Abstract
Cap-Analysis gene expression (CAGE) provides accurate high-throughput measurement of RNA expression. CAGE allows mapping of all the initiation sites of both capped coding and noncoding RNAs. In addition, transcriptional start sites (TSSs) within promoters are characterized at single nucleotide resolution. The latter allows the regulatory inputs driving gene expression to be studied, which in turn enables the construction of transcriptional networks. Here we provide an optimized protocol for the construction of CAGE libraries based on the preparation of 27 nucleotide (nt) long tags corresponding to initial bases at the 5’ ends of capped RNAs. We have optimized the methods using simple steps based on filtration, which altogether takes 4 days to complete. The CAGE tags can be readily sequenced with Illumina sequencers and upon modification, they are also amenable to sequencing using other platforms.
Keywords: Cap-analysis gene expression (CAGE), transcriptome, promoter, sequencing, RNA
Introduction
Methods for genome-wide expression analysis
Genome-wide expression analysis is a key approach to rapidly and systematically interrogate biological systems. Microarrays played a major role since their introduction1 due to their ability to analyze the whole set of transcripts expressed in a cell. However, microarrays are based on hybridization, and thus constitute a closed system. Microarrays are generally designed to detect exons of known genes and may not include novel transcripts, such as non coding RNAs (ncRNA). Genome wide tiling arrays2 largely alleviate these issues but suffer from excluding repetitive sequences such as retrotransposon elements (REs). Given the additional problem of cross-hybridization, microarrays are by design not suitable for the unbiased analysis of the transcriptome. Sequencing based technologies offer the opportunity to overcome these limitations by providing transcript identification together with quantification of expression. Of note, RNA-sequencing (RNAseq) promises to reveal comprehensive splicing variants for the mRNAs present in a given samples, together with their expression levels3. However, obtaining transcript models in higher organisms is proving to be challenging, especially in cases where multiple alternative isoforms are present in the cell. In addition, the coverage of 5’ ends of transcripts is too low to clearly identify TSSs.
An alternative is to sequence only a portion of all expressed RNAs. A notable technology here is the Serial Analysis of Gene Expression4 (SAGE). SAGE captures a sequence from each transcript, generally biased towards its 3’ end, and thus cannot provide additional information on regulatory elements found predominantly at the 5’ end.
Development and previous use of CAGE
We have developed Cap Analysis of Gene Expression (CAGE) for high-throughput gene expression profiling, which focuses on capturing the capped 5’ ends of RNAs5. Sequencing short sequence reads (or tags) taken from the 5’ ends of full-length cDNAs allows TSSs to be mapped and their expression, measured by tag frequency, to be analyzed. The TSS-based approach of CAGE has enabled high-throughput identification of promoters6. CAGE has been instrumental in globally mapping specific TSSs in eukaryotes6,7–10, emphasizing the existence of alternatively regulated TSSs11,12, novel regulatory elements6 and has allowed predictions of transcription factor binding sites13 and other motifs associated with transcription14. In addition, the analysis of 5’ ends by CAGE is particular suitable to infer gene regulatory networks and has provided knowledge of the key transcription factors responsible for the differentiation of monoblasts to monocytes15. The fact that CAGE is not biased towards a known gene model has lead to the observation that retrotransposon elements (REs) are specifically expressed and act as regulators of protein coding RNAs and other ncRNAs16. With this protocol, we have provided more than a billion CAGE reads for ENCODE project. The protocol is also being implemented in other laboratories, which are successfully performing cap-trapper/CAGE library preparation17,18.
Initial CAGE protocols were based on sequencing short reads (21 nt) downstream of the cap-sites. The length was dictated by the availability of only relatively short Class IIS restriction enzymes, like MmeI5,19–21 and required large amount of starting material, in the range of 30–50 µg of total RNA. The protocol we present here profoundly differs from previous ones: optimization of ligations and linker design decreased the starting amount of RNA to 1–5 µg or less, a tenfold decrease compared to the previous protocol5. In case of the ENCyclopedia Of DNA Elements (ENCODE) project, we have routinely used 5 µg of poly-A-minus RNAs, which were depleted of capped RNAs. This demonstrates the potential to further scale down the amount of starting RNA. Next, we introduced the EcoP15I restriction enzyme, which cleaves 27 nt apart, resulting in longer reads thus improving their mapping. Third, the protocols can be applied to 96 well plates or 8 strip tubes using multi channel pipettes for high throughput library preparation, matching the increasing sequencing capacities. Altogether, the reduction of material is useful in applying the method to larger number of samples, while maintaining the number of PCR cycles below 15.
The development of CAGE technology has gone alongside the development of sequencing technology, moving from Sanger to next-generation sequencing, which clearly has the power to characterizeRNA expression. CAGE has been deployed using 454 Life Science19, Applied Biosystems SOLiD21, Heliscope6 and Illumina Genome Analyzer21–23 and Hi-Seq 2000 sequencers (H. Kawaji et al, unpublished). Here, we provide the protocol for the latter two sequencers, although changes in linker sequences will allow adaptation to further sequencing platforms.
Limitations of CAGE
Because CAGE selectively removes non-capped RNAs, small RNAs and other non-capped RNAs transcribed from POLR1 and POLR3, like some SINE derived transcripts, are not detected. Additionally, CAGE is not applicable to prokaryotes, or to RNAs shorter to ~100 nt, which are filtered out during the linker purification procedures.
Alternative methods to CAGE
Alternative methods to CAGE include the 5’-end SAGE16, which is based on selectively ligating de-capped 5’ RNA end with T4 RNA ligase. In contrast to CAGE, RNA ligase may show sequence preferences17. NanoCAGE18 is an alternative that requires less RNA, but requires a much larger number of PCR cycles, which have inherent representation bias. The single molecule Heliscope CAGE5 was also shown to work with less than a microgram of RNA, but the number of reads obtained is often lower than with the method presented here. GIS-PET resembles CAGE for capturing 5’ end or capped RNA, but in addition allows the 3’ end of poly-A RNA to be captured. GIS-PET is thus a very powerful tool to measure coupled initiation-termination events and thus define gene borders19. However, its procedure includes a large number of steps, comprising ligation to plasmids and a plasmid amplification step, which may result in both size and representational bias when it is compared to CAGE.
Experimental Design
cDNA synthesis that efficiently reaches the 5’ ends
For an overview of CAGE library preparation, follow Figure 1. Firstly, cDNA is reverse transcribed by reverse transcriptase using a random primer including EcoP15I sequence and polyadenylated and non polyadenylated RNA as template. Engineered reverse transcriptases, which are devoid of RNaseH activity, are necessary to produce high quality first-strand cDNA from randomly primed RNAs. The absence of RNaseH activity is mandatory, otherwise they will cause nicks on the RNA strand of the RNA/cDNA hybrid. Such nicks could result in the depletion of the capped RNA from the hybrid. Additionally, nicks would expose newly formed 3’ ends. These could be biotinylated and compromise the selectivity of the cap-trapping reaction, which is based on selecting the biotin on the cap site of the RNA (Fig. 1). Random primers (RT-N15-EcoP) are used to prime the reverse transcription, allowing capture of all RNAs, including non polyadenylated capped RNAs as identified in an ongoing study21. Random priming also prevents underrepresentation of very long polyadenylated RNAs, which may occur in oligo-dT primed reactions, since the RT may not be efficient enough to reach the 5’ ends of very large mRNAs (5 ~ 10 Kb). Alternatively, if such very long polyadenylated RNAs are oligo-dT primed, oligo-dT priming could introduce a bias against longer mRNAs in the event of partial RNA degradation. Hence, this method is centered on the use of the random primers, which allow the 5’ end of capped RNAs to be effectively and equally reached regardless the size of the RNAs.
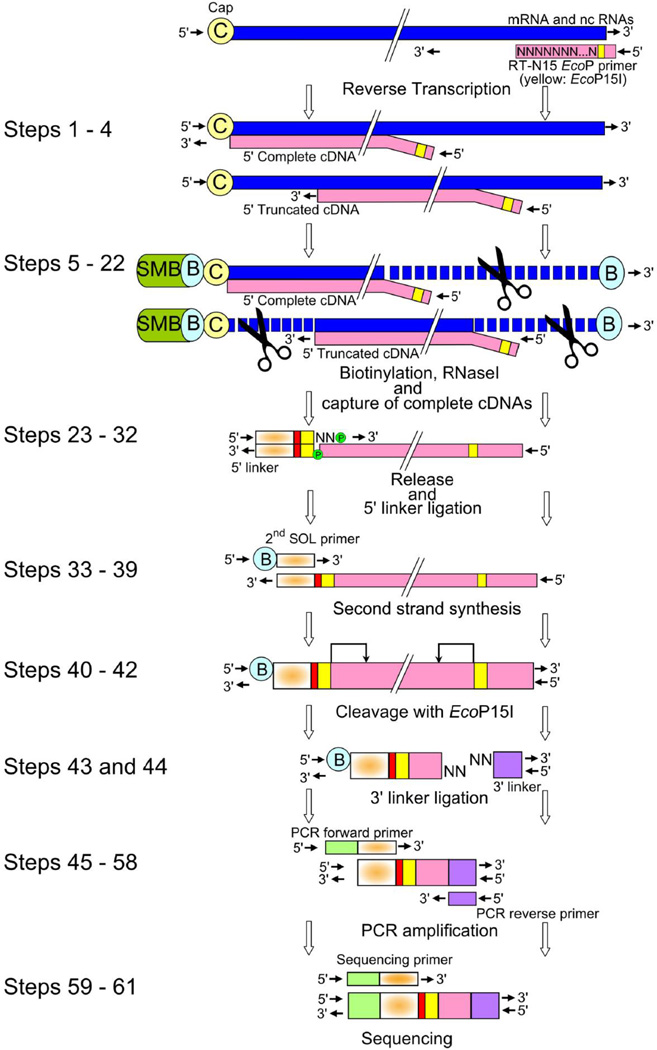
Figure 1. Workflow of CAGE libraries preparation.
cDNA is reverse transcribed by reverse transcriptase using a random primer including EcoP15I sequence (yellow) and polyadenylated and non polyadenylated RNA as template in Steps 1–4. Cap and 3’ end are biotinylated, and after RNAse digestion of non-hybridized single stranded RNA (represented by scissors), 5’ complete cDNAs hybridized to biotinylated capped RNAs are captured by streptavidin coated magnetic beads in Steps 5–22. The cDNA is next released from RNA and ligated to a 5’ linker including a barcode sequence (red) and EcoP15I sequence (yellow) in Steps 23–32. The double strand 5’ linkers is then denatured at 94°C to allow the biotin modified 2nd SOL primer to anneal to the single stranded cDNA and prime second-strand cDNA synthesis in Steps 33–39. Subsequently, cDNA is digested with EcoP15I, which cleaves 27 bp inside the 5’ end of the cDNA in Steps 40–42. Next, a 3’ linker containing the 3’ Illumina primer sequence (purple) is ligated at the 3’ end in Steps 43 and 44. The 96 bp CAGE tags are amplified with the forward primer (green) and reverse primer, which both are compatible with the Illumina flow cell surface, in Steps 45–58. (C) Cap; (B) biotin; (SMB) streptavidin coated magnetic beads.
Besides identifying classic promoters, CAGE also highlights other types of putatively capped transcripts in 3’ UTRs and exons11,24. We have found that the optional inclusion of oligo-dT primer adaptors to the random primers allows better identification of these unconventional TSSs, suggesting that they represent TSS of relatively short polyadenylated RNAs. Addition or removal of the oligo-dT in the priming reaction helps to tune up/down the representation of these putative ncRNAs (Supplementary Fig. 1).
Random priming takes place at 25°C (see Step 4). This low annealing temperature is deliberately used to introduce numerous mismatches on the primer-RNA hybrid that allows subsequent digestion of these non-hybridized ssRNA parts (Fig. 2a) with RNaseI.
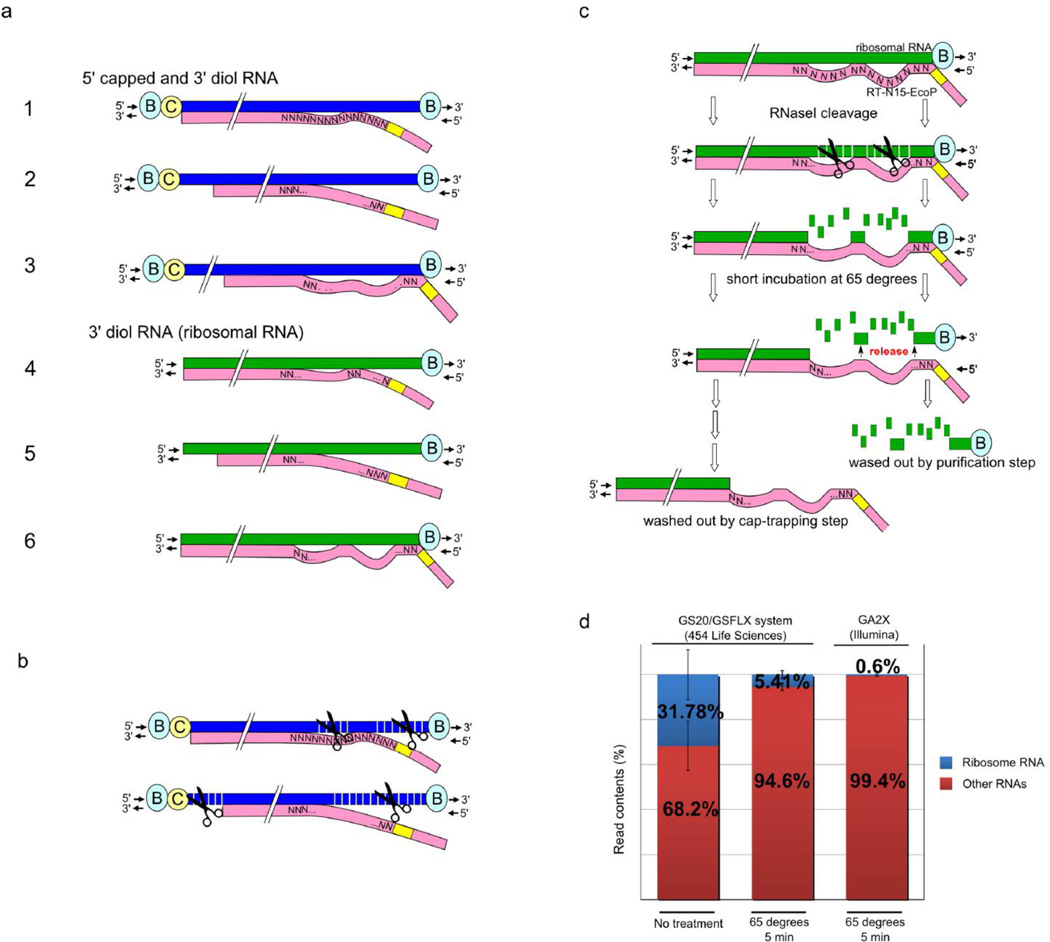
Figure 2. Strategies to eliminate non-capped biotinylated molecules.
(a) As well as at the 5’ end of capped RNAs, biotinylation takes place also on the diol group at the 3’ end of capped RNAs and at the 3’ end of ribosomal/other uncapped RNAs, which must be subsequently eliminated to avoid contamination of 5’ complete cDNA. Careful usage of random primers has been instrumental in achieving this. Blue strand indicates 5’ capped RNA and green strand indicates non capped RNA. Pink strand includes random primer (with restriction enzyme site in yellow) and shows first strand cDNA extension. Examples 1–6 show different potential random priming patterns. (C) Cap; (B) biotin. (b) RNase I is used to cleave single strand mismatched regions produced by cDNA synthesis using random primers. The two examples show different random priming patterns on 5’ capped RN; the upper example (from example 1 in panel a) results in capture of 5’ complete cDNA, while the bottom example (from example 2 in panel a) shows incomplete cDNA that did not extend to the 5’ end. The incomplete cDNA is subsequently eliminated due to RNase I cleavage from the biotinylated cap. (c) Uncapped/incomplete cDNAs derived from primers that perfectly matched the 3’ end of the RNA and biotinylated at the 3’ end need to be eliminated from the library to reduce bias due to ribosomal RNA contamination. Infrequent cases of perfectly aligned random priming at the 3’ end would cause capture through the 3’ end biotin on ribosomal RNA. However, long random primers (N15; pink) leave mismatches that are cleaved by RNAse I treatment, as described in (b). Heating to 65 °C after RNAse treatment releases the biotin at the 3’ end from the cDNA/RNA hybrid, which is then washed out at captrapping step. (d) Dramatic removal of ribosomal cDNA sequence tags by RNAse treatment and heating at 65°C. In a previous protocol using the GS20/GSFLX sequencer (454 Life Sciences)40, ribosomal CAGE tags represented ~30 % of the tags without treatment at 65°C (n = 6). Other samples were incubated 65 °C for 5 min resulting in ribosomal RNA decrease to 5.41 % (n = 6). Error bars indicate a standard deviation of experiments. A third sample shows further decreased ribosomal RNAs with the protocol here presented for the Illumina sequencing instruments. Other RNAs include RNA sequences mappable to the genome (60–89%) or unmapped RNA-derived sequences.
Sorbitol and trehalose addition increase the reverse transcriptase activity at high temperature (55~60°C), further extending the cDNA to the cap site, even for difficult RNA templates, such highly structured, GC-rich 5’ untranslated regions (5’ UTRs)25,26. Cleaning-up the first-strand cDNA reaction is necessary before the chemical biotinylaiton of the cap: the Tris buffer, the saccharides and glycerol present the first-strand cDNA reaction would provide diol groups, which are targets for chemical oxidation (and its subsequent biotinylation), thus competing with the cap for biotinylation. Purification also helps to remove nucleic acids shorter than 100 nt, which include cDNA primers.
Cap-trapping: chemical biotinylation and capture of complete cDNAs
Chemical oxidation with NaIO4 is used to open the RNA diol groups. These include the diol at the 5’ ends, on the cap-structure and the diol-group at the 3’ ends of all RNAs (Fig. 1). The derived oxidized dialdehyde reacts to a long arm biotin hydrazide, which results in biotinylation of the cap-site and the 3’ ends of RNAs (Fig. 2a). Before capturing the biotinylated cap, it is necessary to treat the sample with RNase I, to cleave single stranded RNA (ssRNA) regions that are not protected by newly synthesized cDNAs (Fig. 2), typically when the cDNA did not reach the cap-site. Usually, the 3’ RNA ends are also left single stranded, because random priming seldom primes exactly at the 3’ ends of RNAs. To minimize the chances of capturing cDNAs derived from primers that perfectly matched the 3’ end of the RNA, we use primers with a long random region (15 nt long, or N15). Due to the low priming temperature (RT, which is very efficient to extend N15 partially mismatched primers15) and the complexity of the primer (415 theoretical combinations of sequences), most primers are likely to prime leaving several mismatched bases. This would not be necessarily the case if using a much shorter N6 random primer. More than one random primer could anneal to the same RNA at different positions, producing multiple tandem aligned cDNAs (see Fig. 2a for examples of different random priming patterns). However, only some reach the cap site (as shown in example 1 in Fig. 2a), thus the others must be eliminated to avoid contaminating the library. Mismatches within the random priming sites allow RNase I to nick the RNAs at all the mismatched bases (see Fig. 2b), and the single RNA nucleotides released from the imperfect DNA hybrids are degraded. The next step, where RNase I treatment is followed by a short incubation at 65°C, also releases the biotinylated 3’ end of RNAs from RNA-cDNA hybrids, ensuring that cDNAs derived from primers that perfectly matched the 3’ end of the RNA are not subsequently captured (Fig. 2c). Since we mostly use total RNA as starting material, heating up at 65°C is essential to reduce the final ribosomal contamination to less than 1% (Fig. 2d). The cDNAs including the biotinylated cap site are finally captured with streptavidin coated magnetic beads. cDNAs that did not reach the cap-site are not bound and are eliminated (Fig. 1). To collect 5’-complete cDNA from RNA hybrids, the magnetic beads are treated with alkali, causing denaturation of nucleic acids hybrids and partial hydrolysis of RNAs. RNAse H treatment can be used as an alternative to recover cDNAs6,27.
Providing a priming site for the second strand cDNA synthesis
The recovered cap-selected single-strand cDNA requires the addition of a primable sequence for 2nd strand cDNA synthesis and subsequent sequencing operations. This is achieved with the single-strand linker ligation method (SSLLM), which exploits the ability of DNA ligase to join single-strand cDNAs to a partial double strand linker with a protruding single strand random tail in presence of PEG28. Sequences of 5' linkers with barcodes and their preparation are listed in the REAGENT SETUP section and Table 1. “Phos” and NH2 stand for a phosphate and amino-link modifications respectively (see Table 1). These modifications allow a dramatic reduction of 5’ linker dimers and simultaneously increase the linker–cDNA ligation efficiency (Fig. 3a) because linkers do not ligate to each other but are available only to ligate cDNA, during the whole reaction. The upper linkers (see Table 1) contain a mixture of fully random hexamers at the 3’ end regions, which will anneal to the cDNA at the sequences corresponding to the 5’ end of the original capped RNAs. Additionally, a GN5 is also mixed with the N6 ending linkers, where the first N is substituted with a Guanine. The GN5 linker is used at a ratio of 5:1 with the random N6 purposely, because the majority of the cDNAs carry a Cytosine as the last base. This is often added by the terminal transferase activity of the reverse transcriptase when it reaches the cap site29. Additionally, in mammalian promoters, the first transcribed base of POLR2A promoters is often a G as a part of the initiator element sequence11, resulting often in a C as the last nucleotide of cDNAs.
Table 1.
Primer and linker sequences.
| Step | Primer name | Sequence (5' → 3') | Grade |
|---|---|---|---|
| 1 | RT-N15-EcoP primer | AAGGTCTATCAGCAGNNNNNNNNNNNNNNN | Salt free |
| 28 | ACTB forward (human) |
TATAGCCAGCAGGACCGCC | Salt free |
| ACTB reverse (human) |
ACATGCCGGAGCCGTTGTC | Salt free | |
| ACTB forward (mouse) |
TACCCCCAGCAGGACTGTC | Salt free | |
| ACTB reverse (mouse) |
GTCATCCATGGCGAACTGG | Salt free | |
| Ribosome forward (human) |
CTGGTTGATCCTGCCAGTAG | Salt free | |
| Ribosome reverse (human) |
TCTAGAGTCACCAAAGCCGC | Salt free | |
| Ribosome forward (mouse) |
GCCATGCATGTCTAAGTACGCACG | Salt free | |
| Ribosome reverse (mouse) |
TCAGCGCCCGTCGGCATGTA | Salt free | |
| 30 | 5’SOL-N6-AGA | CCACCGACAGGTTCAGAGTTCTACAGAGACAGCAGNNNN NN Phos |
HPLC |
| 5’SOL-GN5-AGA | CCACCGACAGGTTCAGAGTTCTACAGAGACAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-AGA | Phos CTGCTGTCTCTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 5’SOL- N6-CTT | CCACCGACAGGTTCAGAGTTCTACAGCTTCAGCAGNNNN NN Phos |
HPLC | |
| 5’SOL- GN5-CTT | CCACCGACAGGTTCAGAGTTCTACAGCTTCAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-CTT | Phos CTGCTGAAGCTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 5’SOL- N6-GAT | CCACCGACAGGTTCAGAGTTCTACAGGATCAGCAGNNNN NN Phos |
HPLC | |
| 5’SOL- GN5-GAT | CCACCGACAGGTTCAGAGTTCTACAGGATCAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-GAT | Phos CTGCTGATCCTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 5’SOL-N6-ACA | CCACCGACAGGTTCAGAGTTCTACAGACACAGCAGNNNN NN Phos |
HPLC | |
| 5’SOL- GN5-ACA | CCACCGACAGGTTCAGAGTTCTACAGACACAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-ACA | Phos CTGCTGTGTCTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 5’SOL- N6-ACT | CCACCGACAGGTTCAGAGTTCTACAGACTCAGCAGNNNN NN Phos |
HPLC | |
| 5’SOL- GN5-ACT | CCACCGACAGGTTCAGAGTTCTACAGACTCAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-AGT | Phos CTGCTGAGTCTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 5’SOL- N6-ACG | CCACCGACAGGTTCAGAGTTCTACAGACGCAGCAGNNNN NN Phos |
HPLC | |
| 5’SOL- GN5-ACG | CCACCGACAGGTTCAGAGTTCTACAGACGCAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-ACG | Phos CTGCTGCGTCTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 5’SOL- N6-ATC | CCACCGACAGGTTCAGAGTTCTACAGATCCAGCAGNNNN NN Phos |
HPLC | |
| 5’SOL- GN5-ATC | CCACCGACAGGTTCAGAGTTCTACAGATCCAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-ATC | Phos CTGCTGGATCTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 5’SOL- N6-ATG | CCACCGACAGGTTCAGAGTTCTACAGATGCAGCAGNNNN NN Phos |
HPLC | |
| 5’SOL- GN5-ATG | CCACCGACAGGTTCAGAGTTCTACAGATGCAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-ATG | Phos CTGCTGCATCTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 5’SOL- N6-AGC | CCACCGACAGGTTCAGAGTTCTACAGAGCCAGCAGNNNN NN Phos |
HPLC | |
| 5’SOL- GN5-AGC | CCACCGACAGGTTCAGAGTTCTACAGAGCCAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-AGC | Phos CTGCTGGCTCTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 5’SOL- N6-AGT | CCACCGACAGGTTCAGAGTTCTACAGAGTCAGCAGNNNN NN Phos |
HPLC | |
| 5’SOL- GN5-AGT | CCACCGACAGGTTCAGAGTTCTACAGAGTCAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-AGT | Phos CTGCTGACTCTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 5’SOL- N6-TAG | CCACCGACAGGTTCAGAGTTCTACAGTAGCAGCAGNNNN NN Phos |
HPLC | |
| 5’SOL- GN5-TAG | CCACCGACAGGTTCAGAGTTCTACAGTAGCAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-TAG | Phos CTGCTGCTACTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 5’SOL- N6-TGG | CCACCGACAGGTTCAGAGTTCTACAGTGGCAGCAGNNNN NN Phos |
HPLC | |
| 5’SOL- GN5-TGG | CCACCGACAGGTTCAGAGTTCTACAGTGGCAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-TGG | Phos CTGCTGCCACTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 5’SOL- N6-GTA | CCACCGACAGGTTCAGAGTTCTACAGGTACAGCAGNNNN NN Phos |
HPLC | |
| 5’SOL- GN5-GTA | CCACCGACAGGTTCAGAGTTCTACAGGTACAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-GTA | Phos CTGCTGTACCTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 5’SOL- N6-GAC | CCACCGACAGGTTCAGAGTTCTACAGGACCAGCAGNNNN NN Phos |
HPLC | |
| 5’SOL- GN5-GAC | CCACCGACAGGTTCAGAGTTCTACAGGACCAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-GAC | Phos CTGCTGGTCCTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 5’SOL- N6-GCC | CCACCGACAGGTTCAGAGTTCTACAGGCCCAGCAGNNNN NN Phos |
HPLC | |
| 5’SOL- GN5-GCC | CCACCGACAGGTTCAGAGTTCTACAGGCCCAGCAGGNNN NN Phos |
HPLC | |
| 5’SOL-lower-GCC | Phos CTGCTGGGCCTGTAGAACTCTGAACCTGTCGGTGG NH2 |
HPLC | |
| 35 | 2nd SOL primer | Bio CCACCGACAGGTTCAGAGTTCTACAG | Salt free |
| 43 | 3’ upper linker | NNTCGTATGCCGTCTTCTGCTTG | Cartridge |
| 3’ lower linker | CAAGCAGAAGACGGCATACGA | Cartridge | |
| 51 | PCR Forward primer | AATGATACGGCGACCACCGACAGGTTCAGAGTTC | Salt free |
| PCR Reverse primer | CAAGCAGAAGACGGCATACGA | Salt free | |
| 60 | Sequencing primer | CGGCGACCACCGACAGGTTCAGAGTTCTACAG | Salt free |
Bold indicates barcode sequence. Italics indicate EcoP15I sequence.
5’SOL- N6, 5’SOL- GN5 and 5’SOL-lower indicate 5’- N6 upper linker, 5’- GN5 upper linker and 5’-lower linker respectively.
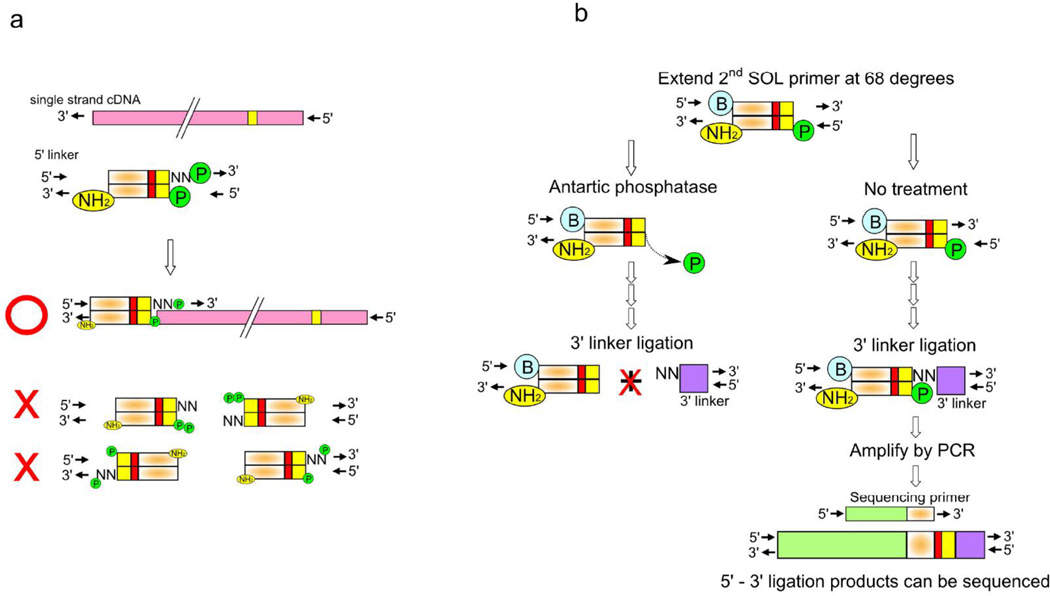
Figure 3. Linker dimer elimination strategies.
(a) Phosphate and NH2 modified 5’ linkers ligate to only cDNA (pink) and not with themselves. (b) Small amounts of linker dimers may form after ligation of the NN single strand of the 3’-linker to the 5’ end linker extended by the 2nd SOL primer (bottom right). This can form artifacts as identified by sequencing if left without treatment. Antarctic phosphatase treatment prevents ligation of the 3’ linker by removing the phosphate group at the end of this artifact, eliminating linker dimers (bottom left).
As this is a critical reaction, linkers should be properly purified by HPLC or by preparative gel electrophoresis. We also recommend synthesizing the linker with fresh reagents. We have noticed that some of the random N nucleotides may be underrepresented when using old reagents, thus introducing ligation biases, because non-random linkers could miss some of the cDNA sequences.
In this step, the linkers ligated to the cDNAs can also be barcoded, in order to pool multiple samples into a single lane to cut down the sequencing costs. We commonly pool 4 to 6 libraries, although progress in sequencing technologies will allow multiplexing with many more samples in the future such an Illumina Multiplex Sequencing system. When barcoding is used, PCR amplification can be applied to all samples simultaneously, which minimizes amplification bias and further aids quantitative comparisons across different conditions30. Conversely, if very deep CAGE sequencing is desired, barcoding and pooling are not necessary.
The second strand cDNA synthesis is initiated by a biotin modified primer, which is used to purify CAGE tags. A thermostable DNA polymerase is used in order to extend at high temperature any potential secondary structure of the cDNA.
Cleavage of the 27 nt CAGE tags and PCR amplification
Before the restriction digestion, the cDNA is treated with Antarctic phospatase, which removes the phosphate modification at the 5’ end of the 5’-linker, as the phosphorylated 5’-linker would otherwise nonspecifically ligate to the 3’-linker during the subsequent 3’-linker ligation step causing artifacts that will contaminate the library (Fig. 3b). The second-strand cDNA is then cleaved by EcoP15I, a type III restriction enzyme that cleaves 27 nt downstream of the enzyme recognition site. Since EcoP15I requires two recognition sequences in opposite (head to head) orientation on the same DNA regardless of their reciprocal distance31, another EcoP15I was included in the first strand cDNA primer (RT-N15-EcoP). Additionally, cleavage is enhanced in presence of AdoMet, an analogue of sinefungin, which stimulates EcoP15I32. The amount of enzyme should be carefully determined. Excess of this enzyme inhibits the cleavage of small cDNA amounts. After cleavage, the 3’ linker (Table 1), ending with two NN protruding nucleotides, is ligated at the EcoP15I cleavage site and provides a priming site for the subsequent PCR amplification. The excess of 3’ linkers should be removed before PCR, to avoid contamination of the library with 3’ linker-dimers. To do so, we take advantage of the Biotin present at the 5’ end on the second strand primer (the 2nd SOL primer); streptavidin-based capture leaves the 3’ linker dimers in solution, allowing their removal.
For the PCR amplification step, the Phusion High-Fidelity DNA Polymerase performs very well allowing reduction in the number of PCR cycles. Additionally, Phusion does not add extra As at the 3’ end of the PCR products, which would cause mismatches with the primer of the Illumina sequencer. Before bulk PCR of the whole CAGE library, we determine the optimal PCR cycle number. This optimization is important to keep the PCR cycle number to a minimum in order to reduce any possible PCR bias. The whole library is finally amplified by PCR and is subsequently sequenced. By using 5 µg of total mammalian RNAs, less than 15 PCR cycles are usually necessary. The CAGE reads are 96 bp long, which include the specified sequences for Illumina sequencing platforms. The excess of PCR primers is degraded with Exonuclease I, which degrades ssDNAs but not the dsDNA CAGE tags.
Library sequencing
We usually apply 5.0 pMoles l−1 of CAGE library to each flow cell of GA2X Illumina (36 cycles protocol), or 4.0 pMoles l−1 of library to the Illumina Hi-Seq 2000 (50 cycles protocol), essentially following the manufacturer’s instructions. The sequencing yield is improving constantly, thanks to continuous advances in technology. This protocol is in principle suitable for use with other sequencing platforms, if the linkers are appropriately modified by the user. CAGE libraries are sequenced as efficiently as other applications (for instance, ChIP-seq, RNAseq) and do not require any further manipulations for sequencing.
Controls
We suggest that inexperienced users of the protocol prepare a control library with an RNA that is commonly used in the laboratory, such as Mouse Embryo 17.5. It is possible to take aliquots of the cDNA to measure the efficiency of cDNA synthesis using quantitative RT-PCR (qRT-PCR) with housekeeping genes such as Actin-β (ACTB; see Step 28) We recommend using genes that show specific transcription starting sites, like ACTB. qRT-PCR primers should be designed to include the borders of cDNA at the points where they ligate with the linker sequences at the 5’ end of the transcript. For example, primers include the 5’ of ACTB and part of the linker (including the EcoP15I site); the other primer is simply designed in the middle of the ACTB. Similarly, we also prepare a set of primers to amplify ribosomal derived cDNA sequence. qRT-PCR primer sequences are shown in Table 1. RT-PCR output is provided in Ct (see ref. 33). The Ct values of ACTB should not be higher than the Ct values for the Ribosome cDNA reaction. The remaining part of the library can be further sequenced as control library.
MATERIALS
REAGENTS
-
·
Trehalose Dihydrate Molecular Biology Grade (Life Sciences Advanced Technologies, cat. no. TDH033)
-
·
D-Sorbitol (Sigma-Aldrich, cat. no. 85529-250G)
-
·
NaIO4 (ICN Bio. Inc, cat. no. 152577)
-
·
Biotin (Long Arm) Hydrazide (VECTOR Lab, cat. no. SP-1100, 50 mg)
-
·
E. coli tRNA (Ribonucleic acid, transfer from Escherichia coli Type XX, Strain W, lyophilized powder; Sigma, cat. no. R1753)
-
·
RQ1 RNase-Free DNase (Promega, cat. no. M6101)
-
·
Protenase K (Invitrogen, cat. no. 25530-049)
-
·
10 mg ml−1 BSA (NEB, cat. no. B9001S)
-
·
10 mM ATP (NEB, cat. no. P0756S)
-
·
Trizol LS (Invitrogen, cat. no. 10296-010) !CAUTION Toxic. This reagent involves phenol solution that may cause a serious health hazard. Handle using appropriate safety measures such as the use of safety goggles, gloves, mask and fume hood.
-
·
RNeasy kit (QIAGEN, cat. no. 74104)
-
·
Poly(A)Purist mRNA Purification Kits (Ambion, cat. no. AM1916)
-
·
PrimeScript Reverse Transcriptase (TAKARA, cat. no. 2680A, 10000 U) ▲CRITICAL Reverse transcribed cDNA yields are best with this enzyme. If other enzymes have to be used, select RT devoid of RNAseH activity.
-
·
Agencourt RNAClean XP Kit (BECKMAN COULTER, cat. no. A63987, 40 ml)
-
·
Agencourt AMPure XP Kit (BECKMAN COULTER, cat. no. A63881, 60 ml)
-
·
RNase ONE Ribonuclease (Promega, cat. no. M4261, 1000 U)
-
·
MPG Streptavidin (TAKARA, cat. no. 6124A, 2 ml)
-
·
Quant-iT™ OliGreen® ssDNA Reagent and Kit (Invitrogen, cat. no. O11492)
-
·
SYBR Premix Ex Taq (TAKARA, cat. no. RR041A)
-
·
Agilent RNA Pico kit (Agilent, cat. no. 5067-1513)
-
·
Agilent DNA1000 kit (Agilent, cat. no. 5067-1504)
-
·
DNA Ligation Kit <Mighty Mix> (TAKARA, cat. no. 6023, 1 kit) ▲CRITICAL This kit contains polyethylene glycol, which increases ligation efficiency.
-
·
T4 DNA ligase (NEB, cat. no. M0202S, 20000 U)
-
·
TaKaRa LA Taq (TAKARA, cat. no. RR002A, 125 U)
-
·
Antarctic Phosphatase (NEB, cat. no. M0289L, 5000 U)
-
·
EcoP15I (NEB, cat. no. R0646S, 500 U)
-
·
Sinefungin (Calbiochem-Novabiochem international, cat. no. 567051, 2 mg)
-
·
Phusion™ High-Fidelity DNA Polymerase (FINNZYMES, cat. no. F-530S, 100 U) ▲CRITICAL This DNA polymerase shows better yield than Taq DNA polymerase, allowing a lower number of PCR cycles.
-
·
Exonuclease I (E. coli) (NEB, cat. no. M0293S, 3000 U)
-
·
MinElute PCR Purification Kit (QIAGEN, cat. no. 28004, 50 columns)
-
·
QIAquick Gel Extraction Kit (QIAGEN, cat. no.28704, 50 columns)
-
·
Ethanol (99.5) (WAKO, 057-00456, 500 ml) !CAUTION Flammable. Handle using appropriate safety precautions.
-
·
SDS (WAKO, cat. no. 196-08678, 500 g)
-
·
10 mM dNTPs (Invitrogen, cat. no. 18427-088)
-
·
NaOAc (WAKO, cat. no. 199-01085, 500 g)
-
·
NaCitrate (MP Biomedicals, Inc., cat. no. 194817, 500 g)
-
·
EDTA (Dojindo, cat. no. 345-01865, 500 g)
-
·
Glycerol (WAKO, cat. no. 075-00616, 500 ml)
-
·
Tris (WAKO, cat. no. 207-06275, 500 g)
-
·
HCl (WAKO, cat. no. 080-01066, 500 ml) !CAUTION Poison. When you adjust pH with HCl solution, handle using appropriate safety equipments such as the use of safety goggles, gloves, mask and fume hood.
-
·
1M Tris-HCl (pH 7.0), Tris (WAKO, cat. no. 207-06275, 500 g), HCl (WAKO, cat. no. 080-01066, 500 ml) !CAUTION Poison. When you adjust pH with HCl solution, handle using appropriate safety equipment.
-
·
10 mM Tris-HCl (pH 8.5), Tris (WAKO, cat. no. 207-06275, 500 g), HCl (WAKO, cat. no. 080-01066, 500 ml) !CAUTION Poison. When you adjust pH with HCl solution, handle using appropriate safety equipment.
-
·
50 mM NaOH, NaOH (WAKO, cat. no. 197-02015, 500 g)
-
·
0.4M MgCl2, MgCl2 (WAKO, cat. no. 135-00165, 500 g)
-
·
Nuclease-free water (Invitrogen Corp, cat. no. 10977-015)
-
·
Standard Cluster Generation Kit v4 (Illumina, cat. no. GD-103-4001, 1 flow cell)
EQUIPMENT
-
·
Micro pipettes
-
·
Multiple channel pipetters
-
·
Pipette Tips (DNase/RNase free, Low binding tips)
-
·
1.5 ml SnapLock Microtube, MaxyClear, Maxymum Recovery (AXYGEN, cat. no. MCT-150-L-C, 250 tube)
-
·
96 well PCR Plate, 0.2 ml, Clear (AXYGEN, cat. no. PCR-96-C, 50 plate)
-
·
0.2 ml 8-Strip PCR Dome Tube cup, for the cup of 96 well PCR Plate, (AXYGEN, cat. no. PCR-02CP-C)
-
·
0.2 ml Thin Wall Clear PCR Strip Tubes and Clease Strip Caps (AXYGEN, cat. no. PCR-0208-CP-C)
-
·
96 well plate and tube centrifuge instrument
-
·
StepOnePlus Real Time PCR system (Applied Bio systems, cat. no. StepOnePlus-01)
-
·
Agilent 2100 Bioanalyzer (Agilent Technologies, cat. no. G2928B)
-
·
NanoDrop 1000 spectrophotometer (Thermo Fisher Inc., cat. no. S09NND360)
-
·
Dynal Magnetic stand (Invitriogen, cat. no. MPC-96S)
-
·
Centrifugal Concentrator (TOMY Digital Biology Co., Ltd., cat. no. 35041048)
-
·
Thermal cycler
-
·
Genome Analyzer IIx (Illumina)
REAGENT SETUP
CRITICAL All regents should be prepared using RNA/DNA free solutions and clean dedicated equipments.
RNA stability check
It is highly recommended to test the stability of RNA in presence of the reagents that are made by operator in REAGENTS and REAGENT SETUP.
Incubate 1 µg of total mammalian RNA (like mouse liver RNA) in 10 µl of reagent at 37°C for 1 h.
Analyze 1 µl of the reaction with the Agilent Bioanalyzer RNA pico/nano kit and measure the RIN value. This should be unchanged (or 1 the original RIN value.)
3.3 M Sorbitol / 0.66 M Trehalose mix
-
Saturate 8.02 g of trehalose and 17.8 g of sorbitol in 30 ml of water and autoclave at 121°C, 30 min. Store at room temperature (15 – 25°C) for up to one year or store at −20°C in aliquots for up to 5 years.
!CAUTION Trehalose and sorbitol are purchased as high-quality essentially free of heavy metals, which would cause nucleic acid degradation.
250 mM NaIO4 for Oxidation of the diol groups
Dissolve 0.053 g NaIO4 in 1 ml of water. !CAUTION Store at room temperature in a dark place. The solution should be freshly prepared before use.
15 mM Biotin (Long Arm) Hydrazide for Biotinylation
-
Dissolve 0.0038g Biotin (Long Arm) Hydrazide in 675 µl of water. Store at room temperature.
!CAUTION Solution should be freshly prepared before use (Step 13). The Biotin will not dissolve immediately in the water, mix continuously until dissolved.
20 µg µl−1E. coli tRNA
Dissolve 30 mg E. coli tRNA lyophilized powder in 400 µl of water and add 45 µl of 10×RQ1 DNase buffer and 30 µl of RQ1 RNase-Free DNase. Incubate at 37 °C, 2 hrs. Add 10 µl of 0.5 M EDTA (pH 8.0), 10 µl of 10% SDS and 10 µl of 10 ng ml−1 Proteinase K to tRNA solution. Incubate at 45 °C, 30 min. Extract with 500 µl of phenol/chloroform and centrifuge at 15,000 rpm for 3 min. at room temperature. Collect the aqueous phase and extract with 500 µl of chloroform. Centrifuge again at 15,000 rpm for 3 min. Collect the aqueous phase and add 25 µl of 5M NaCl, 525 µl of Isopropanol. Centrifuge at 15,000 rpm for 5 min. at room temperature. Remove the supernatant and add 900 µl of 80 % Ethanol to tRNA pellet. Centrifuge at 15,000 rpm for 5 min. at room temperature. Repeat the ethanol wash and centrifugation then discard the supernatant and dissolve the tRNA pellet in 1.5 ml water. Aliquots can be stored at −20 °C for up to 5 years.
Preparation of wash buffers for MPG beads
Wash buffer 1: Mix 45 ml of 5 M NaCl and 5 ml of 0.5 M EDTA (pH 8.0). Store at room temperature for up to one year.
Wash buffer 2: Mix 3 ml of 5M NaCl, 100 µl of 0.5 M EDTA (pH 8.0) and 46.9 ml of water. Store at room temperature for up to one year.
Wash buffer 3: Mix 1 ml of 1M Tris-HCl (pH 8.5), 100 µl of 0.5 M EDTA (pH 8.0), 25 ml of 1 M NaOAc (pH 6.1), 2 ml of 10% SDS and 21.9 ml of water. Store at room temperature for up to one year. !CAUTION If the room temperature drops, 10%SDS in Wash buffer 3 may form crystals. In this case, dissolve crystallized SDS in a water bath at 37°C before usage.
Wash buffer 4: Mix 500 µl of 1M Tris-HCl (pH 8.5), 100 µl of 0.5 M EDTA (pH 8.0), 25 ml of 1M NaOAc (pH 6.1) and 24.4 ml of water. Store at room temperature for up to one year.
Preparation of 3’ linker ligation buffer (5 X)
Mix 50 µl of 1 M Tris-HCl (pH 7.0), 10 µl of 100 mM ATP and 0.5 µl of 10 mg ml−1 BSA and make up to 200 µl with water. Store at room temperature for up to one year.
Preparation of 5’- and 3’-linkers
See Box 1.
Box 1. 5’- and 3’-linker preparation.
Barcoded 5’ linker preparation ● TIMING 1 h
Prepare a 2 µg µl−1 solution of each HPLC-grade 5’ linker (see Table 1) in 1mM Tris-HCl (pH 7.5) and 0.1 mM EDTA (pH 8.0).
For N6 linker reaction solution, mix 1.5 µl of each specific 5’-N6 upper linker (3.0 µg), 1.5 µl of each specific 5’-lower linker (3.0 µg), 0.75 µl of 1 M NaCl and 3.25 µl of water. For GN5 linker reaction solution , mix 6 µl of each specific 5’-GN5 upper linker (12 µg), 6 µl of each specific 5’-lower linker (12 µg), 3 µl of 1 M NaCl and 15 µl of water. CRITICAL STEP Upper and lower linkers with matching specific barcode sequences are combined together to form a double strand with partial single strand random protruding ends, which ligate to the terminal end of the cDNAs.
-
To carry out the annealing reaction, incubate the linker reaction solutions using the following conditions: 95°C, 5 min, −0.1°C/sec down to 83°C, 5 min at 83°C, −0.1°C/sec down to 71°C, 5 min at 71°C, −0.1°C/sec down to 59°C, 5 min at 59°C, −0.1°C/sec, to 47°C, 5 min at 47°C, −0.1°C/sec, to 35°C, 5 min at 35°C, −0.1°C/sec to 23°C, 5 min at 23°C, −0.1°C/sec to 11°C, then hold at 11°C (annealing is considered finished when sample reach 11°C).
CRITICAL STEP Annealing takes place by slowly cooling the linkers at the described temperature.
The final annealed linker solutions can be kept at 4°C for one month, but for long term storage, should be frozen at −20°C and can be kept for up to 5 years.
The “N6” and “GN5” linker solutions carrying the same barcode should be mixed at this stage. The total volume is 37.5 µl (0.8 µg µl−1), using a final ratio of N6:GN5 = 1:4. CRITICAL STEP Using these mixed linkers was found to be effective to maximize the ligation efficiency of the linkers with cap-trapped cDNA ends.
Linkers are used at 200 ng µl−1 (see Step 30), when starting from 5 µg of total RNA.
3’ linker preparation ● TIMING 1 h
Prepare 2 µg µl−1 solutions of each HPLC- or cartridge-grade 3’ linker (see Table 1) in 1mM Tris-HCl (pH 7.5) and 0.1 mM EDTA (pH 8.0).
For 3’-linker reaction solution, mix 2.5 µl of 3’ upper linker (5.0 µg), 2.5 µl of 3’ lower linker (5.0 µg), 1.25 µl of 1 M NaCl and 6.25 µl of water.
To carry out the annealing reaction, incubate the 3’-linker reaction solution using the following conditions: 95°C, 5 min, −0.1°C/sec down to83°C, 5 min at 83°C, −0.1°C/sec down to 71°C, 5 min at 71°C, −0.1°C/sec down to 59°C, 5 min at 59°C, −0.1°C/sec, to 47°C, 5 min at 47°C, −0.1°C/sec, to 35°C, 5 min at 35°C, −0.1°C/sec to 23°C, 5 min at 23°C, −0.1°C/sec to 11°C, then hold at 11°C (annealing is considered finished when sample reaches 11°C).
The final annealed linker concentration is 0.8 µg µl−1. These linkers are used at the concentration of 100 ng µl−1 when starting from 5 µg of total RNA (see Step 43). It can be kept at 4°C for up to one month, but for long term storage should be frozen at −20°C for up to 5 years.
RNA isolation and Preparation (5 µg total RNA, polyA plus RNA or polyA minus RNA)
It is advisable to use RNA that was isolated using Trizol LS (Invitrogen), RNeasy (micro) kit (QIAGEN) or equivalent methods, where obtained RNAs have RIN value above 7 as measured with Agilent RNA nano kit. We generally use 5 µg of total RNA. The protocol can also be used to prepare CAGE libraries from 5 µg polyA minus RNA or 1 µg polyA plus RNA, which can be prepared by Poly(A)Purist mRNA Purification Kits (Ambion) or equivalent kits. As these types of samples contain different amount of capped RNAs, it is likely that this protocol can be used with larger or smaller amount of RNAs.
PROCEDURE
CRITICAL The procedure described here is for a single sample. However, the protocol is commonly performed using multiple samples, including preparation of CAGE libraries with multipipettes. In this case, where appropriate, prepare a master mix of reagents to avoid technical bias.
!CAUTION Wear gloves and lab coat throughout the procedure. Keep samples and reagents under RNase free conditions until the end of the cap-trap procedures, as RNA degradation will interfere with several steps.
Reverse transcription (RT) ● TIMING 1.5 h
-
1
Mix 5 µg of total RNA and 2.2 µl of 210 µM RT-N15-EcoP primer. Adjust volume to 7.5 µl in water. Incubate at 65°C, 5min and then cool on ice immediately.
-
2Mix the following components:
Component Volume Final concentration 5 X PrimeScript buffer 7.5 µl 1 X dNTPs (10 mM each) 1.87 µl 0.5 mM each 3.3 M Sorbitol / 0.66 M Trehalose mix solution 7.5 µl 0.66 M / 0.132 M PrimeScript Reverse
Transcriptase (200 U µl−1)3.75 µl 750 U Water 9.38 µl - Total volume 30 µl -
▲CRITICAL STEP Volumes provided are for a single sample, but can be scaled up for the number of samples used, typically using 8 strip tubes or 96 well plates.
-
3
Add Enzyme mix solution from Step 2 to RNA and primer mix solution from Step 1 then carefully mix by pipetting on ice (total volume 37.5 µl).
-
4
Incubate at following temperatures, for the time indicated; 25°C, 30 sec; 42°C, 30 min; 50°C, 10 min; 56°C, 10 min; 60°C, 10 min; keep on hold on ice.
cDNA purification with the RNAClean XP kit ● TIMING 1.5 h
-
5
Mix 67.5 µl of RNAClean XP and 37.5 µl of RT reaction solution from Step 4 thoroughly by pipetting 10 times. Incubate at room temperature for 30 min, mixing every 10 min by pipetting.
-
6
Place the reaction solution on the magnetic stand and wait for 5 min. Aspirate the cleared solution and discard.
▲CRITICAL STEP Aspirate carefully. Be sure not to aspirate beads in the pipet tip together with the solution, in order to avoid contaminations constituted by residual primers.
-
7
Keep the sample on the magnetic stand and wash the beads with 150 µl of 70% ethanol; wash both the beads and the tube walls. After checking that the beads are settled on the tube wall, aspirate the cleared solution. Repeat this washing step.
-
8
Off the magnetic stand, add 40 µl of 37°C pre-heated water and extensively pipet. The manufacturer suggests pipetting at least 20 times to completely elute the nucleic acids.
-
9
Incubate at 37°C for10 min and then place on the magnetic stand for 5 min to separate the beads. Transfer the eluant to the new tube (40 µl).
-
10
Keep the cDNA on ice until the next step.
Diol-Oxidation with NaIO4 ● TIMING 50 min
-
11Mix the following reagents on ice and incubate on ice for 45 min:
Component Volume Final concentration RNA-cDNA hybrid 40 µl - 1 M NaOAc (pH 4.5) 2 µl 45.7 mM 250 mM NaIO4 2 µl 11 mM Total volume 44 µl -
!CAUTION let the reaction proceed in the dark by putting promptly covering the tube(s) with an aluminum foil.
-
12
After the incubation, add 2 µl of 40% glycerol and mix thoroughly to stop the oxidation reaction. Add 14 µl of 1M Tris-HCl (pH 8.5) to bring the pH above 5.6. (Total volume: 60 µl)
cDNA purification with the RNAClean XP kit ● TIMING 1.5 h
-
13
Mix 108 µl of RNAClean XP and 60 µl of cDNA from the diol-oxidation reaction solution in Step 12 thoroughly by pipetting 10 times. Incubate at room temperature for 30 min, mixing every 10 min by pipetting.
▲CRITICAL STEP It is important to mix the Agencourt RNAClean XP reagent and cDNA solution at a ratio of 1:1.8, which ensures efficient capturing of all nucleic acids longer than 100 nt. In parallel with this procedure, prepare the 15 mM biotin hydrazide (Long Arm) solution.
-
14
Repeat Steps 6–10. Elute in a final volume of 40 µl.
Biotinylation of the RNA diols ● TIMING 16 h (overnight)
-
15Mix the following components by pipetting 10 times and incubate at room temperature (23°C) for 16 h (overnight):
Component Volume Final concentration Purified oxidated cDNA/RNA hybrids from Step 14 40 µl - 1 M Na-Citrate (pH 6.0) 4 µl 70 mM 15 mM biotin hydrazide (Long Arm) 13.5 µl 3.5 mM Total volume 57.5 µl -
CRITICAL STEP Instead of reacting overnight, it is possible perform the reaction at 37°C for 3 h. Although some biotin hydrazide batches have shown to degrade nucleic acids at 37°C, testing new batches of biotin hydrazide solves this problem. To do this, incubate DNA and RNA markers at 37°C with 3.5 mM biotin hydrazide and exclude nucleic acids degradation by gel electrophoresis.
RNase I treatment ● TIMING 45 min
-
16Mix the following reagents by pipetting and incubate at 37°C, 30 min. After the reaction, inactivate at 65°C 5 min and then cool on ice immediately for at least 2 min.
Component Volume Final concentration Biotinylation reaction from Step 15 57.5 µl - 1 M Tris-HCl (pH 8.5) 6 µl 86 mM 0.5 M EDTA (pH 8.0) 1 µl 7.2 mM RNase ONE Ribonuclease ( 10 U µl−1) 5 µl 50 U Total volume 69.5 µl -
cDNA purification with the RNAClean XP kit ● TIMING 1.5 h
-
17
Mix 125 µl of RNAClean XP and 69.5 µl of cDNA from the RNase I reaction solution from Step 16 thoroughly by extensive pipetting. Incubate at room temperature for 30 min, mixing every 10 min by pipetting.
-
18
Repeat Steps 6–10. Elute in a final volume of 40 µl.
Cap-trapping ● TIMING 2.5 h
-
19
Prepare tRNA coated magnetic beads by adding 1.5 µl of 20 µg µl−1 E. coli tRNA mix to 100 µl of MPG beads and incubate at room temperature for 30–60 min, mixing every 10 min by pipetting or moderate vortexing. Separate the beads on a magnetic stand and remove the supernatant. Wash the beads with 50 µl of Wash buffer 1. Repeat this washing step. Resuspend magnetic beads in 80 µl of Wash buffer 1.
▲CRITICAL STEP coating the beads with tRNA before cDNA capture is essential to diminish non-specific cDNA/beads interactions and thus reducing the contamination of cDNAs that did not reach the cap site. Note that E. coli tRNA are added after the RT reaction thus these sequences cannot contaminate the CAGE library.
-
20
Add 40 µl of the purified cDNA from Step 18 to the 80 µl of washed MPG beads from Step 19.
-
21
Incubate at room temperature for 30 min (pipet thoroughly ~10 times or vortex moderately every 5 min.). Place the reaction solution on the magnetic stand and wait for 3 min for the beads to separate. Aspirate and discard the cleared solution.
-
22
Keep the sample on the magnetic stand and wash the beads with 150 µl of the various wash buffers as follows. Wash buffer 1 (1 time), Wash buffer 2 (1 time) Wash buffer 3 (2 times) and Wash buffer 4 (2 times). At each wash, resuspend the beads and let them separate for 3 minutes on the magnetic stand before discarding the washing solution.
▲CRITICAL STEP It is important to wash multiple times. We found that this helps to prevent contamination of non-capped molecules in the obtained CAGE library.
Release 5’-completed cDNAs from magnetic beads ● TIMING 15 min
-
23
Add 60 µl of 50 mM NaOH solution to the beads with the cDNA/RNA bound from Step 22 and incubate at room temperature for 10 min, with occasional mixing by pipetting or vortexing.
-
24
Place the beads on the magnetic stand and wait for 3 min. Transfer the supernatant to a new tube.
-
25
Add to the 60 µl of eluant 12 µl of 1M Tris-HCl (pH 7.0) to neutralize the alkali solution. The total volume is now 72 µl. Store the cDNA on ice before the next step.
cDNA purification with the AMPure XP kit ● TIMING 1.5 h
-
26
Mix 130 µl of AMPure XP ad 72 µl of cDNA from Step 25 thoroughly by pipetting 10 times. Incubate at room temperature for 30 min, mixing every 10 min by pipetting.
-
27
Repeat Steps 6–10. Transfer the 35 µl of eluant to the new tube, and keep 5 µl for quality check.
Quality Check ● TIMING 1.5 h
-
28
Measure the concentration using 2 µl of purified single strand cDNAs by OliGreen (1/10 dilution and measure it in duplicate). Measure the size distribution with 1 µl of the cDNA by Agilent Bioanalyzer RNA pico Kit following the manufacturer’s instructions. Optionally, to analyze the yield of cDNA, the operator can also use 1µl of cDNA and after 10 times dilution, can perform qRT-PCR with ACTB specific primers and ribosomal cDNA primers (see Experimental Design) to monitor specific enrichment of capped molecules using SYBR Premix Ex Taq (TaKaRa) and StepOnePlus Real Time PCR system, follow the manufacturer’s protocol.
▲CRITICAL STEP Ensure that the total amount of obtained cDNA is between 3 ~ 30 ng, when starting from 5 µg of total RNA. The size range should be broad, as shown in Fig. 4.
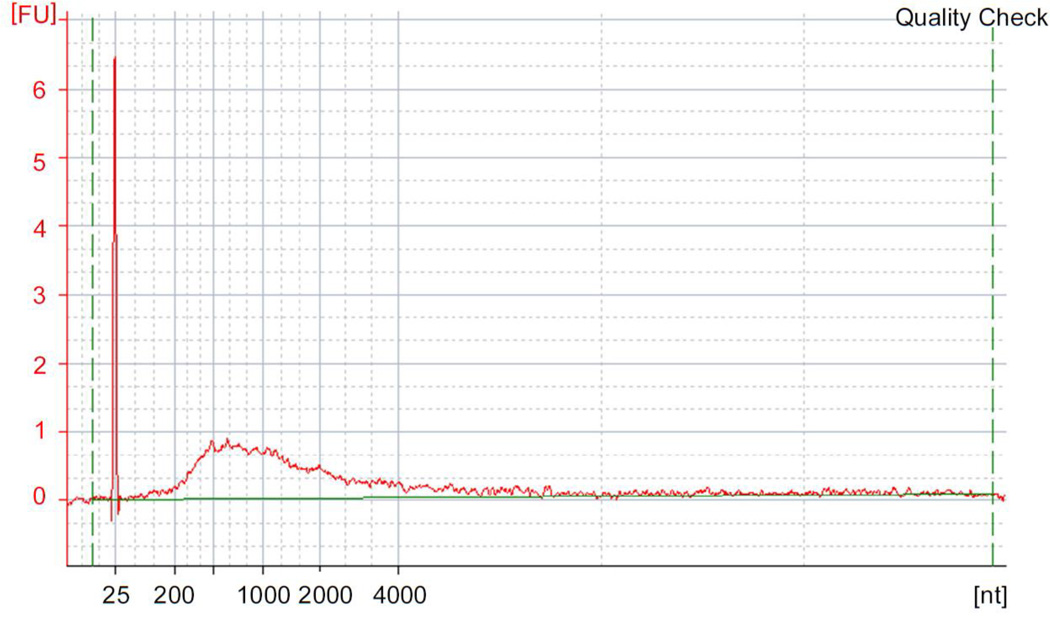
Figure 4. Cap trapped cDNA size distribution.
Quality check result of cap-trapped, single strand cDNA obtained at Step 28. One µl of purified cDNA is measured with the Agilent RNA pico kit. cDNA should range from few hundred base pairs and may reach the length of 4Kb. FU: fluorescence unit, dashed green lines: baseline, 25 nt peak: molecular size marker.
?TROUBLESHOOTING
-
29
Concentrate the cDNA using a centrifugal concentrator at room temperature in a siliconized tube, and then redissolve in 4 µl of water.
▲CRITICAL STEP It may be preferable to avoid complete drying of the pellet by frequently inspecting the remaining volume of water in the sample when drying.
Barcoded 5' linker ligation to the single stranded (ss) cDNA ● TIMING 16 h (overnight)
-
30
Add 1µl of the 5’ linker (from Box 1; 200 ng µl−1) to an empty tube for each cDNA sample and incubate at 37°C, 5 min. At the same time, incubate the 4 µl of redissolved single strand cDNA from Step 29 at 65°C for 5 min. Cool the linker and cDNA on ice for 2 min.
▲CRITICAL STEP It is important to denature the linker and cDNA secondary structure for ligating them efficiency.
-
31
Mix 4 µl of cDNA and 10 µl of DNA ligation Mighty Mix and add the mixture to 1 µl of 5’ linker tubes (total volume 15 µl).
-
32
After extensive mixing, incubate at 16°C, 16 h (overnight).
▲CRITICAL STEP Using differently barcoded 5' linkers it is possible to pool different cDNAs in the following purification step.
cDNA purification with the AMPure XP kit and Pooling samples ● TIMING 1.5h
-
33
Add 55 µl of water to the 15 µl of 5’ linker ligated cDNAs. In case of pooling cDNA, pool ligated cDNAs and make up the volume to 70 µl with water.
▲CRITICAL STEP In this step, due to volume constrains, the maximum number of cDNA samples that can be pooled limited to four. It is possible to pool more cDNAs together in the following, second purification step.
-
34
Mix 126 µl of Agencourt AMPure XP reagent and 70 µl of cDNAs. Purify as in Steps 6–10 and repeat the whole purification process (Steps 6–10) again. The final elution volume is 30.5 µl.
▲CRITICAL STEP Since the concentration of the 5’ linker is high, it is important to perform the purification twice to avoid any linker dimers in the final library. During the second purification step, it is possible to pool together different sets of pooled cDNA samples, after the binding step, just before the 70% ethanol washing step.
Second strand cDNA synthesis ● TIMING 30 min.
-
35Set up the second strand synthesis on ice as described below and gently mix by pipetting.
Component Volume Final concentration 5’ linker ligated 30.5 µl - 10 X LA Taq buffer 5 µl 1 X 25 mM MgCl2 5 µl 2.5 mM dNTPs (2.5 mM each) 8 µl 0.4 mM each 2nd SOL primer (200 ng µl−1, 24 µM) 1 µl 2.4 µM LA Taq (5U µl−1) 0.5 µl 2.5 U Total volume 50 µl - -
36
Run the thermal cycler using the following conditions; 94°C for 3 min, 42°C for 5 min to anneal the primer, 68°C for 20 min, 72°C for 2 min and then hold at 4°C.
Antarctic Phosphatase activation ● TIMING 1.5 h
-
37Add the following reagents to the second strand cDNA reaction solution and gently mix by pipetting for 10 times. Incubate at 37°C for 1 h. Inactivate the enzyme at 65°C for 5 min and then cool on ice for 2 min.
Component Volume Final concentration Second strand cDNA reaction from Step 36 50 µl - Antarctic Phosphatase (5 U µl−1) 4 µl 1 X 10 X Antarctic Phospatase reaction buffer 6 µl 1 X Total volume 60 µl -
cDNA purification with the AMPure XP kit ● TIMING 1.5 h
-
38
Mix 108 µl of AMPure XP and 60 µl of cDNA after the Antarctic Phosphatase treatment thoroughly by pipetting 10 times. Incubate at room temperature for 30 min, mixing every 10 min by pipetting.
-
39
Repeat Steps 6–10. Elute in a final volume of 30 µl.
■ PAUSE POINT The purified cDNA can be frozen for up to a month at −20°C.
EcoP15I digestion ● TIMING 3.5 h
-
40Mix the following reagents and solutions on ice. At the end, incubate at 37°C for 3 h.
Component Volume Final concentration Purified cDNA from Step 39 30 µl - 10 X NEBuffer 4 µl 1 X 10 mg ml−1 (100 X) BSA 0.4 µl 10 X 10 mM (10 X) ATP 4 µl 1 mM 10 mM sinefungin 0.4 µl 0.1 mM EcoP15I (10 U µl−1) 0.1 µl 1 U Water 1.1 µl - Total volume 40 µl - -
41
Add 1 µl of 0.4 M MgCl2 (10 mM final concentration) to EcoP15I digested cDNA to stabilize short tags and to prevent their denaturation.
-
42
Incubate at 65°C, 20 min to inactivate the restriction enzyme. Store on ice until the next step.
Addition of a 3’ linker to the cleaved tags ● TIMING 16 h (overnight)
-
43This step provides to the 5’ cDNA tags with a 3’ end linker. Set up the ligation solution as shown below and mix by pipetting on ice.
Component Volume Final amount cDNA from Step 42 41 µl - 5X 3’ linker ligation buffer 16 µl 1 X 3’ linker (100 ng µl−1) see Box 1 1 µl 100 ng T4 DNA Ligase (400 U µl−1) 3 µl 1200 U Water 19 µl - Total volume 80 µl - -
44
Incubate the reaction solution at 16°C, 16 h (overnight).
Removal of excess 3’ linkers ● TIMING 2.5 h
-
45
Prepare tRNA coated magnetic beads. Add 1 µl of 20 µg µl−1 E. coli tRNA mix to 10 µl of MPG beads and incubate at room temperature for 30–60 min, mixing every 10 min by pipetting. Separate the beads on a magnetic stand and remove the supernatant. Wash the beads with 50 µl of Wash buffer 1. Repeat this washing step. Resuspend magnetic beads in 25 µl of Wash buffer 1.
▲CRITICAL STEP tRNA coating step is essential to diminish non-specific interactions.
-
46
Add 80 µl of 3’ linker ligated cDNA from Step 44 to the 25 µl of washed MPG beads from Step 45.
-
47
Incubate at room temperature for 30 min (pipette thoroughly ~ 10 times or vortex moderately every 5 min.). Place the reaction solution on the magnetic stand and wait for 3 min for the beads to separate. Aspirate and discard the cleared solution.
-
48
Keep the sample on the magnetic stand and wash the beads with 150 µl of the various wash buffers as follows. Wash buffer 1 (1 time), Wash buffer 2 (1 time) Wash buffer 3 (2 times) and Wash buffer 4 (2 times). At each wash, re-suspend the beads and let them separate for 3 minutes on the magnetic stand before discarding the washing solution.
▲CRITICAL STEP It is important to wash multiple times. We found that this helps to prevent contamination of excess 3’ linkers in the final library.
-
49
For the final wash, keep the sample on the magnetic stand and quickly wash with 50 µl of water.
▲CRITICAL STEP To avoid losing cDNAs due to denaturation, do not heat up the sample and perform these steps as quickly as possible
-
50
After removing from the magnetic stand, add 20 µl of water to the magnetic beads. This will be the template for subsequent PCR reactions.
■ PAUSE POINT The purified beads with cDNA can be kept frozen for up to a month at −20°C.
PCR amplification for determination of cycle number ● TIMING 1 h
-
51To decide the optimal number of PCR cycles (e.g. 8, 10, 12 cycles), set up the following PCR premix reaction solution and mix by pipetting on ice.
Component Volume Final concentration 5 X High-Fidelity buffer 10 µl 1 X dNTPs (2.5 mM each) 4 µl 0.2 mM 100 µM PCR Forward
primer0.5 µl 1 µM 100 µM PCR Reverse
primer0.5 µl 1 µM Phusion polymerase (2 U µl−1) 0.5 µl 1 U cDNA with magnetic beads from Step 50 0.5 – 2 µl - Water variable - Total volume 50 µl -
▲CRITICAL STEP The number of PCR tested during the “check cycle” depends also on the amount of cDNAs. For example, barcoded, pooled cDNA may require fewer cycles than non pooled cDNA. Thus, we recommend testing different number of PCR cycles, such as 10, 12 and 14 for non-pooled cDNAs and 8, 10 and 12 for pooled cDNA.
-
52Run the PCR under the following conditions.
Cycle number Denature Anneal/Extend Hold 1 98°C, 30 sec - - 2-8/10/12 98°C, 10 sec 60°C, 10 sec - 9/11/13 - - 4°C
Quality check of PCR products ● TIMING 1 h
-
53
Add 1 µl of PCR product to the Agilent Bioanalyzer DNA1000 tip by pipetting. Measure concentration and confirm the product size. As an example, refer to Fig. 5a–d.
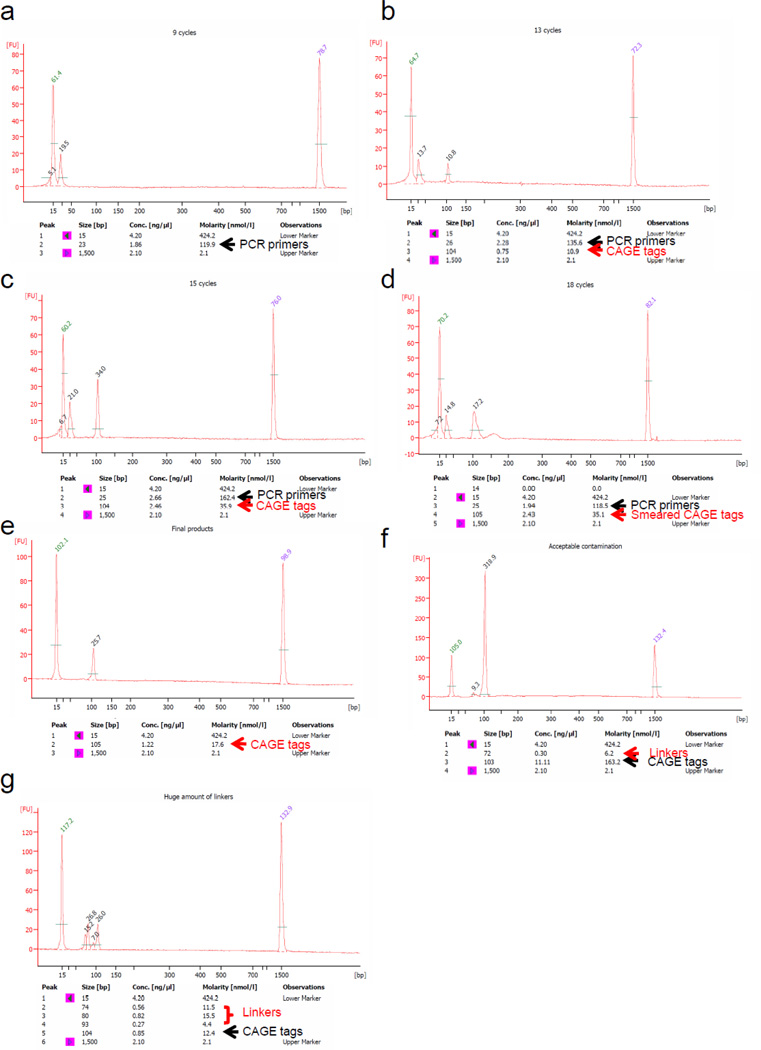
Figure 5. Measurement of PCR products.
Example of PCR cycle optimization by Agilent Bioanalyzer DNA 1000 kit. The amount of applied PCR product is 1 µl. (a) 9 cycles (b) 13 cycles, (c) 15 cycles, (d) 18 cycles. Peak values indicate the height of Fluorescence Units (FU). With only 9 cycles, there is only primer peak (25 bp) and the CAGE peak is not visible. With 13 cycles, there are two peaks, the primer peak and the CAGE peak. The measured size may slightly differ from the actual 96 bp within the inherent instrument error range (103 – 105 bp). CAGE tag peaks with FU values between 5 and 10 (molarity: ~10 nmol l−1) are suitable for bulk PCR. In case of 15 cycles, the FU exceeds 20 (molarity: ~30 nmol l−1) and with 18 cycles the reactions shows a broad peak, due to over cycling (compared to a and b). (e) Final product molarity of the single peak was estimated to be 17.6 nmol l−1 at 13 cycles. PCR primers are subsequently removed during Steps 55 and 57. After Step 58, the single peak products are ready for sequencing. (f) Example of small linker dimer contamination (70 – 80 bp), which does not affect sequencing, and large linker contamination (g), which prevents the usage of the library.
▲CRITICAL STEP The desired product is 96 bp long; a large 30 bp peak constituted by PCR primers is also present. Appearance of other double strand cDNA contaminants around 70 bp, constituted by amplified linkers, should be minimal or absent.
?TROUBLESHOOTING
■ PAUSE POINT The PCR products can be stored at 4°C for 1 week or −20°C for at least 1 month.
Bulk PCR amplification ● TIMING 30 min
-
54
After determining the optimal PCR cycle number, perform bulk PCR (6 PCR tubes) to amplify the remaining part of the library (12 µl of remaining cDNAs from Step 50) as described in Steps 51 and 52, using the optimal cycle number determined in Step 53 (use just enough cycles to detect the desired band but minimize the number of cycles). !CAUTION Do not amplify in a single tube with a large amount of beads, as they may inhibit the PCR reaction (in general, do not amplify more than 2 µl of beads for a 50 µl PCR reaction).
Purification of primers with ExonucleaseI ● TIMING 1.5 h
-
55
Pool 3 of the PCR reactions from Step 54, each one containing 50 µl, into one 1.5 ml siliconized tube. Repeat this for the remaining 3 PCR reactions, to give a total of 2 tubes for each CAGE library.
-
56
Add 1 µl of ExonucleaseI (20 U µl−1) to each of the 150 µl of PCR reaction solutions, mix by pipetting on ice and incubate 37°C for 30 min.
Purification with QIAquick PCR purification kit ● TIMING 30 min
-
57
Purify the 151 µl of Exonuclease I treated CAGE tags using QIAquick PCR purification kit, following the manufacturer’s instructions. At the end, elute in 10 µl EB buffer.
Final product concentration check ● TIMING 1 h
-
58
Use 1 µl of eluant to check the quantity with the Agilent Bioanalyzer DNA 1000 kit. An example result is shown in Fig. 5e.
CRITICAL STEP Extra bands around 80 bp are comprised of linker dimers. If the concentration is negligible compare to desired CAGE reads, this is not going to affect the sequencing outcome (Fig. 5f). However, if the concentration is high, the library could be loaded onto a 8% polyacrylamide gel , and subjected to electrophoresis at 120 V for 1 h and the 96 nt band could be purified by QIAquick Gel Extraction kit (Fig. 5g). Alternatively, one should prepare the library again after troubleshooting the steps to avoid linker dimer formation.
Illumina Cluster generation ● TIMING 0.5 days
-
59To prepare the CAGE tags from Step 58 for Illumina sequencing using the Illumina cluster generation standard protocol (http://www.illumina.com/products/standard_cluster_generation_kit_v4.ilmn), mix the following components and incubate for 5min at room temperature to denature the double stranded CAGE library:
Component Volume Final concentration 10 nM CAGE library 2 µl 1.0 nM 10 mM Tris-HCl (pH 8.5)
(Elution buffer)17 µl 8.5 mM HP3 (2N NaOH) in the cluster
generation kit1 µl 0.1 N Total volume 20 µl -
CRITICAL STEP: This requires a DNA concentration of 10 nM (0.67 ng µl−1, 96 bp CAGE tags). The final DNA concentration in the sequencing reaction should be set accordingly to the sequencer specifications. Currently the Illumina sequencer uses 5 pM as final concentration, although this may be subjected to changes.
-
60Mix the following components, load into the Illumina chip following the manufacturer’s instructions and perform cluster amplification with the Illumina cluster generation standard protocol:
Component Volume Final concentration 1.0 nM Denatured library 5 µl 5 pM Pre-chilled HT1
(Hybrydization buffer in the
cluster generation kit)995 µl - Total volume 1000 µl
CRITICAL STEP The sequencing primer (Table 1) is differs from the standard Illumina provided sequencing primer. If performing the cluster generation in parallel with other template DNAs, which require the standard Illumina sequencing primer, the cluster generation program should be set as “multi hybridization program” among the available instrument options, follow the Illumina cluster generation standard protocol (see STEP 59)
Illumina Sequencing ● TIMING 2.5 days
-
61
Place the flowcell with the bridge PCR products from Step 60 into the sequencing instrument Start a 36 cycle single read run operation program in case of a GAII-X sequencer (http://www.illumina.com/systems/genome_analyzer_iix.ilmn) or a 50 cycles single read run operation program in case of a Hi-Seq 2000 sequencer (http://www.illumina.com/systems/hiseq_2000.ilmn).
Bioinformatic analysis of CAGE tags ● TIMING Vary extensively depending on the computers and servers settings and size of the datasets to be analyzed. Even fast turnaround of the data may take more than a week.
-
62
Extraction of CAGE reads from raw reads. Raw reads obtained after base-calling contain linkers and adaptors in addition to CAGE tags. Simple string matching scripts written in Perl can be used to extract the CAGE tag by using a set of regular expressions for the 5’ and 3’ ends, respectively. Alternatively, use the program fastx_clipper (http://hannonlab.cshl.edu/fastx_toolkit/index.html).
-
63
Removal of artifactual reads. Apply the program TagDust34 together with a list of linkers/adapters used in the library preparation to remove linker dimers.
-
64
Mapping of CAGE reads. Use the mapping programs BWA35 or Delve (T. L., unpublished data) to map the CAGE tags to the genome. Both programs calculate the mapping quality Q that can be used to enrich for correct mappings. The resulting mapping files are in SAM format36 and are converted to BAM files using samtools (http://samtools.sourceforge.net/). Tags can be associated to genome features, such as 5’ UTR, exons, introns, intergenic regions, to assess the quality of the library (as example, see Table 3).
-
65
Generating a CAGE tag start site (CTSS) file. The CTSS file lists the genomic positions where CAGE tags start alongside the number of reads found. Use the script “make_ctss.sh” (Supplementary data 1) to generate this file.
-
66
Generating CAGE tag clusters. Use the program paraclu (http://www.cbrc.jp/paraclu/)14 to aggregate CTSS positions into CAGE clusters. CRITICAL STEP When there are multiple replicas of a given same biological sample/conditions, it is recommended to first merge the CTSS files for all the biological/technical replicas and generate clusters based on the merged data, as explained in details on the paraclu specifications (http://www.cbrc.jp/paraclu).
-
67
Assign expression to clusters. Use BEDtools37 to intersect the CTSS files with the boundaries defined by paraclu to obtain a raw tag count per cluster. To normalize the data, divide the raw tag counts by the total number of mapped tags in the library and multiply by 1 million (tag per million). The quantified clusters can be uploaded to the UCSC browser38 for visual inspection.
-
68
Differential Expression. To compare multiple CAGE samples we recommend defining clusters based on merging all libraries using paraclu as outlined at Step 66. After quantification, each cluster effectively has a vector of expression values from the different samples. The program edgeR39 can be used to detect differentially expressed clusters.
Table 3.
Summary of results from libraries produced for the ENCODE Project
● TIMING
Day 1
Steps 1–4, Reverse transcription: 1.5 h
Steps 5–10, cDNA purification with the RNAClean XP kit: 1.5 h
Steps 11 and 12, Diol-Oxidation with NaIO4: 50 min
Steps 13 and 14, cDNA purification with the RNAClean XP kit: 1.5 h
Step 15, Biotinylation of the RNA: 16 h
Day 2
Step 16, RNaseI treatment: 45 min
Steps 17 and 18, cDNA purification with the RNAClean XP kit: 1.5 h
Steps 19–22, Cap-trapping: 2.5 h
Steps 23–25, Release 5’-Completed cDNAs from magnetic beads: 15 min
Steps 26 and 27, cDNA purification with the AMPure XP kit: 1.5 h
Steps 28 and 29, Quality check: 1.5 h
Steps 30–32, Barcoded 5’ linker Ligation to the single strand (ss) DNA: 16 h
Day 3
Steps 33 and 34, cDNA purification with the AMPure XP kit: 1.5 h
Steps 35 and 36, Second strand cDNA synthesis: 30 min
Step 37, Antarctic Phosphatase activation: 1.5 h
Steps 38 and 39, cDNA purification with the AMPure XP kit: 1.5 h
PAUSE POINT
Steps 40–42, EcoP15I digestion: 3.5 h
Steps 43 and 44, Addition of a 3’ linker to the cleaved tags: 16 h
Day 4
Steps 45–50, Removal of excess 3’ linkers: 2.5 h
PAUSE POINT
Steps 51 and 52, PCR amplification for a cycle determination: 1 h
Step 53, Quality check of PCR products: 1 h
Step54, Bulk PCR amplification: 30 min
Steps 55 and 56, Purification of primers with ExonucleaseI: 1.5 h
Step 57, PCR purification: 30 min
Step 58, Final product concentration check: 1 h
Steps 59 and 60, Illumina Cluster generation: 0.5 d
Step 61, Illumina Sequencing: 2.5 d
Steps 62–68 vary extensively depending on the computers and servers settings and size of the datasets to be analyzed. Even fast turnaround of the data takes usually no less than a week.
? TROUBLE SHOOTING
Troubleshooting advice can be found in Table 2.
Table 2.
Troubleshooting.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 28 | cDNA concentration is lower than ~2.0 ng, or undetectable | Protein or polysaccharide contamination in the starting material or RNA degradation | Check RNA absorbance of 230, 260 and 280. If 260/280 is lower than 1.8, there is possibility of contaminating protein. If 260/230 is lower than 1.8, there is possibility of contaminating polysaccharides. Purify RNA with QIAGEN miRNeasy kit. Check RNA quality before the procedure. Too low a cDNA concentration at this step requires too many PCR cycles (Step 52). |
| Apparent cDNA concentration is higher than 100 ng | cDNA was not purified properly; cDNA was not properly cap-selected | Check Agencourt beads and MPG beads purification protocol. Check if RNAse reaction has worked. | |
| cDNA length is much shorter than 500 bp. In the qRT-PCR analysis, the delta Ct (ACTB-Ct minus ribosomal-Ct) value is often higher than 1. | RNA may be degraded. | Prevent contamination of reagents and instrumentation by RNases. Prepare a laboratory environment suitable to work with RNAs; change and test reagents for being RNAse free. Check if the original RNA are devoid of RNAses. | |
| 53 | The only observed peak is at 25 bp | Insufficient CAGE tags amplification. | Use a larger number of PCR cycles. For example, see result in Fig. 5a. One can amplify up to 25 cycles. However, with more than 25 cycles, we observe a sharp increase of sequencing redundancy, showing molecular bottlenecks. |
| There is a strong peak around 70–80 bp. | Contaminating 5' linker and 3' linker dimers | If the contaminant band molarity is considerably lower than 96 bp, the sequencing yield will not be dramatically affected (Fig. 5f). High molarity contamination will affect sequencing (Fig. 5g), causing sequencing reads to be heavily contaminated by linker sequences. An option to rescue the library is to cut the 96 band from gel, while the linkers should be tested to prevent long-term problems. | |
| Phospate modification remains at 3’ end of excess 5' linker. | Check Antarctic Phosphatase enzyme activity at Step 37. |
Anticipated results
Sequencing coverage
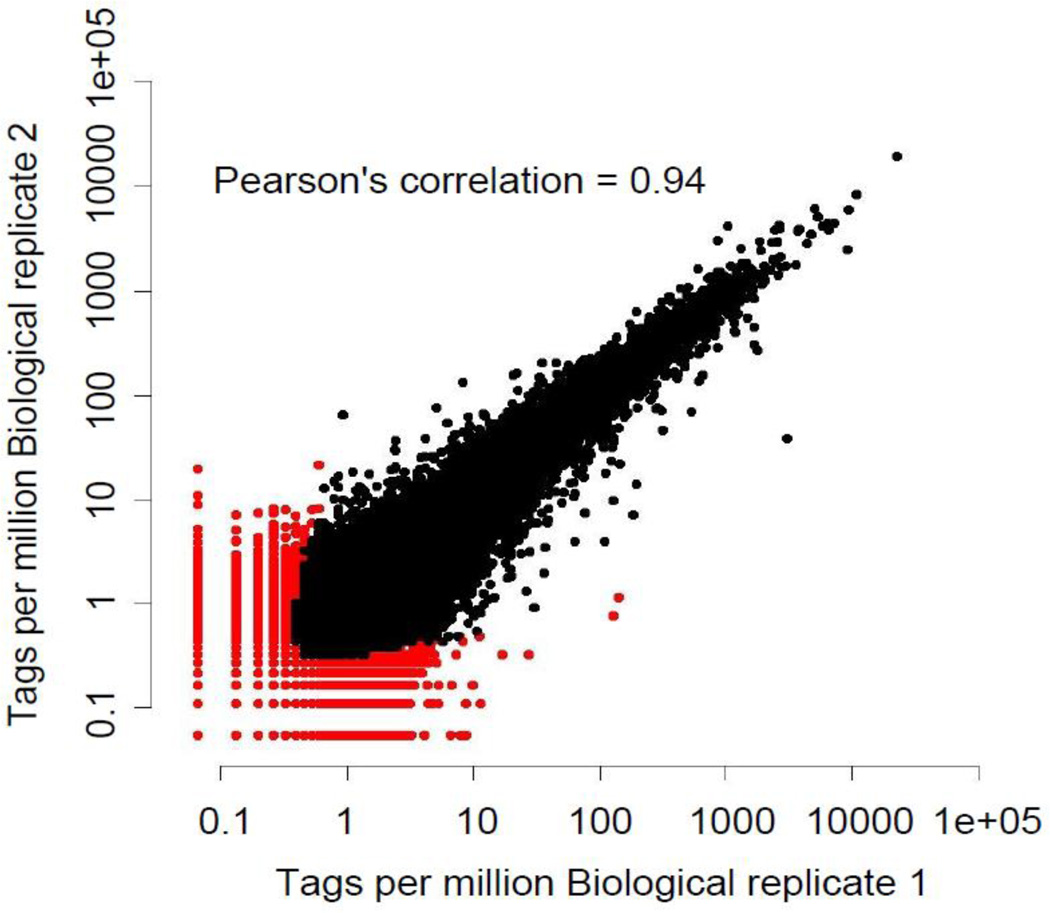
We generate more than 20 million tags per lane with the Illumina GA2X and more than 50 million tags per lane with the Illumina Hi-seq 2000 sequencers. There is not yet consensus on how deep a CAGE sequencing experiment should be but in general, we use 5–10 M reads per sample when performing multiple measurements, like time courses, where the cost of sequencing is an issue. Sequencing multiple barcoded CAGE libraries per one lane makes the process competitive with microarrays. Conversely, 30–50 M tags are desirable for deeper explorations of the transcriptome and identification of rarely expressed transcripts, like in the ENCODE project23 (see also Table 3 for a summary of results from libraries produced for the ENCODE Project and internal control). A full set of CAGE libraries produced for the ENCODE is available at the web site: http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeRikenCage/. Sequencing 50M reads gives a deep coverage of the TSS-ome, including rare RNAs that are present in less than a copy per cell in the analyzed sample. We confirmed the accuracy of the CAGE method by analyzing correlation efficiency; biological replicates, using different RNA replicates, show correlation coefficients as high as 0.94 (Fig. 6).
Figure 6. Scatter plot of cluster expression between 2 biological replicas (K562 whole cell).
Points in red represent measurements above a 0.1 IDR (Irreproducible Discovery Rate)21 threshold. The×axis and y axis indicate log2 of the sequence tags per million.
Bioinformatics analysis of CAGE tags is a rapidly growing area. Major challenges and solutions have been reported elsewhere in detail40. As we have also above outlined (Steps 62–68), there are several necessary key steps, which include tag extraction and separation from the linker, followed by mapping to the genome: at this point one can assess the quality of the library. The RIKEN OSC automated pipeline extracts reads from remaining linker and adapter sequences. Furthermore, the “Tagdust” tool34 identifies reads which can be explained by combinations of sequences used during the library preparation. This estimate of the fraction of non-RNA derived artifacts should be below 10% in a good library. A secondary indicator of library quality is the read redundancy, or how many times the same read is observed in the library on average. A high redundancy might point to a molecular bottleneck introduced by PCR. A typical library will have a redundancy of less than 3 (more than 300,000 different tags identified per million sequences). Although a too low redundancy may imply RNA degradation and failure to aggregate tags to the cap, in reality mammalian11 and other metazoan23 promoters show a distributed shape with multiple starting sites within core promoters that justify a relatively low and spread distribution of tags-identified starting sites. Redundancy above 5 for a mammalian RNA (at a fixed depth of 1M tags) is suggestive of molecular bottlenecks, likely introduced by PCR. It is likely that redundancy/million tags will be higher for other eukaryotes in which the genome is smaller and the complexity of the transcriptome is lower. Mapping the tags to the genome with “Delve” (T. L., unpublished data) provides yet another estimate of the quality of the library. A high mapping rate to known promoter regions is indicative of high library quality. Conversely, a high mapping rate to ribosomal RNAs indicates a high level of contamination of non-capped RNAs. A typical CAGE library produced from total tissue RNA shows minimal ribosomal RNA contamination, often less than 1%, which is remarkable giving the fact that libraries are random primed and rRNA constitutes the vast majority of RNAs. As a quality control measure, we measure the hits of CAGE sequences to various genome features. For instance, in typical libraries like in the last row of Table 3, we observe an average of 61% hit to 5’ UTR and core promoters, 7.2% to exons, 16.2% to introns and 15.6% to intergenic regions, using poly-A minus RNA from HUVEC and HelaS3 cells.
Following the QC, bioinformatics analysis consists of clustering the tags into groups and assigning them to known genes or new clusters. Grouping by clustering has been at first arbitrary, grouping to single units (TSS candidates) tags which 5’ends were mapping within 20 nt on the genome11,24,10. This approach has the disadvantage that at very deep sequencing, low frequency events (including background of the technology) produce increasingly large clusters, devoid of biological significance. Different definitions of clusters have involved restriction of the clusters to windows no larger than 300 nt wide15, or have taken into account the relative density of the peaks identified by CAGE tags14. When applying the latter approach to biological replicates, a high Pearson’s correlation coefficient of 0.94 can be observed (Fig. 6). The recently developed IDR method41 can be used to distinguish highly reproducible from less reproducible peaks which can arise due to biological variation between the samples.
New approaches to clustering are being developed, which will take into account the distribution of the peaks across different tissues and cells in order to decompose multiple transcription contributions of overlapping functional elements of promoters that promote transcription on different genome bases.
After clustering, bioinformatics tools are available to connect the identified promoters to known gene models37, to ultimately provide a gene name to the promoters and the networks that control gene expression15. A full set of freely available tools and resources are available on line (http://genome.gsc.riken.jp/osc/english/dataresource/). In particular, the reader can download the analytical tools like Nexalign (for the alignment of CAGE sequences to the genome), TagDust (to remove linker dimer sequences), MuMRescueLite (a tool to rescue tags that map in multiple location of the genome) among the most commonly used tools, and use EdgeExpress (a CAGE visualization tool).
Supplementary Material
Acknowledgments
This work was founded by a Research Grant for RIKEN Omics Science Center from the Japanese Ministry of Education, Culture, Sports, Science and Technology to Yoshihide Hayashizaki. This project was also supported by the U.S. National Human Genome Research Institute grants U54 HG004557. We thank Sachi Kato for experimental support, Jessica Severin for the genome browser, RIKEN Genome Network Analysis Service (GENAS) for sequencing and basic bioinformatics analysis and all the colleagues at the OSC for the precious feedbacks during the development of the methodology.
Footnotes
Author contributions
HT performed most experiments. MM performed the background reduction experiment. TL performed computations analysis. HT and PC wrote the manuscript. PC designed the project.
Competing interests
PC is an inventor of various patents owned by RIKEN and Dnaform Inc. on the Cap-trapper technology, Full-length cDNA cloning technologies and the CAGE technology.
References
- 1.Schena M, Shalon D, Davis RW, Brown PO. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 2.Cheng J, et al. Transcriptional maps of 10 human chromosomes at 5-nucleotide resolution. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 3.Forrest AR, Carninci P. Whole genome transcriptome analysis. RNA Biol. 2009;6:107–112. doi: 10.4161/rna.6.2.7931. [DOI] [PubMed] [Google Scholar]
- 4.Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995;270:484–487. doi: 10.1126/science.270.5235.484. [DOI] [PubMed] [Google Scholar]
- 5.Kodzius R, et al. CAGE: cap analysis of gene expression. Nat Methods. 2006;3:211–222. doi: 10.1038/nmeth0306-211. [DOI] [PubMed] [Google Scholar]
- 6.Kanamori-Katayama M, et al. Unamplified cap analysis of gene expression on a singlemolecule sequencer. Genome Res. 2011;21:1150–1159. doi: 10.1101/gr.115469.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kawaji H, et al. Dynamic usage of transcription start sites within core promoters. Genome Biol. 2006;7:R118. doi: 10.1186/gb-2006-7-12-r118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponjavic J, et al. Transcriptional and structural impact of TATA-initiation site spacing in mammalian core promoters. Genome Biol. 2006;7:R78. doi: 10.1186/gb-2006-7-8-r78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frith MC, et al. Evolutionary turnover of mammalian transcription start sites. Genome Res. 2006;16 doi: 10.1101/gr.5031006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoskins RA, et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 2011;21:182–192. doi: 10.1101/gr.112466.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carninci P, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nature Genetics. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 12.Gustincich S, et al. The complexity of the mammalian transcriptome. J Physiol. 2006;575:321–332. doi: 10.1113/jphysiol.2006.115568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vitezic M, et al. Building promoter aware transcriptional regulatory networks using siRNA perturbation and deepCAGE. Nucleic Acids Res. 2010;38:8141–8148. doi: 10.1093/nar/gkq729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frith MC, et al. A code for transcription initiation in mammalian genomes. Genome Res. 2008;18:1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suzuki H, et al. The transcriptional network that controls growth arrest and differentiation in a human myeloid leukemia cell line. Nature Genetics. 2009;41:553–562. doi: 10.1038/ng.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faulkner GJ, et al. The regulated retrotransposon transcriptome of mammalian cells. Nature Genetics. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- 17.Hestand MS, et al. Tissue-specific transcript annotation and expression profiling with complementary next-generation sequencing technologies. Nucleic Acids Res. 2010;38:e165. doi: 10.1093/nar/gkq602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wei CL, et al. 5' Long serial analysis of gene expression (LongSAGE) and 3' LongSAGE for transcriptome characterization and genome annotation. Proc Natl Acad Sci U S A. 2004;101:11701–11706. doi: 10.1073/pnas.0403514101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valen E, et al. Genome-wide detection and analysis of hippocampus core promoters using DeepCAGE. Genome Res. 2009;19:255–265. doi: 10.1101/gr.084541.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiraki T, et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc Natl Acad Sci U S A. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Myers RM, et al. A user's guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takahashi H, Kato S, Murata M, Carninci P. CAGE (Cap Analysis of Gene Expression): A Protocol for the Detection of Promoter and Transcriptional Networks. Methods Mol Biol. 2012;786:181–200. doi: 10.1007/978-1-61779-292-2_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoskins RA, et al. Genome-wide analysis of promoter architecture in Drosophila melanogaster. Genome Res. 2011;21:182–192. doi: 10.1101/gr.112466.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carninci P, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 25.Carninci P, et al. Thermostabilization and thermoactivation of thermolabile enzymes by trehalose and its application for the synthesis of full length cDNA. Proc Natl Acad Sci U S A. 1998;95:520–524. doi: 10.1073/pnas.95.2.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carninci P, Shiraki T, Mizuno Y, Muramatsu M, Hayashizaki Y. Extra-long firststrand cDNA synthesis. Biotechniques. 2002;32:984–985. doi: 10.2144/02325bm01. [DOI] [PubMed] [Google Scholar]
- 27.Carninci P, et al. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics. 1996;37:327–336. doi: 10.1006/geno.1996.0567. [DOI] [PubMed] [Google Scholar]
- 28.Shibata K, et al. RIKEN integrated sequence analysis (RISA) system--384-format sequencing pipeline with 384 multicapillary sequencer. Genome Res. 2000;10:1757–1771. doi: 10.1101/gr.152600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Plessy C, et al. Linking promoters to functional transcripts in small samples with nanoCAGE and CAGEscan. Nat Methods. 2010;7:528–534. doi: 10.1038/nmeth.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maeda N, et al. Development of a DNA barcode tagging method for monitoring dynamic changes in gene expression by using an ultra high-throughput sequencer. Biotechniques. 2008;45:95–97. doi: 10.2144/000112814. [DOI] [PubMed] [Google Scholar]
- 31.Janscak P, Sandmeier U, Szczelkun MD, Bickle TA. Subunit assembly and mode of DNA cleavage of the type III restriction endonucleases EcoP1I and EcoP15I. J Mol Biol. 2001;306:417–431. doi: 10.1006/jmbi.2000.4411. [DOI] [PubMed] [Google Scholar]
- 32.Raghavendra NK, Rao DN. Exogenous AdoMet and its analogue sinefungin differentially influence DNA cleavage by R.EcoP15I--usefulness in SAGE. Biochem Biophys Res Commun. 2005;334:803–811. doi: 10.1016/j.bbrc.2005.06.171. [DOI] [PubMed] [Google Scholar]
- 33.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lassmann T, Hayashizaki Y, Daub CO. TagDust--a program to eliminate artifacts from next generation sequencing data. Bioinformatics. 2009;25:2839–2840. doi: 10.1093/bioinformatics/btp527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fujita PA, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carninci P. Cap-analysis gene expression (CAGE) : the science of decoding gene transcription. Pan Stanford; 2010. [Google Scholar]
- 41.Li Q, Brown JB, Huang H, Bickel PJ. Measuring reproducibility of highthroughput experiments. Annals of Applied Statistics. (In press). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.