Africa Consortium and University of Nairobi for supporting A.J.O. We would like to acknowledge the co-operation and assistance of the Africa Swine Fever Project team (International Livestock Research Institute) for their support in sample collection. We thank smallholder pig farmers in Kenya and Uganda for allowing us to sample their pigs for this research study. The samples discussed in this study were sequenced by the Segolip unit of ILRI-BecA hub in Kenya.
Conflict of interest
None.
Contributor Information
Joshua O. Amimo, Email: J.Amimo@cgiar.org
Linda J. Saif, Email: saif.2@osu.edu
References
- 1.Reuter G, Boldizsa′r A, Kiss I, Pankovics P. Candidate new species of Kobuvirus in porcine hosts. Emerg Infect Dis. 2008;14:1968–1970. doi: 10.3201/eid1412.080797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reuter G, Boros A, Pankovics P. Kobuviruses—a comprehensive review. Rev Med Virol. 2011;21:32–41. doi: 10.1002/rmv.677. [DOI] [PubMed] [Google Scholar]
- 3.Reuter G, Boldizsár A, Pankovics P. Complete nucleotide and amino acid sequences and genetic organization of porcine kobuvirus, a member of a new species in the genus Kobuvirus, family Picornaviridae. Arch Virol. 2009;154:101–108. doi: 10.1007/s00705-008-0288-2. [DOI] [PubMed] [Google Scholar]
- 4.Yamashita T, Kobayashi S, Sakae K, Nakata S, Chiba S, Ishihara Y, Isomura S. Isolation of cytopathic small round viruses with BS-C-1 cells from patients with gastroenteritis. J Infect Dis. 1991;164:954–957. doi: 10.1093/infdis/164.5.954. [DOI] [PubMed] [Google Scholar]
- 5.Oh D-Y, Silva PA, Hauroeder B, Diedrich S, Cardoso DDP, Schreier E. Molecular characterization of the first Aichi viruses isolated in Europe and in South America. Arch Virol. 2006;151:1199–1206. doi: 10.1007/s00705-005-0706-7. [DOI] [PubMed] [Google Scholar]
- 6.Reuter G, Boldizsár A, Papp G, Pankovics P. Detection of Aichi virus shedding in a child with enteric and extraintestinal symptoms in Hungary. Arch Virol. 2009;154:1529–1532. doi: 10.1007/s00705-009-0473-y. [DOI] [PubMed] [Google Scholar]
- 7.Pham NTK, Khamrin P, Nguyen TA, Kanti DS, Phan TG, Okitsu S, Ushijima H. Isolation and molecular characterization of Aichi viruses from fecal specimens collected in Japan, Bangladesh, Thailand, and Vietnam. J Clin Microbiol. 2007;45:2287–2288. doi: 10.1128/JCM.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sdiri-Loulizi K, Hassine M, Aouni Z, Gharbi-Khelifi H, Sakly N, Chouchane S, Guédiche MN, Pothier P, Aouni M, Ambert-Balay K. First molecular detection of Aichi virus in sewage and shellfish samples in the Monastir region of Tunisia. Arch Virol. 2010;155:1509–1513. doi: 10.1007/s00705-010-0744-7. [DOI] [PubMed] [Google Scholar]
- 9.Yamashita T, Ito M, Kabashima Y, Tsuzuki H, Fujiura A, Sakae K. Isolation and characterization of a new species of kobuvirus associated with cattle. J Gen Virol. 2003;84:3069–3077. doi: 10.1099/vir.0.19266-0. [DOI] [PubMed] [Google Scholar]
- 10.Reuter G, Boros A′, Pankovics P, Egyed L. Kobuvirus in domestic sheep, Hungary. Emerg Infect Dis. 2010;18:869–870. doi: 10.3201/eid1605.091934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reuter G, Boldizsar A, Boros A, Pankovics P. Detection and characterization of kobuviruses (in family Picornaviridae) in human and in new host species in Hungary. Clin Microbiol Infect. 2010;16:S402. doi: 10.1111/j.1469-0691.2010.03182.x. [DOI] [Google Scholar]
- 12.Khamrin P, Maneekarn N, Hidaka S, Kishikawa S, Ushijima K, Okitsu S, Ushijima H. Molecular detection of kobuvirus sequences in stool samples collected from healthy pigs in Japan. Infect Genet Evol. 2010;10:950–954. doi: 10.1016/j.meegid.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 13.Park S-J, Kim H-K, Moon H-J, Song D-S, Rho S-M, Han J-Y, Nguyen V-G, Park B-K. Molecular detection of porcine kobuviruses in pigs in Korea and their association with diarrhea. Arch Virol. 2010;155:1803–1811. doi: 10.1007/s00705-010-0774-1. [DOI] [PubMed] [Google Scholar]
- 14.Barry AF, Ribeiro J, Alfieri AF, Van Der Poel WHM, Alfieri AA. First detection of kobuvirus in farm animals in Brazil and the Netherlands. Infect Genet Evol. 2011;11:1811–1814. doi: 10.1016/j.meegid.2011.06.020. [DOI] [PubMed] [Google Scholar]
- 15.Sisay Z, Wang Q, Oka T, Saif L. Prevalence and molecular characterization of porcine enteric caliciviruses and first detection of porcine kobuviruses in US swine. Arch Virol. 2013;158:1583–1588. doi: 10.1007/s00705-013-1619-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeoung H-Y, Lim J-A, Jeong W, Oem J-K, An D-J. Three clusters of bovine kobuvirus isolated in Korea, 2008–2010. Virus Genes. 2011;42:402–406. doi: 10.1007/s11262-011-0593-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jonassen CM, Jonassen T, Saif Y, Snodgrass D, Ushijima H, Shimizu M, Grinde B. Comparison of capsid sequences from human and animal astroviruses. J Gen Virol. 2001;82:1061–1067. doi: 10.1099/0022-1317-82-5-1061. [DOI] [PubMed] [Google Scholar]
- 18.Saif LJ, Bohl HE, Theil KW, Kohler EM, Cross RF (1980) 30 nm virus like particles resembling astrovirus in intestinal contents of a diarrheic pig. In: Proceedings of Conf. Res. Work. Anim. Dis. Chicago, Ill, p Abstract #149
- 19.Bridger JC. Detection by electron microscopy of caliciviruses, astroviruses and rotavirus-like particles in the faeces of piglets with diarrhoea. Vet Rec. 1980;107:532–533. [PubMed] [Google Scholar]
- 20.Shimizu M, Shirai J, Narita M, Yamane T. Cytopathic astrovirus isolated from porcine acute gastroenteritis in an established cell line derived from porcine embryonic kidney. J Clin Microbiol. 1990;28:201–206. doi: 10.1128/jcm.28.2.201-206.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mor SK, Chander Y, Marthaler D, Patnayak DP, Goyal SM. Detection and molecular characterization of Porcine astrovirus strains associated with swine diarrhea. J Vet Diagn Investig Off Publ Am Assoc Vet Lab Diagn Inc. 2012;24:1064–1067. doi: 10.1177/1040638712458781. [DOI] [PubMed] [Google Scholar]
- 22.Shan T, Li L, Simmonds P, Wang C, Moeser A, Delwart E. The fecal virome of pigs on a high-density farm. J Virol. 2011;85:JVI05217-11. doi: 10.1128/JVI.05217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan D, Ji W, Shan T, Cui L, Yang Z, Yuan C, Hua X. Molecular characterization of a porcine astrovirus strain in China. Arch Virol. 2011;156:1869–1875. doi: 10.1007/s00705-011-1050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reuter G, Nemes C, Boros A, Kapusinszky B, Delwart E, Pankovics P. Astrovirus in wild boars (Sus scrofa) in Hungary. Arch Virol. 2012;157:1143–1147. doi: 10.1007/s00705-012-1272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laurin MA, Dastor M, L’Homme Y. Detection and genetic characterization of a novel pig astrovirus: relationship to other astroviruses. Arch Virol. 2011;156:2095–2099. doi: 10.1007/s00705-011-1088-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo Z, Roi S, Dastor M, Gallice E, Laurin M-A, L’homme Y. Multiple novel and prevalent astroviruses in pigs. Vet Microbiol. 2011;149:316–323. doi: 10.1016/j.vetmic.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Indik S, Valı′cek L, Smı′d B, Dvora′ kova′ H, Roda′k L. Isolation and partial characterization of a novel porcine astrovirus. Vet Microbiol. 2006;117:276–283. doi: 10.1016/j.vetmic.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 28.Jiang X, Huang PW, Zhong WM, Farkas T, Cubitt DW, Matson DO. Design and evaluation of a primer pair that detects both Norwalk- and Sapporo-like caliciviruses by RT-PCR. J Virol Methods. 1999;83:145–154. doi: 10.1016/S0166-0934(99)00114-7. [DOI] [PubMed] [Google Scholar]
- 29.Le Guyader F, Estes MK, Hardy ME, Neill FH, Green J, Brown DW, Atmar RL. Evaluation of a degenerate primer for the PCR detection of human caliciviruses. Arch Virol. 1996;141:2225–2235. doi: 10.1007/BF01718228. [DOI] [PubMed] [Google Scholar]
- 30.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;113:1530–1534. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khamrin P, Maneekarn N, Kongkaew A, Kongkaew S, Okitsu S, Ushijima H. Porcine kobuvirus in piglets, Thailand. Emerg Infect Dis. 2009;15:2075–2076. doi: 10.3201/eid1512.090724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reuter G, Kecskeme′ti S, Pankovics P. Evolution of porcine kobuvirus infection, Hungary. Emerg Infect Dis. 2010;16:696–698. doi: 10.3201/eid1604.090937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu J, Xu Z, Li B, Zhang Q, Cui S, Jin M, Duan Z. Analysis and characterization of the complete genome of a member of a new species of kobuvirus associated with swine. Arch Virol. 2011;156:747–751. doi: 10.1007/s00705-010-0907-6. [DOI] [PubMed] [Google Scholar]
- 34.Dufkova L, Scigalkova I, Moutelikova R, Malenovska H, Prodelalova J. Genetic diversity of porcine sapoviruses, kobuviruses, and astroviruses in asymptomatic pigs: an emerging new sapovirus GIII genotype. Arch Virol. 2013;158(3):549–558. doi: 10.1007/s00705-012-1528-z. [DOI] [PubMed] [Google Scholar]
- 35.Xiao C-T, Giménez-Lirola LG, Gerber PF, Jiang Y-H, Halbur PG, Opriessnig T. Identification and characterization of novel porcine astroviruses (PAstVs) with high prevalence and frequent co-infection of individual pigs with multiple PAstV types. J Gen Virol. 2013;94:570–582. doi: 10.1099/vir.0.048744-0. [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]


