Abstract
Background
Sulphadoxine-pyrimethamine (SP), an antifolate, was replaced by artemether-lumefantrine as the first-line malaria drug treatment in Kenya in 2004 due to the wide spread of resistance. However, SP still remains the recommended drug for intermittent preventive treatment in pregnant women and infants (IPTP/I) owing to its safety profile. This study assessed the prevalence of mutations in dihydrofolate reductase (Pfdhfr) and dihydropteroate synthase (Pfdhps) genes associated with SP resistance in samples collected in Kenya between 2008 and 2012.
Methods
Field isolates collected from Kisumu, Kisii, Kericho and Malindi district hospitals were assessed for genetic polymorphism at various loci within Pfdhfr and Pfdhps genes by sequencing.
Results
Among the Pfdhfr mutations, codons N51I, C59R, S108N showed highest prevalence in all the field sites at 95.5%, 84.1% and 98.6% respectively. Pfdhfr S108N prevalence was highest in Kisii at 100%. A temporal trend analysis showed steady prevalence of mutations over time except for codon Pfdhps 581 which showed an increase in mixed genotypes. Triple Pfdhfr N51I/C59R/S108N and double Pfdhps A437G/ K540E had high prevalence rates of 86.6% and 87.9% respectively. The Pfdhfr/Pfdhps quintuple, N51I/C59R/S108N/A437G/K540E mutant which has been shown to be the most clinically relevant marker for SP resistance was observed in 75.7% of the samples.
Conclusion
SP resistance is still persistently high in western Kenya, which is likely due to fixation of key mutations in the Pfdhfr and Pfdhps genes as well as drug pressure from other antifolate drugs being used for the treatment of malaria and other infections. In addition, there is emergence and increasing prevalence of new mutations in Kenyan parasite population. Since SP is used for IPTP/I, molecular surveillance and in vitro susceptibility assays must be sustained to provide information on the emergence and spread of SP resistance.
Keywords: Malaria, Plasmodium falciparum dihydrofolate reductase, Plasmodium falciparum dihydropteroate synthase
Background
Sulphadoxine-pyrimethamine (SP) has been widely used as first-line treatment for uncomplicated falciparum malaria in many parts of the world including sub-Saharan Africa. It is used alone or in combination with other anti-malarials which has been effective until recently because of high levels of resistance [1,2]. SP has been replaced with other anti-malarials as the first-line treatment in most of sub-Saharan Africa including Kenya [3]. However, SP is still used as intermittent preventive treatment of malaria in pregnancy (IPTp) in sub-Saharan Africa [4]. It was recently recommended for use as intermittent preventive treatment of malaria in infants (IPTi) by the World Health Organization (WHO) in areas with low SP resistance [5]. There has been continued increase in SP resistance albeit being replaced as the first-line treatment [6], which is a major public health concern especially in sub-Saharan Africa [7].
SP resistance is conferred by mutations in Plasmodium falciparum dihydrofolate reductase (Pfdhfr) and P. falciparum dihydropteroate synthase (Pfdhps) genes [8,9], whose enzymes are target for pyrimethamine and sulphadoxine respectively. Pfdhfr S108N mutation has been linked to pyrimethamine resistance. Additional point mutations A16V, N51I and C59R lead to increased resistance [10,11]. High grade pyrimethamine resistance is linked to the occurrence of the I164L mutation [12] which has been observed together with the Pfdhfr gene triple N51I/C59R/S108N mutant in Southeast Asia and the Americas [12,13]. Pfdhfr I164L mutation, which is shown to cause rapid spread of antifolate resistance, has been observed in the western parts of Kenya [7,14]. Pfdhps A437G mutation is mainly associated with sulphadoxine resistance with increased resistance conferred in the presence of additional point mutations S436A/F/H, A581G, K540E and A613S/T[9,15]. Differing extents of anti-malarial drug resistance to SP are subject to the varying numbers and combinations of mutations present in the Pfdhfr and Pfdhps genes [11,16]. Primarily, the Pfdhfr/Pfdhps N51I, C59R, S108N/A437G, K540E quintuple mutation has strongly been associated with clinical SP treatment failure [11].
A recent meta-analysis of the frequencies of Pfdhfr and Pfdhps mutant genotype in African P. falciparum parasite populations revealed high prevalence of mutant genotypes along the Kenya-Tanzania border and Malawi [6]. To assess the prevalence of mutations in Pfdhfr and Pfdhps genes and the impact of introducing SP as the first-line treatment in Kenya, two studies were conducted. A baseline study was conducted in 1999-2000 [17] and the follow-up study in 2003-2005 [18]. The baseline study coincided with the implementation of SP as the first-line treatment whereas the follow-up study coincided with transition to artemether-lumefantrine (AL) as the first-line treatment. The prevalence of isolates containing the quintuple mutant genotype (Pfdhfr N51I/C59R/S108N/ Pfdhps A437G/K540E) rose from 21% in the baseline study to 53% in the follow-up study (P < 0.001) [17,18]. The Pfdhfr I164L mutation was not present in either of the study. Interestingly, a study that analyzed prevalence of the I164L mutation in sample isolates collected in the same region in 2002-2008 and 2008-2009 reported a prevalence of 0.6% and 0.8% respectively [19]. There were no mutations at Pfdhps codons 581 and 613 at the baseline study but 85% and 61% of the sample isolates in the follow-up study carried mutations at these two codons, respectively [19].
Since SP remains widely used as IPTp and is a possible ACT partner drug as well as IPTi candidate, there is heightened recognition underlying the need to track the prevalence of mutations associated with SP resistance. This study presents the prevalence and trend of mutations in Pfdhfr and Pfdhps genes in sample isolates collected from 4 different sites in Kenya between 2008 and 2012.
Methods
Study sites and sampling collection
This study was conducted after approval by the Kenya Medical Research Institute (KEMRI) and Walter Reed Army Institute of Research (WRAIR) institutional review boards (protocol numbers: KEMRI 1330, WRAIR 1384; Epidemiology of malaria drug sensitivity pattern). The samples used in this study were collected at district hospitals from four regions in Kenya; viz Kisumu District Hospital (KDH), Kisii District Hospital (KSI), Kericho District Hospital (KCH) and Malindi District Hospital (MDH) between the years 2008 and 2012. Kisumu is located in the western region of the country at the shores of Lake Victoria. This is a lowland region with holoendemic malaria. Kisii and Kericho are both highland areas also in the western Kenya regions which experiences hypoendemic malaria. Malindi is at the Kenyan coastal lowland region, but has different malaria episodes from Kisumu, which is also at lowland in the western Kenya region [20,21].
Consenting patients aged six months and above, testing positive for uncomplicated P. falciparum malaria by rapid diagnostic test (RDT; Parascreen® (Pan/Pf), Zephyr Biomedicals, Verna Goa, India) were allowed to participate in the study. Briefly, 2-3 ml of whole blood was collected from the participants. FTA filter paper (Whatman Inc., Bound Brook, New Jersey, USA) was used to collect 3 blood spots of about 100 μl each for DNA extraction.
Genotypic analysis for Pfdhfr and Pfdhps
The DNA was extracted from the FTA filter papers using the QIAamp DNA mini kit; Qiagen protocol (Qiagen, Valencia, CA). Screening for mutations associated with antifolate resistance at codons 16, 50, 51, 59, 108, and 164 of the Pfdhfr gene and codons 436, 437, 540, 581, and 613 of the Pfdhps gene was performed by nested PCR as previously described [22,23]. After successfully amplifying the target regions, the isolates were purified using Exosap-it® (Affymetrix, Santa Clara, CA) as per the manufacturer’s protocol. Sequencing of the target regions was done on the 3500 xL ABI Genetic analyzer using version 3.1 of the big dye terminator method (Applied Biosystems, Foster City, CA). Contig assembly of the generated sequences was performed using DNA Baser version 3× and the sequences aligned and analysed using BioEdit version 7.1.3.0. All sequences were compared against the Pfdhfr (Accession Number; XM_001351443) and Pfdhps (Accession Number; XM_001349382) 3D7 reference sequence published at the NCBI database.
Statistical analysis
Categorical data consisting of genotype polymorphisms within Pfdhfr, Pfdhps genes was analyzed as proportions showing rates of frequency. The differences in frequencies of point mutations in the Pfdhfr, Pfdhps genes and the combined Pfdhfr/Pfdhps genes within and between regions were determined by the Chi-square test. For evaluating trends, the annual frequencies of polymorphisms of individual codons, during the study period were compared. Further, comparison of genotype frequencies of 2003 to 2005 and 2008 to 2012 study periods was performed by Chi-square test. Yates’ correction was applied for the chi-square value, resulting in corrected P values. All statistical analyses were performed at the 5% significance level and the corresponding 95% Confidence Interval (CI). Prism 4.0 software (Graph pad Software, San Diego, California, USA) was used for analyses.
Results
Prevalence of SNPs in Pfdhfr and Pfdhps genes
A total of 869 archival field isolates collected from KDH, KSI, KCH and MDH between 2008 and 2012 were included in the study of which 822 were successfully sequenced. Eleven SNPs were genotyped, six loci in the Pfdhfr gene and five loci in Pfdhps gene as shown in the methods section. There were no mutations in Pfdhfr gene at codons 16 and 50 as well as Pfdhps gene at codon 613. These codons are excluded from any further analysis. Using the criteria that was recently described by Iriemenam et al. [19], SNPs were designated as pure (which contains only either wild type or mutant strains) or as mixed (which contained both wild type and mutant alleles based on presence of two major peaks of the chromatograms). Figure 1 shows the overall prevalence of genotypes at each codon. The prevalence of mutation at Pfdhfr codons 51, 59 and 108 was 95.5%, 84.1% and 98.6% respectively. There were five (0.8%) sample isolates that carried the pure mutant and 2 (0.3%) samples that had the mixed SNP at Pfdhfr codon 164. The prevalence of mutation at Pfdhps codons 436, 437, 540 and 581 was 0.2%, 95.7%, 86.8%, and 0.8%.
Figure 1.
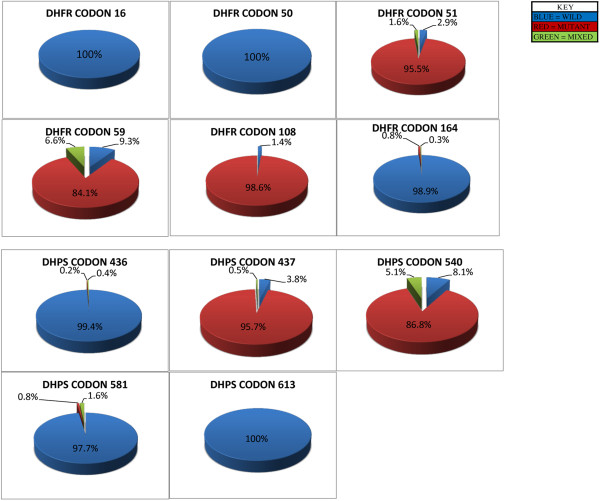
Prevalence of single nucleotide polymorphism in Pfdhfr and Pfdhps. The overall prevalence of genotypes at each codon on the Pfdhfr and Pfdhps genes were determined over the 5 year period, calculated as a percentage of the total number of samples successfully analyzed over the study period per codon.
Prevalence of mutant alleles in Pfdhfr and Pfdhps genes per study site
Table 1 shows the prevalence of Pfdhfr and Pfdhps SNPs per study site for the entire period which isolates were collected. SNPs are classified as wild type, mutant or mixed. The prevalence of mutant allele at Pfdhfr codon 108 was highest, with KSI and KDH having 100% and 99.6% of the sample isolates carrying the mutant allele respectively. MDH had the lowest prevalence of mutations in Pfdhfr gene at codons 51, 59 and 108 compared to the other sites (P = 0.0052, 0.0002 and 0.0066) respectively. Mutation at Pfdhfr codon 164 was present only in samples collected in KDH and KSI. There were five sample isolates which carried the pure mutant (three from KDH and two from KSI), and two sample isolates, one from each site that carried the mixed SNP genotype. In Pfdhps gene, the prevalence of mutant allele at Pfdhps codon 437 was highest, with KDH and KSI having 98.1% and 97.9% of the sample isolates carrying the mutant allele respectively. Mutant and mixed SNPs at Pfdhps codon 581 was present in sample isolates collected in KDH, KSI and KCH. MDH site had the lowest prevalence of mutant alleles at codons 437 and 540 compared to the other sites.
Table 1.
Prevalence of single nucleotide polymorphism in Pfdhfr and Pfdhps per site
| |
|
DHFR prevalence per site |
DHPS prevalence per site |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CODON | 16 | 50 | 51 | 59 | 108 | 164 | S436A/F/H | 437 | 540 | 581 | 613 | |
| |
|
% (n) |
% (n) |
% (n) |
% (n) |
% (n) |
% (n) |
% (n) |
% (n) |
% (n) |
% (n) |
% (n) |
| KDH |
WILD |
100 (266) |
100 (289) |
1.4 (4) |
3.5 (10) |
0.3 (1) |
98.7 (302) |
100 (256) |
1.5 (4) |
7.5 (21) |
97.2 (273) |
100 (284) |
| MUTANT |
0 (0) |
0 (0) |
96.9 (279) |
92.7 (267) |
99.6 (296) |
1 (3) |
0 (0) |
98.1 (264) |
91.1 (256) |
0.4 (1) |
0 (0) |
|
| MIXED |
0 (0) |
0 (0) |
1.7 (5) |
3.8 (11) |
0 (0) |
0.3 (1) |
0.4 (1) |
0.4 (1) |
1.4 (4) |
2.5 (7) |
0 (0) |
|
| KSI |
WILD |
100 (152) |
100 (155) |
1.3 (2) |
11.0 (17) |
0 (0) |
98.1 (155) |
100 (87) |
1.1 (1) |
9.9 (14) |
96.7 (146) |
100 (117) |
| MUTANT |
0 (0) |
0 (0) |
96.9 (152) |
82.5 (127) |
100 (156) |
1.32 (2) |
0 (0) |
97.9 (91) |
89.4 (126) |
2.0 (3) |
0 (0) |
|
| MIXED |
0 (0) |
0 (0) |
1.9 (3) |
6.5 (10) |
0 (0) |
0.6 (1) |
0 (0) |
1.1 (1) |
0.7 (1) |
1.3 (2) |
0 (0) |
|
| KCH |
WILD |
100 (93) |
100 (93) |
2.2 (2) |
7.7 (6) |
2.1 (2) |
100 (93) |
98.9 (89) |
2.2 (2) |
3.8 (4) |
98.1 (102) |
100 (104) |
| MUTANT |
0 (0) |
0 (0) |
97.9 (91) |
73.1 (57) |
97.9 (92) |
0 (0) |
0 (0) |
96.7 (87) |
90.4 (94) |
1.0 (1) |
0 (0) |
|
| MIXED |
0 (0) |
0 (0) |
0 (0) |
19.2 (15) |
0 (0) |
0 (0) |
1.1 (1) |
1.1 (1) |
5.8 (6) |
1.0 (1) |
0 (0) |
|
| MDH | WILD |
100 (99) |
100 (103) |
10.7 (11) |
24.3 (25) |
5.8 (6) |
100 (103) |
98.9 (98) |
13.2 (14) |
11.9 (12) |
100 (104) |
100 (107) |
| MUTANT |
0 (0) |
0 (0) |
87.4 (90) |
70.9 (73) |
94.2 (97) |
0 (0) |
1.0 (1) |
86.8 (92) |
86.1 (87) |
0 (0) |
0 (0) |
|
| MIXED | 0 (0) | 0 (0) | 1.9 (2) | 4.9 (5) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2.0 (2) | 0 (0) | 0 (0) | |
Prevalence of Pfdhfr and Pfdhps wildtype, mutant and mixed genotypes per study site for the entire period was calculated as a percentage of the total number of samples successfully analysed over the study period per codon per the specific site.
Temporal trends of SNPs in Pfdhfr and Pfdhps genes
Additional file 1 shows the temporal trends of mutations in Pfdhfr and Pfdhps genes of samples collected from all the four sites. Samples collected from KDH and KCH displayed steady prevalence over time at all codons except for codon 581 in KDH which the prevalence of mixed genotype increased from 1.2% in 2009 to 15.6% in 2012. However, MDH and to lesser extent KCH showed changing temporal trends. At MDH, samples collected in 2011 had lower prevalence of mutation in Pfdhfr codons 51, 108 and Pfdhps codon 437 compared to samples collected in 2010 and 2012. Chi-square analysis showed that the prevalence of Pfdhfr codon 108 and Pfdhps codon 437 mutant genotypes were significantly different (P = 0.016 and 0.039) respectively. Although it did not reach statistical significance, there was a downward trend of the prevalence of mutation at codon 59 and 540 (P = 0.875 and 0.5146), respectively, for samples collected in KCH and MDH.
Prevalence of Pfdhfr and Pfdhps genotypes
Prevalence of Pfdhfr and Pfdhps genotypes was based on criteria that were originally described by Kublin et al.[11] and recently by Iriemenam et al.[19]. Genotype analysis was done only on those samples that were successfully analysed at all loci. Mixed genotypes containing two alleles at a single locus were pooled with pure mutant (single) alleles for analysis. Pfdhfr genotypes were based on mutations at codons 51, 59, and 108 which were classified as wild type, single, double, and triple. Pfdhps genotypes were based on mutations at codons 437 and 540 which were classified as wild type, single, and double. Combined Pfdhfr (51, 59, and 108) and Pfdhps (437, 540) genotypes were defined as wildtype, single, double, triple, quadruple and quintuple. Quintuple genotype represented infections in which all the five mutations were present. Additional combinations which contained mutation at Pfdhfr codon 164, Pfdhps codons 436 and 581 were genotyped as well.
Additional file 2 shows the prevalence of genotypes in Pfdhfr or Pfdhps genes. In Pfdhfr gene, triple mutant (N51I/C59R/S108N) was the most prevalent at 86.6% in all sites combined whereas single mutant (C59R), double mutant (N51I/C59R) and the quadruple mutant (N51I/C59R/S108N/I164L) were the least prevalent at 0.26% each. When broken further per site, the triple mutant was most prevalent in KDH at 93.7% whereas least prevalent in MDH at 67.9%. There was no genotype observed which contained Pfdhfr N51I single mutant. In the Pfdhps gene, double mutant (A437G/K540E) was the most prevalent at 87.9% whereas single mutant (S436A) was the least prevalent at 0.26% (one sample). Interestingly, two of the four samples in triple mutant 436/437/540 carried S436F and S436H amino acid mutation and not the S436A which has been shown to be more prevalent mutation [19] . These mutations occurred either as a triple S436F/A437G/K540E or S436H/A437G/K540E mutations. Of note is prevalence of A581G mutant which has been rare in the region, but has been seen to emerge in recent years [19]. The A581G mutant appeared as a genotype in combination with the A437G mutation in 0.78% of the isolates analysed. It was also observed as a triple A437G/K540E/A581G mutant genotype in 1.6% of the samples. Interestingly, A581G mutant was not observed as a single mutant, but appeared in the presence of other mutations on the gene.
Additional file 3 shows the prevalence of genotypes in combined Pfdhfr and Pfdhps genes. In all the sites, the quintuple mutant (Pfdhfr/Pfdhps N51I,C59R,S108N/A437G,K540E) was the most prevalent at 75.71% whereas the least prevalent was quadruple mutant (Pfdhfr/Pfdhps N51I,C59R,S108N/S436A) and sextuple mutant (Pfdhfr/Pfdhps N51I,C59R,S108N, I164L/A437G,K540E) at 0.26%. When broken down per site, KCH had the highest prevalence of the quintuple mutant at 89.6% whereas MDH was the lowest at 54.3%.
Temporal trends of genotypes in Pfdhfr and Pfdhps genes
Additional file 4 shows percent temporal prevalence of genotypes from 2008-2012. In Pfdhfr gene, the wildtype of the genotype analysed is N51,C59,S108,I164 (NCSI) and in the Pfdhps gene, the wildtype of the genotype analysed is S436,A437,K540,A581 (SAKA). For Pfdhfr gene, the prevalence of wildtype genotype increased slightly from 2008 to 2011, but was not present in 2012. The IRN I genotype (triple mutant, 51I,59R,108 N) was the most prevalent but decreased from 92.5% in 2008 to 72.3% in 2012. For Pfdhps gene, the prevalence of the wildtype genotype was not steady overtime. The S GE A genotype (double mutant, 437G, 540E) was the most prevalent. However, unlike the Pfdhfr IRN I genotype which declined over time, the Pfdhps S GE A genotype remained relatively stable over time.
Discussion
The Pfdhfr/Pfdhps N51I, C59R, S108N/A437G, K540E quintuple mutant genotype is strongly associated with clinical SP treatment failure [11]. Using sample isolates collected from general patient population, quintuple mutant genotype was shown to be the most prevalent combined genotype at 75.7%. When further broken down per site, KCH and KDH had the highest prevalence at 89.6% and 80.6%, respectively whereas MDH had the lowest prevalence at 54.3% (see Additional file 3). In other studies conducted in Kenya using sample isolates collected in Kisumu, Iriemenam et al. [19] and Spalding et al.[18] showed prevalence of the quintuple mutant genotype to be at 88% and 53% respectively. The difference in prevalence can be explained by the period and patient population which sample isolates were collected from. Iriemenam et al. study used sample isolates collected in 2008-2009 from pregnant patient population whereas Spalding et al. used general patient population sample isolates collected in 2003-2005. In the current study, the prevalence of the quintuple mutant phenotype is similar to that from Iriemenam et al. study most likely because the samples were collected around the same period. The small difference seen can be attributed to the difference in patient population that sample isolates were collected from and is also reflective of the malaria attack rates found in those locations. The attack rate has been significantly lower at the coast over the last five years, due mostly to the use of bed nets and other environmental controls [21].
Although 2003-2005 marked the period which SP was discontinued and AL introduced as first-line treatment for malaria, the prevalence of Pfdhfr/Pfdhps N51I, C59R, S108N/A437G, K540E quintuple mutant genotype which is strongly associated with clinical SP treatment failure continued to increase at an alarming rate during this period. Iriemenam et al. and Spalding et al. studies observed the prevalence of the quintuple mutant genotype increase from 7% and 21% in sample isolates collected in 1996-2000 and 1999-2000 to 88% and 53% in samples collected in 2008-2009 and 2003-2005 respectively. In the current study, the prevalence of quintuple mutant genotype in KDH (western Kenya) was 80.6%. These studies indicate that there is a fixation of the sulphadoxine-pyrimethamine resistant alleles in parasite population from western Kenya. Such fixation has also been observed in Southeast Asia [23], contrary to what has been reported in the Amazon region of Peru where a decline in sulphadoxine-pyrimethamine-resistant alleles was reported five years after change in drug policy [24]. Fixation of the resistance alleles have been attributed to parasites that may have acquired additional compensatory mutations [25]. The fitness cost imposed by the mutations does not impact the parasite and the parasite populations are fixed for resistance with no competing sensitive parasite. There is also the probable presence of additional selective pressure from antifolate drugs such as cotrimoxazole (trimethoprim-sulphamethoxazole) [26,27]. In addition to widespread use of cotrimoxazole, SP continues to be widely available at the local pharmacies (chemists) with self-report SP use still high regardless of AL being the recommended first-line malaria treatment in Kenya. Experimental or epidemiological evidence however will be required to show an association between cotrimoxazole and the occurrence of Pfdhps/Pfdhfr mutations in P. falciparum. In a recently concluded randomized trial, one group of HIV-infected individuals discontinued cotrimoxazole prophylaxis for 12 months whereas the second group continued with cotrimoxazole prophylaxis (unpublished data, personal communication with Dr. Christina Polyak). Comparison of the prevalence of malaria and mutations in Pfdhps/Pfdhfr genes in the group that discontinued cotrimoxazole prophylaxis to the group that continued will provide evidence of any association between cotrimoxazole and the occurrence of Pfdhps/Pfdhfr mutations in P. falciparum. Other factors such as host immunity and transmission intensity might attribute to the fixation of Pfdhps/Pfdhfr mutant alleles in parasites in western Kenya [19].
Mutations at Pfdhps codon 581 and Pfdhfr codon 164 have been linked to high rate of therapeutic failures in Southeast Asia, South America [23,28] and more recently in Africa [29-31]. Recent studies have shown emergence and increase of the triple mutant allele (A437G/K540E/A581G) in Tanzania [29,32]. Alifrangis et al.[32] showed that the prevalence of mutation at Pfdhps codon 581 increased significantly from 12% in 2003 to 56% in 2007, resulting in an increase in the triple mutant Pfdhps haplotype (A437G/K540E/A581G) from 8% to 32%. In studies conducted in Kenya, Iriemenam et al.[19] and Spalding et al.[18] found the prevalence of triple mutant Pfdhps haplotype to be 5.3% and 15% respectively whereas in this study, the prevalence was 1.6% (see Additional file 2). Sample isolates from Iriemenam et al. study and the current study were collected around the same period (2008-2009 vs. 2008-2012) and showed similar prevalence of the triple mutant Pfdhps haplotype. However, sample isolates from Spalding et al. study collected in 2003-2005 showed significantly high prevalence of mutations at Pfdhps codon 581 (85%), only three years after mutations at the Pfdhps codon did not exist [17]. This is unexpected since Iriemenam et al. study and the current study, which used sample isolates collected at later time period showed lower prevalence of the triple mutant Pfdhps haplotype (A437G/K540E/A581G), yet prevalence of this mutation has been on a steady increase during the SP treatment period [19]. However the prevalence of the triple mutant Pfdhps haplotype from Spalding et al. study is similar to that seen in Tanzania [32]. Mutation at Pfdhfr codon 164 remains low. The study done using samples collected from western Kenya between 1992 and1999 by McCollum et al.[33] indicated very low prevalence of Pfdhfr wild type alleles while majority had double Pfdhfr 51/108, 59/108 and Pfdhps 437/540 mutations (50%, 27% and 57% respectively). Pfdhps wild type alleles were relatively high at 34% at that period. This was a period before SP was the first line antimalarial treatment and indicated initial development of resistant traits in the population.
This is the first study which compares prevalence of drug resistance mutant genotypes in Pfdhfr and Pfdhps genes in sample isolates collected from western and coastal regions of Kenya. Sample isolates collected from Malindi (MDH) showed significant difference in prevalence of drug resistance alleles in Pfdhfr and Pfdhps genes compared to sample isolates collected from western Kenya (Table 2). Interestingly, mutation in Pfdhfr codon 51 and 108 as well as Pfdhps codon 437 decreased in 2011 compared to 2010, but increased in 2012. Mutation in Pfdhps codon 540 remained the same in 2010 and 2011, but increased significantly in 2012 (P = 0.0196). To the contrarily, Pfdhfr codon 59 significantly decreased from 2010 to 2012 (P = 0.0064). The prevalence and the temporal trends of mutations in Pfdhfr and Pfdhps genes in Malindi represent interesting dynamics which could be driven by a number of factors including natural phenomenon which is yet to be determined. The prevalence of malaria is generally on a steady decline in the Kenyan coast (personal communication with Dr. Kevin Marsh) [34] which might result to changes in the immunity of the population in this region. Also, the dip in prevalence seen in 2011 might be due to environmental conditions such as weather patterns which can impact transmission intensity hence the prevalence of drug resistance genotype. Interesting however, mutation at Pfdhfr codon 59 showed a steady decline from 2010 to 2012. Further studies are required to further elucidate the dynamics of mutations in Pfdhfr and Pfdhps genes in Kenyan coast.
Table 2.
P values indicating significance in statistical differences, P = 0.05
| Western Kenya (n = 306) | Western Kenya (n = 306) vs MDH (n = 81) | ||
|---|---|---|---|
| Pfdhfr codons |
16 |
1.0000 |
1.0000 |
| 50 |
1.0000 |
1.0000 |
|
| 51 |
0.7762 |
0.0007 |
|
| 59 |
0.0904 |
<0.0001 |
|
| 108 |
0.3642 |
0.0240 |
|
| 164 |
0.3629 |
0.2571 |
|
| Pfdhps codons | 436 |
0.3629 |
0.2964 |
| 437 |
0.4068 |
<0.0001 |
|
| 540 |
0.2323 |
0.2259 |
|
| 581 |
0.7092 |
0.2638 |
|
| 613 | 1.0000 | 1.0000 |
P value of the mutant prevalence of each codon was calculated. Comparison was done in samples from western Kenya study sites (KDH, KSI, and KCH) and also between the western Kenya sites verses the coastal study site (MDH) to determine statistical differences in prevalence between the two geographical regions.
Conclusion
The data presented in this study confirms the fixation of key mutations in the Pfdhfr and Pfdhps genes conferring resistance to SP. It also shows that there is emergence and increasing prevalence of new mutations in the Kenyan parasite population. The adoption of SP in IPTp might only be playing a minor role in these observations. The main contributors of the antifolate drug pressure are likely to be over the counter SP self-medication and the use of cotrimoxazole in management of HIV patients. Apart from discouraging SP use for symptomatic malaria treatment, further evaluation should be undertaken to show effect of the highly prevalent Pfdhfr/Pfdhps quintuple mutant combining with the emerging Pfdhfr 164 and/or the Pfdhps 581 on the efficacy of IPTp or IPTi. A continuous molecular surveillance accompanied by in vitro susceptibility testing will provide valuable information urgently needed to control or reduce spread of SP resistance.
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
DWJ was involved in laboratory oversight, manuscript writing, data analysis and interpretation. AOA carried out molecular assays, drafting the manuscript or revising it critically for important intellectual content. LI and BO participated in data analysis, manuscript revising & molecular assays. ARC was involved in data analysis, revising it critically for important intellectual content. RY participated in data analysis & molecular assays. CO participated in molecular assays, acquisition of data. JC, PM, BN and LC were involved in molecular assays, and data interpretation. FLE was involved in laboratory oversight, revising manuscript critically for important intellectual content. WDB - Manuscript writing, conception and design, data analysis and interpretation. HMA - Data analysis & Manuscript development. JDJ, BA and EK were involved in protocol oversight, Manuscript writing, conception and design, data analysis and interpretation. All authors have read and approved the final version of the manuscript.
Supplementary Material
Prevalence of single nucleotide polymorphism in Pfdhfr and Pfdhps Per year (n = Number of isolates having that genotype). Description: The data shows the frequency of the individual codon genotypes over the study period per site.
Prevalence of combined mutation in the Pfdhfr and Pfdhps genes. Description: The data presented represent prevalence of haplotypes in the Pfdhfr and Pfdhps genes.
Prevalence of Pfdhfr/Pfdhps combined mutations per site (SITE MUTATION/SITE TOTAL*100). Description: The data represents the prevalence of mutation occurring in both Pfdhfr and Pfdhps genes simultaneously.
Percentage temporal prevalence of genotypes. Description: The data shows average genotype prevalence of all the study sites over the period under investigation. The genotypes analysed for Pfdhfr were N51I, C59R, S108N and I164L while for Pfdhps were S436A, A437G, K540E and A581G in that order. Amino acid codes before the loci position indicate the wildtype while the ones indicated after the loci position are the mutations. In the table, the genotypes without neither bold nor underlined amino acid codes are the wild type while the underlined and bold are the mutant alleles.
Contributor Information
Dennis W Juma, Email: Dennis.Juma@usamru-k.org.
Angela A Omondi, Email: Angela.Omondi@usamru-k.org.
Luiser Ingasia, Email: Luiser.Ingasia@usamru-k.org.
Benjamin Opot, Email: Benjamin.Opot@usamru-k.org.
Agnes Cheruiyot, Email: Agnes.Cheruiyot@usamru-k.org.
Redemptah Yeda, Email: Redemptah.Yeda@usamru-k.org.
Charles Okudo, Email: Charles.Okello@usamru-k.org.
Jelagat Cheruiyot, Email: jelagat.cheruiyot@usamru-k.org.
Peninnah Muiruri, Email: peninnah.muiruri@usamru-k.org.
Bidii Ngalah, Email: bidii.ngala@usamru-k.org.
Lorna J Chebon, Email: Lorna.Chebon@usamru-k.org.
Fredrick Eyase, Email: fredrick.eyase@usamru-k.org.
Jacob Johnson, Email: jacob.johnson@usamru-k.org.
Wallace D Bulimo, Email: wallace.bulimo@usamru-k.org.
Hoseah M Akala, Email: hosea.akala@usamru-k.org.
Ben Andagalu, Email: ben.andagalu@usamru-k.org.
Edwin Kamau, Email: edwin.kamau@us.army.mil.
Acknowledgements
We thank the MDR field staff for their contribution in recruitment of participants and the collection of samples. We also thank the Director of the Kenya Medical Research Institute for permission to publish this work. This work was supported by the U.S. Department of Defense, Global Emerging Infections System; Silver Spring, Maryland. The opinions and assertions contained herein are the private opinions of the authors and are not to be construed as reflecting the views of the US Army Medical Research Unit-Kenya or the US Department of Defense.
References
- Bousema JT, Gouagna LC, Meutstege AM, Okech BE, Akim NI, Githure JI, Beier JC, Sauerwein RW. Treatment failure of pyrimethamine-sulphadoxine and induction of Plasmodium falciparum gametocytaemia in children in western Kenya. Trop Med Int Health. 2003;8:427–430. doi: 10.1046/j.1365-3156.2003.01047.x. [DOI] [PubMed] [Google Scholar]
- Mberu EK, Mosobo MK, Nzila AM, Kokwaro GO, Sibley CH, Watkins WM. The changing in vitro susceptibility pattern to pyrimethamine/sulfadoxine in Plasmodium falciparum field isolates from Kilifi, Kenya. Am J Trop Med Hyg. 2000;62:396–401. doi: 10.4269/ajtmh.2000.62.396. [DOI] [PubMed] [Google Scholar]
- Amin AA, Zurovac D, Kangwana BB, Greenfield J, Otieno DN, Akhwale WS, Snow RW. The challenges of changing national malaria drug policy to artemisinin-based combinations in Kenya. Malar J. 2007;6:72. doi: 10.1186/1475-2875-6-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters PJ, Thigpen MC, Parise ME, Newman RD. Safety and toxicity of sulfadoxine/pyrimethamine: implications for malaria prevention in pregnancy using intermittent preventive treatment. Drug Saf. 2007;30:481–501. doi: 10.2165/00002018-200730060-00003. [DOI] [PubMed] [Google Scholar]
- WHO. Report of the Technical Consultation on Intermittent Preventive Treatment in Infants (IPTi), Technical Expert Group on Preventive Chemotherapy. Geneva: World Health Organization; 2009. [Google Scholar]
- Sridaran S, McClintock SK, Syphard LM, Herman KM, Barnwell JW, Udhayakumar V. Anti-folate drug resistance in Africa: meta-analysis of reported dihydrofolate reductase (dhfr) and dihydropteroate synthase (dhps) mutant genotype frequencies in African Plasmodium falciparum parasite populations. Malar J. 2010;9:247. doi: 10.1186/1475-2875-9-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCollum AM, Poe AC, Hamel M, Huber C, Zhou Z, Shi YP, Ouma P, Vulule J, Bloland P, Slutsker L, Barnwell JW, Udhayakumar V, Escalante AA. Antifolate resistance in Plasmodium falciparum: multiple origins and identification of novel dhfr alleles. J Infect Dis. 2006;194:189–197. doi: 10.1086/504687. [DOI] [PubMed] [Google Scholar]
- Peterson DS, Wallikier D, Wellems TE. Evidence that a point mutation in dihydrofolate reductase-thymidylate synthase confers resistance to pyrimethamine in falciparum malaria. Proc Natl Acad Sci U S A. 1988;85:9114–9118. doi: 10.1073/pnas.85.23.9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T, Menting JG, Wilson C, Cowman AF. Mutations in dihydropteroate synthase are responsible for sulfone and sulfonamide resistance in Plasmodium falciparum. Proc Natl Acad Sci U S A. 1997;94:13944–13949. doi: 10.1073/pnas.94.25.13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nzila-Mounda A, Mberu EK, Sibley CH, Plowe CV, Winstanley PA, Watkins WM. Kenyan Plasmodium falciparum field isolates: correlation between pyrimethamine and chlorcycloguanil activity in vitro and point mutations in the dihydrofolate reductase domain. Antimicrob Agents Chemother. 1998;42:164–169. doi: 10.1128/aac.42.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kublin JG, Dzinjalamala FK, Kamwendo DD, Malkin EM, Cortese JF, Martino LM, Mukadam RA, Rogerson SJ, Lescano AG, Molyneux ME, Winstanley PA, Chimpeni P, Taylor TE, Plowe CV. Molecular markers for failure of sulfadoxine-pyrimethamine and chlorproguanil-dapsone treatment of Plasmodium falciparum malaria. J Infect Dis. 2002;185:380–388. doi: 10.1086/338566. [DOI] [PubMed] [Google Scholar]
- Foote SJ, Galatis D, Cowman AF. Amino acids in the dihydrofolate reductase-thymidylate synthase gene of Plasmodium falciparum involved in cycloguanil resistance differ from those involved in pyrimethamine resistance. Proc Natl Acad Sci U S A. 1990;87:3014–3017. doi: 10.1073/pnas.87.8.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowe CV, Cortese JF, Djimde A, Nwanyanwu OC, Watkins WM, Winstanley PA, Estrada-Franco JG, Mollinedo RE, Avila JC, Cespedes JL, Carter D, Doumbo OK. Mutations in Plasmodium falciparum dihydrofolate reductase and dihydropteroate synthase and epidemiologic patterns of pyrimethamine-sulfadoxine use and resistance. J Infect Dis. 1997;176:1590–1596. doi: 10.1086/514159. [DOI] [PubMed] [Google Scholar]
- Oesterholt MJ, Alifrangis M, Sutherland CJ, Omar SA, Sawa P, Howitt C, Gouagna LC, Sauerwein RW, Bousema T. Submicroscopic gametocytes and the transmission of antifolate-resistant Plasmodium falciparum in Western Kenya. PLoS One. 2009;4:e4364. doi: 10.1371/journal.pone.0004364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triglia T, Wang P, Sims PF, Hyde JE, Cowman AF. Allelic exchange at the endogenous genomic locus in Plasmodium falciparum proves the role of dihydropteroate synthase in sulfadoxine-resistant malaria. EMBO J. 1998;17:3807–3815. doi: 10.1093/emboj/17.14.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil I, Ronn AM, Alifrangis M, Gabar HA, Satti GM, Bygbjerg IC. Dihydrofolate reductase and dihydropteroate synthase genotypes associated with in vitro resistance of Plasmodium falciparum to pyrimethamine, trimethoprim, sulfadoxine, and sulfamethoxazole. Am J Trop Med Hyg. 2003;68:586–589. doi: 10.4269/ajtmh.2003.68.586. [DOI] [PubMed] [Google Scholar]
- Mbaisi A, Liyala P, Eyase F, Achilla R, Akala H, Wangui J, Mwangi J, Osuna F, Alam U, Smoak BL, Davis JM, Kyle DE, Coldren RL, Mason C, Waters NC. Drug susceptibility and genetic evaluation of Plasmodium falciparum isolates obtained in four distinct geographical regions of Kenya. Antimicrob Agents Chemother. 2004;48:3598–3601. doi: 10.1128/AAC.48.9.3598-3601.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding MD, Eyase FL, Akala HM, Bedno SA, Prigge ST, Coldren RL, Moss WJ, Waters NC. Increased prevalence of the pfdhfr/phdhps quintuple mutant and rapid emergence of pfdhps resistance mutations at codons 581 and 613 in Kisumu, Kenya. Malar J. 2010;9:338. doi: 10.1186/1475-2875-9-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iriemenam NC, Shah M, Gatei W, van Eijk AM, Ayisi J, Kariuki S, Vanden Eng J, Owino SO, Lal AA, Omosun YO, Otieno K, Desai M, ter Kuile FO, Nahlen B, Moore J, Hamel MJ, Ouma P, Slutsker L, Shi YP. Temporal trends of sulphadoxine-pyrimethamine (SP) drug-resistance molecular markers in Plasmodium falciparum parasites from pregnant women in western Kenya. Malar J. 2012;11:134. doi: 10.1186/1475-2875-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malakooti MA, Biomndo K, Shanks GD. Reemergence of epidemic malaria in the highlands of western Kenya. Emerg Infect Dis. 1998;4:671–676. doi: 10.3201/eid0404.980422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelda AO, Victor AA, Abdisalan MN, Robert WS. Changing malaria intervention coverage, transmission and hospitalization in Kenya. Malar J. 2010;9:285. doi: 10.1186/1475-2875-9-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam MT, de Souza DK, Vinayak S, Griffing SM, Poe AC, Duah NO, Ghansah A, Asamoa K, Slutsker L, Wilson MD, Barnwell JW, Udhayakumar V, Koram KA. Selective sweeps and genetic lineages of Plasmodium falciparum drug -resistant alleles in Ghana. J Infect Dis. 2011;203:220–227. doi: 10.1093/infdis/jiq038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinayak S, Alam MT, Mixson-Hayden T, McCollum AM, Sem R, Shah NK, Lim P, Muth S, Rogers WO, Fandeur T, Barnwell JW, Escalante AA, Wongsrichanalai C, Ariey F, Meshnick SR, Udhayakumar V. Origin and evolution of sulfadoxine resistant Plasmodium falciparum. PLoS Pathog. 2010;6:e1000830. doi: 10.1371/journal.ppat.1000830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Griffing SM, de Oliveira AM, McCollum AM, Quezada WM, Arrospide N, Escalante AA, Udhayakumar V. Decline in sulfadoxine-pyrimethamine-resistant alleles after change in drug policy in the Amazon region of Peru. Antimicrob Agents Chemother. 2008;52:739–741. doi: 10.1128/AAC.00975-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam MT, Vinayak S, Congpuong K, Wongsrichanalai C, Satimai W, Slutsker L, Escalante AA, Barnwell JW, Udhayakumar V. Tracking origins and spread of sulfadoxine-resistant Plasmodium falciparum dhps alleles in Thailand. Antimicrob Agents Chemother. 2011;55:155–164. doi: 10.1128/AAC.00691-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garner P, Gulmezoglu AM, Garner P, Gulmezoglu AM. Drugs for preventing malaria in pregnant women. Cochrane Database Syst. 2006. http://onlinelibrary.wiley.com/doi/10.1002/14651858.CD000169.pub2/full. [DOI] [PubMed]
- Gasaira AF, Kamya MR, Ochong EO, Vora N, Achan J, Charlebois E, Ruel T, Kateera F, Meya DN, Havlir D, Rosenthal PJ, Dorsey G, Gasaira AF, Kamya MR, Ochong EO, Vora N, Achan J, Charlebois E, Ruel T, Kateera F, Meya DN, Havlir D, Rosenthal PJ, Dorsey G. Effect of trimethroprim-sulfamethazole on the risk of malaria in HIV infected Ugandan children living in an area of widespread antifolate resistance. Malar J. 2010;9:177. doi: 10.1186/1475-2875-9-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bras J, Durand R. The mechanisms of resistance to antimalarial drugs in Plasmodium falciparum. Fundam Clin Pharmacol. 2003;17:147–153. doi: 10.1046/j.1472-8206.2003.00164.x. [DOI] [PubMed] [Google Scholar]
- Gesase S, Gosling RD, Hashim R, Ord R, Naidoo I, Madebe R, Mosha JF, Joho A, Mandia V, Mrema H, Mapunda E, Savael Z, Lemnge M, Mosha FW, Greenwood B, Roper C, Chandramohan D. High resistance of Plasmodium falciparum to sulphadoxine/pyrimethamine in northern Tanzania and the emergence of dhps resistance mutation at Codon 581. PLoS One. 2009;4:e4569. doi: 10.1371/journal.pone.0004569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lee CS, Bayoumi R, Djimde A, Doumbo O, Swedberg G, Dao LD, Mshinda H, Tanner M, Watkins WM, Sims PF, Hyde JE. Resistance to antifolates in Plasmodium falciparum monitored by sequence analysis of dihydropteroate synthetase and dihydrofolate reductase alleles in a large number of field samples of diverse origins. Mol Biochem Parasitol. 1997;89:161–177. doi: 10.1016/S0166-6851(97)00114-X. [DOI] [PubMed] [Google Scholar]
- Ishraga A-E, Michael A, Insaf K, Ib B, Emad M, Mustafa E, Hayder G. The implication of dihydrofolate reductase and dihydropteroate synthetase gene mutations in modification of Plasmodium falciparum characteristics. Malar J. 2007;6:108. doi: 10.1186/1475-2875-6-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alifrangis M, Lusingu JP, Mmbando B, Dalgaard MB, Vestergaard LS, Ishengoma D, Khalil IF, Theander TG, Lemnge MM, Bygbjerg IC. Five-year surveillance of molecular markers of Plasmodium falciparum antimalarial drug resistance in Korogwe District, Tanzania: accumulation of the 581G mutation in the P. falciparum dihydropteroate synthase gene. Am J Trop Med Hyg. 2009;80:523–527. [PubMed] [Google Scholar]
- McCollum AM, Schneider KA, Griffing SM, Zhou Z, Kariuki S, Ter-Kuile F, Shi YP, Slutsker L, Lal AA, Udhayakumar V, Escalante AA. Differences in selective pressure on dhps and dhfr drug resistant mutations in western Kenya. Malar J. 2012;11:77. doi: 10.1186/1475-2875-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwangangi JM, Mbogo CM, Orindi BO, Muturi EJ, Midega JT, Nzovu J, Gatakaa H, Githure J, Borgemeister C, Keating J, Beier JC. Shifts in malaria vector species composition and transmission dynamics along the Kenyan coast over the past 20 years. Malar J. 2013;12:13. doi: 10.1186/1475-2875-12-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Prevalence of single nucleotide polymorphism in Pfdhfr and Pfdhps Per year (n = Number of isolates having that genotype). Description: The data shows the frequency of the individual codon genotypes over the study period per site.
Prevalence of combined mutation in the Pfdhfr and Pfdhps genes. Description: The data presented represent prevalence of haplotypes in the Pfdhfr and Pfdhps genes.
Prevalence of Pfdhfr/Pfdhps combined mutations per site (SITE MUTATION/SITE TOTAL*100). Description: The data represents the prevalence of mutation occurring in both Pfdhfr and Pfdhps genes simultaneously.
Percentage temporal prevalence of genotypes. Description: The data shows average genotype prevalence of all the study sites over the period under investigation. The genotypes analysed for Pfdhfr were N51I, C59R, S108N and I164L while for Pfdhps were S436A, A437G, K540E and A581G in that order. Amino acid codes before the loci position indicate the wildtype while the ones indicated after the loci position are the mutations. In the table, the genotypes without neither bold nor underlined amino acid codes are the wild type while the underlined and bold are the mutant alleles.


